Abstract
The blastoderm stage of Drosophila embryogenesis is a time of crucial transitions in RNA transcription, the cell cycle and segment determination. We have previously identified three loci encoding RNAs specific to this stage (Roark et al., Dev. Biol. 109, 476-488, 1985). We present here the complete nucleotide sequence of one of these loci, bsg25D, which encodes a 2.7 kb blastoderm-specific RNA. The primary structure of this RNA, and that of an overlapping 4.5 kb RNA, has been determined. The amino acid sequence of the predicted bsg25D protein has been compared to the NBRF protein database. Structural similarities between domains in the bsg25D, fos, and tropomyosin proteins, and their possible significance for early embryogenesis are discussed.
Full text
PDF


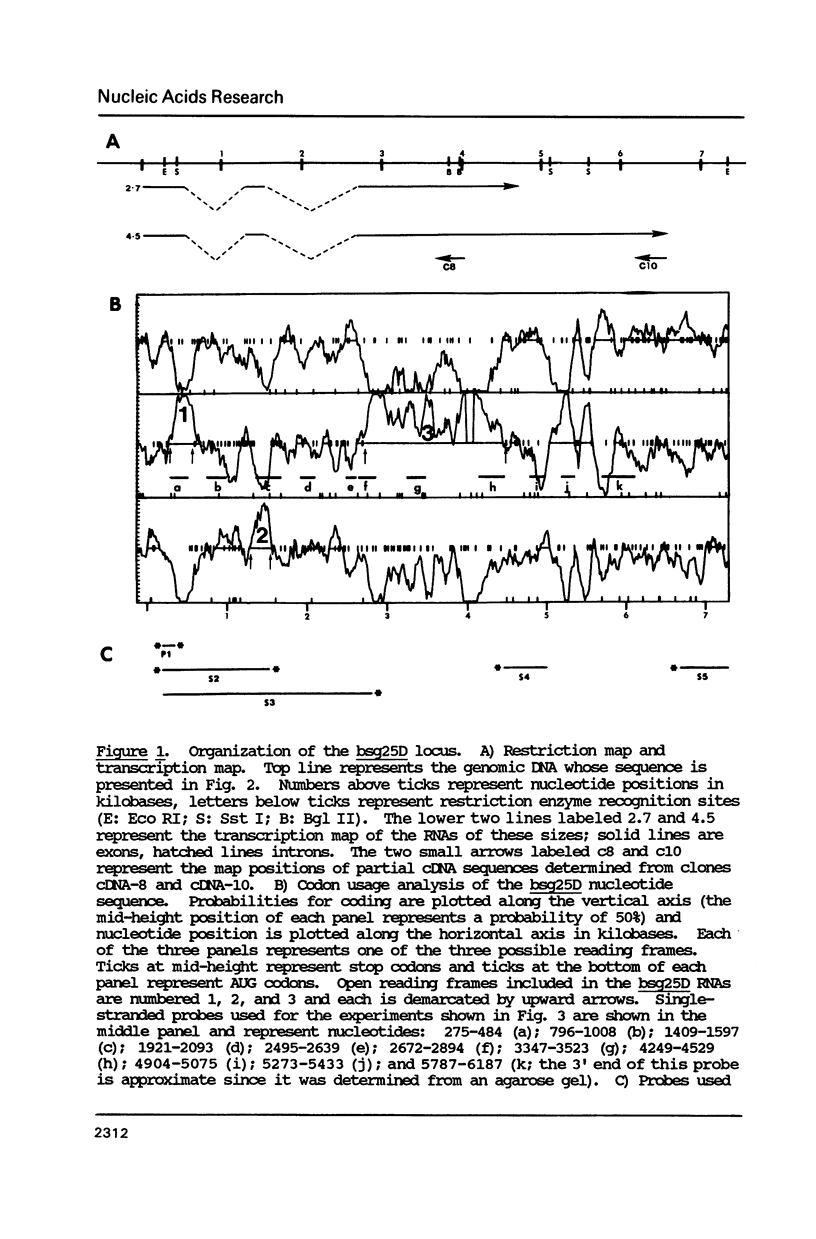





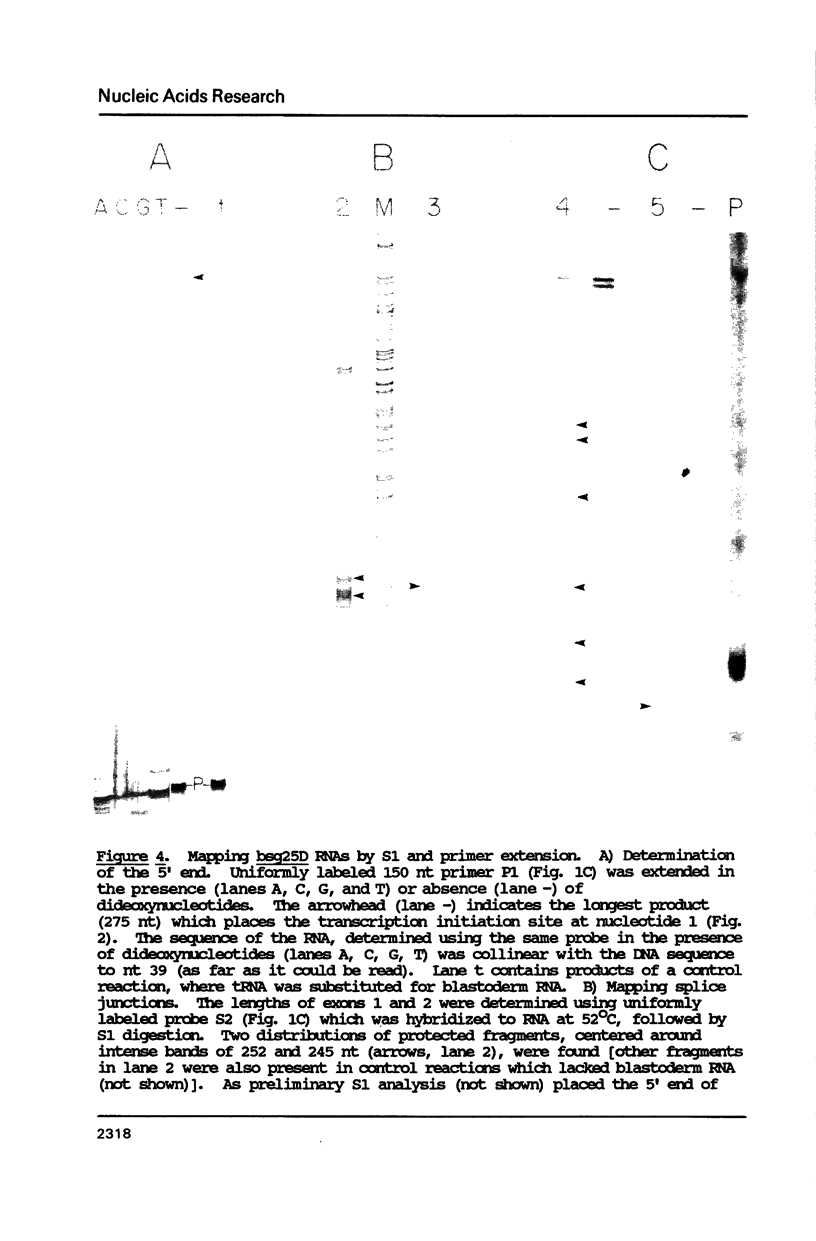







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer P. D. A sensitive technique for detection of RNA with single-stranded probes. Nucleic Acids Res. 1986 Sep 25;14(18):7505–7505. doi: 10.1093/nar/14.18.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Benoist C., O'Hare K., Gannon F., Chambon P. Ovalbumin gene: evidence for a leader sequence in mRNA and DNA sequences at the exon-intron boundaries. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4853–4857. doi: 10.1073/pnas.75.10.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran B. H., Zullo J., Verma I. M., Stiles C. D. Expression of the c-fos gene and of an fos-related gene is stimulated by platelet-derived growth factor. Science. 1984 Nov 30;226(4678):1080–1082. doi: 10.1126/science.6093261. [DOI] [PubMed] [Google Scholar]
- Corden J., Wasylyk B., Buchwalder A., Sassone-Corsi P., Kedinger C., Chambon P. Promoter sequences of eukaryotic protein-coding genes. Science. 1980 Sep 19;209(4463):1406–1414. doi: 10.1126/science.6251548. [DOI] [PubMed] [Google Scholar]
- Deininger P. L. Random subcloning of sonicated DNA: application to shotgun DNA sequence analysis. Anal Biochem. 1983 Feb 15;129(1):216–223. doi: 10.1016/0003-2697(83)90072-6. [DOI] [PubMed] [Google Scholar]
- Doolittle R. F. Similar amino acid sequences: chance or common ancestry? Science. 1981 Oct 9;214(4517):149–159. doi: 10.1126/science.7280687. [DOI] [PubMed] [Google Scholar]
- George D. G., Barker W. C., Hunt L. T. The protein identification resource (PIR). Nucleic Acids Res. 1986 Jan 10;14(1):11–15. doi: 10.1093/nar/14.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hong G. F. A systemic DNA sequencing strategy. J Mol Biol. 1982 Jul 5;158(3):539–549. doi: 10.1016/0022-2836(82)90213-3. [DOI] [PubMed] [Google Scholar]
- Hultmark D., Klemenz R., Gehring W. J. Translational and transcriptional control elements in the untranslated leader of the heat-shock gene hsp22. Cell. 1986 Feb 14;44(3):429–438. doi: 10.1016/0092-8674(86)90464-2. [DOI] [PubMed] [Google Scholar]
- Janin J. Surface and inside volumes in globular proteins. Nature. 1979 Feb 8;277(5696):491–492. doi: 10.1038/277491a0. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Sander C. On the use of sequence homologies to predict protein structure: identical pentapeptides can have completely different conformations. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1075–1078. doi: 10.1073/pnas.81.4.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl N. E., Legouy E., DePinho R. A., Nisen P. D., Smith R. K., Gee C. E., Alt F. W. Human N-myc is closely related in organization and nucleotide sequence to c-myc. Nature. 1986 Jan 2;319(6048):73–77. doi: 10.1038/319073a0. [DOI] [PubMed] [Google Scholar]
- Mason P. J., Jones M. B., Elkington J. A., Williams J. G. Polyadenylation of the Xenopus beta 1 globin mRNA at a downstream minor site in the absence of the major site and utilization of an AAUACA polyadenylation signal. EMBO J. 1985 Jan;4(1):205–211. doi: 10.1002/j.1460-2075.1985.tb02337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K. G., Karr T. L., Kellogg D. R., Mohr I. J., Walter M., Alberts B. M. Studies on the cytoplasmic organization of early Drosophila embryos. Cold Spring Harb Symp Quant Biol. 1985;50:79–90. doi: 10.1101/sqb.1985.050.01.012. [DOI] [PubMed] [Google Scholar]
- Mitchell R. L., Henning-Chubb C., Huberman E., Verma I. M. c-fos expression is neither sufficient nor obligatory for differentiation of monomyelocytes to macrophages. Cell. 1986 May 23;45(4):497–504. doi: 10.1016/0092-8674(86)90281-3. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R., Bravo R., Burckhardt J., Curran T. Induction of c-fos gene and protein by growth factors precedes activation of c-myc. Nature. 1984 Dec 20;312(5996):716–720. doi: 10.1038/312716a0. [DOI] [PubMed] [Google Scholar]
- Poole S. J., Kauvar L. M., Drees B., Kornberg T. The engrailed locus of Drosophila: structural analysis of an embryonic transcript. Cell. 1985 Jan;40(1):37–43. doi: 10.1016/0092-8674(85)90306-x. [DOI] [PubMed] [Google Scholar]
- Roark M., Mahoney P. A., Graham M. L., Lengyel J. A. Blastoderm-differential and blastoderm-specific genes of Drosophila melanogaster. Dev Biol. 1985 Jun;109(2):476–488. doi: 10.1016/0012-1606(85)90473-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Simonsen C. C., Levinson A. D. Analysis of processing and polyadenylation signals of the hepatitis B virus surface antigen gene by using simian virus 40-hepatitis B virus chimeric plasmids. Mol Cell Biol. 1983 Dec;3(12):2250–2258. doi: 10.1128/mcb.3.12.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. A new computer method for the storage and manipulation of DNA gel reading data. Nucleic Acids Res. 1980 Aug 25;8(16):3673–3694. doi: 10.1093/nar/8.16.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Automation of the computer handling of gel reading data produced by the shotgun method of DNA sequencing. Nucleic Acids Res. 1982 Aug 11;10(15):4731–4751. doi: 10.1093/nar/10.15.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Graphic methods to determine the function of nucleic acid sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):521–538. doi: 10.1093/nar/12.1part2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet R. M., Eisenberg D. Correlation of sequence hydrophobicities measures similarity in three-dimensional protein structure. J Mol Biol. 1983 Dec 25;171(4):479–488. doi: 10.1016/0022-2836(83)90041-4. [DOI] [PubMed] [Google Scholar]
- Vincent A., Colot H. V., Rosbash M. Sequence and structure of the serendipity locus of Drosophila melanogaster. A densely transcribed region including a blastoderm-specific gene. J Mol Biol. 1985 Nov 5;186(1):149–166. doi: 10.1016/0022-2836(85)90265-7. [DOI] [PubMed] [Google Scholar]
- Vincent A. TFIIIA and homologous genes. The 'finger' proteins. Nucleic Acids Res. 1986 Jun 11;14(11):4385–4391. doi: 10.1093/nar/14.11.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfenden R. V., Cullis P. M., Southgate C. C. Water, protein folding, and the genetic code. Science. 1979 Nov 2;206(4418):575–577. doi: 10.1126/science.493962. [DOI] [PubMed] [Google Scholar]
- Wolfenden R., Andersson L., Cullis P. M., Southgate C. C. Affinities of amino acid side chains for solvent water. Biochemistry. 1981 Feb 17;20(4):849–855. doi: 10.1021/bi00507a030. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]




