Background: There is an oxidoreductive d-galactose pathway in filamentous fungi.
Results: We identified an l-xylo-3-hexulose reductase that produces d-sorbitol and that is part of this pathway.
Conclusion: This l-xylo-3-hexulose reductase is the missing link in the oxidoreductive d-galactose pathway.
Significance: The alternative pathway for d-galactose catabolism in filamentous fungi is elucidated.
Keywords: Aspergillus, Enzyme Catalysis, Fungi, Galactose, Galactose Metabolism, L-Xylo-3-hexulose, L-Xylo-3-hexulose Reductase, Trichoderma reesei, lxr4, xhrA
Abstract
In addition to the well established Leloir pathway for the catabolism of d-galactose in fungi, the oxidoreductive pathway has been recently identified. In this oxidoreductive pathway, d-galactose is converted via a series of NADPH-dependent reductions and NAD+-dependent oxidations into d-fructose. The pathway intermediates include galactitol, l-xylo-3-hexulose, and d-sorbitol. This study identified the missing link in the pathway, the l-xylo-3-hexulose reductase that catalyzes the conversion of l-xylo-3-hexulose to d-sorbitol. In Trichoderma reesei (Hypocrea jecorina) and Aspergillus niger, we identified the genes lxr4 and xhrA, respectively, that encode the l-xylo-3-hexulose reductases. The deletion of these genes resulted in no growth on galactitol and in reduced growth on d-galactose. The LXR4 was heterologously expressed, and the purified protein showed high specificity for l-xylo-3-hexulose with a Km = 2.0 ± 0.5 mm and a Vmax = 5.5 ± 1.0 units/mg. We also confirmed that the product of the LXR4 reaction is d-sorbitol.
Introduction
There are several pathways for the catabolism of d-galactose. The most studied is the Leloir pathway, which exists in prokaryotic and eukaryotic microorganisms. In this pathway, α-d-galactose is phosphorylated, and in the subsequent steps, converted to d-glucose-6-phosphate in a redox-neutral way (1). The genes of the Leloir pathway and their regulation have been described in yeast and filamentous fungi (2).
An alternative pathway is the oxidative pathway that is sometimes referred to as the De Ley-Doudoroff pathway and was identified in bacteria (3). In this pathway, d-galactose is first oxidized to d-galactonolactone, which is then hydrolyzed by a lactonase to d-galactonate followed by the removal of a water molecule by a dehydratase to form d-threo-3-deoxy-hexulosonate (2-keto-3-deoxy-d-galactonate). d-threo-3-Deoxy-hexulosonate is then phosphorylated to d-threo-3-deoxy hexulosonate 6-phosphate, which is subsequently split by an aldolase, resulting in pyruvate and d-glyceraldehyde-3-phosphate. In a strain of the mold Aspergillus niger, a nonphosphorylated alternative of the De Ley-Doudoroff pathway was described where the d-threo-3-deoxy-hexulosonate is split by an aldolase to pyruvate and d-glyceraldehyde instead of being phosphorylated (4).
Another pathway for d-galactose catabolism has also been demonstrated in some filamentous fungi. It was observed that a Trichoderma reesei (Hypocrea jecorina) strain with a mutation in the galactokinase gene, gal1, is still able to catabolize d-galactose, indicating the existence of an alternative to the Leloir pathway. Because galactitol accumulated in this strain, it was suggested that a pathway exists in T. reesei where d-galactose is reduced in the first step (5). In T. reesei, an aldose reductase, XYL1, was shown to be responsible for the reduction of d-galactose to galactitol as well as for the reduction of d-xylose and l-arabinose (6). A. niger has distinct l-arabinose and d-xylose reductases, LarA and XyrA (7). XyrA had the highest activity with d-galactose and was suggested to be involved in d-galactose catabolism (8).
It was also observed in T. reesei that a double mutant with deletions in gal1 and lad1, coding for galactokinase and l-arabitol-4-dehydrogenase, respectively, was not able to catabolize d-galactose. This suggested that the l-arabitol-4-dehydrogenase that is induced on l-arabitol (9) is also part of this alternative reductive pathway. Pail et al. (10) showed that purified l-arabitol 4-dehydrogenase, the product of lad1, was capable of converting galactitol to l-xylo-3-hexulose. In A. niger, it was shown that a distinct galactitol dehydrogenase, LadB, exists besides the l-arabitol dehydrogenase, LadA, that produces l-xylo-3-hexulose and that it is induced on d-galactose and galactitol (8).
Seiboth and Metz (11) pointed out the possibility that the l-xylo-3-hexulose is reduced to d-sorbitol in a reaction catalyzed by an enzyme related to l-xylulose reductase or by the l-xylulose reductase itself. The complete oxidoreductive d-galactose pathway would then have the intermediates d-galactose, galactitol, l-xylo-3-hexulose, d-sorbitol, and d-fructose (see Fig. 1). d-Sorbitol was shown to be an intermediate in the pathway of A. niger where the d-fructose-forming d-sorbitol dehydrogenase, sdhA, was found to be induced on d-galactose and galactitol and the sdhA deletion mutant had reduced growth on galactitol and was unable to grow on d-sorbitol (12). In T. reesei, xylitol dehydrogenase, xdh1, was proposed to be the enzyme responsible for this reaction.4 Fekete et al. (13) suggested that l-sorbose is produced in Aspergillus nidulans from galactitol by l-arabitol dehydrogenase and that it could be further converted to d-sorbitol. The reaction mechanism of the galactitol to l-sorbose conversion, however, has not been proposed and currently remains elusive. Here we set out to identify the enzyme that catalyzes the conversion of l-xylo-3-hexulose to d-sorbitol (l-gulitol), which is the missing link in the oxidoreductive d-galactose pathway in the molds T. reesei and A. niger.
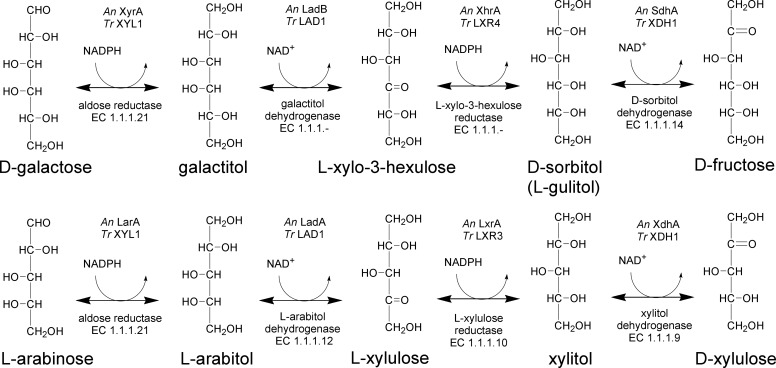
FIGURE 1.
The oxidoreductive d-galactose pathway (upper part) and the eukaryotic pathway for l-arabinose catabolism (lower part). The metabolites are in Fischer projection except for the l-xylo-3-hexulose and l-xylulose, which are oriented so that the C6 and C5, respectively, are at the top to have all molecules in the same orientation. The enzymes for the different reactions in A. niger (An) and T. reesei (Tr) are indicated.
MATERIALS AND METHODS
Strains and Chemicals
The A. niger strain ATCC 1015 (CBS 113.46) was obtained from the Centraalbureau voor Schimmelcultures (Delft, The Netherlands). The T. reesei strain QM9414 (ATCC 26921) was used in this study. For comparison of growth on agar plates, the spores of A. niger and T. reesei strains were applied to agar plates containing 6.7 g of yeast nitrogen base liter−1 (YNB, BD Biosciences), 20 g of agar/liter−1, and 20 g liter−1 of carbon source (see “Results” for details). The l-xylo-3-hexulose used for the in vitro tests was produced as described previously (8).
Transcriptional Analysis
The growth of strains, RNA isolation, and qPCR5 were performed as described previously (12).
Deletion of the xhrA Gene in the A. niger ΔpyrG Strain
Construction of A. niger ATCC 1015 ΔpyrG was described previously (7). The cassette for deletion of the xhrA gene (Table 1) contained 1644 bp from the xhrA promoter region, 1563 bp from the xhrA terminator region, and a 1928-bp fragment containing the pyrG gene flanked by its native promoter and terminator. These fragments were obtained by PCR from the A. niger ATCC 1015 genomic DNA using primers xhrA-5-F, xhrA-5-R, xhrA-3-F, xhrA-3-R, pyrG-del-F_n, and pyrG-del-R_n (Table 2) and the proofreading DNA polymerase Phusion (Finnzymes). The xhrA terminator fragment (xhrA-3) digested with HindIII (New England Biolabs) was inserted into the plasmid pRSET-A (Invitrogen), which was digested with HindIII and PvuII (both New England Biolabs). This intermediary construct was digested with EcoRV and NheI (both New England Biolabs). The resulting fragment was ligated to the NheI-digested promoter fragment (xhrA-5). The resulting vector was digested with EcoRV (New England Biolabs). The pyrG DNA fragment, after digestion with SmaI, was inserted between the two xhrA flanking regions. The resulting plasmid was verified by restriction analysis and sequencing. The deletion cassette, 5097 bp, containing the xhrA flanking regions and the pyrG gene, was released by MluI (New England Biolabs) digestion and transformed into the A. niger ATCC 1015 ΔpyrG strain. Transformants were selected based on their ability to grow in the absence of uracil. Strains with successful deletions were verified by PCR using the primers xhrA_ORF_F and xhrA_ORF_F (Table 2).
TABLE 1.
The transcription of the following genes was tested in A. niger using qPCR
The carbon source was d-galactose or galactitol. The genes can be retrieved from the Aspergillus niger homepage at the DOE Joint Genome Institute, JGI (genome.jgi.doe.gov/Aspni5/Aspni5.home.html) using the JGI identifiers or from the Aspergillus genome database (AspGD) using the identifiers in parentheses.
| JGI: 177738 (An08g01930) lxrA |
| JGI: 184209 (An16g01650) xhrA |
| JGI: 40156 (An07g01830) |
| JGI: 174212 (An02g00220) |
| JGI: 177858 (An06g01980) |
| JGI: 184211 (An16g06440) |
| JGI: 56312 (An15g02280) |
| JGI: 212729 (An09g00620) |
| JGI: 55205 (An04g09990) |
| JGI: 212936 (An05g01210) |
| JGI: 52907 (An11g02460) |
TABLE 2.
Primers
| xhrA-genomic_F | GGCAAGACAACGGACTGAA |
| xhrA-genomic_R | AATTCCTGGTGATTCGGGTT |
| xhrA-ORF_F | ATATGAATTCACAATGTCCCTCAAAGGTAAAGTCG |
| xhrA-ORF_R | ATATGTCGACCTAGATATACAACATCCCACCATT |
| xhrA-N-HIS_F | ATATGAATTCACAATGCATCACCATCACCATCACGGGTCCCTCAAAGGTAAAGTCG |
| xhrA-5_F | TATAGCTAGCAGCTGAACGCCTGATACAAA |
| xhrA-5_R | ATATGATATCGATGGCTTTTGCAGATTGTTTG |
| xhrA-3_F | ATATGATATCCAGAGCCGTGTTAAATAAGGAATAC |
| xhrA-3_F | ATTAAAGCTTACGCGTACGAAGCCGCCGAAGATA |
| act_qPCR_F | CAACATTGTCATGTCTGGTGG |
| act_qPCR_R | GGAGGAGCAATGATCTTGAC |
| xhrA_qPCR_F | GATACAGATATGTACCAGGCAG |
| xhrA_qPCR_R | CTAGATATACAACATCCCACCA |
| galX_qPCR_F | CTGTGAAATGTTTGGGAAGTC |
| galX_qPCR_R | GTTTGTTGGTCGTTCGTAGG |
| pyrG-del-F_n | TATACCCGGGTGATTGAGGTGATTGGCGAT |
| pyrG-del-R_n | TATACCCGGGTTATCACGCGACGGACAT |
| lxr4-HIS-N_F | ATATGAATTCACAATGCATCACCATCACCATCACGGGGCCCGTCCGTATGAAGGC |
| lxr4_ORF_R | TAATGATATCCTACGCAATCGACATGCGCATCC |
| Trire22771_XbaI-Ups-fw | TCTAGACCATTGTCCCAGCCATCTT |
| Trire22771_XhoI-Ups-rev | CTCGAGCGACTTGAGCAATCACCAC |
| Trire22771_XhoI-Dws-fw | CTCGAGGATGTGTACTTGGTGGCTTG |
| Trire22771_Acc-Dws-rev | GGTACCTCTCTGCTCGTTAAATCCCG |
| dlxr4_for3 | GGCGGAGTTTCTATGGAG |
| dlxr4_rev3 | GGGATTGATATTGTTTGC |
Deletion of the lxr4 Gene in T. reesei
A T. reesei Δtku70 strain was used as the parental strain for transformation (14, 15). The cassette for deletion of the lxr4 gene (GenBankTM accession number BK008566) contained 937 bp of the promoter region, 1162 bp of the terminator region, and a fragment containing the pyr4 encoding orotidine-5′-phosphate decarboxylase. The promoter and terminator regions were obtained by PCR from the genomic DNA of the T. reesei strain QM9414 using primers Trire22771_XbaI-Ups-fw, Trire22771_XhoI-Ups-rev, Trire22771_XhoI-Dws-fw, and Trire22771_Acc-Dws-rev (Table 2) and the proofreading DNA polymerase Phusion (Finnzymes). Both fragments were ligated into the vector pBluescript SK(+) (Stratagene) after digestion with XbaI and XhoI (Fermentas) for the promoter fragment and with XhoI and Acc65I (Fermentas) for the terminator fragment using ligation mix III (Takara). The SalI fragment of pyr4 served as the selection marker and was cloned via XhiI restriction between the promoter and terminator regions of lxr4 in pBluescript SK(+). The resulting plasmid (verified by restriction analysis and sequencing) was linearized using XbaI and Acc65I and transformed into the T. reesei Δtku70 strain. Transformants were selected based on their ability to grow in the absence of uridine. Strains with successful deletions were verified by PCR using the primers dlxr4_for3 and dlxr4_rev3 (Table 2).
Reintroduction of xhrA into the ΔxhrA Strain
The xhrA gene, with its native promoter and terminator (2496-bp genomic fragment), was amplified from the A. niger ATCC 1015 genomic DNA using the primers xhrA-genomic_F and xhrA-genomic_R (Table 2). The PCR product was transformed into the ΔxhrA strain, and the transformants were selected on medium with galactitol as a sole carbon source. The resulting strains were tested for the presence of the xhrA gene by PCR and further analyzed for growth on selected carbon sources.
Expression of the Genes in Saccharomyces cerevisiae
For heterologous expression in S. cerevisiae, the open reading frames (ORFs) of xhrA (Table 1) and lxr4 (GenBank accession number BK008566) were amplified from the cDNA of A. niger or T. reesei, respectively, grown in the presence of galactitol with the primers xhrA-H-HIS_F, xhrA-ORF_R, lxr4-HIS-N_F, and lxr4_ORF_R (Table 2). The genes were inserted into the plasmid pYX212 (Ingenius, R&D Systems, Madison, WI) between the EcoRI and SalI sites (in the case of xhrA) or the EcoRI and SmaI sites (in the case of lxr4), allowing the expression to be controlled by the TPI1 promoter. All constructs were verified by sequencing.
The S. cerevisiae strain CEN.PK2-1D was transformed with the pYX212 plasmids containing the xhrA and lxr4 genes as N-terminal His-tagged variants, and the transformants were selected based on their ability to grow in the absence of uracil. The expression of active reductases was tested in crude cell extracts by enzymatic activity measurements. The xhrA was also expressed without a His tag and as a gene that was codon-optimized for expression in S. cerevisiae. The gene was also expressed in the Escherichia coli strain BL21 (DH3) (Invitrogen) using the pBAT4 expression vector (16) and isopropyl-1-thio-β-d-galactopyranoside induction.
Protein Extraction, Enzyme Activity Measurements, and Analysis of the LXR4 Product
For l-xylo-3-hexulose-reductase activity measurements in A. niger extracts, the parent strain ATCC 1015 and the ΔxhrA strains were cultivated overnight in YPG medium (1% yeast extract, 2% Bacto peptone; 3% gelatin). The mycelia were filtered and transferred to fresh medium containing 1% yeast extract, 2% Bacto peptone, and 2% d-glucose, galactitol or l-arabinose and cultivated for 6 h. After incubation in the inducing conditions, the mycelia were isolated by filtration and washed with water, and ∼200 mg of wet mycelia was transferred into 2-ml tubes with 0.6 ml of acid-washed glass beads (Sigma) and 1 ml of lysis buffer containing 50 mm Tris (pH = 7.5) and protease inhibitors (Complete, Roche Applied Science). The cells were disrupted in two 30-s breaking sessions in the Precellys 24 instrument (Bertin Technologies). The cell extracts were clarified by centrifugation, and the supernatants were used in the enzyme assays. The protein concentration was analyzed using the Bio-Rad protein assay kit. In the tests performed with the A. niger protein extracts, 10 mm l-xylo-3-hexulose, 0.5 mm NADPH, and 50 mm Tris-HCl (pH = 7.5) were used. The enzymatic activity was measured at room temperature by monitoring the NADPH disappearance at 340 nm in microtiter plates (Nunc) using the Varioskan spectrophotometer (Thermo Electron). For the protein extractions from S. cerevisiae and the purification of the His-tagged proteins, the same methods were used as described previously (8).
For LXR4 sugar reductase activities, the reactions were set up in 50 mm Tris-HCl (pH = 7.5) with 0.5 mm NADPH and various sugar concentrations (see “Results”). For the LXR4 polyol dehydrogenase activities, the reaction mixtures contained 100 mm Tris-HCl (pH = 8.5), 1 mm NADP+, and various concentrations of polyols (see “Results”). The degradation of NADPH or formation of NADPH was monitored as described above. The Km and Vmax were estimated from the Michaelis-Menten equation fitted to the measured data.
The concentration of l-xylo-3-hexulose produced by E. coli expressing ladB was determined by NMR spectroscopy. 480 μl of D2O containing 0.05% of 3-(trimethylsilyl)propionic-2,2,3,3-d4 acid (TSP, Aldrich) was added to 120 μl of the sample, and a one-dimensional 1H NMR spectrum was acquired on a 600-MHz Bruker Avance III NMR spectrometer equipped with a QCI CryoProbe (Bruker) using the one-dimensional NOESY pulse sequence for presaturation of the water signal. The concentration of the l-xylo-3-hexulose was obtained by comparing the integral over a region of 4.27–4.40 ppm with the integral of TSP.
The in vitro reaction with purified LXR4 for the analysis of the reaction product from d-sorbitol was carried out in 1 ml of reaction mix containing 50 μg of the purified LXR4, 100 mm d-sorbitol, and 5 mm NADP+ in 100 mm Tris-HCl (pH = 9.0). The reaction mix was incubated at room temperature for 16 h. The control reaction was terminated immediately after the components were mixed by incubation at 95 °C for 10 min. The reactions were analyzed by HPLC as described previously (8).
RESULTS
Identification of the l-xylo-3-Hexulose Reductase in A. niger
The suggested oxidoreductive pathway for d-galactose catabolism has similarities with the eukaryotic l-arabinose pathway (Fig. 1). The reductions require NADPH, and the oxidations require NAD+. The reactions that were described are catalyzed by identical or closely related enzymes. In T. reesei, the first and second steps of both pathways are catalyzed by the same enzymes. The T. reesei XYL1 is the major enzyme for l-arabinose and d-galactose reduction (6), and LAD1 is the main enzyme for galactitol and l-arabitol oxidation (9, 10). In A. niger, close homologues of different enzymes are used; XyrA is used for d-galactose reduction, and LarA is used for l-arabinose reduction (7). Also, the second step uses close homologues of different enzymes. Galactitol is oxidized in A. niger by LadB (8), and l-arabitol is oxidized in A. niger by LadA (17). This information suggested that a similar phenomenon may also exist for the third step. Thus, the l-xylo-3-hexulose reductase may be a homologue of the l-xylulose reductase or identical to the l-xylulose reductase. Because LxrA was identified as the l-xylulose reductase in A. niger (18), we examined whether this enzyme could also be the l-xylo-3-hexulose reductase.
The LxrA was purified using the histidine tag as described previously (18). The activity was tested with l-xylo-3-hexulose and NADPH and in the reverse direction with d-sorbitol and NADP+. The enzyme had activity with these substrates, suggesting that this enzyme could be the l-xylo-3-hexulose reductase.
We tested the transcription of this gene by qPCR during growth on galactitol or d-galactose. The lxrA gene was not up-regulated under these conditions, suggesting that although the LxrA has l-xylo-3-hexulose reductase activity, it is not the “true” l-xylo-3-hexulose reductase. Also, the ΔlxrA strain does not confer any growth defect in the presence of galactitol when compared with the wild type strain (data not shown). We then tested the transcription of several close homologues to lxrA on galactitol or d-galactose, which are listed in Table 1. The closest lxrA homologue (E-value = 1.45 × 10−31) JGI: 184209 (An16g01650) was found to be up-regulated on galactitol and d-galactose (Fig. 2A). We called this gene xhrA for l-xylo-3-hexulose reductase.
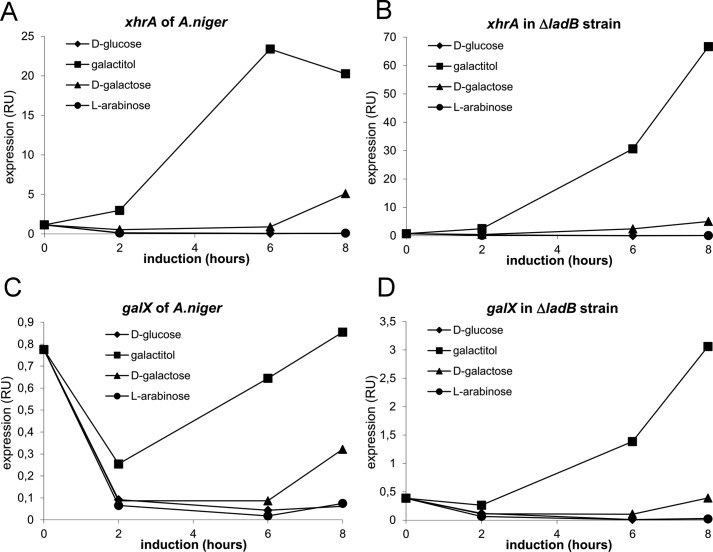
FIGURE 2.
Transcription profiles of the xhrA and galX genes in A. niger. A, the xhrA gene is up-regulated in the presence of galactitol and to a lesser extent and with a delay on d-galactose. The expression on l-arabinose and d-glucose is not affected. RU, response units. B, the transcriptional activation of xhrA is further increased in the ΔladB strain on galactitol. C and D, the galX encoding the transcription factor responsible for control of the d-galactose catabolic genes is up-regulated on galactitol (C), and its transcription is increased in the ΔladB strain (D).
The xhrA gene was also up-regulated on galactitol and d-galactose in a strain where ladB was deleted with the up-regulation being significantly higher (Fig. 2B). The LadB converts galactitol to l-xylo-3-hexulose, and the ladB mutant cannot grow on galactitol (8). This suggests that galactitol, which probably accumulates in the ΔladB strain and not l-xylo-3-hexulose, is required for the induction of xhrA.
A homologue of galX is located next to xhrA on the chromosome. The galX gene was identified in A. nidulans as the gene encoding a regulator that controls the d-galactose utilization, and it was shown to be up-regulated in the presence of galactitol and d-galactose (19). Likewise in A. niger, the transcription of galX was up-regulated in the presence of galactitol and d-galactose. In addition, galX transcription was further enhanced in the ΔladB strain in a similar manner as observed in the case of xhrA expression (Fig. 2, C and D).
Identification of the l-xylo-3-Hexulose Reductase in T. reesei
In T. reesei, several homologues of l-xylulose reductase exist. The product of the lxr1 gene tre74194 (www.jgi.doe.gov) that was described to be active with l-xylulose (20) turned out to be a d-mannitol dehydrogenase (21). Another candidate LXR3 (GenBank accession number BK008567), which was identified and characterized as a gene encoding the true l-xylulose reductase of T. reesei, showed only activity with l-xylulose but not with l-xylo-3-hexulose in vitro.6 Moreover, none of these genes are close homologues of xhrA. By searching the T. reesei genome, we identified a gene that we called lxr4 (GenBank accession number BK008566) as the closest orthologue for lxrA.
Deletion of the l-xylo-3-Hexulose Reductase Genes in A. niger and T. reesei
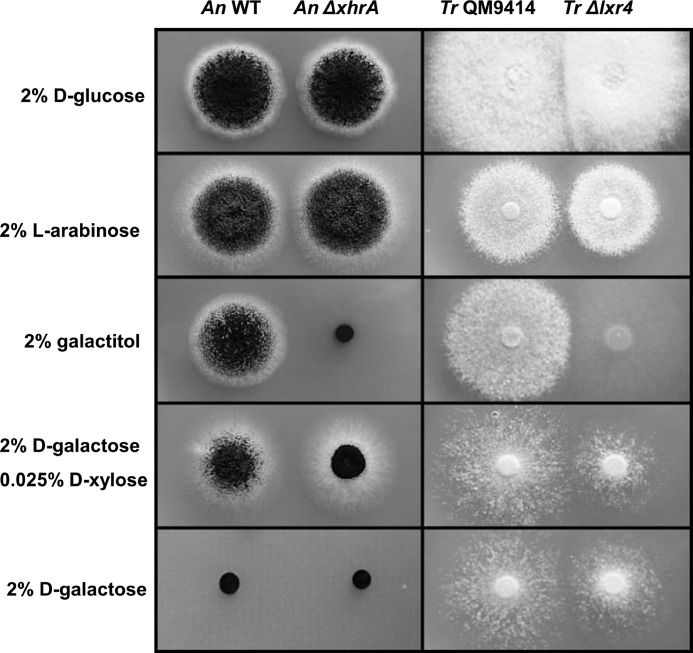
To test whether the xhrA is essential for the pathway, we deleted the gene in A. niger. The resulting strain was then tested for growth on different carbon sources and compared with the ATCC 1015 parent strain (Fig. 3). The mutant showed no growth on galactitol, demonstrating that the xhrA is an essential gene for its utilization. To prove that this phenotype is only related to the deletion of the xhrA, we retransformed the xhrA to the ΔxhrA strain. In the strain expressing the xhrA in the mutant background, growth on galactitol is fully restored (data not shown). The mutant and parent strain do not grow on d-galactose. Only when small amounts of d-xylose (0.025%) are supplemented does the parent strain grow, and the ΔxhrA strain showed reduced growth. 0.025% d-xylose alone does not result in significant growth as described previously (8). The growth on d-glucose and l-arabinose is not affected because the xhrA gene is not required for the metabolism of these sugars.
FIGURE 3.
Growth of the A. niger (An) and T. reesei (Tr) strains with and without a deletion of the gene encoding l-xylo-3-hexulose reductase on solidified medium containing different carbon sources. Equal amounts of spores were applied and grown for 4 days at 28 °C.
The deletion of lxr4 in T. reesei resulted in no growth on galactitol. The phenotype of this mutant is similar to the xhrA deletion in A. niger (Fig. 3). Growth on galactitol was abolished, but growth on d-glucose and l-arabinose was not affected. Growth on d-galactose is significantly slower but not absent. This is expected because it is established that T. reesei has a functional Leloir pathway for d-galactose catabolism and that the oxidoreductive pathway only partially contributes to the d-galactose catabolism. The oxidoreductive pathway only becomes essential if the Leloir pathway is disrupted, for example, when the gene encoding the galactokinase is deleted (5).
Heterologous Expression of the l-xylo-3-Hexulose Reductase
The xhrA of A. niger and the lxr4 of T. reesei were expressed in S. cerevisiae from a multicopy plasmid with a strong constitutive promoter. In the crude S. cerevisiae extracts, we could detect l-xylo-3-hexulose reductase activity when the lxr4 was expressed, but not when the xhrA was expressed. We also tested the expression of a yeast codon-optimized version of the xhrA gene in yeast as well as the expression of cDNA in E. coli using the lac promoter and isopropyl-1-thio-β-d-galactopyranoside induction, but we were not able to detect l-xylo-3-hexulose reductase activity (data not shown).
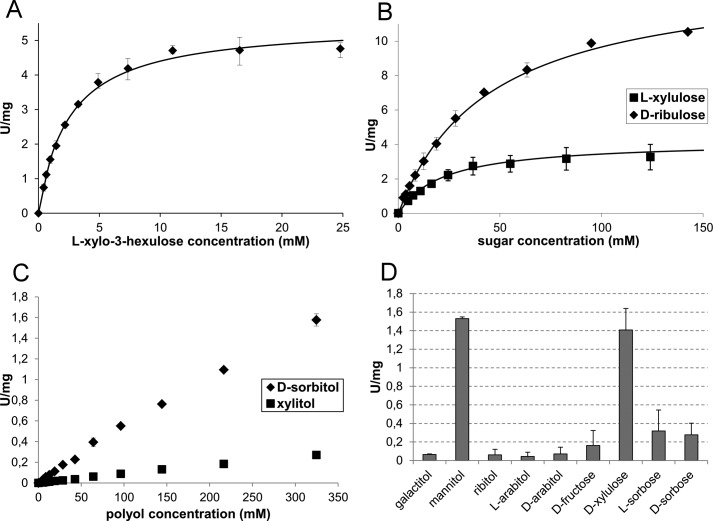
Therefore, we used the T. reesei enzyme for the in vitro characterization of l-xylo-3-hexulose reductase. LXR4 was produced in S. cerevisiae as a recombinant N-terminally His6-tagged protein and subsequently purified. The purified enzyme was NADP(H)-specific and conferred high and specific activity with l-xylo-3-hexulose with a Km = 2.0 ± 0.5 mm and a Vmax = 5.5 ± 1.0 units/mg (Fig. 4A). The enzyme was also active with d-ribulose and l-xylulose, Km = 47 ± 3 mm and Vmax = 14 ± 2 units/mg and Km = 22 ± 3 mm and Vmax = 4.2 ± 1 units/mg, respectively. The enzyme also showed some activity with d-xylulose and very low activity with d-fructose and l- and d-sorbose (Fig. 4, B and D). In the reverse reaction, LXR4 showed activity with d-sorbitol and d-mannitol, low activity with xylitol, and no activity with galactitol, ribitol, and l- and d-arabitol (Fig. 4, C and D).
FIGURE 4.
In vitro activity of purified LXR4. A, initial reaction rate at different l-xylo-3-hexulose concentrations to obtain the Michaelis-Menten-constants: Km = 2.0 ± 0.5 mm and Vmax = 5.5 ± 1 units/mg. B, initial reaction rates of l-xylulose and d-ribulose. The Michaelis-Menten constants are: Km = 22 ± 3 mm and Vmax = 4.2 ± 1 units/mg for l-xylulose and Km = 47 ± 3 mm and Vmax = 14 ± 2 units/mg for d-ribulose. C, the reverse reaction with the polyols d-sorbitol and xylitol. In the concentration range tested, the rate increased linearly with the substrate concentration, so the Michaelis-Menten-constants could not be determined. D, activity with other substrates. The reactions were carried out at room temperature at pH = 7.5 with 0.5 mm NADPH and 50 mm sugars unless otherwise specified. In the reverse direction, 300 mm polyols, 1 mm NADP+, and pH = 8.5 were used. Error bars in panels A–D indicate S.D.
l-xylo-3-Hexulose Reductase Activity in Crude Extracts of A. niger
Because we were not able to produce an active XhrA in a heterologous host, we tested whether this activity could be detected in A. niger. We made a crude cell extract from mycelia that were shifted to different carbon sources. Extracts from mycelia on d-glucose showed the lowest l-xylo-3-hexulose reductase activity. The activity was significantly higher on galactitol and l-arabinose, which is expected because xhrA and lxrA, respectively, are up-regulated under these conditions. In the xhrA deletion mutant, the activity is not increased on galactitol, indicating that the XhrA is mainly contributing to the l-xylo-3-hexulose reductase activity under these conditions. On l-arabinose, the activity is not decreased by the xhrA deletion. This activity is likely due to lxrA, which is up-regulated on l-arabinose and also has high l-xylo-3-hexulose reductase activity (Table 3).
TABLE 3.
l-xylo-3-hexulose activity in crude extracts of A. niger
The activities of the crude extract are given in milliunits/mg of extracted protein. The mycelia were pre-grown in YPG medium and then shifted for 6 h to YP medium supplemented with the 2% carbon sources indicated.
| A. niger (WT) | A. niger ΔxhrA | |
|---|---|---|
| d-Glucose | 7 ± 0.2 | 7 ± 0.8 |
| Galactitol | 25 ± 0.8 | 9 ± 1.0 |
| l-Arabinose | 42 ± 3.4 | 60 ± 2.3 |
l-xylo-3-Hexulose Is Produced from d-Sorbitol in the Reverse Reaction
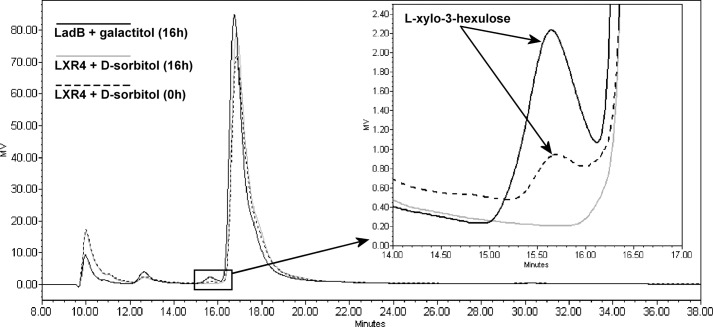
To demonstrate that the l-xylo-3-hexulose reductase is catalyzing the conversion of l-xylo-3-hexulose to d-sorbitol, we tested the reverse reaction with d-sorbitol as the substrate and analyzed the reaction product by HPLC. The product of the reverse reaction of LXR4 with d-sorbitol and NADP+ was identified as l-xylo-3-hexulose. It had the same retention time as the l-xylo-3-hexulose produced from galactitol using LadB (8) (Fig. 5) or LAD1 (not shown). d-Fructose has a different retention time and was not produced.
FIGURE 5.
HPLC elution profile of the reaction products of LadB and LXR4. The reaction product formed from galactitol by LadB is l-xylo-3-hexulose. The reaction product that is formed from d-sorbitol by LXR4 has the same retention time, which is different from the retention time of d-fructose.
The l-xylo-3-hexulose that we had produced from galactitol (8) still contained large amounts of galactitol, which overlaps the d-sorbitol signal in the HPLC. This made it problematic to test whether in the forward reaction d-sorbitol was indeed produced from l-xylo-3-hexulose.
DISCUSSION
In this study, we identified an enzyme with activity for the conversion of l-xylo-3-hexulose to d-sorbitol using NADPH as a cofactor. Such an enzyme activity has not, to the best of our knowledge, been described previously. The enzyme is encoded by xhrA in A. niger and by lxr4 in T. reesei, and deletion mutants are unable to grow on galactitol. This enzyme activity is the missing link in the oxidoreductive pathway for d-galactose catabolism that had been demonstrated to exist in filamentous fungi.
The oxidoreductive d-galactose pathway and the fungal l-arabinose pathway are very similar (Fig. 1). In both cases, the sequence of reactions is reduction, oxidation, reduction and oxidation. In both cases, the reductions are NADPH-linked, and the oxidations are NAD+-linked. In A. niger, d-galactose is reduced by XyrA, and l-arabinose is reduced by LarA. In T. reesei, the same enzyme, XYL1, is used for the reduction of d-galactose and l-arabinose. In A. niger, the second step consisting of the oxidation of galactitol and l-arabitol is carried out by two different enzymes, LadB and LadA, respectively. Again, the same enzyme in T. reesei, LAD1, oxidizes both galactitol and l-arabitol. So far, it seems that in A. niger, two different pathways exist for d-galactose and l-arabinose, whereas in T. reesei, the enzymes of the l-arabinose pathway are also the enzymes of the d-galactose pathway. However, this pattern is different in the third step.
In A. niger, the oxidation of l-xylo-3-hexulose or l-xylulose is performed by two different but highly homologous enzymes, XhrA and LxrA, respectively. However, in this case, T. reesei also uses two different enzymes, LXR4 for the l-xylo-3-hexulose reduction and LXR3 for the l-xylulose reduction.6 The LXR4 has the closest sequence similarity of the A. niger enzymes LxrA and XhrA in T. reesei. The LXR3, however, is more distant in terms of sequence similarity (supplemental Fig. S1), and it also confers considerably different substrate specificity. For example, it shows no activity with l-xylo-3-hexulose, unlike LxrA, whereas it is active with l-sorbose and d-fructose.
In the natural habitats of T. reesei and A. niger, d-galactose is often accompanied by pentose sugars, which indicates that both l-arabinose and d-galactose pathways are active simultaneously in such conditions. In T. reesei, only LXR4 is responsible for the conversion of l-xylo-3-hexulose, whereas in A. niger, XhrA and LxrA both contribute to the reaction.
Surprisingly, we could not obtain an active XhrA after heterologous expression, although the activity was detected in the A. niger crude extracts, and it was reduced in the xhrA deletion mutant. That we could not express the active XhrA in a heterologous host could be due to the protein instability, folding problems, or other issues that we did not pursue as we were able to analyze the T. reesei homologue (LXR4) in vitro.
The last step of the oxidoreductive d-galactose pathway is catalyzed by the d-sorbitol dehydrogenase, SdhA, which was identified in A. niger. This enzyme was shown to be part of the pathway because it is up-regulated on d-galactose and galactitol, and the ΔsdhA strain showed reduced growth on galactitol (12). In A. niger, the corresponding enzyme in the l-arabinose pathway is the xylitol dehydrogenase XdhA (17). In T. reesei, it is apparently a single enzyme, XDH1, that carries out the sorbitol dehydrogenase reaction and the xylitol dehydrogenase reaction. The enzyme is active with d-sorbitol and xylitol, and the xdh1 is up-regulated on the carbon sources l-arabinose and d-galactose (22). Moreover, the Δxdh1 strain fails to grow on galactitol.5
Although the regulation of the Leloir pathway has been studied and the regulation factors have been identified, not much is known about the regulation of the oxidoreductive pathway. We have suggested previously that galactitol might be the inducing compound for the genes of this pathway (8). There are four observations that support this. 1)The expression of ladB is up-regulated sooner and more strongly in the presence of galactitol than in the presence of d-galactose (8). 2) The up-regulation of the sdhA is still observed on galactitol in the ΔladB strain when the pathway is blocked and the production of d-sorbitol, which is the main inducer of sdhA, is reduced or even absent (12). 3) The expression of the xhrA gene is significantly enhanced on galactitol in the ΔladB strain when compared with the wild type strain (Fig. 2B). 4) The expression of the galX gene, which encodes for the transcription factor involved in the regulation of the Leloir pathways genes, and of the ladB gene (19) is significantly more expressed on galactitol in the ΔladB strain (Fig. 2C).
In A. nidulans, the d-galactose catabolism and its regulation seem to be different from other filamentous fungi. In this fungus, the use of different d-galactose pathways is pH-dependent. The Leloir pathway is used between pH 4.0 and 6.5 and via some other route at pH = 7.5 (23). This other route showed oxidation to galactitol, but the subsequent steps were unidentified. It was suggested that the pathway proceeds via l-sorbose and d-sorbitol or even via sorbose 6-phosphate and d-tagatose 1,6-bisphosphate (24). A. nidulans has, in addition to GalX, an additional transcriptional regulator, GalR, which is unique among ascomycetes (19). When comparing the different Aspergillus species using the comparative analysis tool on the JGI A. niger v3.0 database (supplemental Fig. S2), A. nidulans does not have a close homologue of xhrA in the same location as the other Aspergillus species. However, it has a close homologue in a different place. The closest homologue to the xhrA is the gene with the identifier ANID_03400.1 (E-value = 2.93 × 10−41).
Until recently, A. niger was considered unable to use d-galactose as a carbon source (25, 26). In the latest demonstrations of d-galactose utilization in A. niger, the oxidoreductive, but not the Leloir pathway, was shown to be employed, and the d-galactose utilization was enabled with the addition of a small amount of d-xylose (8, 12). Fekete et al. (27) recently suggested that the reason for the inability of A. niger to germinate on d-galactose is due to nonfunctional d-galactose uptake in the conidiospores, whereas the uptake is active in the mycelium. The authors showed that once the spores germinated on a different carbon source, the mycelium continued to grow on d-galactose; however, it was unable to sporulate. In addition, evidence of a functional Leloir pathway was presented. In the work of Fekete et al. (27), the strain N402 (28) was used. The strain ATCC 1015 that we used in our current and previous studies, nonetheless, behaved differently. The pregrown mycelium was not able to continue growth on d-galactose, but the strain can sporulate in its presence. In addition, the Leloir pathway is not active, and the growth on d-galactose is facilitated by the addition of a small amount of d-xylose but not d-glucose (8) (supplemental Fig. S3).
Supplementary Material
Acknowledgments
We thank Dr. Hannu Maaheimo for the quantification of l-xylo-3-hexulose by NMR and Dr. Andrew Conley for critical reading of the manuscript.
This work was supported by the Academy of Finland in the following research programs: Finnish Centre of Excellence in White Biotechnology-Green Chemistry (Grant 118573) and Sustainable Energy (Grant 131869). The work was further supported by a from of the Austrian Science Foundation (Grant P19421) (to B. S.).
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) BK008566 and BK008567.

This article contains supplemental Figs. S1–S3.
B. Seiboth, unpublished observation.
B. Metz, D. Mojzita, S. Herold, C. P. Kubicek, P. Richard, and B. Seiboth, manuscript in preparation.
- qPCR
- quantitative PCR
- TSP
- 3-(trimethylsilyl)propionic-2,2,3,3-d4 acid.
REFERENCES
- 1. Holden H. M., Rayment I., Thoden J. B. (2003) Structure and function of enzymes of the Leloir pathway for galactose metabolism. J. Biol. Chem. 278, 43885–43888 [DOI] [PubMed] [Google Scholar]
- 2. Seiboth B., Pakdaman B. S., Hartl L., Kubicek C. P. (2007) Lactose metabolism in filamentous fungi: how to deal with an unknown substrate. Fungal Biol. Rev. 21, 42–48 [Google Scholar]
- 3. De Ley J., Doudoroff M. (1957) The metabolism of d-galactose in Pseudomonas saccharophila. J. Biol. Chem. 227, 745–757 [PubMed] [Google Scholar]
- 4. Elshafei A. M., Abdel-Fatah O. M. (2001) Evidence for a non-phosphorylated route of galactose breakdown in cell-free extracts of Aspergillus niger. Enzyme Microb. Technol. 29, 76–83 [DOI] [PubMed] [Google Scholar]
- 5. Seiboth B., Hartl L., Pail M., Fekete E., Karaffa L., Kubicek C. P. (2004) The galactokinase of Hypocrea jecorina is essential for cellulase induction by lactose but dispensable for growth on d-galactose. Mol. Microbiol. 51, 1015–1025 [DOI] [PubMed] [Google Scholar]
- 6. Seiboth B., Gamauf C., Pail M., Hartl L., Kubicek C. P. (2007) The d-xylose reductase of Hypocrea jecorina is the major aldose reductase in pentose and d-galactose catabolism and necessary for β-galactosidase and cellulase induction by lactose. Mol. Microbiol. 66, 890–900 [DOI] [PubMed] [Google Scholar]
- 7. Mojzita D., Penttilä M., Richard P. (2010) Identification of an l-arabinose reductase gene in Aspergillus niger and its role in l-arabinose catabolism. J. Biol. Chem. 285, 23622–23628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mojzita D., Koivistoinen O. M., Maaheimo H., Penttilä M., Ruohonen L., Richard P. (2012) Identification of the galactitol dehydrogenase, LadB, that is part of the oxidoreductive d-galactose catabolic pathway in Aspergillus niger. Fungal Genet. Biol. 49, 152–159 [DOI] [PubMed] [Google Scholar]
- 9. Richard P., Londesborough J., Putkonen M., Kalkkinen N., Penttilä M. (2001) Cloning and expression of a fungal l-arabinitol 4-dehydrogenase gene. J. Biol. Chem. 276, 40631–40637 [DOI] [PubMed] [Google Scholar]
- 10. Pail M., Peterbauer T., Seiboth B., Hametner C., Druzhinina I., Kubicek C. P. (2004) The metabolic role and evolution of l-arabinitol 4-dehydrogenase of Hypocrea jecorina. Eur. J. Biochem. 271, 1864–1872 [DOI] [PubMed] [Google Scholar]
- 11. Seiboth B., Metz B. (2011) Fungal arabinan and l-arabinose metabolism. Appl. Microbiol. Biotechnol. 89, 1665–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koivistoinen O. M., Richard P., Penttilä M., Ruohonen L., Mojzita D. (2012) Sorbitol dehydrogenase of Aspergillus niger, SdhA, is part of the oxidoreductive d-galactose pathway and essential for d-sorbitol catabolism. FEBS Lett. 586, 378–383 [DOI] [PubMed] [Google Scholar]
- 13. Fekete E., Karaffa L., Sándor E., Bányai I., Seiboth B., Gyémánt G., Sepsi A., Szentirmai A., Kubicek C. P. (2004) The alternative d-galactose degrading pathway of Aspergillus nidulans proceeds via l-sorbose. Arch. Microbiol. 181, 35–44 [DOI] [PubMed] [Google Scholar]
- 14. Guangtao Z., Hartl L., Schuster A., Polak S., Schmoll M., Wang T., Seidl V., Seiboth B. (2009) Gene targeting in a nonhomologous end joining-deficient Hypocrea jecorina. J. Biotechnol. 139, 146–151 [DOI] [PubMed] [Google Scholar]
- 15. Gruber F., Visser J., Kubicek C. P., de Graaff L. H. (1990) The development of a heterologous transformation system for the cellulolytic fungus Trichoderma reesei based on a pyrG-negative mutant strain. Curr. Genet. 18, 71–76 [DOI] [PubMed] [Google Scholar]
- 16. Peränen J., Rikkonen M., Hyvönen M., Kääriäinen L. (1996) T7 vectors with modified T7lac promoter for expression of proteins in Escherichia coli. Anal. Biochem. 236, 371–373 [DOI] [PubMed] [Google Scholar]
- 17. de Groot M. J., van den Dool C., Wösten H. A., Levisson M., vanKuyk P. A., Ruijter G. J., de Vries R. P. (2007) Regulation of pentose catabolic pathway genes of Aspergillus niger. Food Technol. Biotechnol. 45, 134–138 [Google Scholar]
- 18. Mojzita D., Vuoristo K., Koivistoinen O. M., Penttilä M., Richard P. (2010) The “true” l-xylulose reductase of filamentous fungi identified in Aspergillus niger. FEBS Lett. 584, 3540–3544 [DOI] [PubMed] [Google Scholar]
- 19. Christensen U., Gruben B. S., Madrid S., Mulder H., Nikolaev I., de Vries R. P. (2011) Unique regulatory mechanism for d-galactose utilization in Aspergillus nidulans. Appl. Environ. Microbiol. 77, 7084–7087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Richard P., Putkonen M., Väänänen R., Londesborough J., Penttilä M. (2002) The missing link in the fungal l-arabinose catabolic pathway, identification of the l-xylulose reductase gene. Biochemistry 41, 6432–6437 [DOI] [PubMed] [Google Scholar]
- 21. Metz B., de Vries R. P., Polak S., Seidl V., Seiboth B. (2009) The Hypocrea jecorina (syn. Trichoderma reesei) lxr1 gene encodes a d-mannitol dehydrogenase and is not involved in l-arabinose catabolism. FEBS Lett. 583, 1309–1313 [DOI] [PubMed] [Google Scholar]
- 22. Seiboth B., Hartl L., Pail M., Kubicek C. P. (2003) d-Xylose metabolism in Hypocrea jecorina: loss of the xylitol dehydrogenase step can be partially compensated for by lad1-encoded l-arabinitol-4-dehydrogenase. Eukaryot Cell 2, 867–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roberts C. F. (1970) Enzyme lesions in galactose non-utilizing mutants of Aspergillus nidulans. Biochim. Biophys. Acta 201, 267–283 [DOI] [PubMed] [Google Scholar]
- 24. Flipphi M., Sun J., Robellet X., Karaffa L., Fekete E., Zeng A. P., Kubicek C. P. (2009) Biodiversity and evolution of primary carbon metabolism in Aspergillus nidulans and other Aspergillus spp. Fungal Genet. Biol. 46, Suppl. 1, S19–S44 [DOI] [PubMed] [Google Scholar]
- 25. de Vries R. P. (2008) in Aspergillus in the Genomic Era (Vargas J., Samson R. A., eds), pp. 87–106, Wageningen Academic Publishers, Wageningen [Google Scholar]
- 26. Fekete E., Padra J., Szentirmai A., Karaffa L. (2008) Lactose and d-galactose catabolism in the filamentous fungus Aspergillus nidulans. Acta Microbiol. Immunol. Hung. 55, 119–124 [DOI] [PubMed] [Google Scholar]
- 27. Fekete E., de Vries R. P., Seiboth B., vanKuyk P. A., Sándor E., Fekete E., Metz B., Kubicek C. P., Karaffa L. (2012) d-Galactose uptake is nonfunctional in the conidiospores of Aspergillus niger. FEMS Microbiol Lett. 329, 198–203 [DOI] [PubMed] [Google Scholar]
- 28. Bos C. J., Debets A. J., Swart K., Huybers A., Kobus G., Slakhorst S. M. (1988) Genetic analysis and the construction of master strains for assignment of genes to six linkage groups in Aspergillus niger. Curr. Genet. 14, 437–443 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.