The morning-acting clock factor TIME FOR COFFEE (TIC) was found to work in the jasmonate (JA) hormone pathway through a protein-depletion role at the positive JA regulator MYC2. The Arabidopsis thaliana circadian clock thus allows JA hormone responses to be phased in the morning.
Abstract
Plants are confronted with predictable daily biotic and abiotic stresses that result from the day–night cycle. The circadian clock provides an anticipation mechanism to respond to these daily stress signals to increase fitness. Jasmonate (JA) is a phytohormone that mediates various growth and stress responses. Here, we found that the circadian-clock component TIME FOR COFFEE (TIC) acts as a negative factor in the JA-signaling pathway. We showed that the tic mutant is hypersensitive to growth-repressive effects of JA and displays altered JA-regulated gene expression. TIC was found to interact with MYC2, a key transcription factor of JA signaling. From this, we discovered that the circadian clock rhythmically regulates JA signaling. TIC is a key determinant in this circadian-gated process, and as a result, the tic mutant is defective in rhythmic JA responses to pathogen infection. TIC acts here by inhibiting MYC2 protein accumulation and by controlling the transcriptional repression of CORONATINE INSENSITIVE1 in an evening-phase–specific manner. Taken together, we propose that TIC acts as an output component of the circadian oscillator to influence JA signaling directly.
INTRODUCTION
Plants face predictable daily environmental changes that generate rhythmic biotic and abiotic stresses within a 24-h period. The circadian clock controls the expression of ∼30% of the Arabidopsis thaliana transcriptome, which leads to regulation of diverse metabolic processes and developmental programs in response to daily environmental signals (Davis and Millar, 2001; Covington et al., 2008; Harmer, 2009; Sanchez et al., 2011). The plant circadian clock consists of an input pathway, a core oscillator, and output responses. The components of the input pathway recognize environmental signals, such as light intensity, wavelength and duration, and temperature (Salome and McClung, 2005). Perceived environmental signals are then relayed to the core oscillator for resetting or fine-tuning of the clock. The core oscillator produces a ∼24-h rhythmic periodicity to direct clock output factors. These output factors regulate diverse plant developmental and physiological responses, such as rhythmic hypocotyl growth, photosynthesis, stomata opening, and seasonal flowering-time regulation (Suárez-López et al., 2001; Nozue et al., 2007; Michael et al., 2008; Harmer, 2009). Clock-mediated synchronization of internal growth to external environmental changes is crucial for plant fitness (Dodd et al., 2005).
One important pathway for circadian outputs is rhythmic hormone signaling. A major part of phytohormone production, biosynthesis, and response gene expression oscillates under the control of diurnal signals and/or the circadian clock (Thain et al., 2004; Nováková et al., 2005; Salomé et al., 2006; Covington and Harmer, 2007; Yin et al. 2007; Covington et al., 2008; Michael et al., 2008; Mizuno and Yamashino, 2008; Robertson et al., 2009; Goodspeed et al., 2012). Thereby, an array of hormone-signaling systems function as key circadian output pathways. For example, the clock influences auxin signaling in a time-of-day–specific manner, and this leads to rhythmic hypocotyl growth under diurnal conditions (Covington and Harmer, 2007; Rawat et al., 2009). Another example can be seen with the circadian evening factor, TIMING OF CAB EXPRESSION1 (TOC1). It regulates rhythmic abscisic acid (ABA) responses by repressing transcription of the proposed ABA receptor ABAR; thus, toc1 mutants show altered ABA responses (Legnaioli et al., 2009). Recent studies also revealed that gibberellic acid (GA) signaling is gated by the circadian clock, leading to maximal GA sensitivity during the night phase (Arana et al., 2011). Together, hormone-regulated developmental processes and stress signaling are clear targets of clock outputs.
Hormone signaling can act as a clock input factor. Several hormone-signaling systems modulate the core oscillator. For example, auxin, ABA, and cytokinin alter circadian amplitude, period, and phase, respectively (Hanano et al., 2006). Furthermore, ABA induces the expression of the clock evening element TOC1 in a daytime-specific manner (Legnaioli et al., 2009). Although studies of cross-regulation between the circadian clock and hormone pathways have recently started to emerge, the mechanistic basis for such relationships awaits elucidation.
Jasmonates (JAs) regulate diverse aspects of plant life, from developmental growth processes to physiological responses (Katsir et al., 2008a; Kazan and Manners, 2008; Browse, 2009). Jasmonic acid, its conjugates, and its precursors are oxylipin-signaling molecules. These are collectively termed the JAs. The most biologically active form of JA was reported to be jasmonyl-isoleucine (JA-Ile) (Thines et al., 2007). The bacterial pathogen Pseudomonas syringae produces a functional and structural homolog of JA-Ile called coronatine (COR). This bacterial phytotoxin mimics active JA-Ile in plant tissues and thus alters certain defense responses (Bender et al., 1999; Katsir et al., 2008b). A key player in JA signaling is CORONATINE INSENSITIVE1 (COI1), an F-box protein that forms the E3 ubiquitin ligase SCFCOI1 complex (Xie et al., 1998; Devoto et al., 2002). COI1 binds JA-Ile in cooperation with JAZMONATE ZIM-DOMAIN (JAZ) proteins (Thines et al., 2007; Katsir et al., 2008b). Recent structural studies found that a protein complex of COI1 and JAZ with inositol pentakisphosphate is the complete JA receptor (Sheard et al., 2010; Mosblech et al., 2011). JAZ proteins are JA-signaling repressors, and 12 family members are found in Arabidopsis. In the absence of JA, the JAZs interact with MYC2, a positive transcription factor in JA signaling, and repress MYC2 activity by recruiting the transcription repressors NOVEL INTERACTOR OF JAZ (NINJA) and TOPLESS (TPL)/TPL-RELATED PROTEINS (TPRs) (Pauwels et al., 2010). In the presence of JA, JAZ proteins are ubiquitinated by SCFCOI1 and subsequently degraded by the 26S proteasome (Chini et al., 2007; Thines et al., 2007). Consequently, active MYC2 is released from repressors to induce JA-response gene expression (Lorenzo et al., 2004; Dombrecht et al., 2007).
MYC2 encodes a bHLH motif–containing transcription factor (Abe et al., 2003; Lorenzo et al., 2004). myc2 mutants are less sensitive to JA, in line with MYC2 being a positive regulator in the JA pathway (Lorenzo et al., 2004). MYC2 expression is induced by JA and promotes wound-response gene expression and represses pathogen-response gene expression in a JA-dependent manner (Lorenzo et al., 2004). In Arabidopsis, two genes annotated as MYC3 and MYC4 have sequence and functional similarity to MYC2. Activities of MYC2, MYC3, and MYC4 in JA signaling are partially overlapping (Abe et al., 2003; Fernández-Calvo et al., 2011; Niu et al., 2011). The different spatial expression pattern of these three genes directs distinct regulatory functions. For example, MYC2 is highly expressed in root tissue, whereas the expression of MYC3 and MYC4 is limited in this tissue. Therefore, MYC2 has a dominant role in the JA response in roots (Fernández-Calvo et al., 2011; Niu et al., 2011).
TIME FOR COFFEE (TIC) was identified as a circadian-clock regulator necessary for maintaining circadian period and amplitude (Hall et al., 2003; Ding et al., 2007). tic mutants display a short period with reduced amplitude under free-running conditions (Ding et al., 2007), and TIC was found to reset the core oscillator around the mid–late-night phase (Hall et al., 2003). TIC protein has been shown to localize to the nucleus. The biochemical activity of TIC is currently unknown, because TIC does not have known functional domains in its primary structure to predict its action. To elucidate a functional role and molecular mechanism for TIC, we identified TIC-interacting proteins and explored their functional relationship. Here, we have found MYC2 as a TIC-interacting protein and discovered a previously unknown role of TIC in JA signaling. We established that TIC acts as a negative regulator in JA signaling by repressing MYC2 protein accumulation. In addition, we defined a signaling connection between the circadian clock and the JA pathway and found that TIC plays a key role in clock-gated JA responses.
RESULTS
MYC2 Protein Interacts with TIC in the Nucleus
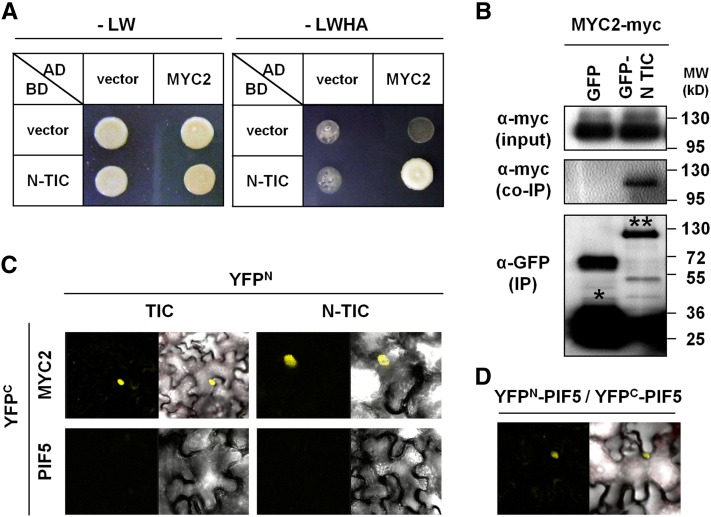
As a first step to investigate the molecular function of TIC, we identified TIC interaction partners in a yeast two-hybrid screen. Because of the protein size of the full-length TIC (1555 amino acids, 165 kDa) and lack of known motifs in its primary structure, we first used its 564-amino acid N terminus as bait. Among several TIC-interacting protein candidates, MYC2 was most frequently identified. To confirm this, we generated constructs with full-length MYC2 and N terminus TIC and repeated the yeast two-hybrid assay. N-TIC and MYC2 constructs enabled yeast growth, suggesting a direct protein–protein interaction (Figure 1A). We next tested the interaction between N-TIC and MYC2 in planta by performing a coimmunoprecipitation assay. Green fluorescent protein (GFP)-fused N terminus TIC (amino acids 1 to 564) and myc-epitope–tagged MYC2 were transiently expressed in Nicotiana benthamiana, and N-TIC was immunoprecipitated with GFP-TRAP beads. MYC2 specifically precipitated with the N terminus of TIC but did not coprecipitate with GFP alone (Figure 1B). For further analysis of protein interaction, we performed a biomolecular fluorescence complementation (BiFC) assay. The N terminus of yellow fluorescent protein (YFP) was fused to full-length TIC (YFPN-TIC) or the N-terminal portion of TIC (YFPN-N TIC), and the YFP C terminus was fused to MYC2 (YFPC-MYC2) or PIF5 (YFPC-PIF5) as a control. Fusion proteins were expressed in N. benthamiana, and interaction was determined by measuring YFP fluorescence under a confocal microscope. Both full-length TIC and N-terminal TIC were found to interact with MYC2, and this was exclusively in nuclei (Figure 1C). A TIC–MYC2 interaction was also found with the reciprocal YFPN-MYC2 and YFPC-TIC combination (see Supplemental Figure 1 online). Association between TIC and MYC2 was specific, because TIC did not interact with another bHLH protein, PIF5 (Figure 1C; see Supplemental Figure 1 online). As a positive experimental control, we confirmed by BiFC assay that PIF5 can successfully form a homodimer under the same experimental conditions (Figure 1D). Taken together, these results demonstrate that TIC physically interacts with MYC2.
Figure 1.
TIC Specifically Interacts with MYC2 in the Nucleus.
(A) Yeast two-hybrid assay with N-TIC and MYC2.
(B) Coimmunoprecipitation assay of N-TIC and MYC2. GFP-fused N-TIC and the myc-epitope–fused MYC2 were transiently expressed in N. benthamiana. GFP-N TIC was immunoprecipitated and detected using an anti-GFP antibody. Coimmunoprecipitated MYC2 was detected using an anti-myc antibody. * indicates GFP protein, and ** indicates GFP-N TIC protein. co-IP, coimmunoprecipitation; IP, immunoprecipitation; MW, molecular weight.
(C) BiFC assay using N-YFP fused with full-length TIC or N terminus TIC, and C-YFP fused with MYC2 or PIF5.
(D) BiFC assay using N-YFP fused with PIF5 and C-YFP fused with PIF5. Proteins were transiently coexpressed in N. benthamiana, and YFP fluorescence was detected under a confocal microscope. BiFC experiments were performed three times, and similar results were observed from each experiment.
TIC Represses JA-Mediated Inhibition of Root Growth in a MYC2-Dependent Manner
For genetic analysis of TIC and MYC2, we obtained two independent T-DNA insertion mutants of MYC2 (Salk_061267, Salk_130877). The Salk_061267 mutant was already characterized and previously named myc2-3/jin1-8 (Lorenzo et al., 2004; Sehr et al., 2010), so we refer to this mutant as myc2-3. We named the Salk_130877 line myc2-4. In both homozygous T-DNA lines, MYC2 mRNA was detected only at background levels by quantitative RT-PCR (qRT-PCR), and MYC2 full-length transcripts were not detected by RT-PCR (see Supplemental Figure 2 online). We also generated MYC2ox plants that express MYC2-myc under control of the CaMV 35S promoter. MYC2ox expressed >10 times more MYC2 transcript compared with the wild type (see Supplemental Figure 3 online). For our analysis, we used the tic-2 (SAIL_753_E03) allele (Ding et al., 2007) and generated the tic-2 myc2-3 double mutant from a cross between tic-2 and myc2-3.
Because MYC2 is an established positive regulator of the JA pathway (Lorenzo et al., 2004; Dombrecht et al., 2007), we asked whether TIC could also be involved in the JA response by testing the inhibition of root growth in response to exogenous methyl jasmonate (MeJA) in Columbia (Col), tic-2, myc2-3, myc2-4, tic-2 myc2-3, and MYC2ox seedlings. All genotypes had similar root length in the absence of exogenous MeJA (Figure 2B). Consistent with previous reports, both myc2-3 and myc2-4 mutants displayed reduced sensitivity to MeJA compared with the wild type; MeJA-treated myc2 mutants displayed 59% inhibition of root growth relative to untreated plants, whereas wild-type plants showed 71% inhibition. Conversely, MYC2ox plants exhibited severely reduced root growth (83% inhibition) on MeJA-containing media. Interestingly, tic-2 displayed significantly shortened primary roots on MeJA-containing media (81% inhibition), similar to the response seen in MYC2ox plants. The response of the tic-2 myc2-3 double mutant (56% inhibition) was comparable with that of the myc2-3 single mutant (Figures 2A and 2B). Therefore, myc2-3 is genetically epistatic to tic-2 in JA-dependent inhibition of root elongation.
Figure 2.
TIC Represses JA-Mediated Inhibition of Root Growth.
(A) Inhibition of root growth of 10-d-old Col (wild type), myc2-3, myc2-4, tic-2, tic-2 myc2-3, and MYC2ox seedlings on medium containing 50 μM MeJA.
(B) Root length of seedlings. The results are the mean of 20 seedlings, and error bars indicate sd.
(C) Relative root length of each genotype grown on MeJA-containing media to root length without MeJA treatment. The results are the mean of 20 seedlings, and error bars indicate sd. Asterisks indicate statistically significant differences compared with Col (Student’s t test, **P < 0.01, and ***P < 0.001).
[See online article for color version of this figure.]
We additionally examined the responsiveness of tic-2 to various concentrations of MeJA. The wild type and tic-2 showed similar responses on 10 μM MeJA-containing medium. tic-2 began to show enhanced sensitivity relative to the wild type on 25 μM MeJA medium, and this hypersensitive phenotype was more pronounced at higher concentrations (Figure 2C).
TIC Promotes Resistance to P. syringae pv tomato DC3000 Infection
To investigate the JA-response phenotype of tic further, we examined JA-dependent susceptibility to bacterial pathogen infection. Col, tic-2, myc2-3, and tic-2 myc2-3 plants were inoculated with the virulent strain P. syringae pv tomato (Pst) DC3000, and in planta bacterial titers were determined on the day of inoculation (day 0) and after 3 d (day 3). Titers of inoculated bacteria at day 0 were similar in all examined genotypes, indicating that Pst DC3000 bacteria enter into tic-2 leaves with the same efficiency as in the wild type. At 3 d after inoculation, tic-2 allowed ∼30 times more bacterial proliferation as compared with the wild type (Figure 3B). The increased susceptibility of tic-2 to Pst DC3000 was also seen at the level of enhanced macroscopic disease symptom development (Figure 3A). Consistent with previous reports (Fernández-Calvo et al., 2011), myc2-3 mutants were more resistant to pathogen infection. tic-2 myc2-3 plants exhibited higher resistance than the tic-2 single mutant; however, this was not as extreme as in myc2-3 (Figure 3B). Because Pst DC3000 produces the JA-Ile structural mimic COR to suppress host defenses by activating JA signaling in a COI1-dependent manner (Bender et al., 1999; Katsir et al., 2008b), these results are consistent with the enhanced sensitivity of tic-2 to JA. We tested this hypothesis by inoculating plants with Pst COR−, a mutant strain deficient in production of COR (Melotto et al., 2006). All examined plant genotypes limited growth of Pst COR− to a similar extent (see Supplemental Figure 4 online). We concluded that TIC serves to restrict Pst DC3000 infection through negative regulation of JA signaling.
Figure 3.
tic-2 Is More Susceptible to Pst DC3000 Infection.
(A) Disease symptoms of Col, tic-2, myc2-3, and tic-2 myc2-3 plants after Pst DC3000 infection. Plants were grown for 5 weeks and sprayed with Pst DC3000. Pictures were taken 4 d after inoculation.
(B) In vivo titers of Pst DC3000 in Col, tic-2,myc2-3, and tic-2 myc2-3 plants on the day of spraying (day 0) or 3 d after inoculation (day 3). Bacterial counts were expressed as log (cfu/cm2). Error bars indicate se. Asterisks indicate statistically significant differences compared with Col (Student’s t test, **P < 0.01).
TIC Regulates JA-Responsive Gene Expression in a MYC2-Dependent Manner
We investigated the JA responses of tic-2 at a molecular level by determining the fold induction of JA-marker gene expression in the presence of exogenous JA relative to untreated tissues. Seedlings were grown under continuous white light (WLc) and treated with MeJA for 1, 3, 6, or 24. For this analysis, we selected three groups of JA-responsive genes: immediate–early-induced genes (JAZ5 and LOX3) (Mandaokar et al., 2006; Chini et al., 2007; Chung et al., 2008), wound-response genes (VSP2 and TAT) (Titarenko et al., 1997; Reymond et al., 2000; Cheong et al., 2002; Farmer et al., 2003; Howe, 2004; Lorenzo et al., 2004), and pathogen-response genes (PDF1.2 and PR4) (Penninckx et al., 1996; Manners et al., 1998; Penninckx et al., 1998; Lorenzo et al., 2003; Lorenzo et al., 2004).
Consistent with a previous report (Lorenzo et al., 2004), we confirmed that myc2-3 displayed lower induction of immediate–early-response genes and wound-responsive genes and enhanced induction of pathogen-response genes (Figures 4A to 4F). The induction of JAZ5 and LOX3 after MeJA treatment was similar in tic-2 and the wild type (Figures 4A and 4B), but levels of VSP2 and TAT were significantly induced in tic-2 relative to the wild type (Figures 4C and 4D). In measuring the expression of pathogen-response marker genes, we found that PR4 and PDF1.2 were only marginally induced by exogenous MeJA treatment in tic-2 compared with the wild type (Figures 4E and 4F). Therefore, TIC represses the wound-response gene expression and promotes the pathogen-response gene expression in a JA-dependent manner. The myc2-3 mutation was either fully (JAZ5, LOX3, TAT) or partially (VSP2, PR4, PDF1.2) epistatic to tic-2 (Figures 4A to 4F). These patterns of gene expression are consistent with our physiological results in Figures 2 and 3 showing a MYC2-dependent repressive function of TIC in JA signaling. Collectively, our data suggest that TIC negatively affects JA signaling and that functional MYC2 is required for this.
Figure 4.
TIC Regulates JA-Responsive Gene Expression in a MYC2-Dependent Manner.
Fold change of JAZ5 (A), LOX3 (B), VSP2 (C), TAT (D), PR4 (E), and PDF1.2 (F) expression by exogenous MeJA treatment relative to mock-treated levels. Plants were grown under WLc for 7 d, and transferred to 50 μM MeJA-containing media for 1, 3, 6, and 24 h, respectively. Gene expression relative to PP2A at each time point was measured by qRT-PCR, and fold induction relative to basal expression level was determined. Fold change indicates a mean of three technical replicates, and error bars indicate sd. qRT-PCR experiments were repeated three times, each with an independent biological replicate, and similar results were obtained. An inset graph in (F) indicates the full scale of the result.
We determined the expression level of several JA biosynthetic genes in the wild type and tic-2 under WLc to test whether the increased JA sensitivity of tic-2 was caused by elevated JA biosynthesis. Transcript levels of various JA biosynthesis genes were not significantly different between tic-2 and wild-type plants (see Supplemental Figure 5 online). Therefore, enhanced responsiveness of tic-2 to JA is unlikely to be caused by elevated JA biosynthesis.
We examined JA responses in the arrhythmic clock mutant elf3-1 (Hicks et al., 1996; McWatters et al., 2000; Reed et al., 2000; Thines and Harmon, 2010; Kolmos et al., 2011) to investigate whether the altered JA sensitivity in tic-2 is caused by general clock defects. Root growth of elf3-1 on MeJA-containing media was similarly inhibited as in the wild type, and MeJA-mediated gene induction of JAZ5 and TAT was comparable between elf3-1 and the wild type (see Supplemental Figure 6 online). Therefore, we concluded that the altered JA response in tic-2 is not the consequence of general clock defects.
MYC2 Protein Accumulation Is Elevated in the tic Mutant
We next investigated whether TIC and MYC2 regulate the expression of each other. We examined transcript accumulation of these genes in the reciprocal mutants. MYC2 transcript levels in tic-2 grown under WLc were similar to those in wild-type plants (see Supplemental Figure 7A online). Also, TIC transcript accumulation was not significantly changed in myc2 mutants or MYC2ox plants (see Supplemental Figure 7B online). Therefore, TIC and MYC2 do not cross-regulate the transcript expression of each other.
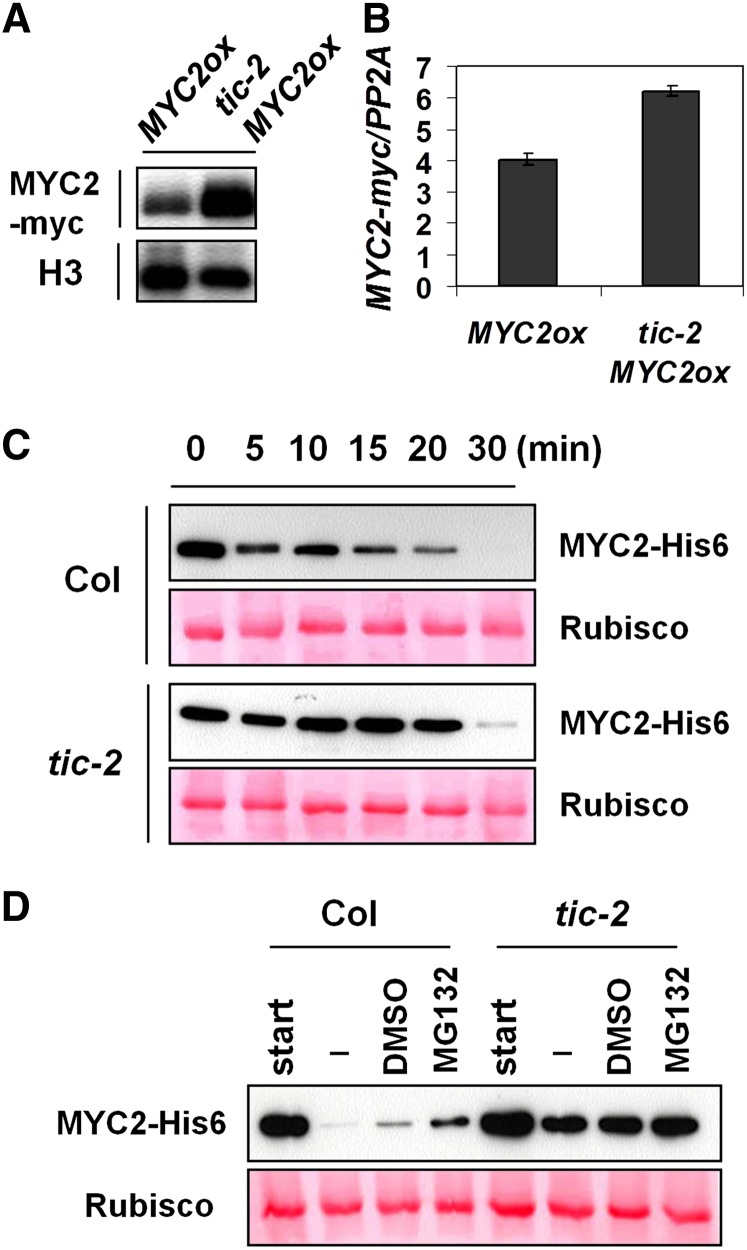
We next tested whether TIC regulates MYC2 protein accumulation. MYC2ox plants were crossed with tic-2 to generate a tic-2 MYC2ox line. Although MYC2-myc transcript levels were slightly higher in tic-2 MYC2ox compared with MYC2ox, MYC2 protein accumulation was dramatically increased in tic-2 MYC2ox compared with MYC2ox (Figures 5A and 5B). We additionally examined MYC2 protein stability in tic-2 by a cell-free protein degradation assay. Recombinant MYC2-His6 protein was purified from Escherichia coli and incubated with a soluble protein extract from Col or tic-2, respectively. The MYC2-His6 protein was rapidly degraded within 15 min in Col extract, whereas it remained more stable for 20 min in tic-2 extract (Figure 5C). We further found that the proteasome is involved in this MYC2 depletion processes, because MYC2 protein degradation was restricted by application of the proteasome inhibitor MG132 in Col extract (Figure 5D). Together, these results indicate that TIC participates in a process leading to MYC2 protein depletion.
Figure 5.
TIC Participates in the MYC2 Protein Depletion.
(A) Immunoblot analysis of MYC2-myc and histone H3 protein in MYC2ox and tic-2 MYC2ox plants.
(B) qRT-PCR of MYC2-myc in MYC2ox and tic-2 MYC2ox plants. Gene expression indicates a mean of three technical replicates, and error bars indicate sd.
(C) Immunoblot analysis of MYC2-His6 protein in a cell-free degradation assay. Recombinant MYC2-His6 protein was incubated in total protein extract of Col or tic-2 for indicated times. MYC2 protein was detected using anti-his6 antibody, and ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) was visualized after Ponceau S staining. This cell-free assay was repeated three times, always with consistent results.
(D) Immunoblot analysis of MYC2-His6 protein in a cell-free degradation assay. Recombinant MYC2-His6 protein was incubated for 20 min with or without DMSO and MG132.
[See online article for color version of this figure.]
MYC2 Expression Oscillates under Control of the Circadian Clock
We next examined whether the circadian clock controls the expression of MYC2. MYC2 transcript profiles were investigated under diurnal and circadian conditions. Plants were entrained under 12-h light/12-h dark diurnal conditions for 7 d and then transferred to WLc free-running conditions for a further 3 d. Total RNA was extracted from replicate samples of plants at defined time points, and MYC2 mRNA levels were measured by qRT-PCR. MYC2 was rhythmically expressed during a 24-h period, with expression highest at dusk (zeitgeber time [ZT] 12) under diurnal conditions (Figure 6A). This rhythmic expression pattern persisted under free-running conditions, with transcripts becoming most abundant at subjective dusk (Figure 6B).
Figure 6.
MYC2 Transcript and Protein Accumulation Are Both Regulated by the Circadian Clock.
qRT-PCR of MYC2 relative to PP2A either under diurnal conditions (A) or under free-running conditions (B). Col plants were grown under 12-h light/12-h dark conditions for 7 d then transferred to continuous light for 3 d. The measurements of gene expression indicate a mean of three technical replicates, and error bars indicate sd. qRT-PCR experiments were repeated three times, each with an independent biological replicate, and similar results were obtained. Immunoblot analysis of MYC2-myc and histone H3 protein in MYC2ox plants, either under diurnal conditions (C) or free-running conditions (D). Immunoblot experiments were repeated three times, each with an independent biological replicate, and similar results were obtained.
We additionally tested the protein accumulation pattern of MYC2 under diurnal and free-running conditions. MYC2ox plants were used for this analysis to remove circadian control of MYC2 transcript accumulation (shown above; Figures 6A and 6B). Whereas MYC2 transcripts were constantly present in MYC2ox, MYC2 protein accumulation varied rhythmically throughout the day (Figure 6C; see Supplemental Figure 8 online). Under diurnal conditions, MYC2 protein was abundant at midday (ZT 4 to ZT 12) and decreased in darkness. This protein destabilization required the proteasome, because application of the MG132 led to increased MYC2 steady state accumulation at night (see Supplemental Figure 9A online). We wanted to establish whether MYC2 protein accumulation was directly regulated by light signaling or by diurnal signaling. For this, MYC2 protein levels were measured in MYC2ox plants grown under continuous darkness, continuous red light, or continuous far-red–light conditions. MYC2 accumulation was not reduced in darkness; rather, its level was comparable with or even higher than those observed under continuous red light or continuous far-red–light conditions (see Supplemental Figure 9B online). Therefore, MYC2 protein does not seem to be stabilized by light signaling. We further determined MYC2 protein levels under free-running conditions and found rhythmic accumulation. MYC2 protein abundance peaked at subjective day and decreased at subjective night (Figure 6D). Taken together, the circadian-clock regulation of MYC2 accumulation uses both transcriptional and posttranslational mechanisms.
JA Response Is Gated by the Circadian Clock
Several studies have reported connections between the circadian clock and various phytohormone-signaling pathways (Hanano et al., 2006; Salomé et al., 2006; Legnaioli et al., 2009; Rawat et al., 2009). This led us to test whether the JA response is gated by the circadian clock and whether TIC is involved in a clock-gating process. We first compared the susceptibility of Col plants with Pst DC3000 at different times of day. Plants grown under short-day conditions (8-h light/16-h dark) were inoculated with Pst DC3000 either at dawn (ZT 0) or at dusk (ZT 8), and bacterial growth was determined 72 h later. Although initial titers of Pst DC3000 bacteria were similar at both examined time points, ∼10 times more Pst DC3000 bacteria were measured in Col at day 3 when plants were inoculated at dawn compared with plants infected at dusk (Figure 7A). Notably, this timing effect was significantly attenuated in tic-2, because Pst DC3000 growth remained high in tic-2 regardless of the inoculation time of day (Figure 7A). We performed the same experiment with the Pst COR− strain and found that Col and tic-2 plants displayed similarly low bacterial titers at day 3 regardless of the inoculation time of day (see Supplemental Figure 10 online). These results suggest that the JA response triggered by COR is higher in the morning than in the evening.
Figure 7.

JA Response Is Gated by the Circadian Clock.
(A) Growth of Pst DC3000 in Col and tic-2 plants 72 h after infection. Plants were grown under short-day (8-h light/16-h dark) conditions for 5 weeks and were inoculated with Pst DC3000 either at ZT 0 or ZT 8. Bacterial counts are expressed as a log scale (cfu/cm2). Error bars indicate se. Asterisks indicate statistically significant differences of ZT 8 compared with ZT 0 of each genotype (Student’s t test, **P < 0.01).
(B) qRT-PCR of JAZ5 expression in Col and tic-2 plants. Plants were grown under 12-h light/12-h dark conditions for 7 d and treated with 50 μM MeJA for 1 h at different times as indicated.
(C) qRT-PCR of JAZ5 expression in Col, tic-2, myc2-3, and tic-2 myc2-3 plants, treated with 50 μM MeJA for 1 h at ZT 0, ZT 12, and ZT 24. The measurements of gene expression indicate a mean of three technical replicates, and error bars indicate sd. qRT-PCR experiments were repeated three times, each with an independent biological replicate, and similar results were obtained.
We next examined JA-responsive gene expression at different times of day. Col plants were grown under 12-h light/12-h dark conditions, and MeJA was applied for 1 h at 4-h intervals over 1 d. Then, immediate–early-induced JA-responsive gene expression was determined. Induction of JAZ5 expression by MeJA treatment was significantly higher at dawn (ZT 0) relative to other time points over a 24-h period (Figure 7B). MYC2 was also rhythmically induced by exogenous MeJA in a similar pattern with JAZ5 (see Supplemental Figure 11 online). Therefore, MYC2 peak-expression phase was shifted from dusk to dawn, in addition to the transcript accumulation being significantly induced by exogenous JA treatment (Figure 6A; see Supplemental Figure 11 online). However, in tic-2, JAZ5 expression on MeJA treatment was not rhythmically regulated, but it was maintained at a high level (Figure 7B). Therefore, the JA response seems to be elevated in tic-2 compared with the wild type, especially at dusk. These data collectively indicate that the circadian clock gates JA responses in a TIC-dependent manner.
We determined JAZ5 induction by MeJA in Col, tic-2, myc2-3, and tic-2 myc2-3 mutants both at dawn (ZT 0, ZT 24) and at dusk (ZT 12). Consistent with the results in Figure 4A, JAZ5 transcript levels were low at all examined time points in myc2-3 as well as in tic-2 myc2-3 (Figure 7C). Interestingly, the JAZ5 level in myc2-3 was similar to that of wild-type plants at dusk (ZT 12). Therefore, JAZ5 is only marginally induced by MeJA at dusk in a wild-type background.
COI1 Transcription Is Controlled by the Circadian Clock in a TIC-Dependent Manner
Our results show that JA-responsive gene induction is differentially regulated depending on the time within diurnal periods (Figure 7B; see Supplemental Figure 11 online). We reasoned that JA signaling, rather than JA biosynthesis, is controlled by the clock for this gating effect. To test this hypothesis, we measured transcript accumulation of the JA receptor COI1 under 12-h light/12-h dark diurnal conditions. COI1 transcript levels oscillated, with expression highest at dawn (ZT 0; Figure 8A). This corresponds to the time when plants most strongly respond to JA (Figure 7). We then measured COI1 transcript accumulation after MeJA application. Although MeJA application did not change the rhythmic expression pattern of COI1, it slightly lowered COI1 expression at all examined time points (Figure 8A). Strikingly, COI1 transcripts did not oscillate in tic-2 and were not reduced by MeJA treatment (Figure 8B). Thus, COI1 expression maintained an elevated peak during 24 h in tic-2 (Figure 8C). COI1 transcript levels in the tic-2 myc2-3 double mutant were similar to those of tic-2 (Figure 8D). Therefore, MYC2 is not required for evening-specific derepression of COI1 in tic-2.
Figure 8.
TIC Is Required for Rhythmic COI1 Expression under Diurnal Conditions.
qRT-PCR of COI1 expression relative to PP2A. Plants were grown under 12-h light/12-h dark conditions for 7 d and treated either with 0.01% ethanol (EtOH) as a mock control ([A] and [B]) or with 50 μM MeJA for 1 h at different times as indicated ([A] to [D]).
(A) COI1 expression in Col plants in response to either ethanol (gray bar) or MeJA (white bar) at different times of day.
(B) COI1 expression in tic-2 plants in response to either ethanol (gray bar) or MeJA (white bar) at different times of day.
(C) Comparison of COI1 expression in Col and tic-2 on MeJA treatment for 1 h under diurnal conditions.
(D) COI1 expression by 50 μM MeJA treatment for 1 h at ZT 0, ZT 12, and ZT 24 in Col, tic-2, myc2-3, and tic-2 myc2-3 plants. The measurements of gene expression indicate a mean of three technical replicates, and error bars indicate sd. qRT-PCR experiments were repeated three times, each with an independent biological replicate, and similar results were obtained.
JA Promotes MYC2 Protein Accumulation in a Circadian Clock–Dependent Manner
We measured MYC2 protein accumulation in the tic-2 background under diurnal conditions. Consistent with the results in Figure 6C, MYC2 accumulated during the day in MYC2ox plants (Figure 9A). In tic-2 MYC2ox plants, a similar rhythmic MYC2 protein accumulation pattern was observed; however, MYC2 protein levels were dramatically elevated during the entire day. This was particularly pronounced at night phases and at dawn (Figure 9B). We then investigated whether MYC2 protein accumulation is regulated by JA in a time-of-day–dependent manner. Plants were grown under 12-h light/12-h dark conditions and treated for 1 h with MeJA at dawn (ZT 0) or at dusk (ZT 12), respectively. MYC2 protein accumulation was increased by MeJA treatment at ZT 0, but not at ZT 12 (Figure 9C). Therefore, MYC2 protein accumulation was more susceptible to the effects of JA in the morning. These data collectively indicate that both TIC and JA-mediated temporal MYC2 stabilization contribute to clock-gated JA responses.
Figure 9.
MYC2 Protein Accumulation Is Promoted by JA Treatment Predominantly at Dawn.
Immunoblot analysis of MYC2-myc and histone H3 protein levels in MYC2ox (A) and tic-2 MYC2ox plants (B). Plants were grown under 12-h light/12-h dark conditions for 7 d.
(C) Immunoblot analysis of MYC2-myc and histone H3 protein levels in MYC2ox and tic-2 MYC2ox plants, with or without 50 μM MeJA treatment for 1 h either at ZT 0 (dawn) or ZT 12 (dusk). Immunoblot experiments were repeated three times, each with an independent biological replicate, and similar results were obtained.
DISCUSSION
Both the circadian clock and hormone signaling are involved in the regulation of plant growth and development by synchronizing the internal growth state to external environmental changes. Therefore, those signaling systems are tightly interconnected and share regulating networks. In this study, we identified signaling crosstalk between the circadian clock and the JA pathway, mediated by TIC. We found that TIC physically interacts with MYC2 to deplete MYC2 protein accumulation. Consequently, TIC acts as a negative regulator of the JA pathway in a MYC2-dependent manner. We further showed that the circadian clock gates JA responses and that TIC is required for evening-specific inhibition of JA signaling.
tic-2 displayed an enhanced responsiveness to JA in the regulation of root growth, susceptibility to Pst DC3000 infection, and JA-dependent wound-response gene expression (Figures 2, 3, 4C, and 4D). By contrast, TIC promotes JA-dependent pathogen-response gene expression (Figures 4E and 4F), and TIC acts as a negative regulator of MYC2 by promoting MYC2 protein degradation (Figure 5). For JA-driven repression of root growth, myc2 was completely genetically epistatic to tic (Figures 2A and 2B), whereas MYC2 was only partially necessary for JA-responsive gene expression and resistance to Pst DC3000 infection (Figures 3 and 4). These data suggest the presence of additional downstream regulators of TIC in JA signaling. MYC2, MYC3, and MYC4 are known to be functionally redundant mainly in aerial tissues in a wide range of JA responses, such as JA-induced gene expression and the pathogen and herbivore infection responses (Fernández-Calvo et al., 2011). MYC family genes also have functional specificity. MYC2 predominantly acts in the root tissue, which supports our root growth results of tic-2 myc2-3 on JA-containing media. It is possible that MYC3 and MYC4 function downstream of TIC in the aerial parts of the plant, and this awaits to be tested.
We showed here cross-regulation of JA signaling and the circadian clock. JA-signaling activity varied depending on the time of day, which is caused by a circadian gating effect. This allows higher JA response in the morning phase. Plants were more susceptible to infection by Pst DC3000 at dawn than at dusk, and JA-responsive gene (JAZ5 and MYC2) induction on JA treatment was highest at dawn (Figure 7; see Supplemental Figure 11 online). TIC was required for the clock gating of JA responses. Susceptibility to Pst DC3000 infection and JA-responsive gene induction were increased in tic-2, regardless of the specific time of a day. Although tic-2 shows only marginally increased sensitivity to JA compared with the wild type at dawn, the difference between the wild type and tic-2 became pronounced at dusk (Figure 7). Therefore, we found that TIC is required at dusk to inhibit JA responses. Because TIC was shown to reset the circadian core clock at the late-night phase (Hall et al., 2003), the function of TIC on the circadian core oscillator and clock output pathways seems to be temporally divided.
In microarray studies, it was reported that clock-regulated JA-responsive genes were mostly expressed in the morning phase (Covington et al., 2008). Some JA-biosynthesis genes were also identified to be rhythmically expressed in a 24-h frame (Mockler et al., 2007). In a recent in planta study, the JA level was found to be rhythmic, with maximum abundance in the middle of the day and lower levels during the night (Goodspeed et al., 2012). Notably, the JA synthesis and signaling pathway is acutely induced by stress signals, such as wound or pathogen infection, whereas it is maintained at only basal levels under nonstressed conditions. Therefore, rhythmic biosynthesis of JA and its signaling gene expression under nonstressed conditions would not likely correlate with the stress-triggered rhythmic JA responses that we have shown in this study.
Daily plant growth is promoted during the night phase and is known to be regulated by rhythmic auxin and GA signaling (Covington and Harmer, 2007; Nozue et al., 2007; Rawat et al., 2009; Arana et al., 2011). Our results support that clock-gated JA responses also contribute to a temporal balance between daily growth and stress responsiveness, because JA response activates stress responses at the expense of energy needed for plant growth (Walters and Heil, 2007). Such a mechanism may allow plants to promote growth in the evening with less-active JA signaling and restrict growth in the morning when JA response is more active. Oxidative stress caused by the dark-to-light transition is one example of a daily event in which JA signaling is vital to plants (Sasaki-Sekimoto et al., 2005; Wolucka et al., 2005; Dombrecht et al., 2007). In previous genomic analysis, clock-regulated genes responsive to the reactive oxygen species or to oxidative damage were shown to be expressed early in the morning phase (Covington et al., 2008). This is consistent with our results showing the clock-gated active JA response in the morning.
The JA pathway is important for defense against necrotrophic pathogen infection and wounding by feeding insects (Howe, 2004; Browse, 2009; Goodspeed et al., 2012). Based on our data, we speculate that plants can predict both the timing of pathogen infection and insect attack by time-specific defense-pathway activation to maximize the defense response against a particular pathogen or pest. Previous studies have shown that plant immune responses against obligate biotrophic pathogens are regulated by the circadian clock (Wang et al., 2011). It is possible that this response allows plants to anticipate pathogen infection in the morning. In particular, the circadian morning element CCA1 was found to direct R gene expression. TIC has been shown to shorten the circadian period and reduce the expression amplitude of the core oscillator, such as CCA1 (Ding et al., 2007). It will be useful to test the functional relationship between TIC and obligate biotrophic pathogen infection in the future.
Here we suggest two components contributing to the JA-gating mechanism: regulation of the expression of the JA receptor COI1 and JA-mediated temporal regulation of MYC2 protein accumulation. We showed here that COI1 transcript accumulation oscillates, with the highest expression at dawn, which corresponds to the time when JA response is high (Figure 8). Previously, the activity of COI1 was shown to correlate with JA responses (Li et al., 2006; Mosblech et al., 2011). This supports our hypothesis that rhythmic COI1 expression would correlate with rhythmic JA responses. We additionally found that the expression of COI1 was modestly downregulated by exogenous JA treatment. This might be caused through transcriptional feedback regulation, which could create homeostasis of JA signaling. In tic-2, COI1 transcript accumulation did not oscillate and was not reduced by JA. The transcriptional regulation of COI1 in tic-2 was not a MYC2-dependent event, because myc2-3 tic-2 double mutants displayed a similar COI1 expression pattern as tic-2 (Figure 8D). We additionally found that JA predominantly promotes MYC2 protein accumulation in the morning. TIC was shown to repress the basal MYC2 protein level during the entire diurnal period; however, its role in the regulation of JA-mediated MYC2 protein accumulation was limited. The underlying molecular mechanism for temporal regulation of JA-mediated MYC2 accumulation should be investigated in future studies.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana Col accession is the genetic background of the wild type and transgenic lines used in this study. Plants were grown on Murashige and Skoog (MS) media (one-half-strength MS [Sigma-Aldrich], 0.8% phytoagar, and 0.05% MES [Duchefa], pH 5.7, 1% Suc) at 22°C under various light conditions. Mutant lines myc2-3 (salk_061267) and myc2-4 (salk_130877) were obtained from the Nottingham Arabidopsis Stock Centre. tic-2 (Ding et al., 2007) and elf3-1 (Hicks et al., 1996; Kolmos et al., 2011) have been previously reported. The tic-2 myc2-3 double mutant was generated by crossing the corresponding parental homozygous lines and genotyping F2 segregating progenies to select homozygous mutations. Primers for genotyping mutants are listed in Supplemental Table 1 online. To generate MYC2ox transgenic plants, full-length MYC2 cDNA was amplified with gene-specific primers (see Supplemental Table 1 online), cloned into the SmaI and AvrII sites of the pBI-HTM vector (kindly provide by Giltsu Choi), and transformed into Col by Agrobacterium tumefaciens–mediated transformation (Davis et al., 2009).
Yeast Two-Hybrid Assay
Full-length MYC2 and N terminus TIC (encoded amino acids 1 to 564) were amplified with Gateway-compatible primers (see Supplemental Table 1 online) and PCR products inserted into pDONR201 with a Gateway BP kit (Invitrogen). A MYC2 construct was used in Gateway LR reactions in combination with the destination yeast-expression vector, pACT-attR (GAL4 AD). A TIC construct was used in Gateway LR reactions in combination with the destination yeast-expression vector, pAS-attR (GAL4 BD). To examine protein–protein interactions in yeast, corresponding plasmids were cotransformed into Saccharomyces cerevisiae AH109 according to the Clontech manual, and transformed cells were spread on selective yeast synthetic dropout media.
Root Measurements
For the JA-mediated root-growth–inhibition assay, seeds were plated on MS agar media with or without 50 μΜ MeJA (Duchefa), placed for 3 d at 4°C in the dark, and subsequently placed vertically under WLc for 10 d. Three independent biological replicates were measured for each sample, and similar results were obtained from each independent experiment. Comparison between Col and mutants (tic-2, myc2-3, myc2-4, MYC2ox, and tic-2 myc2-3) were performed by Student’s t test.
Gene Expression Analysis
Seedlings were grown for 7 d before sampling under various photoperiod conditions, as indicated in the results. For the analysis of JA-induced gene expression, plants were treated with 50 μΜ MeJA (Duchefa) or 0.01% ethanol (mock control) for the time indicated for each experiment. Total RNA was extracted from seedlings using Spectrum Plant Total RNA Kit (Sigma-Aldrich) according to the manufacturer’s instructions. cDNA was synthesized from 4 μg of total RNA with Maxima First Strand cDNA Synthesis Kit (Fermentas). To amplify genes, 5 μL of 1/25 diluted cDNA was used as the template. qRT-PCR analysis was performed using SYBR and iQ5 (Bio-Rad). Primer sequences for qRT-PCR are listed in Supplemental Table 1 online. The resulting gene expression levels were normalized with the level of PP2A as described previously (Czechowski et al., 2005). Data analysis was performed using three technical replicates from each biological sample, and similar results were obtained in three biological replicates.
Protein Extraction and Immunoblotting
For protein extraction, 100 seedlings were grown on MS-agar plates for 7 d. For the analysis of JA-dependent protein accumulation, plants were treated with 50 μΜ MeJA (Duchefa) or 0.01% ethanol (mock control) for 1 h at the indicated time of each experiment. Seedlings were ground in liquid nitrogen and homogenized in a denaturing buffer (100 mM NaH2PO4, 10 mM Tris-Cl, 8 M urea) with vigorous vortexing. Cell debris was removed by centrifugation at 20,817 relative centrifugal force for 15 min at 4°C. For immunoblot analysis, clarified supernatants were separated in a SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore). For detection of GFP-N TIC and MYC2-myc, the membrane was incubated with anti-GFP antibodies (Yau et al. 2008) and anti-myc antibodies (Cell Signaling), respectively, in PBS buffer containing 0.05% Tween-20. For detection of histone H3, the membrane was incubated in the same buffer with antihistone H3 antibodies (Upstate Biotechnology). Bands were visualized with an enhanced chemiluminescence kit (GE Healthcare), according to manufacturer’s instructions.
Coimmunoprecipitation
Four-week-old Nicotiana benthamiana leaves were infiltrated with A. tumefaciens GV3101 strain harboring a vector that generates MYC2 protein fused with myc tag, or N terminus TIC (amino acids 1 to 564) fused with a GFP tag. Briefly, transformed A. tumefaciens were grown at 28°C in yeast-enriched broth media for 2 d, centrifuged, and resuspended with MS media containing 10% Suc, 2.6 mM MES, pH 5.7, and 150 μΜ acetosyringone. Cells were incubated at room temperature in the dark for 3 h, then transformed Agrobacterium were mixed with OD600 ratios as MYC2:N-TIC:p19 = 0.7:0.7:1.0 and infiltrated into the N. benthamiana leaves. At 3 d after infiltration, 100 mg of leaves were collected, ground in liquid nitrogen, and homogenized in a immunoprecipitation buffer (50 mM Tris-Cl, pH, 7.5, 150 mM NaCl, 10% glycerol, 0.1% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride) in addition to 50 μΜ MG132 (Sigma-Aldrich) and 1× complete mini protease inhibitor cocktail (Roche). Resuspended cell lysates were centrifuged at 20,817 relative centrifugal force for 15 min at 4°C. The clarified supernatant was incubated with 20 μL of preequilibrated GFP-Trap-A bead (Chromotek) for 2.5 h at 4°C and washed three times with 1 mL of immunoprecipitation buffer. The immunoprecipitated sample was released by incubation with 2× SDS sample buffer followed by a 95°C treatment for 5 min. These samples were then analyzed by immunoblotting, as described above.
BiFC Assay
Full-length TIC, N terminus TIC, MYC2, and PIF5 were cloned into gateway compatible N-YFP and C-YFP vectors (Kwaaitaal et al., 2010) and transformed into A. tumefaciens GV3101. Different combinations of the A. tumefaciens were infiltrated into N. benthamiana as described above. At 3 d after infiltration, YFP fluorescence was visualized using a Leica confocal microscope. BiFC experiments were performed three times, and the same results were observed from each experiment.
Cell-Free Protein Degradation Assay
For MYC2-His6 protein purification, MYC2 cDNA in pDONR201 was moved into the Gateway-compatible pET-28a vector by Gateway LR reactions. The clone was transformed into the Escherichia coli BL21 strain, and MYC2-His6 protein was isolated using Ni-NTA column (Qiagen).
Cell-free protein degradation assays were performed as described (Chini et al., 2007), with minor modifications. Briefly, total plant protein was extracted with the extraction buffer (20 mM Tris-HCl, pH 7.4, 25 mM NaCl, 0.01% Nonidet P-40, and 1× complete mini protease inhibitor cocktail [Roche]). Protein concentration was determined by the Bradford assay, and 200 μg total protein was added to the reaction mix (20 mM Tris-HCl, pH 7.4, 2 mM MgCl2, 2 mM ATP, 10 μg/μL ubiquitin [Sigma-Aldrich]), and 24 μg of purified recombinant MYC2-His6 protein was added. For proteasome inhibitor assays, either 200 μΜ MG132 or 2% DMSO was added. The protein mixture was incubated at 30°C for different times as indicated, and the reaction was stopped by adding 5× SDS sample buffer.
Bacterial Infection Assays
Plants were grown under short-day (8-h light/16-h dark) conditions for 5 weeks. For spray-infection of Pseudomonas syringae pv tomato (Pst) DC3000 or Pst COR− strains (Melotto et al., 2006), bacteria were adjusted to 108 colony-forming units (cfu)/mL in 10 mM MgCl2 containing 0.04% (v/v) Silwet L-77 (Lehle seeds). In planta bacterial titers were determined 3 to 4 h after spray infection (day 0) and 3 d after infection. For this, the surface of leaves was sterilized with 70% ethanol followed by two washing steps with water, and samples were taken using a cork borer. Bacterial titers were determined by shaking leaf discs in 10 mM MgCl2 with 0.01% Silwet L-77 at 28°C for 1 h, as described previously (Tornero and Dangl, 2001; García et al., 2010). Means and se were calculated from at least three biological replicates per experiment.
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource or GenBank/EMBL databases under the following accession numbers: TIC (locus AT3G22380, GenBank NM_113136), MYC2 (locus AT1G32640, GenBank NM_102998), PIF5 (locus AT3G59060, GenBank NM_180690), JAZ5 (locus AT1G1738, GenBank 0 NM_101599), LOX3 (locus AT1G17420, GenBank NM_101603), VSP2 (locus AT5G24770, GenBank NM_122386), TAT (locus AT2G24850, GenBank NM_128044), PR4 (locus AT3G04720, GenBank NM_111344), PDF1.2 (locus AT5G44420, GenBank NM_123809), PP2A (locus AT1G13320, GenBank NM_001198052).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. TIC Interacts with MYC2 in the Nucleus.
Supplemental Figure 2. Identification of myc2 Mutants.
Supplemental Figure 3. MYC2 Transcript Levels in MYC2ox Plants.
Supplemental Figure 4. Examined Genotypes Are Not Susceptible to Pst COR−.
Supplemental Figure 5. JA Biosynthetic Gene Expression Is Not Altered in tic-2 Relative to the Wild Type.
Supplemental Figure 6. elf3-1 Does Not Show JA-Defective Phenotypes.
Supplemental Figure 7. TIC and MYC2 Do Not Participate in Cross-Regulation of Respective Transcript Accumulation.
Supplemental Figure 8. MYC2 Is Overexpressed without Oscillation in MYC2ox Plants.
Supplemental Figure 9. MYC2 Protein Is Degraded by the Proteasome.
Supplemental Figure 10. Susceptibility to Pst COR− Is Similar Regardless of Genotypes and Infection Time of Day.
Supplemental Figure 11. Circadian Clock Gates the JA-Induced MYC2 Expression.
Supplemental Table 1. Primer Lists.
Acknowledgments
We thank Erich Kombrink, Nora Bujdoso, Amanda Davis, and Giltsu Choi for critical reading of the article and feedback, Giltsu Choi for providing pBI-HTM vector, Amanda Davis and Marc Hallstein for technical support, and Haitao Cui for scientific discussion. This study was supported by the Max Planck Society, a Korea Research Foundation Grant funded by the Korean Government (KRF-2008-357-C00147), an Alexander von Humboldt foundation to J.S., Deutsche Forschungsgemeinschaft grants to S.J.D. (DA1061/4-1) and J.E.P. (Sonderforschungsbereich 670), and Deutsche Forschungsgemeinschaft collaboration funding (Sonderforschungsbereich 635) to S.J.D. and J.E.P.
AUTHOR CONTRIBUTIONS
J.S. and S.J.D. designed the research, J.S., K.H., and A.S.V. performed the research, J.S., K.H., J.E.P., and S.J.D. analyzed data, and J.S., K.H., J.E.P., and S.J.D. wrote the article.
Glossary
- JA
jasmonate
- JA-Ile
jasmonyl-isoleucine
- ABA
abscisic acid
- GA
gibberellic acid
- COR
coronatine
- GFP
green fluorescent protein
- BiFC
biomolecular fluorescence complementation
- YFP
yellow fluorescent protein
- MeJA
methyl jasmonate
- qRT-PCR
quantitative RT-PCR
- Col
Columbia
- Pst
Pseudomonas syringae pv tomato
- WLc
continuous white light
- cfu
colony-forming units
- MS
Murashige and Skoog
- ZT
zeitgeber time
References
- Abe H., Urao T., Ito T., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arana M.V., Marín-de la Rosa N., Maloof J.N., Blázquez M.A., Alabadí D. (2011). Circadian oscillation of gibberellin signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 9292–9297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender C.L., Alarcón-Chaidez F., Gross D.C. (1999). Pseudomonas syringae phytotoxins: Mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 63: 266–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J. (2009). Jasmonate passes muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60: 183–205 [DOI] [PubMed] [Google Scholar]
- Cheong Y.H., Chang H.S., Gupta R., Wang X., Zhu T., Luan S. (2002). Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 129: 661–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Chung H.S., Koo A.J., Gao X., Jayanty S., Thines B., Jones A.D., Howe G.A. (2008). Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 146: 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington M.F., Harmer S.L. (2007). The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol. 5: e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington M.F., Maloof J.N., Straume M., Kay S.A., Harmer S.L. (2008). Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 9: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A.M., Hall A., Millar A.J., Darrah C., Davis S.J. (2009). Protocol: Streamlined sub-protocols for floral-dip transformation and selection of transformants in Arabidopsis thaliana. Plant Methods 5: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S.J., Millar A.J. (2001). Watching the hands of the Arabidopsis biological clock. Genome Biol. 2: 1008.1–1008.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto A., Nieto-Rostro M., Xie D., Ellis C., Harmston R., Patrick E., Davis J., Sherratt L., Coleman M., Turner J.G. (2002). COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 32: 457–466 [DOI] [PubMed] [Google Scholar]
- Ding Z., Millar A.J., Davis A.M., Davis S.J. (2007). TIME FOR COFFEE encodes a nuclear regulator in the Arabidopsis thaliana circadian clock. Plant Cell 19: 1522–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd A.N., Salathia N., Hall A., Kévei E., Tóth R., Nagy F., Hibberd J.M., Millar A.J., Webb A.A. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309: 630–633 [DOI] [PubMed] [Google Scholar]
- Dombrecht B., Xue G.P., Sprague S.J., Kirkegaard J.A., Ross J.J., Reid J.B., Fitt G.P., Sewelam N., Schenk P.M., Manners J.M., Kazan K. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer E.E., Alméras E., Krishnamurthy V. (2003). Jasmonates and related oxylipins in plant responses to pathogenesis and herbivory. Curr. Opin. Plant Biol. 6: 372–378 [DOI] [PubMed] [Google Scholar]
- Fernández-Calvo P., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García A.V., Blanvillain-Baufumé S., Huibers R.P., Wiermer M., Li G., Gobbato E., Rietz S., Parker J.E. (2010). Balanced nuclear and cytoplasmic activities of EDS1 are required for a complete plant innate immune response. PLoS Pathog. 6: e1000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodspeed D., Chehab E.W., Min-Venditti A., Braam J., Covington M.F. (2012). Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc. Natl. Acad. Sci. USA 109: 4674–4677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A., Bastow R.M., Davis S.J., Hanano S., McWatters H.G., Hibberd V., Doyle M.R., Sung S., Halliday K.J., Amasino R.M., Millar A.J. (2003). The TIME FOR COFFEE gene maintains the amplitude and timing of Arabidopsis circadian clocks. Plant Cell 15: 2719–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanano S., Domagalska M.A., Nagy F., Davis S.J. (2006). Multiple phytohormones influence distinct parameters of the plant circadian clock. Genes Cells 11: 1381–1392 [DOI] [PubMed] [Google Scholar]
- Harmer S.L. (2009). The circadian system in higher plants. Annu. Rev. Plant Biol. 60: 357–377 [DOI] [PubMed] [Google Scholar]
- Hicks K.A., Millar A.J., Carré I.A., Somers D.E., Straume M., Meeks-Wagner D.R., Kay S.A. (1996). Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274: 790–792 [DOI] [PubMed] [Google Scholar]
- Howe G.A. (2004). Jasmonates as signals in the wound response. J. Plant Growth Regul. 23: 223–237 [Google Scholar]
- Katsir L., Chung H.S., Koo A.J., Howe G.A. (2008a). Jasmonate signaling: A conserved mechanism of hormone sensing. Curr. Opin. Plant Biol. 11: 428–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L., Schilmiller A.L., Staswick P.E., He S.Y., Howe G.A. (2008b). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 105: 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. (2008). Jasmonate signaling: Toward an integrated view. Plant Physiol. 146: 1459–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmos E., Herrero E., Bujdoso N., Millar A.J., Tóth R., Gyula P., Nagy F., Davis S.J. (2011). A reduced-function allele reveals that EARLY FLOWERING3 repressive action on the circadian clock is modulated by phytochrome signals in Arabidopsis. Plant Cell 23: 3230–3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwaaitaal M., Keinath N.F., Pajonk S., Biskup C., Panstruga R. (2010). Combined bimolecular fluorescence complementation and Forster resonance energy transfer reveals ternary SNARE complex formation in living plant cells. Plant Physiol. 152: 1135–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnaioli T., Cuevas J., Mas P. (2009). TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J. 28: 3745–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Zhao J., Jiang H., Wu X., Sun J., Zhang C., Wang X., Lou Y., Li C. (2006). The wound response mutant suppressor of prosystemin-mediated responses6 (spr6) is a weak allele of the tomato homolog of CORONATINE-INSENSITIVE1 (COI1). Plant Cell Physiol. 47: 653–663 [DOI] [PubMed] [Google Scholar]
- Lorenzo O., Chico J.M., Sánchez-Serrano J.J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Piqueras R., Sánchez-Serrano J.J., Solano R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandaokar A., Thines B., Shin B., Lange B.M., Choi G., Koo Y.J., Yoo Y.J., Choi Y.D., Choi G., Browse J. (2006). Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J. 46: 984–1008 [DOI] [PubMed] [Google Scholar]
- Manners J.M., Penninckx I.A., Vermaere K., Kazan K., Brown R.L., Morgan A., Maclean D.J., Curtis M.D., Cammue B.P., Broekaert W.F. (1998). The promoter of the plant defensin gene PDF1.2 from Arabidopsis is systemically activated by fungal pathogens and responds to methyl jasmonate but not to salicylic acid. Plant Mol. Biol. 38: 1071–1080 [DOI] [PubMed] [Google Scholar]
- McWatters H.G., Bastow R.M., Hall A., Millar A.J. (2000). The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature 408: 716–720 [DOI] [PubMed] [Google Scholar]
- Melotto M., Underwood W., Koczan J., Nomura K., He S.Y. (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980 [DOI] [PubMed] [Google Scholar]
- Michael T.P., Breton G., Hazen S.P., Priest H., Mockler T.C., Kay S.A., Chory J. (2008). A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol. 6: e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T., Yamashino T. (2008). Comparative transcriptome of diurnally oscillating genes and hormone-responsive genes in Arabidopsis thaliana: Insight into circadian clock-controlled daily responses to common ambient stresses in plants. Plant Cell Physiol. 49: 481–487 [DOI] [PubMed] [Google Scholar]
- Mockler T.C., Michael T.P., Priest H.D., Shen R., Sullivan C.M., Givan S.A., McEntee C., Kay S.A., Chory J. (2007). The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb. Symp. Quant. Biol. 72: 353–363 [DOI] [PubMed] [Google Scholar]
- Mosblech A., Thurow C., Gatz C., Feussner I., Heilmann I. (2011). Jasmonic acid perception by COI1 involves inositol polyphosphates in Arabidopsis thaliana. Plant J. 65: 949–957 [DOI] [PubMed] [Google Scholar]
- Niu Y., Figueroa P., Browse J. (2011). Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 62: 2143–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nováková M., Motyka V., Dobrev P.I., Malbeck J., Gaudinová A., Vanková R. (2005). Diurnal variation of cytokinin, auxin and abscisic acid levels in tobacco leaves. J. Exp. Bot. 56: 2877–2883 [DOI] [PubMed] [Google Scholar]
- Nozue K., Covington M.F., Duek P.D., Lorrain S., Fankhauser C., Harmer S.L., Maloof J.N. (2007). Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361 [DOI] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx I.A., Eggermont K., Terras F.R., Thomma B.P., De Samblanx G.W., Buchala A., Métraux J.P., Manners J.M., Broekaert W.F. (1996). Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8: 2309–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx I.A., Thomma B.P., Buchala A., Métraux J.P., Broekaert W.F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10: 2103–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat R., Schwartz J., Jones M.A., Sairanen I., Cheng Y., Andersson C.R., Zhao Y., Ljung K., Harmer S.L. (2009). REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc. Natl. Acad. Sci. USA 106: 16883–16888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.W., Nagpal P., Bastow R.M., Solomon K.S., Dowson-Day M.J., Elumalai R.P., Millar A.J. (2000). Independent action of ELF3 and phyB to control hypocotyl elongation and flowering time. Plant Physiol. 122: 1149–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P., Weber H., Damond M., Farmer E.E. (2000). Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12: 707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson F.C., Skeffington A.W., Gardner M.J., Webb A.A. (2009). Interactions between circadian and hormonal signalling in plants. Plant Mol. Biol. 69: 419–427 [DOI] [PubMed] [Google Scholar]
- Salome P.A., McClung C.R. (2005). What makes the Arabidopsis clock tick on time? A review on entrainment. Plant Cell Environ. 28: 21–38 [Google Scholar]
- Salomé P.A., To J.P., Kieber J.J., McClung C.R. (2006). Arabidopsis response regulators ARR3 and ARR4 play cytokinin-independent roles in the control of circadian period. Plant Cell 18: 55–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A., Shin J., Davis S.J. (2011). Abiotic stress and the plant circadian clock. Plant Signal. Behav. 6: 223–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki-Sekimoto Y., et al. (2005). Coordinated activation of metabolic pathways for antioxidants and defence compounds by jasmonates and their roles in stress tolerance in Arabidopsis. Plant J. 44: 653–668 [DOI] [PubMed] [Google Scholar]
- Sehr E.M., Agusti J., Lehner R., Farmer E.E., Schwarz M., Greb T. (2010). Analysis of secondary growth in the Arabidopsis shoot reveals a positive role of jasmonate signalling in cambium formation. Plant J. 63: 811–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard L.B., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-López P., Wheatley K., Robson F., Onouchi H., Valverde F., Coupland G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Thain S.C., Vandenbussche F., Laarhoven L.J., Dowson-Day M.J., Wang Z.Y., Tobin E.M., Harren F.J., Millar A.J., Van Der Straeten D. (2004). Circadian rhythms of ethylene emission in Arabidopsis. Plant Physiol. 136: 3751–3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B., Harmon F.G. (2010). Ambient temperature response establishes ELF3 as a required component of the core Arabidopsis circadian clock. Proc. Natl. Acad. Sci. USA 107: 3257–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Titarenko E., Rojo E., León J., Sánchez-Serrano J.J. (1997). Jasmonic acid-dependent and -independent signaling pathways control wound-induced gene activation in Arabidopsis thaliana. Plant Physiol. 115: 817–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero P., Dangl J.L. (2001). A high-throughput method for quantifying growth of phytopathogenic bacteria in Arabidopsis thaliana. Plant J. 28: 475–481 [DOI] [PubMed] [Google Scholar]
- Walters D., Heil M. (2007). Costs and trade-offs associated with induced resistance. Physiol. Mol. Plant Pathol. 71: 3–17 [Google Scholar]
- Wang W., Barnaby J.Y., Tada Y., Li H., Tör M., Caldelari D., Lee D.U., Fu X.D., Dong X. (2011). Timing of plant immune responses by a central circadian regulator. Nature 470: 110–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolucka B.A., Goossens A., Inzé D. (2005). Methyl jasmonate stimulates the de novo biosynthesis of vitamin C in plant cell suspensions. J. Exp. Bot. 56: 2527–2538 [DOI] [PubMed] [Google Scholar]
- Yau Y.-Y., Davis S.J., Ipek A., Simon P.W. (2008). Early identification of stable transformation events by combined use of antibiotic selection and vital detection of green fluorescent protein (GFP) in carrot (Daucus carota L.) callus. Agricultural Sciences in China 7: 664–671 [Google Scholar]
- Yin X.-J., et al. (2007). Ubiquitin lysine 63 chain forming ligases regulate apical dominance in Arabidopsis. Plant Cell 19: 1898–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D.X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]