Abstract
Genetic variation and population structure among 1603 soybean accessions, consisted of 832 Japanese landraces, 109 old and 57 recent Japanese varieties, 341 landrace from 16 Asian countries and 264 wild soybean accessions, were characterized using 191 SNP markers. Although gene diversity of Japanese soybean germplasm was slight lower than that of exotic soybean germplasm, population differentiation and clustering analyses indicated clear genetic differentiation among Japanese cultivated soybeans, exotic cultivated soybeans and wild soybeans. Nine hundred ninety eight Japanese accessions were separated to a certain extent into groups corresponding to their agro-morphologic characteristics such as photosensitivity and seed characteristics rather than their geographical origin. Based on the assessment of the SNP markers and several agro-morphologic traits, accessions that retain gene diversity of the whole collection were selected to develop several soybean sets of different sizes using an heuristic approach; a minimum of 12 accessions can represent the observed gene diversity; a mini-core collection of 96 accession can represent a major proportion of both geographic origin and agro-morphologic trait variation. These selected sets of germplasm will provide an effective platform for enhancing soybean diversity studies and assist in finding novel traits for crop improvement.
Keywords: Glycine max, Glycine soja, SNP, Genebank, LD, mini core collection
Introduction
Soybean, Glycine max (L.) Merr., is the most important legume globally and the fourth crop next to rice, wheat and maize in terms of global crop production. Soybean has been used as a major source of nutritious feed for humans and livestock and is an important part of traditional foods in many Asian countries. Soybean seeds contain a high percentage of protein and oil. Soybean is now regarded as a model legume crop owing to the availability of genome sequence information (Schmutz et al. 2010). A large proportion of the soybean genome, 975 Mb out of 1.1 Gb of the estimated size, is available as a chromosome-scale assembly of 20 soybean chromosomes from the phytozome web site (Schmutz et al. 2010, http://www.phytozome.net/soybean.php). Soybean is known as palaeopolyploid species that have undergone whole genome duplication and simultaneous many rearrangements among chromosomes. The complex features in the soybean genome have been reviewed by Cannon and Shoemaker (2012) by comparing genomes across the related pulses. The knowledge and technologies of the newly available genome information are expected to broaden understanding the relationships between genes and agronomically important traits as well as assist in the better use of gene resources for soybean germplasm for breeding.
The recent sequencing of the soybean genome enables molecular tools to be developed for gene discovery, breeding and germplasm characterization. Technological advances in detecting and genotyping of single nucleotide polymorphisms (SNPs) by next generation sequencing and the high throughput genotyping systems has led to a decrease in genotyping cost and time. Simple sequence repeat (SSR) markers have been a conventional tool in soybean genetics because of their high allelic diversity. However, the complexity of the evolutionary process requires estimates of genetic relationships without knowledge of the mutational properties of markers (Ellegren 2004) while SNP markers based on nucleotide change would allow for more exact germplasm characterization. High-throughput genotyping platforms for SNPs can now overcome their biallelic nature and lower allele variability compared with SSR markers. This is due to their vast numbers identified with the most recent generation of sequencers. In soybean, SNP discovery on a large scale identified 5551 SNPs in five genotypes based on gene derived EST assembly (Choi et al. 2007). Hyten et al. (2010) identified 7,108 SNPs between soybean and wild soybean (Glycine soja Sieb. & Zucc.) by next generation sequencing of reduced representation libraries. Recently, 205,614 tag SNPs among Chinese soybean and wild soybeans (Lam et al. 2010), 2.5 million SNPs between Korean wild soybean and the reference sequence (Kim et al. 2010) have been reported.
Wild soybean is presumed to be the direct ancestor of soybean (Hymowitz 1970). It grows in disturbed habitats in the Far East of Russia, eastern China including Taiwan, the Korean peninsula and Japan (Lu 2004). Wild soybeans are important resources of novel alleles to broaden the genetic base of cultivated soybean because diversity in soybean has been greatly reduced by the genetic bottleneck of domestication (Guo et al. 2010, Lam et al. 2010). Domestication of soybean from wild soybean is believed to have occurred around the eleventh century B.C. in the east half of north China and to spread to surrounding countries around the first century A.D. based on various geographical and historical evidences (Hymowitz 1990). Japanese soybeans are thought to have been brought from Korea and China during the Yayoi period (200 B.C. to the first century A.D.) (Hymowiz and Kaizuma 1981). The phylogeny and diversity of cultivated and wild soybeans have been extensively analyzed based on cytoplasmic genome variation and reviewed by Shimamoto (2001). Assessment of nuclear genome diversity based on SSR and other marker system has been conducted on soybean germplasm. The geographical distribution and genetic diversity over 20,000 Chinese soybean accessions (Dong et al. 2004) and 6,000 Chinese wild soybean accessions (Dong et al. 2001) were assessed based on qualitative and quantitative characters. The genetic structure of 1,863 Chinese landraces (Li et al. 2008) and 231 Chinese wild soybean (Guo et al. 2010) were characterized by SSR markers. In Korea, the genetic structure of 260 Korean landraces (Cho et al. 2008) and 210 Korean wild soybean accessions (Lee et al. 2008) and the genetic diversity of 2,758 accessions among approximately 7,000 Korean landraces conserved in the RDA genebank (Yoon et al. 2009) were evaluated by SSR markers.
In Japan, soybean is an important source of traditional foods such as Tofu, natto, miso, soy sauce and edamame and various landraces have been established according to the food usage. Seed quality has been one of conventional breeding targets for these purposes. Recently, the most serious problems affecting soybean production in Japan are the unstable and low yields when soybeans are cultivated in paddy fields in a rotation system with rice. To overcome these problems, flood stress resistance, disease and pest resistance in lowland conditions and adaptation to mechanical harvesting are the main breeding objectives in Japan. Abe et al. (2003) found that the Japanese soybean gene pool is distinct from the Chinese gene pool. Guan et al. (2010) compared seven out of eight different Chinese soybean ecotypes from the three large eco-regions with improved Japanese varieties. They found that Japanese varieties had a lower genetic diversity than Chinese ecotypes but distinct alleles from them and suggested the potential to broaden the genetic base by exchanging germplasm between both countries. Zhou et al. (2000) indicated that the genetic base of Japanese improved varieties was quite distinct from that of China, USA, Canada and also low genetic exchanges has occurred between germplasm from northern, central and southern regions in Japan. Although soybean breeding has mainly focused on seed quality and food processing such as large seed and high protein content, a high degree of diversity among Japanese improved varieties released during 1950 to 1988 was found to be maintained through continual use of local and exotic landraces (Zhou et al. 2002). They suggested that Japanese germplasm has the potential to use genetic diversity of soybean varieties from other countries in Japanese soybean breeding.
Approximately 11,300 soybean accessions are conserved at the National Institute of Agrobiological Sciences (NIAS) Genebank. They include local landraces collected in Japan and overseas, improved varieties and breeding lines developed by regional Japanese agricultural research institutes or introduced from overseas agricultural research institutes and wild soybeans. During the period from 1939 to 2003, 125 soybean varieties with the ‘Soybean Norin’ name have been registered by the Ministry of Agriculture, Forestry and Fisheries (MAFF). Among these improved varieties preserved at the NIAS Genebank, authentic seeds of 91 varieties released during 1939 to 1989 were identified by Miyazaki et al. (1995a, 1995b) based on passport data. At that time, they found many duplicated accessions and homonyms with many ecotypes. Therefore, it is important to eliminate duplicated materials in order to conserve and manage efficiently large germplasm collections and to provide authentic materials to users. However, the identity among landraces with similar names collected by many researchers over a long period cannot be resolved by passport data alone.
In order to improve accessibility of useful accessions from the available large germplasm collection for biologists and breeders, the concept of a core collection, manageable and representative set of the entire collection, was proposed (Frankel and Brown 1984). A large number of core collections have been established in various germplasm so far. A standard figure for core collection is 10% of the whole collection, hence the number of accessions in a core collection varies according to the total number of accessions in the genebank. The number of accessions in the core collection can be too large to handle by researchers in some cases. Recently a collection of reduced size called mini-core subset has been developed in combination with data of molecular markers. In the NIAS Genebank, Japan, a Rice Diversity Research Set of NIAS representing world-wide rice germplasm (Kojima et al. 2005), mini core collection of Japanese rice landrace (Ebana et al. 2008) and Sorghum diversity Research Set (Shehzad et al. 2009) were developed from conserved germplasm based on this strategy. Despite its importance, no genetic characterization has examined soybean germplasm conserved in the NIAS Genebank and no core collection has been developed. A Chinese soybean core collection (943 accessions) and mini-core collection (118 accessions) have been developed from 23,587 accessions conserved in the Chinese National Soybean GeneBank (CNSGB) (Qiu et al. 2009). Cho et al. (2008) developed core set consisted of 260 Korean landraces from approximately 7,000 accessions conserved in the National Genebank of Rural Development Administration, Korea (RDA-Genebank). Recently, a core collection (1600 accessions) was selected from 16,999 accessions in USDA Soybean Germplasm Collection (Oliveira et al. 2010).
The characterization of the population structure of Japanese soybean germplasm is required to provide genetic materials with novel variation to soybean biologists and breeders. This will enable the development of strategies for effective breeding and for the isolation of agronomically important genes in the future. The objectives of this study were to characterize the genetic diversity of Japanese soybean germplasm and develop a mini-core collection.
Materials and Methods
Plant materials
A small set of germplasm accessions was used for SNP marker screening. This set consisted of 96 accessions; 65 Japanese accessions, 26 accessions from outside Japan (considered exotic germplasm here) and 5 wild soybean (Glycine soja) accessions. Japanese accessions were comprised of 44 crossbred varieties, 11 recent varieties developed by pure-line selection and 10 landraces (Supplemental Table 1). Several recent varieties were kindly provided from regional Japanese agricultural research institutes and the rest obtained from the National Institute of Agrobiological Sciences Genebank. Exotic accessions consisted of landraces from China (6 accessions), India (2 acc.), Korea (2 acc.), Vietnam (2 acc.), Myanmar (3 acc.), Nepal (3 acc.), Indonesia (1 acc.), Pakistan (1 acc.), Paraguay (1 acc.), the Philippines (1 acc.) and Thailand (1 acc.). Three major varieties of the U.S.A. (3 acc.) were obtained from the United States Department of Agriculture (USDA).
Based on passport data records, a Japanese landrace subset consisting of 832 accessions was proportionally selected from 3994 accessions of the National Institute of Agrobiological Sciences Genebank so that it reflected the number of samples collected at prefectural level (Fig. 1 and Table 1). Accessions with passport data of collection site and/or local name were preferentially selected. Simultaneously, 109 varieties, which were widely grown before the 1950’s described in the reports issued by the Ministry of Agriculture and Forestry (1952a, 1952b, 1953b, 1954), and 47 crossbred and 10 pure line selected varieties described above were evaluated together (Supplemental Table 1). An exotic land-race subset consisting of 341 accessions from 16 Asian countries was proportionally selected from 2034 landraces in NIAS Genebank. Accessions with passport data for the collection site were preferentially selected. For comparative purposes, wild soybean (Glycine soja) accessions analyzed by SSR marker (Kuroda et al. 2009) were included in the analysis as a reference for wild soybean. The data for seed size, SSR genotype and geographic origin of 690 accessions introduced from USDA were re-examined. Fifty accessions with 100 seed weight more than 2.5 g and/or high heterozygosity were removed and, finally, 190 accessions were proportionally selected again based on their origin so that the samples were from a wide region. For Japanese wild soybeans, 74 accessions different materials from those used by Kuroda et al. (2009) were selected according to their geographic origin based on passport data and with 100 seed weight not greater than 2.5 g. In total, 190 exotic wild accessions from the USDA and 74 Japanese wild accessions (Supplemental Fig. 1) were analyzed in the present study.
Fig. 1.
Geographical distribution of cultivated soybean accessions analyzed in the present study. Japan was subdivided into 11 historic regions.
Table 1.
Number of accessions analyzed in the present study and the gene diversity partitioned by countries and/or regions. ‘Other’ includes cultivars from U.S., Paraguay and Sweden
| Country | Region | Classification | Number of accessions | Gene diversity | Proportion of accessions to the inferred clusters | ||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Japanese cultivated | Exotic cultivated | Wild | |||||
| Japan | A01 | Landrace | 39 | 0.33 | 0.89 | 0.04 | 0.07 |
| Japan | A02 | Landrace | 177 | 0.37 | 0.94 | 0.04 | 0.02 |
| Japan | A03 | Landrace | 128 | 0.37 | 0.92 | 0.05 | 0.02 |
| Japan | A04 | Landrace | 126 | 0.36 | 0.96 | 0.03 | 0.02 |
| Japan | A05 | Landrace | 26 | 0.35 | 0.96 | 0.02 | 0.02 |
| Japan | A06 | Landrace | 35 | 0.37 | 0.92 | 0.04 | 0.04 |
| Japan | A07 | Landrace | 74 | 0.35 | 0.96 | 0.03 | 0.02 |
| Japan | A08 | Landrace | 47 | 0.36 | 0.95 | 0.02 | 0.02 |
| Japan | A09 | Landrace | 53 | 0.34 | 0.94 | 0.05 | 0.01 |
| Japan | A10 | Landrace | 109 | 0.38 | 0.89 | 0.08 | 0.03 |
| Japan | A11 | Landrace | 18 | 0.36 | 0.61 | 0.30 | 0.10 |
| Japan | n.d. | Old varietes | 109 | 0.38 | 0.91 | 0.05 | 0.03 |
| Japan | n.d. | Pure line selection | 10 | 0.35 | 0.83 | 0.12 | 0.05 |
| Japan | n.d. | Crossbred | 47 | 0.39 | 0.84 | 0.11 | 0.05 |
| South Korea | n.d. | Landrace | 52 | 0.36 | 0.64 | 0.32 | 0.03 |
| North Korea | n.d. | Landrace | 11 | 0.37 | 0.64 | 0.30 | 0.07 |
| Taiwan | n.d. | Landrace | 13 | 0.39 | 0.54 | 0.43 | 0.03 |
| China | n.d. | Landrace | 81 | 0.41 | 0.27 | 0.64 | 0.09 |
| Vietnam | n.d. | Landrace | 27 | 0.30 | 0.05 | 0.94 | 0.01 |
| Laos | n.d. | Landrace | 5 | 0.23 | 0.06 | 0.94 | 0.01 |
| Cambodia | n.d. | Landrace | 1 | n.d. | 0.01 | 0.99 | 0.00 |
| Thailand | n.d. | Landrace | 13 | 0.29 | 0.12 | 0.87 | 0.01 |
| Myanmar | n.d. | Landrace | 21 | 0.33 | 0.09 | 0.84 | 0.07 |
| Malaysia | n.d. | Landrace | 1 | n.d. | 0.90 | 0.08 | 0.01 |
| Indonesia | n.d. | Landrace | 17 | 0.31 | 0.06 | 0.89 | 0.06 |
| Philippines | n.d. | Landrace | 4 | 0.32 | 0.36 | 0.63 | 0.01 |
| East Timor | n.d. | Landrace | 4 | 0.20 | 0.15 | 0.82 | 0.03 |
| Nepal | n.d. | Landrace | 42 | 0.33 | 0.07 | 0.74 | 0.20 |
| India | n.d. | Landrace | 33 | 0.38 | 0.11 | 0.80 | 0.09 |
| Pakistan | n.d. | Landrace | 11 | 0.12 | 0.00 | 0.60 | 0.40 |
| Other | n.d. | Landrace and cross bred | 5 | n.d. | 0.28 | 0.67 | 0.05 |
| Japan | A01 | Wild | 3 | 0.13 | 0.05 | 0.01 | 0.95 |
| Japan | A02 | Wild | 12 | 0.28 | 0.13 | 0.03 | 0.84 |
| Japan | A03 | Wild | 7 | 0.29 | 0.18 | 0.02 | 0.79 |
| Japan | A04 | Wild | 8 | 0.28 | 0.18 | 0.04 | 0.78 |
| Japan | A05 | Wild | 3 | 0.20 | 0.07 | 0.05 | 0.89 |
| Japan | A06 | Wild | 4 | 0.24 | 0.06 | 0.06 | 0.88 |
| Japan | A07 | Wild | 11 | 0.29 | 0.14 | 0.02 | 0.85 |
| Japan | A08 | Wild | 7 | 0.28 | 0.08 | 0.03 | 0.89 |
| Japan | A09 | Wild | 6 | 0.22 | 0.08 | 0.02 | 0.90 |
| Japan | A10 | Wild | 13 | 0.27 | 0.06 | 0.03 | 0.92 |
| Korea | n.d. | Wild | 81 | 0.27 | 0.00 | 0.01 | 0.99 |
| China | n.d. | Wild | 72 | 0.23 | 0.02 | 0.01 | 0.97 |
| Russia | n.d. | Wild | 37 | 0.22 | 0.00 | 0.01 | 0.99 |
A single plant except for wild soybean accessions was grown in the field the National Institute of Agrobiological Sciences (NIAS), Tsukuba, Japan, 36°2′N, 140°8′E, from 3 June to the end of November, 2009. The spacing of each plant was 1 m between rows and 0.5 m between plants. Since phenotypic data attached to each accession has been obtained from different geographical regions, the number of days from sowing to first flowering, plant height and 100 seed weight of all accessions were evaluated under the same condition to understand phenotypic variation in the accessions. Other traits, such as color of seed coat, hilum, pod, flower, hair of stem, days to flowering on the top of main stem, days to when several pods start to mature (R7), total seed weight were also recorded.
DNA extraction
Total genomic DNA was extracted from 0.3 g of fresh leaf tissue by a modified method using guanidine hydrochloride and protease (Khosla et al. 1999). DNA concentration was quantified using the fluorescence microplate reader ARVO (Perkin Elmer, Boston, MA, USA) according to the manufacture instructions and was adjusted to 20 ng/μl.
Marker analysis
The chromosome-scale assembly of the U.S. cultivar Williams 82, Glyma 1.0, has been available since December 2008 (Soybean Genome Project, DoE Joint Genome Institute, http://www.phytozome.net/soybean.php). Sequence information for 4240 STS containing SNP including indel (Choi et al. 2007) were retrieved from NCBI database and were blasted using default parameters (0.01; low complexity; 100; 100; -G5-E2) against Glyma 1.0 to find low copy sequences. Multiplex assays for 1136 randomly selected SNP including indel covering the genome widely were designed to the low copy sequences by means of the Sequenom Assay Design 3.1 software. In order to use specific amplicon, sequences of the amplification primer pairs were searched against Glyma 1.0 to examine number of binding sites, amplicon size and location by using Genome tester (Andreson et al. 2006) with default parameter. These processes were repeated until a single amplicon was obtained. For evaluation of a large number of accessions, a new multiplex set based on a reduced number of polymorphic SNP markers was designed by means of Sequenom Assay Design 3.1 software. Nomenclature of marker name was followed by the original name (Supplemental Table 2) but the prefix of chromosome number was added to discriminate from the original marker name because sequences of primer pairs designed here were different from those of Chio et al. (2007).
Genotyping was conducted using Sequenom MassARRAY system (methodology of the system reviewed by Oeth et al. 2009). Multiplex PCR followed by a template-directed single base extension at each SNP or indel site were conducted using MassARRAY iPLEX Gold kit (Sequenom) following the manufacture’s protocol. The reaction mixture was dispensed onto a silicon matrix preloaded SpectroCHIP (Sequenom) using Nanodispenser (Sequenom) and analyzed by Compact MassARRAY MALDI-TOF (Sequenom). The genotypes were determined using MassARRAY Typer4.0 (Sequenom).
Population analysis
Population statistics, gene diversity and population differentiation test, were analyzed using software PowerMarker ver. 3.2.5 (Liu and Muse 2005). Linkage disequilibrium (LD) were estimated with squared allele frequency correlations (r2) between marker loci with MAF (minor allele frequency) >0.1 on the same chromosome by using the software TASSEL ver. 2.0.1 (Bradbury et al. 2007). Since estimate of D′ based on a small number of samples becomes unreliable, only estimate of r2 are presented in the study. The LD decay was estimated following the method of Robbins et al. (2011). A smooth spline was fitted between the r2 and distance of the SNP pairs using loess.smooth function (span = 2/3, 2nd degree of polynomial) in R 2.7.2 (R Development Core Team 2008) and then the distance in bp was determined by cross point of the smooth line with the baseline r2 value of 0.1.
STRUCTURE version 2.1 (Pritchard et al. 2000) was used to assess the extent of genetic admixture among Japanese, exotic and wild germplasm. Estimation of allele frequencies in the presumed number of cluster (K) was simulated based on the independent allele frequency model using a Markov Chain Monte Carlo (MCMC) method. The degree of admixture in each accession was estimated under the conditions of 100,000 burn-in period and 100,000 MCMC replications.
In order to reveal genetic relationships among accessions, a neighbor joining tree was constructed based on shared allele distance (DSA; Chakraborty and Jin 1993) of genotype information using PowerMarker software (Liu and Muse 2005) and visualized with Fig Tree v1.3.1 software (Rambaut 2009). Bootstrap values supporting the branch with higher than 50% are indicated on the branch.
Selection of materials for mini-core collection
PowerCore ver 1.0 (Kim et al. 2007) was used to select a mini-core collection. This program uses advanced M strategy that maximizes the allelic and the phenotypic trait representation by using a modified heuristic algorithm to find out optimum path for sample selection. Initially the minimum number of accessions that can represent the gene diversity of all Japanese germplasm were selected based on genotypic data alone. In those accessions, several important varieties that are a major target for re-sequencing to develop a Japanese soybean SNP panel were preferentially selected and landraces that complement the remaining diversity of all Japanese germplasm were allocated. The accessions having heterozygous loci have been removed from all selection procedures. Then, both SNP and phenotypic data, the number of days from sowing to first flowering (DF), plant height (PH) and 100 seed weight (100SW), were included in the selection procedure of the mini-core collection. Twenty, eleven and twelve phenotypic classes were defined for DF, PH and 100SW based on the observed ranges, respectively. The sample sizes for Japanese and the exotic mini-core collection were limited to 96 accessions. Representative accessions having local name at each branch on the dendrogram were preferentially sampled in the Japanese mini-core collection development. As for selecting the exotic mini-core collection, the proportion of accessions to country was also considered. In order to confirm homogeneity, genetic diversity and statistics for phenotypic trait coverage in the selected mini-core collection were compared with the initial large set.
Results and Discussion
Selection of informative markers for germplasm evaluation
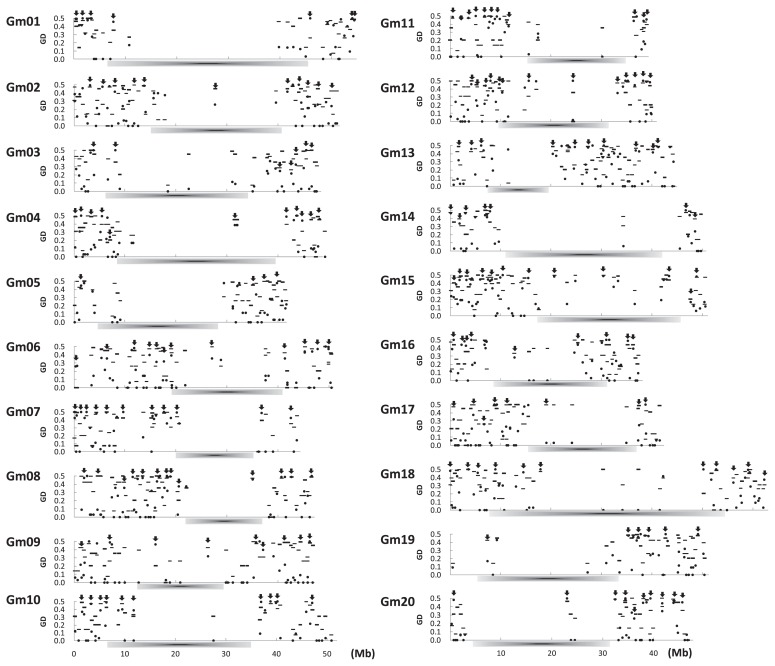
In total, 1057 markers were designed from 4240 STS containing 5551 SNPs (Choi et al. 2007) using the soybean Glyma1.0 chromosome assembly. Polymorphism of these markers was screened using a small set of germplasm consisting of 96 accessions; 65 Japanese and 26 exotic cultivated soybeans and 5 wild soybeans accessions. Out of 1013 SNP markers 963 had a good genotyping quality revealed by polymorphisms between any pair of accessions and those were mainly distributed in euchromatic gene space at a density of 1 marker per 1 Mb (Table 2). The density of markers varied from low (1 markers per 1.6 Mb) on Gm14 to high (1 markers per 690 kb) on Gm13. Among the 963 SNP markers, 145 (15.1%) were markers derived from indel variation. The primers used for marker detection were designed to amplify a single amplicon in order to avoid analytical complexity, especially genotyping, because of the considerable duplication of regions resulting from the ancient poly-ploidization of the soybean genome (Schmutz et al. 2010). Nonetheless, the remaining 20 and 24 markers revealed low genotype call due to signal complexity of multi-copy sequence and low signal, respectively. Biased distribution of the 963 informative SNP markers was observed on each chromosome in spite of designing as many markers as possible to cover the soybean genome evenly (Fig. 2). For example, maximum distance between markers was 29 Mb on chromosome 1. Low marker density on each chromosome corresponded to the pericentromeric genomic region where recombination is highly suppressed in the middle and the extent differed among chromosomes (Schmutz et al. 2010). The repetitive complexity and low gene density at such pericentromeric region may confound the design of SNP markers. The duplicated soybean genome is also problematic for design and detection of functionally important alleles specific SNPs as a single locus and thus a bioinformatic platform to optimize a specific amplicon to soybean gene will be required.
Table 2.
Summary of 963 SNP markers used for screening of 96 accessions
| Chromosome | No. of SNP loci | Average interval distance (bp)a | Gene diversity | No. of markers with minor allele frequency >0.1 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||
| Whole | Japanese | Exotic | Wild | Whole | Japanese | Exotic | Wild | |||
| Gm01 | 38 | 1,460,452 | 0.32 | 0.27 | 0.37 | 0.26 | 28 | 21 | 33 | 24 |
| Gm02 | 58 | 902,512 | 0.29 | 0.22 | 0.34 | 0.22 | 41 | 33 | 49 | 32 |
| Gm03 | 40 | 1,208,800 | 0.25 | 0.15 | 0.37 | 0.24 | 27 | 16 | 36 | 25 |
| Gm04 | 43 | 1,164,079 | 0.27 | 0.19 | 0.32 | 0.22 | 29 | 16 | 35 | 23 |
| Gm05 | 40 | 1,063,030 | 0.27 | 0.18 | 0.36 | 0.27 | 28 | 16 | 37 | 26 |
| Gm06 | 61 | 837,665 | 0.25 | 0.18 | 0.32 | 0.19 | 40 | 25 | 47 | 30 |
| Gm07 | 44 | 1,014,122 | 0.32 | 0.26 | 0.34 | 0.30 | 32 | 26 | 35 | 33 |
| Gm08 | 64 | 715,562 | 0.27 | 0.21 | 0.34 | 0.22 | 41 | 30 | 53 | 36 |
| Gm09 | 56 | 847,321 | 0.22 | 0.13 | 0.33 | 0.17 | 28 | 16 | 49 | 24 |
| Gm10 | 41 | 1,267,401 | 0.26 | 0.19 | 0.32 | 0.20 | 24 | 18 | 34 | 20 |
| Gm11 | 38 | 1,051,934 | 0.31 | 0.26 | 0.34 | 0.20 | 26 | 24 | 32 | 19 |
| Gm12 | 48 | 845,774 | 0.30 | 0.22 | 0.33 | 0.25 | 31 | 25 | 39 | 30 |
| Gm13 | 64 | 690,594 | 0.30 | 0.24 | 0.33 | 0.17 | 45 | 36 | 52 | 27 |
| Gm14 | 32 | 1,600,284 | 0.26 | 0.18 | 0.35 | 0.28 | 21 | 12 | 28 | 22 |
| Gm15 | 57 | 898,824 | 0.34 | 0.26 | 0.39 | 0.26 | 52 | 33 | 53 | 37 |
| Gm16 | 46 | 825,680 | 0.30 | 0.24 | 0.33 | 0.23 | 31 | 27 | 39 | 28 |
| Gm17 | 50 | 841,074 | 0.25 | 0.13 | 0.35 | 0.20 | 32 | 15 | 41 | 25 |
| Gm18 | 57 | 1,110,785 | 0.28 | 0.21 | 0.35 | 0.17 | 39 | 27 | 51 | 26 |
| Gm19 | 42 | 1,215,679 | 0.28 | 0.22 | 0.35 | 0.20 | 29 | 21 | 36 | 23 |
| Gm20 | 44 | 1,073,284 | 0.25 | 0.19 | 0.33 | 0.22 | 24 | 18 | 33 | 24 |
| Average (Total) | (963) | 994,601 | 0.28 | 0.21 | 0.34 | 0.22 | (648) | (455) | (812) | (534) |
the length of each chromosome was divided with the number of intervals between SNP markers.
Fig. 2.
Distribution of 936 SNPs on the chromosome-scale assembly of soybean genome, Glyma 1.0, and their gene diversity values among 69 Japanese accessions (black dot) and 26 exotic accessions (horizontal line). Arrows indicate that the location of SNPs which were used to evaluate a large set of soybean germplasm. Dark bars below each graph indicate presumed pericentromeric regions.
Mean gene diversity over all the 963 informative loci was 0.28 in 96 accessions (Table 2). Among different source of germplasm, mean gene diversity over all the loci of exotic soybeans was the highest (0.34) followed by wild soybeans (0.22) and Japanese soybeans (0.21). Population differentiation between Japanese and exotic cultivated soybeans, and between Japanese cultivated soybeans and wild soybeans were significant at the 1% level based on Mantel’s test of overall loci. Differentiation between exotic cultivated and wild soybeans was not statistically significant. This is because the genotypes of fifteen exotic accessions mainly from South and South-east Asia were relatively similar to wild soybeans (Supplemental Fig. 2). Among exotic germplasm, differentiation of Chinese accession ‘Peking’ from other accessions was prominent. Since the majority of SNPs used in the present study has been identified in re-sequencing the exotic germplasm array including Peking (Choi et al. 2007), the distribution range of the variations is supposed to be limited to those germplasm and possibly explains the low gene diversity of wild soybean as well as Japanese germplasm.
Among 956 polymorphic loci in cultivated soybean, 586 were statistically significant for population differentiation between Japanese and exotic soybeans. Overall, the gene diversity among Japanese accessions was lower than that among exotic accessions across chromosomes and especially on chromosome 3, 9 and 17 (Table 2 and Supplemental Table 2 in detail). In addition, 243 out of 963 loci (25.2%) were found to be monomorphic and not informative in Japanese soybeans compared with only 22 monomorphic loci in exotic germplasm. Thus, re-sequencing using a representative Japanese germplasm array is required in order to capture informative SNPs for Japanese soybean germplasm.
Although the number of informative markers to discriminate Japanese accessions were few, gene diversity of Japanese landraces was generally higher than that of improved varieties. Among 720 polymorphic loci, only 110 were statistically significant at the 1% level to differentiate between Japanese improved varieties and landraces (Supplemental Table 1). A large reduction in gene diversity was found for the markers located in a 8.1–9.8 Mb region of chromosome 8 which is close to seed coat color locus I. This is reasonable because most of Japanese improved varieties have yellow seed color. In contrast, a higher gene diversity of improved varieties than landraces was observed for the markers located in the 9.7–10.4 Mb region of chromosome 18. This region may be involved in the differentiation between cultivated and wild soybean in relation to stem elongation related traits (Lam et al. 2010). Most of the improved varieties having different alleles were breed at Tohoku experimental station. Examination of genetic structure of Japanese improved varieties at fine scale using both more SNP and pedigree information will lead to finding causal factors to explain why such alleles are maintained in those improved varieties and other important genomic regions for Japanese soybean breeding.
Two varieties, ‘Wasesuzunari’ (Hashimoto et al. 1985) and ‘Kosuzu’ (Hashimoto et al. 1988), have been selected for their early maturity by γ-ray irradiation of ‘Okushirome’ and ‘Nattoukotsubu’, respectively. These varieties and their original varieties were expected to have the same genotype for the randomly chosen 963 SNP loci scattered on soybean genome. However, even mutation breeds and their original varieties showed polymorphism; three loci (0.3%) between ‘Wasesuzunari’ and ‘Okushirome’; four loci (0.4%) between ‘Kosuzu’ and ‘Nattoukotsubu’. In contrast, ‘Murayutaka’ (Nakamura et al. 1991) was developed by X-ray irradiation of ‘Fukuyutaka’ and a slightly higher polymorphism between ‘Murayutaka’ and ‘Fukuyutaka’ than the two above-mentioned varieties was observed. Among nine polymorphic loci out of 959 compared (0.9%), three loci were identified to a region between 8.1 Mb to 9.8 Mb on chromosome 8 and two loci to the region between 6.1 Mb to 6.5 Mb. ‘Murayutaka’ was mainly improved to the point of brown hilum color of the major soybean variety ‘Fukuyutaka’ into yellow hilum color. The region on chromosome 8 contains the seed coat color locus I (Bernard and Weiss 1973) and contains the responsible gene (Clough et al. 2004). In the process of mutation breeding of ‘Murayutaka’, backcrossing was not involved. Therefore, it is still unclear why mutations frequently occur around the target locus and larger genomic region with three mutations has been retained by the artificial selection.
Linkage disequilibrium in Japanese germplasm
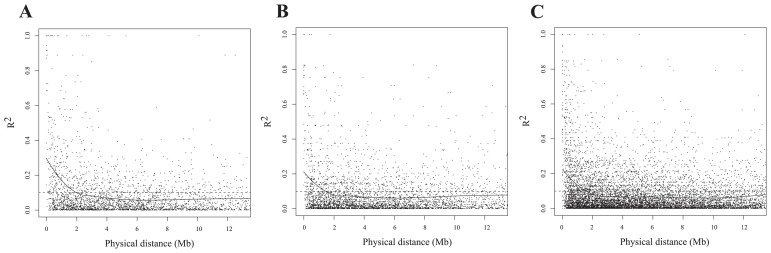
Information on linkage disequilibrium (LD) in Japanese soybean germplasm is important for manipulating strategies to allow genome wide association studies or genomic selection on breeding. Since recombination in each generation leads to break up of allelic associations between neighboring loci, the genetic distance between loci, effective population size and mutation frequency that give rise to new SNPs, largely affect the extent of LD (Hamblin et al. 2011). In the present study, LD was analyzed for the data that partitioned into Japanese crossbred, Japanese landrace and exotic germplasm against physical distance between SNPs. The squared correlation, r2 (Hill and Robertson 1968), between pairs of markers with MAF (minor allele frequency) >0.1 among each categories were calculated. As indicated in Table 2, such markers are limited in Japanese germplasm in spite of greater number of sample analyzed. Hyten et al. (2007) observed that the extent of LD varies depending on the genomic region of the chromosome and between chromosomes. Therefore, decay of LD in the present study was only estimated between marker pairs on the same chromosome. Data for marker pairs across pericentromeric region, where LD at long distance is expected, were excluded. The decline in LD were examined by non-linear curve fitting on r2 estimates of 2841, 3567 and 8669 loci combinations in Japanese cross-bred, Japanese landrace and exotic germplasm, respectively. Decay was declared in regions where the fitted values were above r2 = 0.1 (Fig. 3). Estimates of physical distance in Japanese crossbred, Japanese landrace and exotic germplasm were 1.97 Mb, 1.46 Mb and 1.56 Mb, respectively. Until recently there was no information about genome-wide LD in Japanese soybean germplasm, similar size of decay was observed between Japanese landrace and exotic germplasm whereas slightly slower decay of LD was observed in Japanese improved varieties than that of landraces. Hyten et al. (2007) reported 2~3 fold higher LD in elite northern American cultivars compared with their ancestors. The 1.4-fold increases in LD for Japanese crossbred compared with landraces suggests that effective recombination is restricted under the situation of limited generations and population size in breeding process. The current estimates of LD are likely to be overestimates due to the small sample size, the marker density and evaluation of chromosomes all together in comparison with previously reported estimates of LD ~574 kb (Hyten et al. 2007). Correct estimates of LD are needed in order to provide breeding strategies. Further examination by chromosomes based on a higher density SNP information of Japanese germplasm and each regional set of breeding lines by tracing the pedigree.
Fig. 3.
Decay of the squared correlation of allele frequencies (r2) with physical distance in bp in Japanese crossbred (A), landrace (B) and exotic germplasm (C).
Genetic structure of the soybean germplasm
In order to keep linkage between markers on the same chromosome as low as possible, 197 SNPs were selected at approximately 6 Mb interval apart in order to construct a new SNP multiplex reactions to evaluate a large set of germplasm. Among 197 SNPs analyzed in 1603 accessions, 191 SNPs gave good genotyping calls (99.8%) whereas 6 SNP loci with excess missing genotypes or heterozygous calls (>25% of sample) which may detect signals from paralogous region were removed. In total 97 % of the selected SNPs covering 20 soybean chromosomes were able to provide genetic information of 1603 diverse accessions (Table 3). Average marker interval was 5.2 Mb overall but 2.7 Mb outside the pericentromeric region. The marker density on each chromosome was varied and ranged from four (Gm05) to 13 (Gm08 and Gm12) with an average of 10 markers per chromosome.
Table 3.
Summary of chromosome-wise information of 191 selected SNP markers used for evaluation of 1603 accessions
| Chromosome | No. of SNP loci | Mean distance (bp) | Gene diversity | ||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Whole acc. | Cultivated soybean | Wild soybean | |||||
|
|
|
||||||
| Japanease | Exotic | Japanese | Exotic | ||||
| Gm01 | 7 | 8,691,106 | 0.48 | 0.44 | 0.39 | 0.31 | 0.25 |
| Gm02 | 10 | 5,258,957 | 0.42 | 0.33 | 0.44 | 0.32 | 0.19 |
| Gm03 | 7 | 7,133,078 | 0.39 | 0.29 | 0.44 | 0.16 | 0.15 |
| Gm04 | 11 | 4,783,855 | 0.45 | 0.36 | 0.42 | 0.33 | 0.26 |
| Gm05 | 4 | 12,599,736 | 0.41 | 0.38 | 0.39 | 0.19 | 0.14 |
| Gm06 | 12 | 4,505,019 | 0.44 | 0.37 | 0.44 | 0.33 | 0.27 |
| Gm07 | 11 | 4,234,282 | 0.47 | 0.42 | 0.43 | 0.35 | 0.26 |
| Gm08 | 13 | 3,665,927 | 0.44 | 0.41 | 0.42 | 0.29 | 0.28 |
| Gm09 | 9 | 5,590,517 | 0.41 | 0.32 | 0.42 | 0.20 | 0.22 |
| Gm10 | 9 | 5,693,703 | 0.44 | 0.41 | 0.39 | 0.33 | 0.30 |
| Gm11 | 11 | 3,840,468 | 0.46 | 0.42 | 0.42 | 0.31 | 0.30 |
| Gm12 | 13 | 2,942,636 | 0.45 | 0.41 | 0.44 | 0.30 | 0.21 |
| Gm13 | 10 | 4,391,180 | 0.44 | 0.39 | 0.42 | 0.31 | 0.24 |
| Gm14 | 6 | 9,683,581 | 0.45 | 0.39 | 0.44 | 0.35 | 0.37 |
| Gm15 | 11 | 4,810,803 | 0.47 | 0.42 | 0.44 | 0.41 | 0.39 |
| Gm16 | 8 | 5,067,648 | 0.47 | 0.45 | 0.42 | 0.30 | 0.31 |
| Gm17 | 8 | 5,434,227 | 0.37 | 0.25 | 0.43 | 0.31 | 0.19 |
| Gm18 | 11 | 6,220,394 | 0.47 | 0.42 | 0.43 | 0.34 | 0.24 |
| Gm19 | 10 | 4,581,286 | 0.41 | 0.35 | 0.43 | 0.35 | 0.23 |
| Gm20 | 10 | 5,000,491 | 0.45 | 0.40 | 0.45 | 0.34 | 0.33 |
| Average | 191 in total | 5,176,591 | 0.44 | 0.38 | 0.43 | 0.31 | 0.26 |
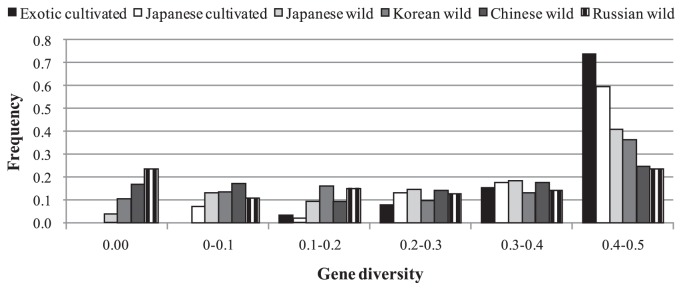
Average gene diversity in Japanese germplasm (0.38) was slightly lower than that of the exotic germplasm (0.43) (Table 3). When partitioned by their locality and breeding method, average gene diversity values from each of the historic regions in Japan, old and recently released varieties were not different from each other (Table 1). Among exotic germplasm, Chinese landraces revealed highest gene diversity and the diversity decreased slightly for countries of South-East Asia. Landraces in India had similar level of diversity to landraces in East Asia. Gene diversity of cultivated soybean was higher than that of wild soybean (Table 3). SNP loci with MAF (minor allele frequency) less than 0.1 was higher in wild soybean (Exotic 37% and Japanese 24%) than in cultivated soybean (Japanese 10% and Exotic 3%). The results are opposite from the diversity estimates between wild and cultivated soybean based on whole genome sequence analysis. Lam et al. (2010) reported that gene diversity of Chinese wild soybean is 1.6 times higher than that of Chinese cultivated soybeans at the whole genome level. Most loci which revealed high gene diversity in exotic and Japanese soybean landraces showed lower diversity in wild soybean (Fig. 4). In addition, the average number of loci that failed to detect expected SNP haplotypes during genotyping was 1.9 in wild soybeans and was >10 times higher that of cultivated soybean (0.13), suggesting that those represent null alleles for detecting SNP due to nucleotide differences at target sites of amplification or extend primers used in MassARRAY system in wild soybeans from the cultivated reference sequence. Lam et al. (2010) found that 35% of SNP alleles were specific to wild soybean. The lower genetic diversity in wild soybean might arise from unbalanced SNP loci selected based on cultivated germplasm characterization and several SNPs in the present study may just occur within the domesticated soybean gene pool.
Fig. 4.
The differences in the frequency of gene diversity for individual SNP locus classified by each germplasm groups.
Based on the Mantel test of overall loci, significant population differentiation at the 1% level was found among four groups; Japanese cultivated soybeans, exotic cultivated soybeans, Japanese wild soybeans and exotic wild soybeans. When population differentiation between these four groups were examined by individual SNP locus, most of the SNP loci were found contribute to differentiation between Japanese and exotic cultivated germplasm (88% of loci), between Japanese cultivated and wild germplasm (80%), between exotic cultivated and wild germplasm (88%). Geographical isolation of Japanese wild soybean populations from populations distributed on mainland Asia were expected to cause clear divergence among populations. However, poor genetic differentiation was observed; only 58% of SNP loci contributed to the population differentiation between Japanese and exotic wild soybean. Since the SNP marker used in the present study may preferentially detect variation within the cultivated soybean gene pool as described above, the variation in wild soybean population may be underestimated. Further population analysis for wild soybean will be needed using SNPs based on the variation within wild soybean populations.
Introgression in soybean germplasm
Using the model-based clustering method, STRUCTURE (Pritchard et al. 2000), population structure in germplasm was inferred and accessions were assigned to the inferred cluster based on 191 multilocus genotype data. At the lower number of clusters, K = 2 and 3, the difference of mean likelihood value, Ln P (D), between the respective K and the successive K were obvious, indicating that there are distinct populations. At K = 2, one cluster corresponds to cultivated soybean while the other corresponds to wild soybean. The majority of Japanese soybeans were grouped into cultivated soybean group while a number of exotic soybean accessions had membership to wild soybean, that is, were genetically close to wild soybean. Further, when three clusters (K = 3) was considered in the whole set of germplasm, cultivated soybeans were subdivided into two groups, Japanese and exotic soybean (Table 1 and Fig. 5). Among Japanese soybeans, the proportion of varieties with a membership of exotic soybeans was relatively high in the varieties developed by crossbreeding and pure line selection. Results suggest that 11–12% of the genetic base of Japanese varieties is derived from exotic germplasm. Zhou et al. (2000) inferred that 91% of the genes in released Japanese varieties between 1950 and 1988 are derived from Japanese ancestors and the remaining proportion from exotic ancestors based on coefficient of parentage analysis using pedigree records. The figure of genetic base of Japanese varieties in the present study was very similar to the previous study. Furthermore, the exotic genetic base in recent Japanese varieties increased by two-fold compared with old varieties. According to the statistics of soybean yield (Statistics Bureau, Ministry of Internal Affairs and Communications of Japan), the average yield of soybean is 144.8 kg/10 a (10 a = 0.1 ha). From 1950 to date yield increased 1.5–2 fold; whereas yield increased only 1.24 fold from 1900 to 1950 from 80.7 to 100 kg/10 a. Recent Japanese breeding program attempt to introgression of exotic genetic base may contributed to this in part.
Fig. 5.
Genetic admixture between Japanese and exotic cultivated soybeans, and wild soybeans. Estimated individual membership to three major groups (K = 3); white (Japanese cultivated soybeans), gray (Exotic cultivated soybeans) and black (wild soybeans).
Among Japanese landraces, approximately 30% of accessions collected from southern island of Japan, ‘A11’ region (Fig. 5, gray), revealed a high membership of exotic soybean class (Table 1); ‘Tashoutou’, ‘Chinpintou’, ‘Tamagomame’ and ‘Aohigu’. Landraces in other groups of ‘old varieties’, ‘A03’ and ‘A10’ regions, were suspected to be recently introduced from other countries; ‘Uda Daizu’, ‘Ippon Sangou’ and ‘Komuta’. Other accessions, with local names including ‘Gedaizu’ from ‘A11’ region, revealed a high membership to wild soybean class. Other landraces which had a membership to the wild soybean class were ‘Madara Ooba Tsurumame’, ‘Tsuru Sengoku’ and ‘Tsunehira Daizu’ from ‘A03’ region and ‘Kuro Sengoku’ from ‘A10’ region. Those accessions could have much of wild soybean genome considering their origin. Although ‘Tsuru Sengoku’ was classified as a landrace in the present study because of the collection record, this accession is a MAFF registered fodder variety selected from hybrid progenies between ‘Kuro Sengoku’ and the wild soybean from Gunma prefecture by National Institute of Animal Industry. ‘Kuro Sengoku’ has been cultivated as a fodder or green manure crop through the ages. ‘Ooba Tsurumame’ was initially collected from wild soybean population in Okayama prefecture because of its larger leaf than common wild soybean and segregated with an intermediate phenotype between cultivated and wild soybean (Sekizuka and Yoshiyama 1960). Although the origin of ‘Tsunehira Daizu’ and ‘Gedaizu’ is still unknown, they contain a proportion of wild soybean genome to a similar level as the other fodder soybeans.
Membership of exotic landraces showed different levels of inclusion in the three categories, Japanese and exotic cultivated soybean and wild soybean. A high proportion of landraces from Korean Peninsula and Taiwan had a membership with Japanese soybean germplasm whereas landraces in the wild soybean class was increased from China to southward. Pakistan and Nepal soybean landraces, 80% and 20%, respectively, had a high membership to the wild soybean class and they revealed morphologically intermediate characteristics between soybean and wild soybean as described below.
Japanese wild soybeans were found to have a higher probability containing cultivated soybean alleles. Average membership of Japanese wild soybeans to Japanese cultivated soybean group was 0.11 (maximum 0.26 in an accession from Nagano Pref. ‘A04’ region) whereas exotic wild soybean revealed very low membership to either exotic or Japanese cultivated soybean groups; Korean 0.01, Chinese 0.01 and Russian 0.01. Kuroda et al. (2006) inferred 6.8% of wild soybean accessions are presumed to have undergone introgression from cultivated soybean. Most of the Japanese wild soybean accessions were different in their study from those used here because the present study intended to capture geographical cline of wild soybean. However, the results obtained here were very similar to their study. One possibility is that the collection site of wild soybean in NIAS genebank tends to be near human disturbed habitats. In modern agro-ecosystem, natural hybridization between wild and cultivated soybean in Japan is thought to be very rare (Kuroda et al. 2010). However, periodical hybridization and introgression from long ago now have the potential of accumulating alleles from cultivated into wild soybean populations and conversely provide the chance to broaden genetic variability of cultivated soybean via gene introgression from wild soybean into cultivated soybean as shown by Xu et al. (2002) based on chloroplast SSR analysis. In the present study, the presumed introgressed alleles, which are frequent in cultivated but not in wild soybean, were not common among accessions that had a scattered distribution in Japan. Abe et al. (1999) reported that wild soybean population with the chloroplast haplotype I, which is the predominant haplotype in cultivated soybean, distribute in Japan. He suggested that the wild type plant with an unusual chloroplast haplotype in natural population could be established by the natural selection on hybrid progenies.
Redundancy and heterogeneities in soybean germplasm
Observed heterozygosity was very high in Japanese wild soybean (mean overall loci, 0.012; range, 0.000–0.068) compared with that of Japanese (0.005; 0.000–0.011) and exotic cultivated soybeans (0.005; 0.000–0.029) and exotic wild soybean (0.001; 0.000–0.049). Since the genotypes for all of accessions was determined from a single plant, the observed high heterozygosity in Japanese wild soybean are reasonable considering natural outcrossing rates range from 2 to 13% in wild soybean populations (Fujita et al. 1997, Kuroda et al. 2008). In contrast, the large difference between Japanese and exotic wild soybean germplasm on observed heterozygosity appear to be related to the interval and frequency of seed propagation after plant introduction. Japanese wild soybean accessions in the NIAS Genebank more or less propagated one or two times after collection whereas exotic wild soybeans which were introduced from USDA have been propagated for very long time and result in high homozygosity. In contrast, several accessions suspected of outcrossing during seed propagation were also identified at the cut off 1% of heterozygous loci against all loci; 5.1% of Japanese and 5.9% of exotic cultivated soybean accessions. At a maximum, one Japanese and one exotic accession had 48.7% and 52.4% heterozygous loci, respectively. Among wild soybean, three individuals were found to be highly heterozygous; JPW11 (12.6% of all loci), JPW17 (14.7%) and JPW26 (23.6%). Therefore, careful seed propagation to prevent out-crossing is necessary even though out-crossing in cultivated soybean is considered to be very low. Nelson (2011) pointed out that germplasm exchange among countries will become more important in the future and high quality materials with their accurate information will be required for exchange. Since phenotype and genotype of only a single plant was evaluated in the present study, there is no data for seed heterogeneity in an accession. However, the finding of heterozygous accessions suggests that heterogeneity exists in seed lots besides accidental seed contamination and for the integrity of accessions in the NIAS genebank it will be necessary with each round of propagation to compare with the original description of the phenotype.
It is expected that duplicates in germplasm collections occur over time after the release of improved varieties. Miyazaki et al. (1995b) traced passport data of 30 accessions with the name ‘Akasaya’ in the NIAS Genebank and found 6 duplicates sent via different institute from the same seed source. They noted that ‘Akasaya’ was developed by pure line selection from a heterogeneous landraces with red pods in Yamanashi in the 1910’s and subsequently numerous ecotypes were developed. There was no way to trace the original seed stock because an accession originating from Yamanashi was not found in the Genebank. In the present study, ten accessions with name of ‘Akasaya’ and having old collection date were examined. Five out of ten were assigned to the completely different varieties from five other ‘Akasaya’ accessions that had identical genotype (Table 4). There were variations on the flower color, the flowering time and the plant height between accessions in the different groups, indicating that genotype information for all of germplasm, would be useful to eliminate the chance of choosing duplicated accessions.
Table 4.
Information of genetic similarity and morphological variations among accessions with name of ‘Akasaya’. Phenotypic data were obtained in the experimental field
| JP No. | Origin | Flower color | Days to flowering | Plant height (cm) | 100 seed weight (g) | Seed coat color | Hilum color | Accession No. | Year of collection |
|---|---|---|---|---|---|---|---|---|---|
| 29265 | Osaka | White | 50 | 78 | 28.8 | Yellowish white | Light buff | 32738 | 1968/5/15 |
| 29346 | Nara | White | 51 | 72 | 29.4 | Yellowish white | Light buff | 32830 | 1972/4/12 |
| 29264 | Shiga | White | 52 | 68 | 26.9 | Yellowish white | Light buff | 32737 | 1968/5/15 |
| 28254 | Toyama | White | 53 | 66 | 33.5 | Yellowish white | Light buff | 31468 | 1969/4/18 |
| 28789 | Toyama | White | 54 | 73 | 35.0 | Yellowish white | Light buff | 32158 | 1972/4/12 |
|
| |||||||||
| 28751 | Nagano | Purple | 48 | 60 | 35.6 | Yellowish white | Dark brown | 32107 | 1972/4/12 |
| 28234 | Aomori | Light purple | 47 | 71 | 39.3 | Yellowish white | Dark brown | 31447 | 1969/4/18 |
| 29428 | Tottori | Purple | 67 | 98 | 35.1 | Yellowish white | Light buff | 32931 | 1968/5/15 |
| 28264 | Ishikawa | Light purple | 70 | 88 | 37.5 | Yellowish white | Dark brown | 31479 | 1969/6/10 |
| 29315 | Kyoto | Purple | 58 | 79 | 30.7 | Yellowish white | Dark brown | 32796 | 1969/6/10 |
On the other hand, 119 accessions were found to be same genotype but having different local names. One case was a breeding variety called by different local names and collected as landrace from different region. This is obvious because the same genotype as a crossbred rarely occur among landraces; four with same genotype to ‘Tamahomare’, five to ‘Enrei’, 10 to ‘Fukuyutaka’. In contrast, landraces and old varieties with different local name revealed sometime large agro-morphological variations in spite of having the same genotype; seed color, days to flowering etc. described individually below. Those variations, yet to be confirmed by replicated field evaluation, in the materials are notable for gene isolation and study for geographical adaptation and human selection of landraces.
Genetic relationships in soybean germplasm
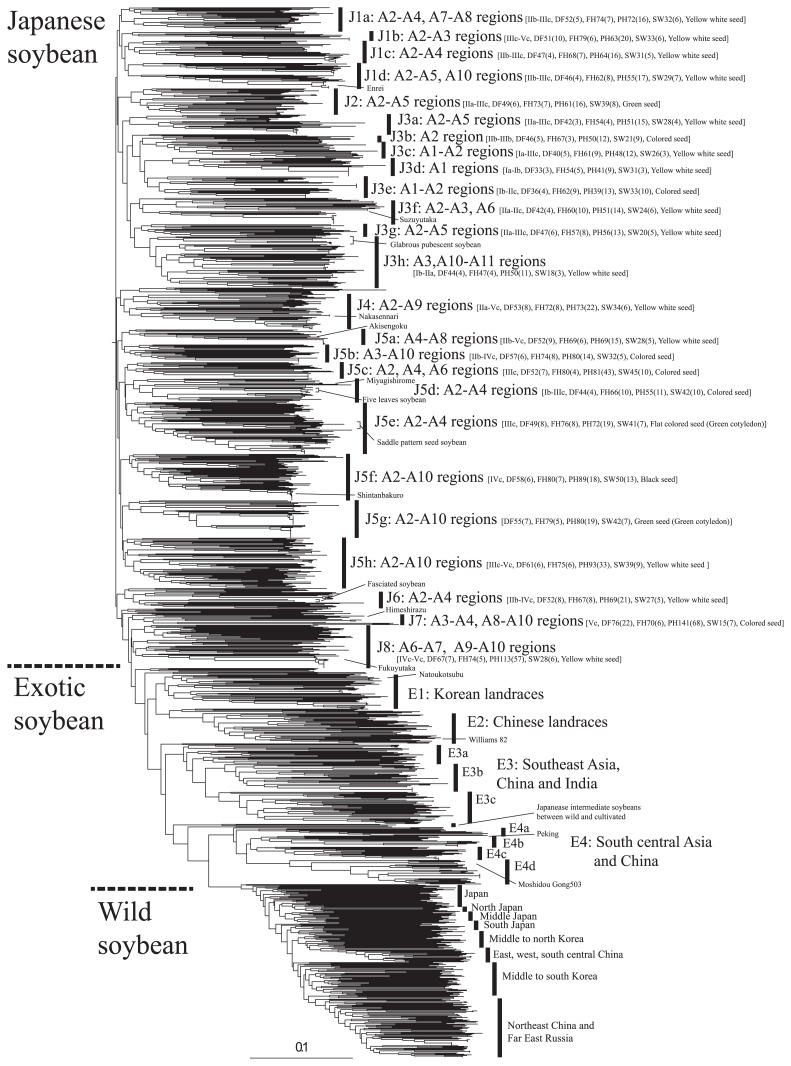
Clustering procedures were used to classify 1603 accessions into groups to examine genetic similarities based on 197 SNP makers. Clear genetic differentiation was observed between Japanese cultivated soybeans, exotic cultivated soybeans and wild soybeans (Fig. 6 and Supplemental Fig. 3 in detail). Exotic cultivated soybeans especially from South Asia and Myanmar were grouped into a distinct cluster (cluster E4) from soybeans from Southeast Asia (cluster E3). Cluster E4 was genetically closer to wild soybean than the other cultivated soybeans. Cluster E4 was found to be highly heterogeneous and comprised of accessions from Nepal, Pakistan, Myanmar and China. These accessions generally flowered very late in the experimental field and had primitive characters such as twining elongated stem and small seeds. Among Chinese accessions, one accession, ‘Peking’, has traditionally been used as a resistant material to soybean cyst nematode in the U.S. breeding program (Ross and Brim 1957) and has other useful characteristics such as resistance to germination under wet conditions (Muramatsu et al. 2008) which is an important traits to have in Japanese soybean cropping systems. Another accession, ‘Moshidou Gong 503’, has been widely used in genetic studies in Japan and recombinant inbred lines derived from a hybrid between Japanese cultivar ‘Misuzudaizu’ and ‘Moshidou Gong 503’ (Watanabe et al. 2004) are available from the National BioResource Project (http://www.legumebase.brc.miyazaki-u.ac.jp/strain/glycineRiLinesList.jsp). Chen and Nelson (2004) classified this accession into semi wild type since it has different phenotype and genotype from cultivated and wild soybeans. Therefore, this group (cluster E4) is expected to be useful for soybean genetic studies and soybean breeding programs. The genetic relationships observed in the present study support well the genetic characteristics of South Asian soybean previously reported (Abe et al. 2003, Xu et al. 2002). They considered the forage type of soybeans with primitive characters as wild soybean. Forage soybeans have a distinct nuclear SSR genotypes and region-specific chloroplast SSR haplotype predominantly in wild soybeans distributed in the Yellow River Valley. Interestingly, several Japanese landraces which had a high membership with wild soybean and intermediate phenotype between cultivated and wild soybean described above, were located outside the clusters of wild soybean and south Asian soybeans. In other words, those South Asian and Chinese soybeans are genotypically more similar to wild soybean than the hybrid progenies between wild and cultivated soybean.
Fig. 6.
A neighbor-joining phylogenetic tree of 1,603 accessions, Japanese (n = 998) and exotic cultivated soybeans (n = 341), Japanese (n = 74) and exotic wild soybeans (n = 190), based on the allele sharing distance. The letters in square bracket for Japanese clusters are phenotype information such as the representative ecological types followed by DF, days to flowering, FH, period from first flowering to start harvest, PH, plant height, SW, 100 seed weight, and seed coat color. Numeric characters after DF, FH, PH and SW and in a round bracket indicate that average and standard deviation among accessions in the cluster, respectively.
Chinese soybeans had the highest gene diversity (Table 1) and were generally either grouped with soybeans from South Asia and Myanmar (cluster E4) or soybeans from Southeast Asia (cluster E3) or grouped into a main cluster E2 unique to Chinese soybeans (Fig. 6). Remaining accessions were scattered widely into various clusters consisting of Japanese accessions. Although Chinese soybean germplasm was randomly chosen, this germplasm exhibits wide genetic and agro-morphologic trait variation. Chinese soybean production is generally divided into three primary regions; northern, Yellow River Valley and southern production regions; and further eight soybean eco-geographical types are classified as Northeastern spring type, Northern spring type, the Yellow River Valley spring and summer types, Yangtze River (Changjiang) spring type, Southern spring, summer and autumn types (Li et al. 2008). Based on the SSR analysis of 1,863 landraces from 29 provinces in China, they found seven genetically distinct clusters; four is generally corresponding to the eco-geographical types but the others had no clear relationships with their geographical origin. Among them, two clusters of landraces from southwestern part of China were reported to be genetically unique and have not been recognized based on the ecotype classification (Li et al. 2008). In the present study, several distinct clusters including Chinese accessions were identified. The cluster of south central Asia (cluster E4) consisted of four sub clusters; E4a, E4b, E4c and E4d. Among them, E4a, and E4d included Chinese accessions and well known accessions for each cluster were ‘Peking’ and ‘Moshidou Gong 503’. In cluster E4d, only Chinese accessions flowered earlier (ave. 46 days after sowing) than other landraces from south Asia (ave. 78 days). Cluster E3 consisted of soybeans from Southeast Asia and were divided into three sub-clusters; E3a, E3b and E3c. Chinese accessions were genetically close to accessions from India and Thailand (E3a) or Vietnam (E3b) rather than accessions from Indonesia and Taiwan (E3c). Most accessions belonging to the main cluster E2 showed very early flowering (ave. 36 days after sowing) and several of them were found to have come from northeast China. However, it was difficult to discuss the consistency of results here with those of Li et al. (2008) because of most Chinese accessions had unknown origin. Abe et al. (2003) found that Japanese soybeans were distinct from soybeans distributed in China whereas soybeans in Korea consisted of accessions similar to Japanese or Chinese germplasm. Similarly, Korean landraces used in the present study were grouped into a main cluster E1 unique to Korean accessions located between the Japanese and Chinese clusters (Fig. 6) and the remaining accessions were scattered into clusters of Japanese germplasm. The level of gene diversity in the Korean accessions was similar to that of Japanese accessions (Table 1). Cho et al. (2008) classified Korean landraces into three groups based on SSR markers and found that each group had different seed related traits and biochemical contents in the seed. Several Japanese small seeded breeding varieties used for ‘natto’ (fermented soybean) were grouped into the main cluster E1 of Korean landraces. Those genotypes were found to be similar with small seeded (Ave. 10g/100SW) Korean landraces. The number of Chinese and Korean accession in the present study was insufficient, further comparison of genotypes and their use by these countries will enable the range of variations to be compared.
Japanese soybean germplasm
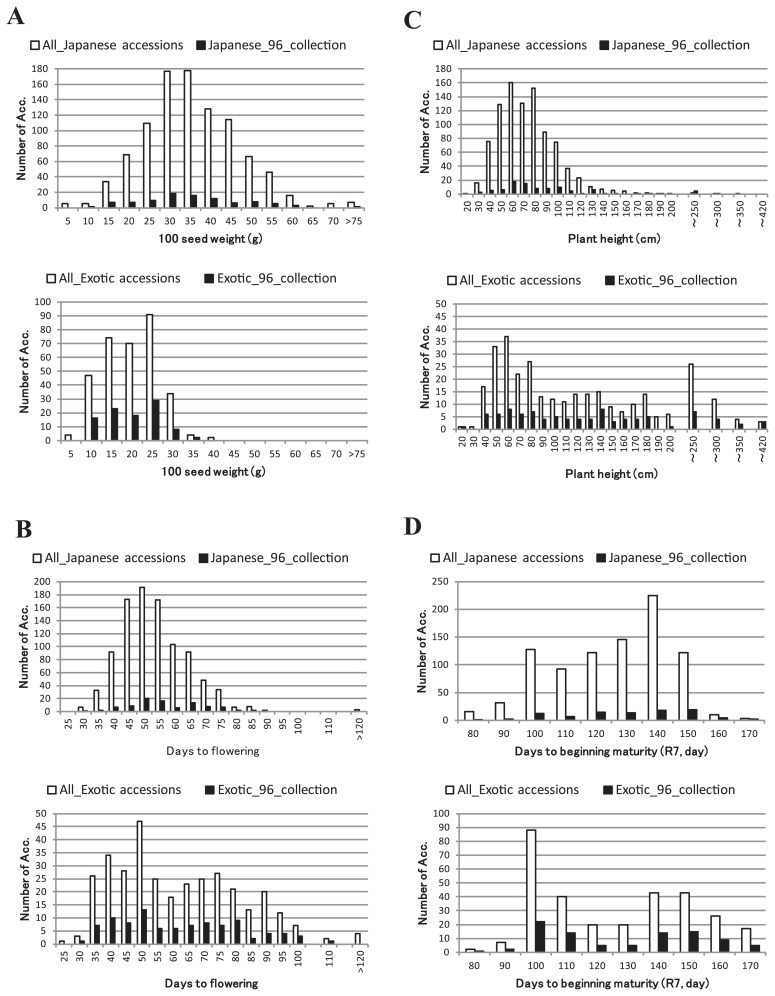
The variation of agro-morphologic characteristics of Japanese soybean germplasm was clearly different from that of exotic germplasm. Compared to exotic germplasm, the range of seed size in Japanese soybean germplasm was about two times greater whereas the range of days to flowering and plant height was less (Fig. 7). Japanese soybean germplasm revealed normal distribution in comparison with bimodal distribution in exotic germplasm for plant height, days to flowering and maturity. Accessions from different geographic region were separated to a certain extent into clusters corresponding to their agro-morphologic characteristics such as seed characteristics and photosensitivity rather than their geographical origin. Since each cluster was not so much homogeneous as a genetic admixed, i.e. branches between clusters were connected continuously. The name of clusters was tentatively assigned as J1 to J8. Lower case letters were appended to clusters and treated as sub-clusters when common agro-morphologic characteristics among accessions were identified in the cluster (Fig. 6 and Supplemental Fig. 3 in detail). In the dendrogram, the genetic relationships among breeding varieties were similar to that based on the 963 SNP markers (Supplemental Fig. 2) while there were some differences on the order of the nodes. Especially, the orders of branch connected at the higher level among the two dendrograms in some parts were inconsistent, suggesting that more SNP loci with variations peculiar to Japanese germplasm is required to divide germplasm into distinct groups. Regardless of the differences, the germplasm in the sub-clusters of which no recent variety was observed; J5b, J5c and J5g, J5h, J6 and J7, could be useful to broaden genetic diversity for breeding.
Fig. 7.
Comparison of phenotypic variation of selected 96 mini-core collection (black) with all accessions (white). A) 100 seed weight; B) days to flowering time; C) plant height; D) days to beginning of maturity (R7); Japanese (upper) and exotic (lower) germplasm comparison.
Cluster J1 covers central regions of Japan from ‘A2’ to ‘A5’ (Fig. 1) and the usages of the germplasm are mainly tofu (soybean curd) and miso (fermented soybean paste), and nimame (boiled soybean). The majority had yellow seed coat color and the Japanese maturity group ranged from ‘IIb’ to ‘IIIc’. Crossbred varieties developed by the public sector in Nagano prefecture located in central Japan were grouped into the cluster J1; ‘Enrei’, ‘Tachinagaha’, ‘Sachiyutaka’, ‘Ohsuzu’ ‘Hatayutaka’, Ootsuru’, ‘Ayakogane’, ‘Suzukogane’, ‘Tanrei’,‘Sayanami’and ‘Norin2’inthe sub-cluster J1d; ‘Misuzudaizu’ in cluster J1c; ‘Chuuteppou’ and ‘Shirorae’ in the sub-cluster J1a. Only the sub-cluster J1b included crossbred varieties, ‘Toyoshirome’ and ‘Nishimusume’, developed by the public sector in Kyushu, south Japan, as well as ‘Tamahomare’ and ‘Ginrei’ from central Japan. The genetic background of the crossbred varieties in the sub-cluster J1d and J1b was very similar to each other nevertheless they had been developed by cross breeding.
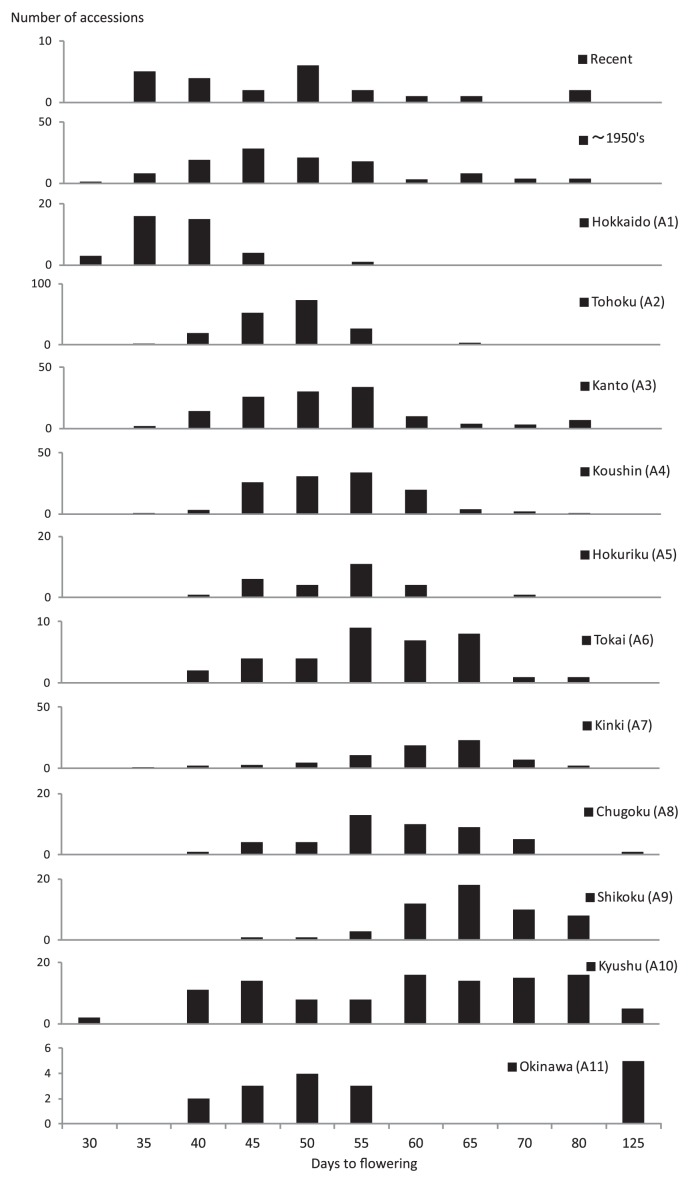
Japan is an archipelago separated from the Eurasian Continent and lie north (46°N, 146°E) and south (24°N, 122°E). Photoperiod and temperature varies greatly from north to south, therefore, the divergence of ecotypes adapted to each regions is expected. Fukui and Arai (1951) classified Japanese germplasm into 9 ecotypes based on days to flowering time and maturity. The system is commonly used in Japan. Hymowitz and Kaizuma (1979) reported that soybean maturity grouping system in North America does not correspond with that of Japanese accessions. The large scale evaluation of germplasm has been reported by Hirata et al. (1995, 1999). However, no marked association between ecotypes and genotype has been reported. In the present study, a single plant from approximately 1,300 soybean accessions were evaluated for several morphological and agronomic traits as well as SNP genotypes. Variation in flowering time of each accessions used here is summarized (Fig. 8) and the variation in each cluster is described (Fig. 6) together with their representative ecotypes. The recent bred varieties revealed biased distribution toward early flowering in spite of their higher gene diversity compared with landraces and old varieties (Table 1). It is likely that modern breeding has shifted to be target specific cultivation regions. Since day length and climate in Japan varies north to south, a geographical cline was observed for days to flowering time in the landraces across ‘A1–A10’ regions. Among them, the similar distribution of flowering time was observed among landraces across the regions of ‘A2’, ‘A3’, ‘A4’ and ‘A5’. This might explain the high genetic similarity among land-races from those regions in the cluster J1 (Fig. 6). After region ‘A1’, frost free period in regions ‘A2’, ‘A4’ and ‘A5’ and mountainous areas of ‘A3’ region is very short, high quality large seeded landraces which can grow throughout the frost free period to attain high yield have been preferentially cultivated rather than short summer season type (Fukui and Arai 1951).
Fig. 8.
Variation of flowering time among Japanese soybeans evaluated at the experimental field in A3 region.
Cluster J3 covers mainly north and central regions from ‘A1’ to ‘A3’ and includes accessions with a wide range of the usages. The cluster can be further sub-divided into eight clusters according to the region and their usage. Since Hokkaido, ‘A1’ region, is the northern limit of soybean cultivation in Japan, the locally adapted landraces and breed varieties of this region are expected to have distinct variation and adaptation to the specific environmental conditions of the region. Soybeans from ‘A1’ region were grouped mainly into three sub-clusters; J3c, J3d and J3e, and most of them were in Japanese maturity group from ‘Ia’ to ‘IIb’ (Fig. 6). Representative accessions of the sub-cluster J3c is ‘Tokachinagaha’ and landraces with narrow leaflet, which is known to be associated with a higher number of seed in pods (Bernard and Weiss 1973), were clustered together. The sub-cluster J3d mainly consisted of crossbreds, ‘Yukihomare’, ‘Tokachikuro’, ‘Toyomusume’, ‘Hayahikari’, ‘Kitamusume’, ‘Toyohomare’, ‘Toyokomachi’ and ‘Wasehadaka’ developed for tofu (soybean curd) and nimame (boiled bean) usages by the public sector in Hokkaido prefecture. In contrast, the sub-cluster J3e consisted of colored seeded landraces and one crossbred ‘Yuuzuru’. Their usages are nimame (boiled bean), edamame (green boiled pod) and kinako (soybean flour). The main difference from other regions was that there were distinct variations in seed color and size despite their similar genetic background and lower photosensitivity reaction. In this region many local landraces with different local names have the same genetic background. Mutations may explain this recent variation because of seed color variation in the same genetic background; a landrace with brown saddle seed (JP28162) in yellow seed color landraces in the sub-cluster J3d and other landraces with black (JP29619), black saddle (‘Sakamotowase’) and green saddle seed (JP27462 and JP27454) in yellow green seeded landraces in the sub-cluster J3e.
There were two distinct groups in southern regions ‘A10’ and ‘A11’ based on the variation of flowering time (Fig. 8). The landraces with early flowering and maturity from ‘A10’ and ‘A11’ regions were grouped with each other in the sub-cluster J3h and had distinct genotypes to the other clusters, J3a-g, that mainly consisted of early flowering soybeans from the northern region (Fig. 6). One exception is that the earliest landrace, ‘Wase-Kuro-daizu’ in region ‘A10’ (Fig. 8) was grouped with landraces from the northern region, ‘A1’, in the sub-cluster J3e. This accession seems to have been introduced as a cover crop from region ‘A1’ (Ministry of Agriculture and Forestry, Japan, 1952b). The majority of sub-cluster J3h consisted of old varieties known as precocious summer type of soybean, Japanese maturity group ‘IIa’, in region ‘A10’ and landraces from the Ryukyu Islands, ‘A11’. Other summer season type from region ‘A3’, included a few Korean and Taiwanese landraces. Fukui and Arai (1951) discuss the relationships of ecotypes with frost free period, crop rotation system and escape from pest damage. In region ‘A3’, summer season type soybean has been preferentially cultivated either as a crop preceding buckwheat and wheat or because of good performance on soil of the region that lacks available phosphate and ground water. In contrast, summer season type had been widely cultivated in ‘A10’ region in order to avoid pod damage by soybean pod gall midge (Asphondylia sp.). The majority of landraces with glabrous leaves were found in this cluster. Although summer season type soybeans in ‘A10’ region are known to be genetically differentiated from autumn season type soybean at many isozyme loci (Hirata et al. 1995), origin of their multi-locus genotype has been unclear. Subsequently, Hirata et al. (1999) suggests that summer season type soybeans are possibly derived from the ‘A2’ region where various multi-locus genotypes consisted of alleles both from summer and autumn season type soybeans based on isozyme variation of a larger number of accessions. Hymowitz and Kaizuma (1979) suggested that the summer season type soybeans in Kyushu region, ‘A10’, originated either in the region or from Korea based on the specific seed protein alleles in the region. The summer season type soybeans in Kyushu region are also known to have the specific cytoplasmic genome type which evolved from the common type of soybean (Shimamoto 2001). A few Taiwanese landraces and land-races from the Ryukyu Islands of Japan clustered together with the precocious summer type of soybean in ‘A10’ region in sub-cluster J3h. In the Ryukyu Islands, the soybean varietal names “Anda” and “Higu” mean for oil and seed coat, respectively, and the soybeans seem to be traditionally recognized as having a different purpose from others in region ‘A10’. The close genetic relationships among very early flowering summer type and landrace from the Ryukyu Islands suggest that some of the very early flowering summer types in region ‘A10’ have been introduced through Taiwan and the Ryukyu Islands from China. It is necessary to investigate how the previously reported specific cytoplasmic genome type is distributed in the cluster and where the genes responsible for early flowering and maturing originated.
In sub-clusters J3a, J3b and J3g, no recent variety was observed and maturity group of the landraces ranged from ‘IIa’ to ‘IIIc’. Sub-cluster J3a consisted of landraces with white flowers and several Chinese landraces. Many of them are reported to have very high protein content (Watanabe and Nagasawa 1990). Sub-cluster J3b consisted of landraces with small green cotyledon and are used for Kinako (soy flour), Nattou (fermented beans) and fodder. The genotype of ‘Wase Kurosengoku’ was clearly different from ‘Kuro sengoku’, described above, in spite of the similar name. In contrast, the sub-cluster J3f consisted of the crossbred varieties with maturity group ‘IIa’ released from the breeding sector in Tohoku region for Tofu usage; ‘Ryuuhou’, ‘Suzuyutaka’, ‘Suzukari’, ‘Wasesuzunari’, ‘Okushirome’, ‘Tachiutaka’ and ‘Fukushirome’. Those varieties had some lineage of ‘Gedenshirazu’, a source of nematode resistance.
The origin of the landraces in cluster J4 and sub-clusters J5a and J5h was not as specific as other clusters and included a very wide range of germplasm. The maturity group in these clusters varied considerably from ‘IIa’ to ‘Vc’, suggesting that selection for maturity has not been as strong as other clusters. Soybeans in these clusters are used for Tofu, edamame (boiled green pods) and nimame (boiled beans). In cluster J4, a landrace ‘Hakkou’, a crossbred ‘Nakasenari’ and its parental landrace ’Houjaku’, and a green seeded landrace ‘Aonyuudo’ were clustered together. A crossbred ‘Akisengoku’ and its parental landrace ‘Akasaya’ were grouped in the sub-cluster J5a.
Three fasciated accessions were found in cluster J6 and two of them had identical genotype with the material used by Onda et al. (2011). Fasciation in soybean is known to be controlled by a recessive gene, f, (Albertsen et al. 1983) and recently the locus has been characterized by molecular markers in detail (Onda et al. 2011). In contrast, one accession ‘JP29177’ showed different genotype and phenotype from those accessions in the cluster J6; early flowering and maturity, purple flower, tall plant, many branches.
Three Japanese accessions, PI171451, PI229358 and PI227687, in the USDA Soybean Germplasm are known to have antixenosis and antibiosis resistances to numerous insect pests (Boerma and Walker 2005). These accessions are ‘Soden daizu’, ‘Kosamame’ and ‘Miyako White’ and were collected in Kanagawa prefecture, unknown and Okinawa prefecture, respectively. In the present study, ‘Soden daizu’ and ‘Kosamame’ in cluster J7 differed only at 1.6% of SNP loci. Komatsu et al. (2006) compared a resistance QTL allele to common cutworm in a fodder breeding variety ‘Himeshirazu’ with that in PI171451 and suggest these alleles have different resistance effects. Interestingly, these germplasm and several accessions with the name ‘Gedaizu’ from ‘A11’ region were genetically similar each other. The other germplasm clustered with these accessions are of interest to examine the level of insect resistances.
Genetically Japanese soybean accessions in cluster J8 are closest to exotic germplasm and mainly consisted of yellow seeded landraces from southern regions and breeding varieties, ‘Fukuyutaka’, ‘Akiyoshi’, ‘Hougyoku’, ‘Hyuuga’, ‘Murayutaka’ and ‘Erusuta’, released from Kyushu region for Tofu. The maturity group ranged from ‘IVc’ to ‘Vc’.
The geographical and historical isolation from the Asian continent has allowed distinctive and diverse soybean cuisine. This unique cuisine has probably shaped Japanese soybean diversity and structure. Japanese landraces with distinguishing seed characteristics, e.g. colored seed coat, seed size and shape, tended to be grouped in separate clusters; J2, J3e, J5b to J5g, from clusters of yellow seeded landraces (Fig. 6). Since those Japanese landraces have been cultivated in mountainous areas, on ridges between rice fields and home gardens, landraces with late flowering and maturity are not a problem within Japanese cropping system. In addition, landraces in these clusters revealed large variation of flowering time and maturity or adapted regions. These facts suggest that the population structures of colored seeded land-races have been strongly influenced by human mediated seed dispersal rather than their maturity.
Distribution of similar genotypes across regions may be related to diversity of local foods and the varieties for food processing. The accessions from north Japan, especially Tohoku region, have a higher level of diversity than soybean landraces from other regions (Table 1). Watanabe and Nagasawa (1990) pointed out that the Japanese soybean germplasm collection in the NIAS genebank was biased towards regions A2–A4 and A10 and did not reflect the proportion of regional landraces, but rather reflected the number of accessions deposited by the regional breeding institutions. Even so, plenty of landraces with different special local names means farmers consciously discriminate characteristics of landraces, that seems to be important for genetic diversification, and this might explain the higher level of genetic diversity in these areas.
The uses of landraces in cluster J2 are various, edamame (boiled green pod), nimame (boiled beans), miso (fermented soybean paste) and tofu (soybean curd). The crossbreds in the cluster were ‘Akishirome’ and ‘Kiyomidori’ released by the public sector in south Japan and ‘Tamamasari’ by that in central Japan. Similarly, the sub-cluster J5d included land-races mainly used for Edamame production but some for nimame and miso. Crossbred in the cluster was only ‘Iwaikuro’anditsparentallandrace‘Banseihikarikuro’ from Hokkaido had similar genotype. Another variety ‘Miyagishirome’ was developed by pure line selection in Miyagi prefecture, northern Japan. In the sub-cluster J5f, black and large seeded landraces from various geographical regions used for nimame were grouped with breeding varieties, ‘Shintanbakuro’, ‘Hyoukei kuro3’ and ‘Kurodamaru’.
Vegetable soybean, edamame, is a popular snack food (Lumpkin et al. 1993) and various varieties have been released mainly from private companies in Japan. Mimura et al. (2007) evaluated 130 varieties for edamame, of which the majority were obtained from Japanese seed companies. It was found that the genetic diversity among Japanese vegetable soybean was lower than that of Chinese vegetable soybean landraces ‘maodou’ whereas the nine distinct clusters different from ‘maodou’ were identified among Japanese edamame varieties based on SSR markers. However, little is known about genetic variation of local Japanese vegetable soybean landraces. Lumpkin and Konovsky (1991) questioned how vegetable soybean varieties differ from grain soybean. Actually, the boundary of soybean variety for cooking between boiled bean and edamame is obscure. Thus, the identification of edamame in local landraces in the present study was based on the local name and its usage described in passport records and old literature. Most of land-races used for edamame fall into nine clusters, J2, J3e and J5b to J5g. These clusters consisted mainly of soybeans with colored and large seed (ranged from 32 g/100 SW to 50 g). Among them, sub-clusters J5c and J5d included many middle maturity landraces with the name including ‘Kaori’ which means good fragrance and ‘Dadacha’ which is famous local name of edamame. Selection for harvest time of vegetable soybean shifted from October to August in order to provide these beans during the Obon Festival in the middle of August. This shift in selection is supposed to have occurred by the middle of Edo period (Lumpkin and Konovsky 1991). In the sub-cluster J5d, days to harvest after flowering in more than half of landraces were approximately 20 days shorter than the others, while the genotype and flowering time were similar. Although landraces with five leaves were scattered to the cluster J2, J3e, J3f and this cluster, interestingly all of black seeded landraces had five leaves are in this cluster, J5d.
The relationship between nine clusters among 130 Japanese edamame breeding varieties and their maturity or seed coat color were discussed by Mimura et al. (2007). They indicated that popular landraces of ‘Tanbakuro’ and ‘Dadachamame’ were grouped into a single cluster. The use of ‘Tanbakuro’ is not only for boiled bean (nimame) but for edamame. In the present study, all of landraces with the name of ‘Tanbakuro’ were grouped into sub-cluster J5f which is different from two sub-clusters, J5c and J5d, including landraces with the name of ‘Dadacha’. In order to use edamame genetic diversity efficiently, further studies to compare the genetic similarity of edamame breeding varieties released from private companies with landraces in the nine clusters is required. The characteristic for sub-cluster J5f soybean was their heavy seed weight and black seed color with wax. The origin was mostly from southern regions and use for boiled soybean. Among them, only landraces having a highly similar genotype with two breeding varieties, ‘Shintanbakuro’ and ‘HyoukeiKuro3’, had heavy (Ave. 70 g/100SW) seed weight as compare with others (Ave. 45 g/100SW). These varieties were developed by pure line selection from landraces with the name of ‘Tanbakuro’ in ‘Tanba’ area where landraces of varied seed shape, seed size and maturity had been cultivated (Yamashita 2003). In ‘Tanba’ area, these landraces are assumed to have been consciously discriminated from other type of soybeans and selected to increase seed size by the local farmers.
The sub-clusters, J5e and J5g, consisted of mainly landraces with the name of ‘Hitashimame’ and green cotyledon from ‘A2’ to ‘A4’ regions. ‘Hitashimame’ is a lightly boiled soybean pickled with salt or vinegar, and two types, green and green with black saddle seed, have been traditionally cultivated in the southern part of ‘A2’, and mountainous area of ‘A3’ and ‘A4’ regions (Maruyama et al. 1987). In the present study, there was a slight genetic difference between the two types whereas flat and round seed types among landraces with the name of ‘Hitashimame’ were grouped into the sub-clusters J5e and J5g, respectively, suggesting that those types are genetically differentiated to some extent. A breeding variety ‘Shinano Kurakake’ in the cluster J5e was developed by pure line selection from one of the black saddle seeded landrace (Maruyama et al. 1987). They also described that another type of miscellaneous landraces with green seed for Kinako have been cultivated in these areas. ‘Aobata’, ‘Sokoshin’ and ‘Kinakomame’ are representative landraces used for ‘Kinako’. They were grouped in cluster J6 while landraces with the same or similar name also appeared in sub-clusters, J5b, J5e and J5g. This supports the view that these landraces have a wide diversity. The protein content in ‘Norimame’ is reported to be very high among large seeded landraces (Watanabe and Nagasawa 1990). The name of ‘Aobata’ comes from the fact that the plant drops its green leaves (Maruyama et al. 1987) and interestingly their cotyledon color was green, which is the frequent color in these clusters. Studies of responsible genes for the large increase in seed size, the flat shape of seed and fragrance in the colored soybeans may be important to understand the relationships between local selection and soybean cuisine in Japan. Although 160 scientific reports related to vegetable soybean were published in the 70 years from 1921 to 1991, the accessibility is restricted to researchers because of limited bibliography and publication in Japanese language (Lumpkin and Konovsky 1991). Further effort to review previous reports on other kinds of soybean and their usage is also required to explain other clusters.
Japanese wild soybean germplasm
All 264 wild soybean accessions were grouped into a single cluster distinct from cultivated soybeans (Fig. 6). With respect to the genetic relationship with cultivated soybean, the wild soybean accessions from Japan was closest to cultivated Japanese soybeans followed by Korean, Chinese and Far East Russian wild soybeans. The population structure analysis described above suggested that approximately 10% of Japanese wild soybeans are presumed to have cultivated soybean alleles (Table 1). These accessions on the phylogenetic tree were close to cultivated soybean cluster and did not reflect geographic origin. The remainder exhibited genetic relatedness to geographic origin in Japan (Fig. 6). A weak geographical cline exists from northern, central to southern part of Japan with several exceptions. Southern Japanese accessions were similar to accessions from north and central parts of the Korean peninsula and north, east and south central regions of China. Two other clusters consisted of (a) accessions from central to southern parts of the Korean peninsula and (b) accessions from northeast China to Far East Russia. The results were generally consistent with the results of structure analysis based on SSR variations of wild soybean populations covering the whole range of distribution (Guo et al. 2010). The difference is that continental coastal populations may constitute a distinct group; accessions from central and southern Korean peninsula, Japanese accessions collected near the Korean Peninsula and accessions from Taiwan were genetically similar and formed a distinct cluster. Unfortunately, no wild soybean accessions from east or southern China was included in the present study, while Guo et al. (2010) reported that wild soybean accession from south China are genetically similar to Korean and southern Japanese accessions. The two accessions from Taiwan, PI245331 and PI518279, were formally classified as G. formosana Hosok. Thseng et al. (2000) suggested that accessions from north Taiwan was taxonomically distinct from G. soja based on various characteristics. The two Taiwanese accessions actually revealed unusual morphological variations with narrow leaves and small sized seed compared with G. soja variation. However, genetically these two accessions were included in the variation of G. soja in the present study.
Gene diversity from the Japanese wild soybean population (0.31) was highest, followed by the Korean population (0.27), Chinese (0.23) and Far East Russian (0.22) populations (Table 1). Cultivated alleles by introgression from soybean may have increased the diversity in Japanese wild soybean population as described above. Nevertheless, the geographical distribution range of wild soybean accessions did not reflect their gene diversity; genetic variation in Chinese accessions was not so high in spite of its wider range, while Korean accessions had similar genetic variation to Japanese accessions of its geographic range is narrow. The result is consistent with previous research in which high genetic diversity in the Korean wild soybean population based on SSR markers (Lee et al. 2008). Also, they found that Korean accessions had lower number of null alleles for SSR markers than those of other countries. Here the average number of null alleles on SNP marker was also lowest in Korean accessions (0.76, n = 81) as compared with that of Japanese (2.47, n = 74), Chinese (2.77, n = 72) and Far East Russian (2.1, n = 37) accessions. It is interesting that the Korean wild soybean accessions have a high similarity to cultivate soybean in respect to lack of null alleles.
A genetic bottleneck has occurred in cultivated soybeans during domestication from wild soybeans therefore wild soybeans are an important source of novel alleles to broaden the genetic base of cultivated soybean (Guo et al. 2010, Hyten et al. 2006, Lam et al. 2010, Stupar 2010). In China, more than 6,000 accessions have already been collected from throughout their range (Dong et al. 2001). In the present study, a weak geographical cline was observed among spatially selected Japanese wild soybean accessions whereas many accessions from the same prefecture had different genotypes. Japanese wild soybean populations that are more than 10 km apart are likely to genetically different (Kuroda et al. 2008). Kim et al. (2010) estimated the divergence of a Korean wild soybean from soybean, based on comparison of the whole genome sequence with the cultivated soybean reference sequence, occurred 270,000 years ago predating archaeological evidence of soybean in China. Even though the wild soybean population used was not the direct ancestor, their study indicates a large genetic variation exists among wild soybean populations. Although 741 and 1159 Japanese wild soybean accessions are conserved in NIAS genebank and in Legume base of National BioResource Project (NBRP), respectively, the present data suggests that many more populations are required to fully understand the genetic variation in wild soybean. In NIAS genebank, there remain gaps in the collection particularly Sanin, Shikoku, Hokuriku and Sanriku regions. Therefore, further countrywide collection efforts for wild soybeans will be required to provide an accurate picture of genetic variation in Japanese wild soybean that should be reflected in the core collection.
Exotic germplasm and implication of future characterization
Incorporation of exotic germplasm into breeding programs is laborious because of the differences in their agronomic traits from domestic germplasm. Therefore, information on genetic similarities among germplasm and evidence of new genes in germplasm are helpful to promote their use. Regarding the clustering pattern, germplasm from South Asia and Myanmar in cluster E4 was most distantly related to Japanese germplasm and are expect to have unique useful alleles. Likewise many important genes have been identified from one of accession ‘Peking’ in the cluster E4a (Fig. 6). For example, protein content of accessions from Myanmar in sub-cluster E4b is very high according to the data in Genebank. The germplasm from Southeast Asia was most genetically different from Japanese germplasm but had more wild soybean like characteristics. Soybeans in Southeast and South Asia are thought to have been repeatedly introduced from the China based on SSR markers (Abe et al. 2003). Since soybeans have spread to Southeast and South Asian countries via land and sea trade routes (Shurtleff and Aoyagi 2010a, 2010b), the observation of genetic similarities between accessions collected from Southeast (excluding Myanmar) and South Asia (including Myanmar) with several Chinese accessions was expected. However, these accessions revealed some genetic and morphologic distinctiveness from Chinese accessions as found in Japanese germplasm and may have accumulated variations required due to adaptation to southern latitudes.
Vietnam: Vietnam is 1650 km from south (8°30′N) to north (23°22′N) and landraces adapted to such a geographic range are expected to have a large genetic variation. Among the 72 accessions preserved in NIAS Genebank from Vietnam, 13 accessions (Kobayashi et al. 1994) analyzed in the present study were grouped into single sub-cluster E3b. Accessions in sub-cluster E3b were genetically slightly different according to their geographical distribution; 1) northeast (representative accession JP78839), 2) south central highland (JP105774) and 3) south central coast of Vietnam (JP78857). Landraces from northeast Vietnam showed early flowering (50–61 days), short stature (60–100 cm) and small seed (9–17 g/100SW). The small seeded landraces can be subdivided into two groups corresponding to their seed color; greenish yellow and yellow. Genotypes with greenish yellow seed color were different from other green seed landraces from the northern border area of China as described below. In contrast, there is differentiation between accessions from southern Vietnam between coastal and highland regions. Landraces from the central highland region revealed relatively later flowering (72–93 days), tall stature (143–270 cm) and small seed (17–20 g/100SW) and were genetically similar to landrace (‘JP226666’) from Laos. Coastal landraces had early flowering (54–61 days), short stem (78–127 cm) and large seed (25–27 g/100SW) and had no genetic variation and had similar genotypes to landraces from central Thailand. Two accessions from south highland region and two accessions collected from the northern border area of China were included in different sub-clusters, E3c and E4b. The two accessions from the south highland region were genetically, as well as phenotypically, similar to landraces from northern Thailand in the sub-cluster E3c. In contrast, a small green seeded landrace, JP207933, collected from a minority ethnic group, H’Mong, Sapa district, Lao Cai province (Shimada et al. 2001) and a yellow seeded landrace, JP105759, from Bạch Thông district were grouped with landraces from several different countries in the cluster E4b.
Myanmar: Myanmar is bounded by China, India, Bangladesh, Laos and Thailand, therefore, a large genetic variation among soybean landrace is expected. Among 43 accessions preserved in the NIAS Genebank, 21 were characterized by SNP markers. Twenty-one accessions collected from central and northern Myanmar were genetically divided into two major clusters regardless of their geographic origin (Takahashi et al. 2002, Tomooka et al. 2003, Watanabe et al. 2007). One sub-cluster E3a was grouped together with Chinese landraces, the other sub-cluster E4b included Nepal, Vietnam, Thailand and Laos accessions. This indicates that soybean germplasm of Myanmar is associated with that of surrounding countries. There was no relationship between genetic differences and their cropping season, cropping system, usage or local names described in the collection reports. Many morpho-agronomic trait differences among the two major groups of Myanmar were observed. Accessions in the sub-cluster E3a had early flowering (47–66 days), short stature (69–128 cm) and middle size seed (11–22 g/100SW) whereas characteristics of soybean in sub-cluster E4b were late flowering (82–122 days), long stature (120–205 cm) and small seeds (8–11 g/100SW). Among accessions in the sub-cluster E4b, the flowering related gene in three accessions, JP217434, JP217458 and JP217507, is of interest because these flowered almost one month later than other genetically similar accessions from Myanmar. As a consequence it was difficult to obtain seeds from the experimental field where they were grown in Japan.
Thailand: Thirteen accessions from Thailand among 40 accessions preserved in the NIAS Genebank were grouped into single cluster E3 and further divided into three sub-clusters. Accessions mainly collected from the northern part of Thailand and registered in the NIAS Genebank around 1968, accessions from northeast Thailand and from central part of Thailand (Sasaki and Shigemori 1986) were grouped into sub-cluster E3c, E3a and E3b, respectively. All accessions have yellow seeds but agro-morphological differences, for example photosensitivity, plant height and seed size, were observed between soybeans in these sub-clusters. Northern accessions in sub-cluster E3c were later flowering (72–97 days), taller (150–370 cm), and had smaller seeds (10–15 g/100 SW). Northeast accessions in sub-cluster E3a had early flowering (60–68 days), short stature (100–170 cm) and large seeds (20–25/100 SW). Genotypes and phenotype of accessions from the central part in sub-cluster E3b were almost the same as accessions from the south central coast of Vietnam (Kobayashi et al. 1994). Chotiyarnwong et al. (2007) reported 149 landrace and 11 improved varieties in Thailand were divided into two major groups and a further 14 minor groups based on 18 SSR markers. Common landraces were limited in the present study, ‘Mae-Jo’ belong to group ‘2e’, ‘Chiangmai Palmetto’ to ‘2b’,’Mae-Rim’ to ‘2c’, ‘Sansai’ to ‘1c’ (Chotiyarnwong et al. 2007). Although results of SNP and SSR markers cannot simply be compared, the high similarity between ’Mae-Rim’ and ‘Sansai’ in the sub-cluster E3c is questionable. Based on the collection record of Sasaki and Shigemori (1986), ‘JP38386’ is suppose to be an improved variety ‘SJ 4’ or ‘SJ 5’ of which one parent is an accession from Taiwan and its similarity with JP30146 from Taiwan in the cluster E3a is understandable. However, the genotype of ‘Mae-Jo’ was the same as accession JP30146 from Taiwan is questionable. Genotype confirmation of JP30187 and JP30179 that were registered with the NIAS Genebank around 1968 should be compared with the original accessions preserved in Thailand.
Indonesia: 21 accessions were analyzed in the present study. The first record of soybean in Indonesia has been found to be around the 13th century and soybean was introduced by traders or merchants from southern China based on chronology of soybean in Southeast Asia (Shurtleff and Aoyagi 2010a). Most of the materials used in this study were registered in the NIAS Genebank around 1973 as local landraces. The twenty-one accessions from Indonesia were grouped into single sub-cluster E3c and were similar to accessions from Taiwan, East Timor, India, Vietnam and Thailand. Banba and Takahashi (1985) reported that most of cultivated soybeans in Java were old varieties which introduced from Taiwan or the Philippines. In contrast, local cultivars ‘Presi’ and ‘Petek’, which were popular until 1980 (Soemarno 1995), and high-yielding soybean varieties ‘Merapi’ and ‘Ringgit’ which were developed by pure line selection from Chinese germplasm and released in the 1930’s (Aman et al. 1988) were grouped with the landraces collected from Java suggests that all of these accessions originated from a similar gene pool.
East Timor: East Timor is located in the eastern end of Lesser Sunda Islands and is surrounded by Indonesia. Four accessions collected in 2005 (Tomooka et al. 2006) were analyzed. Although the soybean germplasm were expected to be genetically similar to germplasm from Indonesia, these four accessions were divided into three clusters. JP226605 and JP226631 had early flowering (74–77 days) and belong to the sub-cluster E3c including Indonesian accessions. In contrast, JP226632 was grouped into the sub-cluster E4b and showed very late flowering (98 days), long stem (350 cm) and was black seeded. Interestingly, the seed size, flowering time and plant height of JP226606 was similar with that of JP226605 and JP226631 but was genetically very similar with the southern Japanese landraces in the cluster J8.
India: Soybean cultivation in south Asia is referred to in the English literature as starting in 1798 (India), 1819 (Nepal) and 1868 (Pakistan) but there appear to be much earlier records in native literature (Shurtleff and Aoyagi 2010b). According to these records, Indian soybeans were supposed to have been introduced through the Himalayan mountain range from China, through Myanmar and from Indonesia by traders. Most of Indian accessions preserved in NIAS Genebank (174 accessions in total) were introduced from Hariyana University, India, in 1978 (Koyama et al. 1983) but unfortunately no passport data for those accessions is available. Thirty-three accessions evaluated were mainly grouped into four sub-clusters; E3a, E3c, E4c and E4d.
Pakistan: Twenty three accessions were introduced by Nakagawara et al. (1990) and Egawa et al. (1992) and are preserved in NIAS Genebank. Eleven accessions from Pakistan were grouped into a single sub-cluster E4d. Soybeans in the northern areas of Pakistan are cultivated as a fodder crop (Nakagawara et al. 1990). In experimental fields, those accessions were genetically similar to 7 accessions from West Nepal in sub-cluster E4d and revealed brachytic stem with many braches. The evolution of brachytic stem phenotype in relation to selection by animal feeding is of interest.
Nepal: Many accessions (336) were introduced between 1984 and 1988 to Japan and are conserved in the NIAS genebank [IBPGR Multicrop collection mission conducted by U. Nare, M. Izuka, A. Ujihara and M. Nakagawara in 1984 and Miyazaki and Adachi (1988)]. Most accessions were grouped into cluster E4 and further subdivided into sub-clusters, E4c or E4d. The remaining two accessions from Nepal, JP40374 and JP100202 (Miyazaki and Adachi 1988) were grouped with Japanese landraces in the cluster J8. According to the collection report (Miyazaki and Adachi 1988), there were variations in growth habit, determinate and indeterminate, and time of maturity, which is different from other Nepal accessions. Therefore, these accessions may be recently introduced from East Asian countries.
Preservation and passport data of soybeans from Laos, Malaysia, Philippines and Cambodia is very limited in the NIAS Genebank. There still needs to be further collection of soybeans from these countries. Nelson (2011) considers that addition of accessions to ex situ collections is unnecessary except for some remote areas where cultivation of newly developed varieties has not yet spread. Therefore, more intensive collection from such areas has a great importance in the conservation of soybean genetic resources.
Development of mini-core collection
Sampling to develop a mini-core core collection followed several steps. Initially, a minimum of 12 representative Japanese accessions were provisionally selected without phenotype information using the PowerCore program. This program used advanced M strategy that maximizes genetic and phenotypic variation using a modified heuristic algorithm to find the optimum path for establishing core sets (Kim et al. 2007). Several accessions with heterozygous loci were removed because those accessions having many more alleles than homozygotes and were preferentially selected by the program. The average gene diversity of the minimum representative Japanese accessions was 0.41 that is almost equivalent to the gene diversity of all Japanese accessions (0.38). These consisted of four Japanese representative varieties, ‘Enrei’, ‘Fukuyutaka’, ‘Tokachinagaha’ and ‘Nattoukotsubu’, and eight local or old varieties that complement this diversity; ‘Miyagishirome’, ‘COL/Tanba/1989/Odagaki/2’, ‘Kurohira’, ‘Kanagawa wase’, ‘Himeshirazu’, ‘Chizuka Ibaraki 1’, ‘Onihadaka’ and ‘Tsurusengoku’. Although several of the varieties were assigned to different germplasm clusters from Japanese germplasm described above, these are a second target for re-sequencing in order to develop Japanese soybean SNP panel.
Two mini-core collections each consisting of 96 accessions from (a) Japanese and (b) exotic germplasm were selected. These were selected to retain 100% of the gene diversity based on the SNP variation considering morpho-agronomic trait variation, population structure and geographic origin in all cultivated soybean accessions. These 192 accessions are approximately 3% of the entire landrace germplasm conserved in the NIAS Genebank. Based on the results of clustering procedures described above, representative accessions having a local name from Japanese clustered branches were preferentially included in the analysis. Simultaneously, the accessions without Japanese breeder’s right were selected. Other accessions that complement for genetic and agro-morphologic diversity of phenotypic classes, described in the material and method section, were further selected and added with the aid of software. Some accessions that seem to be recently introduced from overseas countries were analyzed with exotic germplasm. The country-wise gene diversity and the proportion of accessions across countries were considered to select an exotic mini-core collection.
The resultant mini-core collections of 96 Japanese and 96 exotic accessions should represent a major proportion of gene diversity (0.40 and 0.43, Supplemental Fig. 4 in detail) observed among Japanese and exotic cultivated soybean germplasm analyzed here. In order to confirm homogeneity, statistics for phenotypic trait coverage in the selected Japanese mini-core collection were compared with the entire Japanese cultivated soybeans following Hu et al. (2000). The mean difference (%) was 7.8% and lower than the significance critical value of 20%. The coincidence rate of range (%) was 92%, indicating that the range of variation in the mini-core collection reflects well the variation range of all accessions. Variable rate of coefficient of variance was 123% and higher than the required value, which is more than 100% to represent the variance of all accessions. The graphical distribution of 100 seed weight, days to flowering time, plant height, days to beginning of maturity (R7) of the two selected 96 mini-core collections were found to represent well variations of all Japanese and exotic accessions (Fig. 7).
Screening germplasm on a large scale is difficult for complex quantitative traits which are a main target for breeders. Development of a core collection will enable repeated evaluations; however, even a core collection has limitations to practical application. Therefore, smaller germplasm subsets, called mini-core collections, for Japanese and exotic land-races were developed here. These subsets enable examination of many economically important characteristics. Further effort, for example characterization of maturity related genes in these collections, will be required to achieve a breakthrough in the use of diverse exotic germplasm in Japanese soybean breeding programs.
Seed multiplication of these accessions (Fig. 9) has started from the single genotyped plant. Distribution of the ‘Japanese soybean mini-core collection ver. 1’ and ‘Exotic mini-core collection ver. 1’ is in preparation and will be available soon from NIAS Genebank (http://www.gene.affrc.go.jp/distribution_en.php?section=plant). However, several accessions in ‘Exotic mini-core collection ver. 1’ introduced to Japan after the Convention on Biological Diversity (CBD) are regulated with respect to distribution. Since the mini-core collections have been selected only based on information of a limited number of SNP markers and agro-morphological traits, these collections will be revised when more genomic and morpho-agronomic information becomes available in the future. Both mini-core collections developed in this study will assist in finding novel traits for crop improvement and provide an effective platform for enhancing soybean diversity studies.
Fig. 9.
Morphological variation in the seeds among ‘Japanese soybean mini-core collection ver. 1’ (left) and ‘Exotic mini-core collection ver. 1’ (right). The picture order reflects the genetic relatedness.
Conclusions
To enhance the use of unexploited soybean germplasm in the NIAS Genebank for breeding and diversity research, mini-core collections of Japanese and exotic soybeans have been identified in the present study. Initially, polymorphisms of 963 SNP markers mainly distributed at a density of 1 marker per 1 Mb were evaluated using 96 accessions consisting of 65 Japanese and 26 exotic cultivated soybeans and 5 wild soybeans. Overall, the gene diversity among Japanese accessions (0.21) and wild soybeans (0.22) was lower than that among exotic accessions (0.31). In Japanese soybean 44% of SNP loci were not informative. Thus, re-sequencing using a representative Japanese germplasm array is required in order to capture informative SNPs for Japanese soybean germplasm.
A 1.4-fold increases in linkage disequilibrium (LD) was found in Japanese crossbred germplasm compared with landraces. This was not as great as that observed in elite North American cultivars. The increment of LD in the cross-bred germplasm suggests that effective recombination is restricted under the situation of limited generations and population size in breeding process.
Further, genetic variation and population structure among 1603 soybean accessions were characterized using 191 SNP markers. The soybean accessions consisted of 832 Japanese landraces, 109 old Japanese varieties which were grown before the 1950’s, 57 recent Japanese varieties, 341 landrace from 16 Asian countries and 264 wild soybean accessions. Although gene diversity of Japanese soybean germplasm was slight lower than that of exotic soybean germplasm, analyses for population differentiation and clustering indicated clear genetic differentiation among Japanese cultivated soybeans, exotic cultivated soybeans and wild soybeans. Accessions originating from different geographic region were separated to a certain extent into groups corresponding to their agro-morphologic characteristics such as photosensitivity and seed characteristics rather than their geographical origin. The information of the detailed genetic similarity among germplasm may provide a genetic platform to choose experimental and breeding materials.
Based on this evaluation of soybean germplasm conserved in the NIAS Genebank, exotic germplasm from many Asian countries was found to be poorly represented. To understand the variation in soybean germplasm from various countries, genomic information for accessions will be needed using common genetic markers or sequence information. This is needed not just for exotic germplasm but also Japanese germplasm conserved in public institutes in Japan. Further, germplasm collection in collaboration with other countries and of Japanese wild soybeans are considered necessary.
Based on the assessment of the SNP markers and several agro-morphologic traits, a number of accessions retain gene diversity of the whole collection and several soybean sets of different sizes were selected by using an heuristic approach; a minimum of 12 accessions can represent the observed gene diversity; mini-core collections can represent a major proportion of both geographic origin and agro-morphologic trait variation. These collections will provide an effective platform for enhancing soybean diversity studies and assist in finding novel traits for crop improvement.
Supplementary Material
Acknowledgements
The authors thank the following Japanese institutes for the supply of the materials used in the experiments: National Agricultural Research Center for Kyushu Okinawa Region (KONARC), National Agricultural Research Center for Tohoku Region (NARCT), National Institute of Crop Science, Hokkaido Prefectural Tokachi Agricultural Experimental Station, Nagano Chushin Agricultural Experimental Station, and Saga Prefectural Agricultural Experimental Center. The authors also appreciate the technical support of field materials from the following staff of the National Institute of Agrobiological Sciences: H. Nakazawa, Y. Ito, M. Akiba, T. Misawa J. Inoue and T. Taguchi. This work is supported by grants from the Ministry of Agriculture, Forestry and Fisheries of Japan (Genomics for Agricultural Innovation, DD-1010 and SOY1002) and from the NIAS Genebank Project.
Literature Cited
- Abe J, Hasegawa A, Fukushi H, Ikami TM, Ohara M, Shimamoto Y. Introgression between wild and cultivated soybeans of Japan revealed by RFLP analysis for chloroplast DNAs. Econ Bot. 1999;53:285–291. [Google Scholar]
- Abe J, Xu DH, Suzuki Y, Kanazawa A, Shimamoto Y. Soybean germplasm pools in Asia revealed by SSR. Theor Appl Genet. 2003;106:445–453. doi: 10.1007/s00122-002-1073-3. [DOI] [PubMed] [Google Scholar]
- Albertsen MC, Curry TM, Palmer RG, Lamotte CE. Genetics and comparative growth morphology of fasciation in soybeans (Glycine max (L.) Merr) Bot Gaz. 1983;144:263–275. [Google Scholar]
- Andreson R, Reppo E, Kaplinski L, Remm M. GENOME-MASKER package for designing unique genomic PCR primers. BMC Bioinfo. 2006;7:172. doi: 10.1186/1471-2105-7-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aman D, Sultoni A, Hidajat N, Dauphin F, Morooka Y, Rachim A. Government Policy, Regulations and Support Programs. Research for varietal improvement. In: Bottemat T, editor. The Soybean Commodity System in Indonesia, CGPRT No. 3, Bogor. CGPRT. 1988. pp. 84–85. [Google Scholar]
- Banba K, Takahashi N. Report on exploration to collect local soybean cultivars in Indonesia. Ann Rep Explor Intro Plant Genet Resourc. 1985;1:82–91. [Google Scholar]
- Bernard RL, Weiss MG. Qualitative genetics. Soybeans Improvement, Production, and Uses. In: Caldwell BE, editor. Agronomy Monograph. Vol. 16. American Society of Agronomy; Madison, WI: 1973. pp. 117–154. [Google Scholar]
- Boerma HR, Walker DR. Discovery and utilization of QTLs for insect resistance in soybean. Genetica. 2005;123:181–189. doi: 10.1007/s10709-004-2741-9. [DOI] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang ZW, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- Cannon SB, Shoemaker RC. Evolutionary and comparative analyses of the soybean genome. Breed Sci. 2012;61:437–444. doi: 10.1270/jsbbs.61.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty R, Jin L. Determination of relatedness between individuals using DNA fingerprinting. Hum Biol. 1993;65:875–895. [PubMed] [Google Scholar]
- Chen Y, Nelson RL. Genetic variation and relationships among cultivated, wild, and semiwild Soybean. Crop Sci. 2004;44:316–325. [Google Scholar]
- Cho GT, Yoon MS, Lee J, Baek HJ, Kang JH, Kim TS, Paek NC. Development of a core set of Korean soybean landraces [Glycine max (L.) Merr.] J Crop Sci Biotech. 2008;11:157–162. [Google Scholar]
- Choi IY, Hyten DL, Matukumalli LK, Song Q, Chaky JM, Quigley CV, Chase K, Lark KG, Reiter RS, Yoon MS, et al. A soybean transcript map: gene distribution, haplotype and single-nucleotide polymorphism analysis. Genetics. 2007;176:685–696. doi: 10.1534/genetics.107.070821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotiyarnwong O, Chatwachirawong P, Chanprame S, Sriniv P. Evaluation of genetic diversity in Thai indigenous and recommended soybean varieties by SSR markers. Thai J Agri Sci. 2007;40:119–126. [Google Scholar]
- Clough SJ, Tuteja JH, Li M, Marek LF, Shoemaker RC, Vodkin LO. Features of a 103-kb gene-rich region in soybean include an inverted perfect repeat cluster of CHS genes comprising the I locus. Genome. 2004;47:819–831. doi: 10.1139/g04-049. [DOI] [PubMed] [Google Scholar]
- Dong YS, Zhuang BC, Zhao LM, Sun H, He MY. The genetic diversity of annual wild soybeans grown in China. Theor Appl Genet. 2001;103:98–103. [Google Scholar]
- Dong YS, Zhao LM, Liu B, Wang ZW, Jin ZQ, Sun H. The genetic diversity of cultivated soybean grown in China. Theor Appl Genet. 2004;108:931–936. doi: 10.1007/s00122-003-1503-x. [DOI] [PubMed] [Google Scholar]
- Ebana K, Kojima Y, Fukuoka S, Nagamine T, Kawase M. Development of mini core collection of Japanese rice landrace. Breed Sci. 2008;58:281–291. [Google Scholar]
- Egawa Y, Nakano H, Bhatti MS. 1. Grain legumes. In: Kawase M, Okuno K, Egawa Y, Katsuta-Seki M, Nakano H, Nagamine T, Anwar R, Bhatti MS, Ahmad Z, Afzal M, editors. A Report of IBPGR Exploration in Northern Pakistan. National Institute of Agrobiological Resources, MAFF, Japan, and International Board for Plant Genetic Resources; Rome, Italy: 1992. 1991. [Google Scholar]
- Ellegren H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 2004;5:435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- Frankel OH, Brown AHD. Plant genetic resources today: a critical appraisal. In: Holden JHW, Williams JT, editors. Crop Genetic Resources: Conservation and Evaluation. Allen and Unwin; London: 1984. pp. 249–257. [Google Scholar]
- Fujita R, Ohara M, Okazaki K, Shimamoto Y. The extent of natural cross-pollination in wild soybean (Glycine soja) J Heredity. 1997;88:124–128. [Google Scholar]
- Fukui J, Arai M. Ecological studies on Japanese soybean varieties. I. Classification of soybean varieties on the basis of the days from germination to blooming and from blooming to ripening with special refference to their geographical differentiation. Jpn J Breed. 1951;1:27–39. [Google Scholar]
- Guan R, Chang R, Li Y, Wang L, Liu Z, Qiu L. Genetic diversity comparison between Chinese and Japanese soybeans (Glycine max (L.) Merr.) revealed by nuclear SSRs. Genet Resour Crop Evol. 2010;57:229–242. [Google Scholar]
- Guo J, Wang Y, Song C, Zhou J, Qiu L, Huang H, Wang Y. A single origin and moderate bottleneck during domestication of soybean (Glycine max): implications from microsatellites and nucleotide sequences. Ann Bot. 2010;106:505–514. doi: 10.1093/aob/mcq125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamblin MT, Buckler ES, Jannink JL. Population genetics of genomics-based crop improvement methods. Trends in Genet. 2011;27:98–106. doi: 10.1016/j.tig.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Nagasawa T, Murakami S, Kokubun K, Nakamura S, Koyama T, Matsumoto S, Sasaki K, Okabe A. A new soybean variety “Wasesuzunari”. Bull Tohoku Natl Agric Exp Stn. 1985;71:23–42. [Google Scholar]
- Hashimoto K, Nagasawa T, Murakami S, Watanabe I, Kokubun K, Sakai S, Igita K, Okabe A. A new soybean variety “Kosuzu”. Bull Tohoku Natl Agric Exp Stn. 1988;77:45–61. [Google Scholar]
- Hill WG, Robertson A. Linkage disequilibrium in finite populations. Theor Appl Genet. 1968;33:54–78. doi: 10.1007/BF01245622. [DOI] [PubMed] [Google Scholar]
- Hirata T, Kaneko M, Misawa T, Abe J, Shimamoto Y. Regional differentiation in agricultural characters of soybean landraces. Res Bull Univ Farm Hokkaido Univ. 1995;29:41–54. [Google Scholar]
- Hirata T, Abe J, Shimamoto Y. Genetic structure of the Japanese soybean population. Genet Resour Crop Evol. 1999;46:441–453. [Google Scholar]
- Hu J, Zhu J, Xu HM. Methods of constructing core collections by stepwise clustering with three sampling strategies based on the genotypic values of crops. Theor Appl Genet. 2000;101:264–268. [Google Scholar]
- Hymowitz T. On the domestication of the soybean. Econ Bot. 1970;24:408–421. [Google Scholar]
- Hymowitz T. Soybeans: The success story. In: Janick J, Simon JE, editors. Advances in New Crops. Timber Press; Portland: 1990. pp. 159–163. [Google Scholar]
- Hymowitz T, Kaizuma N. Dissemination of soybean (Glycine max): seed protein electrophoresis profiles among Japanese cultivar. Econ Bot. 1979;33:311–319. [Google Scholar]
- Hymowitz T, Kaizuma K. Soybean seed protein electrophoresis profiles from 15 Asian countries or regions: hypotheses on paths of dissemination of soybeans from China. Econ Bot. 1981;35:10–23. [Google Scholar]
- Hyten DL, Song Q, Zhu Y, Choi IY, Nelson RL, Costa JM, Specht JE, Shoemaker RC, Cregan PB. Impacts of genetic bottlenecks on soybean genome diversity. Proc. Natl. Acad. Sci USA. 2006;103:16666–16671. doi: 10.1073/pnas.0604379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyten DL, Choi IY, Song Q, Shoemaker RC, Nelson RL, Costa JM, Specht JE, Cregan PB. Highly variable patterns of linkage disequilibrium in multiple soybean populations. Genetics. 2007;175:1937–1944. doi: 10.1534/genetics.106.069740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyten DL, Cannon SB, Song Q, Weeks N, Fickus EW, Shoemaker RC, Specht JE, Farmer AD, May GD, Cregan PB. High-throughput SNP discovery through deep resequencing of a reduced representation library to anchor and orient scaffolds in the soybean whole genome sequence. BMC Genomics. 2010;11:38. doi: 10.1186/1471-2164-11-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosla S, Augustus M, Brahmachari V. Sex-specific organisation of middle repetitive DNA sequences in the mealybug Planococcus lilacinus. Nucleic Acids Res. 1999;27:3745–3751. doi: 10.1093/nar/27.18.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KW, Chung HK, Cho GT, Ma KH, Chandrabalan D, Gwag JG, Kim TS, Cho EG, Park YJ. PowerCore: a program applying the advanced M strategy with a heuristic search for establishing core sets. Bioinformatics. 2007;23:2155–2162. doi: 10.1093/bioinformatics/btm313. [DOI] [PubMed] [Google Scholar]
- Kim MY, Lee S, Van K, Kim TH, Jeong SC, Choi IY, Kim DS, Lee YS, Park D, Ma J, et al. Whole-genome sequencing and intensive analysis of the undomesticated soybean (Glycine soja Sieb. and Zucc.) genome. Proc. Natl. Acad. Sci USA. 2010;107:22032–22037. doi: 10.1073/pnas.1009526107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Shimada H, Thang NQ, Tung LT. Exploration and Collection of Grain Legumes Germplasm in Vietnam. Ann Rep Explor Intro Plant Genet Resourc. 1994;10:141–169. [Google Scholar]
- Kojima Y, Ebana K, Fukuoka S, Nagamine T, Kawase M. Development of an RFLP-based rice diversity research set of germplasm. Breed Sci. 2005;55:431–440. [Google Scholar]
- Komatsu K, Takahashi M, Nakazawa Y. Antibiosis resistance of QTL introgressive soybean lines to common cutworm (Spodoptera litura Fabricius) Crop Sci. 2006;48:527–532. [Google Scholar]
- Koyama T, Sasaki K, Watanabe I. Characteristics of soybean varieties introduced from India and Nepal. Tohoku J Crop Sci. 1983;26:67–68. [Google Scholar]
- Kuroda Y, Kaga A, Guaf J, Vaughan DA, Tomooka N. Exploration, collection and monitoring of wild soybean, cultivated soybean and hybrid derivatives between wild soybean and cultivated soybean: based on field surveys at Akita, Ibaraki, Kochi and Saga Prefectures. Ann Rep Explor Intro Plant Genet Resourc. 2006;22:1–12. [Google Scholar]
- Kuroda Y, Kaga A, Tomooka N, Vaughan DA. Gene flow and genetic structure of wild soybean (Glycine soja) in Japan. Crop Sci. 2008;48:1071–1079. [Google Scholar]
- Kuroda Y, Tomooka N, Kaga A, Wanigadeva SMSW, Vaughan DA. Genetic diversity of wild soybean (Glycine soja Sieb. et Zucc.) and Japanese cultivated soybeans [G. max (L.) Merr.] based on microsatellite (SSR) analysis and the selection of a core collection. Genet Resour Crop Evol. 2009;56:1045–1055. [Google Scholar]
- Kuroda Y, Kaga A, Tomooka N, Vaughan DA. The origin and fate of morphological intermediates between wild and cultivated soybeans in their natural habitats in Japan. Mol Ecol. 2010;19:2346–2360. doi: 10.1111/j.1365-294X.2010.04636.x. [DOI] [PubMed] [Google Scholar]
- Lam HM, Xu X, Liu X, Chen W, Yang G, Wong FL, Li MW, He W, Qin N, Wang B, et al. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat Genet. 2010;42:1053–1059. doi: 10.1038/ng.715. [DOI] [PubMed] [Google Scholar]
- Lee JD, Yu JK, Hwang YH, Blake S, So YS, Lee GJ, Nguyen HT, Shannon JG. Genetic diversity of wild Soybean (Glycine soja Sieb. and Zucc.) accessions from South Korea and other countries. Crop Sci. 2008;48:606–616. [Google Scholar]
- Li Y, Guan R, Liu Z, Ma Y, Wang L, Li L, Lin F, Luan W, Chen P, Yan Z, et al. Genetic structure and diversity of cultivated soybean (Glycine max (L.) Merr.) landraces in China. Theor Appl Genet. 2008;117:857–871. doi: 10.1007/s00122-008-0825-0. [DOI] [PubMed] [Google Scholar]
- Liu K, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- Lu BR. Conserving biodiversity of soybean gene pool in the biotechnology era. Plant Spec Biol. 2004;19:115–125. [Google Scholar]
- Lumpkin TA, Konovsky JC. A critical analysis of vegetable soybean production, demand, and research in Japan. In: Shanmugasundaram S, editor. Vegetable Soybean: Research Needs for Production and Quality Improvement. Asian Vegetable Res. Dev. Center; Taiwan: 1991. pp. 120–140. [Google Scholar]
- Lumpkin TA, Konovsky JC, Larson KJ, McClary DC. Potential new specialty crops from Asia: Azuki bean, edamame soybean, and astragalus. In: Janick J, Simon JE, editors. New Crops. Wiley; New York: 1993. pp. 45–51. [Google Scholar]
- Maruyama N, Matsuzawa H, Hiroma K, Horiuchi J, Hagiwara H, Mikoshiba K. New soybean varieties for special uses: Black seeded variety “Shinano-kuro”, green-seeded variety “Shinano-aomame”, saddle pattern-seeded variety “Shinano-kurakake” and flat-seeded variety “Shinano-hiramame”. Bull Nagano Chushin Agr Ex Sta. 1987;5:19–28. [Google Scholar]
- Ministry of Agriculture and Forestry. A Study on Distribution of Soybean Varieties in Japan. Japan Ministry of Agriculture and Forestry, Agricultural Improvement Bureau, Research Division; Tokyo: 1952a. Kanto and Tosan Region [translated title] p. 33. [Google Scholar]
- Ministry of Agriculture and Forestry. A Study on Distribution of Soybean Varieties in Japan. Japan Ministry of Agriculture and Forestry, Agricultural Improvement Bureau, Research Division; Tokyo: 1952b. Hokkaido and Tohoku Region [translated title] p. 34. [Google Scholar]
- Ministry of Agriculture and Forestry. A Study on Distribution of Soybean Varieties in Japan. Japan Ministry of Agriculture and Forestry, Agricultural Improvement Bureau, Research Division; Tokyo: 1953. Tokai, Shikoku and Kyushu Region [translated title] p. 46. [Google Scholar]
- Ministry of Agriculture and Forestry. A Study on Distribution of Soybean Varieties in Japan. Japan Ministry of Agriculture and Forestry, Agricultural Improvement Bureau, Research Division; Tokyo: 1954. Hokuriku, Kinki and Chugoku Region [translated title] [Google Scholar]
- Mimura M, Coyne CJ, Bambuck MW, Lumpkin TA. SSR diversity of vegetable soybean [Glycine max (L.) Merr.] Genet Resour Crop Evol. 2007;54:441–453. [Google Scholar]
- Miyazaki S, Adachi D. Exploration of legumes in East Nepal. Ann Rep Explor Intro Plant Genet Resourc. 1988;4:87–107. [Google Scholar]
- Miyazaki S, Carter TE, Jr, Hattori S, Nemoto H, Shiina T, Yamaguchi E, Miyashita S, Kunihiro Y. Identification of representative accessions of Japanese soybean [Glycine max] varieties registered by the Ministry of Agriculture, Forestry and Fisheries, based on passport data analysis. Misc Publ of Nat Ins Agrobiol Resourc. 1995a;8:1–17. [Google Scholar]
- Miyazaki S, Carter TE, Jr, Shiina T, Chibana T, Miyashita S, Kunihiro Y. Identification of representative accessions of old cultivars that contribute to the pedigree of modern Japanese soybean varieties, based on passport data analysis. Misc Publ of Nat Ins Agrobiol Resourc. 1995b;8:18–37. [Google Scholar]
- Muramatsu N, Kokubun M, Horigane A. Relation of seed structures to soybean cultivar difference in pre-germination flooding tolerance. Plant Prod Sci. 2008;11:434–439. [Google Scholar]
- Nakagahra M, Kawase M, Nagamine T, Anwar R, Bhatti MS, Ahmad Z, Afzal M. Nat. Ins. Agrobiol. Res. MAFF; Japan: 1990. A Report of PARK/NIAR cereal collecting expedition in Pakistan, 1989. [Google Scholar]
- Nakamura D, Yokoh H, Hirota Y, Nonaka Shigyo, Ohishi H, Shigetomi O, Tagagi Y, Kishikawa H. A new soybean cultivar “Murayutaka”. Bull Saga Pref Agric Exp Stn. 1991;27:21–42. [Google Scholar]
- Nelson RL. Managing self-pollinated germplasm collections to maximize utilization. Plant Genet Res. 2011;9:123–133. [Google Scholar]
- Oeth P, del Mistro G, Marnellos G, Shi T, van den Boom D. Qualitative and quantitative genotyping using single base primer extension coupled with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MassARRAY) Methods Mol Biol. 2009;578:307–343. doi: 10.1007/978-1-60327-411-1_20. [DOI] [PubMed] [Google Scholar]
- Oliveira MF, Nelson RL, Geraldi IO, Cruz CD, de Toledo JF. Establishing a soybean germplasm core collection. Field Crops Res. 2010;119:277–289. [Google Scholar]
- Onda R, Watanabe S, Sayama T, Komatsu K, Okano K, Ishimoto M, Harada K. Genetic and molecular analysis of fasciation mutation in Japanese soybeans. Breed Sci. 2011;61:26–34. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu LJ, Li YH, Guan RX, Liu ZX, Wang LX, Chang RZ. Establishment, representative testing and research progress of soybean core collection and mini core collection. Acta Agron Sin. 2009;35:571–579. [Google Scholar]
- Rambaut A. FigTree v1.3.1. 2009 available http://tree.bio.ed.ac.uk/software/figtree/
- Robbins MD, Sim SC, Yang W, Deynze AV, van der Knaap E, Joobeur T, Francis DM. Mapping and linkage disequilibrium analysis with a genome-wide collection of SNPs that detect polymorphism in cultivated tomato. J Exp Bot. 2011;62:1831–1845. doi: 10.1093/jxb/erq367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J, Brim CA. Resistance of soybean to the soybean-cyst nematode as determined by a double-row method. Plant Dis Rep. 1957;41:923–924. [Google Scholar]
- Sasaki K, Shigemori I. Exploration and introduction of genetic resources of soybeans in Thailand. Ann Rep Explor Intro Plant Genet Resourc. 1986;2:134–142. [Google Scholar]
- Schmutz J, Cannon S, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. Genome sequence of the paleopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Sekizuka S, Yoshiyama T. Studies on the native wild grasses for fodder. (IV) crop-scientific studies on wild species of Glycine soja. J. Kanto-Tosan Agr Exp Stn. 1960;15:57–73. [Google Scholar]
- Shehzad T, Okuizumi H, Kawase M, Okuno K. Development of SSR-based sorghum (Sorghum bicolor (L.) Moench) diversity research set of germplasm and its evaluation by morphological traits. Genet Resour Crop Evol. 2009;56:809–827. [Google Scholar]
- Shimada H, Kasahara Y, Chi VL, Ut NT. Collaborative exploration for collecting legume genetic resources in Vietnam, 2000. Ann Rep Explor Intro Plant Genet Resourc. 2001;17:81–104. [Google Scholar]
- Shimamoto Y. Polymorphism and phylogeny of soybean based on chloroplast and mitochondrial DNA analysis. JARQ. 2001;35:79–84. [Google Scholar]
- Shurtleff W, Aoyagi A. History of soybeans and soyfoods in Southeast Asia (13th Century to 2010) Soyinfo Center; CA, USA: 2010a. [Google Scholar]
- Shurtleff W, Aoyagi A. History of soybeans and soyfoods in South Asia/Indian subcontinent (1656–2010) Soyinfo Center; CA, USA: 2010b. [Google Scholar]
- Soemarno . Soybean variety development in Indonesia. In: van Amstel H, Bottema JWT, Sidik M, van Santen CE, editors. Integrating Seed Systems for Annual Food Crops, CGPRT No. 32, Bogor. CGPRT. 1995. pp. 207–214. [Google Scholar]
- Stupar RM. Into the wild: The soybean genome meets its undomesticated relative. Proc. Natl. Acad. Sci USA. 2010;107:21947–21948. doi: 10.1073/pnas.1016809108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Ishii T, Pe S, Htay MT. Collaborative exploration for collecting legume germplasm in Myanmar. Ann Rep Explor Intro Plant Genet Resourc. 2002;18:93–113. [Google Scholar]
- Thseng F, Lin TK, Wu ST. The relations of genus Glycine subgenus soja and Glycine formosana Hosok. collected from Taiwan: Revealed by RAPD Analysis. J Jpn Bot. 2000;75:270–279. [Google Scholar]
- Tomooka N, Abe K, Thein MS, Twat W, Maw JB, Vaughn D, Kaga A. Collaborative exploration of cultivated and wild legume species in Myanmar (Oct. 15th–Nov.15th, 2002) Ann Rep Explor Intro Plant Genet Resourc. 2003;19:67–83. [Google Scholar]
- Tomooka N, Abe K, Vaughan DA, Kaga A, Isemura T, Kuroda Y. Conservation of legume—symbiotic rhizobia genetic diversity in East Timor, 2005. Ann Rep Explor Intro Plant Genet Resourc. 2006;22:135–148. [Google Scholar]
- Watanabe I, Nagasawa T. Appearance and chemical composition of soybean [Glycine max] seeds in germplasm collection of Japan. Jpn J Crop Sci. 1990;59:649–660. [Google Scholar]
- Watanabe K, Tun YT, Kawase M. Field survey and collection of traditionally grown crops in northern areas in Myanmar. Ann Rep Explor Intro Plant Genet Resourc. 2007;23:161–175. [Google Scholar]
- Watanabe S, Tajuddin T, Yamanaka N, Hayashi M, Harada K. Analysis of QTLs for reproductive development and seed quality traits in soybean using recombinant inbred lines. Breed Sci. 2004;54:399–407. [Google Scholar]
- Xu D, Abe J, Gai J, Shimamoto Y. Diversity of chloroplast DNA SSRs in wild and cultivated soybeans: evidence for multiple origins of cultivated soybean. Theor Appl Genet. 2002;105:645–653. doi: 10.1007/s00122-002-0972-7. [DOI] [PubMed] [Google Scholar]
- Yamashita M. Nougyou Gijyutu Taikei: Crop: no. 6: Soybean, Azuki and Peanut. Nousangyoson Bunka Kyokai; Tokyo: 2003. Kurodaizu no raireki to hinshu seitai. [Google Scholar]
- Yoon MS, Lee J, Kim CY, Kang JH, Cho EG, Baek HJ. DNA profiling and genetic diversity of Korean soybean (Glycine max (L.) Merrill) landraces by SSR markers. Euphytica. 2009;165:69–77. [Google Scholar]
- Zhou XL, Jr, Carter TE, Cui ZL, Miyazaki S, Burton JW. Genetic base of Japanese soybean cultivars released during 1950 to 1988. Crop Sci. 2000;40:1794–1802. [Google Scholar]
- Zhou XL, Jr, Carter TE, Cui ZL, Miyazaki S, Burton JW. Genetic diversity patterns in Japanese soybean cultivars based on coefficient of parentage. Crop Sci. 2002;42:1331–1342. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.