Abstract
The acquisition of multiple genes from long terminal repeat (LTR) retrotransposons occurred in mammals. Genes belonging to a sushi-ichi-related retrotransposon homologs (SIRH) family emerged around the time of the establishment of two viviparous mammalian groups, marsupials and eutherians. These genes encode proteins that are homologous to a retrotransposon Gag capsid protein and sometimes also have a Pol-like region. We previously demonstrated that PEG10 (SIRH1) and PEG11/RTL1 (SIRH2) play essential but different roles in placental development. PEG10 is conserved in both the marsupials and the eutherians, while PEG11/RTL1 is a eutherian-specific gene, suggesting that these two domesticated genes were deeply involved in the evolution of mammals via the establishment of the viviparous reproduction system. In this review, we introduce the roles of PEG10 and PEG11/RTL1 in mammalian development and evolution, and summarize the other genes domesticated from LTR retrotransposons and endogenous retroviruses (ERVs) in mammals. We also point out the importance of DNA methylation in inactivating and neutralizing the integrated retrotransposons and ERVs in the process of domestication.
Keywords: domesticated genes, LTR retrotransposons and ERVs, mammals, development and evolution
INTRODUCTION
Domestication (exaptation, co-option) is an extended mode of restricting the retrotransposons, endogenous retroviruses (ERVs), and DNA transposons that are integrated into host genomes. It has been proposed that host organisms make use of such transposable DNA elements as a genetic resource of genes for novel purposes (Brosius and Gould, 1992; Smit, 1999). Telomerase, which maintains the telomere end repeats in chromosomes in eukaryotes, and two recombination activating genes (RAG1 and RAG2) that are essential for producing the vast diversity of immunoglobulin types by V(D)J recombination in vertebrates, are good examples. The former was derived from a reverse transcriptase of a long terminal repeat (LTR) retrotransposon or retrovirus (Nakamura and Cech, 1998) and the latter from a transposase of a DNA transposon (Agrawal et al., 1998; Hiom et al., 1998). Mammalian centromere-associated protein B (CENP-B) facilitates centromere formation and is a DNA-binding protein derived from a transposase of the pogo-like DNA transposon family (Tudor et al., 1992; Casola et al., 2008). Although it bears considerable similarity to three fission yeast proteins, ARS-binding protein (Abp1), CENP-B homologs 1 and 2 (Cbh1 and Cbh2), which also exhibit centromere binding, it was recently reported that the origin of mammalian CENP-B is different from that of the three fission yeast proteins. That is, they are all derived from distinct pogo-like DNA transposons, indicating that convergent domestication occurred in the mammalian and fission yeast lineages. In plants, the Arabidopsis far-red elongated hypocotyls 3 (FHY3) and far-red-impaired response (FAR1) genes are derived from an ancient Mutator-like transposase, a kind of DNA transposons. They encode transcription factors essential for the light response via phytochrome A signaling (Lisch et al., 2001; Hudson et al., 2003; Lin et al., 2007). From these data, it is clear that the domestication of transposable elements had a profound effect on quite a large numbers of animals and plants during the course of biological evolution, even though only few cases are currently known.
The retrotransposons, ERVs, and their remnant DNA sequences occupy approximately 40% of the mammalian genome and they have long been thought to be either “selfish” genes or useless “junk.” Is it thus the case that domestication events are very rare in mammals? Are there any domesticated genes which are present in a mammalian-, therian-, and eutherian-specific manner? Alternatively, are there domesticated genes which have been conserved in a more restricted manner, i.e., as species- and strain-specific genes? If so, it would be highly probable that they have contributed to mammalian evolution in various ways and to different degrees.
The recent availability of mammalian genome sequence information enabled us to identify dozens of novel domesticated genes from LTR retrotransposons/ERVs. In 2000, human SYNCYTIN (ERVWE1) was identified as the first candidate domesticated gene derived from ERVs in mammals (Blond et al., 2000; Mi et al., 2000). As shown in Figure 1, it derives from an envelope (Env) gene of a human-specific endogenous retrovirus, HERV-W, and was suggested by in vitro study to mediate placental cytotrophoblast fusion so as to produce syncytiotrophoblast cells in human placental morphogenesis. Interestingly, humans have two SYNCYTIN genes, but they are primate-specific genes (Blaise et al., 2003). Similar genes (also called Syncytin) were also discovered in several mammalian lineages that were independently acquired from Env genes from different ERVs (Dupressoir et al., 2005; Heidmann et al., 2009). Finally, mouse SyncytinA and B have been to be essential placental genes using knockout mice (Dupressoir et al., 2009, 2011).
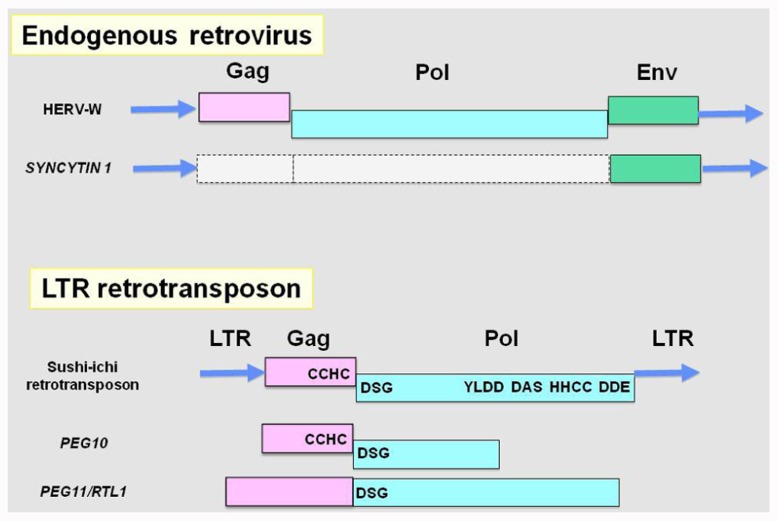
FIGURE 1.
Endogenous retrovirus, LTR retrotransposon and their domesticated genes. Top: an endogenous retrovirus, HERV-W, and SYNCYTIN 1. SYNCYTIN 1 retains LTRs at both ends and the Env gene, while the Gag and Pol genes do not support ORFs corresponding functional proteins because of stop mutations. Bottom: an LTR retrotransposon, sushi-ichi, and the domesticated PEG10 and PEG11/RTL1 genes. Both PEG10 and PEG11/RTL1 have lost LTRs while entire regions exhibit significant homologies to the Gag and Pol genes remaining in some of the retrotransposon domains. The -1 frameshift mechanism is conserved in PEG10. LTR, long terminal repeat; Gag, group-specific antigen; Pol, polymerase; Env, envelope; CCHC, RNA-binding motif; DSG, protease active site; YYDD, reverse transcriptase; DAS, RNase highly conserved motif; HHCC, integrase DNA binding motif; DDE, strongly conserved integrase.
In 2001, the first and second candidate domesticated genes from a sushi-ichi-related LTR retrotransposon were identified as paternally expressed 10 (PEG10; Ono et al., 2001) and paternally expressed 11/retrotransposon-like 1 (PEG11/RTL1; Charlier et al., 2001). They encode proteins homologous to a retrotransposon Gag and a Pol protein, respectively (Figure 1). Combined with definitive genetic studies using knockout mice, PEG10 and PEG11/RTL1 have been shown to be essential for mammalian development via placenta formation and the subsequent maintenance of its placental function, respectively (Ono et al., 2006; Sekita et al., 2008). As PEG10 is conserved in all the eutherian and marsupial species, it is a therian-specific gene (Suzuki et al., 2007), while PEG11/RTL1 is eutherian-specific (Edwards et al., 2008). All these findings demonstrated that these two domesticated genes are essential in the current mammalian developmental system and indicate that they have been critically involved in the establishment and diversification of viviparous mammals. In other words, these domesticated genes could be major players in the macroevolution of mammals (Kaneko-Ishino and Ishino, 2010).
The concept of macroevolution by such domesticated genes from the LTR retrotransposons/ERVs and the DNA transposons, as well as rewiring gene regulatory networks by non-LTR retrotransposons (Kuwabara et al., 2009; Lynch et al., 2011; Schmidt et al., 2012) is a subject of interest not only to biologists, but also to those in the general public who are interested in biological evolution and the origin of human beings. It is of special interest because it implies the existence of a unique long-term relationship between the transposable elements and the emergence of mammals.
In this review, we introduce the essential role played by PEG10 and PEG11/RTL1 in mammalian development via placenta formation, and summarize the current understanding of domesticated genes from the LTR retrotransposons/ERVs, especially those in the mammalian lineages. We also discuss the critically important role of DNA methylation in the process of retrotransposon domestication.
PEG10 AND PEG11/RTL1 IN MAMMALIAN DEVELOPMENT AND EVOLUTION
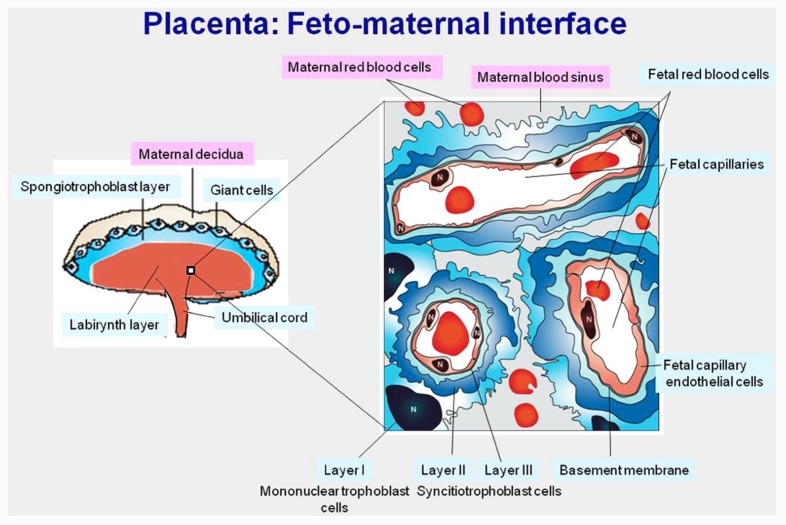
PEG10 and PEG11/RTL1 were identified as paternally expressed genes in the course of an investigation on genomic imprinting (Charlier et al., 2001; Ono et al., 2001). Genomic imprinting is a mammalian-specific epigenetic mechanism regulating the parent-of-origin expression of a subset of specific genes. For these imprinted genes, the two parental alleles are not equivalent: some of the genes are transcribed only from maternally transmitted alleles (maternally expressed genes, MEGs) and the others are transcribed only from paternally transmitted alleles (paternally expressed genes, PEGs; Kaneko-Ishino et al., 2006). Then, genomic imprinting plays an essential role in mammalian development, growth, and behavior via the activity of certain critically important imprinted genes. In mice, there are more than 10 imprinted regions which have been identified, consisting of both PEGs and MEGs. Among them, a proximal region of chromosome 6 is known to cause early embryonic lethality upon maternal duplication, while maternal duplication of a distal region of chromosome 12 causes late embryonic/neonatal lethality associated with growth retardation (Cattanach and Beechey, 1990; see also Genomic imprinting map: http://www.har.mgu.ac.uk/research/genomic_imprinting/). Mouse Peg10 and Peg11/Rtl1 are the major genes responsible for the lethal phenotypes observed in these imprinted regions, respectively (Ono et al., 2006; Sekita et al., 2008). Using knockout mice, we demonstrated that Peg10 and Peg11/Rtl1 play essential roles in early placenta formation and maintenance of the placenta in the mid-to-late stages of gestation, respectively. No labyrinth or spongiotrophoblast formation was observed in the placenta of Peg10 knockout mice. The labyrinth layer is a central part of the mouse placenta in which feto-maternal interactions take place. A large portion of the fetal capillaries exist in the labyrinth layer and allow an exchange of nutrients and gases between maternal and fetal blood cells (Figure 2). Mouse embryos require nutrient supply from the placenta starting on day 9.5 of gestation, therefore, Peg10 KO embryos do not survive beyond this stage.
FIGURE 2.
Mouse mature placenta. Left: mouse placenta is composed of labyrinth, spongiotrophoblast, and giant cell layers. Right: magnified view of the labyrinth layer. In the labyrinth layers, fetal capillaries are surrounded by three layers of trophoblast cells and are bathed in maternal blood, functioning as a site of nutrient and gas exchange between the fetal and maternal blood.
PEG10 encodes two open reading frames exhibiting the highest homology to the Gag and Pol proteins of the sushi-ichi retrotransposon, respectively, and produce two types of proteins, one derived from ORF1 and the other from both ORF1 and 2 (Ono et al., 2001; Volff et al., 2001; Figure 1). The PEG10 protein retains a CCHC RNA-binding motif in the Gag protein and there is a DSG protease domain in the Pol protein. The -1 frameshift mechanism which produces a Gag–Pol fusion protein that is unique to LTR retrotransposons and ERVs is conserved in PEG10, providing strong evidence for its origin from an LTR retrotransposon (Ono et al., 2001; Shigemoto et al., 2001; Manktelow et al., 2005). The biochemical function of the PEG10 protein has yet to be elucidated. However, it was reported that PEG10 is highly expressed in a great majority of hepatocellular carcinomas and confers oncogenic activity. Furthermore, the PEG10 protein is reportedly associated with a member of the “seven in absentia homolog” family (SIAH1 protein) that acts as a mediator of apoptosis. Overexpression of PEG10 decreased the cell death mediated by SIAH1, suggesting that PEG10 has a growth promoting function related to apoptosis in somatic cells (Okabe et al., 2003).
The genomic record shows that PEG10 is conserved in the eutherian and marsupial mammals among the vertebrates (Suzuki et al., 2007). As the placenta is an organ unique to the viviparous reproduction system in these two mammalian groups, it is clearly evident that this gene domesticated from the LTR retrotransposon contributed to the establishment of the current developmental systems of viviparous mammalian groups as a positively selected gene (Suzuki et al., 2007; Kaneko-Ishino and Ishino, 2010). Thus, PEG10 is a very good example of Darwinian evolution and natural selection at work in a macroevolutionary process beyond the individual species which led to the establishment of a subclass of mammals, the therians (Figure 3).
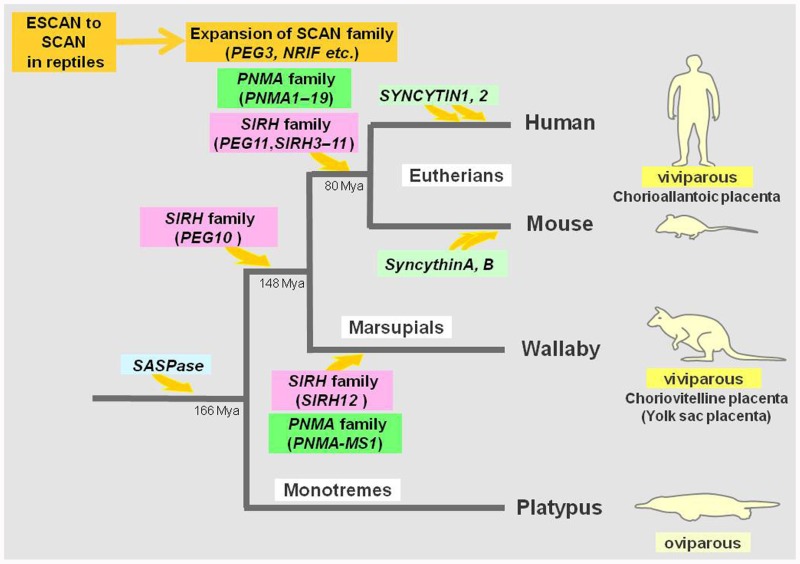
FIGURE 3.
Domestication from LTR retrotransposons and ERVs in mammals. The acquisition of SASPase occurred in a common mammalian ancestor. PEG10 was domesticated in a common therian ancestor while PEG11/RTL1, SIRH3–11, and PNMA1–19 were domesticated in a common eutherian ancestor with subsequent loss of some of the PNMA genes in rodents. The ESCAN domain was domesticated in lower vertebrates and its transition to the SCAN domain took place by combining with the zinc finger and/or KRAB motifs which had already occurred in certain reptiles. In any event, the expansion of SCAN family is obvious in the eutherians. SIRH12 and PNMA-MS1 are derived from marsupialspecific domestication events. The SYNCYTINs were independently recruited in several different mammalian lineages. The eutherians and marsupials are viviparous, having chorioallantoic and choriovitelline placentas (yolk sac placentas), respectively. Both PEG10 and PEG11/RTL1 are essential for the proper formation of efficient chorioallantoic placentas.
The PEG11/RTL1 protein also possesses homology to both the Gag and Pol proteins, including the DSG protease domain in the latter, although no frameshift is required in this case (Charlier et al., 2001). The amino acid sequence homology between the PEG10 and PEG11/RTL1 proteins is approximately 20–30%, indicating their different functions. Mouse Peg11/Rtl1 knockout clearly showed that Peg11/Rtl1 has both a different role from Peg10 and is essential for the maintenance of placental function in the mid-to-late fetal stages (Sekita et al., 2008). Peg11/Rtl1 is expressed in endothelial cells (of extraembryonic mesoderm lineage) of the fetal capillaries in the labyrinth layer, in contrast with Peg10, which is expressed in the labyrinth and spongiotrophoblast cells (of extraembryonic endoderm lineage; Figure 2). As mentioned above, the fetal capillary is the place where feto-maternal interaction occurs. The loss of Peg11/Rtl1 causes clogging in many of the fetal capillaries in the labyrinth layer because of the phagocytosis of endothelial cells carried out by the surrounding trophoblast cells. The Peg11/Rtl1 protein may protect endothelial cells against placental trophoblast cells, which have a highly invasive and hence dangerous nature, although its biochemical function awaits demonstration. It should be noted that the loss and overexpression of PEG11/RTL1 are thought to attribute to the etiology of two different human imprinted diseases, maternal and paternal disomies of human chromosome 14 (matUPD14 and patUPD14), where PEG11/RTL1 is located, respectively (Kagami et al., 2008). In these cases, PEG11/RTL1 plays a major role, not only in placental function, but also in fetal and postnatal growth.
PEG11/RTL1 is conserved in all eutherian mammals but is absent from marsupial mammals, and is, therefore, a eutherian-specific gene (Edwards et al., 2008). Marsupials use a choriovitelline placenta (yolk sac placenta), which is different from the eutherian chorioallantoic placenta and give birth to their young after a very short gestation period compared to the eutherians (Renfree, 2010). PEG11/RTL1 function is necessary for the latter to complete their longer gestational period. Therefore, it is probable that PEG11/RTL1 has a role in the reproduction system of eutherians, which have the chorioallantoic placenta and that it thus contributed to the diversification of these two viviparous mammalian groups. We can say that PEG11/RTL1 provides another good example of macroevolution in mammals (i.e., the establishment of an infraclass of mammals, the eutherians) by domesticated genes from LTR retrotransposons (Kaneko-Ishino and Ishino, 2010; Figure 3).
OTHER SIRH FAMILY GENES DERIVING FROM THE SUSHI-ICHI-RELATED RETROTRANSPOSON
PEG10 and PEG11/RTL1 belong to a sushi-ichi-related retrotransposon homolog (SIRH) family consisting of 12 genes, including these two genes as SIRH1 and SIRH2, respectively (Figure 2; Ono et al., 2006). It is also called the mammalian-specific retrotransposon transcripts (MART; Brandt et al., 2005a,b) or SUSHI family (Youngson et al., 2005). The SIRH1–11 genes are conserved in the eutherian species but no marsupial orthologs have been found, yet nevertheless, SIRH12 is derived from a marsupial-specific insertion event (Ono et al., 2011; Figure 3). Among the SIRH family genes, PEG10 (SIRH1), PEG11/RTL1 (SIRH2), and SIRH9 share homology to both the Gag and Pol proteins, while all the others bear homology only to the Gag protein, but encode proteins of more than 100 amino acid sequences (Brandt et al., 2005a,b; Youngson et al., 2005; Campillos et al., 2006; Ono et al., 2006).
SIRH12 is present in the tammar wallaby, an Australian marsupial species, but its amino acid sequence is degenerated in the gray short-tailed opossum, a South American marsupial species, suggesting that it is only functional in the former (Ono et al., 2011). No ortholog has been reported in any eutherian species in the corresponding region between the ectodermal-neural cortex (ENC1) and rho-guanine nucleotide exchange factor (RGNEF) genes where wallaby SIRH12 and opossum pseudo SIRH12 are located in the marsupial genome. These findings demonstrate that the SIRH family of genes appeared mainly around the establishment of the therian mammals, one (PEG10) before and all the others after the split of the marsupials and eutherians (Figure 3). Consequently, the eutherians and the marsupials have a different set of SIRH family genes except for PEG10. It is likely that SIRH3–11 and SIRH12 also have roles in the eutherian and marsupial developmental and reproductive systems as well as PEG10 and PEG11. As they are expressed in the brain, ovary, and testis as well as the placenta, they may be related to ovulation, gestation, delivery, and/or maternal nursing behaviors, including lactation, as well as placenta formation. Their respective functions are now under investigation using knockout mice.
PNMA-FAMILY GENES FROM THE Gypsy12_DR-RELATED LTR RETROTRANSPOSON
A paraneoplastic Ma antigen (PNMA) family is another large family consisting of eutherian- and marsupial-specific genes (Schüller, 2005; Iwasaki et al., in preparation; Figure 3). PNMA-family genes (PNMA1–3) were first identified as genes encoding neuronal auto-antigens using sera from human patients with paraneoplastic neurological syndromes (Voltz et al., 1999; Rosenfeld et al., 2001). By comprehensive search of a protein database, Schüller (2005) identified additional human PNMA genes, MOAP1/PNMA4, PNMA5, and PNMA6, among which PNMA6 has no mouse ortholog. Campillos et al. (2006) performed a genome-wide search for PNMA genes and identified a total 15 genes and 1 pseudogene in humans. They also showed that all of the PNMA genes were related to a Gypsy12_DR-related Gag protein group of the Ty3/Gypsy LTR retrotransposons isolated from zebrafish (Danio rerio) and that there was no Gypsy12_DR-derived sequences in birds. Recently, Iwasaki and colleagues identified novel PNMA genes by a re-examination of the entire mouse and human entire genome sequences and the PNMA-family genes found thus far number 15 and 19 in mice and humans, respectively; all of these genes have Gag-like proteins, but none are homologous to the Pol protein. The difference in number may be due to the rodent-specific deletion of the PNMA6A–6D genes on X chromosome (Schüller, 2005; Iwasaki et al., in preparation).
No knockout mouse studies on PNMA genes have been reported, but there are reports indicating that these genes are involved in important biological pathways and related to human diseases. PNMA4-deficient cells exhibit aggressive anchorage-independent growth, suggesting that PNMA4 has an important role in regulating apoptosis signaling in a strict temporal manner in mammalian cells, because the PNMA4/MOAP-1 protein is short-lived and constitutively degraded by the ubiquitin–proteasome system (Lee et al., 2009).
Cho et al. (2008a), 2011 reported PNMA10 to be a candidate gene for X-linked mental retardation (XLMR) in humans. In mice, Pnma10/Zcchc12 is expressed in the embryonic ventral forebrain in a cholinergic–neuron-specific manner (Cho et al., 2011), and is known to act as a transcriptional co-activator for bone morphogenic protein (BMP) signaling by binding to the SMAD family of proteins (Cho et al., 2008b). Therefore, it is likely that PNMA10 is related to the evolution of brain function in mammals.
Recently, Pnma14/CCDC8 was suggested to be one of the genes responsible for 3-M syndrome (Hanson et al., 2011a,b). 3-M syndrome is an autosomal-recessive disease characterized by severe postnatal growth restriction, leading to a significantly diminished stature. CULLIN7 (CUL7) and Obsculin-like 1 (OBSL1) are both related to the transcription of insulin-like growth factor-binding protein (IGFBP) genes and have been identified as two of the genes involved in 3-M syndrome (Huber et al., 2005; Hanson et al., 2009). Importantly, the OBSL1 protein interacts with both the PNMA14/CCDC8 and CUL7 proteins, indicating that this protein complex is responsible for the growth retardation observed in 3-M patients. These findings suggest that PNMA genes play an important role in the eutherian development and growth that are impacted by human diseases.
We have recently identified two marsupial-specific PNMA genes, PNMA-MS1 and -MS2 (Iwasaki et al., in preparation; Figure 3). PNMA-MS1 exists in the same genome location in both the Australian (tammar wallaby) and South American (gray short-tailed opossum) marsupial species, but no orthologs exist in the eutherians, suggesting that they are derived from a marsupial-specific domestication event similar to that of SIRH12. PNMA-MS2 was only found in the opossum because there is a gap in the corresponding region of the wallaby genome sequence. However, it is clear that no ortholog exists in any eutherian species. Thus, it is also clear that PNMA-family genes were independently domesticated in the eutherian and the marsupial lineages, and may have certain eutherian- and marsupial-specific functions, respectively.
THE RETROVIRAL-LIKE ASPARTIC PROTEASE SASPase IS CONSERVED IN MAMMALS
Skin aspartic protease (SASPase), which is known a retroviral-like aspartic protease (Bernard et al., 2005), plays a key role in determining the texture of skin by modulating the degree of hydration via the processing of profilaggrin (Matsui et al., 2006, 2011; Barker et al., 2007). SASPase is a single gene conserved in the eutherians, marsupials, and presumably the monotremes (Matsui, personal communication), and thus is a mammalian-specific gene (Figure 3). The profilaggrin protein comprises a tandem array of filaggrin monomers and the SASPase is its specific protease which produces the filaggrin monomer (Matsui et al., 2011). The Filaggrin gene has recently been identified to be etiologically responsible for atopic dermatitis (Barker et al., 2007). Interestingly, both SASPase and profilaggrin are unique to mammals and expressed exclusively in the stratified epithelia in skin. Therefore, it is highly likely that they contributed to the establishment of the mammalian-specific skin barrier system. Aberrant SASPase expression in transgenic mice reportedly leads to impaired skin regeneration and skin remodeling after cutaneous injury or chemically induced hyperplasia (Hildenbrand et al., 2010), and SASPase-deficient mice exhibit fine wrinkles on the sides of the adult body (Matsui et al., 2006).
SCAN-FAMILY GENES RAPIDLY EXPANDED IN THE COURSE OF EUTHERIAN EVOLUTION
The SCAN-family is not a mammalian-specific gene family because its ancestral form exists in non-mammalian vertebrates, but nevertheless, an enormous expansion occurred in the eutherian species (Figure 3). The SCAN motif consists of only a C-terminal portion of the Gag capsid (CA) protein and, in mammals, it always accompanied by multiple C2H2 zinc finger motifs and/or Krüppel-associated box (KRAB) domains neither of which is of retrotransposon origin. It is suggested that the former part was already domesticated at or near the root of the tetrapod animal branch from a full-length CA gene derived from a Gmr1-like retrotransposon. This is called the extended SCAN (ESCAN) domain and that either it or its truncated SCAN motif combined with the zinc finger and/or KRAB motifs in the Anolis lizard (Emerson and Thomas, 2011). Approximately, 60 and 40 genes are known in humans and mice, respectively, and some of them are involved in development and differentiation as transcription factors, such as ZNF202, ZNF197, ZNF444, ZNF274 (neurotrophin receptor interacting factor, NRIF), Zfp496 (NSD1-interacting zinc finger protein 1, Nizp1) and Zfp263 (NT2; Edelstein and Collins, 2005).
Therefore, it is highly likely that some of the SCAN-family genes are related to certain eutherian-specific functions. One example is paternally expressed gene 3 (PEG3; Kuroiwa et al., 1996) that was reported to be essential for maternal nursing behavior as well as promoting embryonic growth (Li et al., 1999). The PEG3 protein has very unique structural features among C2H2 zinc finger proteins, such as amino acid sequences for 11 C2H2 zinc finger motifs and a wider spacing of these motifs. The C2H2 zinc finger proteins comprise the largest class of eukaryotic transcription factors, yet no other C2H2 zinc finger proteins have such features (Kuroiwa et al., 1996). PEG3 is widely expressed during fetal development of mice, and strongly in adult neurons and skeletal muscle. The Peg3 KO offspring are approximately 20% smaller at birth, with markedly reduced nursing behavior and a reduced number of oxytocin-positive neurons in the hypothalamus of Peg3 KO females (Li et al., 1999). Human PEG3 has tumor-suppressing activity in glioma cell lines by its capacity to inhibit Wnt signaling, and the loss of its expression is reportedly observed in gliomas (Kohda et al., 2001; Maekawa et al., 2004; Jiang et al., 2010).
INDEPENDENT DOMESTICATION EVENTS OF THE SYNCYTINs IN DIFFERENT LINEAGES IN EUTHERIANS
As mentioned in Section “Introduction,” SYNCYTIN was first discovered in humans (Blond et al., 2000; Mi et al., 2000). Although there are many Env-related DNA sequences in the human genome, only two exhibit fusogenic activity in cell fusion assays and now these are called SYNCYTIN1 and 2 (Blaise et al., 2003). They are derived from different human-specific ERVs, HERV-W, and HERV-FRD, and became integrated into a primate lineage 25 and >40 MYA, respectively (Figure 3). Recent studies demonstrated that similar genes exist in an order- or family-specific manner in several mammalian lineages, i.e., producing syncytiotrophoblast cells by cell fusion in the placenta. Mice also have two Syncytin genes, SyncytinA and B, derived from Muridae family-specific integrations of HERV-F/H-related ERV(s) approximately 20 MYA (Dupressoir et al., 2005; Figure 3), and rabbits (Oryctolagus cuniculus) have another SYNCYTIN-Ory1 from Leporidae family-specific integration of a different type-D retrovirus 12–30 MYA (Heidmann et al., 2009). Therefore, at least three independent domestication events have been confirmed in the eutherians, indicating that domestication from ERVs which were actively functioning during the time of mammalian radiation.
SyncytinA knockout mice exhibit mid-fetal lethality because of the structural abnormality of the placenta (Dupressoir et al., 2009), and double knockout of both SyncytinA and B causes an even more severe phenotype, early embryonic lethality (Dupressoir et al., 2011). Among the eutherians, placental morphology and functions are quite substantially diverged. Therefore, it is very interesting that the SYNCYTINs from the ERVs appear to have important roles in the placenta that they play in an order- or family-specific manner, while PEG10 and PEG11/RTL1 from the LTR retrotransposons are conserved in the therians and eutherians, respectively, and presumably have contributed to the establishment of the basic structure of viviparous reproductive systems in the current eutherian species.
RESISTANCE TO VIRAL INFECTION BY DOMESTICATED VIRAL GENES
ERVs have long been thought to confer resistance to infection by exogenous retroviruses. Well-known examples are Friend virus susceptibility 1 and 4 (Fv1 and Fv4) and resistance to mink cell focus-forming (MCF) virus (Rmcf) genes, which exhibit resistance to murine leukemia viruses (MuLVs) in mice (Pincus et al., 1971; Suzuki, 1975; Hartley et al., 1983). Fv1 is derived from the Gag region of an ancient MERV-L element (Best et al., 1996; Bénit et al., 1997), whereas Fv4 and Rmcf consist of intact Env genes, the expression of which prevents infection via receptor interference (Ikeda et al., 1985; Lyu and Kozak, 1996; Taylor et al., 2001; Jung et al., 2002). Endogenous betaretroviruses (enJSRVs) in sheep are another example (Dunlap et al., 2005). The ovine genome possesses approximately 20 copies of enJSRVs that are highly related to two exogenous oncogenic viruses, Jaagsiekte sheep retrovirus (JSRV) and enzootic nasal tumor virus. It has been proposed that the enJSRVs Env genes are beneficial to the host and help protect of the uterus from viral infection and act as regulators of placental morphogenesis and function. They exist as species- or strain-specific genes, meaning that they are derived from recent domestication events compared to the SIRH-, PNMA-, and SCAN-family genes as well as the SASPase gene.
Therefore, it is clear that the domestication from LTR retrotransposons and ERVs has a very long history, dating from around the time of the establishment of vertebrates, on through the establishment and diversification of mammals and ultimately to the radiation of each mammalian species. Koala retrovirus (KoRV) has recently been reported to cause leukemia, lymphoma, and immunosuppression in the Australian Koala population (Tarlinton et al., 2006, 2008; Stoye, 2006). Interestingly, KoRV is currently undergoing endogenization and it is likely that it entered the koala genome within the last 200 years. Therefore, retrotransposon endogenization may be a fairly ordinary process in the long course of evolution, and novel genes may continue to appear by this mechanism in the future.
GENE DUPLICATION OF DOMESTICATED GENES
Although dozens of domesticated genes have been found in mammals, this does not necessarily mean that independent domestication events have happened as often as the number of domesticated genes. Certain domesticated genes have apparently been produced by the gene duplication of an originally domesticated gene, such as in the SCAN family of genes. The SCAN domain was domesticated long before the emergence of mammals in the lower vertebrates (ESCAN) and then a new combination of this domain and zinc finger and/or KRAB motifs produced the SCAN-family gene prototype in a reptile, and its expansion occurred during radiation of the eutherians (Emerson and Thomas, 2011; Figure 3).
SIRH4, 5, and 6 as well as PNMA6A, 6B, 6C, and 6D, are other clear examples of gene duplication. The domestication of the original gene must have occurred in the ancestral eutherian mammals, but these clusters were produced by gene duplication because they encode very nearly the same coding frames. It is interesting to elucidate whether they are in the process of diversifying into genes with different functions or there is some as yet unknown reason for them to multiply and increase their copy numbers in this way. Nevertheless, as discussed above, at least two independent domestication events occurred in the eutherians and the marsupials in the SIRH and PNMA families, and at least four independent domestications have been confirmed in three different eutherian lineages in the case of the SYNCYTINs (Figure 3).
THE ESSENTIAL ROLE OF DNA METHYLATION IN THE DOMESTICATION PROCESS
Retrotransposons are potentially harmful to host organisms because their integration not only causes genetic diseases by disrupting essential genes, but also induces chromosomal deletion as well as recombination by DNA homologous recombination between the two of them. Their integration could also disturb transcription of neighboring genes. Thus, host organisms must prevent any further propagation that would result in an accumulation of new insertion events by regulating their transcription. How are they critically silenced and yet stably inherited from generation to generation in a manner similar to endogenous DNA sequences in the host genome?
Mammals have adopted certain defense mechanisms against them, such as DNA methylation and histone modifications (Rowe and Trono, 2011). The integrated retrotransposons are usually heavily DNA methylated and transcriptionally silenced in almost all somatic cells. They have the character of being neutral genes in the mammalian genome. According to the neutral theory of molecular evolution proposed by Kimura (1968), 1983, such neutral mutations are fixed in a population by the mechanism of random drift. Ohta (2002) proposed in her “nearly neutral theory” as an extension of neutral evolution that less harmful mutations could become fixed in a population if the population size were sufficiently small). We previously proposed the hypothesis that in the course of retrotransposon domestication the neutral or nearly neutral evolution preceded Darwinian evolution and helped supply novel genes for novel purposes from the integrated retrotransposons (Kaneko-Ishino et al., 2006; Kaneko-Ishino and Ishino, 2010). In brief, we assume that either neutral or nearly neutral evolution played essential background roles by both inactivating and neutralizing integrated retrotransposons. Subsequently, their gradual conversion from silenced harmful genes to slightly advantageous genes took place as the result of multiple mutations. A loss of such silencing, at least in a subset of tissues, was ultimately required for the “new gene” to have a certain function. Darwinian forces then came into play, and by natural selection certain of these genes became more usefully functional and thus advantageous for the host organisms. It should be noted that in extraembryonic organs, such as the yolk sac and placenta in mammals, the DNA methylation levels are lower than those in other embryonic and adult tissues. Therefore, a leaky expression of retrotransposons and retroviruses constantly occurs. In this situation, the integrated retrotransposons and their subsequent mutated forms would be less harmful. However, in the case of advantageous mutations, a swift transition from the state of nearly neutral evolution to that of Darwinian evolution would take place. In this regard, the extraembryonic tissues might have been a site of retrotransposon domestication during the course of mammalian evolution, which is consistent with the fact that the domesticated PEG10, PEG11/RTL1, and SYNCYTIN genes play essential roles in the placenta (Kaneko-Ishino and Ishino, 2010).
In this hypothetical scenario, various epigenetic mechanisms, such as DNA methylation and/or histone modification, might have played a critical role. In mammals, DNMT1 is the essential maintenance DNA methyltransferase and the loss of its activity causes early embryonic lethality associated with overexpression of IAP retrovirus (Li et al., 1992; Walsh et al., 1998). The two de novo DNA methyltransferase DNMT3A and DNMT3B are also essential for mammalian development, and the loss of their activities causes lethality in the postnatal and embryonic period in mice, respectively (Okano et al., 1999). Overexpression of IAP retrovirus was also observed in Dnmt3a and 3b double knockout mice, although to a lesser degree than Dnmt1 KO mice (Okano et al., 1999). DNMT3L does not have DNA methyltransferase activity itself, but has an essential function of producing a different DNA methylation status in female- and male-derived genomic DNA in the process of establishing the genomic imprinting memories associated with DNMT3A (Bourc’his et al., 2001; Hata et al., 2002). It is known that this complex is also essential for retrotransposon methylation in the paternal germ line (Bourc’his et al., 2001; Bourc’his and Bestor, 2004). The coincident emergence of DNMT3L in the therian mammals is highly suggestive, both for the origin of the genomic imprinting mechanism as well as the abundance of LTR retrotransposons/ERVs, each of which is specific to the therian genome (Yokomine et al., 2006). It should be noted that H3K9 methyltransferase ERG-associated protein with SET domain (ESET, also called SETDB1) coupled with KRAB-associated protein 1 (KAP1, also called TRIM28) and zinc finger protein ZFP806 is required for H3K9 trimethylation as well as the repression of the retrotransposons and ERVs in undifferentiated mouse ES cells (Wolf and Goff, 2007, 2009; Matsui et al., 2010). Such a DNA methylation-independent pathway may be necessary, because DNA methylation is dynamically reprogrammed during the early embryonic period in mammals.
Finally, we would like to consider how the mammalian viviparous reproductive system originally started using the retrotransposon-derived PEG10 gene. If this new reproductive system first happened in a single individual, was it possible for such an individual to survive and propagate his or her offspring? It is worth mentioning that the nearly neutral theory of molecular evolution can also explain how new species originated not from a single individual, but rather from a population subset (Kimura, 1983). Preadaptive mutations were already distributed in a neutral manner. Adaptive functions emerged under the selective pressures of a new environment. This suggests the neutral evolution process could also play a role as an “evolutionary capacitor,” as predicted in the case of heat shock protein (Hsp) 90, where genotypic variations in other genes are masked and therefore are accumulated without causing any evident phenotypic changes in the chaperone activity of Hsp90 per se (Rutherford and Lindquist, 1998; Bergman and Siegel, 2003). However, this original scenario has recently come under challenge because Hsp90 also acts as a suppressor of retrotransposons and its mutation induces retrotransposon transposition, thus causing a number of secondary mutations (Specchia et al., 2010; Gangaraju et al., 2011).
DNA methylation is commonly observed in a wide range of organisms, from bacteria to plants and animals, although certain model organisms do in fact lack this feature. We propose that changes in DNA methylation in genome regulation systems gave rise to the great diversity of the organisms across the earth. In particular, as mammals developed their particularly specialized DNA methylation system, mammalian evolution was advanced by a series of retrotransposon domestication events. Retrotransposons serve as a double-edged sword in development and evolution, i.e., either harmful or beneficial depending on which time scale is used. The domestication of retrotransposons seems likely to be a very rare event, but once it has taken place, its impact is profound, which is especially the case in mammalian evolution. That may provide the raison d’etre for the LTR retrotransposons in the mammalian genome.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all the collaborators and all the laboratory members, especially, Ryuichi Ono, Yoichi Sekita, Shunsuke Suzuki, and Takashi Kohda for analyzing Peg10 and Peg11 KO mice and marsupial PEG10 and Marilyn Renfree and Jeniffer Graves for several marsupial researches. The work has long been supported by a number of grants, Grants-in-Aid for Scientific Research (S) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan, Funding Program for Next Generation World-Leading Researchers (NEXT Program; to Tomoko Kaneko-Ishino), Bilateral Program on Joint Research Project (to Fumitoshi Ishino), and Creative Science Research (to Fumitoshi Ishino and Tomoko Kaneko-Ishino) from the Japan Society for the Promotion of Science (JSPS), Asahi Glass Foundation (to Tomoko Kaneko-Ishino) and Mitsubishi foundation (to Fumitoshi Ishino). Pacific Edit reviewed the manuscript prior to submission.
REFERENCES
- Agrawal A., Eastman Q. M., Schatz D. G. (1998). Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 394 744–751 [DOI] [PubMed] [Google Scholar]
- Barker J. N., Palmer C. N., Zhao Y., Liao H., Hull P. R., Lee S. P., Allen M. H., Meggitt S. J., Reynolds N. J., Trembath R. C., McLean W. H. (2007). Null mutations in the filaggrin gene (FLG) determine major susceptibility to early-onset atopic dermatitis that persists into adulthood. J. Invest. Dermatol. 127 564–567 [DOI] [PubMed] [Google Scholar]
- Bénit L., De Parseval N., Casella J. F., Callebaut I., Cordonnier A., Heidmann T. (1997). Cloning of a new murine endogenous retrovirus, MuERV-L, with strong similarity to the human HERV-L element and with a gag coding sequence closely related to the Fv1 restriction gene. J. Virol. 71 5652–5657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman A., Siegel M. L. (2003). Evolutionary capacitance as a general feature of complex gene networks. Nature 424 549–552 [DOI] [PubMed] [Google Scholar]
- Bernard D., Méhul B., Thomas-Collignon A., Delattre C., Donovan M., Schmidt R. (2005). Identification and characterization of a novel retroviral-like aspartic protease specifically expressed in human epidermis. J. Invest. Dermatol. 125 278–287 [DOI] [PubMed] [Google Scholar]
- Best S., Le Tissier P., Towers G., Stoye J. P. (1996). Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382 826–829 [DOI] [PubMed] [Google Scholar]
- Blaise S., de Parseval N., Bénit L., Heidmann T. (2003). Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl. Acad. Sci. U.S.A. 100 13013–13018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blond J.-L., Lavillette D., Cheynet V., Bouton O., Oriol G., Chapel-Fernandes S., Mandrand B., Mallet F., Cosset F. L. (2000). An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J. Virol. 74 3321–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc’his D., Bestor T. H. (2004). Transposon silencing and imprint establishment in mammalian germ cells. Cold Spring Harb. Symp. Quant. Biol. 69 381–388 [DOI] [PubMed] [Google Scholar]
- Bourc’his D., Xu G. L., Lin C. S., Bollman B., Bestor T. H. (2001). Dnmt3L and the establishment of maternal genomic imprinting. Science 294 2536–2539 [DOI] [PubMed] [Google Scholar]
- Brandt J., Veith A. M., Volff J. N. (2005a). A family of neofunctionalized Ty3/gypsy retrotransposon genes in mammalian genomes. Cytogenet. Genome Res. 110 307–317 [DOI] [PubMed] [Google Scholar]
- Brandt J., Schrauth S., Veith A. M., Froschauer A., Haneke T., Schultheis C., Gessler M., Leimeister C., Volff J. N. (2005b). Transposable elements as a source of genetic innovation: expression and evolution of a family of retrotransposon-derived neogenes in mammals. Gene 345 101–111 [DOI] [PubMed] [Google Scholar]
- Brosius J., Gould S. J. (1992). On “genomenclature”: a comprehensive (and respectful) taxonomy for pseudogenes and other “junk DNA”. Proc. Natl. Acad. Sci. U.S.A. 89 10706–10710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campillos M., Doerks T., Shah P. K., Bork P. (2006). Computational characterization of multiple Gag-like human proteins. Trends Genet. 22 585–589 [DOI] [PubMed] [Google Scholar]
- Casola C., Hucks D., Feschotte C. (2008). Convergent domestication of pogo-like transposases into centromere-binding proteins in fission yeast and mammals. Mol. Biol. Evol. 25 29–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattanach B. M., Beechey C. V. (1990). Autosomal and X-chromosome imprinting. Dev. Suppl. 1990 63–72 [PubMed] [Google Scholar]
- Charlier C., Segers K., Wagenaar D., Karim L., Berghmans S., Jaillon O., Shay T., Weissenbach J., Cockett N., Gyapay G., Georges M. (2001). Human–ovine comparative sequencing of a 250-kb imprinted domain encompassing the callipyge (clpg) locus and identification of six imprinted transcripts: DLK1, DAT, GTL2, PEG11, antiPEG11, and MEG8. Genome Res. 11 850–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho G., Bhat S. S., Gao J., Collins J. S., Rogers R. C., Simensen R. J., Schwartz C. E., Golden J. A., Srivastava A. K. (2008a). Evidence that SIZN1 is a candidate X-linked mental retardation gene. Am. J. Med. Genet. A 146A 2644–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho G., Lim Y., Zand D., Golden J. A. (2008b). Sizn1 is a novel protein that functions as a transcriptional coactivator of bone morphogenic protein signaling. Mol. Cell. Biol. 28 1565–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho G., Lim Y., Golden J. A. (2011). XLMR candidate mouse gene, Zcchc12 (Sizn1) is a novel marker of Cajal–Retzius cells. Gene Expr. Patterns 11 216–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap K. A., Palmarini M., Adelson D. L., Spencer T. E. (2005). Sheep endogenous betaretroviruses (enJSRVs) and the hyaluronidase 2 (HYAL2) receptor in the ovine uterus and conceptus. Biol. Reprod. 73 271–279 [DOI] [PubMed] [Google Scholar]
- Dupressoir A., Marceau G., Vernochet C., Bénit L., Kanellopoulos C., Sapin V., Heidmann T. (2005). Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc. Natl. Acad. Sci. U.S.A. 102 725–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A., Vernochet C., Bawa O., Harper F., Pierron G., Opolon P., Heidmann T. (2009). Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc. Natl. Acad. Sci. U.S.A. 106 12127–12132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A., Vernochet C., Harper F., Guégan J., Dessen P., Pierron G., Heidmann T. (2011). A pair of co-opted retroviral envelope syncytin genes is required for formation of the two-layered murine placental syncytiotrophoblast. Proc. Natl. Acad. Sci. U.S.A. 108 1164–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein L. C., Collins T. (2005). The SCAN domain family of zinc finger transcription factors. Gene 359 1–17 [DOI] [PubMed] [Google Scholar]
- Edwards C. A., Mungall A. J., Matthews L., Ryder E., Gray D. J., Pask A. J., Shaw G., Graves J. A., Rogers J., SAVOIR Consortium, Dunham I., Renfree M. B., Ferguson-Smith A. C. (2008). The evolution of the DLK1-DIO3 imprinted domain in mammals. PLoS Biol. 6 e135 10.1371/journal.pbio.0060135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson R. O., Thomas J. H. (2011). Gypsy and the birth of the SCAN domain. J. Virol. 85 12043–12053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangaraju V. K., Yin H., Weiner M. M., Wang J., Huang X. A., Lin H. (2011). Drosophila Piwi functions in Hsp90-mediated suppression of phenotypic variation. Nat. Genet. 43 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson D., Murray P. G., O’Sullivan J., Urquhart J., Daly S., Bhaskar S. S., Biesecker L. G., Skae M., Smith C., Cole T., Kirk J., Chandler K., Kingston H., Donnai D., Clayton P. E., Black G. C. (2011a). Exome sequencing identifies CCDC8 mutations in 3-M syndrome, suggesting that CCDC8 contributes in a pathway with CUL7 and OBSL1 to control human growth. Am. J. Hum. Genet. 89 148–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson D., Murray P. G., Black G. C., Clayton P. E. (2011b). The genetics of 3-m syndrome: unravelling a potential new regulatory growth pathway. Horm. Res. Paediatr. 76 369–378 [DOI] [PubMed] [Google Scholar]
- Hanson D., Murray P. G., Sud A., Temtamy S. A., Aglan M., Superti-Furga A., Holder S. E., Urquhart J., Hilton E., Manson F. D., Scambler P., Black G. C., Clayton P. E. (2009). The primordial growth disorder 3-M syndrome connects ubiquitination to the cytoskeletal adaptor OBSL1. Am. J. Hum. Genet. 84 801–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley J. W., Yetter R. A, Morse H. C. III. (1983). A mouse gene on chromosome 5 that restricts infectivity of mink cell focus-forming recombinant murine leukemia viruses. J. Exp. Med. 158 16–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata K., Okano M., Lei H., Li E. (2002). Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferase to establish maternal imprints in mice. Development 129 1983–1993 [DOI] [PubMed] [Google Scholar]
- Heidmann O., Vernochet C., Dupressoir A., Heidmann T. (2009). Identification of an endogenous retroviral envelope gene with fusogenic activity and placenta-specific expression in the rabbit: a new “syncytin” in a third order of mammals. Retrovirology 6 107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildenbrand M., Rhiemeier V., Hartenstein B., Lahrmann B., Grabe N., Angel P., Hess J. (2010). Impaired skin regeneration and remodeling after cutaneous injury and chemically induced hyperplasia in taps-transgenic mice. J. Invest. Dermatol. 130 1922–1930 [DOI] [PubMed] [Google Scholar]
- Hiom K., Mele M., Gellert M. (1998). DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell 94 463–470 [DOI] [PubMed] [Google Scholar]
- Huber C., Dias-Santagata D., Glaser A., O’Sullivan J., Brauner R., Wu K., Xu X., Pearce K., Wang R., Uzielli M. L., Dagoneau N., Chemaitilly W., Superti-Furga A., Dos Santos H., Mégarbané A., Morin G., Gillessen-Kaesbach G., Hennekam R., Van der Burgt I., Black G. C., Clayton P. E., Read A., Le Merrer M., Scambler P. J., Munnich A., Pan Z. Q., Winter R., Cormier-Daire V. (2005). Identification of mutations in CUL7 in 3-M syndrome. Nat. Genet. 37 1119–1124 [DOI] [PubMed] [Google Scholar]
- Hudson M. E., Lisch D. R., Quail P. H. (2003). The FHY3 and FAR1 genes encode transposase-related proteins involved in regulation of gene expression by the phytochrome A-signaling pathway. Plant J. 34 453–471 [DOI] [PubMed] [Google Scholar]
- Ikeda H., Laigret F., Martin M. A., Repaske R. (1985). Characterization of a molecularly cloned retroviral sequence associated with Fv-4 resistance. J. Virol. 55 768–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Yu Y., Yang H. W., Agar N. Y., Frado L., Johnson M. D. (2010). The imprinted gene PEG3 inhibits Wnt signaling and regulates glioma growth. J. Biol. Chem. 285 8472–8480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y. T., Lyu M. S., Buckler-White A., Kozak C. A. (2002). Characterization of a polytropic murine leukemia virus proviral sequence associated with the virus resistance gene Rmcf of DBA/2 mice. J. Virol. 76 8218–8224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagami M., Sekita Y., Nishimura G., Irie M., Kato F., Okada M., Yamamori S., Kishimoto H., Nakayama M., Tanaka Y., Matsuoka K., Takahashi T., Noguchi M., Tanaka Y., Masumoto K., Utsunomiya T., Kouzan H., Komatsu Y., Ohashi H., Kurosawa K., Kosaki K., Ferguson-Smith A. C., Ishino F., Ogata T. (2008). Deletions and epimutations affecting the human 14q32.2 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat. Genet. 40 237–242 [DOI] [PubMed] [Google Scholar]
- Kaneko-Ishino T., Ishino F. (2010). Retrotransposon silencing by DNA methylation contributed to the evolution of placentation and genomic imprinting in mammals. Dev. Growth Differ. 52 533–543 [DOI] [PubMed] [Google Scholar]
- Kaneko-Ishino T., Kohda T., Ono R., Ishino F. (2006). Complementation hypothesis: the necessity of a monoallelic gene expression mechanism in mammalian development. Cytogenet. Genome Res. 113 24–30 [DOI] [PubMed] [Google Scholar]
- Kimura M. (1968). Evolutionary rate at the molecular level. Nature 217 624–626 [DOI] [PubMed] [Google Scholar]
- Kimura M. (1983). The Neutral Theory of Molecular Evolution. Cambridge: Cambridge University Press; 305–327 [Google Scholar]
- Kohda T., Asai A., Kuroiwa Y., Kobayashi S., Aisaka K., Nagashima G., Yoshida M. C., Kondo Y., Kagiyama N., Kirino T., Kaneko-Ishino T., Ishino F. (2001). Tumour suppressor activity of human imprinted gene PEG3 in a glioma cell line. Genes Cells 6 237–247 [DOI] [PubMed] [Google Scholar]
- Kuroiwa Y., Kaneko-Ishino T., Kagitani F., Kohda T., Li L-L., Tada M., Suzuki R., Yokoyama M., Shiroishi T., Wakana S., Barton S. C., Ishino F., Surani M. A. (1996). Peg3 imprinted gene on proximal chromosome 7 encodes for a zinc finger protein. Nat. Genet. 12 186–190 [DOI] [PubMed] [Google Scholar]
- Kuwabara T., Hsieh J., Muotri A., Yeo G., Warashina M., Lie D. C., Moore L., Nakashima K., Asashima M., Gage F. H. (2009). Wnt-mediated activation of NeuroD1 and retro-elements during adult neurogenesis. Nat. Neurosci. 12 1097–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. S., Fu N. Y., Sukumaran S. K., Wan K. F., Wan Q., Yu V. C. (2009). TRIM39 is a MOAP-1-binding protein that stabilizes MOAP-1 through inhibition of its poly-ubiquitination process. Exp. Cell Res. 315 1313–1325 [DOI] [PubMed] [Google Scholar]
- Li E., Bestor T. H., Jaenisch R. (1992). Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69 915–926 [DOI] [PubMed] [Google Scholar]
- Li L., Keverne E. B., Aparicio S. A., Ishino F., Barton S. C., Surani M. A. (1999). Regulation of maternal behavior and offspring growth by paternally expressed Peg3. Science 284 330–333 [DOI] [PubMed] [Google Scholar]
- Lin R., Ding L., Casola C., Ripoll D. R., Feschotte C., Wang H. (2007). Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318 1302–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D. R., Freeling M., Langham R. J., Choy M. Y. (2001). Mutator transposase is widespread in the grasses. Plant Physiol. 125 1293–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch V. J., Leclerc R. D., May G., Wagner G. P. (2011). Transposon-mediated rewiring of gene regulatory networks contributed to the evolution of pregnancy in mammals. Nat. Genet. 43 1154–1159 [DOI] [PubMed] [Google Scholar]
- Lyu M. S., Kozak C. A. (1996). Genetic basis for resistance to polytropic murine leukemia viruses in the wild mouse species Mus castaneus. J. Virol. 70 830–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa S., Itaba N., Otsuka. S. Kamitani, H., Watanabe T., Tahimic C. G., Nanba E., Oshimura M. (2004). Coordinate downregulation of a novel imprinted transcript ITUP1 with PEG3 in glioma cell lines. DNA Res. 11 37–49 [DOI] [PubMed] [Google Scholar]
- Manktelow E., Shigemoto K., Brierley I. (2005). Characterization of the frameshift signal of Edr, a mammalian example of programmed -1 ribosomal frameshifting. Nucleic Acids Res. 33 1553–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T., Kinoshita-Ida Y., Hayashi-Kisumi F., Hata M., Matsubara K., Chiba M., Katahira-Tayama S., Morita K., Miyachi Y., Tsukita S. (2006). Mouse homologue of skin-specific retroviral-like aspartic protease involved in wrinkle formation. J. Biol. Chem. 281 27512–27525 [DOI] [PubMed] [Google Scholar]
- Matsui T., Leung D., Miyashita H., Maksakova I. A., Miyachi H., Kimura H., Tachibana M., Lorincz M. C., Shinkai Y. (2010). Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464 927–931 [DOI] [PubMed] [Google Scholar]
- Matsui T., Miyamoto K., Kubo A., Kawasaki H., Ebihara T., Hata K., Tanahashi S., Ichinose S., Imoto I., Inazawa J., Kudoh J., Amagai M. (2011). SASPase regulates stratum corneum hydration through profilaggrin-to-filaggrin processing. EMBO Mol. Med. 3 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S., Lee X., Li X.-P., Veldman G. M., Finnerty H., Racie L., LaVallie E., Tang X.-Y., Edouard P., Howes S., Keith J. C., Jr., McCoy J. M. (2000). Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403 785–789 [DOI] [PubMed] [Google Scholar]
- Nakamura T. M., Cech T. R. (1998). Reversing time: origin of telomerase. Cell 92 587–590 [DOI] [PubMed] [Google Scholar]
- Ohta T. (2002). Near-neutrality in evolution of genes and gene regulation. Proc. Natl. Acad. Sci. U.S.A. 99 16134–16137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe H., Satoh S., Furukawa Y., Kato T., Hasegawa S., Nakajima Y., Yamaoka Y., Nakamura Y. (2003). Involvement of PEG10 in human hepatocellular carcinogenesis through interaction with SIAH1. Cancer Res. 63 3043–3048 [PubMed] [Google Scholar]
- Okano M., Bell D. W., Haber D. A., Li E. (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99 247–257 [DOI] [PubMed] [Google Scholar]
- Ono R., Kobayashi S., Wagatsuma H., Aisaka K., Kohda T., Kaneko-Ishino T., Ishino F. (2001). A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics 73 232–237 [DOI] [PubMed] [Google Scholar]
- Ono R., Kuroki Y., Naruse M., Ishii M., Iwasaki S., Toyoda A., Fujiyama A., Shaw G., Renfree M. B., Kaneko-Ishino T., Ishino F. (2011). Identification of SIRH12, a retrotransposon-derived gene specific to marsupial mammals. DNA Res. 18 211–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono R., Nakamura K., Inoue K., Naruse M., Usami T., Wakisaka-Saito N., Hino T., Suzuki-Migishima R., Ogonuki N., Miki H., Kohda T., Ogura A., Yokoyama M., Kaneko-Ishino T., Ishino F. (2006). Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat. Genet. 38 101–106 [DOI] [PubMed] [Google Scholar]
- Pincus T., Hartley J. W., Rowe W. P. (1971). A major genetic locus affecting resistance to infection with murine leukemia viruses. I. Tissue culture studies of naturally occurring viruses. J. Exp. Med. 133 1219–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfree M. B. (2010). Marsupials: placental mammals with a difference. Placenta 31(Suppl.), 24 S21–S26 [DOI] [PubMed] [Google Scholar]
- Rosenfeld M. R., Eichen J. G., Wade D. F., Posner J. B., Dalmau J. (2001). Molecular and clinical diversity in paraneoplastic immunity to Ma proteins. Ann. Neurol. 50 339–348 [PubMed] [Google Scholar]
- Rowe H. M., Trono D. (2011). Dynamic control of endogenous retroviruses during development. Virology 411 273–287 [DOI] [PubMed] [Google Scholar]
- Rutherford S. L., Lindquist S. (1998). Hsp90 as a capacitor for morphological evolution. Nature 396 335–342 [DOI] [PubMed] [Google Scholar]
- Schmidt D., Schwalie P. C., Wilson M. D., Ballester B., Gonçalves A., Kutter C., Brown G. D., Marshall A., Flicek P., Odom D. T. (2012). Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell 148 335–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüller M., Jenne D., Voltz R. (2005). The human PNMA family: novel neuronal proteins implicated in paraneoplastic neurological disease. J. Neuroimmunol. 169 172–176 [DOI] [PubMed] [Google Scholar]
- Sekita Y., Wagatsuma H., Nakamura K., Ono R., Kagami M., Wakisaka N., Hino T., Suzuki-Migishima R., Kohda T., Ogura A., Ogata T., Yokoyama M., Kaneko-Ishino T., Ishino F. (2008). Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nat. Genet. 40 243–248 [DOI] [PubMed] [Google Scholar]
- Shigemoto K., Brennan J., Walls E., Watson C. J., Stott D., Rigby P. W., Reith A. D. (2001). Identification and characterisation of a developmentally regulated mammalian gene that utilises -1 programmed ribosomal frameshifting. Nucleic Acids Res. 29 4079–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A. F. (1999). Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 9 657–663 [DOI] [PubMed] [Google Scholar]
- Specchia V., Piacentini L., Tritto P., Fanti L., D’Alessandro R., Palumbo G., Pimpinelli S., Bozzetti M. P. (2010). Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature 463 662–665 [DOI] [PubMed] [Google Scholar]
- Stoye J. P. (2006). Koala retrovirus: a genome invasion in real time. Genome Biol. 7 241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S. (1975). Fv-4: a new gene affecting the splenomegaly induction by Friend leukemia virus. Jpn. J. Exp. Med. 45 473–478 [PubMed] [Google Scholar]
- Suzuki S., Ono R., Narita T., Pask A. J., Shaw G., Wang C., Kohda T., Alsop A. E., Graves M. J. A., Kohara Y., Ishino F., Renfree M. B., Kaneko-Ishino T. (2007). Retrotransposon silencing by DNA methylation can drive mammalian genomic imprinting. PLoS Genet. 3 e55 10.1371/journal.pgen.0030055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarlinton R., Meers J., Young P. (2008). Biology and evolution of the endogenous koala retrovirus. Cell. Mol. Life. Sci. 65 3413–3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarlinton R. E., Meers J., Young P. R. (2006). Retroviral invasion of the koala genome. Nature 442 79–81 [DOI] [PubMed] [Google Scholar]
- Taylor G. M., Gao Y., Sanders D. A. (2001). Fv-4: identification of the defect in Env and the mechanism of resistance to ecotropic murine leukemia virus. J. Virol. 75 11244–11248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudor M., Lobocka M., Goodell M., Pettitt J, O’Hare K. (1992). The pogo transposable element family of Drosophila melanogaster. Mol. Gen. Genet. 232 126–134 [DOI] [PubMed] [Google Scholar]
- Volff J., Körting C., Schartl M. (2001). Ty3/Gypsy retrotransposon fossils in mammalian genomes: did they evolve into new cellular functions? Mol. Biol. Evol. 18 266–270 [DOI] [PubMed] [Google Scholar]
- Voltz R., Gultekin S. H., Rosenfeld M. R., Gerstner E., Eichen J., Posner J. B., Dalmau J. (1999). A serologic marker of paraneoplastic limbic and brain-stem encephalitis in patients with testicular cancer. N. Engl. J. Med. 340 1788–1795 [DOI] [PubMed] [Google Scholar]
- Walsh C. P., Chaillet J. R., Bestor T. H. (1998). Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20 116–117 [DOI] [PubMed] [Google Scholar]
- Wolf D., Goff S. P. (2007). TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131 46–57 [DOI] [PubMed] [Google Scholar]
- Wolf D., Goff S. P. (2009). Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458 1201–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomine T., Hata K., Tsudzuki M., Sasaki H. (2006). Evolution of the vertebrate DNMT3 gene family: a possible link between existence of DNMT3L and genomic imprinting. Cytogenet. Genome Res. 113 75–80 [DOI] [PubMed] [Google Scholar]
- Youngson N. A., Kocialkowski S., Peel N., Ferguson-Smith A. C. (2005). A small family of sushi-class retrotransposon-derived genes in mammals and their relation to genomic imprinting. J. Mol. Evol. 61 481–490 [DOI] [PubMed] [Google Scholar]