Abstract
Herein a photon manipulated mesoporous release system was constructed based on azobenzene-modified nucleic acids. In this system, the azobenzene-incorporated DNA double strands were immobilized at the pore mouth of meso-porous silica nanoparticles. The photo-isomerization of azobenzene induced dehybridization/hybridization switch of complementary DNA, causing uncapping/capping of pore gates of mesoporous silica. This nanoplatform permits holding of guest molecules within the nanopores under visible light but release them when light wavelength turns to UV range. These DNA/mesoporous silica hybrid nanostructures were exploited as carriers for cancer cell chemotherapy drug doxorubicin (DOX) due to its stimuli-responsive property as well as good biocompatibility via MTT assay. It is found that the drug release behavior is light wavelength sensitive. Switching of the light from visible to UV range uncapped the pores causes the release of DOX from the mesoporous silica nanospheres and an obvious cytotoxic effect on cancer cells. We envision that this photo-controlled drug release system could find potential applications in cancer therapy.
Keywords: azobenzene, photoregulation, mesoporous silica, nucleic acids, drug delivery
Nucleic acids have been recognized as important building blocks for nanotechnology by virtue of their remarkable molecular self-recognition capabilities and unique structural motif. Fabrication and characterization of DNA nanomachines that respond to external stimuli have indicated that these structures have practical applications in molecular sensing, logic gate operation, and nanomedicine.1–4 However, few studies have investigated the use of photon-manipulated DNA nanomachines for drug delivery applications.
The features of mesoporous silica, such as its stable mesostructure, lack of cytotoxicity, large surface areas, and tailorable pore sizes, make it an excellent candidate for drug release. In conventional mesoporous systems, the release of guest molecules usually follows a sustained kinetic mechanism that can be expressed in terms of diffusion of adsorbed molecules throughout the mesopore channels. Release kinetics, therefore, can be interpreted in terms of the Fickian diffusion coefficients, which depend on the characteristics of both the molecules and the silica particles.5,6
For certain applications, such as those involving highly toxic antitumor drugs, zero release before the targeted cells or tissues is required. As a result, together with the development of mesoporous silica for drug-delivery, the stimuli-responsive ability of this system has been a point at issue in current research. Most of previously reported cases have been limited to the use of nanoparticles and polymer molecules for controllable nanovalves on mesoporous surfaces.7–14 Recently, DNA molecules have been exploited to control the pore opening/closing due to their unique complementary properties and flexible structures. Bein et al. first attached biotin-labeled DNA double strands on the pores of mesoporous nanoparticles,15 and they used temperature changes to control the hybridization of DNA double strands, corresponding to valve opening and closing. Ren et al. attached quadruplex DNA to mesoporous silica and achieved controlled release by changing the solution pH.16 They further capped pores by the DNA duplexes and used endonuclease to degrade the DNA for pore opening.17 However, compared to temperature, pH, or other external stimuli, photoregulation has significant advantages for easy control of movement and conformation. In addition to the capability for remote control, irradiation is an accurate and simple method, which does not require any additional solution components. Moreover, it is a clean source of energy, and as demonstrated in this study, it can be applied repeatedly and rapidly without loss of efficiency.
In this study, we report a new photo-responsive DNA/mesoporous silica hybrid formed by introducing azobenzene moieties into the DNA molecule. As the most popular phototransformable molecules, azobenzene and its derivatives can undergo reversible isomerization from the trans to cis forms under irradiation at 300–380 nm and from cis to trans using light at wavelengths > 400 nm.18 Applications of stereoisomerization in the development of nonlinear optical materials,19 molecular motors,20,21 and even in peptide engineering22,23 have been reported. With azobenzene-tethered DNA, formation and dissociation of a double-stranded structure can be reversibly photo-regulated by cis-trans isomerization of the azobenzene upon UV-Vis light irradiation.24,25 Immobilization of the azobenzene-incorporated DNA double strands at the pore mouth of mesoporous silica provides a novel type of reversible photo-regulated release platform. This nanoplatform permits holding of guest molecules within the nanopores under visible light, with controlled release when the wavelength is switched to the UV range.
In this work, Rhodamine 6G (Rh6G) was used as a model guest molecule to evaluate the loading and controlled releasing behavior of the composite nanospheres. The photo-isomerization of azobenzene results in dehybridization/hybridization of complementary DNA, causing uncapping/capping of pore gates of mesoporous silica. After loading with the anticancer drug doxorubicin (DOX) in the interior pores, this nanostructure was utilized for chemotherapy of cancer cells. The cytotoxic effect of the DOX-loaded nanospheres to CEM cells and A549 cells was examined, showing effective light controlled therapy after release of DOX from the pore structure.
Materials and Methods
Synthesis of azobenzene phosphoramidite
Azobenzene phosphoramidite was synthesized according to the protocol reported by Asanuma et al.30 Compound 1. 1H NMR (CDCl3): δ 7.96-7.38 (m, 9H), δ 7.12 (d, 1H), δ 4.33 (m, 1H), δ 4.09 (m, 1H), δ 3.98 (d, 2H), δ 1.29 (d, 3H). Compound 2. 1H NMR (CDCl3): δ 8.00-6.78 (m, 23H), δ 4.25 (m, 1H), δ 4.17 (m, 1H), δ 3.77 (s, 6H), δ 3.60 and 3.42 (dd, 2H), 1.23 (d, 3H). Compound 3. 1H NMR (CDCl3): δ 8.00-6.79 (m, 22H), δ 6.62 (d, 1H), δ 4.48 (m, 1H), δ 4.39 (m, 1H), δ 4.21-4.10 (m, 2H), δ 3.77 (s, 6H), δ 3.57-3.34 (m, 4H), δ 2.76-2.72 (m, 2H), δ 1.30-1.25 (m, 15H). 31P (CDCl3): δ 149.
Synthesis of azobenzene-modified-DNA
The azobenzene-modified DNA sequences were synthesized using an ABI3400 DNA/RNA synthesizer (Applied Biosystems). The synthesis started with a controlled pore glass (CPG) column at the 1 µmole scale. The azobenzene coupling was realized using the azobenzene phosphoramidite synthesized by the protocol mentioned above. A proper amount of azobenzene phosphoramidite was dissolved in dry acetonitrile in a vial connected to the synthesizer. A ProStar HPLC (Varian) with a C18 column (Econosil, 5µ, 250 mm × 4.6 mm) from Alltech (Deerfield, IL) was used for DNA purification. The arm-DNA (5’-T-A-C-C-T-A-NH2-3’) and normal DNA (5’-T-A-G-G-T-A-CCC-CCC-CCC-CCC-T-A-G-G-T-A-3’) were synthesized by the same protocol.
Synthesis of mesoporous silica MCM-41
CTAB (1.00 g, 2.74 mmol) was dissolved in 480 mL of nanopure water. Sodium hydroxide aqueous solution (2.00 M, 3.50 mL) was introduced to the CTAB solution and the temperature of the mixture was adjusted to 80 °C. TEOS (5.00 mL, 22.4 mmol) was added dropwise to the surfactant solution under vigorous stirring. The mixture was allowed to react for 2 h to give a white precipitate. This solid crude product was filtered, washed with deionized water and methanol, and dried in air to yield the as-synthesized mesoporous silica nanoparticles (denoted as MSN). To remove the surfactant template (CTAB), 1.50 g of the as-synthesized MSN was refluxed for 24 h in a methanolic solution of 9.00 mL of HCl (37.4%) in 160.00 mL methanol. The resulting material was filtered and extensively washed with deionized water and methanol.
Functionalization of mesoporous silica with DNA
MSN (1.00 g) was refluxed for 20 h in 80.00 mL of anhydrous toluene with 0.25 mL (1.00 mmol) of 3-isocyanatopropyltriethoxysilane to yield the 3-isocyanatopropyl-functionalized MSN (denoted as ICP-MSN) material. After removal of toluene by a rotary evaporator, the purified ICP-MSN (100 mg) was redispersed in 2 mL of anhydrous acetonitrile. The amine-modified oligonucleotides (5’-T-A-C-C-T-A-NH2-3’) with a solution concentration of 200 µM were then added. The amino groups of DNA were allowed to react with the ICP functional groups present on the surface of MSN overnight at room temperature to obtain the DNA modified MSN.
Rhodamine 6G loading and DNA capping
The functionalized MSN (50 mg) was added to 5 mL of Rh6G (5.00 mM) in PBS buffer (100 mM, pH = 7.4) solution. After the mixture was stirred for 24 h in a dark area, the final mixture was then centrifuged and washed with a PBS buffer solution three times. The azobenzene-modified-ssDNA (1 mL, 120 µM) was added to complement to DNA linkers functionalized on MSN and cap the pores on the mesoporous particles. The surface-adsorbed dye was washed with PBS buffer and removed by centrifugation. The amount of Rh6G loaded on the MSN was determined by analyzing the supernatant solution spectrophotometrically. All the washing solutions were collected, and the loading amount was calculated from the difference in number of moles of the initial and uncapped dye to be approximately 47 µmol/g MSNs.
Model drug release test
DNA-modified-MSN loaded with Rh6G was dispersed in 6 mL of Tris buffer (20 mM Tris-HCl, 20 mM NaCl, 2 mM MgCl2, pH = 7.0). The release kinetics study was carried out using a mini dialysis tube (MW = 7,000). The released Rh6G could cross the dialysis membrane, but not the MSN. Aliquots were taken from the solution outside the dialysis membrane, and the delivery of rhodamine dye from the pores to the buffer solution was monitored via the fluorescence peak of the dye centered at 560 nm.
Cytotoxicity Assay
The cytotoxicity study was performed using the MTT assay for CEM and A549 cell lines. Cells were seeded into 96-well cell-culture plate at 105/well. The cells were incubated for 48 h at 37 °C under 5% CO2. The mesoporous silica particles at concentrations of 100 µg·mL−1 in medium were added to the wells of the treatment group. Six-fold diluted MTT (120 µL/well) in PBS solution was added to each well and incubated at 37 °C for 2 h. The cell viability was determined colorimetrically by using a microplate reader (VERSA), which was used to measure the OD490 nm (Absorbance value).
Internalization Study
To investigate the internalization of DOX-loaded azo-DNA MSNs into cells, samples containing A549 cells with a concentration of 105 cells/well were incubated with the desired concentrations of DOX-loaded azo-DNA MSNs at 37 °C in a volume of 200 µL of binding buffer for 2 h with 5% CO2 atmosphere. The cells were then subjected to confocal fluorescence microscopy analysis. A 488 nm argon laser was used for the excitation of DOX.
Results and Discussion
Details of azobenzene-modified DNA (denoted as azo-DNA) strand synthesis are shown in the Supporting Information. The single-stranded DNA arms (denoted as arm-DNA) were designed having a complementary sequence to azo-DNA. Duplex formation between azo-DNA strand and arm-DNA was tested using free DNA strands in solution by fluorescence resonance energy transfer (FRET), as reported in our previous works.25,26 The azo-DNA and arm-DNA were labelled with FAM (fluorophore) and Dabcyl (quencher) respectively as shown below: azo-DNA labelled with FAM: 5’-T-Azo-A-G-Azo-G-T-Azo-A-CCC-CCC-CCC-CCC-T-Azo-A-G-Azo-G-T-Azo-A -FAM-3’ arm-DNA labelled with Dabcyl: 5’-Dabcyl-T-A-C-C-T-A-3’
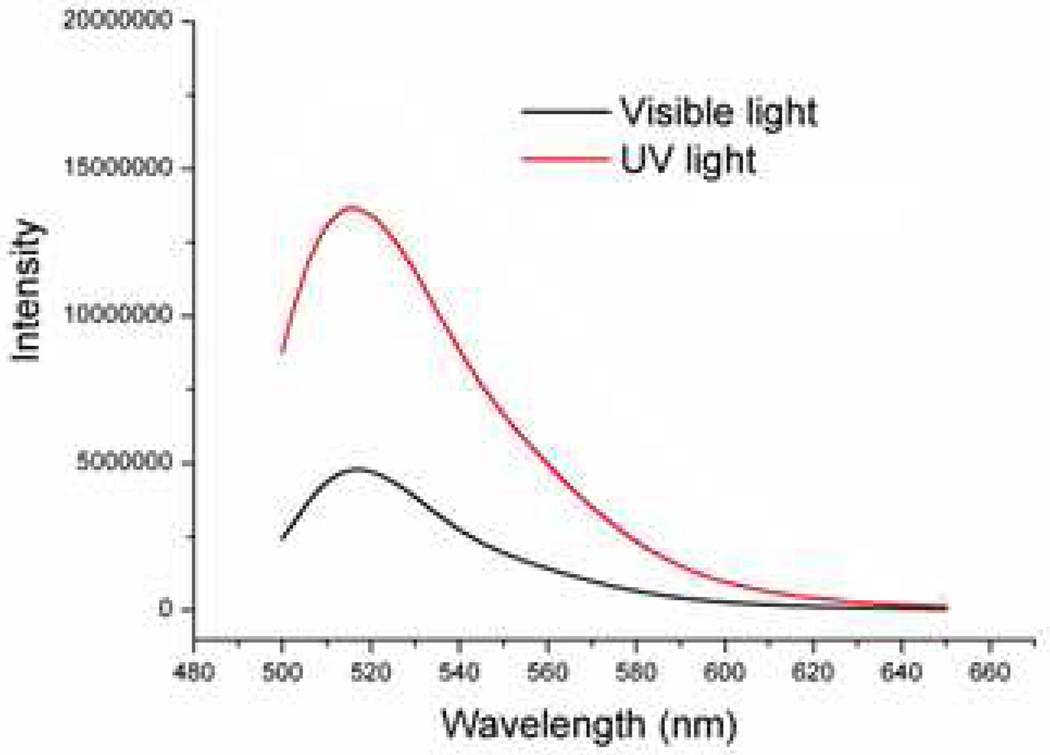
The fluorescence signal of FAM is quenched because of FRET between FAM and Dabcyl when azo-DNA hybridizes with arm-DNA. However, when the azo-DNA dehybridizes from the arm DNA, the fluorescence intensity will recover due to the separation between FAM and Dabcyl. As shown in Figure 1, the fluorescence intensity was low under visible light (λ = 450 nm), indicating the duplex formation between azo-DNA and arm-DNA strands. However, upon irradiation with UV light (λ = 365 nm), a significant increase of fluorescence intensity was detected due to the dehybridization between azo-DNA and arm-DNA induced by UV light. This result demonstrated the obvious hybridization behavior difference between the trans and cis forms of the azo-DNA sequence.
Figure 1.
Fluorescence spectra of buffer solution containing FAM labeled azo-DNA and Dabcyl labeled arm-DNA. Black line represents DNA under visible light and red line represents DNA after UV light irradiation. Buffer: 20 mM Tris-HCl, 20 mM NaCl, 2 mM MgCl2, pH = 7.0, [azo-DNA] = 2 µM, [arm-DNA] = 4 µM.
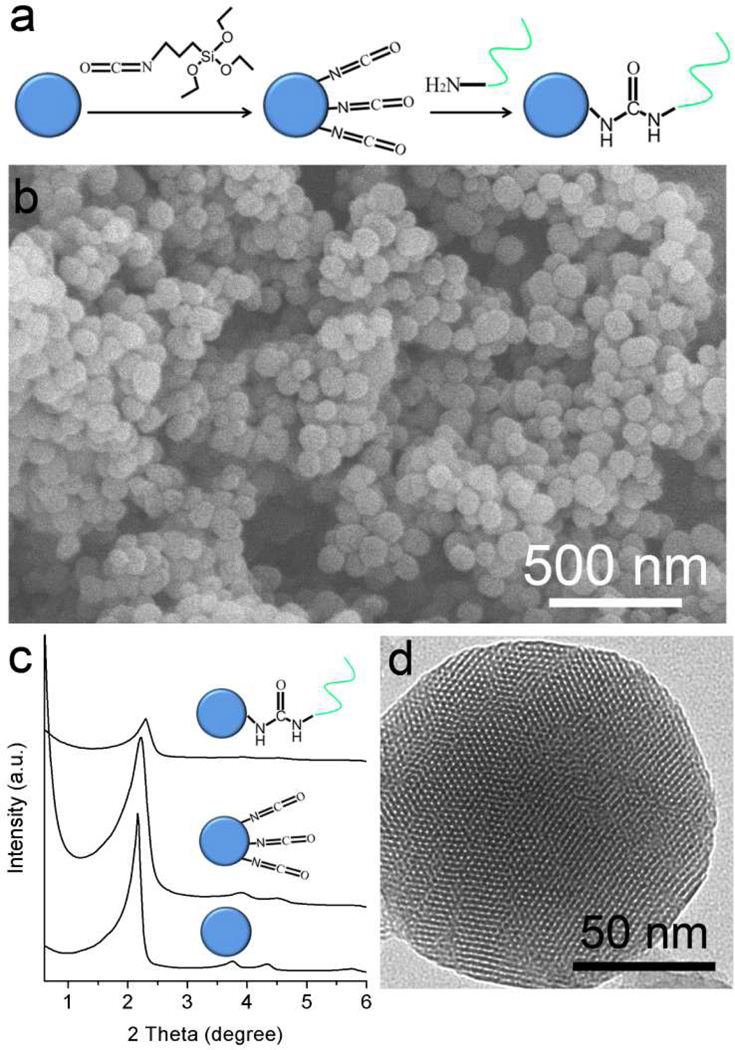
The synthesis of mesoporous silica nanoparticles (MSNs) was carried out following a base-catalyzed sol-gel procedure.27 The surface was first functionalized with cyanato groups via post-modification with 3-isocyanatopropyltriethoxysilane, followed by treatment with the arm-DNA modified with an amino group at the 3’ end (5’-T-A-C-C-T-A-NH2-3’) for conjugation to the MSN surface (Figure 2a).28 The scanning electron microscopy (SEM) image in Figure 2b shows that the resulting silica particles have uniform size of about 100 nm. The ordered two-dimensional hexagonal mesoporous structure was confirmed by small-angle X-ray diffraction (XRD) (Figure 2c) and transmission electron microscopy (TEM) characterization (Figure 2d), which indicated that the morphology and structure of mesoporous silica nanoparticles were not affected by DNA modification. The immobilization efficiency was determined to be 4.3 µmol DNA arm per gram SiO2 by UV/Vis spectroscopy. The N2 adsorption–desorption isotherm recorded at 77 K shows a typical type IV curve and a narrow pore size distribution centered at 2.5 nm (Figure S1). Using the BJH model on the adsorption branch of the isotherm, the surface area and pore volume of mesoporous silica were calculated to be up to 842 m2 g−1 and 1.032 cm3 g−1, respectively. After modification with arm-DNA, the surface area changed to 721 m2 g−1 while pore volume was 0.894 cm3 g−1.
Figure 2.
(a) Synthetic strategy for arm-DNA grafting onto the surface of MSNs. (b) Representative SEM image of DNA-functionalized MSN. (c) Small-angle XRD of MSNs, cyanato-functioned-MSN, and DNA-modified-MSN. (d) TEM image of DNA-modified-MSNs.
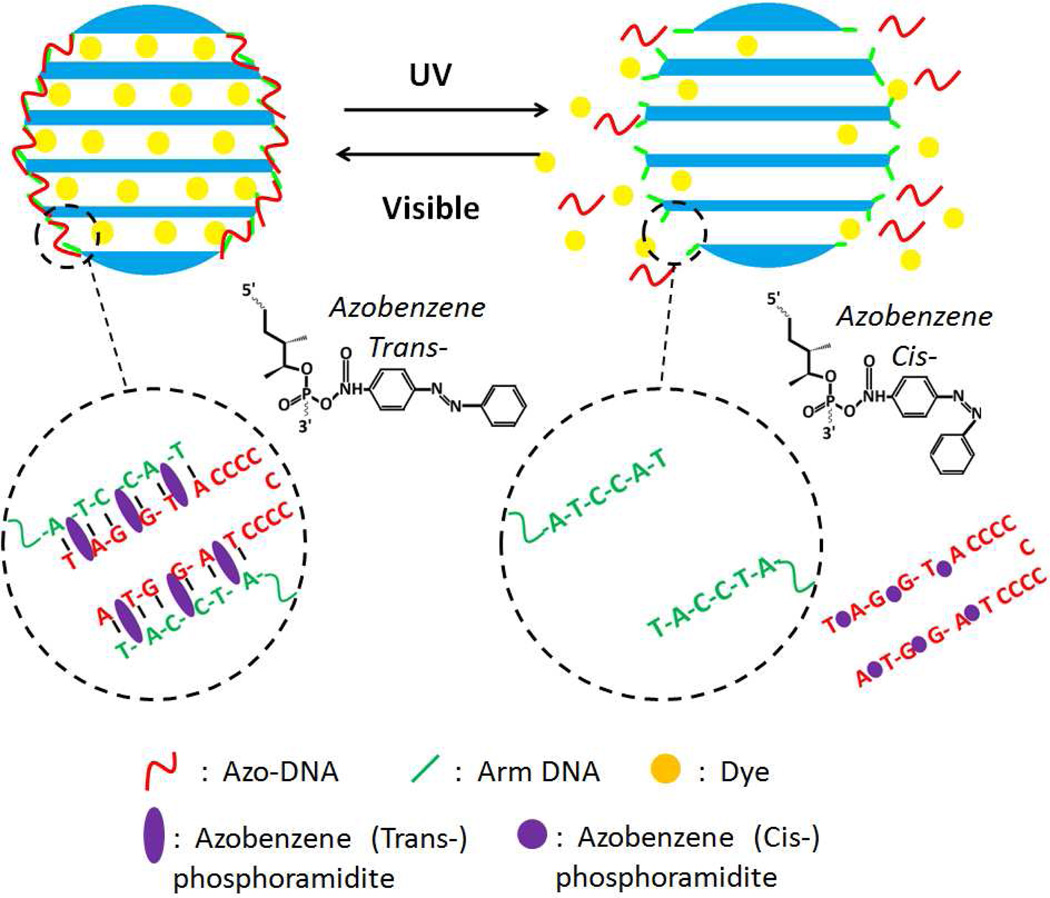
The opening of the valve is achieved by DNA duplex dehybridization caused by UV light irradiation, as shown in Scheme 1. Amino-modified arm-DNA strands are attached to the pore opening at their 3’ ends. The linker azo-DNA strands contain azobenzene moieties and segments complementary to the arms, as shown in Table 1. Under visible light irradiation (λ = 450 nm), the azobenzene moieties are in the trans form, and the linkers can hybridize to the arms, as shown in Scheme 1, to form a cap over the pore mouth. Upon exposure to UV (λ = 365 nm), the azobenzenes are converted to the cis form. This causes dehybridization and the pore opens as the linker is released into solution. The sensitive and rapid response, as well as precise recognition and positioning capability of the designed azo-DNA nanoswitch, make it possible to achieve effective photo-controlled release. This new design represents a major step toward the development of more highly efficient carrier and release systems. Furthermore, rapid switching between the open and closed states can be used to control drug dose, thereby avoiding side effects caused by excessive doses.
Table 1.
The sequences of azobenzene-modified DNA and normal DNA.
| Name | Sequence |
|---|---|
| Azo-DNA-1 | 5’-T-A-G-G-Azo-T-A-CCC-CCC-CCC-CCC-T-A-G-G- Azo-T-A-3’ |
| Azo-DNA-2 | 5’-T-A-Azo-G-G-Azo-T-A-CCC-CCC-CCC-CCC-T-A-Azo-G-G-Azo-T-A-3’ |
| Azo-DNA-3 | 5’-A-Azo-T-A-Azo-G-G-Azo-T-A-CCC-CCC-CCC-CCC-A-Azo-T-A-Azo-G-G-Azo-T-A-3’ |
| Azo-DNA-4 | 5’-T-Azo-A-G-Azo-G-T-Azo-A-CCC-CCC-CCC-CCC-T-Azo-A-G-Azo-G-T-Azo-A-3’ |
| Normal DNA | 5’-T-A-G-G-T-A-CCC-CCC-CCC-CCC-T-A-G-G-T-A-3’ |
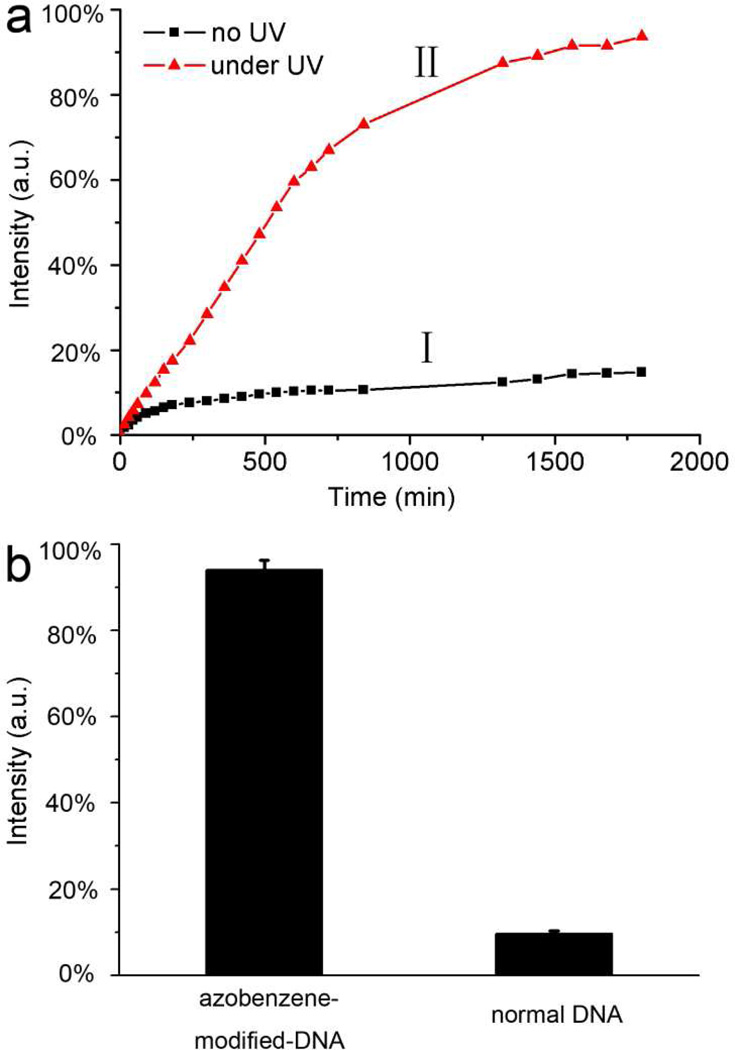
Rh6G was loaded into the pore structures to verify the photo-sensitive release mechanism. After loading the pores with Rh6G molecules, a significant decrease of surface area to 502 m2 g−1 was observed (Table S1), indicating that the pores have been filled by the dye molecules. Fluorescence spectroscopy was used to quantitate the release of Rh6G monitored at the emission maximum (560 nm) as a function of time. To optimize the design of azobenzene incorporated DNA oligomer, DNA strands (azo-DNA-1 through azo-DNA-4, Table 1) were prepared with different numbers and positions of azobenzene moieties. Azo-DNA-4 gave the best release results, as shown in Figure 3a. Under visible light (450 nm), no noticeable release was observed (curve I), indicating good capping efficiency. When the wavelength was changed to 365 nm, Rh6G escaped into solution as confirmed by the fluorescence intensity increase. The emission intensity of Rh6G in solution increased rapidly and 91% release was obtained after 1500 min (curve II). This result showed that the azobenzene-incorporated-DNA linker dehybridized from the arm-DNA on the silica surface and unblocked the pore openings. For comparison, a normal DNA strand without azobenzene modification was also tested for release of rhodamine molecules under photo-irradiation. However, no significant fluorescence signal could be detected under UV light for 1500 min, as shown in Figure 3b. These data clearly demonstrated that we were able to both close the MSN pore system with complementary azobenzene-incorporated DNA linkers, and subsequently to release the loaded molecules by dehybridizing the azo-DNA in response to UV light stimulus. As a result of diffusion-controlled kinetics, the dye release process is very slow. These slow release properties will result in optimum therapeutic effect without overdose. In contrast, the rapid capping/uncapping response to light provided by the MSN system allows exact point-to-point drug release.
Figure 3.
(a) Time course of Rh6G release from MSN modified by azobenzene-incorporated-DNA (azo-DNA-4). Data have been normalized to the maximum level of dye released in the experiment. (b) UV-light responsive release from MSN system modified with azobenzene-incorporated-DNA (azo-DNA-4) and normal DNA for 1500 min, as determined by fluorescence intensity. Data have been normalized to the loading amount of dye in the experiment.
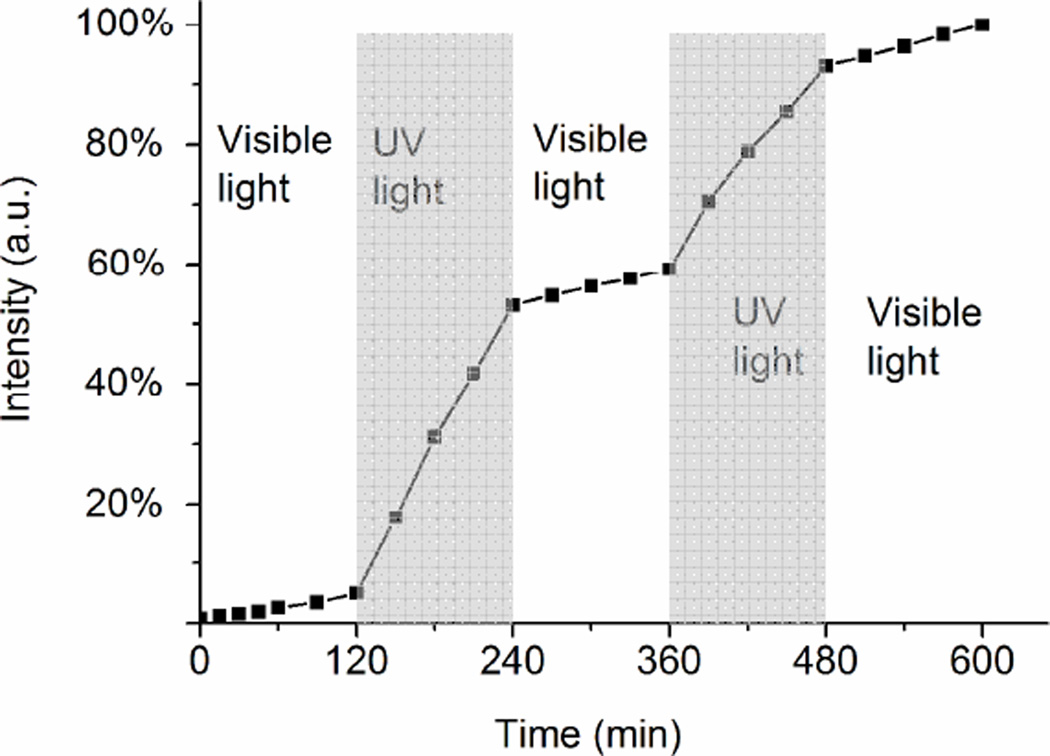
A distinctive advantage of this system lies in the reversible capping of pores, thereby enabling more complicated on-demand cargo delivery. As a proof of concept, the release of trapped dye molecules was regulated with open-close cycles via alternating irradiation wavelengths. As demonstrated in Figure 4, the closed state with visible irradiation strongly constrained the delivery of the Rh6G molecules. On the other hand, a distinct release of the entrapped rhodamine dye was triggered in the open state as a result of dehybridization when the wavelength was suddenly changed to 365 nm at 120 min. After 240 min the release of the entrapped dye was again restricted by changing to visible light at 450 nm. At 360 min, UV irradiation was repeated, and further delivery of the entrapped dye occurred. Subsequent irradiation with visible light again inhibited the release of the dye. The decreased dye release rate in each open segment is attributed to the reduced amount of dye to be delivered from the pores in each cycle. The release process shows that the on-off switching is reversible and can be repeated several times. Based on the photosensitive DNA, the delivery of small molecules by mesoporous silica nanoparticles can be achieved automatically by sequential adjustment of light. This smart stimuli-responsive behavior should be useful for on-demand dosing in clinical situations.
Figure 4.
Rh6G release profile from MSN as a function of time, showing the reversible dehybridization/rehybridization switching by changing the light wavelength. Data have been normalized to the largest amount of released dye at 600 min in the experiment.
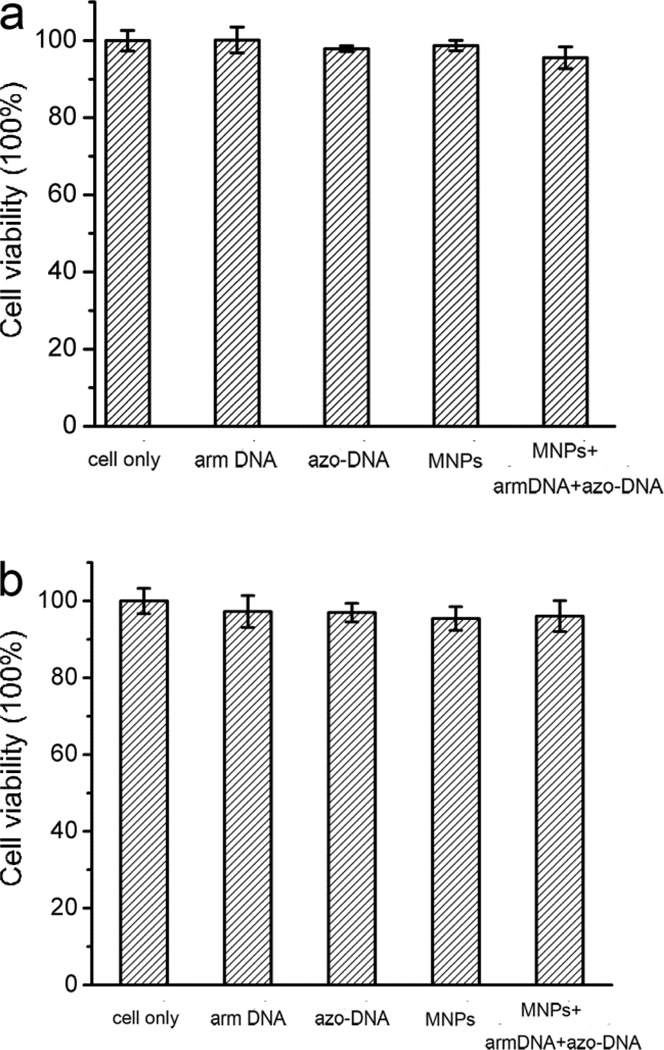
The cytotoxicity of the obtained azo-DNA/mesoporous-silica nanoparticles was investigated to examine its feasibility in bio-related fields. The effect of this kind of mesoporous silica particles on cell proliferation was assessed with human acute lymphoblastic leukemic CCRF-CEM cells and human lung adenocarcinoma A549 cells respectively by means of a methyl thiazolyl tetrazolium (MTT) assay. The viability of untreated cells was assumed to be 100%. Upon incubating the CEM cells with the arm-DNA (0.2 mM) and the azo-DNA (0.1 mM), less than 2.1% and 1.3% of the cells died after a 2 h exposure (Figure 5a). In addition, the CEM cell viability still remained above 98% when they were incubated with mesoporous silica particles for 2 h. The cytotoxicity of the azo-DNA MSNs to CEM cells was tested, respectively. Although the concentration was as high as 100 µg·mL−1, the cell viability still remained above 95%. For A549 cells with the arm-DNA (0.2 mM) and the azo-DNA (0.1 mM), the cell viabilities are 97.3% and 97.0%, respectively (Figure 5b). After incubation with azo-DNA MSNs for 2 h, only 4% of the cells died. These results clearly indicate that our synthesized mesoporous particles have low in vitro cytotoxicities, thereby demonstrating the excellent biocompatibility of this kind of vehicle.29
Figure 5.
The in vitro cell viability of (a) CEM cells and (b) A549 cells incubated with arm-DNA (0.2 mM), azo-DNA (0.1 mM), MSNs (100 µg mL−1), and azo-DNA/MSNs (100 µg mL−1) for 2 h.
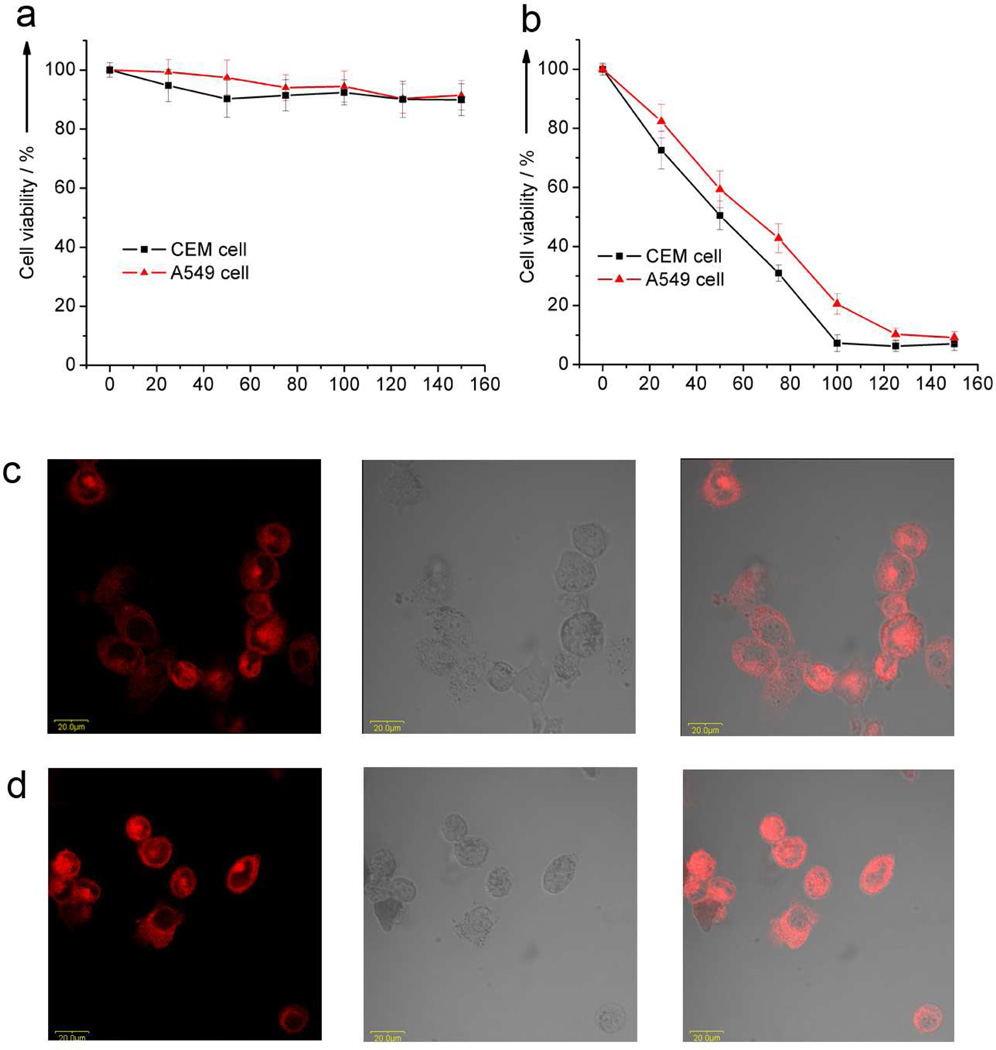
To evaluate the capacity as release-controlled drug delivery vehicles of azo-DNA/MSNs, a typical chemotherapeutic agent, DOX was selected and loaded into it to prepare DOX-loaded azo-DNA/MSNs. As demonstrated by the release kinetics study, these modified MSNs showed light-responsive release of guest Rh6G molecules. To determine whether DOX-loaded azo-DNA/MSNs could show a similar light-responsive function, the in vitro cytotoxicity of DOX-loaded azo-DNA/MSNs was evaluated for both CEM and A549 cells. DOX-loaded azo-DNA/MSNs of different concentrations were incubated with cells for 2 h. Without UV light treatment, the particles showed very low cytotoxicity to both CEM cells and A549 cells, even at concentrations as high as 150 µg·mL−1, because DOX was imprisoned in the pores of MSNs by azo-DNA blocking (Figure 6a). After UV irradiation for 30 min, the particles showed dose-dependent cytotoxicity to CEM as well as A549 cells (Figure 6b). The IC50 of DOX-loaded azo-DNA/MSNs was 49.2 µg·mL−1 for CEM cells and 62.5 µg·mL−1 for A549 cells, showing the obviously high killing efficacy due to DOX release from azo-DNA/MSNs under UV light. In addition, the confocal fluorescence microscopy images (Figure 6c,d) showed that the intracellular distribution of DOX increased with increased azo-DNA/MSN concentration, suggesting an efficient cellular uptake of DOX-loaded azo-DNA/MSNs and gradual release of DOX from azo-DNA/MSNs.
Figure 6.
Cytotoxicity assay of CEM cells and A549 cells treated with DOX-loaded azo-DNA/MSNs (a) without UV light illumination and (b) after UV light illumination for 30 min. Confocal microscopic images of A549 cells after incubating with DOX-loaded azo-DNA/MSNs of (c) 50 µg mL−1, and (d) 100 µg mL−1 after treatment with UV light.
Conclusions
In summary, we have demonstrated, as a proof-of-concept, how the incorporation of azobenzene into a novel biocompatible DNA/MSN molecular valve can impart controllable photo-responsive release behavior. The successful assembly of this multifunctional nanodevice is made possible by the novel multifunctional mesoporous silica host, free available pore volume inside the host, azobenzene-modified-dsDNA caps, and photo-controllable azobenzene isomerization induced opening/closing of pore gates. This photo-sensitive “on-off” release system exhibits highly efficient operation at different light wavelengths. Under visible light irradiation (λ = 450 nm), the linker DNA strand can hybridize to the arm-DNA to form a cap over the pore mouth. Upon exposure to UV (λ = 365 nm), the pore opens as the linker DNA is released into solution. We exploited this azo-DNA/MSN nanoplatform as a carrier for the anticancer drug DOX because of its good biocompatibility via MTT assay. The endocytosis of DOX-loaded azo-DNA/MSN by A549 cells was demonstrated through confocal laser microscopy. Efficient photon controlled release of DOX was confirmed for cancer therapy. Therefore, we envision that this intelligent nanodevice, which allows the release of incorporated guest molecules to be adjusted precisely, can lead to a wide range of desired applications, especially for targeted drug delivery.
Supplementary Material
Scheme 1.
Schematic of azobezene-modified DNA controlled reversible release system. Visible irradiation at 450 nm (azobenzene trans) leads to hybridization of the linker and the complementary DNA arm. Irradiation with UV (365 nm) converts azobenzene to the cis form, leading to dehybridization and pore opening.
Acknowledgement
This work was also supported by the National Natural Science Foundation of China (20975034) and the National Key Scientific Program of China (2011CB911001, 2011CB911003). This work was also supported by grants awarded by the National Institutes of Health (GM066137, GM079359 and CA133086). The authors gratefully thank Professor Xiaoxiao He and Mr. Lvan Yan for their help.
Footnotes
Supporting Information Available: Additional information on the synthesis of azobenzene phosphoramidite, pore structure of DNA-functionalized MSN, set-up for Rh6G release test and UV light responsive release of Rh6G toward MSN functionalized with different azobenzene incorporated DNA. This material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.Beissenhirtz MK, Willner I. DNA-Based Machines. Org. Biomol. Chem. 2006;4:3392–3401. doi: 10.1039/b607033g. [DOI] [PubMed] [Google Scholar]
- 2.Liedl T, Sobey TL, Simmel FC. DNA-Based Nanodevices. Nano Today. 2007;2:36–41. [Google Scholar]
- 3.Bath J, Turberfield AJ. DNA Nanomachines. Nat. Nanotechnol. 2007;2:275–284. doi: 10.1038/nnano.2007.104. [DOI] [PubMed] [Google Scholar]
- 4.Liu HJ, Liu DS. DNA Nanomachines and Their Functional Evolution. Chem. Commun. 2009:2625–2636. doi: 10.1039/b822719e. [DOI] [PubMed] [Google Scholar]
- 5.Balas F, Manzano M, Horcajada P, Vallet-Regí M. Confinement and Controlled Release of Bisphosphonates on Ordered Mesoporous Silica-Based Materials. J. Am Chem. Soc. 2006;128:8116–8117. doi: 10.1021/ja062286z. [DOI] [PubMed] [Google Scholar]
- 6.Vallet-Regí M, Rámila A, del Real RP, Pérez-Pariente J. A New Property of MCM-41: Drug Delivery System. Chem. Mater. 2001;13:308–311. [Google Scholar]
- 7.Mal NK, Fujiwara M, Tanaka Y. Photocontrolled Reversible Release of Guest Molecules from Coumarin-modified Mesoporous Silica. Nature. 2003;421:350–353. doi: 10.1038/nature01362. [DOI] [PubMed] [Google Scholar]
- 8.Vallet-Regi M, Balas F, Arcos D. Mesoporous Materials for Drug Delivery. Angew. Chem. Int. Ed. 2007;46:7548–7758. doi: 10.1002/anie.200604488. [DOI] [PubMed] [Google Scholar]
- 9.Angelos S, Yang YW, Patel K, Stoddart JF, Zink JI. pH-Responsive Supramolecular Nanovalves Based on Cucurbit[6]uril Pseudorotaxanes. Angew. Chem. Int. Ed. 2008;47:2222–2226. doi: 10.1002/anie.200705211. [DOI] [PubMed] [Google Scholar]
- 10.Vivero-Escoto JL, Slowing II, Wu CW, Lin VSY. Photoinduced Intracellular Controlled Release Drug Delivery in Human Cells by Gold-Capped Mesoporous Silica Nanosphere. J. Am. Chem. Soc. 2009;131:3462–3463. doi: 10.1021/ja900025f. [DOI] [PubMed] [Google Scholar]
- 11.Park C, Kim H, Kim S, Kim C. Enzyme Responsive Nanocontainers with Cyclodextrin Gatekeepers and Synergistic Effects in Release of Guests. J. Am. Chem. Soc. 2009;131:16614–16615. doi: 10.1021/ja9061085. [DOI] [PubMed] [Google Scholar]
- 12.Aznar E, Marcos MD, Martínez-Máñez, Sancenón F, Soto J, Amorós P, Guillem C. pH- and Photo-Switched Release of Guest Molecules from Mesoporous Silica Supports. J. Am. Chem. Soc. 2009;131:6833–6843. doi: 10.1021/ja810011p. [DOI] [PubMed] [Google Scholar]
- 13.Liu R, Zhao X, Agarwal A, Mueller LJ, Feng PY. pH-Responsive Nanogated Ensemble Based on Gold-Capped Mesoporous Silica through an Acid-Labile Acetal Linker. J. Am. Chem. Soc. 2010;132:1500–1501. doi: 10.1021/ja907838s. [DOI] [PubMed] [Google Scholar]
- 14.Kim H, Kim S, Park C, Lee H, Park H, Kim C. Glutathione-Induced Intracellular Release of Guests from Mesoporous Silica Nanocontainers with Cyclodextrin Gatekeepers. Adv. Mater. 2010;22:4280–4283. doi: 10.1002/adma.201001417. [DOI] [PubMed] [Google Scholar]
- 15.Schlossbauer A, Warncke S, Gramlich PME, Kecht J, Manetto A, Carell T, Bein T. A Programmable DNA-Based Molecular Valve for Colloidal Mesoporous Silica. Angew. Chem. Int. Ed. 2010;49:4734–4737. doi: 10.1002/anie.201000827. [DOI] [PubMed] [Google Scholar]
- 16.Chem C, Pu F, Huang ZZ, Liu Z, Ren JS, Qu XG. Stimuli-responsive Controlled-release System Using Quadruplex DNA-Capped Silica Nanocontainers. Nucleic Acids Res. 2011;39:1638–1644. doi: 10.1093/nar/gkq893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen C, Geng J, Pu F, Yang X, Ren J, Qu X. Polyvalent Nucleic Acid/Mesoporous Silica Nanoparticle Conjugates: Dual Stimuli-Responsive Vehicles for Intracellular Drug Delivery. Angew. Chem. Int. Ed. 2011;50:882–886. doi: 10.1002/anie.201005471. [DOI] [PubMed] [Google Scholar]
- 18.Yager KG, Barrett CJ. Novel Photo-switching Using Azobenzene Functional Materials. J. Photochem Photobio. A. 2006;182:250–261. [Google Scholar]
- 19.Yesodha SK, Pillai CKS, Tsutsumi N. Prog. Polym. Sci. 2004;29:45–74. [Google Scholar]
- 20.Kay ER, Leigh DA, Zerbetto F. Synthetic Molecular Motors and Mechanical Machines. Angew. Chem. Int. Ed. 2007;46:72–191. doi: 10.1002/anie.200504313. [DOI] [PubMed] [Google Scholar]
- 21.Liang XG, Nishioka H, Takenaka N, Asanuma H. A DNA Nanomachine Powered by Light Irradiation. ChemBioChem. 2008;9:702–705. doi: 10.1002/cbic.200700649. [DOI] [PubMed] [Google Scholar]
- 22.Bose M, Groff D, Xie J, Brustad E, Schultz PG. The Incorporation of a Photoisomerizable Amino Acid into Proteins in. E. Coli J. Am. Chem. Soc. 2006;128:388–389. doi: 10.1021/ja055467u. [DOI] [PubMed] [Google Scholar]
- 23.Woolley GA. Photocontrolling Peptide α Helices. Acc. Chem. Res. 2005;38:486–493. doi: 10.1021/ar040091v. [DOI] [PubMed] [Google Scholar]
- 24.Asanuma H, Liang X, Yoshida T, Komiyama M. Photocontrol of DNA Duplex Formation by Using Azobenzene-Bearing Oligonucleotides. ChemBioChem. 2001;2:39–44. doi: 10.1002/1439-7633(20010105)2:1<39::AID-CBIC39>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 25.Kang HZ, Liu HP, Phillips JA, Cao ZH, Kim YM, Chen Y, Yang ZY, Li JW, Tan WH. Single-DNA Molecule Nanomotor Regulated by Photons. Nano Lett. 2009;9:2690–2696. doi: 10.1021/nl9011694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan Q, Zhang YF, Chen Y, Wang RW, Du CL, Yasun E, Tan WH. Using Silver Nanowire Antennas to Enhance the Conversion Efficiency of Photoresponsive DNA Nanomotors. Proc. Natl. Acad. Sci. U S A. 2011;108:9331–9336. doi: 10.1073/pnas.1018358108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai CY, Trewyn BG, Jeftinija DM, Jeftinija K, Xu S, Jeftinija S, Lin VSY. A Mesoporous Silica Nanosphere-Based Carrier System with Chemically Removable CdS Nanoparticle Caps for Stimuli-Responsive Controlled Release of Neurotransmitters and Drug Molecules. J. Am Chem. Soc. 2003;125:4451–4459. doi: 10.1021/ja028650l. [DOI] [PubMed] [Google Scholar]
- 28.Radu DR, Lai C-Y, Jeftinija K, Rowe EW, Jeftinija S, Lin VS-Y. A Polyamidoamine Dendrimer-Capped Mesoporous Silica Nanosphere-Based Gene Transfection Reagent. J. Am Chem. Soc. 2004;126:13216–13217. doi: 10.1021/ja046275m. [DOI] [PubMed] [Google Scholar]
- 29.Babincová M, Sourvong P, Leszczynska D, Babinec P. Blood-specific Whole-body Electromagnetic Hyperthermia. Med. Hypotheses. 2000;55:459–460. doi: 10.1054/mehy.2000.1089. [DOI] [PubMed] [Google Scholar]
- 30.Asanuma H, Liang X, Nishioka H, Matsunaga D, Liu M, Komiyama M. Synthesis of Azobenzene-tethered DNA for Reversible Photo-regulation of DNA Functions: Hybridization and Transcription. Nat. Protoc. 2007;2:203–212. doi: 10.1038/nprot.2006.465. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.