Abstract
Background
Prediction of prognosis remains a major unmet need in new-onset heart failure (HF). Although several clinical tests are in use, none accurately distinguish between patients with poor versus excellent survival. We hypothesized that a transcriptomic signature, generated from a single endomyocardial biopsy, could serve as a novel prognostic biomarker in HF.
Methods and Results
Endomyocardial biopsy samples and clinical data were collected from all patients presenting with new-onset HF from 1997 to 2006. Among a total of 350 endomyocardial biopsy samples, 180 were identified as idiopathic dilated cardiomyopathy. Patients with phenotypic extremes in survival were selected: good prognosis (event-free survival for at least 5 years; n=25) and poor prognosis (events [death, requirement for left ventricular assist device, or cardiac transplant] within the first 2 years of presentation with HF symptoms; n=18). We used human U133 Plus 2.0 microarrays (Affymetrix) and analyzed the data with significance analysis of microarrays and prediction analysis of microarrays. We identified 46 overexpressed genes in patients with good versus poor prognosis, of which 45 genes were selected by prediction analysis of microarrays for prediction of prognosis in a train set (n=29) with subsequent validation in test sets (n=14 each). The biomarker performed with 74% sensitivity (95% CI 69% to 79%) and 90% specificity (95% CI 87% to 93%) after 50 random partitions.
Conclusions
These findings suggest the potential of transcriptomic biomarkers to predict prognosis in patients with new-onset HF from a single endomyocardial biopsy sample. In addition, our findings offer potential novel therapeutic targets for HF and cardiomyopathy.
Keywords: biopsy, genes, heart failure, prognosis, transcriptome
The clinical course of patients with newly diagnosed heart failure varies drastically, with some patients recovering and returning to completely normal levels of ejection fraction,1,2 whereas others develop severe symptoms of cardiac decompensation that require insertion of a left ventricular assist device (LVAD) or a heart transplant.3,4 Accurate risk assessment and prediction of prognosis at first presentation are crucial for appropriate allocation of therapy,4,5 monitoring,3,6 and patient management.1,7 Prediction tools based on standard criteria have had limited accuracy.8–11 In the present study, we present a transcriptome-based biomarker (TBB),5,7 which has been derived from a single endomyocardial biopsy sample (EMB) and which predicts the long-term clinical outcome of patients with idiopathic dilated cardiomyopathy (IDCM) with very high accuracy.
Methods
Patients
EMBs were collected from patients who were referred to Johns Hopkins Hospital between 1997 and 2006 for evaluation of cardiomyopathy (n=350).3 A clinical database of patient outcome was maintained concurrently for a 10-year period beginning in 1997. All patients gave written informed consent for sample collection and medical chart abstraction. Transvenous EMBs from the right ventricular septum were obtained as described previously3 for subsequent microarray analysis. To avoid possible disease-specific confounding factors, only samples from patients with IDCM were selected. IDCM was a diagnosis of exclusion after extensive histological workup without any detectable pathological signs.4 Within a repository of 180 IDCM samples, biopsy samples were selected in a case-control fashion based on the phenotypic extremes in survival of the cohort.12 A group with good prognosis (n=25) was defined as having event-free survival for at least 5 years after initial presentation with heart failure symptoms; a group with poor prognosis (n=18) experienced an event within the first 2 years. Events included death (n=14), requirement for an LVAD (n=2), or cardiac transplantation (n=2).
Processing of Biopsy Samples
EMBs were immediately flash-frozen in liquid nitrogen for storage in a biorepository. All steps of RNA isolation and processing were performed according to MIAME guidelines (Minimum Information About a Microarray Experiment). Tissue samples (average diameter 2 mm) were homogenized with the MM 301 Mixer Mill (Retsch, Inc, Newtown, Pa; catalog No. 85120). Trizol reagent together with the Micro-to-Midi Total RNA Purification System (Invitrogen, Carlsbad, Calif; catalog No. 12183-018) was used for extraction of total RNA (success rate 97% of samples). Concentration and integrity of total RNA were measured with the Agilent 2100 Bioanalyzer (Agilent Technologies, Inc, Santa Clara, Calif). All RNA samples exhibited intact 28S and 18S ribosomal RNA on denaturing agarose gel electrophoresis, and the 260/280-nm absorbance readings fell within the acceptable range of 1.8 to 2.1. An average of 568±92 ng (mean±SEM) of total RNA was isolated and preprocessed with the Ovation Biotin RNA Amplification and Labeling System (NuGEN Technologies, Inc, San Carlos, Calif; catalog No. 2300-12).13
Microarray Hybridization
Samples were hybridized to the Human Genome U133 Plus 2.0 Array from Affymetrix (Santa Clara, Calif) without additional amplification steps. We judged the microarray experiments to be successful when RNA isolation and microarray hybridization met all the indices of quality control as specified in the Affymetrix guidelines for assessing sample and array quality.7 Average background and noise of all chips registered within acceptable ranges, and hybridization efficiencies were similar for all samples.
Statistical Analysis
Raw intensity values from microarray hybridization were normalized with the robust multiarray average implemented in the R package for statistical computing (available at www.R-project.org). In the next step, significance analysis of microarrays14 was used to identify phenotype-specific differences in gene expression. Significance analysis of microarrays defines significance with the q value, an adjusted probability value for multiple comparisons. For the development of a TBB, we used prediction analysis of microarrays15 to create a classifier in a training set (containing two thirds of the data, n=29), with subsequent validation in a test set (containing one third of the data, n=14).16 Prediction analysis of microarrays is software that enables one to find the minimum number of genes necessary to create a phenotype-specific “nearest shrunken centroid” for classification.15 This is done by a balanced 10-fold cross-validation in a training set, which enables one to choose a threshold that minimizes classification errors.15 Overall accuracy of the discovered transcriptomic biomarker was assessed after 50 random partitions. To test for balanced baseline conditions of the cohort with good versus poor prognosis, we used Student’s t test for numerical variables and Fisher’s exact test for categorical variables.
The authors had full access to the data and take full responsibility for its integrity. All authors have read and agree to the manuscript as written.
Results
We sought to test the hypothesis that the transcriptome, derived from a single EMB, contains sufficient information to develop a biomarker for prognosis in heart failure. Although substantial research has demonstrated the prognostic value of a variety of biomarkers and clinical prediction algorithms in heart failure,17,18 the ability to distinguish individual patients who will improve their functional status from those who will go on to circulatory collapse and require cardiac transplantation or LVAD placement remains an important clinical challenge.1,3
Patient Characteristics
A total of 43 EMBs were analyzed with microarray technology to identify phenotype-specific differences in gene expression and to develop a prognostic TBB. Table 1 contains the baseline conditions of both cohorts, patients with good prognosis and those with poor prognosis. Importantly, there were no significant differences in age, gender, ventricular function, hemodynamics, or drug therapy between the 2 groups. The overall population with IDCM presented at an average age of 46±15 years, with slight overrepresentation of males (67%). Most patients were at an advanced New York Heart Association classification stage (at least stage II), with severely compromised ejection fraction of 23±13%, average left ventricular internal diastolic dimension of 6.1±1.5 cm, and pulmonary capillary wedge pressure of 15±9 mm Hg.
Table 1.
Baseline Conditions
| Good Prognosis (n=25) | Poor Prognosis (n=18) | |
|---|---|---|
| Age, y | 46±15 | 48±17 |
| Male, n (%) | 17 (68) | 12 (67) |
| NYHA, n (%) | ||
| I | 1 (4) | 1 (5) |
| II | 13 (52) | 6 (33) |
| III | 10 (40) | 8 (44) |
| IV | 1 (4) | 4 (22) |
| LVEF, % | 24±13 | 23±13 |
| LVIDD, cm | 6.4±1 | 6.3±2 |
| PAP, mm Hg | ||
| Systolic | 36±13 | 41±13 |
| Diastolic | 16±6 | 20±11 |
| Pulmonary capillary wedge pressure, mm Hg | 13±7 | 18±10 |
| Medications, n (%) | ||
| β-Antagonist | 17 (68) | 13 (72) |
| ACE inhibitor | 17 (68) | 12 (67) |
| Aldosterone antagonist | 4 (16) | 4 (22) |
| Diuretic | 17 (68) | 15 (83) |
| Intravenous inotropic therapy | 0 | 0 |
NYHA indicates New York Heart Association classification; LVEF, left ventricular ejection fraction; LVIDD, left ventricular internal diastolic dimension; PAP, pulmonary artery pressure; and ACE, angiotensin-converting enzyme inhibitor.
Microarray Analysis
An average of 568±92 ng (mean±SEM) of total RNA was isolated from all EMBs and tested with the Agilent 2100 Bioanalyzer, which revealed high integrity and purity of RNA of all samples with consistent bands of 18S and 28S RNA (Figure 1). There were 46 significantly overexpressed genes in patients who recovered from heart failure (q<5%, fold change >1.2; Table 2), as determined with significance analysis of microarrays. Prediction analysis of microarrays was used to evaluate the predictive value of this set of genes. To achieve this and to test for validity, patients were allocated into training sets that consisted of two thirds of samples (n=29) and test sets that contained one third of samples (n=14; Figure 2). This approach resulted in a “nearest shrunken centroid” of 45 genes, which distinguished with high accuracy between high-risk patients and those with an excellent prognosis.
Figure 1. Analysis of extracted total RNA with Agilent 2100 Bioanalyzer.
Every sample was tested for its integrity and purity before microarray hybridization. The graph depicts a gel of 12 samples with consistent bands of 18S and 28S RNA. The left lane contains the reference marker.
Table 2.
Forty-Six Significantly Overexpressed Genes in Patients With Heart Failure and Good Prognostic Outcome (q<5%, Fold Change >1.2)
| Affymetrix ID | Gene Title | Fold Change |
|---|---|---|
| 232669_at | Hypoxia-inducible factor 3, α-subunit | 1.8 |
| 214951_at | Solute carrier family 26, member 10 | 1.8 |
| 243482_at | Epidermal growth factor receptor pathway substrate 15-like 1 | 1.8 |
| 226210_s_at | Maternally expressed 3 | 1.7 |
| 232159_at | Epidermal growth factor receptor pathway substrate 15-like 1 | 1.7 |
| 233026_s_at | PDZ domain containing 2 | 1.6 |
| 211996_s_at | KIAA0220-like protein hypothetical gene LOC 283846 | 1.6 |
| 243774_at | Mucin 20, cell surface associated | 1.6 |
| 242551_at | Chromosome 18 open reading frame 1 | 1.6 |
| 244548_at | Rho GTPase activating protein 26 | 1.6 |
| 244208_at | Checkpoint suppressor 1 | 1.6 |
| 239984_at | Sodium channel, voltage-gated, type VII, alpha | 1.6 |
| 230683_at | CDNA:FLJ20892 fis, clone ADKA03430 | 1.5 |
| 241869_at | Apolipoprotein L, 6 | 1.5 |
| 241597_at | Arginine-glutamic acid dipeptide (RE) repeats | 1.5 |
| 235887_at | Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) | 1.5 |
| 229957_at | Transmembrane protein 91 | 1.5 |
| 223546_x_at | LUC7L-like (S. cerevisiae) | 1.5 |
| 239567_at | Rho GTPase activating protein 10 | 1.5 |
| 242194_at | Cullin 4A | 1.5 |
| 1558525_at | Hypothetical protein LOC283901 | 1.4 |
| 227178_at | CUG triplet repeat, RNA binding protein 2 | 1.4 |
| 228198_s_at | Mitochondrial ribosomal protein S9 | 1.4 |
| 202379_s_at | Natural killer-tumor recognition sequence | 1.4 |
| 224260_at | CDNA clone | 1.4 |
| 238643_at | Neuroblastoma, suppression of tumorigenicity 1 | 1.4 |
| 232253_at | RAD50 homolog (S. cerevisiae) | 1.4 |
| 227968_at | Parkinson disease 7 domain containing 1 | 1.4 |
| 233197_at | Kelch-like 9 (Drosophila) | 1.4 |
| 244512_at* | Transcribed locus, strongly similar to XP_001081342.1 | 1.4 |
| 233443_at | Hypothetical protein LOC389362 | 1.4 |
| 231275_at | FLJ42875 protein | 1.4 |
| 226419_s_at | Hypothetical protein LOC645460 | 1.4 |
| 201221_s_at | Small nuclear ribonucleoprotein 70-kDa polypeptide | 1.4 |
| 209354_at | Tumor necrosis factor receptor family member 14 | 1.4 |
| 226571_s_at | Protein tyrosine phosphatase receptor type, S | 1.4 |
| 220728_at | EST | 1.3 |
| 203071_at | Sema domain, immunoglobulin domain (Ig), short basic domain | 1.3 |
| 213946_s_at | Obscurin-like 1, similar to titin isoform N2-B | 1.3 |
| 201394_s_at | RNA binding motif protein 5 | 1.3 |
| 203748_x_at | RNA binding motif, single-stranded interacting protein 1 | 1.3 |
| 223147_s_at | WD repeat domain 33 | 1.3 |
| 213773_x_at | NOL1/NOP2/Sun domain family, member 5 | 1.3 |
| 1560049_at | CUG triplet repeat, RNA binding protein 2 | 1.3 |
| 243974_at | CDNA clone IMAGE:4821815 | 1.2 |
| 201510_at | E74-like factor 3(ets domain transcription factor, epithelial specific) | 1.2 |
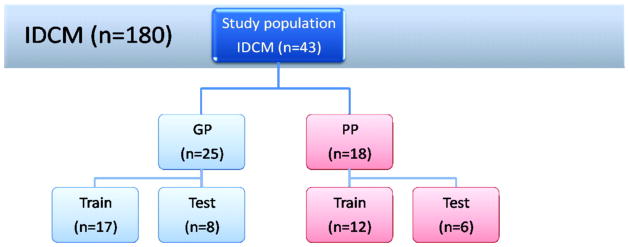
Figure 2. Scheme of training and test sets as used for the development of a classifier with prediction analysis of microarrays.
All samples obtained from patients with poor prognosis (PP, n=18) were selected from a biorepository (n=180), and those with good prognosis (GP, n=25) were chosen in a case-control fashion (see text for definitions of PP and GP). The classifier, a nearest shrunken centroid, was developed in two thirds of the data (17 samples with GP, 12 samples with PP) with subsequent validation in the remaining one third of data (8 samples with GP, 6 samples with PP). The overall test accuracy of the TBB was calculated from 50 random partitions into training and test sets.
Validation
To evaluate the overall performance of the transcriptomic biomarker, we performed 50 random partitions into training and test sets. This approach revealed an overall sensitivity of 74% (95% CI 69% to 79%) and an overall specificity of 90% (95% CI 87% to 93%). The positive predictive value was 85% (95% CI 80% to 89%), whereas the negative predictive value was 82% (95% CI 78% to 86%). The log OR for having an event if classified with poor prognosis was 3.3.
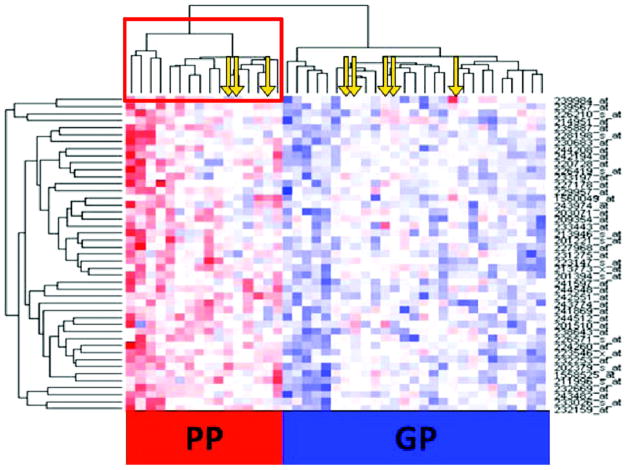
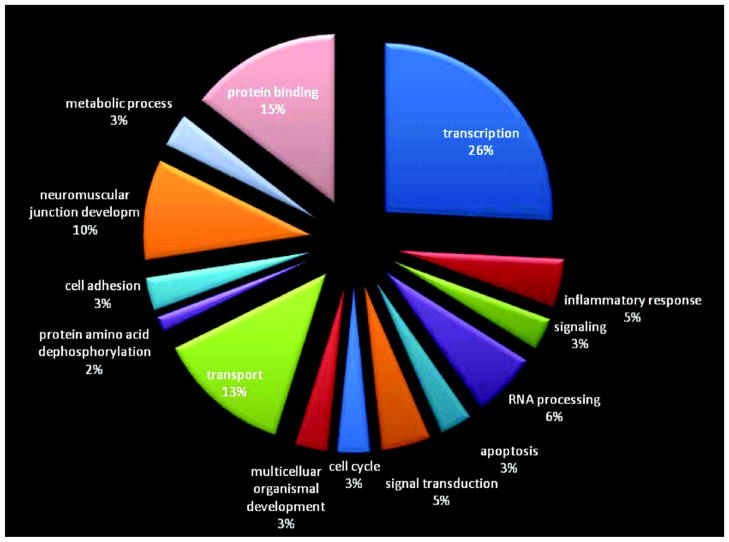
The molecular signature was illustrated in a heat map, which was created by euclidean distance (Figure 3). This independent approach of unsupervised clustering additionally confirmed the robustness of the discovered set of genes, with very clear distinction of samples with poor prognosis from those with good prognosis. Pathways with major involvement were ion transport mechanisms (13%), neuromuscular development (10%), protein binding (15%), and transcription (26%; www.geneontology.org; Figure 4).
Figure 3. Heat map of samples from all patients with IDCM (n=43).
Each column corresponds to a patient sample, and each row represents a gene. Samples classified as having poor prognosis (PP) form a distinct cluster and are highlighted in a red square. Downregulated genes are depicted with red, whereas upregulated genes are labeled blue. Yellow arrows denote misclassified samples. GP indicates good prognosis.
Figure 4. Pie chart illustrating involved pathways within the prognostic biomarker.
Major pathways overexpressed in patients with good prognosis included transcription (26%), protein binding (15%), ion transport (13%), and neuromuscular development (10%). Developm indicates development.
Prediction of Recovery in Left Ventricular Function
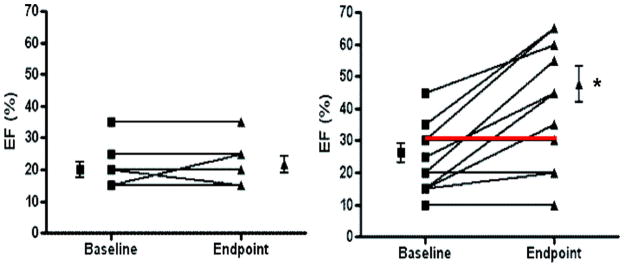
Improved clinical outcome is often associated with recovery in left ventricular function. Accordingly, we tested the hypothesis that the good prognosis signature would be associated with improved ejection fraction. Among the study sample (n=43), we selected all subjects in whom paired echocardiography data from baseline and follow-up were available (n=17) and characterized them as good prognosis or poor prognosis according to their TBB (Figure 5). Patients classified as having good prognosis experienced a substantial improvement in ejection fraction, from 23±3% to 42±5% (P=0.0009), whereas patients with a poor prognosis molecular signature continued to have a depressed ejection fraction, with 20±3% and 21±3% at baseline and follow-up, respectively.
Figure 5. Functional improvement of ejection fraction (EF) from baseline to end point.
Within all enrolled cases of idiopathic cardiomyopathy (n=43), we further analyzed those for whom echocardiographic measurements at baseline and end point were available (n=17). Samples were classified into good prognosis (right panel) or poor prognosis (left panel) based on TBB prediction. Patients classified as having a good prognosis (n=11, average follow-up 49.9±21 months) experienced improvement of EF (*P=0.0009), whereas those with a poor prognosis (n=6, average follow-up 6.2±2.9 months) did not. Red line depicts 1 misclassified sample. Error bars represent SEM.
Discussion
One of the most valuable applications of genomic information has proven to be clinical prediction.5 Whereas the pattern of differentially expressed genes provides insight into disease origin,19 it has also been used for the development of biomarkers.20 This approach has been highly successful in neoplastic disease15,20,21 and is emerging in a variety of other disease processes.6,16 Here, we sought to identify a TBB that predicted the clinical outcome of patients with new-onset heart failure. Using a repository of EMBs collected over a 10-year period,3 we have developed a highly accurate prognostic biomarker that distinguishes patients with very poor trajectory from those with excellent long-term prognosis. Importantly, the present study used biopsy samples from patients presenting at an early disease stage, without additional amplification of RNA.
The present findings address a major unmet need in the management of heart failure: the ability to accurately assess patient prognosis.1 Although there are emerging biomarkers17,18 and clinical prediction algorithms9,11 of prognostic value, these markers do not dichotomize substantially between patients with highly variable outcomes.3,8–11 It has long been known that patients with near-identical features at presentation, receiving identical therapy, can have dramatically differing prognoses.1,2,4 Although some patients undergo complete recovery of their heart function within an average of 5 years, others progress into circulatory collapse within the first 2 years of presentation and require aggressive therapeutic interventions, such as mechanical circulatory assistance or cardiac transplantation.3 The ability to predict these patients has great clinical value in an era of multiple therapeutic options.
Previous transcriptomic studies16,22–24 investigated heart tissue from explanted hearts or LVAD. The results of these studies continue to be controversial and have led to the questioning of the feasibility of TBBs in cardiovascular disorders.23 Many previous studies evaluated gene expression changes during left ventricular functional recovery with mechanical circulatory assistance to discover genes associated with recovery. This approach is inherently limited, because gene expression changes could be either causative or due to a bystander effect of volume unloading.23,25 Although Hall et al.22 discovered a molecular signature of recovery that was specific for patients who recovered from heart failure during LVAD placement versus patients who continued to be dependent on a device, Margulies et al23 concluded from a similarly designed study that transcriptomic changes in recovering LVAD patients would not consistently explain their functional improvement. These controversial findings motivated the present study with biopsy samples obtained at the time of patient presentation.
Other investigators have attempted TBB studies from explanted hearts rather than LVAD patients. Steenman et al.24 investigated samples with ischemic cardiomyopathy versus IDCM and failed to find significant differences in gene expression. However, that study was limited by an extremely small sample size (n=2 in each group), and data were evaluated by pooling the RNA from these samples. This negative finding highlights the importance of sample size in microarray experiments. Learning-curve constructions provided evidence that 20 to 30 samples are required per training set to achieve significance in a classification problem.26 In a previous study that involved a larger number of samples (n=48), we discovered a cluster of genes that accurately distinguished idiopathic from ischemic cardiomyopathy.16
Several features of this approach may have contributed to the present results. First, the present study used biopsy samples obtained at the time of patient presentation, not taken from explanted hearts, a time at which disease could be considered end-stage. Arguably this approach reduces transcriptomic changes that could be considered compensatory and that could have been activated during disease progression.23,25 A second key issue results from our technical approach: We used a highly efficient RNA processing technique that allowed microarray analysis without additional amplification and thus avoided possible bias due to different binding preferences of primers and reduction of time and costs with regard to a later clinical application.27
The transcriptomic biomarker reported here for cardiomyopathy has a similar degree of accuracy as those developed for cancer.15,20,21 Previous proof-of-concept studies suggested the feasibility of obtaining clinically useful TBBs from cardiac biopsy samples.16 In this regard, we previously demonstrated in explanted heart tissue obtained at the time of heart transplantation that a transcriptomic signature could be developed to delineate between ischemic and nonischemic cardiomyopathy.16 Additional support for the concept of TBBs for heart muscle disease arises from the successful development of expression profiles to detect cardiac inflammation.6,28
The developed TBB, which contains a molecular signature of 45 genes, predicted the clinical phenotype of samples with very high specificity (90%), which suggests its utility, particularly as a rule-out test in addition to current standard risk assessment. Furthermore, results of the TBB might be included in the assessment of a priority score in transplant waiting lists. In combination with clinical data and laboratory values, the TBB has the potential to offer guidance in patient management.6 Furthermore, the present data offer insights into novel molecular pathways involved in the recovery of patients with heart failure.29 Of the 46 differentially expressed genes (Table 2), all were overexpressed in patients who recovered from heart failure. Many overexpressed genes in these patients have RNA or DNA binding functions, eg, RBMS1 and WDR33, playing a crucial role in DNA replication, gene transcription, cell cycle progression, and apoptosis.30–32 Various genes with critical regulatory functions were identified, ie, SNRP70; the nuclear transcription factors ELF3, CHES1, and RERE; NSUN4, a gene with methyltransferase activity; and the transcription factor HIF3A. HIF3A has been discussed as an inductor of glucose transporters, vascular endothelial growth factor, and erythropoietin, similar to HIF1A and HIF2A, whereas others postulated its counteraction with the other 2 subunits.33,34
The genes CUGBP2, LUC7-like, and SEMA3B are involved in neuromuscular development, axon guidance, and regulation of heart contractility (http://www.geneontology.org/). Closely related in its function is the overexpressed obscurin-like 1 gene (OBSL1), a linker that stabilizes cell contacts and organelles within the cytoskeleton35 and that is located at the intercalated discs in the adult cardiomyocyte.35 OBSL1 is supposed to have similar functions to obscurin, a multifunctional protein responsible for assembly of myofibrils and myocyte cellular organization.36 By its interaction with titin and ankyrin,36 as well as by linking sarcomere and sarcoplasmic reticulum,36 it provides the required alignments for contraction.36
Furthermore, we discovered 2 genes previously implicated in regenerative pathways, Rad50 and SMG6, important key regulators in telomerase activity and DNA repair.37 Rad50 is part of the Mre11/Rad50/Nbs1 (MRN) complex,37 a functional unit that generates t loops by inserting 3′ G overhangs at human telomere ends.37 These t loops prevent chromosome ends from being recognized as damaged DNA and provide a template for telomerase and preservation of genome stability.38 The increase in telomerase activity might explain a protective effect against degenerative processes and aging in the hearts of patients with good prognosis. It is attractive to speculate that the reason for better recovery might be increased cellular regenerative capacity,39,40 given observations that Rad50 is essential for viability of stem cells.38 Of note, 3 genes (SNRP70, OBSL1, and RBMS1) reported here were also reported in a study of 199 human LVAD samples that suggested genes involved in recovery.23 There was no overlap with results published by Lowes et al,41,42 who investigated genes that were overexpressed during recovery from heart failure. This may suggest that there are important differences between gene expression changes caused by therapy and characteristics of gene expression that may predict clinical outcome.
Validation of microarray experiments is an issue of considerable importance. In this context, it remains highly controversial whether independent RNA quantification is essential. Given the technical approach and motivation to avoid RNA amplification, we lacked sufficient RNA for independent reverse-transcription polymerase chain reaction quantification. Recent technical advances, however, obviate the need for polymerase chain reaction–based validation in clinical prediction studies. Indeed, the HG-U133 Plus 2.0 Gene Chip has been shown to perform with high consistency compared with established quantitative gene expression platforms.43,44 Moreover, numerous published studies have not used independent RNA quantification for validation. In this regard, recent TBB studies in oncology45,46 have used split-sample validation, which was the approach used in the present study. Additional validation of selected transcripts by reverse-transcription polymerase chain reaction was not considered mandatory, because the individual expression value of a single gene has less predictive power than the averaged overall gene expression pattern of a molecular signature.7,47
Although endomyocardial biopsy is a low-risk procedure,48 alternative methods for obtaining transcriptomic biomarkers might be developed.5 There is evidence that affected tissue and circulating blood cells share a high percentage of common genes.6,49 Easy accessibility of peripheral blood mononuclear cells by a simple venous puncture makes those cells very attractive for clinical application.5,6,47 Several studies have proven a remarkably high correlation between the gene expression profiles of peripheral blood mononuclear cells and various tissues,6,49 and first diagnostic kits, based on blood samples, are commercially available.6 Future studies will show whether the biopsy-derived biomarkers are also present in the corresponding peripheral blood mononuclear cells of patients, whether peripheral blood mononuclear cell–derived markers perform even better, or whether a combination of both can potentiate the overall accuracy. Future studies will be also necessary to compare the utility of the transcriptomic biomarker in combination with existing biomarkers (e.g., brain natriuretic peptide18 uric acid17) and established prediction algorithms (artificial neural networks, the Seattle Heart Failure Model9). As a result of frequent comorbidity in patients with cardiomyopathy, a comprehensive profile of laboratory values and clinical data may be necessary for accurate prediction of an individual’s prognostic outcome.
Although the present study is one of the most extensive transcriptomic studies of heart tissue, the fact that the patient sample size was much smaller than the measured markers (>40 000 transcripts per sample) raises important statistical challenges.7,26,50 Power calculations are particularly challenging in this scenario, largely because the variance of each gene contained within the biomarker is impossible to assess, but learning-curve modulations26 have led to a general recommendation that samples of 20 to 30 in the training set are adequate. Nevertheless, logistic regression models indicate that under these circumstances, the probability of developing a nonrepresentative prediction algorithm rises.7 Sophisticated statistical modeling, however, indicates that several issues and approaches mitigate this problem.50 First, if the number of genes included in the predictive biomarker as predictors can be reduced to <50 and can be chosen according to evidence of differences in gene expression, methods of classification may be applied successfully.50 Critical to this approach is the practice of validating the biomarker obtained in the training set in an independent test set. The ultimate validation of a successful TBB requires prospective study in an independent clinical trial, an approach highly successful in various cancers.7,26
In addition to the sample size issue, another study limitation that warrants mention is that some patients were lost to follow-up, and prognostic information was retrieved solely from the Social Security Death Index. Consequently, we were not able to assess echocardiographic data in all patients. Prospective studies are under way to address this issue.
In summary, we have developed a novel transcriptomic biomarker for prognosis in heart failure that is obtainable from a single EMB with potential for a direct clinical application. This approach should improve individualization of cardiac care and help identify patients at highest risk for circulatory collapse within the first years of presentation with heart failure.
CLINICAL PERSPECTIVE.
New technologies that measure expression levels of the entire complement of messenger RNAs in a cell or tissue have become highly useful for clinical prediction of disease origin, prognosis, and therapeutic response. Because they are highly comprehensive, they have the potential to be highly accurate. The present study shows that this approach could be very important to fulfill an unmet need in the field of heart failure: accurate prediction of the long-term clinical course of a patient. New-onset heart failure is very common and has a highly variable outcome; thus, the ability to accurately assess individual patient risk is of major significance. Using endomyocardial biopsy tissue obtained at the time of clinical presentation, we developed a molecular signature comprising 45 genes that predicted long-term clinical outcome in patients with new-onset heart failure. This transcriptomic biomarker distinguished patients who survived at least 5 years after first diagnosis from those who did poorly and required left ventricular mechanical assistance or cardiac transplantation or who died. These findings may provide the physician with important prognostic information about individual patients and could provide tools for personalized treatment or monitoring. Importantly, the biomarker can be obtained from a single endomyocardial biopsy and therefore is clinically practical. The biomarker contained biologically relevant genes, including those involved in regeneration and angiopoiesis, which suggests possible novel therapeutic targets.
Acknowledgments
The authors acknowledge the efforts of Gina Edness, RN, Elayne Breton, RN, and the staff of the Johns Hopkins Hospital cardiac catheterization laboratory for assistance and support in the collection of patient samples. We thank Francisco Martinez Murillo, PhD, Linda Dorsch, BS, and Ira Maine, PhD, from the Johns Hopkins Microarray Core Facility for consultation and their assistance with sample processing.
Sources of Funding
This work was supported by National Institutes of Health grants HL-084275, AG-02017, and HL-65455 (Dr Hare).
Footnotes
Disclosures
None.
Guest Editor for this article was Roberto Bolli, MD.
Presented in part for the Samuel A. Levine Young Clinical Investigator Award at the meeting of the American Heart Association in Orlando, Fla, November 4, 2007, and published in abstract form (Circulation. 2007;116[suppl II]:II-370).
References
- 1.Felker GM, Adams KF, Jr, Konstam MA, O’Connor CM, Gheorghiade M. The problem of decompensated heart failure: nomenclature, classification, and risk stratification. Am Heart J. 2003;145:S18–S25. doi: 10.1067/mhj.2003.150. [DOI] [PubMed] [Google Scholar]
- 2.Diaz RA, Obasohan A, Oakley CM. Prediction of outcome in dilated cardiomyopathy. Br Heart J. 1987;58:393–399. doi: 10.1136/hrt.58.4.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Felker GM, Thompson RE, Hare JM, Hruban RH, Clemetson DE, Howard DL, Baughman KL, Kasper EK. Underlying causes and long-term survival in patients with initially unexplained cardiomyopathy. N Engl J Med. 2000;342:1077–1084. doi: 10.1056/NEJM200004133421502. [DOI] [PubMed] [Google Scholar]
- 4.Hare JM. The dilated, restrictive and infiltrative cardiomyopathies. In: Zipes DP, Libby P, Bonow R, Braunwald E, editors. Braunwald’s Heart Disease. 8. Philadelphia, Pa: Elsevier; 2007. [Google Scholar]
- 5.Heidecker B, Hare JM. The use of transcriptomic biomarkers for personalized medicine. Heart Fail Rev. 2007;12:1–11. doi: 10.1007/s10741-007-9004-7. [DOI] [PubMed] [Google Scholar]
- 6.Deng MC, Eisen HJ, Mehra MR, Billingham M, Marboe CC, Berry G, Kobashigawa J, Johnson FL, Starling RC, Murali S, Pauly DF, Baron H, Wohlgemuth JG, Woodward RN, Klingler TM, Walther D, Lal PG, Rosenberg S, Hunt S. Noninvasive discrimination of rejection in cardiac allograft recipients using gene expression profiling. Am J Transplant. 2006;6:150–160. doi: 10.1111/j.1600-6143.2005.01175.x. [DOI] [PubMed] [Google Scholar]
- 7.Kittleson MM, Irizarry RA, Heidecker B, Hare JM. Transcriptomics: translation of global expression analysis to genomic medicine. In: Willard HF, Ginsburg GS, editors. Genomic and Personalized Medicine. 1. Philadelphia, Pa: Elsevier; In press. [Google Scholar]
- 8.Lee DS, Austin PC, Rouleau JL, Liu PP, Naimark D, Tu JV. Predicting mortality among patients hospitalized for heart failure: derivation and validation of a clinical model. JAMA. 2003;290:2581–2587. doi: 10.1001/jama.290.19.2581. [DOI] [PubMed] [Google Scholar]
- 9.Levy WC, Mozaffarian D, Linker DT, Sutradhar SC, Anker SD, Cropp AB, Anand I, Maggioni A, Burton P, Sullivan MD, Pitt B, Poole-Wilson PA, Mann DL, Packer M. The Seattle Heart Failure Model: prediction of survival in heart failure. Circulation. 2006;113:1424–1433. doi: 10.1161/CIRCULATIONAHA.105.584102. [DOI] [PubMed] [Google Scholar]
- 10.Aaronson KD, Schwartz JS, Chen TM, Wong KL, Goin JE, Mancini DM. Development and prospective validation of a clinical index to predict survival in ambulatory patients referred for cardiac transplant evaluation. Circulation. 1997;95:2660–2667. doi: 10.1161/01.cir.95.12.2660. [DOI] [PubMed] [Google Scholar]
- 11.Smits JM, Deng MC, Hummel M, De MJ, Schoendube F, Scheld HH, Persijn GG, Laufer G, Van Houwelingen HC. A prognostic model for predicting waiting-list mortality for a total national cohort of adult heart-transplant candidates. Transplantation. 2003;7:1185–1189. doi: 10.1097/01.TP.0000091171.82384.33. [DOI] [PubMed] [Google Scholar]
- 12.Schork NJ, Nath SK, Fallin D, Chakravarti A. Linkage disequilibrium analysis of biallelic DNA markers, human quantitative trait loci, and threshold-defined case and control subjects. Am J Hum Genet. 2000;67:1208–1218. doi: 10.1086/321201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dafforn A, Chen P, Deng G, Herrler M, Iglehart D, Koritala S, Lato S, Pillarisetty S, Purohit R, Wang M, Wang S, Kurn N. Linear mRNA amplification from as little as 5 ng total RNA for global gene expression analysis. Biotechniques. 2004;37:854–857. doi: 10.2144/04375PF01. [DOI] [PubMed] [Google Scholar]
- 14.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kittleson MM, Ye SQ, Irizarry RA, Minhas KM, Edness G, Conte JV, Parmigiani G, Miller LW, Chen Y, Hall JL, Garcia JG, Hare JM. Identification of a gene expression profile that differentiates between ischemic and nonischemic cardiomyopathy. Circulation. 2004;110:3444–3451. doi: 10.1161/01.CIR.0000148178.19465.11. [DOI] [PubMed] [Google Scholar]
- 17.Kittleson MM, St John ME, Bead V, Champion HC, Kasper EK, Russell SD, Wittstein IS, Hare JM. Increased levels of uric acid predict haemodynamic compromise in patients with heart failure independently of B-type natriuretic peptide levels. Heart. 2007;93:365–367. doi: 10.1136/hrt.2006.090845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maisel A. B-type natriuretic peptide levels: diagnostic and prognostic in congestive heart failure: what’s next? Circulation. 2002;105:2328–2331. doi: 10.1161/01.cir.0000019121.91548.c2. [DOI] [PubMed] [Google Scholar]
- 19.Chakravarti A, Little P. Nature, nurture and human disease. Nature. 2003;421:412–414. doi: 10.1038/nature01401. [DOI] [PubMed] [Google Scholar]
- 20.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 21.Marchionni L, Wilson RF, Wolff AC, Marinopoulos S, Parmigiani G, Bass EB, Goodman SN. Systematic review: gene expression profiling assays in early-stage breast cancer. Ann Intern Med. 2008;148:358–369. doi: 10.7326/0003-4819-148-5-200803040-00208. [DOI] [PubMed] [Google Scholar]
- 22.Hall JL, Birks EJ, Grindle S, Cullen ME, Barton PJ, Rider JE, Lee S, Harwalker S, Mariash A, Adhikari N, Charles NJ, Felkin LE, Polster S, George RS, Miller LW, Yacoub MH. Molecular signature of recovery following combination left ventricular assist device (LVAD) support and pharmacologic therapy. Eur Heart J. 2007;28:613–627. doi: 10.1093/eurheartj/ehl365. [DOI] [PubMed] [Google Scholar]
- 23.Margulies KB, Matiwala S, Cornejo C, Olsen H, Craven WA, Bednarik D. Mixed messages: transcription patterns in failing and recovering human myocardium. Circ Res. 2005;96:592–599. doi: 10.1161/01.RES.0000159390.03503.c3. [DOI] [PubMed] [Google Scholar]
- 24.Steenman M, Chen YW, Le CM, Lamirault G, Varro A, Hoffman E, Leger JJ. Transcriptomal analysis of failing and nonfailing human hearts. Physiol Genomics. 2003;12:97–112. doi: 10.1152/physiolgenomics.00148.2002. [DOI] [PubMed] [Google Scholar]
- 25.Depre C, Shipley GL, Chen W, Han Q, Doenst T, Moore ML, Stepkowski S, Davies PJ, Taegtmeyer H. Unloaded heart in vivo replicates fetal gene expression of cardiac hypertrophy. Nat Med. 1998;4:1269–1275. doi: 10.1038/3253. [DOI] [PubMed] [Google Scholar]
- 26.Mukherjee S, Tamayo P, Rogers S, Rifkin R, Engle A, Campbell C, Golub TR, Mesirov JP. Estimating dataset size requirements for classifying DNA microarray data. J Comput Biol. 2003;10:119–142. doi: 10.1089/106652703321825928. [DOI] [PubMed] [Google Scholar]
- 27.van Haaften RI, Schroen B, Janssen BJ, van Erk A, Debets JJ, Smeets HJ, Smits JF, van den Wijngaard A, Pinto YM, Evelo CT. Biologically relevant effects of mRNA amplification on gene expression profiles. BMC Bioinformatics. 2006;7:200. doi: 10.1186/1471-2105-7-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morgun A, Shulzhenko N, Perez-Diez A, Diniz RV, Sanson GF, Almeida DR, Matzinger P, Gerbase-DeLima M. Molecular profiling improves diagnoses of rejection and infection in transplanted organs. Circ Res. 2006;98:e74–e83. doi: 10.1161/01.res.0000228714.15691.8a. [DOI] [PubMed] [Google Scholar]
- 29.Hajjar RJ, del Monte F, Matsui T, Rosenzweig A. Prospects for gene therapy for heart failure. Circ Res. 2000;86:616–621. doi: 10.1161/01.res.86.6.616. [DOI] [PubMed] [Google Scholar]
- 30.Ito S, Sakai A, Nomura T, Miki Y, Ouchida M, Sasaki J, Shimizu K. A novel WD40 repeat protein, WDC146, highly expressed during spermatogenesis in a stage-specific manner. Biochem Biophys Res Commun. 2001;280:656–663. doi: 10.1006/bbrc.2000.4163. [DOI] [PubMed] [Google Scholar]
- 31.Negishi Y, Nishita Y, Saegusa Y, Kakizaki I, Galli I, Kihara F, Tamai K, Miyajima N, Iguchi-Ariga SM, Ariga H. Identification and cDNA cloning of single-stranded DNA binding proteins that interact with the region upstream of the human c-myc gene. Oncogene. 1994;9:1133–1143. [PubMed] [Google Scholar]
- 32.Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, Kimura K, Makita H, Sekine M, Obayashi M, Nishi T, Shibahara T, Tanaka T, Ishii S, Yamamoto J, Saito K, Kawai Y, Isono Y, Nakamura Y, Nagahari K, Murakami K, Yasuda T, Iwayanagi T, Wagatsuma M, Shiratori A, Sudo H, Hosoiri T, Kaku Y, Kodaira H, Kondo H, Sugawara M, Takahashi M, Kanda K, Yokoi T, Furuya T, Kikkawa E, Omura Y, Abe K, Kamihara K, Katsuta N, Sato K, Tanikawa M, Yamazaki M, Ninomiya K, Ishibashi T, Yamashita H, Murakawa K, Fujimori K, Tanai H, Kimata M, Watanabe M, Hiraoka S, Chiba Y, Ishida S, Ono Y, Takiguchi S, Watanabe S, Yosida M, Hotuta T, Kusano J, Kanehori K, Takahashi-Fujii A, Hara H, Tanase TO, Nomura Y, Togiya S, Komai F, Hara R, Takeuchi K, Arita M, Imose N, Musashino K, Yuuki H, Oshima A, Sasaki N, Aotsuka S, Yoshikawa Y, Matsunawa H, Ichihara T, Shiohata N, Sano S, Moriya S, Momiyama H, Satoh N, Takami S, Terashima Y, Suzuki O, Nakagawa S, Senoh A, Mizoguchi H, Goto Y, Shimizu F, Wakebe H, Hishigaki H, Watanabe T, Sugiyama A, Takemoto M, Kawakami B, Yamazaki M, Watanabe K, Kumagai A, Itakura S, Fukuzumi Y, Fujimori Y, Komiyama M, Tashiro H, Tanigami A, Fujiwara T, Ono T, Yamada K, Fujii Y, Ozaki K, Hirao M, Ohmori Y, Kawabata A, Hikiji T, Kobatake N, Inagaki H, Ikema Y, Okamoto S, Okitani R, Kawakami T, Noguchi S, Itoh T, Shigeta K, Senba T, Matsumura K, Nakajima Y, Mizuno T, Morinaga M, Sasaki M, Togashi T, Oyama M, Hata H, Watanabe M, Komatsu T, Mizushima-Sugano J, Satoh T, Shirai Y, Takahashi Y, Nakagawa K, Okumura K, Nagase T, Nomura N, Kikuchi H, Masuho Y, Yamashita R, Nakai K, Yada T, Nakamura Y, Ohara O, Isogai T, Sugano S. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- 33.Heidbreder M, Frohlich F, Johren O, Dendorfer A, Qadri F, Dominiak P. Hypoxia rapidly activates HIF-3alpha mRNA expression. FASEB J. 2003;17:1541–1543. doi: 10.1096/fj.02-0963fje. [DOI] [PubMed] [Google Scholar]
- 34.Semenza GL. Pulmonary vascular responses to chronic hypoxia mediated by hypoxia-inducible factor 1. Proc Am Thorac Soc. 2005;2:68–70. doi: 10.1513/pats.200404-029MS. [DOI] [PubMed] [Google Scholar]
- 35.Geisler SB, Robinson D, Hauringa M, Raeker MO, Borisov AB, Westfall MV, Russell MW. Obscurin-like 1, OBSL1, is a novel cytoskeletal protein related to obscurin. Genomics. 2007;89:521–531. doi: 10.1016/j.ygeno.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raeker MO, Su F, Geisler SB, Borisov AB, Kontrogianni-Konstantopoulos A, Lyons SE, Russell MW. Obscurin is required for the lateral alignment of striated myofibrils in zebrafish. Dev Dyn. 2006;235:2018–2029. doi: 10.1002/dvdy.20812. [DOI] [PubMed] [Google Scholar]
- 37.Chai W, Sfeir AJ, Hoshiyama H, Shay JW, Wright WE. The involvement of the Mre11/Rad50/Nbs1 complex in the generation of G-overhangs at human telomeres. EMBO Rep. 2006;7:225–230. doi: 10.1038/sj.embor.7400600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo G, Yao MS, Bender CF, Mills M, Bladl AR, Bradley A, Petrini JH. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc Natl Acad Sci U S A. 1999;96:7376–7381. doi: 10.1073/pnas.96.13.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boyle AJ, Schulman SP, Hare JM, Oettgen P. Is stem cell therapy ready for patients? Stem cell therapy for cardiac repair: ready for the next step. Circulation. 2006;114:339–352. doi: 10.1161/CIRCULATIONAHA.105.590653. [DOI] [PubMed] [Google Scholar]
- 40.Moore KA, Lemischka IR. Stem cells and their niches. Science. 2006;311:1880–1885. doi: 10.1126/science.1110542. [DOI] [PubMed] [Google Scholar]
- 41.Lowes BD, Gilbert EM, Abraham WT, Minobe WA, Larrabee P, Ferguson D, Wolfel EE, Lindenfeld J, Tsvetkova T, Robertson AD, Quaife RA, Bristow MR. Myocardial gene expression in dilated cardiomyopathy treated with beta-blocking agents. N Engl J Med. 2002;346:1357–1365. doi: 10.1056/NEJMoa012630. [DOI] [PubMed] [Google Scholar]
- 42.Lowes BD, Zolty R, Minobe WA, Robertson AD, Leach S, Hunter L, Bristow MR. Serial gene expression profiling in the intact human heart. J Heart Lung Transplant. 2006;25:579–588. doi: 10.1016/j.healun.2006.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Canales RD, Luo Y, Willey JC, Austermiller B, Barbacioru CC, Boysen C, Hunkapiller K, Jensen RV, Knight CR, Lee KY, Ma Y, Maqsodi B, Papallo A, Peters EH, Poulter K, Ruppel PL, Samaha RR, Shi L, Yang W, Zhang L, Goodsaid FM. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat Biotechnol. 2006;24:1115–1122. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- 44.Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F, Kawasaki ES, Lee KY, Luo Y, Sun YA, Willey JC, Setterquist RA, Fischer GM, Tong W, Dragan YP, Dix DJ, Frueh FW, Goodsaid FM, Herman D, Jensen RV, Johnson CD, Lobenhofer EK, Puri RK, Schrf U, Thierry-Mieg J, Wang C, Wilson M, Wolber PK, Zhang L, Amur S, Bao W, Barbacioru CC, Lucas AB, Bertholet V, Boysen C, Bromley B, Brown D, Brunner A, Canales R, Cao XM, Cebula TA, Chen JJ, Cheng J, Chu TM, Chudin E, Corson J, Corton JC, Croner LJ, Davies C, Davison TS, Delenstarr G, Deng X, Dorris D, Eklund AC, Fan XH, Fang H, Fulmer-Smentek S, Fuscoe JC, Gallagher K, Ge W, Guo L, Guo X, Hager J, Haje PK, Han J, Han T, Harbottle HC, Harris SC, Hatchwell E, Hauser CA, Hester S, Hong H, Hurban P, Jackson SA, Ji H, Knight CR, Kuo WP, LeClerc JE, Levy S, Li QZ, Liu C, Liu Y, Lombardi MJ, Ma Y, Magnuson SR, Maqsodi B, McDaniel T, Mei N, Myklebost O, Ning B, Novoradovskaya N, Orr MS, Osborn TW, Papallo A, Patterson TA, Perkins RG, Peters EH, Peterson R, Philips KL, Pine PS, Pusztai L, Qian F, Ren H, Rosen M, Rosenzweig BA, Samaha RR, Schena M, Schroth GP, Shchegrova S, Smith DD, Staedtler F, Su Z, Sun H, Szallasi Z, Tezak Z, Thierry-Mieg D, Thompson KL, Tikhonova I, Turpaz Y, Vallanat B, Van C, Walker SJ, Wang SJ, Wang Y, Wolfinger R, Wong A, Wu J, Xiao C, Xie Q, Xu J, Yang W, Zhang L, Zhong S, Zong Y, Slikker W., Jr The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bueno-de-Mesquita JM, van Harten WH, Retel VP, van’t Veer LJ, van Dam FS, Karsenberg K, Douma KF, van Tinteren H, Peterse JL, Wesseling J, Wu TS, Atsma D, Rutgers EJ, Brink G, Floore AN, Glas AM, Roumen RM, Bellot FE, van Krimpen C, Rodenhuis S, van de Vijver MJ, Linn SC. Use of 70-gene signature to predict prognosis of patients with node-negative breast cancer: a prospective community-based feasibility study (RASTER) Lancet Oncol. 2007;8:1079–1087. doi: 10.1016/S1470-2045(07)70346-7. [DOI] [PubMed] [Google Scholar]
- 46.van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 47.Barth AS, Hare JM. The potential for the transcriptome to serve as a clinical biomarker for cardiovascular diseases. Circ Res. 2006;98:1459–1461. doi: 10.1161/01.RES.0000231257.15059.d7. [DOI] [PubMed] [Google Scholar]
- 48.Cooper LT, Baughman KL, Feldman AM, Frustaci A, Jessup M, Kuhl U, Levine GN, Narula J, Starling RC, Towbin J, Virmani R. The role of endomyocardial biopsy in the management of cardiovascular disease: a scientific statement from the American Heart Association, the American College of Cardiology, and the European Society of Cardiology. Circulation. 2007;116:2216–2233. doi: 10.1161/CIRCULATIONAHA.107.186093. [DOI] [PubMed] [Google Scholar]
- 49.Liew CC, Ma J, Tang HC, Zheng R, Dempsey AA. The peripheral blood transcriptome dynamically reflects system wide biology: a potential diagnostic tool. J Lab Clin Med. 2006;147:126–132. doi: 10.1016/j.lab.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 50.Dudoit S, Fridlyand J, Speed TP. Comparison of discrimination methods for the classification of tumors using gene expression data. J Am Stat Assoc. 2002;97:77–87. [Google Scholar]