Abstract
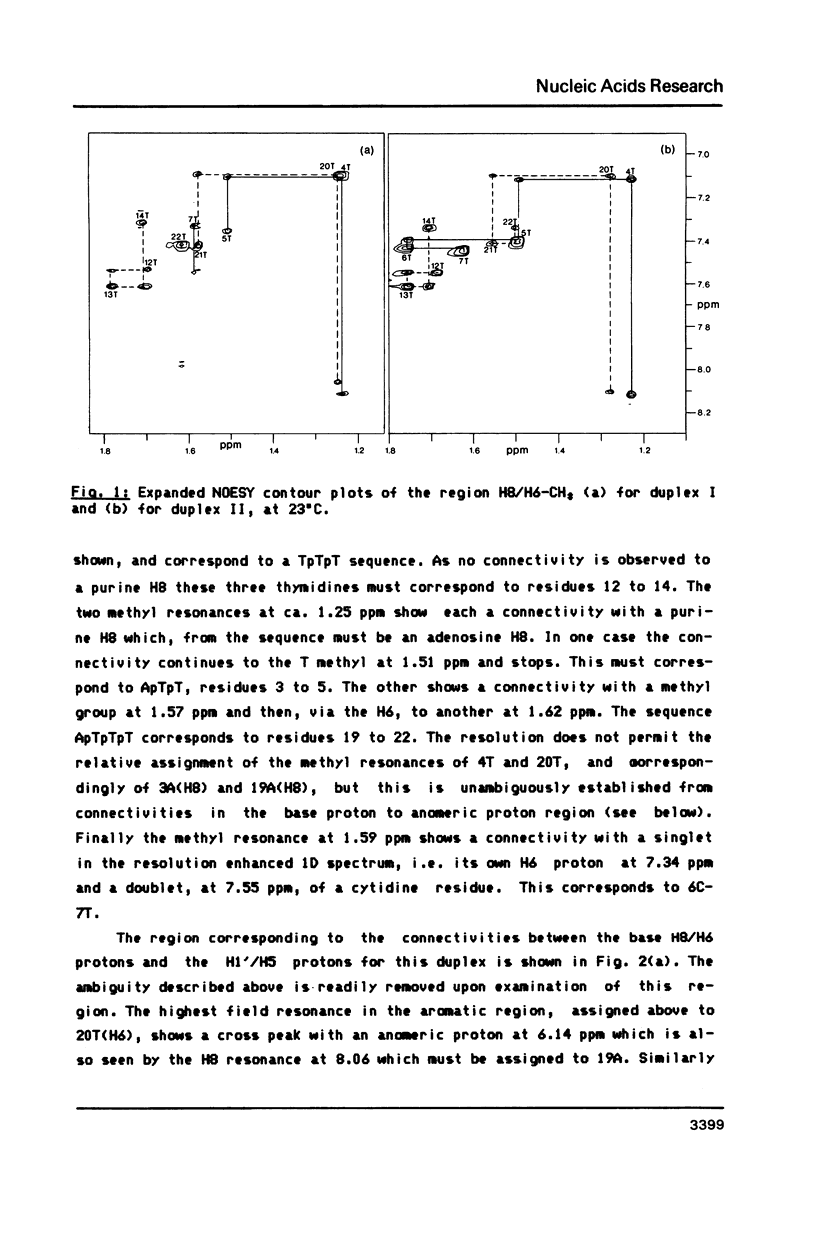
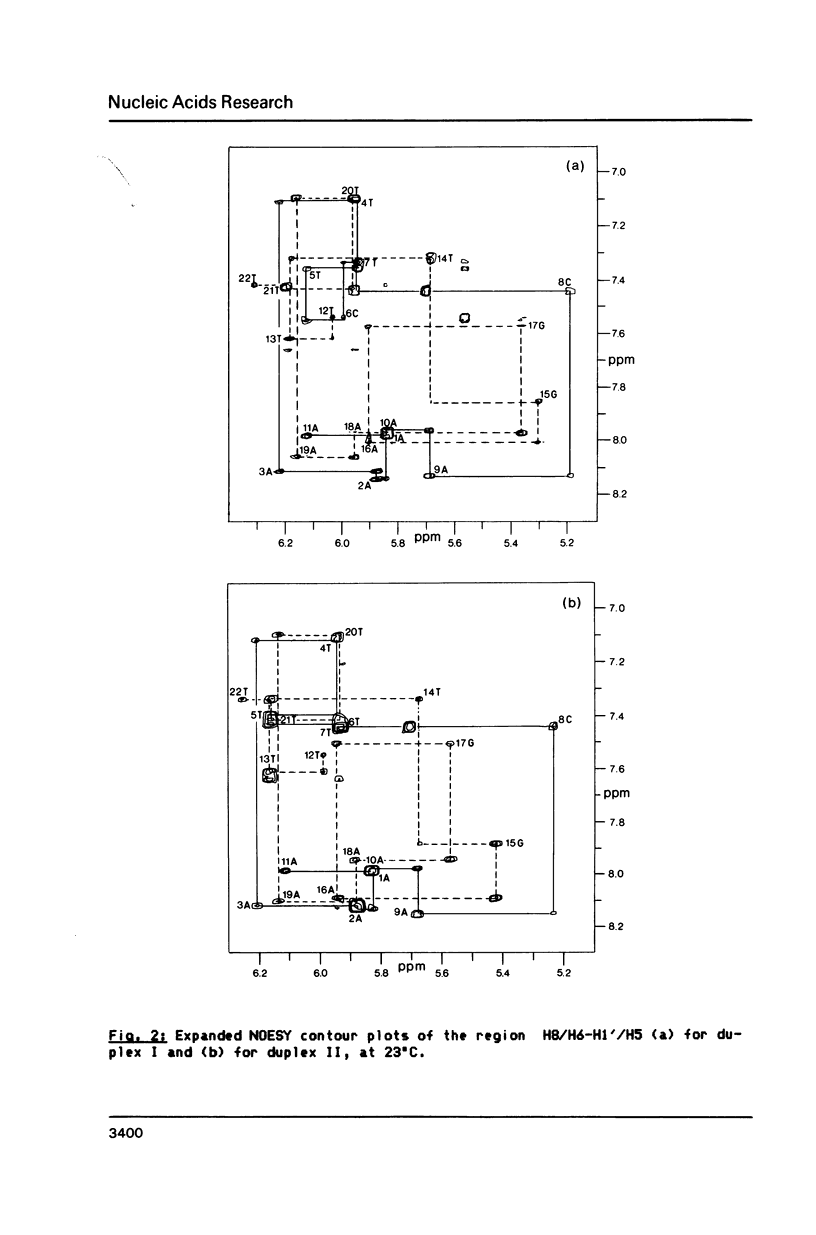
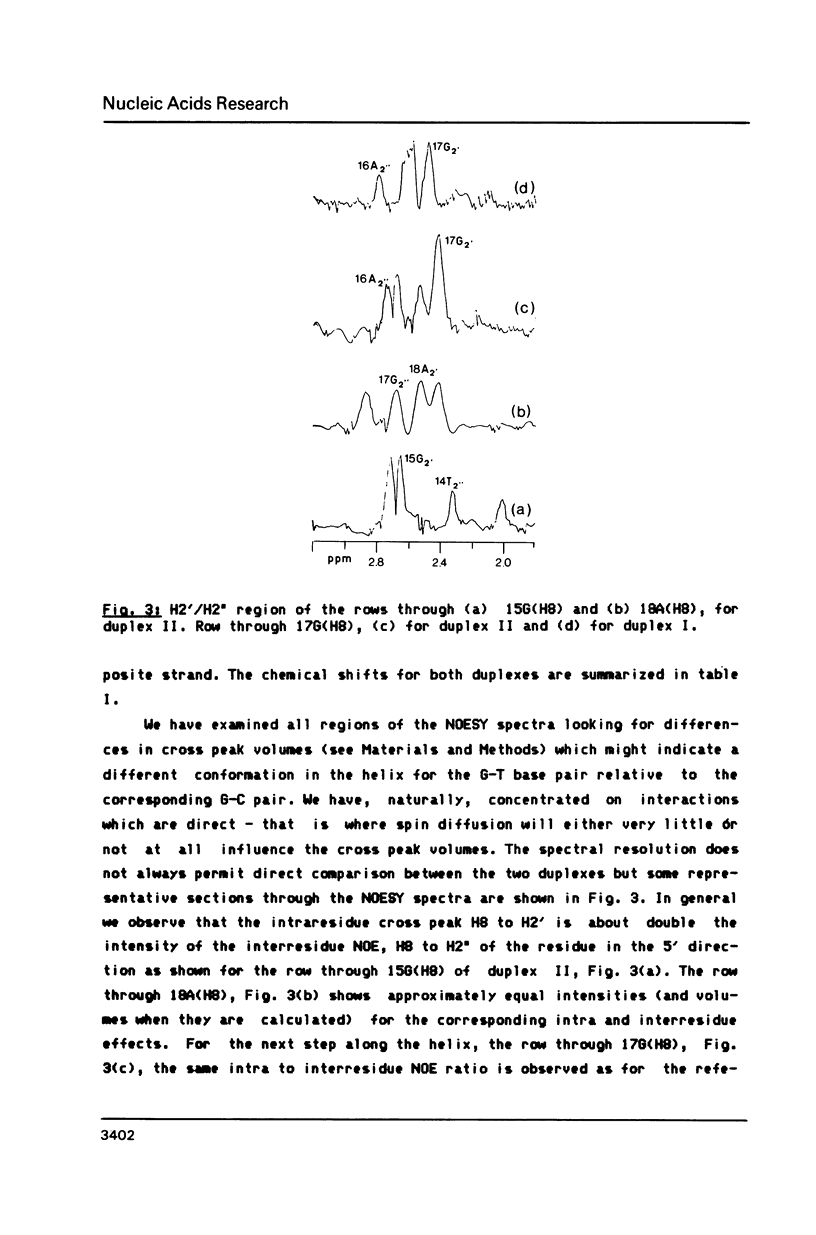
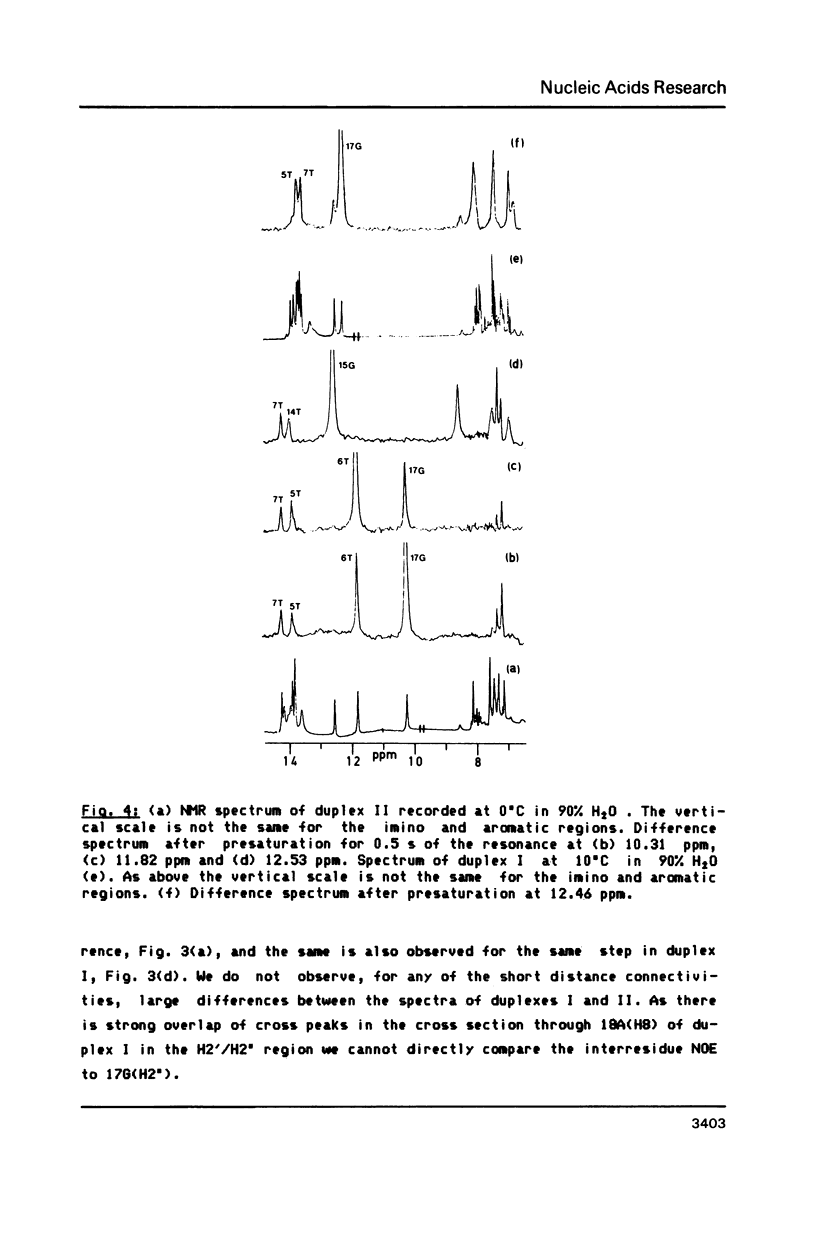
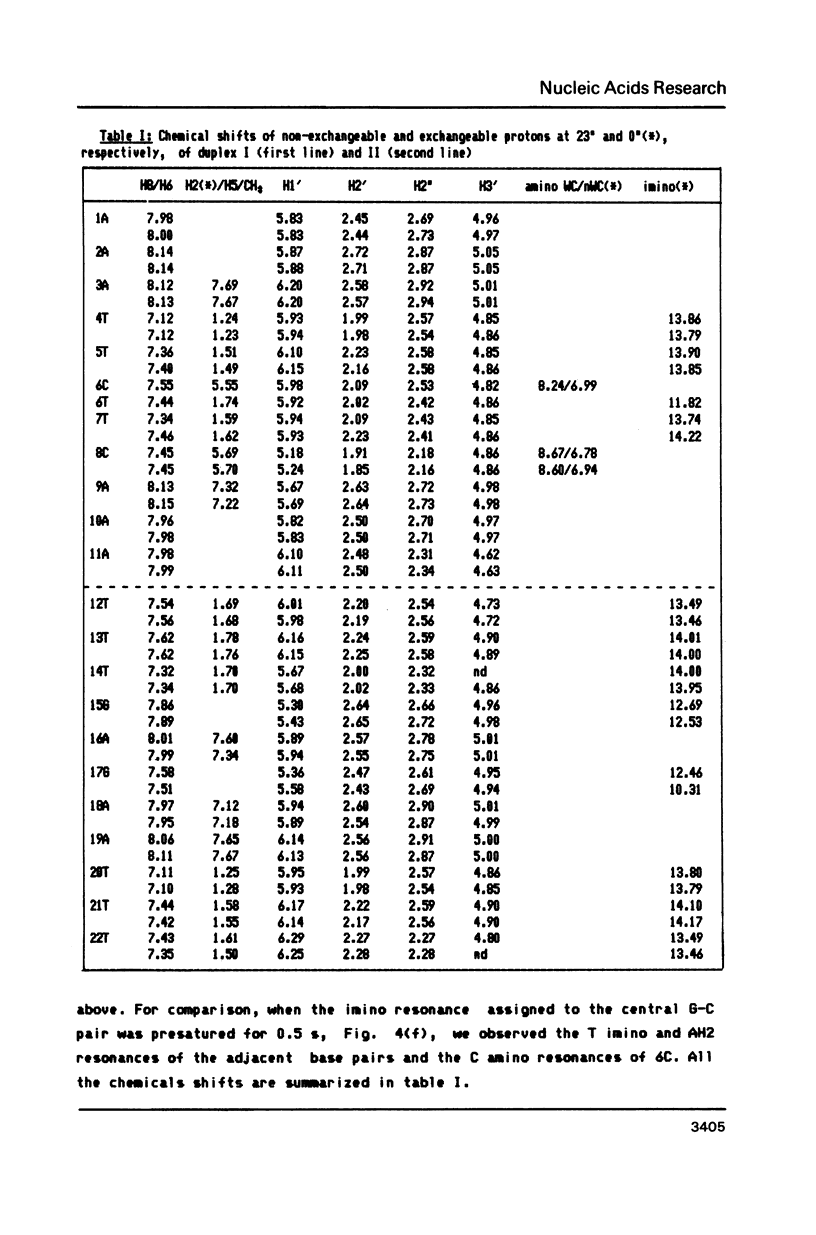
We have recorded NOESY spectra of two non-selfcomplementary undecanucleotide duplexes. From the observed NOEs we do not detect any significant distortion of the helix when a G-C pair is replaced by a G-T pair and the normal interresidue connectivities can be followed through the mismatch site. We conclude that the 2D spectra of the non-exchangeable protons do not allow differentiation between a wobble or rare tautomer form for the mismatch. NOE measurements in H2O, however, clearly show that the mismatch adopts a wobble structure and give information on the hydration in the minor groove for the G-T base pair which is embedded between two A-T base pairs in the sequence.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Crick F. H. Codon--anticodon pairing: the wobble hypothesis. J Mol Biol. 1966 Aug;19(2):548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- Dohet C., Wagner R., Radman M. Repair of defined single base-pair mismatches in Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jan;82(2):503–505. doi: 10.1073/pnas.82.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R., Dickerson R. E. Structure of a B-DNA dodecamer. III. Geometry of hydration. J Mol Biol. 1981 Sep 25;151(3):535–556. doi: 10.1016/0022-2836(81)90009-7. [DOI] [PubMed] [Google Scholar]
- Early T. A., Olmsted J., 3rd, Kearns D. R., Lezius A. G. Base pairing structure in the poly d(G-T) double helix: wobble base pairs. Nucleic Acids Res. 1978 Jun;5(6):1955–1970. doi: 10.1093/nar/5.6.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazakerley G. V., Quignard E., Woisard A., Guschlbauer W., van der Marel G. A., van Boom J. H., Jones M., Radman M. Structures of mismatched base pairs in DNA and their recognition by the Escherichia coli mismatch repair system. EMBO J. 1986 Dec 20;5(13):3697–3703. doi: 10.1002/j.1460-2075.1986.tb04702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazakerley G. V., Téoule R., Guy A., Fritzsche H., Guschlbauer W. NMR studies on oligodeoxyribonucleotides containing the dam methylation site GATC. Comparison between d(GGATCC) and d(GGm6ATCC). Biochemistry. 1985 Aug 13;24(17):4540–4548. doi: 10.1021/bi00338a009. [DOI] [PubMed] [Google Scholar]
- Fazakerley G. V., van der Marel G. A., van Boom J. H., Guschlbauer W. Helix opening in deoxyribonucleic acid from a proton nuclear magnetic resonance study of imino and amino protons in d(CG)3. Nucleic Acids Res. 1984 Nov 12;12(21):8269–8279. doi: 10.1093/nar/12.21.8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigon J., Leupin W., Denny W. A., Kearns D. R. Two-dimensional proton nuclear magnetic resonance investigation of the synthetic deoxyribonucleic acid decamer d(ATATCGATAT)2. Biochemistry. 1983 Dec 6;22(25):5943–5951. doi: 10.1021/bi00294a038. [DOI] [PubMed] [Google Scholar]
- Frechet D., Cheng D. M., Kan L. S., Ts'o P. O. Nuclear Overhauser effect as a tool for the complete assignment of nonexchangeable proton resonances in short deoxyribonucleic acid helices. Biochemistry. 1983 Oct 25;22(22):5194–5200. doi: 10.1021/bi00291a020. [DOI] [PubMed] [Google Scholar]
- Hare D. R., Wemmer D. E., Chou S. H., Drobny G., Reid B. R. Assignment of the non-exchangeable proton resonances of d(C-G-C-G-A-A-T-T-C-G-C-G) using two-dimensional nuclear magnetic resonance methods. J Mol Biol. 1983 Dec 15;171(3):319–336. doi: 10.1016/0022-2836(83)90096-7. [DOI] [PubMed] [Google Scholar]
- Hare D., Shapiro L., Patel D. J. Wobble dG X dT pairing in right-handed DNA: solution conformation of the d(C-G-T-G-A-A-T-T-C-G-C-G) duplex deduced from distance geometry analysis of nuclear Overhauser effect spectra. Biochemistry. 1986 Nov 18;25(23):7445–7456. doi: 10.1021/bi00371a029. [DOI] [PubMed] [Google Scholar]
- Hunter W. N., Kneale G., Brown T., Rabinovich D., Kennard O. Refined crystal structure of an octanucleotide duplex with G . T mismatched base-pairs. J Mol Biol. 1986 Aug 20;190(4):605–618. doi: 10.1016/0022-2836(86)90246-9. [DOI] [PubMed] [Google Scholar]
- Kneale G., Brown T., Kennard O., Rabinovich D. G . T base-pairs in a DNA helix: the crystal structure of d(G-G-G-G-T-C-C-C). J Mol Biol. 1985 Dec 20;186(4):805–814. doi: 10.1016/0022-2836(85)90398-5. [DOI] [PubMed] [Google Scholar]
- Lu A. L., Clark S., Modrich P. Methyl-directed repair of DNA base-pair mismatches in vitro. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4639–4643. doi: 10.1073/pnas.80.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A. L., Welsh K., Clark S., Su S. S., Modrich P. Repair of DNA base-pair mismatches in extracts of Escherichia coli. Cold Spring Harb Symp Quant Biol. 1984;49:589–596. doi: 10.1101/sqb.1984.049.01.066. [DOI] [PubMed] [Google Scholar]
- Pardi A., Morden K. M., Patel D. J., Tinoco I., Jr Kinetics for exchange of the imino protons of the d(C-G-C-G-A-A-T-T-C-G-C-G) double helix in complexes with the antibiotics netropsin and/or actinomycin. Biochemistry. 1983 Mar 1;22(5):1107–1113. doi: 10.1021/bi00274a018. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Marky L. A., Rice J. A., Broka C., Dallas J., Itakura K., Breslauer K. J. Structure, dynamics, and energetics of deoxyguanosine . thymidine wobble base pair formation in the self-complementary d(CGTGAATTCGCG) duplex in solution. Biochemistry. 1982 Feb 2;21(3):437–444. doi: 10.1021/bi00532a003. [DOI] [PubMed] [Google Scholar]
- Pukkila P. J., Peterson J., Herman G., Modrich P., Meselson M. Effects of high levels of DNA adenine methylation on methyl-directed mismatch repair in Escherichia coli. Genetics. 1983 Aug;104(4):571–582. doi: 10.1093/genetics/104.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman M., Wagner R. Effects of DNA methylation on mismatch repair, mutagenesis, and recombination in Escherichia coli. Curr Top Microbiol Immunol. 1984;108:23–28. doi: 10.1007/978-3-642-69370-0_3. [DOI] [PubMed] [Google Scholar]
- Scheek R. M., Boelens R., Russo N., van Boom J. H., Kaptein R. Sequential resonance assignments in 1H NMR spectra of oligonucleotides by two-dimensional NMR spectroscopy. Biochemistry. 1984 Mar 27;23(7):1371–1376. doi: 10.1021/bi00302a006. [DOI] [PubMed] [Google Scholar]
- Tibanyenda N., De Bruin S. H., Haasnoot C. A., van der Marel G. A., van Boom J. H., Hilbers C. W. The effect of single base-pair mismatches on the duplex stability of d(T-A-T-T-A-A-T-A-T-C-A-A-G-T-T-G) . d(C-A-A-C-T-T-G-A-T-A-T-T-A-A-T-A). Eur J Biochem. 1984 Feb 15;139(1):19–27. doi: 10.1111/j.1432-1033.1984.tb07970.x. [DOI] [PubMed] [Google Scholar]
- Topal M. D., Fresco J. R. Base pairing and fidelity in codon-anticodon interaction. Nature. 1976 Sep 23;263(5575):289–293. doi: 10.1038/263289a0. [DOI] [PubMed] [Google Scholar]
- WATSON J. D., CRICK F. H. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953 May 30;171(4361):964–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- Wagner R., Dohet C., Jones M., Doutriaux M. P., Hutchinson F., Radman M. Involvement of Escherichia coli mismatch repair in DNA replication and recombination. Cold Spring Harb Symp Quant Biol. 1984;49:611–615. doi: 10.1101/sqb.1984.049.01.069. [DOI] [PubMed] [Google Scholar]


