Here, antisense inhibition of the E1 subunit of the 2-oxoglutarate dehydrogenase complex in tomato was associated with generally accelerated development. This report reveals the importance of this mitochondrial enzyme in both photosynthetic and respiratory metabolism, highlighting its significance in programs of plant development connected to carbon–nitrogen interactions.
Abstract
Transgenic tomato (Solanum lycopersicum) plants expressing a fragment of the gene encoding the E1 subunit of the 2-oxoglutarate dehydrogenase complex in the antisense orientation and exhibiting substantial reductions in the activity of this enzyme exhibit a considerably reduced rate of respiration. They were, however, characterized by largely unaltered photosynthetic rates and fruit yields but restricted leaf, stem, and root growth. These lines displayed markedly altered metabolic profiles, including changes in tricarboxylic acid cycle intermediates and in the majority of the amino acids but unaltered pyridine nucleotide content both in leaves and during the progression of fruit ripening. Moreover, they displayed a generally accelerated development exhibiting early flowering, accelerated fruit ripening, and a markedly earlier onset of leaf senescence. In addition, transcript and selective hormone profiling of gibberellins and abscisic acid revealed changes only in the former coupled to changes in transcripts encoding enzymes of gibberellin biosynthesis. The data obtained are discussed in the context of the importance of this enzyme in both photosynthetic and respiratory metabolism as well as in programs of plant development connected to carbon–nitrogen interactions.
INTRODUCTION
In recent years, a number of studies have focused on reverse genetic analysis of the function of the enzymes of the plant mitochondrial tricarboxylic acid (TCA) cycle (Nunes-Nesi et al., 2005, 2007a; Studart-Guimarães et al., 2007; Sienkiewicz-Porzucek et al., 2008, 2010; van der Merwe et al., 2009, 2010; Araújo et al., 2011a). These studies have revealed important roles for the enzymes of the TCA cycle in root metabolism and morphology, with all transgenic plants characterized thus far exhibiting decreased rates of respiration and reduced root biomass (reviewed in Nunes-Nesi et al., 2011). By contrast, the situation in photosynthetic tissues is more complicated, with antisense inhibition of mitochondrial malate dehydrogenase or succinate dehydrogenase and a deleterious mutation of aconitase leading to elevated rates of photosynthesis (Carrari et al., 2003; Nunes-Nesi et al., 2005; Araújo et al., 2011a), whereas antisense lines in which fumarase was targeted exhibited decreased rates of photosynthesis and stunted growth (Nunes-Nesi et al., 2007a). Deficiency in the expression of succinyl-CoA ligase and mitochondrial isoforms of citrate synthase and isocitrate dehydrogenase, however, had little effect on the photosynthetic rate (Studart-Guimarães et al., 2007; Sienkiewicz-Porzucek et al., 2008, 2010). It is important to mention that the effects of the various genetic manipulations within the TCA cycle are distinctive for each enzyme and are qualitatively similar over a range of reduced levels for a given enzyme (Nunes-Nesi et al., 2011). Accordingly, the results of recent genome-scale models of Arabidopsis thaliana metabolism (Poolman et al., 2009; de Oliveira Dal’Molin et al., 2010), coupled with the lack of congruence between the expression levels (Fait et al., 2008) and maximum catalytic activities of the constituent enzymes of the TCA cycle (Gibon et al., 2004), provide further support for the contention that, as in bacteria, the TCA cycle of plants operates in a modular manner (Tcherkez et al., 2009; Gauthier et al., 2010; Sweetlove et al., 2010).
Studies in a broad range of organisms have pointed to an essential role of the 2-oxoglutarate dehydrogenase reaction in overall metabolic activity (reviewed in Bunik and Fernie, 2009). Using specific chemical inhibitors, we have previously demonstrated that the 2-oxoglutarate dehydrogenase complex (OGDH) is limiting for respiration in potato (Solanum tuberosum) tubers and plays an important role in nitrogen assimilation (Araújo et al., 2008). Moreover, a study in Arabidopsis mutants of cytosolic pyruvate,orthophosphate dikinase has implicated OGDH in nitrogen remobilization during leaf senescence (Taylor et al., 2010). During senescence, leaf cells undergo massive changes in cellular metabolism and a progressive degeneration of cellular structures (Bleecker and Patterson, 1997; Wingler and Roitsch, 2008). The induced metabolic changes include a loss of photosynthetic activity and the hydrolysis of macromolecules, the products of which are remobilized to still-growing parts of the plant. Senescence is affected by endogenous factors such as aging and hormones but also by environmental factors such as stress and nutrient supply (Gan and Amasino, 1997; Jing et al., 2002; Schippers et al., 2008). Specifically, the hormones ethylene and abscisic acid (ABA) have long been recognized as endogenous positive regulators of senescence (Hung and Kao, 2004; Lim et al., 2007), whereas gibberellin (GA) delays senescence in several species (Whyte and Luckwill, 1966; Goldthwaite and Laetsch, 1968; Chin and Beevers, 1970; Back and Richmond, 1971; Aharoni and Richmond, 1978; Kappers et al., 1998; Rosenvasser et al., 2006). Similarly, the supply of nutrients such as nitrogen, phosphorus, and metals is important during the senescence process (Lim et al., 2007; Wingler and Roitsch, 2008). Recent work indicates that the levels of pyridine nucleotides also play a critical role in this process (Schippers et al., 2008). However, the relative importance of these cues remains unknown, despite the fact that, although deteriorative in nature, the process of senescence is crucial for plant fitness (Lim et al., 2003).
In this study, we describe the generation of transgenic tomato (Solanum lycopersicum) plants deficient in the expression of OGDH. The transformants were comprehensively analyzed with respect to their physiology and metabolism. These studies reveal a clear reduction in respiration and considerable shifts in metabolism but little difference in the photosynthetic capacity of young plants. By contrast, at later stages of development, the plants displayed reduced photosynthetic capacity due to the early onset of senescence. Analysis of microarray expression data and selected hormones indicated that the senescence phenotype observed was independent of the ethylene and ABA signal transduction pathway but rather linked to an inhibition of GA biosynthesis. These data are discussed both in the context of the role of 2-oxoglutarate in general and with respect to current models of the metabolic shifts occurring during leaf senescence.
RESULTS
Generation of OGDH Antisense Tomato Lines
OGDH operates as a heterodimer of OGDH E1 and E2 subunits. Searches of tomato EST collections (van der Hoeven et al., 2002) revealed the presence of 11 tentative consensus sequences encoding subunits of the OGDH, five of them with similarity to the E1 subunit of OGDH and six similar to E2. Full-length cDNAs encoding subunits E1 and E2 of OGDH were isolated by a PCR-based approach. Using this strategy, one clone per subunit was isolated, having 3.5 and 1.9 kb in length for E1 and E2, respectively. When the sequences of the isolated clones were BLASTed against the Sol Genomics Network (SGN) tomato database (http://solgenomics.net), 99 and 98% of similarity was found to SGN-U579159 and SGN-U568073 for E1 and E2 OGDH subunits, respectively. Further assembly and sequence analysis of the isolated clones indicated that the whole coding region had been amplified and revealed open reading frames encoding proteins of 1020 and 288 amino acids for E1 and E2 subunits, respectively. In silico analysis suggest that both Sl OGDH E1 and E2 subunits bear characteristics of a mitochondrial transit peptide sequence, indicating a mitochondrial location for the protein encoded by the Sl OGDH E1 and E2 cDNA. Comparison between the E1 and E2 subunit sequences revealed no significant similarity between these subunits. Comparison at the protein level between the tomato OGDH E1 amino acid sequences and the sequences from other species revealed relatively high identity (82%) with the maize (Zea mays; Zm OGDH E1), soybean (Glycine max; 82%), rice (Oryza sativa; 82%), and grapevine (Vitis vinifera; 86%) proteins (see Supplemental Figure 1 online; see Supplemental Data Set 1 online). Additionally, comparison between the OGDH E2 and the amino acid sequences from other species revealed relatively high identity (82 to 90%) with the potato protein, while it also showed high identity to the grapevine protein (86%). There is lower identity to proteins from Arabidopsis (69 to 71%), rice (58% [Os OGDH E2a] and 70% [Os OGDH E2b]), and soybean and Brassica napus (70%) and weak similarity to maize (65%) and a homologous Chlamydomonas reinhardtii protein (50%). Analysis of mRNA level, using a quantitative RT-PCR protocol (Czechowski et al., 2004), indicates a nearly constitutive expression of both genes, with the transcripts present at approximately equivalent levels in leaves, stems, roots, and fruits but only at very low levels in flowers (see Supplemental Figure 1 online). In addition, the transcripts for both subunits showed increasing expression during fruit development, with the highest expression in red fruits (see Supplemental Figure 1 online).
A 3050-bp fragment of the cDNA encoding E1 OGDH was cloned in the antisense orientation into the transformation vector pART27 between the cauliflower mosaic virus (CaMV) promoter and the ocs terminator (see Supplemental Figure 1 online). Similarly, a 1400-bp fragment of the cDNA encoding E2 OGDH was cloned in the antisense orientation into the transformation vector pBinAR between the CaMV promoter and the ocs terminator (see Supplemental Figure 1 online). Transgenic tomato lines harboring the resulting constructs were screened at the level of fruit enzyme activity, revealing three lines expressing the E1 OGDH antisense construct that exhibited suitably reduced enzyme activity (lines OGDH14, OGDH36, and OGDH37; Figure 1A). We were unable to detect any enzyme activity reduction in the E2 OGDH antisense lines (Figure 1A) and therefore did not study these further. The selected E1 OGDH lines were clonally propagated in tissue culture and then transferred to the greenhouse for further analyses.
Figure 1.
Characterization and Expression of Tomato OGDH.
OGDH activity was determined in fruits at 35 DAA (A) and in 4-week-old leaves (B) of tomato plants. ODGH E1 (C) and OGDH E2 (D) expression was determined in leaves of 4-week-old plants. The abundance of OGDH mRNAs was measured by quantitative RT-PCR. Values are presented as means ± se of six individual plants per line. Asterisks indicate values that were determined by Student’s t test to be significantly different from the wild type (WT; P < 0.05). FW, Fresh weight.
We also analyzed the activity in leaves of these propagants, confirming our previous screening result (Figure 1B). It is important to notice, however, that the activity in leaves was only around 20% of that observed in fruits. To verify the specificity of the constructs as well as to ensure that no compensatory effect occurred in the expression of the other subunits, a secondary screen was performed at the mRNA level. This revealed that only E1 OGDH expression was significantly reduced in leaves of the transgenic lines used (Figure 1C). Additionally, these analyses showed that the expression of the other subunit was not altered in the respective transformants (Figure 1D). Off-target effects of RNA interference constructs in plants have been suggested for fragments of 21 to 24 nucleotides or more (Watson et al., 2005; Rossel et al., 2007). Furthermore, it has been computationally predicted that between 50 and 70% of gene transcripts in plants have potential off targets when used for posttranscriptional gene silencing (Xu et al., 2006). Therefore, we decided to confirm that nonspecific gene silencing had not taken place in our studies. The fragment used for the antisense construct was designed to have minimal complementarity to other genes; thus, a BLAST query against the SGN database (SGN unigenes) revealed the absence of identical regions of at least 20 nucleotides. Therefore, it can be anticipated that there were no regions of identity to any other members of the OGDH family with the exception of the already tested E2 OGDH (Figure 1D) or, indeed, to any other transcript that could potentially be responsible for the phenotypes observed here.
Visual Phenotype of the Antisense Plants
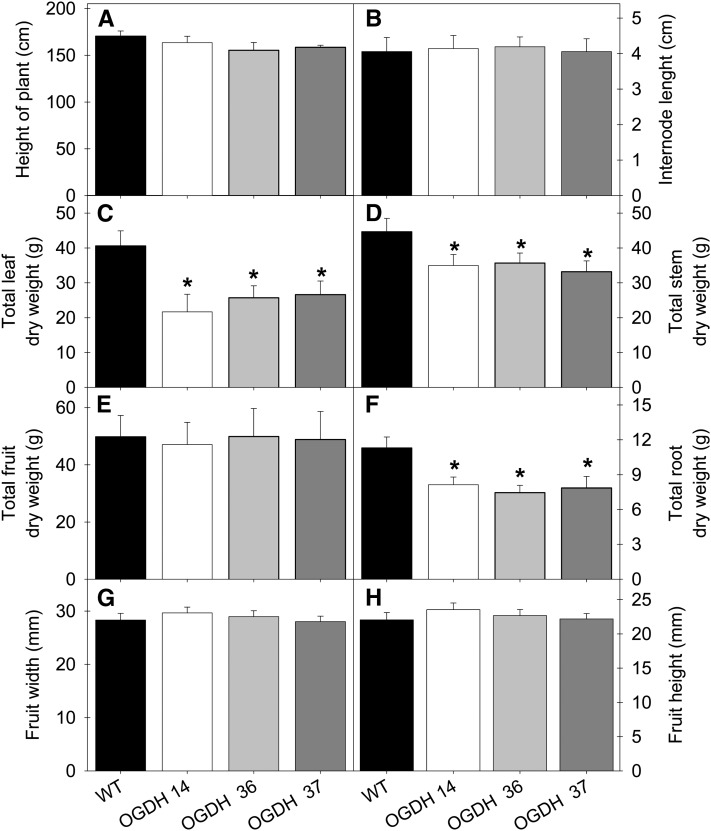
The antisense lines displayed early flowering (Figures 2A and 2B) but no difference in total flower number per plant (Figure 2C). We also observed early senescence in the antisense plants and accelerated fruit ripening (Figure 3). The flowers of antisense lines developed normally; thus, this accelerated fruit ripening was further confirmed using fruits of similar ages. Transgenic fruits exhibited a similar phenotype to wild-type fruits at 10 d after anthesis (DAA), while fruits at 35 and 55 DAA already demonstrated early ripening (see Supplemental Figure 2 online). A close inspection of 55-DAA fruits showed that a considerable proportion of wild-type fruits were orange whereas all fruits from the transgenics were red ripe (see Supplemental Figure 2 online). After 11 weeks of growth, the transgenic lines displayed decreased root, shoot, and leaf dry weight but no difference from the wild type in plant height or fruit biomass or size (Figure 4).
Figure 2.
Early-Flowering Phenotype of Antisense OGDH Tomato Plants.
(A) Cumulative number of flowers determined after the first flower was observed.
(B) Number of days needed for tomato plants to start to flower, determined as days after planting until the first flower appears.
(C) Total number of flowers per plant determined in 10-week-old plants.
Similar results were observed in three independent experiments. Values are presented as means ± se of determinations on at least six individual plants per line. Asterisks indicate values that were determined by Student’s t test to be significantly different (P < 0.05) from the wild type (WT).
Figure 3.
Growth Phenotype of Antisense OGDH Tomato Plants.
Transgenic plants showed early flowering (A), early senescence (B), and early fruit ripening (C) with respect to the wild type (WT). Representative plants after 5 and 8 weeks of growth as well as branches with fruits of a similar age are shown.
Figure 4.
Growth Phenotype of Antisense OGDH Tomato Plants.
Transgenic plants showed an enhanced aerial biomass with respect to the wild type (WT) in the later stages of growth (10-week-old plants). Data shown are for plant height (A), internode length (B), total leaf dry weight (C), total stem dry weight (D), total fruit dry weight (E), total root dry weight (F), mean fruit width (G), and mean fruit height (H). The lines used were as follows: the wild type, black bars; OGDH14, white bars; OGDH36, light gray bars; OGDH37, dark gray bars. Values are presented as means ± se of six individual plants per line. Asterisks indicate values that were determined by Student’s t test to be significantly different (P < 0.05) from the wild type.
Inhibition of OGDH Results in a Reduced Flux through the TCA Cycle
In a number of our previous studies (Carrari et al., 2003; Nunes-Nesi et al., 2005, 2007a), the analysis of the incorporation and subsequent metabolism of 14CO2 in genotypes deficient in the expression of TCA cycle enzymes suggested a reduction in flux through this cycle. However, analysis of the OGDH antisense plants revealed no such change in the incorporation of radiolabel in TCA cycle intermediates, or downstream metabolites thereof, under CO2-saturating conditions (Table 1).
Table 1. Effect of Decreased OGDH Activity on Photosynthetic Carbon Partitioning at the Onset of Illumination of 4-Week-Old Fully Expanded Source Leaves.
| Parameter | Wild Type | OGDH14 | OGDH36 | OGDH37 |
|---|---|---|---|---|
| Label incorporated (Bq) | ||||
| Total uptake | 305.6 ± 42.9 | 283.0 ± 53.1 | 278.5 ± 26.2 | 254.9 ± 34.5 |
| Organic acids | 13.2 ± 2.3 | 12.4 ± 2.3 | 11.6 ± 1.1 | 10.9 ± 1.8 |
| Amino acids | 12.4 ± 2.5 | 10.5 ± 1.5 | 10.6 ± 1.4 | 10.8 ± 4.8 |
| Soluble sugars | 114.2 ± 26.5 | 111.8 ± 44.2 | 110.7 ± 10.2 | 92.1 ± 30.5 |
| Starch | 165.8 ± 19.6 | 148.3 ± 20.5 | 145.6 ± 13.8 | 141.1 ± 14.5 |
| Redistribution of radiolabel (% of total assimilated) | ||||
| Organic acids | 4.5 ± 0.7 | 4.3 ± 0.1 | 4.2 ± 0.2 | 4.4 ± 0.8 |
| Amino acids | 4.3 ± 0.7 | 3.9 ± 0.4 | 3.7 ± 0.2 | 4.5 ± 0.5 |
| Soluble sugars | 35.7 ± 3.8 | 35.6 ± 6.8 | 39.8 ± 0.4 | 33.6 ± 7.8 |
| Starch | 55.6 ± 3.9 | 56.1 ± 6.6 | 52.3 ± 0.3 | 57.5 ± 6.9 |
Leaf discs were cut from separate plants of each genotype at the end of the night and illuminated at 700 µmol photons m−2 s−1 photosynthetically active radiation in an oxygen electrode chamber containing air saturated with 14CO2. After 30 min, the leaf discs were extracted and fractionated. Values presented are means ± se of measurements from six individual plants per genotype.
Given that we have previously documented that chemical inhibition of the OGDH reaction dramatically affects the rate of respiration (Araújo et al., 2008; van der Merwe et al., 2010), we next assessed the rate of respiration more directly using two complementary approaches. First, we measured the rate of dark respiration via infrared gas-exchange analyses (Figure 5A). These measurements revealed a reduction in the rate of carbon dioxide production, with that of the transformants being less than 60% of that observed in the wild type. We next directly evaluated the rate of light respiration in the transformants. For this purpose, we recorded the evolution of 14CO2 following the incubation of leaf discs in positionally labeled [14C]Glc molecules to assess the relative rate of flux through the TCA cycle. To do so, we incubated leaf discs taken from plants in the light and supplied these with [1-14C]Glc, [2-14C]Glc, [3,4-14C]Glc, or [6-14C]Glc over a period of 5 h. During this time, we collected the 14CO2 evolved at hourly intervals. Carbon dioxide can be released from the C1 position by the action of enzymes that are not associated with mitochondrial respiration, but carbon dioxide released from the C3/C4 positions of Glc cannot (Nunes-Nesi et al., 2005). Thus, the ratio of carbon dioxide evolution from C1 to C3/C4 positions of Glc provides a strong indication of the relative rate of the TCA cycle with respect to other processes of carbohydrate oxidation. When we compared the relative 14CO2 release of the transgenic and wild-type lines for the various fed substrates, the absolute release was always higher in the wild type than in the antisense lines (Figure 5B). In addition to these changes in absolute release, there was a shift in the evolution of 14CO2 from the variously labeled Glc, with the relative release from the C3/C4 positions being much lower in the transgenic lines OGDH14, OGDH36, and OGDH37 than in the wild type (the C3,4/C1 ratios were as follows: wild type = 0.98 ± 0.14; OGDH14 = 0.68 ± 0.08; OGDH36 = 0.69 ± 0.04; OGDH37 = 0.63 ± 0.07). Thus, these data reveal that a lower proportion of carbohydrate oxidation is performed by the TCA cycle in the transgenic lines, which is in keeping with the observation of reduced dark respiration in these plants. The differences in release from C2 and C6 positions were less marked, suggesting that there were no major alterations in metabolic fluxes involved in cycling through the pentose phosphate pathway or in pentan synthesis (Keeling et al., 1988).
Figure 5.
Respiratory Parameters in Leaves of Antisense OGDH Tomato Plants.
(A) Dark respiration (Rd) measurements performed in 4-week-old plants.
(B) Evolution of 14CO2 from isolated leaf discs in the light.
The leaf discs were taken from 4-week-old plants and incubated in 10 mM MES-KOH solution, pH 6.5, and 0.3 mM Glc supplemented with 2.32 kBq mL−1 [1-14C]Glc, [2-14C]Glc, [3,4-14C]Glc, or [6-14C]Glc at an irradiance of 200 μmol m−2 s−1. The 14CO2 liberated was captured (at hourly intervals) in a KOH trap, and the amount of radiolabel released was subsequently quantified by liquid scintillation counting. Values are presented as means ± se of determinations on six individual plants per line. Asterisks indicate values that were determined by Student’s t test to be significantly different (P < 0.05) from the wild type (WT).
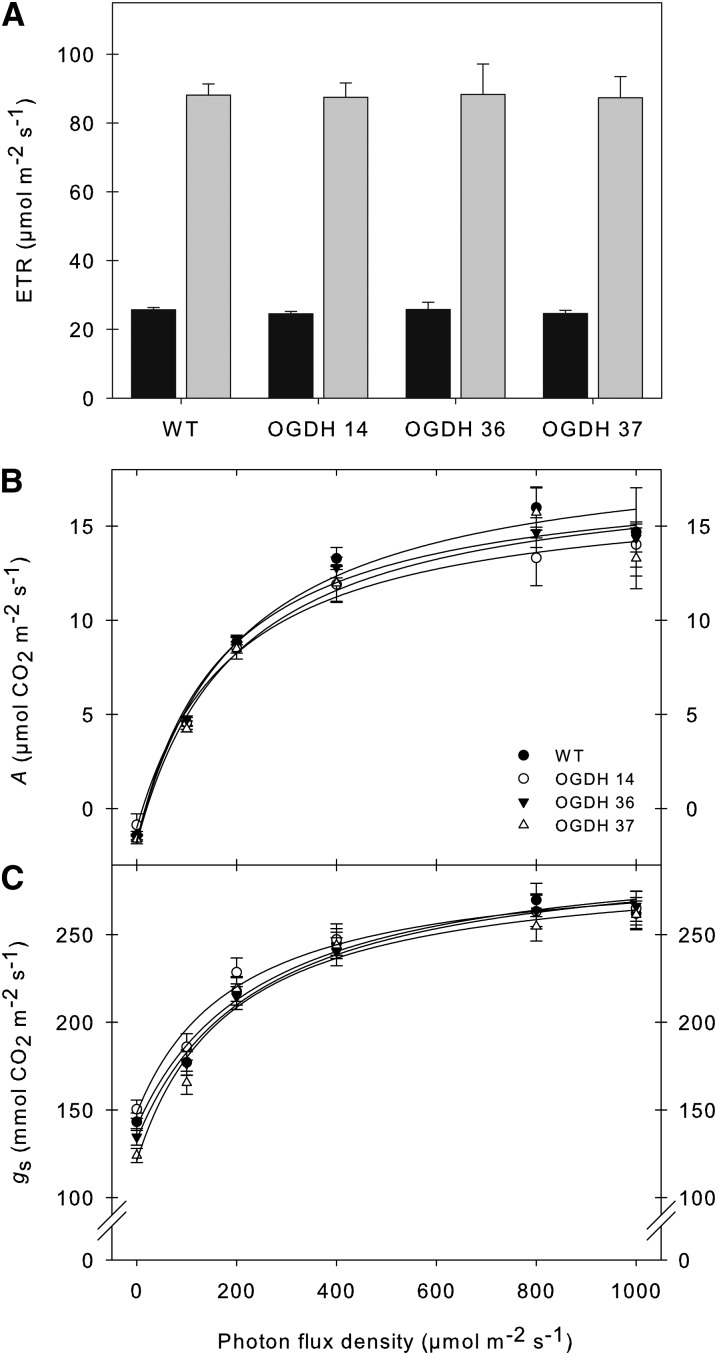
Antisense Inhibition of OGDH Has Little Effect on Photosynthesis during Early Development
Since our previous work has indicated a close link between TCA cycle activity and photosynthesis, we next studied this process in the transgenic lines. For this purpose, we determined chlorophyll fluorescence in vivo using a pulse amplitude modulation (PAM) fluorimeter in order to calculate relative electron transport rates (ETRs) at both high (700 μmol m−2) and low (100 μmol m−2) irradiances. These experiments revealed that the transformants displayed similar chloroplastic ETRs irrespective of the irradiance (Figure 6A). Additionally, gas exchange was measured directly, in 4-week-old plants, under photon flux densities (PFDs) that ranged from 100 to 1000 μmol m−2 s−1. The transformants exhibited unaltered assimilation rates irrespective of the irradiance (Figure 6B). Analysis of other parameters of gas exchange revealed that the transformants were also characterized by unaltered stomatal conductance regardless of the irradiance level (Figure 6C). When taken together, alongside the results of the 14CO2 feeding experiments, these data strongly suggest that these lines are characterized by unaltered photosynthesis at this time point (4-week-old plants). Additionally, these data indicate that the photosynthetic machinery is not compromised in the transformants.
Figure 6.
Effect of Decreased OGDH Activity on Photosynthetic Parameters.
(A) In vivo chlorophyll a fluorescence was measured as an indicator of the ETR by use of a PAM fluorometer at PFDs of 100 (black bars) and 700 (gray bars) μmol m−2 s−1.
(B) Assimilation rate as a function of PFD.
(C) Stomatal conductance as a function of PFD.
Values are presented as means ± se of six individual determinations per line. All measurements were performed in 4- to 5-week-old plants. The lines used were as follows: the wild type (WT), black circles; OGDH14, white circles; OGDH36, black triangles; OGDH37, white triangles.
Pigment Contents in the Transgenic Lines
Given the above results, we next decided to evaluate the levels of the photosynthetic pigments, since these compounds have often been reported as important indicators of nitrogen deficiencies (Gaude et al., 2007). Analysis of pigment content revealed that chlorophylls a and b were both significantly reduced in the transformants, as was β-carotene, while lutein was decreased only in the most strongly repressed line, OGDH37 (Table 2). Most of our results were obtained in 4- to 5-week-old tomato plants, but the most clearly visible phenotype (that of early senescence) was observed in 10-week-old plants. Thus, we measured two parameters related to the function of chloroplasts, chlorophyll content and photochemical efficiency (maximum variable fluorescence/maximum yield of fluorescence [Fv/Fm]), as diagnostics of leaf senescence (Oh et al., 1996). During normal leaf development of 4- to 10-week-old tomato plants, the chlorophyll content declined more rapidly in the transformants than in the wild type (see Supplemental Figure 3 online). This decline was linked with a minor increase in the chlorophyll a/b ratio, a typical feature of senescence-related chlorophyll breakdown in Arabidopsis (Pružinská et al. 2005; Araújo et al., 2011b) (see Supplemental Figure 3 online). Accordingly, these results were associated with a rapid decline in the photochemical efficiency of photosystem II (Fv/Fm) in the transformants (see Supplemental Figure 3 online).
Table 2. Pigment Contents in Antisense OGDH Tomato Plants.
| Pigment | Wild Type |
OGDH14 |
OGDH36 |
OGDH37 |
|---|---|---|---|---|
| μmol kg-1 fresh wt | ||||
| Neoxanthin | 40.9 ± 9.7 | 36.9 ± 7.9 | 35.4 ± 9.3 | 34.4±7.7 |
| Violaxanthin | 73.7 ± 9.3 | 74.3 ± 9.1 | 65.9 ± 6.1 | 69.5 ± 9.01 |
| Antheraxanthin | 4.1 ± 0.7 | 4.3 ± 0.5 | 3.5 ± 0.4 | 3.6 ± 0.6 |
| Zeaxanthin | 33.1 ± 11.1 | 29.4 ± 7.2 | 38.5 ± 11.2 | 25.8 ± 6.9 |
| β-Carotene | 93.4 ± 16.8 | 41.1 ± 4.5 | 46.3 ± 5.4 | 45.3 ± 6.2 |
| Lutein | 596.4 ± 41.1 | 516.6 ± 59.2 | 471.9 ± 92.1 | 445.4 ± 51.5 |
| Chlorophyll a | 4629.5 ± 166.2 | 3894.6 ± 158.8 | 3816.5 ± 189.5 | 3735.9 ± 186.4 |
| Chlorophyll b | 1641.8 ± 131.9 | 1382.4 ± 87.1 | 1475.1 ± 121.7 | 1387.1 ± 119.5 |
| Chlorophyll a/b | 2.9 ± 0.3 | 2.8 ± 0.2 | 2.7 ± 0.4 | 2.9 ± 0.4 |
Pigments were determined in 4-week-old fully expanded source leaves harvested 6 h into the photoperiod. Values presented are means ± se of six individual plants per line. Values in boldface were determined by Student’s t test to be significantly different (P < 0.05) from the wild type.
In order to gain a deeper comprehension of the reasons underlying the observed leaf and fruit phenotypes, we further analyzed the seed yield of nonstressed plants. This was largely unaltered (see Supplemental Figure 4 online) in both number of seeds per fruit and total mass per fruit. Tests of germination efficiency confirmed that the germination rate of the transgenic seeds was also unaffected (see Supplemental Figure 4 online). These results suggest that the early leaf senescence and fruit maturation may represent a strategy to facilitate full seed development. Indeed, the sacrifice of nutrients from the leaves and a slight acceleration of fruit development are seemingly able to compensate for the reduced energy capacity of the transgenic fruit (described in detail below).
Metabolite Profile of Leaves of the Antisense Lines
We next decided to extend this study to the major primary pathways of plant photosynthetic metabolism by utilizing an established gas chromatography–mass spectrometry (GC-MS) protocol for metabolic profiling (Fernie et al., 2004a; Lisec et al., 2006). These studies revealed considerable changes in the levels of a wide range of organic acids, amino acids, and sugars (Figure 7; see Supplemental Table 1 online). As would be anticipated, the level of 2-oxoglutarate was significantly increased in all antisense lines. The levels of succinate, γ-amino butyric acid (GABA), and ascorbate all increased also. Interestingly, these metabolites have previously been demonstrated to be increased as a strategy to bypass blocks in the TCA cycle and thus augment respiration (Nunes-Nesi et al., 2005; Studart-Guimarães et al., 2007). By contrast, the levels of the TCA cycle intermediates isocitrate, fumarate, and glycerate were significantly decreased in all antisense lines. Additionally, citrate (OGDH14 and OGDH37) and lactate (OGDH14 and OGDH36) were significantly decreased. The majority of the amino acids were decreased, most notably those derived from 2-oxoglutarate (Pro, Orn, Glu, and Gln) but also pyruvate-derived Ala, oxaloacetate-derived homoserine, Thr, and Met, as well as 3-phosphoglycerate-derived Ser and Gly. By contrast, Trp levels were significantly increased in all antisense lines. In addition, the levels of Arg (OGDH36 and OGDH37), Asn (OGDH14 and OGDH36), and the branched chain amino acids Ile (OGDH14 and OGDH36), Val (OGDH14 and OGDH36), as well as Lys (OGDH14 and OGDH37) were significantly decreased. Furthermore, the levels of sugars were elevated, with Fru, Glc, maltose, and Suc all increasing. Another change of note was the increase in the levels of the polyamine spermine.
Figure 7.
Relative Metabolite Content of Fully Expanded Leaves from 4-Week-Old Antisense OGDH Plants.
Amino acids (A) and organic acids (B) were determined as described in Methods. The full data sets from these metabolic profiling studies are available in Supplemental Table 1 online. Data are normalized with respect to the mean response calculated for the wild type (WT); to allow statistical assessment, individual plants from this set were normalized in the same way. The lines used were as follows: the wild type, black bars; OGDH14, white bars; OGDH36, light gray bars; OGDH37, dark gray bars. Values are presented as means ± se of six individual plants per line. Asterisks indicate values that were determined by Student’s t test to be significantly different (P < 0.05) from the wild type.
Given the considerable changes in amino acid metabolism, we next performed complementary measurements to determine the total cellular amino acid, protein, nitrate, and starch levels. While the levels of amino acids, protein, and nitrate were significantly reduced in the transgenics (Figures 8A to 8C, respectively), starch, malate, and fumarate levels remained unaltered (Figures 8D to 8F, respectively). Additional analysis of the carbohydrate content of leaves from 4-week-old plants revealed that the transformants were characterized by significant increases in the levels of Suc (Figure 8G), Glc (Figure 8H), and Fru (Figure 8I).
Figure 8.
Metabolite Levels in Leaves of OGDH Transgenic Tomato Plants.
Total amino acid (A), protein (B), nitrate (C), starch (D), malate (E), fumarate (F), Suc (G), Glc (H), and Fru (I) levels were measured using leaf material harvested in the middle of the light period from 4-week-old plants. Values are means ± se of six independent samplings. The lines used were as follows: the wild type (WT), black bars; OGDH14, white bars; OGDH36, light gray bars; OGDH37, dark gray bars. Asterisks indicate values that were determined by Student’s t test to be significantly different (P < 0.05) from the wild-type. FW, Fresh weight.
To provide compelling evidence of the reasons behind the early senescence observed, we additionally determined the levels of these metabolites during normal plant development of leaves from 4- to 10-week-old tomato plants. This analysis revealed that nitrate, amino acid, and protein contents declined more rapidly in the transformants than in the wild type (see Supplemental Figure 3 online), in accordance with the substantial alteration of nitrogen metabolism already documented in the OGDH antisense lines.
Metabolite Profile of Tomato Fruits during Normal Development
To follow the repertoire of metabolic changes that occur during tomato fruit development in the OGDH transformants, we performed extensive metabolic profiling. For these analyses, we harvested fruit samples at 10, 35, and 55 DAA (see Supplemental Figure 3 online) and evaluated their metabolic composition by GC-MS. These experiments revealed large changes in the levels of a range of sugars, organic acids, and amino acids across fruit development in all genotypes (see Supplemental Figure 5 online; see Supplemental Data Set 2 online). Perhaps surprisingly, the metabolite profiles of the transgenic lines followed a remarkably similar pattern to those of the wild type. Thus, as expected, the level of 2-oxoglutarate was significantly higher in all antisense lines during fruit development and ripening, with a clear trend of reduction during ripening. Comparison of metabolite levels revealed that the amounts of several compounds that normally increase during fruit ripening, such as Phe, Asp, Ile, and Glu, were higher in the OGDH transformants than in the wild type across fruit development.
Suc decreased to less than 50% between 10 and 55 DAA, with no difference between transgenic and wild-type fruits, whereas Glc and Fru increased in an essentially linear manner, with significantly higher levels in all OGDH transformants. The changes in major sugars during ripening largely mirror those reported previously for these carbohydrates and for the enzymes involved in their interconversion (Carrari et al., 2006). Accordingly cell wall–related metabolites, namely Man, Rha, Xyl, and Gal, increased during normal tomato ripening, and these were higher in all antisense lines. During ripening, we observed a strong increase in the level of succinate, with higher values detected in all OGDH transformants. These changes were coupled with a much faster decline in GABA levels in all antisense lines (see Supplemental Figure 5 online; see Supplemental Data Set 2 online). By contrast, the levels of malate and fumarate were strongly decreased during normal fruit ripening, reaching lower levels in all antisense lines, while citrate and isocitrate increased during normal fruit ripening, reaching higher values in all antisense lines than in the wild type during normal ripening.
The levels of amino acids were also highly variable during fruit ripening. Declines (more rapid in all OGDH antisense lines) in metabolite levels were observed for GABA, Ala, β-Ala, Arg, Asn, Gln, Ile, Ly, Ser, Thr, and Val across normal fruit ripening. By contrast, Phe, Glu, Asp, Met, and putrescine increased, peaking at the onset of fruit ripening. One of the most prominent changes associated with these processes in ripening tomatoes is the increase in Glu content, although the values observed for OGDH transformants were always lower than in wild-type fruits of the same age.
We next determined the levels of chlorophyll, total amino acids, protein, nitrate, sugars, and starch in fruits at 10, 35, and 55 DAA. No significant major changes in those metabolites were observed in fruits at 10 DAA when comparing wild-type and OGDH antisense fruits (see Supplemental Table 2 online). Similarly no changes in protein, total amino acid, starch, or Suc levels were observed across fruit development (see Supplemental Table 2 online). Interestingly, a reduction in chlorophyll content linked to a decline in the chlorophyll a/b ratio was observed in ODGH fruits at both 35 and 55 DAA. Accordingly, higher levels of nitrate, Fru, and Glc were observed in all antisense lines during normal fruit development.
Pyridine Nucleotide Content in the Transgenic Plants
Since the oxidative decarboxylation of 2-oxoglutarate to succinyl-CoA catalyzed by OGDH also generates the reduced coenzyme NADH, it is reasonable to anticipate that reduction in the activity of this enzyme may affect the redox balance in the transformants. Therefore, we decided to assay the levels of pyridine dinucleotides in the leaves of wild-type and transformant plants. Surprisingly, alterations in neither pyridine nucleotide level per se nor in the NADH/NAD and NADPH/NADP ratios were observed (Figure 9). Similarly, during normal fruit development, no changes between wild-type and antisense lines were apparent when comparing both pyridine nucleotide levels and ratios across fruit ripening (see Supplemental Table 2 online)
Figure 9.
Pyridine Nucleotide Levels and Ratios in Leaves of OGDH Transgenic Tomato Plants.
The leaf material was harvested in the middle of the light period from 4-week-old plants. Values are means ± se of six independent samplings. The lines used were as follows: the wild type (WT), black bars; OGDH14, white bars; OGDH36, light gray bars; OGDH37, dark gray bars. Asterisks indicate values that were determined by Student’s t test to be significantly different (P < 0.05) from the wild-type. FW, Fresh weight.
Effect of the Reduction of OGDH on the Activities of Other Enzymes of Primary Metabolism
To understand the above-described changes in metabolites better, we next analyzed the maximal activities of a wide range of key enzymes of carbon and nitrogen metabolism (Table 3). Our results suggested a substantial suppression of nitrogen metabolism in the OGDH antisense lines. However, analyses of the maximal catalytic activities of important enzymes of nitrogen metabolism (nitrate reductase, ferredoxin-dependent glutamate synthase, and Glu dehydrogenase) were not significantly altered in leaves of the transgenic plants. Furthermore, analysis of key enzymes of photosynthesis (ribulose-1,5-bis-phosphate carboxylase/oxygenase, transketolase, transaldolase, Fru bisphosphatase, and glyceraldehyde 3-phosphate dehydrogenase) and starch synthesis (AGPase) revealed few consistent changes. Considering the TCA cycle and associated enzymes, there were no significant differences in the total activities of aconitase, succinyl-CoA ligase, cytosolic NADP-dependent isocitrate dehydrogenase, fumarase, and pyruvate dehydrogenase. Evaluation of the NAD-dependent malate dehydrogenase, mitochondrial NAD-dependent isocitrate dehydrogenase, and phosphoenolpyruvate carboxylase activities revealed isolated changes; however, these did not change in a manner consistent with the altered activity and expression of OGDH. In addition, there were no consistent changes in either the initial or total activities of the NADP-dependent malate dehydrogenase of the chloroplast, a commonly used diagnostic marker for alterations in plastidial redox status (Scheibe et al., 2005).
Table 3. Enzyme Activities in Leaves of OGDH Transgenic Tomato Lines.
| Enzymes | OGDH36 | OGDH37 | Parameter | OGDH36 |
|---|---|---|---|---|
| Phosphoenolpyruvate carboxylase | 253.3 ± 11.5 | 258.5 ± 24.2 | 308.4 ± 15.2 | 268.4 ± 17.5 |
| AGPase | 1598.5 ± 265.1 | 1662.4 ± 217.8 | 1478.9 ± 210.6 | 1423.5 ± 127.6 |
| Transketolase | 208.9 ± 12.1 | 215.8 ± 12.1 | 227.9 ± 15.7 | 234.0 ± 12.4 |
| Transaldolase | 21.2 ± 2.4 | 24.8 ± 1.6 | 24.2 ± 1.8 | 24.9 ± 2.0 |
| FBPase | 178.6 ± 39.7 | 203.7 ± 27.9 | 191.4 ± 53.2 | 180.8 ± 52.1 |
| NADP-glyceraldehyde 3-phosphate dehydrogenase | 70.9 ± 5.7 | 56.0 ± 4.4 | 58.2 ± 2.2 | 65.2 ± 3.1 |
| Aconitase | 34.2 ± 8.7 | 36.4 ± 2.3 | 35.5 ± 4.7 | 30.6 ± 3.9 |
| Succinyl-CoA ligase | 13.1 ± 0.9 | 13.6 ± 1.5 | 13.3 ± 2.2 | 15.8 ± 4.0 |
| Fumarase | 3431.1 ± 256.1 | 23.1 ± 226.7 | 3585.5 ± 192.9 | 3223.1 ± 226.7 |
| NAD-malate dehydrogenase | 56.4 ± 4.2 | 56.2 ± 5.4 | 65.6 ± 2.8 | 55.9 ± 4.5 |
| NADP-malate dehydrogenase initial | 90.3 ± 4.2 | 103.5 ± 7.9 | 105.4 ± 6.5 | 102.3 ± 8.6 |
| NADP-malate dehydrogenase total | 234.7 ± 8.1 | 223.6 ± 22.9 | 241.2 ± 12.2 | 240.4 ± 1.3 |
| NADP-malate dehydrogenase activation state (%) | 38.5 ± 4.2 | 47.3 ± 6.2 | 43.8 ± 5.3 | 42.7 ± 2.6 |
| NADP-isocitrate dehydrogenase | 1572.9 ± 154.2 | 1615.0 ± 247.4 | 1493.2 ± 247.3 | 1425.7 ± 217.9 |
| NAD-isocitrate dehydrogenase | 29.2 ± 1.3 | 26.6 ± 1.9 | 27.1 ± 2.9 | 27.9 ± 1.6 |
| Pyruvate dehydrogenase | 9.6 ± 2.2 | 11.1 ± 1.5 | 11.3 ± 1.5 | 10.8 ± 0.9 |
| Ferredoxin-dependent glutamate synthase | 3697.4 ± 423.3 | 3261.5 ± 338.6 | 3399.1 ± 431.9 | 3257.3 ± 351.7 |
| Nitrate reductase | 352.7 ± 30.4 | 316.7 ± 37.6 | 307.6 ± 69.7 | 328.3 ± 33.4 |
| Glu dehydrogenase | 270.5 ± 24.5 | 258.5 ± 24.2 | 257.3 ± 15.2 | 248.4 ± 17.5 |
NAD-malate dehydrogenase activity is expressed in mmol min−1 g−1 fresh weight.
Activities were determined in 4-week-old fully expanded source leaves harvested 6 h into the photoperiod. Data presented are means ± se (n = 6). Values in boldface were determined by Student’s t test to be significantly different (P < 0.05) from the wild type.
Involvement of 2-Oxoglutarate Metabolism in the GABA Shunt
To elucidate further the changes in primary metabolism and the connection between the 2-oxoglutarate complex and the GABA shunt, we next evaluated the relative isotope redistribution in leaves excised from wild-type and transformant plants using a combination of feeding 13C-labeled substrate to the leaf via the transpiration stream and a modified GC-MS protocol that facilitates the estimation of intracellular fluxes (Roessner-Tunali et al., 2004). We supplied either 13C-labeled Glc (Table 4) or Glu (Table 5) for a period of 4 h and evaluated the redistribution of 13C to TCA cycle, GABA shunt, and photorespiratory pathways. In both instances, the rate of isotope redistribution to 2-oxoglutarate was significantly increased in all antisense lines, as was that to Glu. The GABA shunt can bypass the reaction catalyzed by succinyl-CoA ligase and sustain succinate supply to the TCA cycle (Studart-Guimarães et al., 2007). Given this fact, we additionally quantified the label redistribution through the GABA shunt, observing a significant increase in the label redistribution to both GABA and succinate from either 13C-labeled Glc (Table 4) or Glu (Table 5). Interestingly, however, label redistribution to malate and fumarate was unaltered in both cases. In contrast to the general increase in label redistribution to GABA shunt intermediates, when looking at photorespiratory pathway intermediates, a clearly decreased label redistribution was observed to Gly (significant in all antisense lines and in both feeding experiments) and Ser (significant in all antisense lines in the Glu feeding). Additionally, a decreased label redistribution was observed for a range of other amino acids, namely Asp (significant in all antisense lines), Met (significant in all antisense lines), Orn (significant in all antisense lines), and Pro (significant in all antisense lines). Interestingly, the changes in redistribution of isotope were essentially conserved between the transgenics, with results from these experiments in close agreement with the observed alteration in the steady state levels of sugars, organic acids, and amino acids (Figure 7; see Supplemental Table 1 online). Taken together, these data, and those of the respiration measurements described above, indicate a clear imbalance of the TCA cycle coupled with an upregulation of the GABA shunt and also suggest a reduction in the in vivo activity of mitochondrial photorespiratory enzymes.
Table 4. Redistribution of Heavy Label following Glc Feeding of Tomato OGDH Antisense Lines and Wild-Type Leaves.
| Metabolites | Wild Type |
OGDH14 |
OGDH36 |
OGDH37 |
|---|---|---|---|---|
| μmol C1 equivalent g−1 fresh wt h−1 | ||||
| Ala | 4.8687 ± 1.2559 | 4.0151 ± 0.5819 | 4.4456 ± 0.7116 | 4.4152 ± 0.6440 |
| Asn | 0.1591 ± 0.0449 | 0.1250 ± 0.0046 | 0.1319 ± 0.0145 | 0.1655 ± 0.0160 |
| Asp | 0.1355 ± 0.0213 | 0.0636 ± 0.0152 | 0.0641± 0.0156 | 0.0723 ± 0.0131 |
| Fru | 0.5056 ± 0.0339 | 0.4099 ± 0.1132 | 0.6478 ± 0.0786 | 0.7956 ± 0.1236 |
| Fumarate | 0.1308 ± 0.0685 | 0.1143 ± 0.0128 | 0.1627 ± 0.0662 | 0.1367 ± 0.0334 |
| GABA | 0.0770 ± 0.0061 | 0.1161 ± 0.0063 | 0.9772 ± 0.0074 | 0.1021 ± 0.0157 |
| Glu | 1.2066 ± 0.1731 | 1.7232 ± 0.1640 | 1.6014 ± 0.1346 | 1.9555 ± 0.2489 |
| Gln | 0.9369 ± 0.1349 | 0.9130 ± 0.0956 | 1.1067 ± 0.2001 | 1.4611 ± 0.2310 |
| Gly | 0.7898 ± 0.1448 | nd | nd | nd |
| His | 1.0570 ± 0.1482 | 0.9155 ± 0.0479 | 0.9450 ± 0.1252 | 0.9976 ± 0.2123 |
| Ile | 0.0190 ± 0.0048 | 0.0164 ± 0.0032 | 0.0126 ± 0.0030 | 0.0318 ± 0.0090 |
| Lys | 1.5608 ± 0.2336 | 1.4119 ± 0.1509 | 1.4434 ± 0.1961 | 1.4965 ± 0.4709 |
| Malate | 2.1743 ± 0.1658 | 1.9227 ± 0.1583 | 1.8409 ± 0.2056 | 2.0265 ± 0.3760 |
| Met | 0.0437 ± 0.0013 | 0.0068 ± 0.0006 | 0.0023 ± 0.0011 | 0.0139 ± 0.0045 |
| Orn | 0.3665 ± 0.1085 | 0.0736 ± 0.0155 | 0.0633 ± 0.0202 | 0.1186 ± 0.0454 |
| 2-Oxoglutarate | 0.3223 ± 0.1687 | 1.1038 ± 0.0349 | 0.8570 ± 0.1264 | 0.8754 ± 0.0837 |
| Phe | 0.0600 ± 0.0122 | 0.0464 ± 0.0113 | 0.0722 ± 0.0107 | 0.0850 ± 0.0136 |
| Pro | 0.0308 ± 0.0124 | 0.0011 ± 0.0003 | 0.0017 ± 0.0010 | 0.0044 ± 0.0026 |
| Ser | 0.1215 ± 0.0235 | 0.0918 ± 0.0144 | 0.0926 ± 0.0186 | 0.1032 ± 0.0330 |
| Succinate | 1.3544 ± 0.3976 | 3.4968 ± 0.4090 | 3.0023 ± 0.3375 | 3.2060 ± 0.3398 |
| Suc | 1.3267 ± 0.1347 | 1.1869 ± 0.3273 | 1.1997 ± 0.2562 | 1.1500 ± 0.3886 |
| Trp | 0.5457 ± 0.0913 | 0.7438 ± 0.0643 | 0.5941 ± 0.2260 | 0.6641 ± 0.0481 |
| Val | 0.4520 ± 0.1098 | 0.3271 ± 0.0543 | 0.4440 ± 0.0617 | 0.4246 ± 0.0398 |
Fully expanded leaves of 4-week-old plants were harvested at the middle of the light period and fed via the petiole with [U-13C]Glc solution. Values represent absolute redistribution of the label and are given as means ± se of determinations on six independent plants. Those in boldface were determined by Student’s t test to be significantly different (P ≤ 0.05) from the wild-type. nd, not determined.
Table 5. Redistribution of Heavy Label following Glu Feeding of Tomato OGDH Antisense Lines and Wild-Type Leaves.
| Metabolites | Wild Type |
OGDH14 |
OGDH36 |
OGDH37 |
|---|---|---|---|---|
| μmol C1 equivalent g−1 fresh wt h−1 | ||||
| Ala | 3.2768 ± 0.6249 | 3.1572 ± 0.4030 | 2.9734 ± 0.5652 | 3.2731 ± 0.9014 |
| Asn | 0.1759 ± 0.0354 | 0.1405 ± 0.0169 | 0.1021 ± 0.0255 | 0.1840 ± 0.0241 |
| Asp | 0.2161 ± 0.4212 | 0.1663 ± 0.3179 | 0.1780 ± 0.0508 | 0.1184 ± 0.1818 |
| Fumarate | 0.3507 ± 0.0585 | 0.2827 ± 0.0708 | 0.3366 ± 0.0964 | 0.3287 ± 0.0100 |
| GABA | 0.1949 ± 0.0370 | 0.2981 ± 0.0351 | 0.2814 ± 0.0054 | 0.2614 ± 0.0411 |
| Glu | 2.0356 ± 0.2106 | 2.9169 ± 0.1512 | 2.7413 ± 0.3287 | 2.8551 ± 0.4369 |
| Gln | 4.1566 ± 0.8797 | 4.9032 ± 1.2392 | 4.0780 ± 0.4483 | 4.2725 ± 1.7787 |
| Gly | 2.3895 ± 0.5198 | 1.0203 ± 0.1515 | 1.1861 ± 0.2726 | 1.0856 ± 0.1305 |
| Malate | 2.9408 ± 0.4378 | 3.2693 ± 0.5074 | 2.8744 ± 0.2350 | 2.9035 ± 0.1853 |
| 2-Oxoglutarate | 0.5529 ± 0.1176 | 0.9963 ± 0.0775 | 1.4167 ± 0.2268 | 1.0377 ± 0.1008 |
| Pro | 0.2248 ± 0.0843 | 0.0870 ± 0.0143 | 0.0165 ± 0.0695 | 0.0592 ± 0.0150 |
| Ser | 0.2266 ± 0.0266 | 0.1711 ± 0.0111 | 0.1630 ± 0.0216 | 0.1538 ± 0.0214 |
| Succinate | 1.7913 ± 0.5689 | 3.7237 ± 0.4290 | 3.8405 ± 0.3592 | 3.9789 ± 0.3777 |
Fully expanded leaves of 4-week-old plants were harvested at the middle of the light period and fed via the petiole with [U-13C]Glu solution. Values represent absolute redistribution of the label and are given as means ± se of determinations on six independent plants. Those in boldface were determined by Student’s t test to be significantly different (P ≤ 0.05) from the wild-type.
Antisense Inhibition of OGDH Affects Hormone Metabolism and Gene Expression in Illuminated Leaves
Given that several of the observed phenotypes cannot be directly explained by the changes in OGDH activity in the transformants, we next decided to evaluate other potential mechanisms that are more directly related to plant morphology. Given the growing body of evidence suggesting sugar–hormone crosstalk (León and Sheen, 2003; Moore et al., 2003; Carrari et al., 2004; Loreti et al., 2008) as well as the recognized impact of hormones on senescence (Hung and Kao, 2004; Rosenvasser et al., 2006; Lim et al., 2007), we decided to focus on hormones. As a first experiment, we used the Affymetrix Tomato Genome Array to profile the transcript levels of a wide number of genes in leaf material of line OGDH36 and the wild type. Our studies revealed relatively few significant changes after adjusting for multiple testing. ANOVA of the microarray results indicated that the expression of 103 genes (33 upregulated and 70 downregulated) showed a significant change between the wild type and OGDH36, with a detection false discovery rate value of P < 0.005 in all replicates (see Supplemental Data Sets 3 and 4 online).
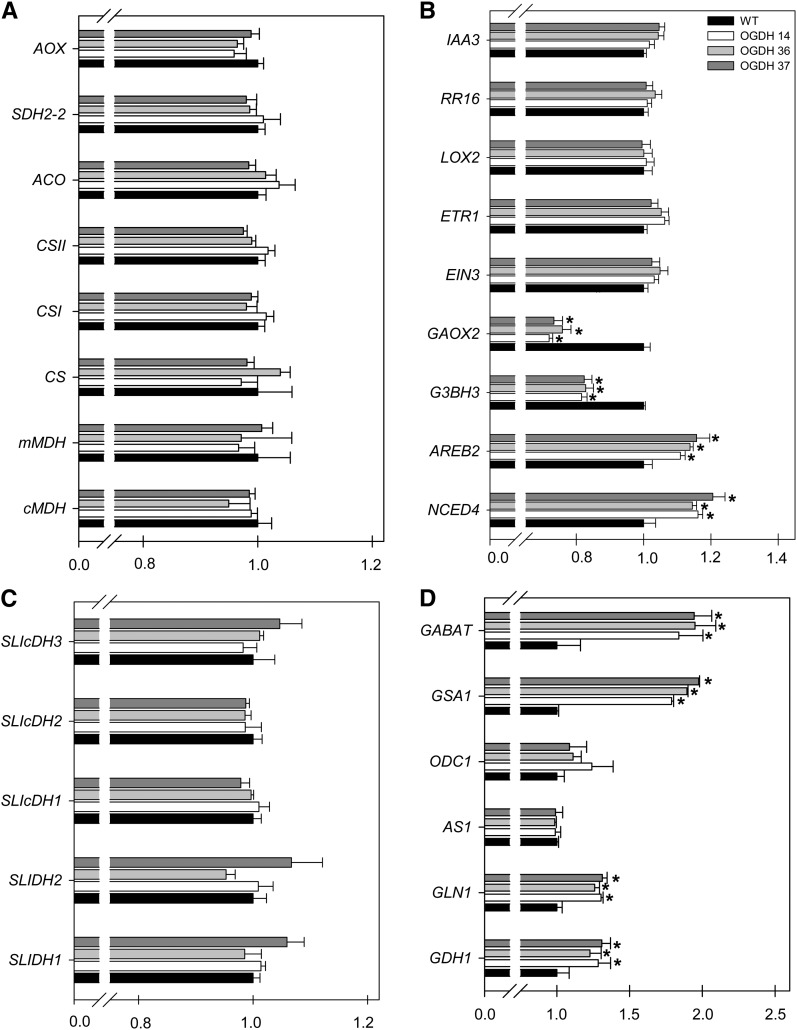
Overrepresentation analysis revealed a range of transcripts that were either upregulated or downregulated (see Supplemental Data Set 5 online). Among the transcripts that were upregulated in the OGDH samples, we observed transcripts associated with hormone metabolism, cell wall metabolism (pectin degradation [glycosyl hydrolases, pectate lyase, and polygalacturonase]), stress response (osmotic stress-responsive Pro dehydrogenase), and amino acid degradation (see Supplemental Data Sets 3 and 4 online). Among those that were downregulated were transcripts encoding proteins associated with hormone metabolism (GA-responsive protein, 2-oxoglutarate-dependent dioxygenase, ethylene-responsive proteinase inhibitor 1, allene oxide synthase, and auxin-induced protein) and transcripts associated with photosynthesis (including the Calvin cycle, light reactions, photosystem II, and photorespiration) (see Supplemental Data Sets 3 and 4 online). In addition to the changes in the expression of transcripts associated with the above processes, the OGDH antisense inhibition also seems to have mixed effects (induction/repression) on small sets of transcripts involved in several metabolic processes as well as those associated with RNA transcription and processing, posttranslational modifications, and protein turnover (see Supplemental Data Sets 3 and 4 online). The general picture, however, is relatively similar to that found following transcript profiling for the fumarase (Nunes-Nesi et al., 2007a) and succinate dehydrogenase (Araújo et al., 2011a) antisense lines. That said, it is important to note that the microarray falls well short of whole-genome coverage, and this may mask specific changes in certain key transcript levels. To gain a broader overview of the changes occurring following the genetic perturbation introduced, we additionally evaluated the relative levels of a wide range of transcripts by comparing their abundance using a more sensitive quantitative RT-PCR platform (Zanor et al., 2009). We identified the tomato homologs of signature genes from the literature, including mitochondrion-related genes, hormone-related genes, and GABA shunt– and amino acid–related genes (Figure 10). The primer information for these studies is given in Supplemental Data Set 6 online. As can be seen in Figure 10, the majority of the transcripts evaluated showed similar patterns when compared with wild-type plants. In fact, there were no significant changes in any of the TCA cycle or mitochondrial electron transport chain transcripts analyzed. When the transcripts associated with hormone biosynthesis or function were analyzed, changes were observed, with increased expression in the ABA-related transcripts AREB2 (for ABA-responsive element binding protein 2) and NCED4 (for 9-cis-epoxy carotenoid dioxygenase; a key regulated step in ABA biosynthesis in plants, which cleaves 9-cis-xanthophylls to xanthoxin, a precursor of ABA). In addition, the expression of the GA-related transcripts Gibberellin 2-Oxidase4 and Gibberellin 3β-Hydroxylase3 was were significantly reduced. Notwithstanding transcripts associated with auxin, ethylene and jasmonate showed relatively similar values in all genotypes analyzed here. Furthermore, the levels of transcripts involved in the GABA shunt, namely γ-Aminobutyrate Transaminase and Glutamate-1-Semialdehyde2,1-Aminomutase, were significantly increased in all antisense lines, in good agreement with the documented upregulation of the GABA shunt in these lines (Tables 4 and 5). Furthermore, elevated levels of the transcripts for two senescence marker genes (Dubois et al., 1996; Masclaux et al., 2000; Masclaux-Daubresse et al., 2002), Cytosolic Glutamine Synthetase and Glutamate Dehydrogenase, were also observed (Figure 10).
Figure 10.
Transcript Responses Involved in Regulatory and/or Signaling Responses in Plant Growth and Development.
Relative transcript abundance is shown for mitochondrion-related genes (A), hormone-related genes (B), various cellular isoforms of isocitrate dehydrogenase (C), and GABA shunt– and amino acid–related genes (D). The transcripts analyzed here were as follows: alternative oxidase (AOX), iron–sulfur subunit of succinate dehydrogenase (SDH2-2), aconitase hydratase (ACO), peroxisomal isoforms of citrate synthase (CSI and CSII), mitochondrial isoform of citrate synthase (CS), mitochondrial NAD-dependent malate dehydrogenase (mMDH), cytosolic malate dehydrogenase (cMDH), indole-3-acetic acid induced–related protein (IAA3), response regulator 16 (cytokinin, signal transduction; RR16), lipoxygenase 2 (LOX2), ethylene response 1 (ETR1), ethylene-insensitive 3 protein (EIN3), gibberellin 2-oxidase 4 (GAOX2), gibberellin 3β-hydroxylase 3 (G3BH3), ABA-responsive element binding protein 2 (AREB2), 9-cis-epoxycarotenoid dioxygenase (NCED4), most likely mitochondrial NADP-ICDH (SlICDH3), most likely cytosolic NADP-ICDH (SlICDH2), cytosolic NADP-ICDH (SlICDH1), mitochondrial regulatory NAD-IDH (SIIDH2), mitochondrial regulatory NAD-IDH (SIIDH1), γ-aminobutyrate transaminase (GABAT), glutamate-1-semialdehyde 2,1-aminomutase (GSA1), ornithine decarboxylase (ODC1), asparagine synthetase 1 (AS1), glutamine synthetase 1 (GLN1), and glutamate dehydrogenase (GDH1). Analyses were done in fully expanded leaves of 4-week-old plants. Values are presented as means ± se of six individual plants. The lines used were as follows: the wild type (WT), black bars; OGDH14, white bars; OGDH36, light gray bars; OGDH37, dark gray bars. Asterisks indicate values that were determined by Student’s t test to be significantly different (P < 0.05) from the wild type.
Given the suggestion that hormone metabolism is somehow altered, we next evaluated the levels of the hormones ABA and GA themselves. Interesting, there was a clear trend toward reduced levels of bioactive GAs, namely GA1, GA3, and GA7 (Figures 11A to 11C, respectively); however, it is important to note that a dramatic decrease was observed only for the levels of GA3 (which was reduced to ∼40% of the wild type levels; Figure 11B). This demonstrates that the levels of transcripts of the GA biosynthetic pathway are indeed reflected in an altered content of the phytohormone. However, the levels of the phytohormone ABA (Figure 11D) were invariant between genotypes, revealing that the increased transcript levels of AREB2 and NCED4 in the transgenics do not translate to altered levels of ABA itself (Figure 11D). We next extended the analysis of GA to fruits, where, although a strong variation was observed, we also found a clear trend toward decreased values of bioactive GAs (see Supplemental Figure 6 online). This is even more evident in the case of GA3, although the reduction was not of a similar magnitude to that observed for leaf material.
Figure 11.
Hormone Contents of the Wild-Type and Tomato OGDH Antisense Lines.
Data shown are GA1 (A), GA3 (B), GA7 (C), and ABA (D) contents of wild-type (WT) and OGDH antisense lines. Values are presented as means ± se of six individual determinations per line. All measurements were performed in 4-week-old plants. The lines used were as follows: the wild type, black bars; OGDH14, white bars; OGDH36, light gray bars; OGDH37, dark gray bars. Asterisks indicate values that were determined by Student’s t test to be significantly different (P < 0.05) from the wild type. FW, Fresh weight.
DISCUSSION
The TCA cycle is a fundamental component of mitochondrial respiration, linking glycolysis and/or extramitochondrial malate synthesis to the mitochondrial electron transport chain (Fernie et al., 2004b; Millar et al., 2004; Sweetlove et al., 2010). Unsurprisingly, then, intense efforts are currently devoted to elucidating the metabolic basis of the regulation of the TCA cycle (Sweetlove et al., 2007, 2010). As part of an ongoing project to elucidate the role of the TCA cycle in illuminated leaves, we have characterized plants deficient in the expression of enzymes of the TCA cycle (Sweetlove et al., 2010; Nunes-Nesi et al., 2011; Araújo et al., 2012). Here, we complete the characterization of the TCA cycle by characterizing the metabolic impact of antisense inhibition of OGDH in tomato plants. This allows us to assess whether long-term manipulation of its activity produces additional changes to those observed following its pharmacological inhibition (Araújo et al., 2008). In recent years, a considerable amount of research in nonplant systems has pointed to an essential role of OGDH in overall metabolic activity (reviewed in Bunik and Fernie, 2009). Here, we provide experimental evidence that the inhibition of the OGDH complex results in altered plant development. This occurs despite the fact that the lines were characterized by a compensatory increased flux through the GABA shunt, presumably in an attempt to maintain a supply of succinate for the mitochondrial electron transport chain. However, it is important to note that this compensatory response causes significant shifts in cellular pools of amino acids and nitrate. Recently, Sweetlove et al. (2010) combined evidence from labeling studies and metabolic network models to suggest that under certain conditions, the TCA cycle can function in a noncyclic manner with an absence of flux between 2-oxoglutarate and fumarate. In this study, we did not observe any alteration in the label redistribution to fumarate. However, we did observe an increase in label redistribution to Glu and GABA, providing further evidence for the occurrence of a functional bypass of 2-oxoglutarate to the succinate section of the TCA cycle. This raises the possibility that carboxylic acid metabolism may be driven to provide carbon skeletons for nitrogen assimilation (the reactions from acetyl-CoA to 2-oxoglutarate) and Asp biosynthesis (the conversion of malate to oxaloacetate) rather than the synthesis of ATP (Sweetlove et al., 2010). The reduced pool of total amino acids and nitrate we observed could thus be directly connected to the well-characterized involvement of 2-oxoglutarate and oxaloacetate in providing carbon skeletons for nitrogen assimilation (Hodges, 2002), since it clearly implies the importance of the OGDH, in addition to the well-documented isocitrate dehydrogenases (Dry and Wiskich, 1985; Nichols et al., 1994; Cornu et al., 1996; Gálvez et al., 1999; Igamberdiev and Gardeström, 2003), in both the TCA cycle and nitrogen assimilation. Moreover, bioinformatic analyses have revealed that the TCA cycle and the GABA shunt are differentially regulated at the level of gene expression (Fait et al., 2008). It thus seems reasonable to assume that the increased GABA levels represent an important adaptive mechanism for maintaining the rate of respiration. However, perhaps this is unsurprising, given that recent studies have demonstrated that there are several functionally active, alternative donors to the plant mitochondrial electron transport chain (Ishizaki et al., 2005, 2006; Nunes-Nesi et al. 2005; Araújo et al., 2010). When taken together, our results suggest that the activity of the OGDH is a highly sensitive metabolic juncture, and as such, they reveal the importance of mitochondrial reactions involving 2-oxoglutarate and by implication also of the mitochondrial membrane protein that mediates its transport between the cytosol and mitochondria (Picault et al., 2002).
Previous studies have demonstrated that interactions between photosynthesis and respiration induce changes in metabolic pathways of primary carbon metabolism (Hurry et al., 2005; Nunes-Nesi et al., 2007b) and that, in illuminated leaves, intracellular metabolism is dynamically modulated as a function of environmental changes (Noguchi and Yoshida, 2008). Historically speaking, photosynthesis and respiration have been considered as independent pathways; however, a growing body of evidence suggests that the functions of both chloroplasts and mitochondria are closely coordinated and tightly interact through intracellular metabolite pools (Carrari et al., 2003; Raghavendra and Padmasree, 2003; Nunes-Nesi et al., 2005, 2007b; Noguchi and Yoshida, 2008).That said, as described in the Introduction, alteration of the activities of the various enzymes of the mitochondrial TCA cycle resulted in diverse photosynthetic and phenotypic phenotypes. The results presented here suggest that despite displaying marked metabolic shifts, a deficiency of OGDH does not result in major changes in photosynthesis or growth rates. However, the results presented here, coupled with our previous studies, where OGDH was chemically inhibited (Araújo et al., 2008), suggest a greater impact of the OGDH on the respiration rate than previously observed for other genotypes deficient in the expression of TCA cycle enzyme activities. Indeed, evaluation of the control coefficient of OGDH for respiration from these data reveals it to exhibit a value of 0.736, which is largely similar to that estimated from the potato tuber data (0. 786; Araújo et al., 2012). This value suggests that the control of leaf respiration is largely shared between malate dehydrogenase and OGDH, with OGDH harboring the second greatest share of the control. Furthermore, our results clearly indicate that OGDH plays an important role in modulating the rate of flux from 2-oxoglutarate into amino acid metabolism. This conclusion is further endorsed by the altered levels of several amino acids, including those derived from 2-oxoglutarate (Pro, Orn, Glu, and Gln), supporting the growing evidence for strong network behavior in the coordination of plant amino acid metabolism (Coruzzi and Last, 2000; Foyer et al., 2003; Zhu and Galili, 2003; Sweetlove and Fernie, 2005; Less and Galili, 2008; Gu et al., 2010).
Mitochondrial effects on plant development have mainly been associated with plants having growth or floral development defects (Gutierres et al., 1997; Brangeon et al., 2000; Sabar et al., 2000; Pineau et al., 2005; León et al., 2007; Garmier et al., 2008; Roschzttardtz et al., 2009). Cytoplasmic male sterility traits involve chimeric mitochondrial genes that lead to altered expression of certain nuclear genes and, ultimately, to modified stamen phenotypes without affecting the vegetative plant phenotype in general (Schnable and Wise, 1998; Linke and Börner, 2005; Carlsson et al., 2008). By contrast, Nicotiana sylvestris and maize genotypes in which subunits of the mitochondrial electron transport chain are absent were characterized by alterations in photosynthesis, growth rate, leaf shape, and greening (Marienfeld and Newton, 1994; Gutierres et al., 1997; Karpova et al., 2002; Dutilleul et al., 2003; Noctor et al., 2004). In addition, the repression of TCA cycle enzymes has produced yet more variation. For instance, phenotypes observed in tomato include impairment in stomatal function and growth rate in fumarase antisense plants (Nunes-Nesi et al., 2007a), while increases of photosynthesis and growth rate were observed in plants deficient in the expression of mitochondrial malate dehydrogenase (Nunes-Nesi et al., 2005) and succinate dehydrogenase (Araújo et al., 2011a). Furthermore, physiological and genetic studies in Arabidopsis have uncovered the importance of the uncoupling protein UCP1 for the maintenance of optimal photosynthetic performance and growth rate (Sweetlove et al., 2006). There is also strong evidence that external mitochondrial NADPH dehydrogenase, NDB1, present in the mitochondrial electron transport chain, can, by modifying cell redox levels, specifically affect developmental processes via a modulation of bolting, resulting in an altered flowering time (Liu et al., 2009). To summarize, many plants with alterations in the expression and activity of enzymes of the mitochondrial respiratory pathways have displayed phenotypes of modified photosynthesis and growth, but not specific developmental changes, which culminated in a generally accelerated development, as evidenced by their early flowering, accelerated fruit ripening, and markedly earlier onset of leaf senescence, such as those described in this study (Figure 3; see Supplemental Figure 3 online). Our results revealed that the phenotype observed in OGDH antisense plants cannot be associated with alterations in the redox state, since no significant differences in pyridine dinucleotide levels were observed between leaves or fruits of wild-type and transformant plants (Figure 9; see Supplemental Table 2 online). Accordingly, these results suggest that the OGDH is important per se for normal plant development and performance. However, it is highly interesting that, despite these changes, the transgenic lines were characterized by largely unaltered fruit set, seed yield, and germination rates, suggesting the presence of sophisticated signaling mechanisms by which the plant uses the senescence process to prioritize the supply of nutrients to reproductive tissues.
Leaf senescence is a highly regulated developmental process that involves orderly, sequential changes in cellular physiology, biochemistry, and gene expression; as such, it has an essential role in plant survival and productivity (Hörtensteiner, 2006). However, despite its clear economic importance, the metabolic network that allows carbon and nitrogen to be remobilized from senescent leaves is yet poorly understood. Here, we demonstrate that the antisense inhibition of OGDH accelerates plant development in a manner that was coupled to reduced levels of nitrate as well as protein and amino acids (Figure 8). This likely indicates increased rates of nutrient remobilization from leaves to the seeds of the transformants. Additionally, the senescence phenotype observed is supported by several other results obtained in leaf material from 4-week-old plants, such as an increase in the hexose:Suc ratio (as deduced from the results shown in Figure 8) and an increase in transcripts for the senescence marker genes (Dubois et al., 1996; Masclaux et al., 2000; Masclaux-Daubresse et al., 2002) Cytosolic Glutamine Synthetase and Glutamate Dehydrogenase (Figure 10), concomitant with the reduced levels of total amino acids, particularly Gln and Asn (Figure 7). Additionally, high sugar levels (Figure 8) can be associated with senescence (van Doorn, 2008) and thus are likely to be an important factor involved in the process, especially given that they are subsequently processed via glycolysis to provide the pyruvate required to support mitochondrial respiration through the TCA cycle (Fernie et al., 2004b). Despite these findings, it is important to note that the role of sugars in plant senescence is still unclear, and the question of whether leaf senescence is induced or repressed by sugars has not yet been fully answered (van Doorn, 2004, 2008; Wingler et al., 2006).
Interestingly, several lines of evidence revealed that ethylene-mediated pathways leading to leaf senescence in Arabidopsis depend on age-dependent factors (Grbic and Bleecker, 1995); thus, ethylene can induce senescence only after leaves reach a certain developmental stage (reviewed in Lim et al., 2003). Similarly, tomato fruit ripening was induced by exogenously applied ethylene in mature green fruit but not in immature fruit (Yang, 1987). In addition, several aspects of plant growth and development, including fruit ripening and leaf and petal senescence, are controlled by ethylene (for details, see Lin et al., 2009; Bapat et al., 2010). However, we did not observe any increase in transcript levels of ethylene-related genes (Figure 10), indicating that ethylene is unlikely to be of major mechanistic importance with respect to the phenotypes observed in this study. Nevertheless, due to the fact that increased levels of ascorbate were observed and that this metabolite serves as an important cofactor in the biosynthesis of many plant hormones, including ethylene, gibberellic acid, and ABA, one has to assume that the endogenous level of ascorbate will affect not only the biosynthesis but also the levels and therefore the signaling of these molecules (for details, see Barth et al., 2006). Indeed, there is evidence for an important role of 2-oxoglutarate in GA biosynthesis (Lange et al., 1994; Hedden and Kamiya, 1997). Consequently, it is perhaps unsurprising that we have seen alterations in the transcript levels specifically for GA- and ABA-associated genes. ABA is commonly regarded as a senescence promoter (Barth et al., 2006); therefore, it could be predicted that a consequent elevation in ABA levels, which would be coupled to the alteration of ascorbate, would lead to an early induction of senescence. However, despite the observation that supposed key genes involved in ABA biosynthesis were increased, these were not supported by increased levels of this phytohormone itself. That said, high levels of GAs have been documented to inhibit senescence (Goldwaithe, 1972; Manos and Goldwaithe, 1975); therefore, interactions of GAs with the senescence inhibitor cytokinin and other regulatory genes in the GA response pathway also must be considered (Greenboim-Wainberg et al., 2005). This is indeed realistic when considering the reduced expression of GA-related genes coupled to the significant drop in the level of active GAs. Consequently, we can suggest not only that a single hormonal regulatory mechanism is responsible for the phenotype observed in OGDH transformants but that a correct interaction and balance of hormones is required for normal plant development. This conclusion is further supported by recent evidence that the inhibitory effect on tomato ovary growth by indole-3-acetic acid transported from the apex seems to be mediated by preventing the synthesis of active GAs (Serrani et al., 2010). While the precise nature of the interaction between energy metabolism and hormone-mediated regulation of growth and senescence could not be fully resolved in this study, it remains an exciting topic for future research.
In conclusion, we present here compelling evidence of the importance of the OGDH in plant development and metabolism. This enzyme exhibited a greater influence on respiration rate than we have observed previously, suggesting that 2-oxoglutarate plays a critical regulatory role in the rate of respiration (Araújo et al., 2008). The specific developmental change that culminated in a generally accelerated development of the transformants was involved in carbon and nitrogen remobilization pathways during natural senescence, revealing OGDH as a new possible target for the manipulation of these traits in plants. This observation additionally supports the recently proposed pathway of carbon–nitrogen metabolism during senescence suggested by Taylor et al. (2010). The somewhat surprising absence of alterations in fruit yield and seed production and germination makes it tempting to speculate that pathways of energy metabolism are tightly interregulated at the whole-plant level in a manner that allows the plant to prioritize reproductive organs during senescence.
METHODS
Materials
Tomato (Solanum lycopersicum) cv Moneymaker was obtained from Meyer Beck. Plants were handled as described in the literature (Carrari et al., 2003; Nunes-Nesi et al., 2005). Briefly, plants were grown side by side in the greenhouse with a minimum of 250 µmol photons m−2 s−1, 22°C, under a 16-h-light/8-h-dark regime. Except where mentioned, leaf experiments were performed on mature fully expanded source leaves from 4- to 5-week-old plants; fruit samples were harvested as indicated. All chemicals and enzymes used in this study were obtained from Roche Diagnostics, with the exception of radiolabeled sodium bicarbonate and d-[1-14C]Glc, d-[2-14C]Glc, d-[3,4-14C]Glc, and d-[6-14C]Glc, from American Radiolabeled Chemicals; [U-13C]Glc and [U-13C]-Glu were both from Cambridge Isotope Laboratories.
cDNA Cloning and Expression
The 3050-bp fragment of the E1 subunit of tomato OGDH was amplified using the primers 35S-OGDH E1 forward (5′-CACGCACGAGGAGGAAATGTAGG-3′) and 35S-OGDH E1 reverse (5′-AGGGCGACTAGTTTAATCCGACG-3′) and cloned in the antisense orientation into the vector pART27 (Liu et al., 1990) between the CaMV 35S promoter and the ocs terminator. This construct was introduced into plants by an Agrobacterium tumefaciens–mediated transformation protocol, and plants were selected and maintained as described in the literature (Tauberger et al., 2000). Initial screening of 16 lines was performed by enzymatic assay. These screens allowed the selection of three lines, which were taken to the next generation. Total OGDH activity was confirmed in the second harvest of these lines, after which three lines were chosen for detailed physiological and biochemical analyses.
Phylogenetic Analysis
Protein sequences were retrieved from GenBank through the BLASTp algorithm using the OGDH E1 and E2 amino acid sequences as queries. Sequences were aligned using the ClustalW software package (Higgins and Sharp, 1988) using default parameters. Neighbor-joining trees (Saitou and Nei, 1987) were constructed with MEGA4.1 Beta 2 software (Tamura et al., 2007) using midpoint rooting. Distances were calculated using pairwise deletion and Poisson correction for multiple hits, and bootstrap values were obtained with 1000 pseudoreplicates.
Analysis of Enzyme Activities
Enzyme extracts were prepared as described previously (Gibon et al., 2004), except that Triton X-100 was used at a concentration of 1% and glycerol at 20%. Enzymes were assayed exactly as described in the literature (Gibon et al., 2004; Nunes-Nesi et al., 2005; Studart-Guimarães et al., 2007), with the exception of the maximum activity of OGDH, which was determined as described by Araújo et al. (2008) with a few modifications. Briefly, the OGDH activity was measured by determining NADH formation at 340 nm in a reaction medium containing 75 mM TES-KOH (pH 7.5), 0.05% (w/v) Triton X-100, 0.5 mM MgCl2, 2 mM NAD+, 0.12 mM lithium-CoA, 0.5 mM thiamine pyrophosphate, 2.5 mM Cys-HCl, 1 mM AMP, 1 mM sodium-2-oxoglutarate, and 5 units of lipoamide dehydrogenase.
Determination of Metabolite Levels
Leaf and fruit pericarp samples were taken at the time points indicated, immediately frozen in liquid nitrogen, and stored at −80°C until further analysis. Extraction was performed by rapid grinding of tissue in liquid nitrogen and immediate addition of the appropriate extraction buffer. The levels of starch, Suc, Fru, and Glc in the leaf tissue were determined exactly as described previously (Fernie et al., 2001). The levels of nitrate, total amino acids, and protein were measured as described by Sienkiewicz-Porzucek et al. (2008); NAD(H) and NADP(H) were determined as described by Schippers et al. (2008). The levels of all other metabolites were quantified by GC-MS as described by Roessner et al. (2001), with the exception that the peak identification was optimized to tomato tissues (Roessner-Tunali et al., 2003), and the metabolites studied included recent additions to our mass spectral libraries (Schauer et al., 2005). Photosynthetic pigments were determined exactly as described by Bender-Machado et al. (2004). Full documentation of metabolite profiling data acquisition and interpretation is provided in Supplemental Data Set 7 online following recommended guidelines (Fernie et al., 2011).
Determination of Hormone Levels
The levels of ABA and GA were determined exactly as described previously (van der Merwe et al., 2009; Csukasi et al., 2011).
Measurements of Photosynthetic Parameters
The 14C-labeling pattern of Suc, starch, and other cellular constituents was determined by illuminating leaf discs (10 mm diameter) in a leaf disc oxygen electrode (Hansatech) in saturating 14CO2 at a PFD of 700 μmol m−2 s−1 photosynthetically active radiation at 22°C for 30 min, and subsequent fractionation was performed exactly as detailed by Lytovchenko et al. (2002). Fluorescence emission was measured in vivo using a PAM fluorometer (Walz) on 4-week-old plants maintained at fixed irradiance (250 and 700 µmol photons m−2 s−1) for 30 min prior to measurement of chlorophyll fluorescence yield and relative ETR, which were calculated using the WinControl software package (Walz). The CO2 response curves were measured at saturating irradiance with an open-flow gas-exchange system (LI-COR; model LI-6400). Four- to five-week-old plants maintained at a fixed irradiance of 250 μmol photons m−2 s−1 were used for gas exchange using a LI-6400 gas-exchange system (www.licor.com/) under different light intensities (as described in Results) and 400 µmol CO2 mol−1 air. All measurements were performed at 25°C, vapor pressure deficit was kept at 2.0 ± 0.2 kPa, while the amount of blue light was set to 10% PFD to optimize stomatal aperture. In addition, using a dark-adaptation leaf clip, minimum (F0) and maximum (Fm) dark-adapted (30 min) fluorescence were measured, from which Fv/Fm, in which Fv = Fm − F0, was calculated. This ratio has been used as a measure of the potential photochemical efficiency of photosystem II.
Measurement of the Redistribution of Isotope
The fate of 13C-labeled Glc and 13C-labeled Glu was traced following the feeding of leaves excised from 4-week-old plants via the petiole placed in a solution containing 10 mM MES-KOH (pH 6.5) and either 20 mM [U-13C]Glc or 20 mM [U-13C]Glu for 4 h. Fractional enrichment of metabolite pools was determined exactly as described previously (Roessner-Tunali et al., 2004; Tieman et al., 2006), and label redistribution was expressed as per Studart-Guimarães et al. (2007).
Measurement of Respiratory Parameters
Dark respiration was measured using the same gas-exchange system as defined above. Estimations of the TCA cycle flux on the basis of 14CO2 evolution were performed following the incubation of isolated leaf discs in 10 mM MES-KOH, pH 6.5, containing 2.32 kBq mL−1 [1-14C]Glc, [2-14C]Glc, [3,4-14C]Glc, or [6-14C]Glc. 14CO2 evolved was trapped in KOH and quantified by liquid scintillation counting. The results were interpreted following ap Rees and Beevers (1960).
Microarray Data Analysis
All experiments analyzing RNA expression levels were performed using four independent replicates of leaf materials, obtained from whole leaves from 36 wild-type and OGDH plants. Four independent replicates were harvested, and the material was immediately snap frozen in liquid nitrogen. Leaf material was homogenized using a ball mill (Retsch). Immediately afterward, RNA was extracted from the homogenized leaf material using Trizol (Invitrogen) exactly following the manufacturer’s suggestions. After quality control using an Agilent Bioanalyzer, at least 10 µg of total RNA was sent to ATLAS Biolabs for microarray analysis. At the service provider, cDNA synthesis, biotinylation, and microarray hybridization were performed exactly as specified by Affymetrix (http://media.affymetrix.com/support/downloads/manuals/gcas_ht_plate_manual.pdf).
The raw microarray data were processed using the Robin application (Lohse et al., 2010) to check the hybridization quality and identify significantly differentially expressed genes. Two samples (control sample 1 and OGDH36 sample 4) were excluded from downstream analyses because they showed strong outlier behavior in the quality checks. Statistical assessment of differential gene expression was performed using default settings in Robin. Briefly, raw chip data were normalized by applying the Robust Multichip Average method (Irizarry et. al., 2003), and subsequently, differentially expressed genes were identified using the limma R package (Smyth, 2004). Genes showing an absolute log2-fold change greater than 1 and a corrected P value (using the method developed by Benjamini and Hochberg, 1995) less than 0.05 were accepted as significantly differentially expressed. The significantly changed genes were divided into upregulated and downregulated genes and separately examined for functional category enrichment using the PageMan tool (http://mapman.mpimp-golm.mpg.de/general/ora/ora.shtml; Usadel et al., 2006).
The microarray data were submitted in MIAME-compliant (for minimum information about a microarray experiment) format to the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) and have been assigned accession number GSE35618. For a complete description of the experimental design of the microarray experiment and the submission details, readers are referred to this accession number.
Quantitative Real-Time PCR
Quantitative real-time PCR was performed exactly as described by Zanor et al. (2009). The primers that were used here are described in Supplemental Data Set 6 online, and a detailed experimental description is available in Supplemental Table 3 online. RNA was extracted from at least six biological replicates and at least two technical replicates. Analyses were done in fully expanded leaves of 4-week-old plants by harvesting and immediately snap-freezing the samples in liquid nitrogen. Extraction of total RNA was made by phenol separation and LiCl precipitation as described by Bugos et al. (1995). Digestion with DNase I (Ambion; http://www.ambion.com/) was performed according to the manufacturer’s instructions. To confirm the absence of genomic DNA contamination, a quantitative PCR analysis using three primer pairs was performed. The primer pairs were designed to amplify intron sequences of three tomato genes (AJ224356, forward [5′-CGAAGCAAGCGTGAACAAAT-3′] and reverse [5′-TGCGGAGATTAGGATGGACA-3′]; X58273, forward [5′-TCTCTTCTTTTCGTCGCTCTTG-3′] and reverse [5′-TTGCAACTTGGCAGTTGAATTA-3′]; Y00478, forward [5′-CTGCGAACTTCATTCAACAGC-3′] and reverse [5′-GCTCCACCATCGATCAAAAC-3′]). The integrity of the RNA was checked on 1% (w/v) agarose gels, and the concentration was measured before and after DNase I digestion using a Nanodrop ND-1000 spectrophotometer (http://www.nanodrop.com/). cDNA was synthesized from 2 μg of total RNA using SuperScript III reverse transcriptase (Invitrogen; http://www.invitrogen.com/) according to the manufacturer’s instructions. The efficiency of cDNA synthesis was estimated by quantitative PCR using two primer pairs amplifying the 5′ and 3′ regions of glyceraldehyde 3-phosphate dehydrogenase (AB110609; forward [5′-GATATCCCATGGGGTGAAGC-3′] and reverse [5′-CACAACCTTCTTGGCACCAC-3′]; forward [5′-GGCTGCAATCAAGGAGGAAT-3′] and reverse [5′- CAGCCTTGGCATCAAAAATG-3′]). PCR was performed using an ABI PRISM 7900 HT sequence detection system (Applied Biosystems; http://www.appliedbiosystems.com/). PCR was performed in a 5-μL volume as described by Caldana et al. (2007). Data analysis was performed using SDS software version 2.3 (Applied Biosystems). The reference gene was measured using two replicates in each PCR run, and their mean cycle threshold was used for relative normalized expression analyses. To normalize gene expression, we used the constitutively expressed Ubiquitin3 with the following primers, forward (5′-AGGTTGATGACACTGGAAAGGTT-3′) and reverse (5′-ATCGCCTCCAGCCTTGTTGTA-3′) (Wang et al., 2008).
Statistical Analysis
The term significant is used here only when the change in question has been confirmed to be significant (P < 0.05) with Student’s t test. All statistical analyses were performed using the algorithm embedded into Microsoft Excel.
Accession Numbers
Sequence data from this article for the major genes discussed can be found in the GenBank/EMBL databases under the following accession numbers: OGDH E1, SGN-U579159; OGDH E2a, SGN-U568073; and OGDH E2b, SGN-U568072. Additional accession numbers for other genes discussed in this article are shown in Supplemental Data Set 6 online. The microarray data from this article can be found in the National Center for Biotechnology Information Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE35618.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Characterization and Expression of the Tomato OGDH.
Supplemental Figure 2. Accelerated Fruit-Ripening Phenotype of Antisense OGDH Tomato Plants.
Supplemental Figure 3. Metabolic Phenotype of OGDH Transgenic Tomato Plants during Normal Plant Development.
Supplemental Figure 4. Seed Phenotype of the Wild Type and Tomato OGDH Antisense Lines.
Supplemental Figure 5. Heat Map Representing the Changes in Relative Metabolite Contents of the Wild Type and Tomato OGDH Antisense Lines during Fruit Development.
Supplemental Figure 6. Hormone Content Phenotype of the Wild Type and Tomato OGDH Antisense Lines during Fruit Development.
Supplemental Table 1. Relative Metabolite Content of Fully Expanded Leaves from 4-Week-Old Plants of the Antisense OGDH Plants.
Supplemental Table 2. Metabolite Content during Fruit Development of OGDH Tomato Plants.
Supplemental Table 3. Checklist Features To Be Followed for the Analyses Done by Quantitative RT-PCR.
Supplemental Data Set 1. Text File Corresponding to the Alignment Shown in Supplemental Figure 1 Online.
Supplemental Data Set 2. Relative Metabolite Content during Fruit Development of the Antisense OGDH Plants.
Supplemental Data Set 3. Expression Values of Upregulated Genes between Wild-Type and OGDH36 Samples.
Supplemental Data Set 4. Expression Values of Downregulated Genes between Wild-Type and OGDH36 Samples.
Supplemental Data Set 5. Functional Categories That Are Overrepresented in the List of Genes That Exhibit Significant Changes in Expression Levels between the Wild Type and OGDH36 and Were Consistently Upregulated (33) and Downregulated (70) according to ANOVA.
Supplemental Data Set 6. Primers Used in the RT-PCR Analyses Performed in This Study.
Supplemental Data Set 7. Overview and Checklist Features of the Metabolite Reporting List.
Acknowledgments
We acknowledge the excellent care of the plants by Helga Kulka (Max-Planck-Institut für Molekulare Pflanzenphysiologie) and the excellent photographic work by Josef Bergstein (Max-Planck-Institut für Molekulare Pflanzenphysiologie). This work was supported by grants from the Max Planck Gesellschaft (to W.L.A. and A.N.-N.), by the ERAnet Plant Genomics projects TRIPTOP funded by the German Research Council (to A.R.F.) and the Deutsche Forschungsgemeinschaft within the research unity FOR 1186-Promics Photorespiration Network (to A.R.F.), and by the National Council for Scientific and Technological Development, Brazil (to W.L.A. and A.N.-N.).
AUTHOR CONTRIBUTIONS
W.L.A., A.N.-N., and A.R.F. designed the research; W.L.A., T.T., S.O., M.L., I.B., I.K., A.S.-P., and A.N.-N. performed the research. W.L.A., B.U., and A.R.F. analyzed the data, and W.L.A. and A.R.F. wrote the article, which was later approved by all the other authors.
Glossary
- TCA
tricarboxylic acid
- OGDH
2-oxoglutarate dehydrogenase complex
- ABA
abscisic acid
- GA
gibberellin
- SGN
Sol Genomics Network
- CaMV
cauliflower mosaic virus
- DAA
d after anthesis
- PAM
pulse amplitude modulation
- ETR
electron transport rate
- PFD
photon flux density
- Fv/Fm
maximum variable fluorescence/maximum yield of fluorescence
- GC-MS
gas chromatography–mass spectrometry
- GABA
γ-amino butyric acid
References
- Aharoni N., Richmond A.E. (1978). Endogenous gibberellin and abscisic acid content as related to senescence of detached lettuce leaves. Plant Physiol. 62: 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ap Rees T.A., Beevers H. (1960). Pathways of glucose dissimilation in carrot slices. Plant Physiol. 35: 830–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo W.L., Ishizaki K., Nunes-Nesi A., Larson T.R., Tohge T., Krahnert I., Witt S., Obata T., Schauer N., Graham I.A., Leaver C.J., Fernie A.R. (2010). Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 22: 1549–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo W.L., Ishizaki K., Nunes-Nesi A., Tohge T., Larson T.R., Krahnert I., Balbo I., Witt S., Dörmann P., Graham I.A., Leaver C.J., Fernie A.R. (2011b). Analysis of a range of catabolic mutants provides evidence that phytanoyl-coenzyme A does not act as a substrate of the electron-transfer flavoprotein/electron-transfer flavoprotein:ubiquinone oxidoreductase complex in Arabidopsis during dark-induced senescence. Plant Physiol. 157: 55–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo W.L., Nunes-Nesi A., Nikoloski Z., Sweetlove L.J., Fernie A.R. (2012). Metabolic control and regulation of the tricarboxylic acid cycle in photosynthetic and heterotrophic plant tissues. Plant Cell Environ. 35: 1–21 [DOI] [PubMed] [Google Scholar]
- Araújo W.L., et al. (2011a). Antisense inhibition of the iron-sulfur subunit of succinate dehydrogenase enhances photosynthesis and growth in tomato via an organic acid-mediated effect on stomatal aperture. Plant Cell 23: 600–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo W.L., Nunes-Nesi A., Trenkamp S., Bunik V.I., Fernie A.R. (2008). Inhibition of 2-oxoglutarate dehydrogenase in potato tuber suggests the enzyme is limiting for respiration and confirms its importance in nitrogen assimilation. Plant Physiol. 148: 1782–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Back A., Richmond A.E. (1971). Interrelations between gibberellic acid, cytokinins and abscisic acid in retarding leaf senescence. Physiol. Plant. 24: 76–79 [Google Scholar]
- Bapat V.A., Trivedi P.K., Ghosh A., Sane V.A., Ganapathi T.R., Nath P. (2010). Ripening of fleshy fruit: Molecular insight and the role of ethylene. Biotechnol. Adv. 28: 94–107 [DOI] [PubMed] [Google Scholar]
- Barth C., De Tullio M., Conklin P.L. (2006). The role of ascorbic acid in the control of flowering time and the onset of senescence. J. Exp. Bot. 57: 1657–1665 [DOI] [PubMed] [Google Scholar]
- Bender-Machado L., Bäuerlein M., Carrari F., Schauer N., Lytovchenko A., Gibon Y., Kelly A.A., Loureiro M., Müller-Röber B., Willmitzer L., Fernie A.R. (2004). Expression of a yeast acetyl CoA hydrolase in the mitochondrion of tobacco plants inhibits growth and restricts photosynthesis. Plant Mol. Biol. 55: 645–662 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300 [Google Scholar]
- Bleecker A.B., Patterson S.E. (1997). Last exit: Senescence, abscission, and meristem arrest in Arabidopsis. Plant Cell 9: 1169–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangeon J., Sabar M., Gutierres S., Combettes B., Bove J., Gendy C., Chétrit P., Des Francs-Small C.C., Pla M., Vedel F., De Paepe R. (2000). Defective splicing of the first nad4 intron is associated with lack of several complex I subunits in the Nicotiana sylvestris NMS1 nuclear mutant. Plant J. 21: 269–280 [DOI] [PubMed] [Google Scholar]
- Bugos R.C., Chiang V.L., Zhang X.H., Campbell E.R., Podila G.K., Campbell W.H. (1995). RNA isolation from plant tissues recalcitrant to extraction in guanidine. Biotechniques 19: 734–737 [PubMed] [Google Scholar]
- Bunik V.I., Fernie A.R. (2009). Metabolic control exerted by the 2-oxoglutarate dehydrogenase reaction: A cross-kingdom comparison of the crossroad between energy production and nitrogen assimilation. Biochem. J. 422: 405–421 [DOI] [PubMed] [Google Scholar]
- Caldana C., Scheible W.R., Mueller-Roeber B., Ruzicic S. (2007). A quantitative RT-PCR platform for high-throughput expression profiling of 2500 rice transcription factors. Plant Methods 3: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson J., Leino M., Sohlberg J., Sundström J.F., Glimelius K. (2008). Mitochondrial regulation of flower development. Mitochondrion 8: 74–86 [DOI] [PubMed] [Google Scholar]
- Carrari F., Baxter C., Usadel B., Urbanczyk-Wochniak E., Zanor M.I., Nunes-Nesi A., Nikiforova V., Centero D., Ratzka A., Pauly M., Sweetlove L.J., Fernie A.R. (2006). Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiol. 142: 1380–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrari F., Fernie A.R., Iusem N.D. (2004). Heard it through the grapevine? ABA and sugar cross-talk: The ASR story. Trends Plant Sci. 9: 57–59 [DOI] [PubMed] [Google Scholar]
- Carrari F., Nunes-Nesi A., Gibon Y., Lytovchenko A., Loureiro M.E., Fernie A.R. (2003). Reduced expression of aconitase results in an enhanced rate of photosynthesis and marked shifts in carbon partitioning in illuminated leaves of wild species tomato. Plant Physiol. 133: 1322–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin T.Y., Beevers L. (1970). Changes in endogenous growth regulators in nasturtium leaves during senescence. Planta 92: 178–188 [DOI] [PubMed] [Google Scholar]
- Cornu S., Pireaux J.C., Gerard J., Dizengremel P. (1996). NAD(P)-dependent isocitrate dehydrogenases in mitochondria purified from Picea abies seedlings. Physiol. Plant. 96: 312–318 [Google Scholar]
- Coruzzi G., Last R. (2000). Amino acids. In Biochemistry and Molecular Biology of Plants, B.B. Buchanan, W. Gruissem, and R.L. Jones, eds (Rockville, MD: American Society of Plant Biologists), pp. 358–410
- Csukasi F., Osorio S., Gutierrez J.R., Kitamura J., Giavalisco P., Nakajima M., Fernie A.R., Rathjen J.P., Botella M.A., Valpuesta V., Medina-Escobar N. (2011). Gibberellin biosynthesis and signalling during development of the strawberry receptacle. New Phytol. 191: 376–390 [DOI] [PubMed] [Google Scholar]
- Czechowski T., Bari R.P., Stitt M., Scheible W.R., Udvardi M.K. (2004). Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: Unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J. 38: 366–379 [DOI] [PubMed] [Google Scholar]
- de Oliveira Dal’Molin C.G., Quek L.E., Palfreyman R.W., Brumbley S.M., Nielsen L.K. (2010). AraGEM, a genome-scale reconstruction of the primary metabolic network in Arabidopsis. Plant Physiol. 152: 579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dry I.B., Wiskich J.T. (1985). Inhibition of 2-oxoglutarate oxidation in plant mitochondria by pyruvate. Biochem. Biophys. Res. Commun. 133: 397–403 [DOI] [PubMed] [Google Scholar]
- Dubois F., Brugière N., Sangwan R.S., Hirel B. (1996). Localization of tobacco cytosolic glutamine synthetase enzymes and the corresponding transcripts shows organ- and cell-specific patterns of protein synthesis and gene expression. Plant Mol. Biol. 31: 803–817 [DOI] [PubMed] [Google Scholar]
- Dutilleul C., Lelarge C., Prioul J.L., De Paepe R., Foyer C.H., Noctor G. (2005). Mitochondria-driven changes in leaf NAD status exert a crucial influence on the control of nitrate assimilation and the integration of carbon and nitrogen metabolism. Plant Physiol. 139: 64–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fait A., Fromm H., Walter D., Galili G., Fernie A.R. (2008). Highway or byway: The metabolic role of the GABA shunt in plants. Trends Plant Sci. 13: 14–19 [DOI] [PubMed] [Google Scholar]
- Fernie A.R., Aharoni A., Willmitzer L., Stitt M., Tohge T., Kopka J., Carroll A.J., Saito K., Fraser P.D., DeLuca V. (2011). Recommendations for reporting metabolite data. Plant Cell 23: 2477–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie A.R., Carrari F., Sweetlove L.J. (2004b). Respiratory metabolism: Glycolysis, the TCA cycle and mitochondrial electron transport. Curr. Opin. Plant Biol. 7: 254–261 [DOI] [PubMed] [Google Scholar]
- Fernie A.R., Roscher A., Ratcliffe R.G., Kruger N.J. (2001). Fructose 2,6-bisphosphate activates pyrophosphate:fructose-6-phosphate 1-phosphotransferase and increases triose phosphate to hexose phosphate cycling in heterotrophic cells. Planta 212: 250–263 [DOI] [PubMed] [Google Scholar]
- Fernie A.R., Trethewey R.N., Krotzky A., Willmitzer L. (2004a). Metabolic profiling: From diagnostics to systems biology. Nat. Rev. Mol. Cell Biol. 5: 1–7 [DOI] [PubMed] [Google Scholar]
- Foyer C.H., Parry M., Noctor G. (2003). Markers and signals associated with nitrogen assimilation in higher plants. J. Exp. Bot. 54: 585–593 [DOI] [PubMed] [Google Scholar]
- Gálvez S., Lancien M., Hodges M. (1999). Are isocitrate dehydrogenases and 2-oxoglutarate involved in the regulation of glutamate synthesis? Trends Plant Sci. 4: 484–490 [DOI] [PubMed] [Google Scholar]
- Gan S., Amasino R.M. (1997). Making sense of senescence: Molecular genetic regulation and manipulation of leaf senescence. Plant Physiol. 113: 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmier M., Carroll A.J., Delannoy E., Vallet C., Day D.A., Small I.D., Millar A.H. (2008). Complex I dysfunction redirects cellular and mitochondrial metabolism in Arabidopsis. Plant Physiol. 148: 1324–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaude N., Bréhélin C., Tischendorf G., Kessler F., Dörmann P. (2007). Nitrogen deficiency in Arabidopsis affects galactolipid composition and gene expression and results in accumulation of fatty acid phytyl esters. Plant J. 49: 729–739 [DOI] [PubMed] [Google Scholar]
- Gauthier P.P.G., Bligny R., Gout E., Mahé A., Nogués S., Hodges M., Tcherkez G.G.B. (2010). In folio isotopic tracing demonstrates that nitrogen assimilation into glutamate is mostly independent from current CO2 assimilation in illuminated leaves of Brassica napus. New Phytol. 185: 988–999 [DOI] [PubMed] [Google Scholar]
- Gibon Y., Blaesing O.E., Hannemann J., Carillo P., Höhne M., Hendriks J.H.M., Palacios N., Cross J., Selbig J., Stitt M. (2004). A robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: Comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16: 3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldwaithe J.J. (1972). Further studies of hormone regulated senescence in Rumex leaf tissue. In Plant Growth Substances, D.J. Carr, ed (Berlin: Springer), pp. 581–588
- Goldthwaite J.J., Laetsch W.M. (1968). Control of senescence in Rumex leaf discs by gibberellic acid. Plant Physiol. 43: 1855–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbic V., Bleecker A.B. (1995). Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J. 8: 595–602 [Google Scholar]
- Greenboim-Wainberg Y., Maymon I., Borochov R., Alvarez J., Olszewski N., Ori N., Eshed Y., Weiss D. (2005). Cross talk between gibberellin and cytokinin: The Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell 17: 92–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L., Jones A.D., Last R.L. (2010). Broad connections in the Arabidopsis seed metabolic network revealed by metabolite profiling of an amino acid catabolism mutant. Plant J. 61: 579–590 [DOI] [PubMed] [Google Scholar]
- Gutierres S., Sabar M., Lelandais C., Chetrit P., Diolez P., Degand H., Boutry M., Vedel F., de Kouchkovsky Y., De Paepe R. (1997). Lack of mitochondrial and nuclear-encoded subunits of complex I and alteration of the respiratory chain in Nicotiana sylvestris mitochondrial deletion mutants. Proc. Natl. Acad. Sci. USA 94: 3436–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P., Kamiya Y. (1997). Gibberellin biosynthesis: Enzymes, genes and their regulation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 431–460 [DOI] [PubMed] [Google Scholar]
- Higgins D.G., Sharp P.M. (1988). CLUSTAL: A package for performing multiple sequence alignment on a microcomputer. Gene 73: 237–244 [DOI] [PubMed] [Google Scholar]
- Hodges M. (2002). Enzyme redundancy and the importance of 2-oxoglutarate in plant ammonium assimilation. J. Exp. Bot. 53: 905–916 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S. (2006). Chlorophyll degradation during senescence. Annu. Rev. Plant Biol. 57: 55–77 [DOI] [PubMed] [Google Scholar]
- Hung K.T., Kao C.H. (2004). Hydrogen peroxide is necessary for abscisic acid-induced senescence of rice leaves. J. Plant Physiol. 161: 1347–1357 [DOI] [PubMed] [Google Scholar]
- Hurry V., Igamberdiev A.U., Keerberg O., Pärnik T., Atkin O.K., Zaragoza-Castells J., Gardeström P. (2005). Respiration in photosynthetic cells: Gas exchange components, interactions with photorespiration and the operation of mitochondria in the light. In Plant Respiration: From Cell to Ecosystem, Vol. 18, Advances in Photosynthesis and Respiration Series, H. Lambers and M. Ribas-Carbo, eds (Dordrecht, The Netherlands: Springer), pp. 43–61
- Igamberdiev A.U., Gardeström P. (2003). Regulation of NAD- and NADP-dependent isocitrate dehydrogenases by reduction levels of pyridine nucleotides in mitochondria and cytosol of pea leaves. Biochim. Biophys. Acta 1606: 117–125 [DOI] [PubMed] [Google Scholar]
- Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Ishizaki K., Larson T.R., Schauer N., Fernie A.R., Graham I.A., Leaver C.J. (2005). The critical role of Arabidopsis electron-transfer flavoprotein:ubiquinone oxidoreductase during dark-induced starvation. Plant Cell 17: 2587–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki K., Schauer N., Larson T.R., Graham I.A., Fernie A.R., Leaver C.J. (2006). The mitochondrial electron transfer flavoprotein complex is essential for survival of Arabidopsis in extended darkness. Plant J. 47: 751–760 [DOI] [PubMed] [Google Scholar]
- Jing H.C., Sturre M.J., Hille J., Dijkwel P.P. (2002). Arabidopsis onset of leaf death mutants identify a regulatory pathway controlling leaf senescence. Plant J. 32: 51–63 [DOI] [PubMed] [Google Scholar]
- Kappers I.F., Jordi W., Maas F.M., Stoopen G.M., van der Plas L.H.W. (1998). Gibberellin and phytochrome control senescence in Alstromeria leaves independently. Physiol. Plant. 103: 91–98 [Google Scholar]
- Karpova O.V., Kuzmin E.V., Elthon T.E., Newton K.J. (2002). Differential expression of alternative oxidase genes in maize mitochondrial mutants. Plant Cell 14: 3271–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling P.L., Wood J.R., Tyson R.H., Bridges I.G. (1988). Starch biosynthesis in developing wheat grain: Evidence against the direct involvement of triose phosphates in the metabolic pathway. Plant Physiol. 87: 311–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange T., Schweimer A., Ward D.A., Hedden P., Graebe J.E. (1994). Separation and characterization of three 2-oxoglutarate-dependent dioxygenases from Cucurbita maxima L. endosperm involved in gibberellin biosynthesis. Planta 195: 98–107 [Google Scholar]
- León G., Holuigue L., Jordana X. (2007). Mitochondrial complex II is essential for gametophyte development in Arabidopsis. Plant Physiol. 143: 1534–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- León P., Sheen J. (2003). Sugar and hormone connections. Trends Plant Sci. 8: 110–116 [DOI] [PubMed] [Google Scholar]
- Less H., Galili G. (2008). Principal transcriptional programs regulating plant amino acid metabolism in response to abiotic stresses. Plant Physiol. 147: 316–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim P.O., Kim H.J., Nam H.G. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58: 115–136 [DOI] [PubMed] [Google Scholar]
- Lim P.O., Woo H.R., Nam H.G. (2003). Molecular genetics of leaf senescence in Arabidopsis. Trends Plant Sci. 8: 272–278 [DOI] [PubMed] [Google Scholar]
- Lin Z., Zhong S., Grierson D. (2009). Recent advances in ethylene research. J. Exp. Bot. 60: 3311–3336 [DOI] [PubMed] [Google Scholar]
- Linke B., Börner T. (2005). Mitochondrial effects on flower and pollen development. Mitochondrion 5: 389–402 [DOI] [PubMed] [Google Scholar]
- Lisec J., Schauer N., Kopka J., Willmitzer L., Fernie A.R. (2006). Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat. Protoc. 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Liu X.J., Prat S., Willmitzer L., Frommer W.B. (1990). cis regulatory elements directing tuber-specific and sucrose-inducible expression of a chimeric class I patatin promoter/GUS-gene fusion. Mol. Gen. Genet. 223: 401–406 [DOI] [PubMed] [Google Scholar]
- Liu Y.J., Nunes-Nesi A., Wallström S.V., Lager I., Michalecka A.M., Norberg F.E., Widell S., Fredlund K.M., Fernie A.R., Rasmusson A.G. (2009). A redox-mediated modulation of stem bolting in transgenic Nicotiana sylvestris differentially expressing the external mitochondrial NADPH dehydrogenase. Plant Physiol. 150: 1248–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M., et al. (2010). Robin: An intuitive wizard application for R-based expression microarray quality assessment and analysis. Plant Physiol. 153: 642–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E., Povero G., Novi G., Solfanelli C., Alpi A., Perata P. (2008). Gibberellins, jasmonate and abscisic acid modulate the sucrose-induced expression of anthocyanin biosynthetic genes in Arabidopsis. New Phytol. 179: 1004–1016 [DOI] [PubMed] [Google Scholar]
- Lytovchenko A., Sweetlove L.J., Pauly M., Fernie A.R. (2002). The influence of cytosolic phosphoglucomutase on photosynthetic carbohydrate metabolism. Planta 215: 1013–1021 [DOI] [PubMed] [Google Scholar]
- Manos P.J., Goldwaithe J.J. (1975). A kinetic analysis of the effects of gibberellic acid, zeatin, and abscisic acid on leaf tissue senescence in Rumex. Plant Physiol. 55: 192–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marienfeld J.R., Newton K.J. (1994). The maize NCS2 abnormal growth mutant has a chimeric nad4-nad7 mitochondrial gene and is associated with reduced complex I function. Genetics 138: 855–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux C., Valadier M.H., Brugière N., Morot-Gaudry J.F., Hirel B. (2000). Characterization of the sink/source transition in tobacco ( Nicotiana tabacum L.) shoots in relation to nitrogen management and leaf senescence. Planta 211: 510–518 [DOI] [PubMed] [Google Scholar]
- Masclaux-Daubresse C., Valadier M.-H., Carrayol E., Reisdorf-Cren M., Hirel B. (2002). Diurnal changes in the expression of glutamate dehydrogenase and nitrate reductase are involved in the C/N balance of tobacco source leaves. Plant Cell Environ. 25: 1451–1462 [Google Scholar]
- Millar A.H., Eubel H., Jänsch L., Kruft V., Heazlewood J.L., Braun H.-P. (2004). Mitochondrial cytochrome c oxidase and succinate dehydrogenase complexes contain plant specific subunits. Plant Mol. Biol. 56: 77–90 [DOI] [PubMed] [Google Scholar]
- Moore B., Zhou L., Rolland F., Hall Q., Cheng W.H., Liu Y.X., Hwang I., Jones T., Sheen J. (2003). Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300: 332–336 [DOI] [PubMed] [Google Scholar]
- Nichols B.J., Rigoulet M., Denton R.M. (1994). Comparison of the effects of Ca2+, adenine nucleotides and pH on the kinetic properties of mitochondrial NAD(+)-isocitrate dehydrogenase and oxoglutarate dehydrogenase from the yeast Saccharomyces cerevisiae and rat heart. Biochem. J. 303: 461–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G., Dutilleul C., De Paepe R., Foyer C.H. (2004). Use of mitochondrial electron transport mutants to evaluate the effects of redox state on photosynthesis, stress tolerance and the integration of carbon/nitrogen metabolism. J. Exp. Bot. 55: 49–57 [DOI] [PubMed] [Google Scholar]
- Noguchi K., Yoshida K. (2008). Interaction between photosynthesis and respiration in illuminated leaves. Mitochondrion 8: 87–99 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A., Araújo W.L., Fernie A.R. (2011). Targeting mitochondrial metabolism and machinery as a means to enhance photosynthesis. Plant Physiol. 155: 101–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A., Carrari F., Gibon Y., Sulpice R., Lytovchenko A., Fisahn J., Graham J., Ratcliffe R.G., Sweetlove L.J., Fernie A.R. (2007a). Deficiency of mitochondrial fumarase activity in tomato plants impairs photosynthesis via an effect on stomatal function. Plant J. 50: 1093–1106 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A., Carrari F., Lytovchenko A., Smith A.M., Loureiro M.E., Ratcliffe R.G., Sweetlove L.J., Fernie A.R. (2005). Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol. 137: 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A., Sweetlove L.J., Fernie A.R. (2007b). Operation and function of the tricarboxlic acid cycle in the illuminated leaf. Physiol. Plant. 129: 45–56 [Google Scholar]
- Oh S.A., Lee S.Y., Chung I.K., Lee C.H., Nam H.G. (1996). A senescence-associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant Mol. Biol. 30: 739–754 [DOI] [PubMed] [Google Scholar]
- Picault N., Palmieri L., Pisano I., Hodges M., Palmieri F. (2002). Identification of a novel transporter for dicarboxylates and tricarboxylates in plant mitochondria. Bacterial expression, reconstitution, functional characterization, and tissue distribution. J. Biol. Chem. 277: 24204–24211 [DOI] [PubMed] [Google Scholar]
- Pineau B., Mathieu C., Gérard-Hirne C., De Paepe R., Chétrit P. (2005). Targeting the NAD7 subunit to mitochondria restores a functional complex I and a wild type phenotype in the Nicotiana sylvestris CMS II mutant lacking nad7. J. Biol. Chem. 280: 25994–26001 [DOI] [PubMed] [Google Scholar]
- Poolman M.G., Miguet L., Sweetlove L.J., Fell D.A. (2009). A genome-scale metabolic model of Arabidopsis and some of its properties. Plant Physiol. 151: 1570–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pružinská A., Tanner G., Aubry S., Anders I., Moser S., Müller T., Ongania K.-H., Kräutler B., Youn J.-Y., Liljegren S.J., Hörtensteiner S. (2005). Chlorophyll breakdown in senescent Arabidopsis leaves: Characterization of chlorophyll catabolites and chlorophyll catabolic enzymes involved in the degreening reaction. Plant Physiol. 139: 52–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra A.S., Padmasree K. (2003). Beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Trends Plant Sci. 8: 546–553 [DOI] [PubMed] [Google Scholar]
- Roessner U., Luedemann A., Brust D., Fiehn O., Linke T., Willmitzer L., Fernie A.R. (2001). Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13: 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner-Tunali U., Hegemann B., Lytovchenko A., Carrari F., Bruedigam C., Granot D., Fernie A.R. (2003). Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol. 133: 84–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner-Tunali U., Liu J., Leisse A., Balbo I., Perez-Melis A., Willmitzer L., Fernie A.R. (2004). Kinetics of labelling of organic and amino acids in potato tubers by gas chromatography-mass spectrometry following incubation in (13)C labelled isotopes. Plant J. 39: 668–679 [DOI] [PubMed] [Google Scholar]
- Roschzttardtz H., Fuentes I., Vásquez M., Corvalán C., León G., Gómez I., Araya A., Holuigue L., Vicente-Carbajosa J., Jordana X. (2009). A nuclear gene encoding the iron-sulfur subunit of mitochondrial complex II is regulated by B3 domain transcription factors during seed development in Arabidopsis. Plant Physiol. 150: 84–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenvasser S., Mayak S., Friedman H. (2006). Increase in reactive oxygen species (ROS) and in senescence-associated gene transcript (SAG) levels during dark-induced senescence of Pelargonium cuttings, and the effect of gibberellic acid. Plant Sci. 170: 873–879 [Google Scholar]
- Rossel J.B., Wilson P.B., Hussain D., Woo N.S., Gordon M.J., Mewett O.P., Howell K.A., Whelan J., Kazan K., Pogson B.J. (2007). Systemic and intracellular responses to photooxidative stress in Arabidopsis. Plant Cell 19: 4091–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabar M., De Paepe R., de Kouchkovsky Y. (2000). Complex I impairment, respiratory compensations, and photosynthetic decrease in nuclear and mitochondrial male sterile mutants of Nicotiana sylvestris. Plant Physiol. 124: 1239–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Schauer N., Steinhauser D., Strelkov S., Schomburg D., Allison G., Moritz T., Lundgren K., Roessner-Tunali U., Forbes M.G., Willmitzer L., Fernie A.R., Kopka J. (2005). GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett. 579: 1332–1337 [DOI] [PubMed] [Google Scholar]
- Scheibe R., Backhausen J.E., Emmerlich V., Holtgrefe S. (2005). Strategies to maintain redox homeostasis during photosynthesis under changing conditions. J. Exp. Bot. 56: 1481–1489 [DOI] [PubMed] [Google Scholar]
- Schippers J.H.M., Nunes-Nesi A., Apetrei R., Hille J., Fernie A.R., Dijkwel P.P. (2008). The Arabidopsis onset of leaf death5 mutation of quinolinate synthase affects nicotinamide adenine dinucleotide biosynthesis and causes early ageing. Plant Cell 20: 2909–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P.S., Wise R.P. (1998). The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 3: 175–180 [Google Scholar]
- Serrani J.C., Carrera E., Ruiz-Rivero O., Gallego-Giraldo L., Peres L.E.P., García-Martínez J.L. (2010). Inhibition of auxin transport from the ovary or from the apical shoot induces parthenocarpic fruit-set in tomato mediated by gibberellins. Plant Physiol. 153: 851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienkiewicz-Porzucek A., Nunes-Nesi A., Sulpice R., Lisec J., Centeno D.C., Carillo P., Leisse A., Urbanczyk-Wochniak E., Fernie A.R. (2008). Mild reductions in mitochondrial citrate synthase activity result in a compromised nitrate assimilation and reduced leaf pigmentation but have no effect on photosynthetic performance or growth. Plant Physiol. 147: 115–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienkiewicz-Porzucek A., Sulpice R., Osorio S., Krahnert I., Leisse A., Urbanczyk-Wochniak E., Hodges M., Fernie A.R., Nunes-Nesi A. (2010). Mild reductions in mitochondrial NAD-dependent isocitrate dehydrogenase activity result in altered nitrate assimilation and pigmentation but do not impact growth. Mol Plant 3: 156–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. (2004). Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol 3: Article3 [DOI] [PubMed] [Google Scholar]
- Studart-Guimarães C., Fait A., Nunes-Nesi A., Carrari F., Usadel B., Fernie A.R. (2007). Reduced expression of succinyl-coenzyme A ligase can be compensated for by up-regulation of the gamma-aminobutyrate shunt in illuminated tomato leaves. Plant Physiol. 145: 626–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove L.J., Beard K.F.M., Nunes-Nesi A., Fernie A.R., Ratcliffe R.G. (2010). Not just a circle: Flux modes in the plant TCA cycle. Trends Plant Sci. 15: 462–470 [DOI] [PubMed] [Google Scholar]
- Sweetlove L.J., Fait A., Nunes-Nesi A., Williams T., Fernie A.R. (2007). The mitochondrion: An integration point in cellular metabolism and signalling. Crit. Rev. Plant Sci. 26: 17–43 [Google Scholar]
- Sweetlove L.J., Fernie A.R. (2005). Regulation of metabolic networks: Understanding metabolic complexity in the systems biology era. New Phytol. 168: 9–24 [DOI] [PubMed] [Google Scholar]
- Sweetlove L.J., Lytovchenko A., Morgan M., Nunes-Nesi A., Taylor N.L., Baxter C.J., Eickmeier I., Fernie A.R. (2006). Mitochondrial uncoupling protein is required for efficient photosynthesis. Proc. Natl. Acad. Sci. USA 103: 19587–19592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tauberger E., Fernie A.R., Emmermann M., Renz A., Kossmann J., Willmitzer L., Trethewey R.N. (2000). Antisense inhibition of plastidial phosphoglucomutase provides compelling evidence that potato tuber amyloplasts import carbon from the cytosol in the form of glucose-6-phosphate. Plant J. 23: 43–53 [DOI] [PubMed] [Google Scholar]
- Taylor L., Nunes-Nesi A., Parsley K., Leiss A., Leach G., Coates S., Wingler A., Fernie A.R., Hibberd J.M. (2010). Cytosolic pyruvate,orthophosphate dikinase functions in nitrogen remobilization during leaf senescence and limits individual seed growth and nitrogen content. Plant J. 62: 641–652 [DOI] [PubMed] [Google Scholar]
- Tieman D., Taylor M., Schauer N., Fernie A.R., Hanson A.D., Klee H.J. (2006). Tomato aromatic amino acid decarboxylases participate in synthesis of the flavor volatiles 2-phenylethanol and 2-phenylacetaldehyde. Proc. Natl. Acad. Sci. USA 103: 8287–8292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkez G., Mahé A., Gauthier P., Mauve C., Gout E., Bligny R., Cornic G., Hodges M. (2009). In folio respiratory fluxomics revealed by 13C isotopic labeling and H/D isotope effects highlight the noncyclic nature of the tricarboxylic acid “cycle” in illuminated leaves. Plant Physiol. 151: 620–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B., Nagel A., Steinhauser D., Gibon Y., Bläsing O.E., Redestig H., Sreenivasulu N., Krall L., Hannah M.A., Poree F., Fernie A.R., Stitt M. (2006). PageMan: An interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoeven R., Ronning C., Giovannoni J., Martin G., Tanksley S. (2002). Deductions about the number, organization, and evolution of genes in the tomato genome based on analysis of a large expressed sequence tag collection and selective genomic sequencing. Plant Cell 14: 1441–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Merwe M.J., Osorio S., Araújo W.L., Balbo I., Nunes-Nesi A., Maximova E., Carrari F., Bunik V.I., Persson S., Fernie A.R. (2010). Tricarboxylic acid cycle activity regulates tomato root growth via effects on secondary cell wall production. Plant Physiol. 153: 611–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Merwe M.J., Osorio S., Moritz T., Nunes-Nesi A., Fernie A.R. (2009). Decreased mitochondrial activities of malate dehydrogenase and fumarase in tomato lead to altered root growth and architecture via diverse mechanisms. Plant Physiol. 149: 653–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn W.G. (2004). Is petal senescence due to sugar starvation? Plant Physiol. 134: 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn W.G. (2008). Is the onset of senescence in leaf cells of intact plants due to low or high sugar levels? J. Exp. Bot. 59: 1963–1972 [DOI] [PubMed] [Google Scholar]
- Wang S., Liu J., Feng Y., Niu X., Giovannoni J., Liu Y. (2008). Altered plastid levels and potential for improved fruit nutrient content by downregulation of the tomato DDB1-interacting protein CUL4. Plant J. 55: 89–103 [DOI] [PubMed] [Google Scholar]
- Watson J.M., Fusaro A.F., Wang M.B., Waterhouse P.M. (2005). RNA silencing platforms in plants. FEBS Lett. 579: 5982–5987 [DOI] [PubMed] [Google Scholar]
- Whyte P., Luckwill L.C. (1966). A sensitive bioassay for gibberellins based on retardation of leaf senescence in Rumex obtusifolious. Nature 210: 1360 [Google Scholar]
- Wingler A., Purdy S., MacLean J.A., Pourtau N. (2006). The role of sugars in integrating environmental signals during the regulation of leaf senescence. J. Exp. Bot. 57: 391–399 [DOI] [PubMed] [Google Scholar]
- Wingler A., Roitsch T. (2008). Metabolic regulation of leaf senescence: Interactions of sugar signalling with biotic and abiotic stress responses. Plant Biol (Stuttg) 10 (suppl. 1): 50–62 [DOI] [PubMed] [Google Scholar]
- Xu P., Zhang Y.J., Kang L., Roossinck M.J., Mysore K.S. (2006). Computational estimation and experimental verification of off-target silencing during posttranscriptional gene silencing in plants. Plant Physiol. 142: 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.F. (1987). The role of ethylene and ethylene biosynthesis in fruit ripening. In Plant Senescence: Its Biochemistry and Physiology, W.W. Thomson, , E.A. Nothnagel, and R.C. Huffaker, eds (Rockville, MD: American Society of Plant Physiologists), pp. 156–165
- Zanor M.I., et al. (2009). RNA interference of LIN5 in tomato confirms its role in controlling Brix content, uncovers the influence of sugars on the levels of fruit hormones, and demonstrates the importance of sucrose cleavage for normal fruit development and fertility. Plant Physiol. 150: 1204–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Galili G. (2003). Increased lysine synthesis coupled with a knockout of its catabolism synergistically boosts lysine content and also transregulates the metabolism of other amino acids in Arabidopsis seeds. Plant Cell 15: 845–853 [DOI] [PMC free article] [PubMed] [Google Scholar]