Background: Light is an important factor for plant growth and development.
Results: GBF1 interacts with two other bZIP proteins, HY5 and HYH, in light signaling.
Conclusion: This work demonstrates the functional interconnections of GBF1, HY5, and HYH.
Significance: The knowledge gained in this study will help to understand light-controlled plant growth and development.
Keywords: Development, Photomorphogenesis, Plant Molecular Biology, Transcription Factors, Transcription Regulation, HY5, HYH, ZBF2/GBF1, Gene Regulation, Photomorphogenesis
Abstract
Arabidopsis bZIP transcription factor, GBF1, acts as a differential regulator of cryptochrome-mediated blue light signaling. Whereas the bZIP proteins, HY5 (elongated hypocotyl 5) and HYH (HY5 homologue), are degraded by COP1-mediated proteasomal pathways, GBF1 is degraded by a proteasomal pathway independent of COP1. In this study, we have investigated the functional interrelations of GBF1 with HY5 and HYH in Arabidopsis seedling development. The genetic studies using double and triple mutants reveal that GBF1 largely acts antagonistically with HY5 and HYH in Arabidopsis seedling development. Further, GBF1 and HY5 play more important roles than HYH in blue light-mediated photomorphogenic growth. This study reveals that GBF1 is able to form a G-box-binding heterodimer with HY5 but not with HYH. The in vitro and in vivo studies demonstrate that GBF1 co-localizes with HY5 or HYH in the nucleus and physically interacts with both of the proteins. The protein-protein interaction studies further reveal that the bZIP domain of GBF1 is essential and sufficient for the interaction with HY5 or HYH. Taken together, these data demonstrate the functional interrelations of GBF1 with HY5 and HYH in Arabidopsis seedling development.
Introduction
In unfavorable environmental conditions, an intact and healthy seed remains dormant in a dry state. In Arabidopsis thaliana, seed dormancy is terminated by environmental signals, such as light, temperature, nutrient availability, and duration of storage in the dried state (1). After germination, seedlings are able to follow two developmental patterns, depending upon the presence or absence of light. Skotomorphogenesis (or etiolation) in the dark is characterized by long hypocotyl, closed cotyledons protected by apical hooks, and the development of proplastids into etioplasts. By contrast, growth in the light results in photomorphogenesis (or de-etiolation), characterized by short hypocotyl, open cotyledons, and the development of mature green chloroplasts (2–4).
A wide spectrum of light, in particular far red, red, blue, and ultraviolet (UV) light, induces photomorphogenesis (5). It is therefore not surprising that plants have adopted the ability to sense multiple parameters of ambient light signals, including light quantity (fluence), quality (wavelength), direction, and duration. Light signals are perceived through at least four different types of photoreceptors, which include phytochromes, cryptochromes, phototropins, and recently identified UV-B photoreceptor, UVR8 (5–9). The dramatic morphological change from skotomorphogenic to photomorphogenic growth involves a regulated change in the expression of an estimated one-third of the genes in Arabidopsis (10). The results indicate that the massive change in gene expression is mediated by several transcriptional cascades (11). The genomic expression profiles show that a large percentage of early responsive genes (induced in <1 h) are transcription factors, whereas it takes several h before many of the genes associated with photomorphogenic development are highly expressed (11). These results suggest that photoreceptors transduce the signal to a set of key transcription factors that in turn rapidly induce expression of a new set of transcriptional regulators. This results in an extensive branching of the signal and the promotion of photomorphogenic development.
Transcriptional regulatory networks have a key role in mediating light signaling through the coordinated activation and repression of specific downstream genes. So far, HY5 and CAM7/ZBF3 are two positive regulators known of light signaling pathways that act downstream of multiple photoreceptors, including phytochromes and cryptochromes (5, 12–14). HY5 has been genetically defined as a positive regulator of photomorphogenesis based on its partially etiolated phenotype in light-grown mutant seedlings (15–18). HY5 encodes a bZIP4 protein that can physically interact with COP1 (12). DNA-protein interaction studies have revealed that HY5 specifically interacts with the G-box and is required for the proper activation of G-box-containing promoters in light (12, 19, 20). Recent ChIP-on-chip studies have shown that HY5 binds to the promoters of a large number of regulatory genes in Arabidopsis (21, 22).
HYH, a bZIP protein and a close homologue of HY5, works as a positive regulator of photomorphogenesis in blue light (23). HY5 and HYH proteins have an overlapping yet non-redundant function in blue light-mediated inhibition of hypocotyl elongation. Furthermore, HY5 and HYH form heterodimers and mediate light-regulated expression of overlapping as well as distinct target genes (23). The synergism between red and blue light for control of gene expression and seedling development has also been reported where HY5 and HYH were found to enhance the phyB signaling output (24). Both of these proteins are degraded by COP1-mediated proteasomal pathways in the dark (13, 23).
GBF1 (G-box-binding factor 1)/ZBF2 (Z-box-binding factor 2), a bZIP transcription factor, has been shown to bind to the G-box of RBCS-1A or the Z-box of the CAB1 promoter and regulate the expression of these genes (25–27). It has been further reported that the G- and Z-box are functionally equivalent with context to GBF1 (27). The examination of physiological functions of GBF1 has revealed that it functions in cryptochrome-mediated blue light signaling and plays a differential regulatory role in Arabidopsis seedling development (27). Further studies have revealed that GBF1 protein is degraded by a proteasomal pathway in the dark, which is independent of COP1 and SPA1 (28). COP1 physically interacts with GBF1 and is required for the optimum accumulation of GBF1 protein in light-grown seedlings (28).
Transcription factors are capable of activating or repressing gene expression by binding to specific cis-elements in the promoter. The bZIP transcription factors have a basic region that binds DNA and a leucine zipper region. The leucine zipper region is α-helical and can form dimer via a coiled-coil structure (29). Plant bZIP proteins preferentially bind to the cis-elements that have an ACGT core sequence, such as G-box (CACGTG), C-box (GACGTC), Z-box (ATACGTGT), and A-box (TACGTA). The formation of homo- and heterodimers offers a combinatorial flexibility to the bZIP proteins to regulate transcription. Although there are several transcription factors known to function in light signaling pathways, the genetic and molecular interrelations among them are largely unknown. In fact, such interrelations among the handful of bZIP proteins known in light signaling pathways are not clearly understood. The genetic and molecular relationships between HY5 and HYH have been investigated (23); however, the interrelations of GBF1 with HY5 and HYH are unknown. In this study, we have investigated the genetic and molecular relationships of GBF1 with HY5 and HYH in Arabidopsis seedling development.
EXPERIMENTAL PROCEDURES
Plant Materials and Growth Conditions
The wild type Arabidopsis thaliana used in this study is the segregated wild type obtained from the genetic crosses of various mutants in Col-0 or Wassilewskija (WS) background. The gbf1–1 mutant (27) is in Col-0 accession, whereas hy5-ks50 (18), hyh (23), and GBF1OE (27) are in WS. The hy5 hyh double mutant lines are as described previously (23). The gbf1 hy5, gbf1 hyh, and gbf1 hy5 hyh mutants are constructed by genetic crosses of gbf1-1 individually with hy5-ks50, hyh, and hy5 hyh, respectively. The hy5 GBF1OE and hyh GBF1OE mutant transgenic lines were generated by genetic crosses of GBF1OE individually with hy5-ks50 and hyh, respectively. Unless stated otherwise, seeds were surface-sterilized and plated on Murashige and Skoog medium supplemented with 0.8% Bactoagar (Difco) and 1% sucrose. The plates were then cold-treated at 4 °C for 4 days and transferred to light chambers maintained at 22 °C with the desired light intensities. Hypocotyl length measurements were performed with the help of ImageJ 1.41 software (National Institutes of Health, Bethesda, MD).
Electrophoretic Mobility Shift (Gel Shift) Assay
GST-GBF1 and GST-HY5 were induced and overexpressed in Escherichia coli using 1 and 0.5 mm isopropyl thio-β-galactoside, respectively. Both of the overexpressed fusion proteins were affinity-purified following the manufacturer's protocol (Amersham Biosciences). The 196-bp fragment of native RBCS-1A promoter (from −320 to −125) (19) was digested with HindIII and XhoI, purified, and 3′-end-labeled by filling 3′-recessed ends for use as probe for the DNA binding assays. One ng of labeled DNA was used for each binding reaction. Both of the proteins were mixed and incubated at 55 °C for 5 min. The DNA binding assays were performed at room temperature in a final volume of 30 μl with a binding buffer of 15 mm Hepes, pH 7.5, 35 mm KCl, 1 mm EDTA, pH 8.0, 6% glycerol, 1 mm DTT, 1 mm MgCl2, and 1 μg of poly(dI-dC). The samples were incubated at room temperature for 30 min and then run on the 4% polyacrylamide gel containing acrylamide/bisacrylamide (29:1) at 10 mA. After drying, the gels were autoradiographed.
Chromatin Immunoprecipitation (ChIP) Assays
The ChIP assays were carried out as described previously (30) with some modifications. Briefly, a 4-day-old constant WL or BL (20 μmol/m2/s)-grown 1.5-mg seedling was harvested and immersed in buffer A (0.4 m sucrose, 10 mm Tris-HCl pH 8.0, 1 mm EDTA pH 8.0, 1 mm PMSF, 1% formaldehyde) and kept under vacuum for 10 min. Glycine was added to a final concentration of 0.25 m to stop the cross-linking. Seedlings were rinsed, excess water was dried off using tissue paper, and seedlings were frozen in liquid nitrogen. Then the tissues were ground into fine powder and suspended in lysis buffer (50 mm HEPES, pH 7.5, 150 mm NaCl, 1 mm EDTA, pH 8.0, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 1 mm PMSF, 10 mm sodium butyrate, and 1× plant protease inhibitor (Sigma)). Chromatin was sheared to about 0.5–1.0-kb fragments by sonication. After centrifugation, the supernatant was precleared with 60 μl of salmon sperm DNA/protein A-agarose for 60 min at 4 °C. An appropriate amount (5–10 μl) of anti-GBF1 polyclonal antibody was added to the supernatant and was further incubated overnight at 4 °C. 60 μl of salmon sperm DNA/protein A-agarose was added the next day and incubated further for 2 h. The agarose beads were then washed with 1 ml of the following buffers for 3 min at 4 °C: two times with lysis buffer, one time with LNDET buffer (0.25 m LiCl, 1% Nonidet P-40, 1% sodium deoxycholate, 1 mm EDTA, pH 8.0, 10 mm Tris-HCl, pH 8.0), and two times with TE buffer (10 mm Tris-HCl, pH 8.0, 1 mm EDTA, pH 8.0). The immunocomplexes were collected from beads with 250 μl of elution buffer (1% SDS, 0.1 m NaHCO3), incubated at 65 °C for 20 min with agitation. 0.3 m NaCl was added to each tube, and cross-linking was reversed by incubation at 65 °C for 6–8 h. Residual protein was degraded by the incubation with 20 mg of Proteinase K in 10 mm EDTA and 40 mm Tris-HCl, pH 8.0, at 45 °C for 1 h, followed by phenol/chloroform/isoamyl alcohol extraction and ethanol precipitation. DNA pellets were washed with 70% ethanol and suspended in 30 μl of sterile water. About 10% of non-immunoprecipitated sonicated chromatin was reverse cross-linked and used as input DNA control. Both immunoprecipitated DNA and input DNA were analyzed by real-time PCR (Light Cycler; Roche Applied Science). Otherwise, after reverse cross-linking by boiling for 25 min, it was resolved on SDS-PAGE. Both the input and immunoprecipitation were probed with anti-GBF1 antibodies. The sequence of the primer pair (resulting products of 135 bp) used was as follows: RBCS-1A PROMO FP, 5′-TAAGATTCATGGAATTATC-3′; RBCS-1A PROMO RP, 5′-GGATTTTGAGTGTGGATA-3′.
In Vitro Binding Assays
GST and GST fusion proteins were expressed in E. coli strain BL21/DE3 after cloning full-length coding sequence (CDS) in pGEX-4T-2 vector (Amersham Biosciences). Proteins were purified using glutathione-Sepharose 4B beads (GE Healthcare) as described previously (31). HY5-His6 and HYH-His6 recombinant proteins were obtained by cloning full-length CDS in pET-20b (+) (Novagen) vector and were purified using Ni-NTA-agarose beads (Qiagen) after overexpressing in E. coli strain BL21/DE3. For in vitro pull-down assays, 2 μg of GST and GST fusion proteins were bound to glutathione-Sepharose 4B beads. 2 μg of precleared HY5-His6 and HYH-His6 was incubated with fusion proteins bound to glutathione-Sepharose 4B beads for 2 h at 4 °C in in vitro pull-down buffer (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 0.2% (v/v) glycerol, 0.1% (v/v) Triton X-100, 1 mm EDTA, pH 8.0, 1 mm PMSF, 0.1% (v/v) Nonidet P-40). After washing three times with the same buffer, pulled-down proteins were separated on 10% SDS-polyacrylamide gels and were detected by Western blot using anti-HY5 and anti-His antibodies.
For pull-down assays using plant protein extract, HY5-His6 and HYH-His6 proteins were bound to the Ni-NTA beads, followed by a 6-h incubation at 4 °C with precleared plant protein extract in a pull-down buffer consisting of 50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 10% (v/v) glycerol, 1 mm PMSF, 0.1% (v/v) Tween 20, and 1× plant protease inhibitor mixture (Sigma). After washing three times with the same buffer, pulled down proteins were separated on 10% SDS-polyacrylamide gel and were detected by Western blot using anti-GBF1 and anti-actin antibodies.
Co-immunoprecipitation Assays
4-day-old BL (20 μmol/m2/s) grown seedlings of wild type and various mutants were harvested for total protein extraction in Arabidopsis total protein extraction buffer (400 mm sucrose, 50 mm Tris-Cl, pH 7.5, 10% glycerol, 2.5 mm EDTA). 500 mg of total protein of each line was used for co-immunoprecipitation in co-immunoprecipitation buffer (50 mm Tris-HCl, 100 mm NaCl, 10% glycerol, 5 mm EDTA, 0.1% Triton X-100, 0.2% Nonidet P-40) using 10–15 μl of affinity-purified HY5 polyclonal antibody for 6 h at 4 °C. Then 30 μl of preblocked protein A-agarose beads (Sigma) were added and further incubated for 2 h at 4 °C. After the beads were washed three times with co-immunoprecipitation buffer, they were kept in a boiling water bath in 2× protein-loading dye for 10 min and then run in SDS-PAGE and probed with anti-GBF1 antibodies.
Co-localization Experiments
For co-localization studies, full-length CDS of GBF1 and HYH were cloned at EcoRI and NcoI sites of pAM-PAT-35SS-CFP vector (32) to produce GBF1-CFP and HYH-CFP fusion proteins. Similarly, GBF1-YFP and HY5-YFP fusion proteins were obtained by cloning full-length CDS of GBF1 or HY5 at EcoRI and NcoI sites of pAM-PAT-35SS-YFP vector (32). For transient expression, particle bombardment was performed using onion epidermal cells. In brief, gold particles (1.0-μm diameter) were washed with 100% ethanol and coated with 10 μg of each plasmid. DNA-coated gold-particles were bombarded using the Biolistic particle delivery system (Bio-Rad) PDS-1000. 0.5–0.6 mg of gold particles/shot were used with a chamber vacuum of 27 inches Hg. Particles were accelerated with a pressure of 1100 p.s.i. The distance between the projectile source and the samples was 6 cm. After bombardment, samples were incubated for 24–48 h at room temperature in the dark and then visualized under a confocal microscope (Leica). Both CFP and YFP fluorescence were visualized at the same parameters using respective lasers. Red false color was given to CFP fluorescence images, and green false color was given to YFP fluorescence images to obtain a different color (yellow) of the merged image.
Bimolecular Fluorescence Complementation (BiFC) Experiments
For BiFC experiments, full-length CDS of GBF1 was cloned in the vectors pUC-SPYNE and pUC-SPYCE (33) at BamHI and SalI sites to obtain GBF1-YFPN-ter and GBF1-YFPC-ter fusion proteins, respectively. To obtain HY5-YFPC-ter and HYH-YFPN-ter fusion proteins, full-length CDS of HY5 and HYH was cloned at BamHI and SalI restriction sites of pUC-SPYCE and pUC-SPYNE vectors, respectively. BiFC experiments were carried out in onion epidermal cells using a PDS-1000 Gene-gun (Bio-Rad), as described under “Co-localization Experiments.” Either empty vectors or BiFC constructs were bombarded along with pCAMBIA-1302 vector (CAMBIA, Canberra, Australia). The GFP fluorescence of pCAMBIA-1302 vector was used as a marker for transformed cells.
For the domain-wise study, both the split domains of HY5, ΔZIP (aa 1–115) and bZIP (aa 77–168), were cloned into the pUC-SPYCE vector at SalI and BamH1 sites, whereas the ΔZIP (aa 1–100) and bZIP (aa 74–149) domains of HYH were cloned into the pUC-SPYNE vector at SalI and BamHI sites. The ΔZIP (aa 1–243) and bZIP (aa 221–315) domains of GBF1 were cloned in both the BiFC vectors at SalI and BamHI sites. BiFC analysis of these truncated domains was done similarly to full-length BiFC constructs.
Gene Expression Analysis
Wild type and different mutant seedlings were grown in the dark for 5 days and then transferred to continuous blue light at 30 μmol/m2/s for 12 or 24 h, and total RNA was extracted using the RNeasy plant minikit (Qiagen). cDNA was synthesized from 1 μg of the total RNA using RT-AMV reverse transcriptase (Titan One Tube RT-PCR System, Roche Applied Science). Real-time PCR analyses were carried out by using the Thermal Cycler Applied Biosystem StepOneTM and Light Cycler Faststart DNA Masterplus SYBR Green 1 systems (Roche Applied Science). The transcript levels of CAB1 were determined by using FP CAB1 (5′-GTTAACAACAACGCATGGC-3′) and RP CAB1 (5′-CCTCTCACACTCACGAAGCA-3′), and transcript levels of RBCS-1A were determined using FP RBCS-1A (5′-TCGGATTCTCAACTGTCTGATG-3′) and RP RBCS-1A (5′-ATTTGTAGCCGCATTGTCCT-3′). The transcript levels were normalized with the level of actin2 transcript abundance using FP Actin2 (5′-TGATGCACTTGTGTGTGACAA-3′) and RP Actin2 (5′-GGGACTAAAACGCAAAACGA-3′).
Chlorophyll and Anthocynin Estimations
For chlorophyll estimation, 6-day-old WL-grown seedlings were immediately frozen in liquid nitrogen. The frozen tissue was ground in a 1.5-ml microcentrifuge tube with a disposable pestle, and chlorophyll was extracted repeatedly into 80% acetone in the dark until the pellet appeared colorless. Chlorophyll a and b contents were calculated using MacKinney's specific absorption coefficients, in which chlorophyll a = 12.7(A663) − 2.69(A645), and chlorophyll b = 22.9(A645) − 4.48(A663). The total specific chlorophyll content is expressed as μg of chlorophyll a and b normalized by g, fresh weight, of tissue.
For anthocyanin estimation, frozen plant tissues were ground in a 1.5-ml microcentrifuge tube with a disposable pestle, and total plant pigments were extracted overnight in 0.3 ml of 1% HCl in methanol. After the addition of 0.2 ml of H2O, chlorophyll was separated from the anthocyanin by extraction with an equal volume of chloroform. The quantity of anthocyanin was determined by spectrophotometric measurements by taking readings at wavelengths of 530 and 657 nm. The total anthocyanin content was calculated with the help of the formula, (A530 − 0.33A657)/g of fresh weight.
Transient Expression Analysis in Arabidopsis Protoplasts
Protoplast isolation and transformation was performed essentially as described previously (34). For construction of RBCS-1A-NAN construct, a 355-bp region of RBCS-1A promoter upstream of the start codon was amplified from genomic DNA of WT (Col-0) using FP RBCS-1A Pro HindIII (5′-CCCAAGCTTGGGCCAAGTCCACCAGGCAAG-3′) and RP RBCS-1A Pro XbaI (5′-GCTCTAGATGTTCTTCTTTACTCTTTG-3′). pBIB-HYG vector, which has a CaMV 35S promoter fused to the NAN reporter (35), was digested with HindIII and XbaI to remove the CaMV 35S promoter, and then the amplified RBCS-1A promoter was ligated in place of it. The digested CaMV 35S promoter was used to replace GAL-UAS in pGAL-UAS:GUS vector (36), which resulted in p35S:GUS construct, which was used as an internal control to normalize the transfection efficiencies. For construction of effector constructs, full-length CDS of GBF1, HY5, and HYH was individually cloned into Gateway®-compatible (GW) vector p35S-HA-GW (36) under the control of the CaMV 35S promoter. For reporter gene assays, 16 μg of reporter, 20 μg of each effector, and 4 μg of internal control plasmid, p35S:GUS, were transformed in the Arabidopsis protoplasts. The overall amount of DNA was set to 80 μg by adding p35S-HA-GW plasmid DNA. After transformation, protoplast samples were incubated in continuous BL (40 μmol/m2/s) for 12–14 h. After incubation, half of the protoplasts were harvested for total RNA extraction, and the rest were used for NAN and GUS activity measurements. NAN and GUS enzyme assays were performed essentially as described previously (35).
RESULTS
GBF1 and HY5 Form DNA-binding Heterodimer at RBCS-1A Promoter
The b-ZIP proteins bind as dimers to their palindromic DNA targets. The spatial orientation resulting from leucine zipper dimerization leads two opposed protein monomers to interact symmetrically at the site of the DNA palindrome (37). GBF1, HY5, and HYH have previously been shown to bind to the G-box light-responsive element present in the RBCS-1A minimal promoter, and furthermore HY5 and HYH form a heterodimer at this element (19, 23, 26). Considering their comparable affinity to the RBCS-1A minimal promoter, we wanted to investigate whether GBF1 heterodimerizes with HY5 or HYH and binds to the G-box light-responsive element present in the RBCS-1A minimal promoter.
We carried out electrophoretic mobility shift assays (gel shift) using GST fusion proteins and a 196-bp minimal promoter fragment of RBCS-1A (19). Pre-existing homodimeric complexes were dissociated by incubation of an equimolar mixture of GST-GBF1 and GST-HY5 at 50 °C for 5 min prior to its addition to the DNA. Although added individually, GBF1 and HY5 form discrete protein-DNA complexes that are readily resolved, as shown in Fig. 1A (lanes 3 and 4). It is worth mentioning here that the HY5 monomer is predicted to migrate at ∼18 kDa; however, it migrates at the ∼30 kDa region (13). This may be the reason, while individually incubated with DNA, GST-GBF1 runs much faster than GST-HY5 (Fig. 1A). When GBF1 and HY5 were mixed together, a complex with intermediary mobility was detected (Fig. 1A and supplemental Fig. 1A). Because the DNA probe contains only one copy of the G-box motif, our result suggests the formation of GBF1-HY5 heterodimers. We carried out similar experiments to determine the possible heterodimer formation between GBF1 and HYH during DNA binding; however, no such heterodimerization was observed.
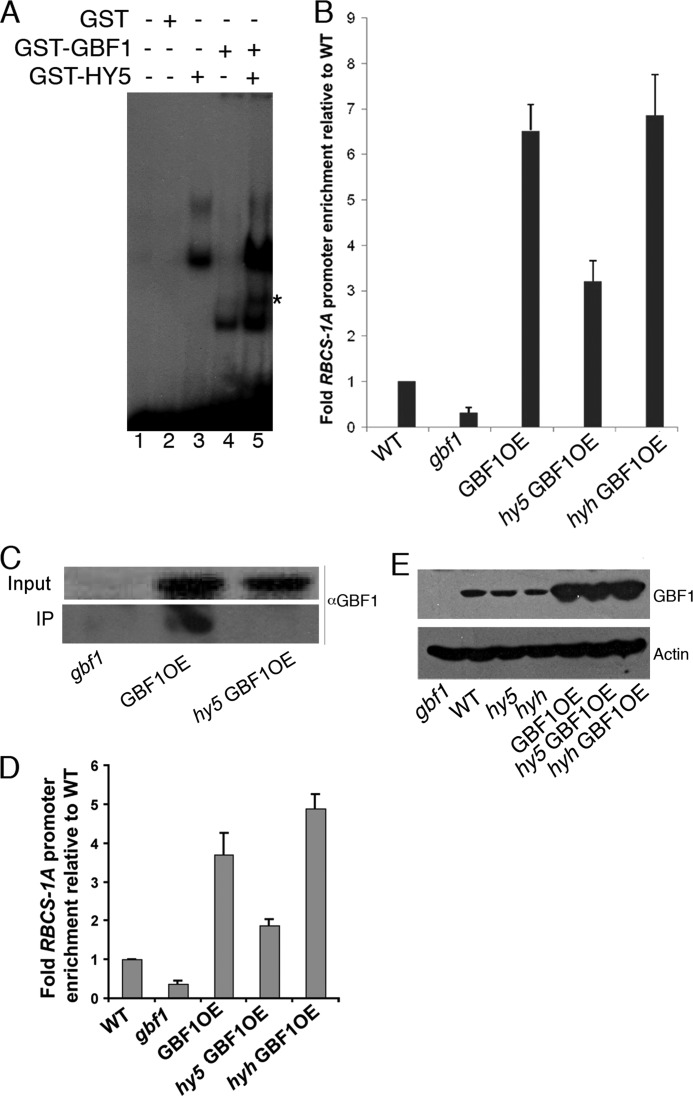
FIGURE 1.
GBF1 and HY5 form a G-box-binding heterodimer. A, electrophoretic mobility shift assays (EMSAs) using recombinant GBF1 and HY5 proteins bind to the G-box of the RBCS-1A minimal promoter. Approximately 100 ng of each of the proteins was mixed and incubated at 50 °C for 5 min to dissociate pre-existing dimers prior to their addition to radioactively labeled probe. Approximately 200 ng of GST protein was added in lane 2. The protein-DNA complexes were resolved on a 4% native 0.5× TBE polyacrylamide gel. Plus and minus signs show the presence and absence of the components in respective lanes. The star indicates the heterodimer complex. B, ChIP assays of RBCS-1A promoter of white light-grown WT, mutant, and overexpresser transgenic seedlings using antibodies to GBF1. The result of the real-time PCR is presented as the ratio of immunoprecipitated DNA to input DNA from various backgrounds. Error bars, S.D. n ≥ 3 independent experiments with similar results. C, in vivo interactions of GBF1 and HY5 proteins in ChIP assays. The cross-linked complex of GBF1-HY5 with the RBCS-1A promoter was pulled down by antibodies to HY5. The complex was reverse cross-linked and resolved onto SDS-PAGE. Both the input and immunoprecipitates were probed with antibodies to GBF1. D, ChIP assays of the RBCS-1A promoter of blue light-grown WT, mutant, and overexpresser transgenic seedlings using antibodies to GBF1. The result of the real-time PCR is presented as the ratio of immunoprecipitated DNA to input DNA from various backgrounds. Error bars, S.D. n ≥ 3 independent experiments with similar results. E, analysis of GBF1 protein levels in wild type, mutant, and overexpresser transgenic lines by Western blots using antibodies to GBF1.
To further substantiate the above result, we carried out ChIP experiments. We used Arabidopsis transgenic lines overexpressing GBF1 (27). We introduced the 35S-GBF1 transgene individually into hy5 or hyh mutant backgrounds by genetic crosses and obtained the homozygous transgenic mutant lines. The GBF1 protein was immunoprecipitated using affinity-purified antibody to GBF1 (28). The co-immunoprecipitated genomic DNA fragments with GBF1 were then analyzed by real-time PCR. The amount of DNA fragment of RBCS-1A promoter co-immunoprecipitated from the GBF1 overexpresser transgenic seedlings (GBF1OE) was found to be about 14-fold enriched as compared with the gbf1 mutant background (Fig. 1B). Whereas the level of enrichment of RBCS-1A promoter fragment was reduced to about 6-fold in the transgenic hy5 mutant background, it remained at the similar level in transgenic hyh mutants. Examination of the GBF1 protein level in various mutants and overexpresser transgenic backgrounds revealed that the accumulation of the protein remained at similar levels in wild type, hy5, and hyh mutants and at higher and comparable levels in GBF1OE, hy5 GBF1OE, and hyh GBF1OE transgenic lines (Fig. 1E). Taken together, these results demonstrate the preferred G-box-binding heterodimer formation of GBF1 and HY5 proteins.
The in vivo heterodimerization of GBF1 and HY5 was further examined by ChIP assays. For this experiment, gbf1 mutants, GBF1OE and hy5 GBF1OE transgenic lines were individually used to immunoprecipitate the RBCS-1A promoter-GBF1-HY5 complex by using antibody to HY5. The immunoprecipitated complex was processed and subjected to Western blot analyses using antibodies to GBF1. Although no GBF1 protein was detected in hy5 GBF1OE transgenic lines, a significant amount of GBF1 was detected in the GBF1OE transgenic line (Fig. 1C). These results in combination with the ChIP-quantitative PCR results clearly demonstrate the preferred heterodimer formation of GBF1 and HY5 proteins at the RBCS-1A promoter.
To determine whether GBF1, a negative regulator of hypocotyl growth in BL (27), and HY5 form a heterodimer at the RBCS-1A promoter in BL, we performed ChIP assays using wild type, gbf1 mutants, GBF1OE, hy5 GBF1OE, and hyh GBF1OE transgenic lines grown in BL. As shown in Fig. 1D, the amount of the DNA fragment of RBCS-1A promoter co-immunoprecipitated from the GBF1OE transgenic seedlings was about 10-fold higher than the gbf1 mutant background. The level of enrichment of the RBCS-1A promoter fragment was reduced to about 5-fold in the transgenic hy5 mutant background; however, it remained at a similar level in transgenic hyh mutants. Taken together, these results demonstrate that GBF1 and HY5 proteins form a heterodimer at the G-box of RBCS-1A promoter in BL-grown Arabidopsis seedlings.
GBF1 Physically Interacts with HY5 and HYH
Because the DNA-protein interaction studies suggest the formation of heterodimer between GBF1 and HY5, we examined the possible physical interactions of GBF1 with both HY5 and HYH proteins in the absence of DNA, through protein-protein interaction studies. First, we carried out in vitro binding assays, in which we used poly-His or GST fusion proteins. For these experiments, HY5-His6 protein was separately passed through columns containing glutathione-Sepharose 4B beads attached to GST, GST-GBF1, or GST-COP1 (used as a positive control) proteins. The α-HY5 immunoblot showed that the amount of HY5-His6 retained by GST-GBF1 was comparable with GST-COP1 and significantly higher than the background level retained by GST alone (Fig. 2, A and E). In a similar experiment, HYH-His6 protein was separately passed through columns containing glutathione-Sepharose 4B beads bound to GST, GST-GBF1, or GST-HY5 (used as positive control) proteins. The immunoblot analyses using anti-His antibodies showed that GST-GBF1 retained HYH-His6 comparable with GST-HY5, whereas no detectable HYH-His6 protein was retained by GST alone (Fig. 2C). Taken together, these results indicate that GBF1 physically interacts with HY5 and HYH proteins.
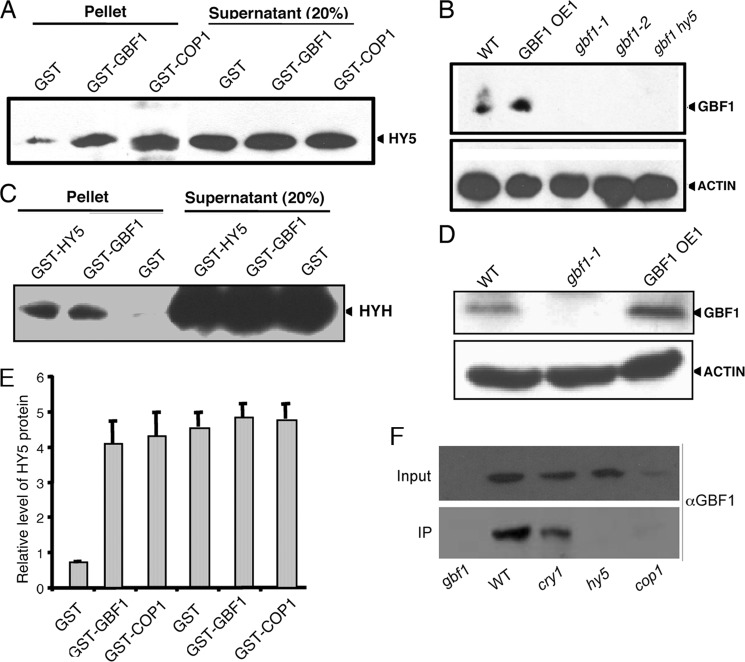
FIGURE 2.
GBF1 Interacts with HY5 and HYH. A, in vitro binding assays using 2 μg of bacterially expressed HY5-His6 separately incubated with 2 μg of glutathione-Sepharose-bound GST-COP1 (used as control), GST-GBF1, and GST proteins. The blot was probed with anti-HY5 antibodies. The signal in supernatant serves as a loading control. B, in vitro pull-down assays using 100 mg of total plant protein extract of WT, gbf1-1, gbf1-2, gbf1 hy5, and GBF1 OE1 separately incubated with Ni-NTA-agarose-bound HY5-His6. The blot was probed with anti-GBF1 antibodies. The anti-actin immunoblot serves as a loading control. The arrowheads show the bands respective to proteins detected in immunoblot. C, in vitro binding assays using 2 μg of bacterially expressed HYH-His6 separately incubated with 2 μg of glutathione-Sepharose-bound GST-HY5 (used as a control), GST-GBF1, and GST proteins. The blot was probed with anti-HIS antibodies. Signal in supernatant serves as loading control. D, in vitro pull-down assays using 100 mg of total plant protein extract of WT, gbf1, and GBF1 OE1 separately incubated with Ni-NTA-agarose-bound HYH-His6. The blot was probed with anti-GBF1 antibodies. An anti-actin immunoblot serves as loading control. The arrowheads show the bands respective to proteins detected in the immunoblot. E, quantification of the results in A. The error bars represent S.E. of three independent experiments with similar results. F, co-immunoprecipitation (CoIP) experiments using 500 μg of total protein from WT, cry1, gbf1, hy5, and cop1 mutants grown in BL. The co-immunoprecipitation experiments were carried out with antibodies to HY5, and then input (10%) and the immunoprecipitated (IP) fractions were fractionated onto SDS-PAGE and probed with antibodies to GBF1.
To further substantiate the above results, we carried out in vitro pull-down assays using the total plant protein extracts. In this experiment, HY5-His6 recombinant fusion protein was bound to Ni-NTA-agarose beads and incubated with total protein extracts of wild type, GBF1OE, or various mutant lines. As shown in Fig. 2B, when protein extract from wild type and GBF1OE lines was passed through an HY5-His6/Ni-NTA column, GBF1 protein was retained in the column. A higher level of retention was also observed in GBF1OE lines compared with wild type background while probed with αGBF1. However, the Western blot analyses using αGBF1 did not show any detectable band, whereas the protein extracts from gbf1-1, gbf1-2, or gbf1 hy5 were passed through the column. These results suggest that GBF1 and HY5 proteins physically interact with each other. Similarly, to determine the physical interaction between GBF1 and HYH, HYH-His6 recombinant fusion protein was bound to the Ni-NTA-agarose beads and separately incubated with the total protein extracts of wild type, gbf1, or GBF1OE seedlings. As shown in Fig. 2D, when crude protein extract from wild type or the GBF1OE line was passed through the HYH-His6/Ni-NTA column, GBF1 protein was retained in the column; however, no protein was detected from the protein extract of gbf1 mutant seedlings. Taken together, these results strongly suggest that GBF1 physically interacts with HY5 and HYH proteins.
GBF1 is known to work downstream from the cry1 photoreceptor (27). It has also been shown that the optimum accumulation of GBF1 required COP1 (28). To examine whether GBF1 is able to interact with HY5 in the absence of cry1 or COP1, we carried out co-immunoprecipitation experiments using cry1, cop1, and hy5 (as control) mutant seedlings grown in BL. The co-immunoprecipitation experiment was carried out using antibody to HY5. The complex was processed and subjected to Western blot analyses using antibodies to GBF1. Whereas no GBF1 protein was detected in hy5 mutant lines, GBF1 protein was detected in the cry1 mutant background (Fig. 2F). Because GBF1 protein accumulates at a very low level in cop1 mutants (28), very faint, if any, GBF1 protein band was detected in the cop1 mutant background (Fig. 2F). Taken together, these results demonstrate that GBF1 and HY5 physically interact in vivo in the absence of CRY1.
GBF1 Co-localizes with HY5 and HYH in Onion Cells
HY5 and HYH display diffused and uniform nuclear fluorescence when expressed in onion epidermal cells (12, 23). In order to determine subcellular localization of interaction events of GBF1 with HY5 and HYH, we prepared cyan fluorescent protein (CFP) or yellow fluorescent protein (YFP) fusion proteins of GBF1, HY5, and HYH and co-expressed GBF1 with HY5 or HYH in onion epidermal cells. Similar to HY5 and HYH, GBF1 was also uniformly localized in the nucleus (Fig. 3, B and C). Superimposition of the images of GBF1-CFP with HY5-YFP or of GBF1-YFP with HYH-CFP showed obvious color changes in fluorescence (Fig. 3, B and C). The images of empty vectors are shown in Fig. 3A. Taken together, these results suggest that GBF1 co-localizes with HY5 or HYH in the nucleus of onion epidermal cells.
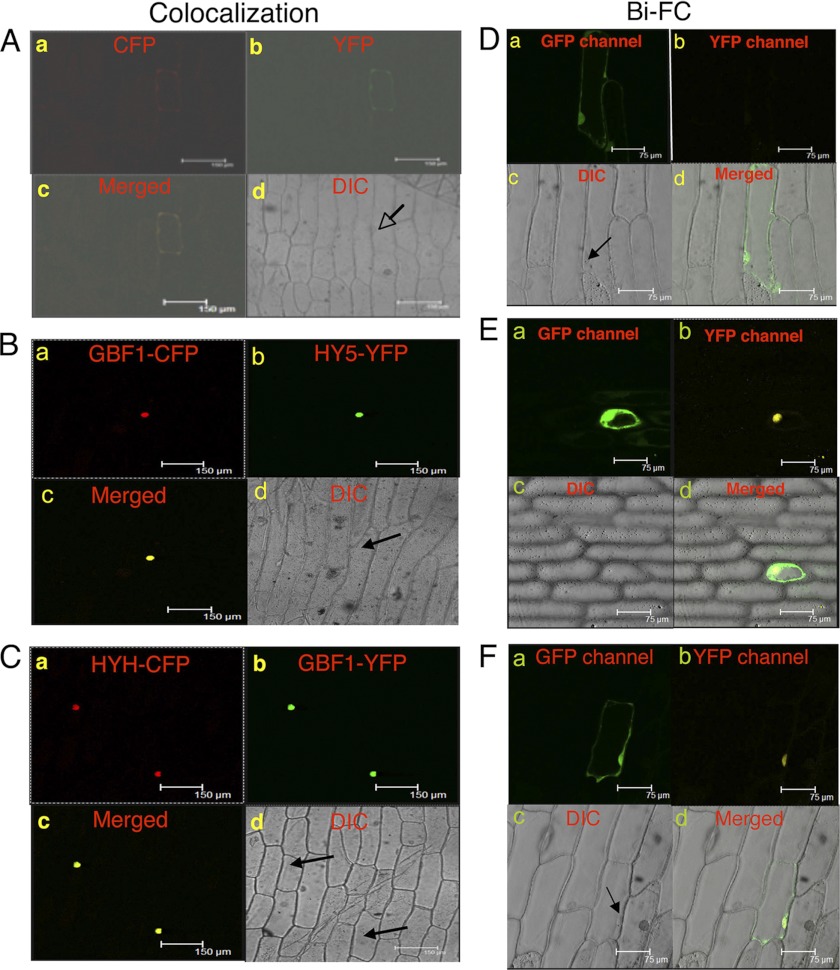
FIGURE 3.
GBF1 co-localizes and interacts with HY5 and HYH in the nucleus of onion epidermal cells. In A–C, a shows CFP channel fluorescence, changed to a red color; b shows YFP channel fluorescence, changed to a green color; c shows a merged image of a and b; and d shows the corresponding bright field image. The arrowheads indicate the positions of the nuclei. A, pAM-PAT-35SS-CFP and pAM-PAT-35SS-YFP vectors were co-transformed into onion epidermal cells. B, pAM-PAT-35SS-GBF1-CFP and pAM-PAT-35SS-HY5-YFP constructs were co-transformed into onion epidermal cells. C, pAM-PAT-35SS-HYH-CFP and pAM-PAT-35SS-GBF1-YFP constructs were co-transformed into onion epidermal cells. In D–F, a shows GFP channel fluorescence produced by GFP of pCAMBIA-1302 vector, serving as a control of transformation; b shows the YFP channel image produced by reconstruction of YFP; c shows the respective bright field image; and d shows a merged image of a, b, and c. The arrowheads indicate the position of the nuclei. D, empty BiFC vectors and pCAMBIA-1302 vector (GFP) were co-transformed into onion epidermal cells. E, GBF1-YFPN-ter and HY5-YFPC-ter constructs were co-expressed into onion epidermal cells along with GFP. F, HYH-YFPN-ter and GBF1-YFPC-ter constructs were co-expressed into onion epidermal cells along with GFP. DIC, differential interference contrast.
GBF1 Physically Interacts in Vivo with HY5 and HYH
To confirm the physical interactions of GBF1 with HY5 and HYH in vivo conditions, a BiFC experiment was performed. For this, GBF1 full-length (FL) CDS was fused to the N terminus of YFP in the pUC-SPYNE vector, and HY5 FL CDS was fused to the C terminus of YFP in the pUC-SPYCE vector (33). These constructs were co-bombarded into the onion epidermal cells along with pCAMBIA-1302 vector, which has a GFP tag and was used as a positive control for transformation. Although empty vectors did not give any YFP fluorescence (Fig. 3D), interaction of GBF1 and HY5 produced strong YFP fluorescence in the nucleus (Fig. 3E). Similarly, to confirm the in vivo physical interactions between GBF1 and HYH, GBF1 FL CDS was fused to the C terminus of YFP in pUC-SPYCE vector, and HYH FL CDS was fused to the N terminus of YFP in the pUC-SPYNE vector. Upon co-bombardment, interaction of GBF1 and HYH produced strong YFP fluorescence in the nucleus (Fig. 3F). The bright field image and image merged with fluorescence confirm the nuclei positions. Taken together, these results demonstrate the in vivo physical interactions of GBF1 with HY5 and HYH proteins.
The bZIP Domain of GBF1 Is Necessary and Sufficient for Its Interaction with Full-length HY5 and HYH Proteins
In the bZIP proteins, the basic region is usually responsible for DNA sequence recognition, binding, and nuclear localization, and the leucine zipper is important for the creation of hetero- or homodimers (37–39). To determine the specific domains of GBF1, HY5, and HYH that are mediating the direct protein-protein interaction, we split each protein into two domains: 1) the N-terminal domain (ΔZIP), which contains only the N-terminal part of the protein but excludes the leucine zipper domain; 2) the C-terminal domain (bZIP), which contains only the C-terminal bZIP part of the protein. Because the basic region is important for nuclear localization, and the proteins were found to be interacting in the nucleus, the basic region was included in both of the truncated domains. To analyze the HY5 domain that mediates its interaction with GBF1, the ΔZIP (aa 1–115) and bZIP (aa 77–168) domains of HY5 were individually fused with YFPC-ter and were subsequently co-expressed with full-length GBF1-YFPN-ter in onion epidermal cells along with pCAMBIA-1302 vector. Cells positive for GFP fluorescence were further examined for BiFC fluorescence. As shown in Fig. 4, A and B, both ΔZIP and bZIP domains of HY5 were unable to produce BiFC fluorescence with full-length GBF1. To examine whether any specific HYH domain could interact with full-length GBF1, the ΔZIP (aa 1–100) and bZIP (aa 74–149) domains of HYH were individually fused with YFPN-ter and subsequently coexpressed with full-length GBF1-YFPC-ter in onion epidermal cells along with pCAMBIA-1302 vector. As shown in Fig. 4, C and D, neither of the truncated domains of HYH was able to produce BiFC fluorescence with full-length GBF1. The lack of interaction of truncated versions of HY5 and HYH with full-length GBF1 protein suggests that the overall structure of HY5 and HYH is important for interaction with GBF1.
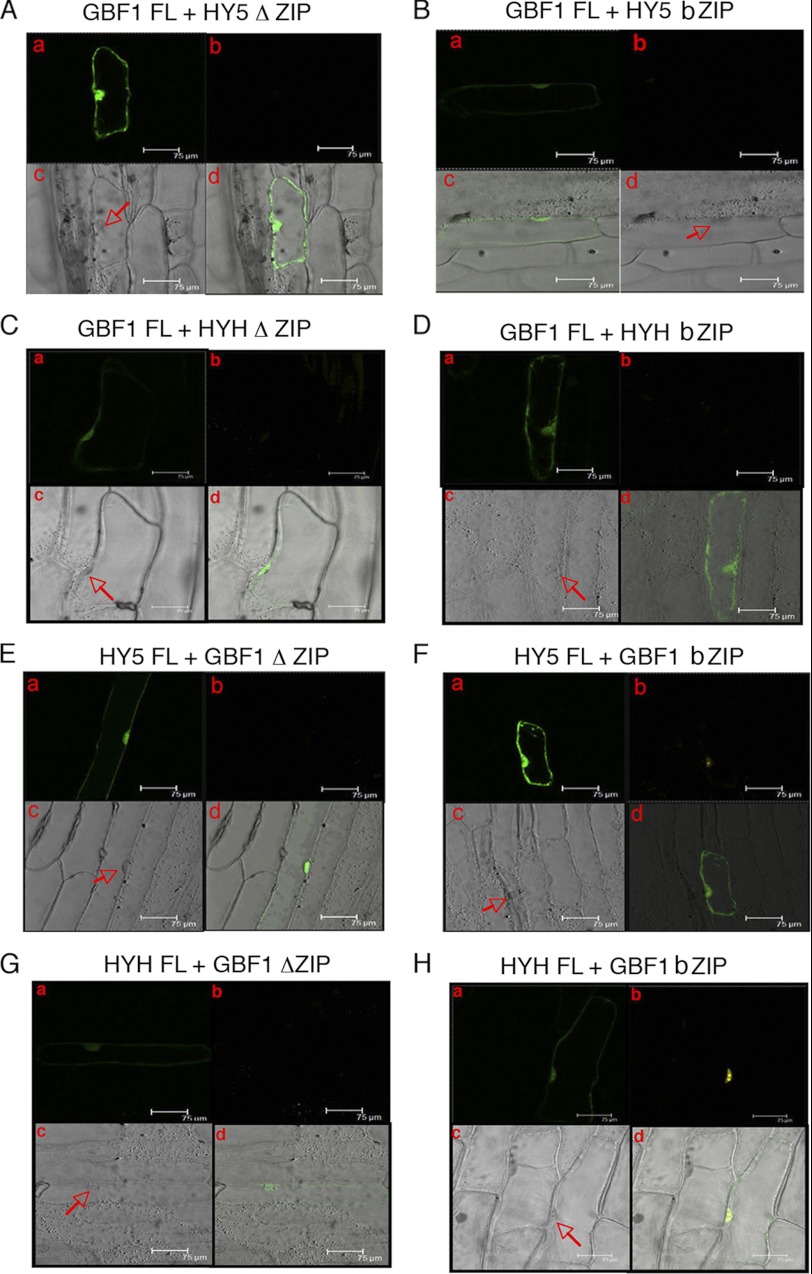
FIGURE 4.
The bZIP domain of GBF1 interacts with full-length HY5 and HYH proteins. In A–H, a shows GFP channel fluorescence produced by GFP of pCAMBIA-1302 vector, serving as a control of transformation; b shows the YFP channel image produced by reconstruction of YFP; c shows the respective bright field image; and d shows a merged image of a, b, and c. A and B, full-length GBF1 (fused to YFPN-ter) and domains of HY5 (fused to YFPC-ter) co-expressed in onion epidermal cells along with GFP. C and D, full-length GBF1 (fused to YFPC-ter) and domains of HYH (fused to YFPN-ter) co-expressed in onion epidermal cells along with GFP. E and F, full-length HY5 (fused to YFPC-ter) and domains of GBF1 (fused to YFPN-ter) co-expressed in onion epidermal cells along with GFP. G and H, full-length HYH (fused to YFPN-ter) and domains of GBF1 (fused to YFPC-ter) co-expressed in onion epidermal cells along with GFP.
In parallel experiments, to dissect the structural requirement of GBF1 protein for interaction with full-length HY5 or HYH, the ΔZIP (aa 1–243) and bZIP (aa 220–315) domains of GBF1 were individually fused with YFPC-ter and YFPN-ter and then co-expressed with FL HYH-YFPN-ter and FL HY5-YFPC-ter, respectively, along with pCAMBIA-1302 vector, in onion epidermal cells. As shown in Fig. 4, E–H, although the ΔZIP domain of GBF1 was unable to produce any BiFC fluorescence with both FL HY5 and FL HYH proteins, the bZIP domain of GBF1 produced the yellow fluorescence of reconstructed YFP. Altogether, these results suggest that the bZIP domain of GBF1 is necessary and sufficient for mediating its interaction with the FL HY5 and FL HYH proteins.
To examine the formation of homodimer of GBF1, we used FL GBF1 and also the bZIP domain of GBF1 in BiFC experiments. As shown in supplemental Fig. 1, B and C, the FL as well as the bZIP domain of GBF1 produced the yellow fluorescence of reconstructed YFP. These results suggest that FL GBF1 protein and the bZIP domains are able to form the homodimer.
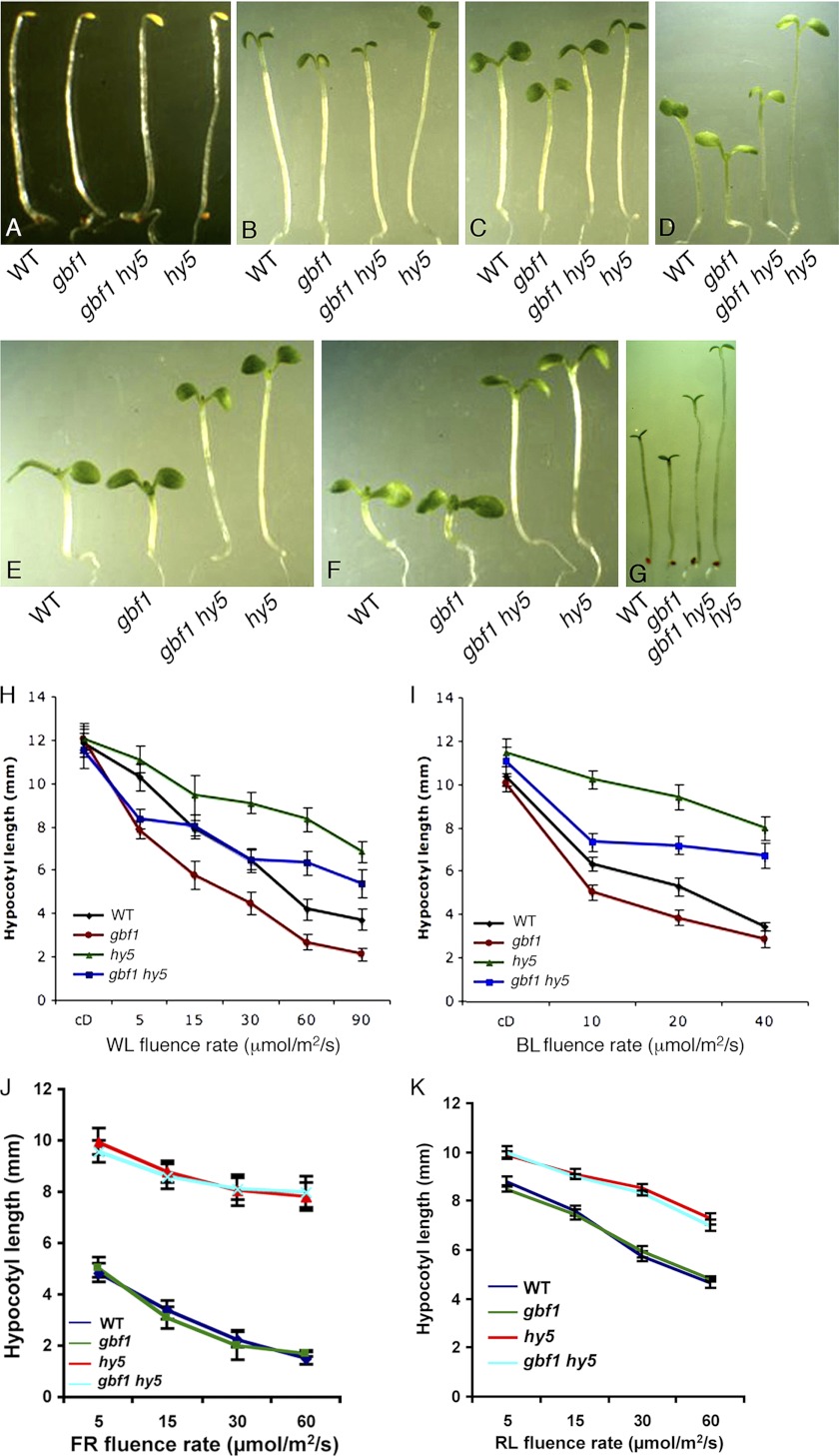
GBF1 and HY5 Show Light Intensity-dependent Epistatic Relationship for White Light-mediated Hypocotyl Elongation
Earlier studies have shown that GBF1 plays a negative regulatory role in WL- and BL-mediated inhibition of hypocotyl elongation, whereas HY5 exhibits a positive regulatory role for hypocotyl elongation at various wavelengths of light (16, 27). Because GBF1 physically interacts with HY5, we wanted to examine the physiological significance of such interactions during Arabidopsis seedling development. To determine that, we constructed gbf1 hy5 double mutants and investigated the morphology of the seedlings in dark and various light conditions. In darkness, gbf1, hy5, and gbf1 hy5 exhibited skotomorphogenic growth similar to wild type seedlings (Fig. 5, A and H). While examining the growth of gbf1 hy5 double mutants at various intensities of WL, it was observed that the inhibition of hypocotyl elongation in gbf1 hy5 double mutants was similar to gbf1 at lower fluence rates (Fig. 5, B and H). However, at moderate light intensities of WL, gbf1 hy5 displayed hypocotyl length similar to wild type (Fig. 5, C, D, and H). Furthermore, at higher fluence rates of WL, gbf1 hy5 double mutants exhibited hypocotyl length similar to hy5 mutants (Fig. 5, E, F, and H). These results revealed altered functional relationships between GBF1 and HY5, depending upon light intensity. Whereas gbf1 is epistatic to hy5 at lower fluences of WL, gbf1 and hy5 work antagonistically at moderate fluence rates of WL. At higher fluence rates, hy5 is epistatic to gbf1 in the regulation of hypocotyl growth.
FIGURE 5.
GBF1 genetically interacts with HY5. The visible phenotypes of the seedlings grown in constant dark (A), WL (5 μmol/m2/s) (B), WL (15 μmol/m2/s) (C), WL (30 μmol/m2/s) (D), WL (60 μmol/m2/s) (E), WL (90 μmol/m2/s) (F), and BL (15 μmol/m2/s) (G). In each panel, 6-day-old segregated wild type (WT) and various mutant lines are shown. H, quantification of hypocotyl length of 6-day-old seedlings grown in constant dark or at various fluences of WL. I, quantification of hypocotyl length of 6-day-old seedlings grown in constant dark or at various fluences of BL. J, quantification of hypocotyl length of 6-day-old seedlings grown at various fluences of FR. K, quantification of hypocotyl length of 6-day-old seedlings grown at various fluences of RL. Approximately 30 seedlings were used for the measurement of hypocotyl length. Error bars, S.D. n ≥ 3 independent experiments with similar results.
To examine whether the light intensity-dependent genetic interaction between GBF1 and HY5 was also present in BL, we monitored the growth of gbf1 hy5 double mutants in BL. As shown in Fig. 5, G and I, gbf1 hy5 seedlings displayed hypocotyl length in between gbf1 and hy5 single mutants at various fluence rates of BL, suggesting that gbf1 and hy5 function antagonistically to each other in BL-mediated inhibition of hypocotyl elongation. To further test the genetic interaction of GBF1 and HY5 in red light (RL) and far red (FR), we monitored the growth of gbf1 hy5 double mutants in RL and FR. As shown in Fig. 5, J and K, gbf1 hy5 seedlings displayed hypocotyl length similar to hy5, suggesting that the additional loss of function of GBF1 does not affect the hypocotyl growth of hy5 mutants in RL and FR.
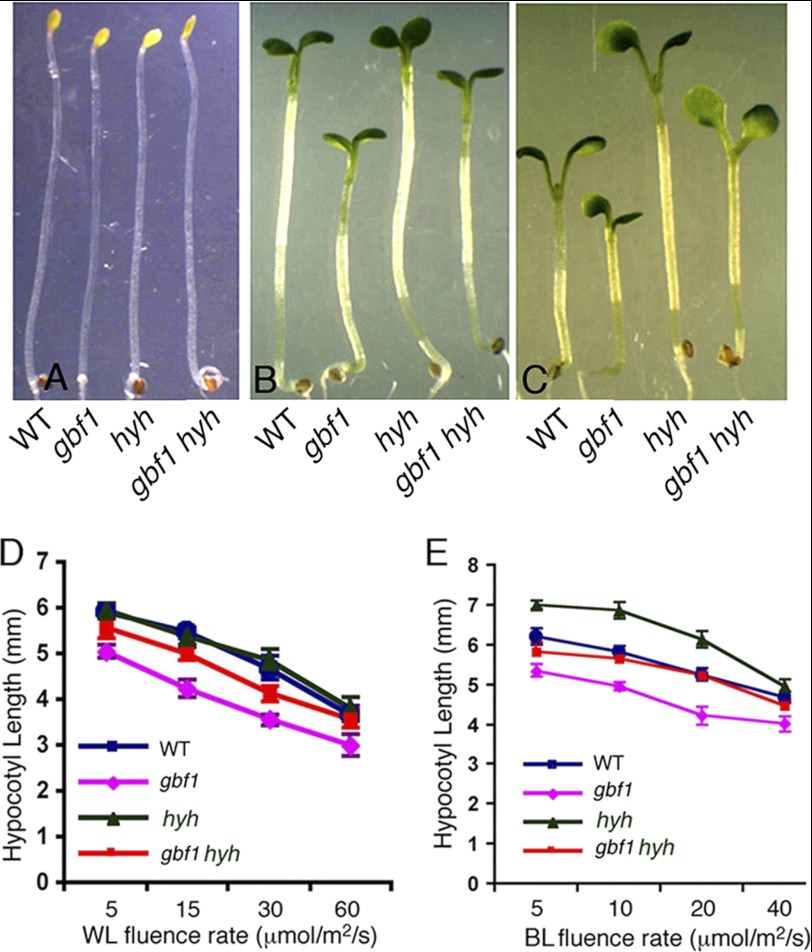
The Additional Mutation in HYH Alters Hypocotyl Length of gbf1 Mutants
HYH, a close homologue of HY5, acts as a positive regulator of hypocotyl growth specifically in BL (23). To determine the possible physiological relationship between GBF1 and HYH, gbf1 hyh double mutants were generated through genetic crosses, and the hypocotyl length of gbf1 hyh double mutants was examined in the dark and at various fluences of WL and BL. The gbf1, hyh, and gbf1 hyh seedlings displayed skotomorphogenic growth similar to wild type seedlings in the darkness (Fig. 6A). As reported earlier, gbf1 mutants showed shorter hypocotyl than wild type, whereas hyh mutants showed hypocotyl length similar to wild type in WL (Fig. 6, B and D) (23, 27). The hypocotyl length of gbf1 hyh double mutants was found to be significantly higher than gbf1 mutants, suggesting that hyh partially suppresses the gbf1 phenotype under WL conditions (Fig. 6D). The gbf1 hyh double mutant seedlings displayed hypocotyl length similar to wild type in BL, suggesting that GBF1 and HYH play antagonistic roles in light-mediated inhibition of hypocotyl elongation in BL (Fig. 6, C and E).
FIGURE 6.
GBF1 Genetically Interacts with HYH. The visible phenotypes of seedlings grown in constant dark (A), WL (15 μmol/m2/s) (B), and BL (30 μmol/m2/s) (C). In each panel, 6-day-old segregated wild type (WT) and various mutant lines are shown. D, quantification of hypocotyl length of 6-day-old seedlings grown at various fluences of WL. E, quantification of hypocotyl length of 6-day-old seedlings grown at various fluences of BL. Approximately 30 seedlings were used for the measurement of hypocotyl length. Error bars, S.D. n ≥ 3 independent experiments with similar results.
Genetic Interactions of GBF1 with HY5 and HYH Modulate Physiological Responses and Light-regulated Gene Expression
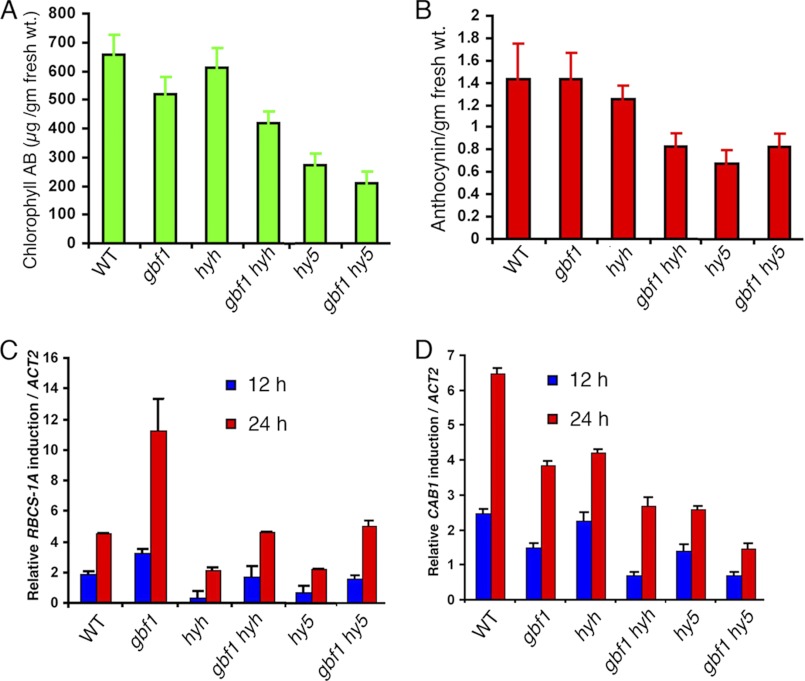
Earlier studies have shown that hyh mutant seedlings accumulate a reduced level of anthocyanin as compared with wild type, although there is no defect in chlorophyll accumulation (23). The hy5 mutants accumulate lower levels of anthocyanin and chlorophyll than the wild type seedlings (16). On the other hand, it has been shown that gbf1 mutant seedlings accumulate a reduced level of chlorophyll compared with wild type; however, there is no significant change in anthocyanin accumulation (27). To determine the genetic interactions of GBF1 with HY5 and HYH for anthocyanin and chlorophyll accumulation, we examined the chlorophyll and anthocyanin levels in various single and double mutant backgrounds. The anthocyanin content in gbf1 hy5 double mutants was found to be similar to hy5 seedlings, suggesting that the additional loss of GBF1 function does not alter the anthocyanin accumulation in hy5 mutants (Fig. 7B). On the other hand, whereas the accumulation of anthocyanin was found to be similar to wild type in gbf1 and slightly reduced in hyh single mutants, there was significant reduction in the accumulation of anthocyanin in gbf1 hyh double mutants. These results suggest a synergistic effect of GBF1 and HYH in anthocyanin accumulation (Fig. 7B).
FIGURE 7.
Modulation of physiological responses and light-induced gene expression in gbf1 hy5 and gbf1 hyh double mutants. A, quantification of chlorophyll content in 6-day-old WT (segregated wild type) and various mutant seedlings grown in WL (60 μmol/m2/s). B, quantification of anthocyanin accumulation in 6-day-old WT and various mutant seedlings grown in WL (60 μmol/m2/s). C and D, real-time PCR analysis of RBCS-1A and CAB1 transcript levels, respectively, from 5-day-old constant dark grown seedlings transferred to BL (30 μmol/m2/s) for 12 and 24 h. ACT2, actin 2, used as control. Error bars, S.D. n ≥ 3 independent experiments with similar results.
Examination of chlorophyll content revealed that gbf1 mutants had a lower level of chlorophyll accumulation than wild type, and the chlorophyll accumulation was drastically reduced in the hy5 mutant. Measurements of chlorophyll contents in gbf1 hy5 double mutants revealed that the chlorophyll accumulation was further reduced in gbf1 hy5 double mutants as compared with either of the single mutants (Fig. 7A). These results indicate an additive effect of HY5 and GBF1 in chlorophyll accumulation. The accumulation of chlorophyll was found to be similar to the wild type level in hyh mutants; however, the chlorophyll accumulation was further reduced in gbf1 hyh double mutants as compared with gbf1 alone (Fig. 7A). These results suggest that HYH and GBF1 work synergistically to regulate the accumulation of chlorophyll.
The light-mediated induction of CAB1 and RBCS-1A gene expression is differentially regulated by GBF1 (27). GBF1 acts as a positive regulator of CAB1 but negatively regulates RBCS-1A gene expression. HY5 acts as a positive regulator for induction of both CAB1 and RBCS-1A gene expression (19–21, 40). HYH directly binds to the G-box of the RBCS-1A promoter (23), although the effect of HYH on RBCS-1A expression is not clear. To determine the effect of genetic interactions of GBF1 with HY5 and HYH on light-regulated expression of CAB1 and RBCS-1A, 5-day-old dark-grown seedlings were transferred to BL for 12 and 24 h, and real-time PCR was performed. As expected, an elevated level of induction of RBCS-1A expression in gbf1 mutants was observed as compared with wild type. The hy5 and hyh mutants showed a lower level of induced expression of RBCS-1A than wild type. The expression of RBCS-1A in gbf1 hy5 and gbf1 hyh double mutants was found to be between the corresponding single mutants (Fig. 7C). These results indicate that GBF1 acts antagonistically to HY5 and HYH in the regulation of RBCS-1A expression. The induction of CAB1 expression was found to be less than that of wild type in gbf1, hy5, and hyh mutants. In gbf1 hy5 or gbf1 hyh double mutants, the induction of CAB1 was further reduced as compared with corresponding single mutants, suggesting that GBF1 additively functions with HY5 and HYH to regulate CAB1 gene expression (Fig. 7D).
Combinatorial Regulation of Photomorphogenic Growth and Gene Expression by GBF1, HY5, and HYH
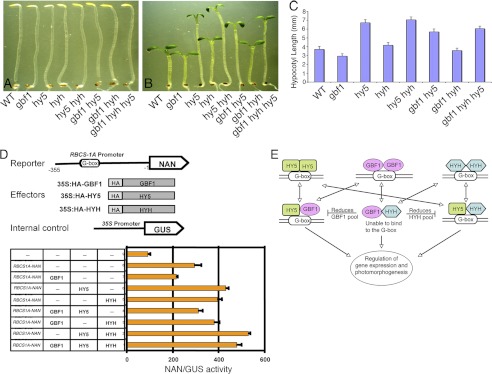
Based on the findings that GBF1 plays an antagonistic role with HY5 and HYH in photomorphogenic growth, we wanted to examine the effect of loss of function of GBF1 on hy5 hyh double mutants. It has been shown that additional loss of HYH function enhances the hy5 phenotype in BL (23). We constructed gbf1 hy5 hyh triple mutants and examined the growth of the seedlings in the dark and BL. The triple mutants displayed skotomorphogenic growth similar to wild type seedlings in the darkness (Fig. 8A). While examining the growth of the seedlings in BL, hypocotyl length of the gbf1 hy5 hyh triple mutant seedlings was found to be similar to that of the gbf1 hy5 double mutants (Fig. 8, B and C). These results together with the DNA-protein interaction studies suggest that GBF1 and HY5 are likely to play more important roles than HYH in BL-mediated inhibition of hypocotyl elongation.
FIGURE 8.
Concerted function of GBF1, HY5, and HYH in light-regulated gene expression and photomorphogenic growth. A, representative picture of dark-grown seedlings. Shown from the left are segregated wild type (WT), gbf1, hy5, hyh, hy5 hyh, gbf1 hy5, gbf1 hyh, and gbf1 hy5 hyh. B, representative picture of 6-day-old BL (30 μmol/m2/s)-grown seedlings. Shown from the left are segregated wild type (WT), gbf1, hy5, hyh, hy5 hyh, gbf1 hy5, gbf1 hyh, and gbf1 hy5 hyh. C, quantification of hypocotyl length of 6-day-old BL-grown seedlings. Approximately 30 seedlings were used for the measurement of hypocotyl length. The error bars indicate S.D. D (top), schematic representation of promoter-reporter, effectors, and internal control construct used in transient expression analysis in Arabidopsis protoplasts. The 355-bp RBCS-1A promoter upstream from the start codon was fused to the NAN reporter. Effector constructs are under the control of the CaMV 35S promoter. A Pro35S:GUS construct was used as an internal control to normalize the differences in the transfection efficiencies. D (bottom), quantification of the RBCS-1A promoter-reporter activity in the presence of given combinations of effectors in transiently transformed Arabidopsis leaf protoplasts. After transfection, protoplast samples were incubated in continuous BL (40 μmol/m2/s) for 12–14 h. Half of the protoplast samples were harvested for total RNA extraction, and the other half were used for the reporter activity measurements. The NAN/GUS activity was then normalized against the relative transcript of the effectors in each combination of the effectors. The x axis values are expressed as NAN activity relative to GUS. Given are mean and S.D. values of three replicates. n ≥ 3 independent experiments with similar results. E, a model of interaction of GBF1 with HY5 and HYH. GBF1, HY5, and HYH homodimers bind to the G-box light-responsive element. GBF1 and HYH heterodimerize with HY5 and bind to the G-box. Physical interaction of GBF1 with HYH does not lead to their heterodimerization at the G-box; however, the GBF1-HYH pool results in the reduction of GBF1 and HYH pools available for the formation of GBF1-HY5 and HY5-HYH heterodimers, respectively.
In order to study the interplay of these three bZIP proteins in gene expression, we analyzed the blue light-regulated RBCS-1A expression by multiple combinations of these proteins. For BL-mediated RBCS-1A expression analysis, we studied transient gene expression in Arabidopsis protoplasts using the RBCS-1A promoter-reporter construct. As shown in Fig. 8D, the RBCS-1A promoter construct driving the expression of a NAN reporter, was co-transformed with effector plasmids of GBF1, HY5, and HYH. NAN is a synthetic sialidase reporter gene that possesses kinetic properties similar to those of the GUS reporter; however, it is more sensitive than GUS (35). Because GBF1 and HYH are BL-specific transcription factors and the gene expression analysis in single and double mutants was performed in BL (Fig. 7), we analyzed the promoter-reporter activity in BL conditions. As shown in Fig. 8D, expression of 35S:GBF1 resulted in the reduction of activity of RBCS-1A-NAN, consistent with the negative regulatory role of GBF1 in RBCS-1A expression. Similarly, the expression of 35S:HY5 and 35S:HYH resulted in an increase of reporter activity, suggesting the positive regulatory roles of HY5 and HYH, respectively, in RBCS-1A expression. Co-expression of both 35S:HY5 and 35S:HYH resulted in further enhancement of the reporter activity compared with either 35S:HY5 or 35S:HYH, indicating an additive effect of HY5 and HYH on RBCS-1A expression. Co-expression of 35S:GBF1 and 35S:HY5 showed intermediate reporter activity, demonstrating the antagonistic effect of GBF1 and HY5 on RBCS-1A expression. However, the antagonistic effect on RBCS-1A expression observed in gbf1 hyh double mutant (Fig. 7) was not found upon co-expression of 35S:GBF1 with 35S:HYH (Fig. 8D). Given the fact that GBF1 and HYH physically interact and do not form the G-box-binding heterodimer (Figs. 1–3), it might be possible that their physical interaction oppositely affects the negative regulation of RBCS-1A expression by GBF1. Upon co-expression of all three bZIP proteins, the RBCS-1A-NAN activity was found to be higher than for individual expression of either 35S:HY5 or 35S:HYH; however, it was less than for co-expression of 35S:HY5 and 35S:HYH. These results indicate that GBF1 antagonistically regulates HY5-HYH heterodimer-mediated RBCS-1A expression. Altogether, these results, along with real-time gene expression analysis, strongly suggest that the observed antagonistic interaction between GBF1 and HY5 for RBCS-1A expression is due to their direct co-binding to the RBCS-1A promoter; however, the observed antagonistic effect between GBF1 and HYH seems to be indirect.
DISCUSSION
Earlier studies have suggested that the massive change in gene expression during photomorphogenic development involves multiple transcriptional cascades (5). Although several transcription factors involved in light-mediated seedling development have been studied in fair detail, functional interactions among themselves are not well established to provide sufficient insights into these transcriptional cascades. In this study, we have analyzed molecular and functional interrelationships of GBF1 with two other bZIP proteins, HY5 and HYH, involved in light-signaling pathways. The DNA-protein interaction studies suggest that although GBF1 does not form the G-box-binding heterodimer with HYH, it does so with HY5 protein. Furthermore, our results show that when both GBF1 and HY5 are incubated together with the DNA, their homodimeric forms bind to DNA more strongly compared with when they are individually incubated with DNA. The ChIP data also support these results. In the absence of functional HY5 protein, the binding of GBF1 to RBCS-1A promoter is decreased to about 2-fold compared with when functional HY5 is present. Altogether, these results suggest that HY5 not only forms a DNA-binding heterodimer with GBF1; it also increases the GBF1 binding to the RBCS-1A promoter. Similarly, GBF1 was also found to increase the binding of HY5 to the RBCS-1A promoter. However, when examined for GBF1-HYH heterodimer, no such regulation was found. The domain-wise BiFC experiments suggest that the N-terminal part of GBF1 is dispensable for dimerization with HY5. Thus, considering the gel shift data together, it is tempting to speculate that whereas one full-length protein increases the DNA-binding ability of the other, the N-terminal part of GBF1 may have some inhibitory role in dimerization with HY5.
Several lines of experimental results, including protein-protein interactions through BiFC experiments and in vitro pull-down assays using total plant protein extract, demonstrate the physical interactions of GBF1 with both HY5 and HYH proteins. One plausible mechanism of physical interaction of GBF1 and HYH may be that it controls the heterodimer pools of HY5-GBF1 and HY5-HYH and thereby regulates gene expression and photomorphogenic growth in BL (Fig. 8E). This notion is supported by the protoplast experiments (Fig. 8D), where we have observed that when all three heterodimers (GBF1-HY5, HY5-HYH, and GBF1-HYH) are present, the activity of the RBCS-1A reporter is between those of the GBF1-HY5- and HY5-HYH-mediated RBCS-1A reporter expression, suggesting that the GBF1-HYH heterodimer pool negatively regulates both the GBF1-HY5 and HY5-HYH heterodimer pools. It is reasonable to speculate that GBF1 or HY5 homodimers should also be present to finely tune the regulation of gene expression and seedling development. For example, in the absence of GBF1 (in a null mutant background), HY5 homodimer and the HY5-HYH heterodimer pool predominate and bind to the G-box and thereby increase the transcript level of RBCS-1A. Similarly, in the hy5 mutant background, thew GBF1 homodimer level goes up, down-regulating the expression of RBCS-1A. Therefore, in wild type seedlings, the pool of homo- and heterodimers of GBF1 and HY5 should be present in an appropriate proportion, which results in the fine-tuning of gene expression. The heterodimerization of GBF1-HY5 or HY5-HYH thus might be a potential mechanism to generate positive or negative regulatory responses specifically downstream from cryptochromes.
The co-localization experiment reveals that GBF1 co-localizes with HY5 or HYH in the nucleus. These observations are further substantiated by BiFC experiments. The b-ZIP factors form dimers, involving leucine zipper domain, and bind to DNA with their basic domain as a dimer (37). Our domain-wise interaction study reveals that the bZIP domain of GBF1 is necessary and sufficient to mediate its interaction with HY5 and HYH proteins. However, in the case of HY5 and HYH, we found that the bZIP domain alone is not sufficient to mediate its interaction with FL GBF1. It is possible that in addition to the bZIP domain, other motifs are also involved in their interaction with FL GBF1. The bZIP domains of HY5 and HYH share the highest sequence similarity (23), and our results show that the nature of interaction of GBF1 with these two close homologous proteins, HY5 and HYH, is very similar.
In order to analyze the physiological significances of the observed protein-protein interactions, gbf1 hy5 and gbf1 hyh double mutants were generated. Analysis of the gbf1 hy5 double mutant revealed that it exhibits altered photomorphogenic growth in a light intensity-dependent manner in WL. At lower intensity of WL, gbf1 hy5 showed hypocotyl length similar to that of gbf1, suggesting that GBF1 is functionally downstream from HY5. However, moderate light intensities promoted the antagonistic interaction between these two transcription factors. At higher intensities, however, the hypocotyl length of gbf1 hy5 is closer to that of hy5 seedlings. These altered morphologies of gbf1 hy5 could be attributed to altered interaction of GBF1 and HY5 at the transcriptional level. In a recent chromatin immunoprecipitation study, the GBF1 upstream region was found to be enriched in the immunoprecipitates with αHA antibody from 35S-HA:HY5 transgenic lines (21). These results indicate that HY5 binds to the promoter of GBF1 and thereby suggest that GBF1 may act downstream from HY5. On the other hand, antagonistic interaction between gbf1 and hy5 at moderate light intensities indicates that these two proteins may physically interact and form heterodimers to regulate light-dependent gene expression. At higher intensities of WL, long hypocotyl phenotype of gbf1 hy5 reveals the increasing essentiality of functional HY5. It is worth mentioning here that earlier studies have revealed more prominent morphological defects of gbf1 at lower fluence rates, whereas hy5 mutants display the strong phenotype at higher fluence rates of WL (13, 27). In BL, the hypocotyl elongation of gbf1 hy5 is found to be largely the outcome of antagonistic interactions of gbf1 and hy5. However, in RL and FR, the gbf1 hy5 double mutants displayed a phenotype similar to that of hy5 mutants. Consistent with these results, Bhatia et al. (41) have previously shown that mutation in SHW1 (short hypocotyl in white light 1) causes altered hypocotyl elongation in WL, whereas no such effect was found in RL, FR, and BL. It could be envisioned that because the wild type plants in an ambient environment are exposed to various intensities of light, altered genetic and molecular interactions are required for optimum use of the available light sources.
The antagonistic effect of GBF1 and HYH for hypocotyl growth is observed in BL. Furthermore, although hyh mutants do not display any morphological defects in WL (23), the hypocotyl length of gbf1 hyh double mutants indicates that hyh partially suppresses the phenotype of gbf1 mutants in WL. Such redundant functional relationships have also been observed with HY5 and CAM7 (14). Furthermore, BL-mediated induction of RBCS-1A was also found to be antagonistically regulated by GBF1 and HY5 or HYH. The transient gene expression analyses further suggest that the observed antagonistic interactions between GBF1 and HY5 is direct; however, in the case of GBF1-HYH, the antagonistic relationship is likely to be due to an indirect effect. Analysis of CAB1 expression revealed that GBF1, which acts as a positive regulator of CAB1, acts additively with both HY5 and HYH in its regulation.
Considering the heterodimer formation of HY5 with HYH and GBF1 and the strong phenotype of hy5 mutants in WL and in multiple wavelengths of light as compared with the BL-specific relatively weak phenotype of gbf1 and hyh, it is likely that HY5 plays the major role and functions in concert with wavelength-specific regulators, such as GBF1 or HYH, in Arabidopsis seedling development. Furthermore, whereas gbf1 mutants display the hypocotyl phenotype in BL and WL, the phenotype of hyh mutants is restricted to BL only. Thus, taken together, HY5 seems to be functionally more important than GBF1, and furthermore, GBF1 appears to be functionally more important than HYH in Arabidopsis seedling development. This notion is further supported by the analyses of gbf1 hy5 hyh triple mutants.
Temporal and spatial availability of the proteins is important for their combinatorial function. GBF1, HY5, and HYH proteins have been shown to accumulate in light-grown seedlings and to be degraded in the dark through the 26S proteosome-mediated pathway. However, the role of COP1 has been found to be opposite in the stability of GBF1 versus HY5 and HYH proteins. COP1 destabilizes HY5 and HYH proteins in the dark, whereas it stabilizes the GBF1 protein in the light (13, 23, 28, 42). Furthermore, it has been reported that COP1 is not involved in the proteasome-mediated degradation of GBF1 protein in the darkness (28). GBF1, HY5, and HYH may be directly related to the subcellular shuttling of COP1 from nucleus to cytoplasm in response to the onset of light signal (23, 43, 44). Light might also regulate the subcellular localization of transcription factors through phosphorylation (45). One plausible mechanism might be that COP1 stabilizes GBF1 in the cytoplasm of light-grown seedlings, and nuclear translocation of this pool of GBF1 may be followed by GBF1-HY5 heterodimer formation based upon the light conditions and physiological necessities. This supports the notion that in the absence of functional COP1, GBF1 gets destabilized, and a reduced pool of GBF1 in the nucleus may give rise to HY5 homodimer formation and up-regulation of RBCS1A promoter in cop1 seedlings grown in light (16). Positive signals from the photoreceptors received by downstream transcription factors are thus balanced and thereby result in finely tuned gene expression and photomorphogenic growth.
Supplementary Material
Acknowledgment
We thank Wolfgang Dröge-Laser for providing vectors for transient expression analysis.
This work is supported by a J. C. Bose National Fellowship Award Grant and in part by a research grant from the Department of Science and Technology, Government of India (to S. C.).

This article contains supplemental Fig. 1.
- bZIP
- basic leucine zipper
- WL
- white light
- FR
- far red light
- RL
- red light
- BL
- blue light
- FL
- full-length
- FP
- forward primer
- RP
- reverse primer
- GW
- Gateway®-compatible
- AMV
- avian myeloblastosis virus
- CFP
- cyan fluorescent protein
- Ni-NTA
- nickel-nitrilotriacetic acid
- BiFC
- bimolecular fluorescence complementation
- CDS
- coding sequence
- aa
- amino acids.
REFERENCES
- 1. Koornneef M., Bentsink L., Hilhorst H. (2002) Seed dormancy and germination. Curr. Opin. Plant Biol. 5, 33–36 [DOI] [PubMed] [Google Scholar]
- 2. Nagatani A., Reed J. W., Chory J. (1993) Isolation and initial characterization of Arabidopsis mutants that are deficient in phytochrome A. Plant Physiol. 102, 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Whitelam G. C., Johnson E., Peng J., Carol P., Anderson M. L., Cowl J. S., Harberd N. P. (1993) Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5, 757–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Neff M. M., Fankhauser C., Chory J. (2000) Light. An indicator of time and place. Genes Dev. 14, 257–271 [PubMed] [Google Scholar]
- 5. Jiao Y., Lau O. S., Deng X. W. (2007) Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8, 217–230 [DOI] [PubMed] [Google Scholar]
- 6. Cashmore A. R., Jarillo J. A., Wu Y. J., Liu D. (1999) Cryptochromes. Blue light receptors for plants and animals. Science 284, 760–765 [DOI] [PubMed] [Google Scholar]
- 7. Lin C. (2002) Phototropin blue light receptors and light-induced movement responses in plants. Sci. STKE 2002, pe5. [DOI] [PubMed] [Google Scholar]
- 8. Schepens I., Duek P., Fankhauser C. (2004) Phytochrome-mediated light signaling in Arabidopsis. Curr. Opin. Plant Biol. 7, 564–569 [DOI] [PubMed] [Google Scholar]
- 9. Rizzini L., Favory J. J., Cloix C., Faggionato D., O'Hara A., Kaiserli E., Baumeister R., Schäfer E., Nagy F., Jenkins G. I., Ulm R. (2011) Perception of UV-B by the Arabidopsis UVR8 protein. Science 332, 103–106 [DOI] [PubMed] [Google Scholar]
- 10. Ma L., Li J., Qu L., Hager J., Chen Z., Zhao H., Deng X. W. (2001) Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13, 2589–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tepperman J. M., Zhu T., Chang H. S., Wang X., Quail P. H. (2001) Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc. Natl. Acad. Sci. U.S.A. 98, 9437–9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ang L. H., Chattopadhyay S., Wei N., Oyama T., Okada K., Batschauer A., Deng X. W. (1998) Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1, 213–222 [DOI] [PubMed] [Google Scholar]
- 13. Osterlund M. T., Hardtke C. S., Wei N., Deng X. W. (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466 [DOI] [PubMed] [Google Scholar]
- 14. Kushwaha R., Singh A., Chattopadhyay S. (2008) Calmodulin7 plays an important role as transcriptional regulator in Arabidopsis seedling development. Plant Cell 20, 1747–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Koornneef M., van der Veen J. H. (1980) Induction and analysis of gibberellin-sensitive mutants in Arabidopsis thaliana (L.) Heyne. Theor. Appl. Genet. 58, 257–263 [DOI] [PubMed] [Google Scholar]
- 16. Ang L. H., Deng X. W. (1994) Regulatory hierarchy of photomorphogenic loci. Allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell 6, 613–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pepper A. E., Chory J. (1997) Extragenic suppressors of the Arabidopsis det1 mutant identify elements of flowering-time and light-response regulatory pathways. Genetics 145, 1125–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oyama T., Shimura Y., Okada K. (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11, 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chattopadhyay S., Ang L. H., Puente P., Deng X. W., Wei N. (1998) Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10, 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yadav V., Kundu S., Chattopadhyay D., Negi P., Wei N., Deng X. W., Chattopadhyay S. (2002) Light-regulated modulation of Z-box-containing promoters by photoreceptors and downstream regulatory components, COP1 and HY5, in Arabidopsis. Plant J. 31, 741–753 [DOI] [PubMed] [Google Scholar]
- 21. Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., Tongprasit W., Zhao H., Lee I., Deng X. W. (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19, 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang H., He H., Wang X., Wang X., Yang X., Li L., Deng X. W. (2011) Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 65, 346–358 [DOI] [PubMed] [Google Scholar]
- 23. Holm M., Ma L. G., Qu L. J., Deng X. W. (2002) Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16, 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sellaro R., Hoecker U., Yanovsky M., Chory J., Casal J. J. (2009) Synergism of red and blue light in the control of Arabidopsis gene expression and development. Curr. Biol. 19, 1216–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Giuliano G., Pichersky E., Malik V. S., Timko M. P., Scolnik P. A., Cashmore A. R. (1988) An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc. Natl. Acad. Sci. U.S.A. 85, 7089–7093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schindler U., Menkens A. E., Beckmann H., Ecker J. R., Cashmore A. R. (1992) Heterodimerization between light-regulated and ubiquitously expressed Arabidopsis GBF bZIP proteins. EMBO J. 11, 1261–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mallappa C., Yadav V., Negi P., Chattopadhyay S. (2006) A basic leucine zipper transcription factor, G-box-binding factor 1, regulates blue light-mediated photomorphogenic growth in Arabidopsis. J. Biol. Chem. 281, 22190–22199 [DOI] [PubMed] [Google Scholar]
- 28. Mallappa C., Singh A., Ram H., Chattopadhyay S. (2008) GBF1, a transcription factor of blue light signaling in Arabidopsis, is degraded in the dark by a proteasome-mediated pathway independent of COP1 and SPA1. J. Biol. Chem. 283, 35772–35782 [DOI] [PubMed] [Google Scholar]
- 29. Ellenberger T., Fass D., Arnaud M., Harrison S. C. (1994) Crystal structure of transcription factor E47. E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 8, 970–980 [DOI] [PubMed] [Google Scholar]
- 30. Johnson L., Cao X., Jacobsen S. (2002) Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 12, 1360–1367 [DOI] [PubMed] [Google Scholar]
- 31. Yadav V., Mallappa C., Gangappa S. N., Bhatia S., Chattopadhyay S. (2005) A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell 17, 1953–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Datta S., Johansson H., Hettiarachchi C., Irigoyen M. L., Desai M., Rubio V., Holm M. (2008) LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell 20, 2324–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438 [DOI] [PubMed] [Google Scholar]
- 34. Yoo S. D., Cho Y. H., Sheen J. (2007) Arabidopsis mesophyll protoplasts. A versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 [DOI] [PubMed] [Google Scholar]
- 35. Kirby J., Kavanagh T. A. (2002) NAN fusions. A synthetic sialidase reporter gene as a sensitive and versatile partner for GUS. Plant J. 32, 391–400 [DOI] [PubMed] [Google Scholar]
- 36. Ehlert A., Weltmeier F., Wang X., Mayer C. S., Smeekens S., Vicente-Carbajosa J., Dröge-Laser W. (2006) Two-hybrid protein-protein interaction analysis in Arabidopsis protoplasts. Establishment of a heterodimerization map of group C and group S bZIP transcription factors. Plant J. 46, 890–900 [DOI] [PubMed] [Google Scholar]
- 37. Sibéril Y., Doireau P., Gantet P. (2001) Plant bZIP G-box binding factors. Modular structure and activation mechanisms. Eur. J. Biochem. 268, 5655–5666 [DOI] [PubMed] [Google Scholar]
- 38. Schindler U., Terzaghi W., Beckmann H., Kadesch T., Cashmore A. R. (1992) DNA binding site preferences and transcriptional activation properties of the Arabidopsis transcription factor GBF1. EMBO J. 11, 1275–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hardtke C. S., Gohda K., Osterlund M. T., Oyama T., Okada K., Deng X. W. (2000) HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 19, 4997–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chattopadhyay S., Puente P., Deng X. W., Wei N. (1998) Combinatorial interaction of light-responsive elements plays a critical role in determining the response characteristics of light-regulated promoters in Arabidopsis. Plant J. 15, 69–77 [DOI] [PubMed] [Google Scholar]
- 41. Bhatia S., Gangappa S. N., Kushwaha R., Kundu S., Chattopadhyay S. (2008) Plant Physiol. 147, 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Saijo Y., Sullivan J. A., Wang H., Yang J., Shen Y., Rubio V., Ma L., Hoecker U., Deng X. W. (2003) The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 17, 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. von Arnim A. G., Deng X. W. (1994) Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79, 1035–1045 [DOI] [PubMed] [Google Scholar]
- 44. Osterlund M. T., Deng X. W. (1998) Multiple photoreceptors mediate the light-induced reduction of GUS-COP1 from Arabidopsis hypocotyl nuclei. Plant J. 16, 201–208 [DOI] [PubMed] [Google Scholar]
- 45. Harter K., Kircher S., Frohnmeyer H., Krenz M., Nagy F., Schäfer E. (1994) Light-regulated modification and nuclear translocation of cytosolic G-box binding factors in parsley. Plant Cell 6, 545–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.