Abstract
Adult skeletal muscle stem cells are a heterogeneous cell population characterized by a small subset of undifferentiated cells that express at high level the paired/homeodomain gene Pax7. This category of satellite cells divides predominantly by asymmetric chromatid segregation generating a daughter cell that carries the mother DNA and retains stem cell property, and a daughter cell that inherits the newly-synthesized DNA and acquires the myocyte lineage.1
Recently, the established modalities of stem cell self-renewal, i.e., generation of a daughter stem cell and a daughter committed cell, have been questioned and an old theory that may have important implications in stem cell function and senescence has been reconsidered. The possibility that non-random chromatid segregation regulates stem cell division has gained interest, promoting intense discussion in the field.2-6 The immortal DNA strand hypothesis advanced by John Cairns in 19752 suggests that stem cell division is characterized by asymmetric segregation of chromatids so that one daughter cell contains only the old intact DNA and the other carries chromatids composed exclusively of the newly synthesized DNA (Figure 1). This process would attenuate the accumulation of replication errors in the parental DNA strand.2,7 In the event that deleterious mutations are acquired during DNA replication, they would be transmitted to daughter cells, which may undergo senescence and apoptosis,8-11 having a reduced capacity to repair DNA damage.2,12,13 Moreover, telomeric shortening dictated by DNA synthesis would affect only partly the actual stem cells retaining the old DNA, and telomere attrition would be largely restricted to the newly synthesized strands when become templates in subsequent descendants.6,14
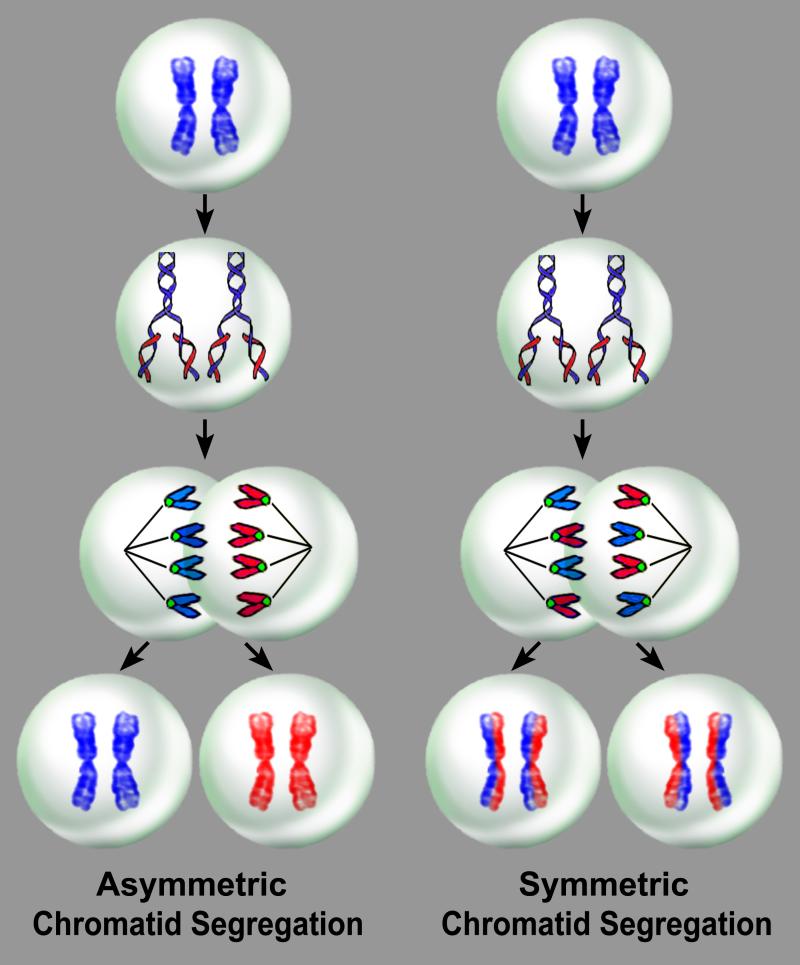
Figure 1. Non-random and random chromatid segregation.
With asymmetric chromatid segregation, a dividing mother stem cell synthesizes new DNA during S-phase and generates two daughter stem cells, one carrying only the mother DNA, which is the true stem cell, and the other only the newly-synthesized DNA. With symmetric chromatid segregation, a dividing stem cell synthesizes new DNA and generates two daughter stem cells, each carrying the mother DNA and the newly-synthesized DNA.
For historical accuracy, experimental evidence of non-random segregation of sister chromatids during cell division was obtained in the 1960s, shortly after the discovery of the double helix structure of the DNA.15 These pioneering studies were performed in mouse fibroblasts and human HeLa cells,16 and in plants, such as beans, Vicia faba, and wheat, Triticum boeoticum.17 However, no emphasis on stem cells was placed at this early time, even though the importance of the problem at hand was apparent.
Unquestionably, the immortal DNA strand theory5-7,14,18-20 has added a level of difficulty to the recognition and understanding of stem cell growth and lineage specification. Although Cairns’ theory is fascinating and potentially very important, the actual behavior of stem cells in self-renewing organs is significantly more complex. According to the immortal DNA strand hypothesis, cells carrying the old DNA maintain their undifferentiated state and preserve the stem cell compartment within the organ, while cells containing the new DNA acquire specialized functions; chromosomes are segregated asymmetrically in stem cells which represent the most primitive pool that controls cell turnover in organs in a steady state. Whether these cells are involved in the expansion of the stem cell population during rapid physiological organ growth and the formation of a large committed progeny required for tissue repair following injury is uncertain. Based on Cairns’ principle, asymmetric segregation of chromatids would be equivalent to asymmetric stem cell division, challenging the ability of stem cells to undergo symmetric division with the formation of two indistinguishable sibling cells. Theoretically, these cells cannot divide and form two daughter stem cells, or two daughter parenchymal cells.
Asymmetric and symmetric chromatid segregation of replicating satellite cells has been convincingly documented by clonal assay in vitro, as the recognition of the daughter stem cell by the expression of the endocytic protein Numb.20 For example, in the heart, the presence of Numb is associated with degradation of the Notch receptor and preservation of cell stemness.21 However, an opposite effect is observed in the brain.22 The uneven distribution of the thymidine analog, an indicator of the newly-synthesized DNA, together with the presence of the intermediate filament desmin, has led to the identification of the daughter cell committed to the myocyte phenotype in vitro.7 Based on this approach, Rocheteau and collaborators have provided strong evidence in favor of the notion that a class of satellite cells in the skeletal muscle divides by template DNA strand segregation, a pattern of stem cell growth apparently restricted to highly primitive cells. Two thymidine analogs were delivered sequentially in vivo, a protocol that allows the temporal identification of DNA strands being formed.1 The biased partitioning of these two types of DNA labeling suggests that the newly-synthesized DNA was limited to one set of duplicated chromosomes, leaving the mother DNA unlabeled. Chromosome orientation-fluorescence in situ hybridization (CO-FISH) of metaphase spreads, or tissue sections led to the discrimination of the pattern of DNA segregation at the single chromatid level,1,23 but the presence or absence of markers of stemness and commitment remained to be defined. Importantly, all chromosomes were found to be involved in the process, excluding that sister chromatid exchange occurred, or that only a unique subset of chromosomes divided by non-random chromatid segregation. These mechanisms have repeatedly been raised as alternatives to the immortal DNA strand hypothesis.4
However, in vivo studies attempting to document the modality of chromosome segregation by consecutive injections of distinct thymidine analogs pose questions on interpretation. Activated cells may enter the S-phase of the cell cycle shortly after the first pulse of DNA labeling, but traverse S-phase during the second administration of the halogenated nucleotide. Adult stem cells, including hematopoietic stem cells and cardiac stem cells (CSCs), are highly heterogeneous cell populations and the length of their telomeres and the activity of telomerase vary dramatically in stem cell subsets,8,10,24 suggesting that the telomere-telomerase axis and the length of the cell cycle are intimately linked.24 A similar condition may be encountered in skeletal muscle satellite cells which exhibit a wide range of Pax7 expression, possibly indicating several stem cell compartments with variable growth characteristics. Slowly dividing stem cells may have a significantly longer G1-phase than rapidly replicating cells, making the timing of delivery of different thymidine analogs an impossible guess. The complexity in defining precisely the length of the cell cycle in dividing stem cells in vivo has to be carefully considered in the analysis of asymmetric chromatid segregation of any class of resident adult stem cells. This information is critical for an accurate correlation between the distribution of DNA labeling and the fate of the target cell.
The possibility that symmetric and asymmetric chromatid segregation during cell division is not restricted to the satellite cells of the skeletal muscle but applies to CSCs as well poses some interesting questions concerning the organization of the cardiac niches and the role that these two types of stem cells may have in niche homeostasis and cardiac cell turnover. The long-term label-retaining assay has been employed for the identification of stem cells in various organs including the heart.25-27 This protocol was established on the assumption that stem cells divide rarely and have a very long cell cycle time. Therefore, the long-term label-retaining property of a cell would document its stemness, while the progressive dilution of the label would identify the generated progeny.
If the immortal DNA strand theory is correct, this view would not be valid. Stem cells which incorporate BrdU lose the labeled DNA by the second division,2,3 challenging the recognition and quantification of stem cells by this methodology. However, the shortcomings of Cairns’ model of stem cell self-renewal suggest that each CSC niche is composed of a dominant cell carrying the old DNA and a cluster of CSCs containing the new DNA. The brightly BrdU-labeled CSC found within each niche structure25 most likely corresponds to the BrdU-labeled parent CSC, which did not divide subsequently (Figure 2). Only CSCs formed by random chromatid segregation may leave the niche area and differentiate into specialized cardiomyocytes and vascular structures. This pattern of stem cell growth would protect the pool size of CSCs harboring the old DNA within the niches. Importantly, the presence of CSCs brightly labeled by the thymidine analog would allow the recognition of cardiac niches which contain a true stem cell that controls the destiny of the less commanding cells.
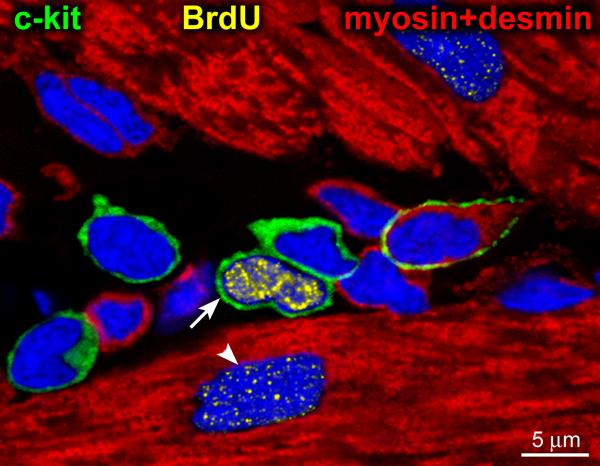
Figure 2. Cardiac stem cell niche.
Five c-kit positive cells (green) surrounded by cardiomyocytes (myosin and desmin, red). One of the 5 c-kit-positive cells is brightly labeled by BrdU (yellow, arrow). A dimly labeled cardiomyocyte nucleus (arrowhead) is also present. The brightly BrdU-labeled c-kit-positive CSC may reflect a parent stem cell derived from a BrdU-negative grandparent stem cell that incorporated the halogenated nucleotide during the first division. Subsequently, the BrdU-labeled parent stem cell entered the G0/G1 phase. Adapted from reference 25.
Antagonism between cells is not new. Competitive mechanisms are known to be critical for the modulation of organ homeostasis and regeneration.28 c-myc has been linked to competition of cells in multicellular structures where some cells survive while other cells die.29 In physiological conditions, an upregulation of c-myc in a group of cells transform these cells in “supercompetitors” capable of clonal expansion.28,29 The cluster of supercompetitor cells influences the behavior of the more vulnerable surrounding cells, which are at a growth disadvantage because of the low level of c-myc.28,29 In the mouse heart, the expression of c-myc has been found in one or two CSCs within niches composed of pockets of c-myc negative cells (Figure 3). Whether a similar mechanism is operative in the human heart and defines a cross-talk between human CSCs carry the old or new DNA is currently unknown and an important question. The presence of supercompetitor cells within niches may regulate niche function, and the absence of supercompetitor cells may alter the preservation of stem cell self-renewal, leading to the generation of old non-functional niches.10
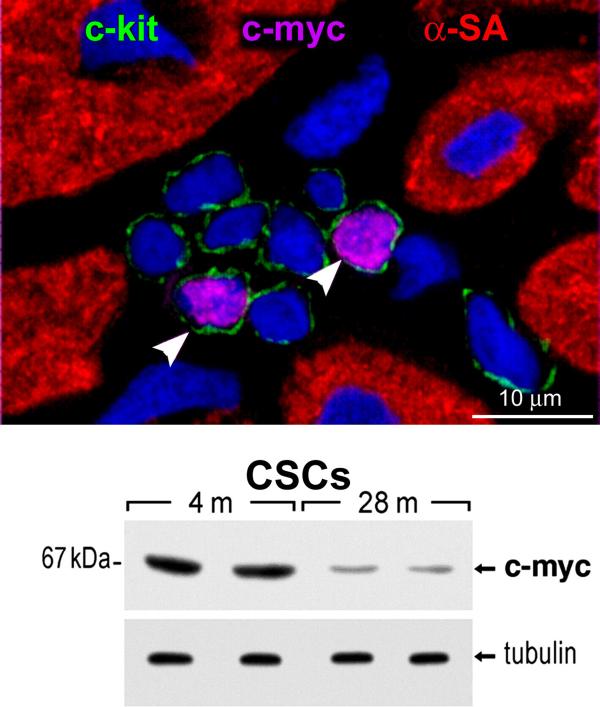
Figure 3. c-myc and CSCs.
Cardiac stem cell niche in a young rat heart contains eight c-kit-positive CSCs (green); two express c-myc (magenta, arrowheads). Nuclei are stained by DAPI. The attenuation in the expression of c-myc in CSCs with age is apparent by Western blotting. m, months. Tubulin, loading conditions.
As emphasized by Lansdorp,4 several questions have to be raised concerning asymmetric segregation of chromatids during stem cell division:30-32 they include the extremely high number of DNA lesions occurring every day in both DNA strands which are successfully repaired;33,34 the low rate of stem cell turnover in organs such as the bone marrow and the gut;35,36 the non-primitive progenitor state of cells showing immortal strand segregation due to their high rate of division;4 the presence of epigenetic marks promoting in one sister chromatid and suppressing in the other the expression of selective genes; 4,37 the impossibility to prevent telomeric shortening at the 5’ end of the immortal DNA template;4,28 and the lack of estimates of the fraction of cells showing asymmetric versus symmetric chromatid segregation.4,30-32 These variables impose a reevaluation of the strategies required for the identification, characterization and quantification of satellite cells or CSCs dividing by non-random or random chromatid segregation.39-45
The immortal DNA strand theory, if valid, would have implications in understanding the biology of myocardial aging and chronic heart failure. Whether the pattern of stem cell division and lineage specification are causally related to organ and organism aging and lifespan is presently unknown.4,24,46-50 The mystery of aging has not been resolved and the etiology of the aging myopathy is unclear although defects in CSC function may condition the senescent cardiac phenotype9,10,51-57 (Figure 4). Based on this premise, the role of symmetric stem cell division and fate, i.e., random template segregation, and asymmetric stem cell division and fate, i.e., non-random template segregation, will have to be determined to define the contribution of CSC growth and differentiation to the development of the old failing heart.
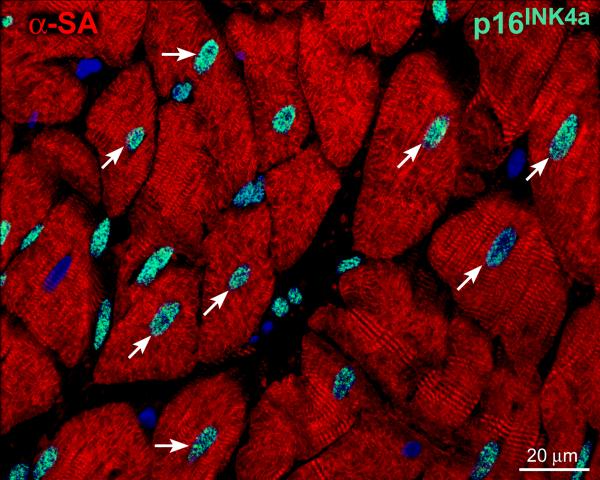
Figure 4. Senescent human heart.
The myocardium of a 79 years old man is characterized by cardiomyocytes (α-sarcomeric actin, α-SA, red) that express the senescence-associated protein p16INK4a (green, arrows).
Whether these patterns of stem cell division are an all-or-none phenomenon that applies to all organs and the heart in particular is unknown. This is biologically and clinically relevant because the growth potential of stem cells possessing the old DNA is theoretically superior to that of stem cells inheriting only the newly synthesized DNA. Throughout life, human CSCs may consist of two distinct subsets and their relative contribution may change dramatically in the developing, adult, and senescent failing heart.
In dividing stem cells, the asymmetric segregation of chromatids has been interpreted as a general process of stem cell growth and differentiation,2,6,7,20 but this may not be the case. Three patterns of stem cell division have to be considered. Non-random chromatid segregation may not necessarily lead to the formation of a daughter stem cell and a daughter committed cell (a); two daughter stem cells (b) or two daughter committed cells (c) may be generated. According to the theory of non-random chromatid segregation, when two daughter stem cells are created, one will carry the old DNA and the other the young DNA, and when two daughter committed cells are formed, the pool size of stem cells with immortal DNA will be reduced. Moreover, environmental factors and cardiac diseases, which increase with age, decrease the number of functionally-competent CSCs,8,58,59 contributing to the onset of the aging myopathy and heart failure. However, a small number of human CSCs harboring the immortal DNA or dividing by random segregation of chromatids may be preserved and employed therapeutically to reverse the old cardiac phenotype and chronic heart failure.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rocheteau P, Gayraud-Morel B, Siegl-Cachedenier I, Blasco MA, Tajbakhsh S. A subpopulation of adult skeletal muscle stem cells retains all template DNA strands after cell division. Cell. 2012;148:112–125. doi: 10.1016/j.cell.2011.11.049. [DOI] [PubMed] [Google Scholar]
- 2.Cairns J. Cancer and the immortal strand hypothesis. Genetics. 2006;174:1069–1072. doi: 10.1534/genetics.104.66886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rando TA. The immortal strand hypothesis: segregation and reconstruction. Cell. 2007;129:1239–1243. doi: 10.1016/j.cell.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 4.Lansdorp PM. Immortal strands? Give me a break. Cell. 2007;129:1244–1247. doi: 10.1016/j.cell.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 5.Merok JR, Lansita JA, Tunstead JR, Sherley JL. Cosegregation of chromosomes containing immortal DNA strands in cells that cycle with asymmetric stem cell kinetics. Cancer Res. 2002;62:6791–6795. [PubMed] [Google Scholar]
- 6.Potten CS, Owen G, Booth D. Intestinal stem cells protect their genome by selective segregation of template DNA strands. J Cell Sci. 2002;115:2381–2388. doi: 10.1242/jcs.115.11.2381. [DOI] [PubMed] [Google Scholar]
- 7.Conboy MJ, Karasov AO, Rando TA. High incidence of non-random template strand segregation and asymmetric fate determination in dividing stem cells and their progeny. PLoS Biol. 2007;5:e102. doi: 10.1371/journal.pbio.0050102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Urbanek K, Torella D, Sheikh F, De Angelis A, Nurzynska D, Silvestri F, Beltrami CA, Bussani R, Beltrami AP, Quaini F, Bolli R, Leri A, Kajstura J, Anversa P. Myocardial regeneration by activation of multipotent cardiac stem cells in ischemic heart failure. Proc Natl Acad Sci U S A. 2005;102:8692–8697. doi: 10.1073/pnas.0500169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torella D, Rota M, Nurzynska D, Musso E, Monsen A, Shiraishi I, Zias E, Walsh K, Rosenzweig A, Sussman MA, Urbanek K, Nadal-Ginard B, Kajstura J, Anversa P, Leri A. Cardiac stem cell and myocyte aging, heart failure, and insulin-like growth factor-1 overexpression. Circ Res. 2004;94:514–524. doi: 10.1161/01.RES.0000117306.10142.50. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez A, Rota M, Nurzynska D, Misao Y, Tillmanns J, Ojaimi C, Padin-Iruegas ME, Müller P, Esposito G, Bearzi C, Vitale S, Dawn B, Sanganalmath SK, Baker M, Hintze TH, Bolli R, Urbanek K, Hosoda T, Anversa P, Kajstura J, Leri A. Activation of cardiac progenitor cells reverses the failing heart senescent phenotype and prolongs lifespan. Circ Res. 2008;102:597–606. doi: 10.1161/CIRCRESAHA.107.165464. [DOI] [PubMed] [Google Scholar]
- 11.Rota M, LeCapitaine N, Hosoda T, Boni A, De Angelis A, Padin-Iruegas ME, Esposito G, Vitale S, Urbanek K, Casarsa C, Giorgio M, Lüscher TF, Pelicci PG, Anversa P, Leri A, Kajstura J. Diabetes promotes cardiac stem cell aging and heart failure, which are prevented by deletion of the p66shc gene. Circ Res. 2006;99:42–52. doi: 10.1161/01.RES.0000231289.63468.08. [DOI] [PubMed] [Google Scholar]
- 12.Potten CS. Extreme sensitivity of some intestinal crypt cells to X and gamma irradiation. Nature. 1977;269:518–521. doi: 10.1038/269518a0. [DOI] [PubMed] [Google Scholar]
- 13.Ijiri K, Potten CS. Further studies on the response of intestinal crypt cells of different hierarchical status to eighteen different cytotoxic agents. Br J Cancer. 1987;55:113–123. doi: 10.1038/bjc.1987.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karpowicz P, Morshead C, Kam A, Jervis E, Ramunas J, Cheng V, van der Kooy D. Support for the immortal strand hypothesis: neural stem cells partition DNA asymmetrically in vitro. J Cell Biol. 2005;170:721–732. doi: 10.1083/jcb.200502073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watson JD, Crick FH. A structure for deoxyribose nucleic acid. 1953. Nature. 2003:421397–421398. [PubMed] [Google Scholar]
- 16.Lark KG, Consigli RA, Minocha HC. Segregation of sister chromatids in mammalian cells. Science. 1966;154:1202–1205. doi: 10.1126/science.154.3753.1202. [DOI] [PubMed] [Google Scholar]
- 17.Lark KG. Nonrandom segregation of sister chromatids in Vicia faba and Triticum boeoticum. Proc Natl Acad Sci U S A. 1967;58:352–359. doi: 10.1073/pnas.58.1.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rambhatla L, Ram-Mohan S, Cheng JJ, Sherley JL. Immortal DNA strand cosegregation requires p53/IMPDH-dependent asymmetric self-renewal associated with adult stem cells. Cancer Res. 2005;65:3155–3161. doi: 10.1158/0008-5472.CAN-04-3161. [DOI] [PubMed] [Google Scholar]
- 19.Smith GH. Label-retaining epithelial cells in mouse mammary gland divide asymmetrically and retain their template DNA strands. Development. 2005;132:681–687. doi: 10.1242/dev.01609. [DOI] [PubMed] [Google Scholar]
- 20.Shinin V, Gayraud-Morel B, Gomès D, Tajbakhsh S. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nat Cell Biol. 2006;8:677–687. doi: 10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- 21.Boni A, Urbanek K, Nascimbene A, Hosoda T, Zheng H, Delucchi F, Amano K, Gonzalez A, Vitale S, Ojaimi C, Rizzi R, Bolli R, Yutzey KE, Rota M, Kajstura J, Anversa P, Leri A. Notch1 regulates the fate of cardiac progenitor cells. Proc Natl Acad Sci USA. 2008;105:15529–15534. doi: 10.1073/pnas.0808357105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Ables JL, Breunig JJ, Eisch AJ, Rakic P. Not(ch) just development: Notch signalling in the adult brain. Nat Rev Neurosci. 2011;12:269–283. doi: 10.1038/nrn3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baerlocher GM, Vulto I, de Jong G, Lansdorp PM. Flow cytometry and FISH to measure the average length of telomeres (flow FISH). Nat Protoc. 2006;1:2365–2376. doi: 10.1038/nprot.2006.263. [DOI] [PubMed] [Google Scholar]
- 24.Aubert G, Lansdorp PM. Telomeres and aging. Physiol Rev. 2008;88:557–579. doi: 10.1152/physrev.00026.2007. [DOI] [PubMed] [Google Scholar]
- 25.Urbanek K, Cesselli D, Rota M, Nascimbene A, De Angelis A, Hosoda T, Bearzi C, Boni A, Bolli R, Kajstura J, Anversa P, Leri A. Stem cell niches in the adult mouse heart. Proc Natl Acad Sci U S A. 2006;103:9226–9231. doi: 10.1073/pnas.0600635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braun KM, Niemann C, Jensen UB, Sundberg JP, Silva-Vargas V, Watt FM. Manipulation of stem cell proliferation and lineage commitment: visualisation of label-retaining cells in wholemounts of mouse epidermis. Development. 2003;130:5241–5255. doi: 10.1242/dev.00703. [DOI] [PubMed] [Google Scholar]
- 27.Potten CS. Keratinocyte stem cells, label-retaining cells and possible genome protection mechanisms. J Investig Dermatol Symp Proc. 2004;9:183–195. doi: 10.1111/j.1087-0024.2004.09305.x. [DOI] [PubMed] [Google Scholar]
- 28.Abrams JM. Competition and compensation: coupled to death in development and cancer. Cell. 2002;110:403–406. doi: 10.1016/s0092-8674(02)00904-2. [DOI] [PubMed] [Google Scholar]
- 29.Moreno E, Basler K. dMyc transforms cells into super-competitors. Cell. 2004;117:117–129. doi: 10.1016/s0092-8674(04)00262-4. [DOI] [PubMed] [Google Scholar]
- 30.Kuroki T, Murakami Y. Random segregation of DNA strands in epidermal basal cells. Jpn J Cancer Res. 1989;80:637–642. doi: 10.1111/j.1349-7006.1989.tb01690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiel MJ, He S, Ashkenazi R, Gentry SN, Teta M, Kushner JA, Jackson TL, Morrison SJ. Haematopoietic stem cells do no asymmetrically segregate chromosomes or retain BrdU. Nature. 2007;449:238–242. doi: 10.1038/nature06115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waghmare SK, Bansal R, Lee J, Zhang YV, McDermitt DJ, Tumbar T. Quantitative proliferation dynamics and random chromosome segregation of hair follicle stem cells. EMBO J. 2008;27:1309–1320. doi: 10.1038/emboj.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfeiffer P. The mutagenic potential of DNA double-strand break repair. Toxicol Lett. 1998;96-97:119–129. doi: 10.1016/s0378-4274(98)00058-7. [DOI] [PubMed] [Google Scholar]
- 34.Rajski SR, Jackson BA, Barton JK. DNA repair: models for damage and mismatch recognition. Mutat Res. 2000;447:49–72. doi: 10.1016/s0027-5107(99)00195-5. [DOI] [PubMed] [Google Scholar]
- 35.Lansdorp PM. Self-renewal of stem cells. Biol Blood Marrow Transplant. 1997;3:171–178. [PubMed] [Google Scholar]
- 36.Barker N, van de Wetering M, Clevers H. The intestinal stem cell. Genes Dev. 2008;22:1856–1864. doi: 10.1101/gad.1674008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 38.Lingner J, Cooper JP, Cech TR. Telomerase and DNA end replication: no longer a lagging strand problem? Science. 1995;269:1533–1534. doi: 10.1126/science.7545310. [DOI] [PubMed] [Google Scholar]
- 39.Beltrami AP, Barlucchi L, Torella D, Baker M, Limana F, Chimenti S, Kasahara H, Rota M, Musso E, Urbanek K, Leri A, Kajstura J, Nadal-Ginard B, Anversa P. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–776. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 40.Oh H, Bradfute SB, Gallardo TD, Nakamura T, Gaussin V, Mishina Y, Pocius J, Michael LH, Behringer RR, Garry DJ, Entman ML, Schneider MD. Cardiac progenitor cells from adult myocardium: homing, differentiation, and fusion after infarction. Proc Natl Acad Sci U S A. 2003;100:12313–12318. doi: 10.1073/pnas.2132126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfister O, Mouquet F, Jain M, Summer R, Helmes M, Fine A, Colucci WS, Liao R. CD31- but Not CD31+ cardiac side population cells exhibit functional cardiomyogenic differentiation. Circ Res. 2005;97:52–61. doi: 10.1161/01.RES.0000173297.53793.fa. [DOI] [PubMed] [Google Scholar]
- 42.Tomita Y, Matsumura K, Wakamatsu Y, Matsuzaki Y, Shibuya I, Kawaguchi H, Ieda M, Kanakubo S, Shimazaki T, Ogawa S, Osumi N, Okano H, Fukuda K. Cardiac neural crest cells contribute to the dormant multipotent stem cell in the mammalian heart. J Cell Biol. 2005;170:1135–1146. doi: 10.1083/jcb.200504061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith RR, Barile L, Cho HC, Leppo MK, Hare JM, Messina E, Giacomello A, Abraham MR, Marbán E. Regenerative potential of cardiosphere-derived cells expanded from percutaneous endomyocardial biopsy specimens. Circulation. 2007;115:896–908. doi: 10.1161/CIRCULATIONAHA.106.655209. [DOI] [PubMed] [Google Scholar]
- 44.Bearzi C, Rota M, Hosoda T, Tillmanns J, Nascimbene A, De Angelis A, Yasuzawa-Amano S, Trofimova I, Siggins RW, Lecapitaine N, Cascapera S, Beltrami AP, D'Alessandro DA, Zias E, Quaini F, Urbanek K, Michler RE, Bolli R, Kajstura J, Leri A, Anversa P. Human cardiac stem cells. Proc Natl Acad Sci USA. 2007;104:14068–14073. doi: 10.1073/pnas.0706760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oyama T, Nagai T, Wada H, Naito AT, Matsuura K, Iwanaga K, Takahashi T, Goto M, Mikami Y, Yasuda N, Akazawa H, Uezumi A, Takeda S, Komuro I. Cardiac side population cells have a potential to migrate and differentiate into cardiomyocytes in vitro and in vivo. J Cell Biol. 2007;176:329–341. doi: 10.1083/jcb.200603014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blasco MA. Telemoere length, stem cells and aging. Nat Chem Biol. 2007;3:640–649. doi: 10.1038/nchembio.2007.38. [DOI] [PubMed] [Google Scholar]
- 47.Dimmeler S, Leri A. Aging and disease as modifiers of efficacy of cell therapy. Circ Res. 2008;102:1319–1330. doi: 10.1161/CIRCRESAHA.108.175943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Janzen V, Forkert R, Fleming HE, Saito Y, Waring MT, Dombkowski DM, Cheng T, DePinho RA, Sharpless NE, Scadden DT. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443:421–426. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- 49.Beausejour CM, Campisi J. Ageing: balancing regeneration and cancer. Nature. 2006;443:404–405. doi: 10.1038/nature05221. [DOI] [PubMed] [Google Scholar]
- 50.Molofsky AV, Slutsky SG, Joseph NM, He S, Pardal R, Krishnamurthy J, Sharpless NE, Morrison SJ. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443:448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chimenti C, Kajstura J, Torella D, Urbanek K, Heleniak H, Colussi C, Di Meglio F, Nadal-Ginard B, Frustaci A, Leri A, Maseri A, Anversa P. Senescence and death of primitive cells and myocytes lead to premature cardiac aging and heart failure. Circ Res. 2003;93:604–613. doi: 10.1161/01.RES.0000093985.76901.AF. [DOI] [PubMed] [Google Scholar]
- 52.Capasso JM, Palackal T, Olivetti G, Anversa P. Severe myocardial dysfunction induced by ventricular remodeling in aging rat hearts. Am J Physiol. 1990;259:H1086–H1096. doi: 10.1152/ajpheart.1990.259.4.H1086. [DOI] [PubMed] [Google Scholar]
- 53.Olivetti G, Melissari M, Capasso JM, Anversa P. Cardiomyopathy of the aging human heart. Myocyte loss and reactive cellular hypertrophy. Circ Res. 1991;68:1560–1568. doi: 10.1161/01.res.68.6.1560. [DOI] [PubMed] [Google Scholar]
- 54.Capasso JM, Fitzpatrick D, Anversa P. Cellular mechanisms of ventricular failure: myocyte kinetics and geometry with age. Am J Physiol. 1992;262:H1770–H1781. doi: 10.1152/ajpheart.1992.262.6.H1770. [DOI] [PubMed] [Google Scholar]
- 55.Kajstura J, Pertoldi B, Leri A, Beltrami CA, Deptala A, Darzynkiewicz Z, Anversa P. Telomere shortening is an in vivo marker of myocyte replication and aging. Am J Pathol. 2000;156:813–819. doi: 10.1016/S0002-9440(10)64949-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leri A, Franco S, Zacheo A, Barlucchi L, Chimenti S, Limana F, Nadal-Ginard B, Kajstura J, Anversa P, Blasco MA. Ablation of telomerase and telomere loss leads to cardiac dilatation and heart failure associated with p53 upregulation. EMBO J. 2003;22:131–139. doi: 10.1093/emboj/cdg013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rota M, Hosoda T, De Angelis A, Arcarese ML, Esposito G, Rizzi R, Tillmanns J, Tugal D, Musso E, Rimoldi O, Bearzi C, Urbanek K, Anversa P, Leri A, Kajstura J. The young mouse heart is composed of myocytes heterogeneous in age and function. Circ Res. 2007;101:387–399. doi: 10.1161/CIRCRESAHA.107.151449. [DOI] [PubMed] [Google Scholar]
- 58.Kajstura J, Leri A, Finato N, Di Loreto C, Beltrami CA, Anversa P. Myocyte proliferation in end-stage cardiac failure in humans. Proc Natl Acad Sci U S A. 1998;95:8801–8805. doi: 10.1073/pnas.95.15.8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beltrami AP, Urbanek K, Kajstura J, Yan SM, Finato N, Bussani R, Nadal-Ginard B, Silvestri F, Leri A, Beltrami CA, Anversa P. Evidence that human cardiac myocytes divide after myocardial infarction. N Engl J Med. 2001;344:1750–1757. doi: 10.1056/NEJM200106073442303. [DOI] [PubMed] [Google Scholar]