Abstract
We previously reported a human-specific gene conversion of SIGLEC11 by an adjacent paralogous pseudogene (SIGLEC16P), generating a uniquely human form of the Siglec-11 protein, which is expressed in the human brain. Here, we show that Siglec-11 is expressed exclusively in microglia in all human brains studied—a finding of potential relevance to brain evolution, as microglia modulate neuronal survival, and Siglec-11 recruits SHP-1, a tyrosine phosphatase that modulates microglial biology. Following the recent finding of a functional SIGLEC16 allele in human populations, further analysis of the human SIGLEC11 and SIGLEC16/P sequences revealed an unusual series of gene conversion events between two loci. Two tandem and likely simultaneous gene conversions occurred from SIGLEC16P to SIGLEC11 with a potentially deleterious intervening short segment happening to be excluded. One of the conversion events also changed the 5′ untranslated sequence, altering predicted transcription factor binding sites. Both of the gene conversions have been dated to ∼1–1.2 Ma, after the emergence of the genus Homo, but prior to the emergence of the common ancestor of Denisovans and modern humans about 800,000 years ago, thus suggesting involvement in later stages of hominin brain evolution. In keeping with this, recombinant soluble Siglec-11 binds ligands in the human brain. We also address a second-round more recent gene conversion from SIGLEC11 to SIGLEC16, with the latter showing an allele frequency of ∼0.1–0.3 in a worldwide population study. Initial pseudogenization of SIGLEC16 was estimated to occur at least 3 Ma, which thus preceded the gene conversion of SIGLEC11 by SIGLEC16P. As gene conversion usually disrupts the converted gene, the fact that ORFs of hSIGLEC11 and hSIGLEC16 have been maintained after an unusual series of very complex gene conversion events suggests that these events may have been subject to hominin-specific selection forces.
Keywords: pseudogene, gene conversion, human evolution, human brain, microglia
Introduction
Sialic acids (Sias) are nine-carbon backbone monosaccharides found at the outer ends of glycan chains on cell surfaces, secreted glycoproteins, and glycolipids of all vertebrates and play important roles as ligands in intercellular communication and in host–pathogen interactions (Angata and Varki 2002; Varki and Schauer 2009; Chen and Varki 2010). Siglecs (Sialic acid binding Ig-like Lectins) are single-pass transmembrane cell surface proteins that recognize Sias as ligands and typically send signals through tyrosine-based signaling motifs in their cytosolic C-terminal domains (Varki and Angata 2006; Crocker et al. 2007; von Gunten and Bochner 2008; Cao and Crocker 2011). A subset of CD33-related Siglecs can negatively regulate cellular responses by recognizing host Sias as “self-associated molecular patterns” (Varki 2011) and via recruitment of tyrosine phosphatases to their cytosolic tails (Whitney et al. 2001; Angata et al. 2002; Zhang et al. 2007). However, surface Sia-expressing (sialylated) pathogens can subvert this self-recognition process and dampen innate immune responses (Carlin et al. 2009).
The SIGLEC11 and SIGLEC16 genes are located in a head-to-head orientation ∼1 Mb away from the CD33-related SIGLEC gene cluster, both in the human and chimpanzee genomes. Human SIGLEC16 lies ∼9 kb 5′ of SIGLEC11 on chromosome 19q13.3. Analysis of the locus in rhesus monkeys, dogs, and cows has shown the presence of a single SIGLEC gene encoding a single ITIM (Immunoreceptor Tyrosine-based Inhibitory Motif), that is, an ancestral SIGLEC11 gene (Cao et al. 2008). Further analysis in the same study shows that this gene was evidently lost in rodents but duplicated in a hominoid common ancestor ∼20 Ma, creating two head-to-head ITIM containing genes. One of these two genes then underwent a gene conversion in the 3′ portion of the gene, resulting in the loss of its ITIM domain and gain of a sequence capable of recruiting an ITAM domain (Immunoreceptor Tyrosine-based Activation Motif). This newly formed gene is SIGLEC16, which then underwent pseudogenization in the human lineage (Cao et al. 2008).
Previously, we reported that human SIGLEC11 appeared to be an unusual gene, generated via human-specific gene conversion by the SIGLEC16 locus (Hayakawa et al. 2005). The converted region was noted to encompass 5′ sequences upstream of the transcription start site and exons encoding the N-terminal region, including the domain mediating Sia recognition. While the new human gene product Siglec-11 showed reduction in overall Sia-binding abilities in comparison with the ancestral ape form, it could recognize oligosialic acids (Hayakawa et al. 2005), which are enriched in the brain (Inoko et al. 2010). Notably, in humans, the antibody against Siglec-11 stained brain cells corresponding in appearance and distribution to brain microglia, and such staining was absent or rare in brains from chimpanzees and orangutans, indicating human-specificity of this expression (Hayakawa et al. 2005). Microglia are cells derived from the yolk sac (Guillemin and Brew 2004; Hanisch and Kettenmann 2007; Ransohoff and Perry 2009), which populate the brain as immature macrophages during early development, with a subsequent turnover rate much lower than that of nonneural tissue macrophages (Ginhoux et al. 2010) and are well known to play a central role in brain immunity with functions such as sending “danger signals,” cytokine secretion, and phagocytosis (Guillemin and Brew 2004).
Although the gene conversion is associated with uniquely human Siglec-11 expression in brains, this did not have a major effect on the expression of Siglec-11 in extraneural tissue macrophages, as shown by expression in both human and chimpanzee tonsillar macrophages (Hayakawa et al. 2005). More recent studies have shown that Siglec-11 is also expressed in stromal fibroblasts of the ovary, which are cells of mesenchymal origin (Wang et al. 2011).
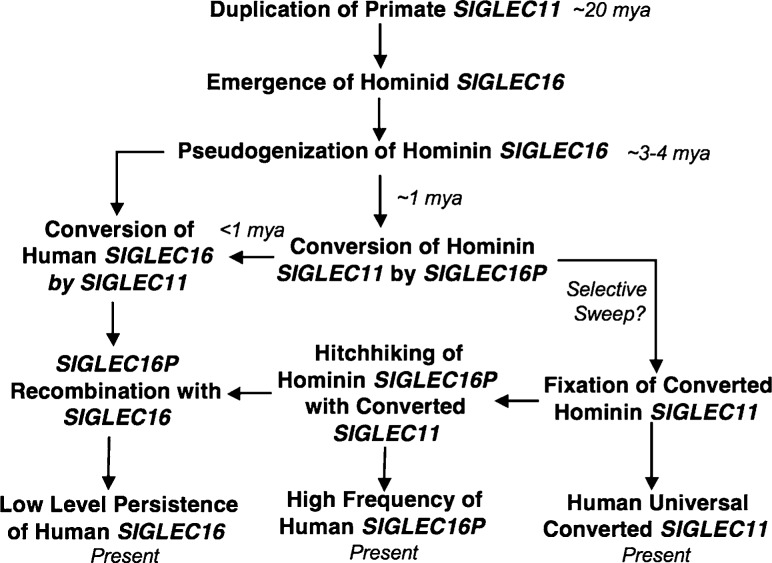
Following our report on the SIGLEC11 gene conversion event, SIGLEC16P was recognized to actually be a null allele of a functional SIGLEC16 gene that exists in some humans (Cao et al. 2008). Here, we report an unusual series of gene conversion events among these loci, including tandem and likely simultaneous gene conversions from SIGLEC16P to SIGLEC11 with a potentially deleterious intervening short segment happening to be excluded; and a more recent gene conversion from SIGLEC11 to SIGLEC16, with the latter gene showing a low allele frequency of ∼0.1–0.3 in a worldwide population study. The potential pathological and evolutionary significance of these gene conversion events are discussed.
Materials and Methods
Siglec-11 Immunostaining
Paraffin sections of eleven human, four chimpanzee, and two orangutan brain samples were deparaffinized, rehydrated, treated to remove endogenous peroxidases, endogenous biotin and nonspecific binding to extracellular matrix. The sections were subjected to heat-induced antigen retrieval in pH = 6.0 citrate buffer and overlaid with anti-Siglec-11 antibody 4C4 (which was used in the biotinylated form for double fluorescence studies) or mouse IgG (X63 supernatant from mouse myeloma cell line as negative control) overnight at 4 °C. The slides were then washed and specific binding was detected using components of the CSA kit (DAKO) following established protocols. For fluorescent labeling to detect binding of biotinylated 4C4, the last step used Alexa-Fluor 488 labeled Streptavidin (Molecular Probes). In the double labeling studies, we used a PE conjugated CD68 macrophage marker for microglia (BD Pharmingen, with PE conjugated IgG as negative control). The double fluorescence assays were viewed by epifluorescence under a Nikon microscope, photographed using a Sony CCD camera with filters for detecting fluorescence. Images were captured using the Scion Image program and overlaid using Adobe Photoshop. The bright field assays with horseradish peroxidase (HRP) and substrate development and hematoxylin nuclear counterstain were viewed using an Olympus BH2 microscope equipped with an Olympus Magnafire image capture system and the photomicrographs were analyzed using Adobe Photoshop.
Sequencing of SIGLEC16 Locus in HEL Cells
HEL (human erythroleukemia cell line), the cell line which was reported to carry a functional SIGLEC16 allele (Cao et al. 2008), was obtained from Health Science Research Resources Bank (Osaka, Japan) and cultured as specified by the distributor. Genomic DNA was purified from the cells by a conventional method, and a segment of SIGLEC16 locus encompassing the exons 1 through 6 (3.2 kb) was amplified by a polymerase chain reaction (PCR). The reaction mixture containing 100 ng of genomic DNA from HEL cells, 300 nM (each) forward and reverse primers (forward primer sequence: CTGCATGGAGTAGATTTTGCCTG; reverse primer sequence: CAGGCCCTTCCTACATCCCTG), 200 μM (each) dNTPs, and two units of Expand High Fidelity Enzyme Mix (Roche Applied Science) in 50 μl of 1× reaction buffer (Roche Applied Science) was subjected to the thermal cycling reaction as follows: 94 °C, 2 min; (94 °C, 15 s—63 °C, 30 s—72 °C, 2 min) × 10 cycles; (94 °C, 15 s—63 °C, 30 s—72 °C, 2 min + n × 5 s) × 20 cycles (where n represents the cycle number); 72 °C, 7 min. The amplified DNA fragment (3.2 kb) was purified by agarose gel electrophoresis and cloned into pCR2.1 TOPO TA cloning vector (Life Technologies). The clones were sequenced at the Support Unit for Bio-material Analysis in RIKEN BSI Research Resources Center. Four independent clones were sequenced and the majority consensus sequence was deduced. The sequence was deposited to GenBank (accession number: JQ045129).
Analysis of Genomic DNA Sequences
Sequences of Chimpanzee SIGLEC11 and SIGLEC16 were obtained from the chimpanzee genome NCBI Build 2.1 http://www.ncbi.nlm.nih.gov/genome/guide/chimp/. The human pseudogene allele of SIGLEC16 was acquired from the human genome NCBI Build 37.2. Genetic distances were calculated with multiple-hit corrections (MEGA4) (Tamura et al. 2007) using the genomic sequence alignment shown in supplementary figure S1 (Supplementary Material online). Phylogenetic trees were constructed using the Neighbor-Joining method (Saitou and Nei 1987)(MEGA4). DnaSP version 3 (Rozas J and Rozas R 1999) was used to obtain a sliding window plot representing the nucleotide differences between human SIGLEC11 and SIGLEC16P, as well as chimpanzee SIGLEC11 and SIGLEC16.
Genotyping of SIGLEC16/P Alleles in Human Populations
Genotyping of SIGLEC16/P was conducted in 12 genomic DNA samples of sub-Saharan Africans obtained from Coriell Institute for Medical Research (Camden, NJ), as well as a HapMap sample set of 27 individuals, including 8 North Europeans, 10 Africans, and 9 Asians (Chinese and Japanese) (table 1). To determine the genomic status of SIGLEC16, we designed two sets of PCR primers: CTGGGAGCCTGGTGTAAGCTGCAG and ATTCCATACCTGTTACTTTTAGAAAG gave a 1,000 bp product only when the SIGLEC16 allele was present. To detect the null allele, we used the primer sets CTGTGTGTCATCGTGTCTTG and ATTCCATACCTGTTACTTTTAGAGC as described earlier (Cao et al. 2008). A 300 bp product was obtained when the SIGLEC16P allele was present. PCR Supermix (Invitrogen) was used with conditions as follows: 1 cycle (96 °C–2 min), 3 cycles (96 °C–24 s, 65 °C–45 s, and 72 °C–30 s), 25 cycles (96 °C–25 s, 62 °C–45 s, and 72 °C–30 s), 4 cycles (96 °C–25 s, 55 °C–1 min, and 72 °C–2 min), and 1 cycle (72 °C–10 min).
Table 1.
SIGLEC16/P Allele Frequencies in a Random Sampling of Human Populations Obtained by Genomic PCR or HapMap SNPs Analysis.
| Homozygous SIGLEC16 (+/+) | Homozygous SIGLEC16P (−/−) | Heterozygous (+/−) | Allele Frequency of SIGLEC16 | Percentage Capable of SIGLEC16 Expression (+/+) and (+/−) | |
|---|---|---|---|---|---|
| African samples (12) | 0 | 10 | 2 | 0.08 | 16.7 |
| Northern European (8) NHGRI | 0 | 5 | 3 | 0.19 | 37.5 |
| Yoruba (10) NHGRI | 0 | 7 | 3 | 0.15 | 30 |
| Chinese (4) NHGRI | 0 | 2 | 2 | 0.25 | 50 |
| Japanese (5) NHGRI | 1 | 3 | 1 | 0.3 | 40 |
| HapMap database (993) | 44 | 608 | 341 | 0.22 | 38.7 |
| All African samples database and NHGRI (368) | 13 | 247 | 108 | 0.18 | 32.8 |
Human SIGLEC16/P Haplotype Analysis
In total, 3,122 bp of human SIGLEC16/P covering the 3′ end of the gene containing the last intron and the last exon (using NR_002825 as a reference) was sequenced in 27 HapMap human samples. This part of the genome is specific to human SIGLEC16/P and not involved in the gene conversion with SIGLEC11. PHASE2.1.1 (Stephens et al. 2001; Stephens and Scheet 2005) was used to reconstruct the haplotypes for the SIGLEC16 locus. The branch length tests using Lintre program (Takezaki et al. 1995) were performed to examine the rate constancy between haplotypes and chimpanzee SIGLEC16 sequence. In these tests, the gorilla sequence was used as an outgroup. Haplotypes showing rate constancy were used in the calculation of time back to the most recent common ancestor (TMRCA) and divergence times. TMRCA was calculated using the Genetree program (Griffiths and Tavare 1994; Griffiths 2002). In the TMRCA calculation, haplotypes showing parallel mutations were eliminated.
Immunohistochemistry and Western Blots for Siglec-11 Ligands in Human Brain
Paraffin sections of two human and two chimpanzee brain samples were prepared for staining and overlaid with 1% BSA and then with recombinant Siglec-11-FLAG-Fc chimera (Angata et al. 2002) at 1 μg/ml. Siglec-11-Fc is a recombinant soluble fusion protein combining the first two domains of human Siglec-11 with human IgG Fc, including a FLAG tag at the point of fusion. Control sections were overlaid with Siglec-6-FLAG-Fc chimera at 1 μg/ml or with blocking buffer alone. Slides were incubated overnight at 4 °C, followed by washes before incubation with mouse anti-FLAG (Sigma, St. Louis, MO) at 1:500, further washes, and then incubated with peroxidase-labeled goat antimouse Ig at 1:500 (Jackson laboratories). After additional washes, substrate was added to develop color (Vector laboratories, Burlingame, CA), washed, counterstained, and aqueous mounted for viewing, using an Olympus BH2 microscope, fitted with Magnafire digital camera.
For western blots, human brain slices in bacterial culture tubes were minced and crushed using a polytron in 1.2 ml of phosphate buffer with protease inhibitor cocktail set III (EMD Biosciences). The sample was spun at 75 g for 15 min to eliminate nuclei and unbroken cells, and the supernatant was spun again at 100,000 × g for 1 h at 4 °C. The second pellet was solubilized in 500 μl of lysis buffer (0.5% TX-100 in PBS with protease inhibitor) for 30 min on ice. The sample was spun at 100,000 × g for 1 h. Protein concentration of the supernatant was estimated using the BCA Protein Assay kit (Thermo Fisher). The sample was biotinylated using EZ-Link Sulfo-NHS-Biotin (Thermo Fisher) and remaining biotinylation reagent was quenched using excess glycine. The membrane protein was divided into 100 μg aliquots. Five micrograms of Siglec-11-FLAG-Fc chimera, precomplexed with M2-Agarose (Sigma) was added to one aliquot of the membrane protein. Final volume was raised to 300 μl by adding more lysis buffer. The tubes were rotated end-over-end overnight in the cold room. Next morning, the samples were spun down, and the M2-agarose beads were washed with wash buffer (10 mM Tris, 0.1% Triton X-100, 150 mM NaCl, and 1 mM ethylenediaminetetraacetic acid, pH7.4). The beads were then soaked in 100 μl, pH = 3.0 glycine buffer for 5 min at room temperature to dissociate bound material. The beads were spun down and the supernatant was resolved on an sodium dodecyl sulfate polyacrylamide gel electrophoresis followed by western blot with streptavidin-HRP.
Results
Exclusive Expression of Siglec-11 in Adult Human Brain Microglia
As reported earlier (Angata et al. 2002), brain cells corresponding in appearance and distribution to human brain microglia showed strong Siglec-11 expression (detected with 4C4 antibody in eight human brains), and such staining has been found to be absent or rare in brains from chimpanzees and orangutans (Hayakawa et al. 2005). Here, we show exclusive microglial expression of Siglec-11 in eleven additional human brains by double-color immunofluorescence analysis, staining with biotinylated 4C4 antibody and an anti-CD68 antibody (macrophage-specific) and using human intestine and liver as positive controls. It can be seen that there is an essentially complete overlap of staining with the two markers (supplementary fig. S2, Supplementary Material online). This fits with a recent study, which also described human microglial expression of Siglec-11 (Wang and Neumann 2010). Based on the human-specific gene conversion events reported earlier, it is likely that this antibody cross-reacts with the extracellular domain of Siglec-16 (cross-reactivity of the antibody 4C4 between Siglec-11 and Siglec-16 was confirmed using flow cytometry). However, as discussed later, most humans are homozygous for a null allele of SIGLEC16 (SIGLEC16P). Thus, the fact that all human brain samples stained positively with 4C4 indicates that Siglec-11 is universally found in human microglia. Since the antibody will stain both Siglec-11 and any functional Siglec-16, and exclusively stains microglial cells in the human brain, it is safe to predict that any functional Siglec-16 expressed in the brain is also found only in microglia.
Close Evolutionary Relationship between Human SIGLEC11 and the Pseudogene Allele SIGLEC16P
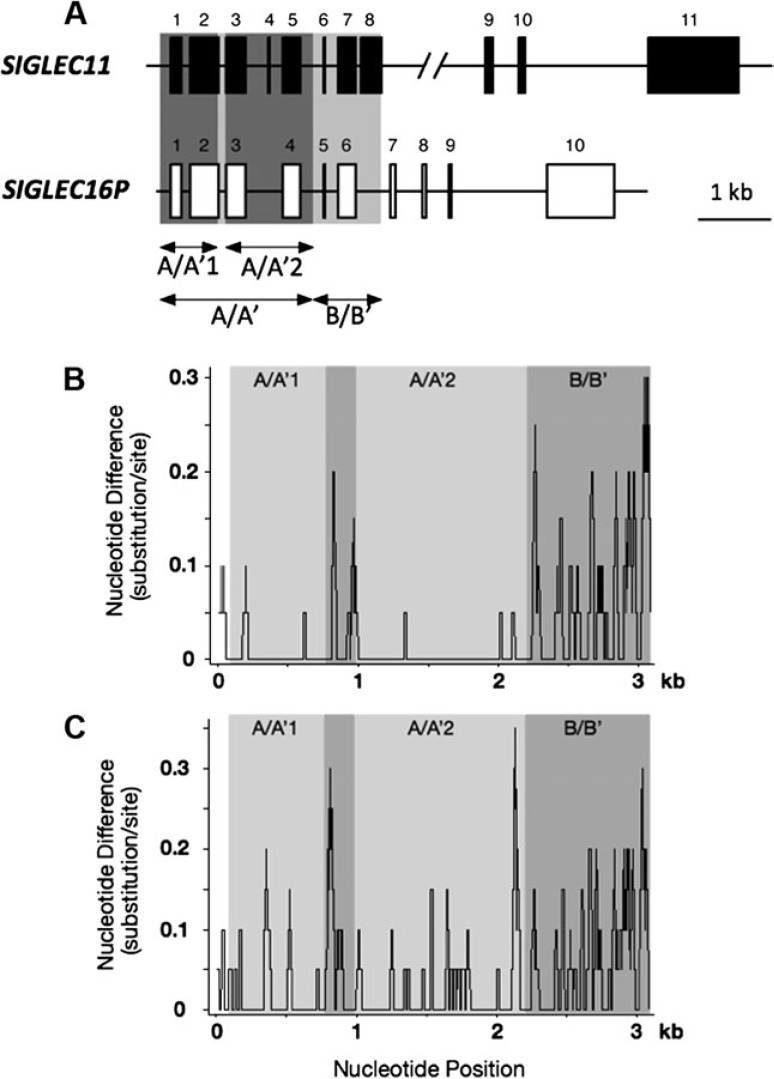
We had originally reported the human-specific gene conversion in the A/A′ region, assuming that SIGLEC16P is a fixed pseudogene in humans (Hayakawa et al. 2005). The ∼2-kb portion (designated A/A′), which includes the first five exons encoding the signal peptide, V-set, and two C2-set domains, shows 99.3% identity between human SIGLEC11 (hSIGLEC11) and human SIGLEC16P (hSIGLEC16P) (Hayakawa et al. 2005). In striking contrast, the rest of the region (designated B/B′, see fig 1A and B) has a much lower degree of identity (94.6%). The corresponding genomic regions from chimpanzee did not show this remarkably high sequence identity in their A/A′ region (97.8%)(fig. 1C). We also found that the relatively higher identity in chimpanzee A/A′ region compared with its B/B′ region (93.9%) could result from an ancient gene conversion before the human–chimpanzee divergence (data not shown, this matter is not pursued further here). Due to the subsequently discovered functional allele of SIGLEC16 gene in human populations (Cao et al. 2008), we needed to completely revisit our previous analysis of human-specific gene conversion.
Fig. 1.
Comparison of SIGLEC11 and SIGLEC16/P sequences. (A) Gene structures of SIGLEC11 and SIGLEC16P. Exons are represented by solid and open boxes. (B) Sliding window analysis of the conservation profile of human SIGLEC16P versus SIGLEC11 (window size 20 bp; step size 1 bp). (C) Sliding window analysis of the conservation profile of chimpanzee SIGLEC16 versus SIGLEC11 (window size 20 bp; step size 1 bp).
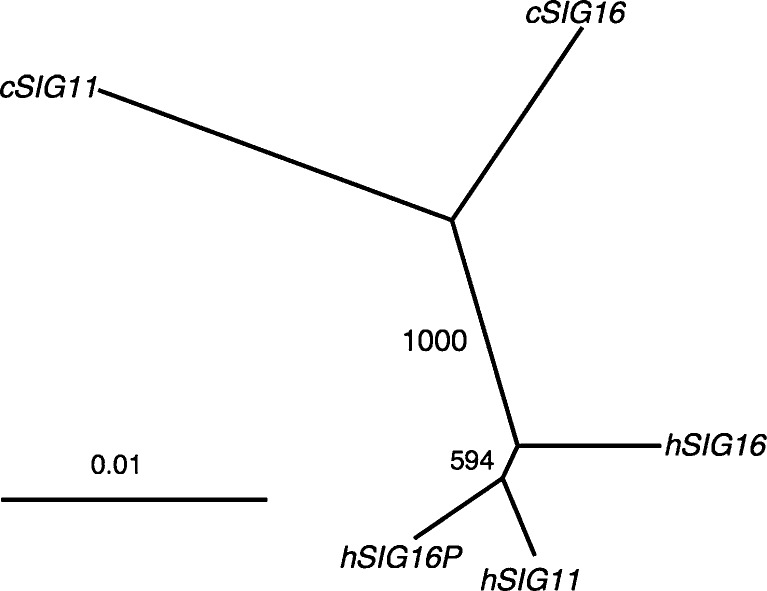
With the newly acquired human SIGLEC16 allele sequence (hSIGLEC16), we examined the phylogenetic relationship of the A/A′ region among hSIGLEC16, hSIGLEC16P, hSIGLEC11, and chimpanzee SIGLEC11 (cSIGLEC11) and chimpanzee SIGLEC16 (cSIGLEC16). The phylogenetic tree of A/A′ showed that hSIGLEC11 is more closely related to hSIGLEC16P than to hSIGLEC16 (fig. 2), which indicates that gene conversion indeed occurred between hSIGLEC11 and hSIGLEC16P.
FIG. 2.
Phylogenetic relationship between the human (h) SIGLEC16P, hSIGLEC16, hSIGLEC11 and chimpanzee (c) SIGLEC11 and cSIGLEC16. Phylogenetic tree of the A/A′ region is constructed using Neighbor-Joining method. The label at the internode represents bootstrap support for 1,000 replications.
Human SIGLEC11 and SIGLEC16P Are Involved in Two Tandem Gene Conversion Events
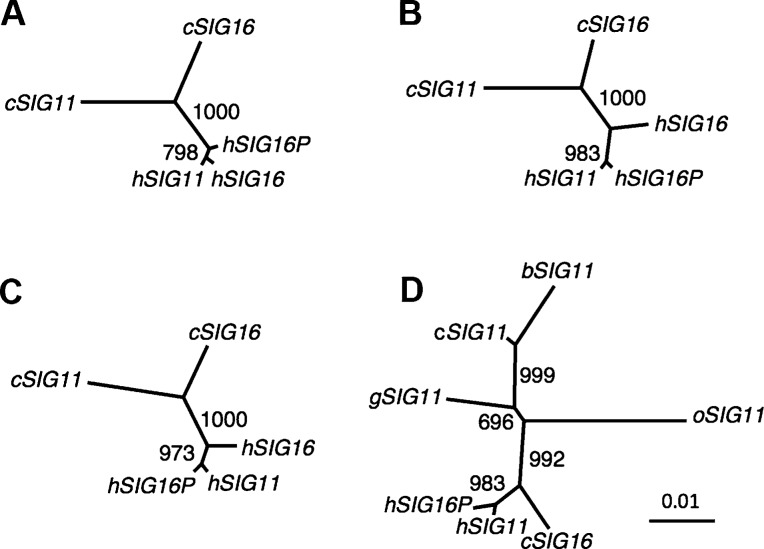
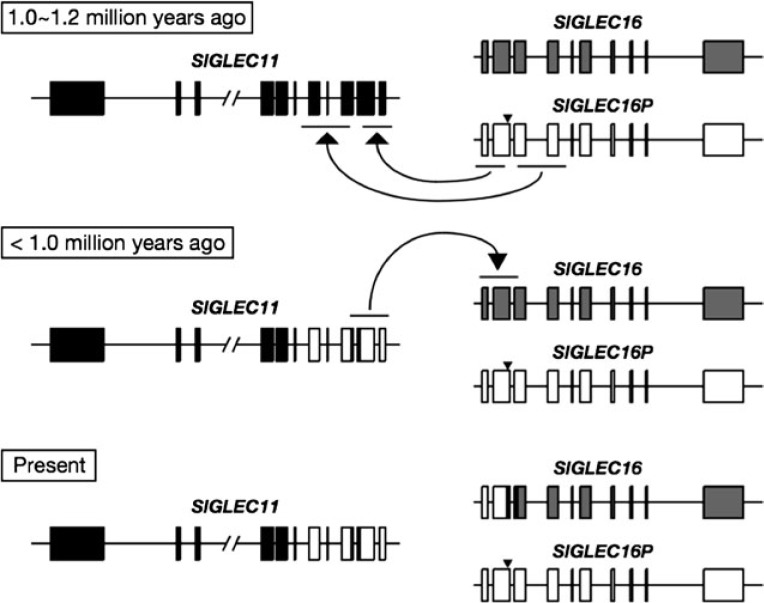
Sequence comparison of the A/A′ region between hSIGLEC11 and hSIGLEC16P showed that the A/A′ region can be divided into two even more highly similar parts A/A′1 (706 bp) and A/A′2 (1,226 bp), located upstream and downstream of a 170-bp divergent region which includes intron 2 that contains 9 of the 15 differences found in the A/A′ region between hSIGLEC11 and hSIGLEC16P (see fig. 1A and B) (Hayakawa et al. 2005). Interestingly, a 4-bp deletion, which caused the frame shift in hSIGLEC16P is coincidently located within this 170-bp divergent region. We calculated the pairwise genetic distances of the A/A′1 together with the A/A′2 regions using introns combined with synonymous sites from hSIGLEC11, hSIGLEC16, hSIGLEC16P, cSIGLEC11, and cSIGLEC16 genes. For comparison, the standard human–chimpanzee genetic distance (0.0166) was calculated using intronic portions downstream of SIGLEC11 A/A′ region. The genetic distance of the A/A′1 + A/A′2 regions between hSIGLEC11 and hSIGLEC16P (0.0031) is much smaller than either the standard human–chimpanzee genetic distance (0.0166) or that between cSIGLEC11 and cSIGLEC16 (0.0239) (table 2). Separate calculations for A/A′1 and A/A′2 regions gave the same result (table 2). The relationship among these genes was also supported by the reconstructed phylogenetic trees (fig. 3A–C). In contrast, the hSIGLEC11–hSIGLEC16P genetic distance of intron 2 (0.0314) is similar to the B/B′ intronic region (0.0434). A phylogenetic tree reconstructed from the 170-bp divergent region also indicates that hSIGLEC11 is more closely related to cSIGLEC11 than to hSIGLEC16P (supplementary fig. S3, Supplementary Material online). These data support the concept that there were actually two tandem gene conversions between hSIGLEC16P and hSIGLEC11, and the 170-bp divergent portion in the A/A′ region appeared to be excluded from these gene conversions. We also noticed that hSIGLEC11 is more closely related to hSIGLEC16 than to hSIGLEC16P in the A/A′1 region (fig. 3A and table 2) and this closer relationship is driven by a single informative site. This phylogenetic pattern actually leads to our finding of another second-round gene conversion, occurring from the initially converted hSIGLEC11 to hSIGLEC16, with the A/A′1 region involved (details shown later).
Table 2.
Genetic Distances of Noncoding Region Sequences Together with Synonymous Sites from Human and Chimpanzee Loci.
| cSIGLEC11 | hSIGLEC16 | hSIGLEC16P | cSIGLEC16 | |
|---|---|---|---|---|
| A/A′1 + A/A′2 (∼1,140 bp) | ||||
| hSIGLEC11 | 0.0329 ± 0.0052 | 0.0087 ± 0.0026 | 0.0031 ± 0.0016 | 0.0199 ± 0.0040 |
| cSIGLEC11 | 0.0321 ± 0.0051 | 0.0346 ± 0.0053 | 0.0239 ± 0.0044 | |
| hSIGLEC16 | 0.0087 ± 0.0026 | 0.0207 ± 0.0041 | ||
| hSIGLEC16P | 0.0215 ± 0.0042 | |||
| A/A′1 (∼320 bp) | ||||
| hSIGLEC11 | 0.0275 ± 0.0080 | 0.0023 ± 0.0023 | 0.0045 ± 0.0032 | 0.0159 ± 0.0060 |
| cSIGLEC11 | 0.0252 ± 0.0076 | 0.0275 ± 0.0080 | 0.0159 ± 0.0060 | |
| hSIGLEC16 | 0.0023 ± 0.0023 | 0.0136 ± 0.0056 | ||
| hSIGLEC16P | 0.0159 ± 0.0060 | |||
| A/A′2 (∼820 bp) | ||||
| hSIGLEC11 | 0.0358 ± 0.0067 | 0.0122 ± 0.0039 | 0.0024 ± 0.0017 | 0.0220 ± 0.0052 |
| cSIGLEC11 | 0.0358 ± 0.0067 | 0.0384 ± 0.0069 | 0.0283 ± 0.0059 | |
| hSIGLEC16 | 0.0122 ± 0.0039 | 0.0245 ± 0.0055 | ||
| hSIGLEC16P | 0.0245 ± 0.0055 | |||
Note.—Genetic distances within the region A/A′ (see also fig. 1) were calculated, with multiple-hit corrections (Jukes and Cantor 1969). h, human; c, chimpanzee. Synonymous sites were represented by the third codon positions of the coding region sequences.
FIG. 3.
Phylogenetic analyses of the A/A′ regions of SIGLEC11 and SIGLEC16/P. Phylogenetic relationships between human (h) and chimpanzee (c) SIGLEC11 and SIGLEC16/P for (A) region A/A′1, (B) region A/A′2, and (C) region A/A′1 + A/A′2. (D). Phylogenetic tree of region A/A′1 + A/A′2 of human, chimpanzee, bonobo (b), gorilla (g), and orangutan (o) SIGLEC11 and human SIGLEC16P and chimpanzee SIGLEC16. The label at the internode represents bootstrap support for 1,000 replications. The GenBank accession numbers of bonobo, gorilla, and orangutan SIGLEC11 sequences are AB211392–AB211394.
Direction of the Gene Conversion between hSIGLEC11 and hSIGLEC16P
We also verified the direction of the two tandem gene conversions is from hSIGLEC16P to hSIGLEC11 by analyzing both the A/A′1 and A/A′2 regions. Table 2 shows that both hSIGLEC11 and hSIGLEC16P are more closely related to cSIGLEC16 than to cSIGLEC11, and that the genetic distance between hSIGLEC11 and cSIGLEC16 is nearly the same as that between SIGLEC16 orthologs. A similar pattern was seen when comparing A/A′1, A/A′2, or A/A′1 + A/A′2 (table 2). Moreover, a phylogenetic tree of A/A′1 + A/A′2 from multiple hominids showed that bonobo, gorilla, and orangutan SIGLEC11 are more closely related to cSIGLEC11 than to cSIGLEC16 (fig. 3D). Phylogenetic analysis of individual A/A′1 or A/A′2 region also showed the same result as figure 3D (data not shown). These data indicated that SIGLEC16P converted SIGLEC11 in both A/A′1 and A/A′2 regions (SIGLEC16P → SIGLEC11).
SIGLEC16P to SIGLEC11 Conversion Events Occurred after the Emergence of the Genus Homo but before the Emergence of Modern Homo sapiens
Analysis of the converted SIGLEC11 sequence in the current Human Single Nucleotide Polymorphism (SNP) database showed no evidence for gene conversion polymorphisms (data not shown). Taken together with the finding that Siglec-11 is expressed in all human brains studied, the gene conversion of SIGLEC11 is apparently universal to modern humans (i.e., was fixed prior to the emergence of modern humans in Africa ∼100–200,000 years ago). We took advantage of the evolutionary neutrality of the pseudogene allele hSIGLEC16P to date the gene conversion events. After the SIGLEC16P → SIGLEC11 gene conversions, mutations in SIGLEC16P should have accumulated at a neutral rate. The timing (T) of gene conversion can therefore be roughly calculated by T = d/λ, where d is the branch length of hSIGLEC16P and λ is the neutral mutation rate of genomic region containing SIGLEC11 locus. Under the assumption of 6 myr of human–chimpanzee divergence, λ was calculated as (1.4 ± 0.1) × 10−9/site/year using the standard human and chimpanzee genetic distance (0.0166). The d value is estimated as 0.0016 in A/A′1 + A/A′2 (based on the number of changes that occurred only in human SIGLEC16P). Thus, T is estimated as 1.1 myr. The d values are also calculated as 0.0014 and 0.0017 for A/A′1 and A/A′2, respectively. Thus, the A/A′1 and A/A′2 gene conversions can be dated back to 1 and 1.2 Ma, respectively. These results support the notion that two SIGLEC16P → SIGLEC11 tandem gene conversions occurred during the same time period. It is possible that one complex gene conversion event occurred in region A/A′ ∼1 Ma, containing two tandem events, or that two gene conversions occurred separately and sequentially in the A/A′1 and A/A′2 regions. In either scenario, the potentially deleterious 170-bp intervening segment was excluded.
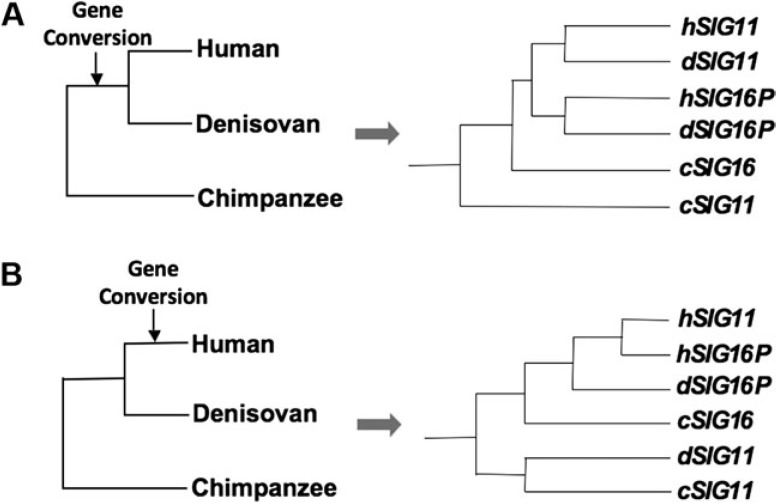
To improve the precision in timing, we examined the recently published genomes of the extinct hominins, Neanderthals, and Denisovans (UCSC Genome Browser, http://genome.ucsc.edu/). Unfortunately, the region containing SIGLEC11 is missing in the Neanderthal genome (Green et al. 2010). However, sufficient sequence was available from the Denisovan genome (Reich et al. 2010) to confirm that the Denisovan SIGLEC11 was much more similar to the modern human SIGLEC11 than the ancestral version represented by chimpanzee SIGLEC11 (see fig. 4 and supplementary fig. S4, Supplementary Material online). This result supports that the gene conversions occurred prior to the divergence of the human and Denisovan. As the common ancestor of humans and Denisovans probably existed about 800,000 years ago (Reich et al. 2010), this is consistent with our estimation that the SIGLEC11 conversion events occurred between 1 and 1.2 Ma.
FIG. 4.
Two possible scenarios for gene conversions from SIGLEC16P to SIGLEC11 during hominin evolution. (A). If the gene conversion occurred in the common ancestor of humans and Denisovans, the inferred phylogeny would then show that Denisovan SIGLEC11 is more closely related to human SIGLEC11 (supported). (B) If the gene conversion occurred only in the human lineage after the human and Denisovan divergence, the inferred phylogeny would then show that Denisovan SIGLEC11 is more closely related to chimpanzee SIGLEC11 (unsupported).
Specific Changes in the 5′ Untranslated Region of the New SIGLEC11 Gene Alter Predicted Transcription Factor Binding Sites
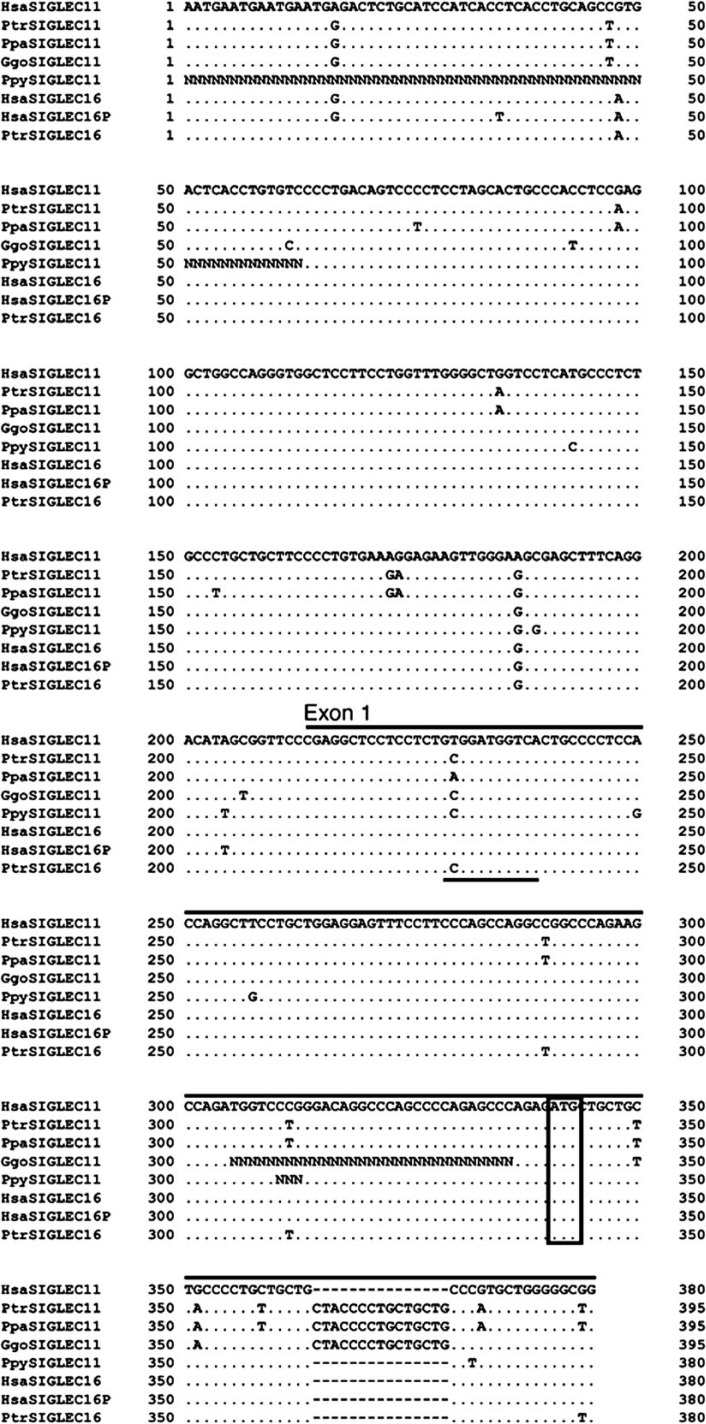
In addition to coding sequences, the human-specific A/A′1 gene conversion event transferred ∼240 bp of additional SIGLEC16P 5′-UTR sequences into the new SIGLEC11 genomic region. Analysis of this ∼240-bp region identified a GATA-1-binding sequence modified by the human-specific gene conversion event (fig. 5 and supplementary fig. S1, Supplementary Material online). A GATA-1-binding sequence acts as a repressor of BACE1 transcription in rat microglial cells (Lange-Dohna et al. 2003). The human-specific gene conversion made the sequence a potentially weaker GATA-1 binding site because thymine is a rare nucleotide at the 5′ second position of GATA-1-binding sequence (analysis done using TransFac Matys et al. 2006) (fig. 5). A modification at the same position is also found in the bonobo, but would not have the same effect, because adenine is a major nucleotide at that position (Matys et al. 2006). Thus, the human-specific gene conversion may have allowed a release from normal suppression of SIGLEC11 transcription in microglia by altering the GATA-1-binding sequence. This is also supported by the presence of the weak GATA-1 binding site found in hSIGLEC16 (fig. 5) and the fact that the SIGLEC16 transcript was also detected in human brain mRNA samples (Cao et al. 2008).
FIG. 5.
Genomic alignments of 5′ noncoding region and exon 1 of SIGLEC11 and SIGLEC16 from multiple hominids. Hsa, Homo sapiens; Ptr, Pan troglodytes; Ppa, Pan paniscus; Ggo, Gorilla gorilla; Ppy, Pongo pygmaeus. Exon 1 is represented by the bars lying on top of the alignment. The open box indicates the ATG start codon. The putative GATA-1-binding sequence is underlined. The “N”s in gorilla and orangutan sequences represent the uncertain sites in sequencing.
Detection of Siglec-11 Ligands in the Human Brain
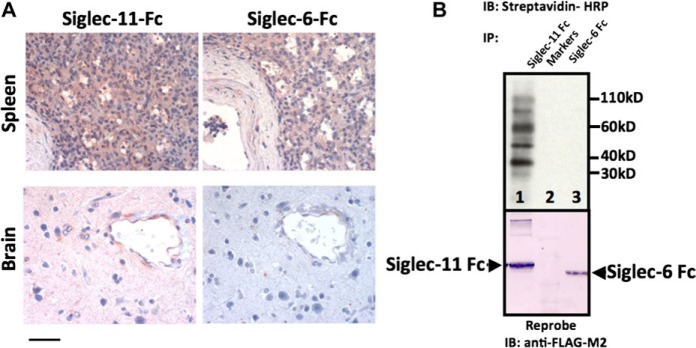
Our previous study using an in vitro Sia-binding assay with semisynthetic glycan ligands reported that chimpanzee Siglec-11 showed much more robust binding than human Siglec-11 to every sialylated glycan probe studied (Hayakawa et al. 2005). This binding difference is likely caused by the SIGLEC16P → SIGLEC11 gene conversions, which involved regions encoding the sialic acid binding domain. One may argue that the decreased Sia-binding of human Siglec-11 was simply a relative loss of binding ability to natural tissue ligands due to amino acid changes that is a “functional pseudogenization.” On the other hand, there could have been a gain of function resulting from expression in human brain microglia. To address the latter possibility, we looked for endogenous ligands in the brain by examining binding of recombinant soluble Siglec-11-Fc protein (containing the extracellular domains of human Siglec-11) to human brain tissue sections. As shown in figure 6A, human brain was strongly stained by human Siglec-11-Fc protein (extraneural tissues like spleen also showed staining). Siglec-6-Fc protein (Patel et al. 1999) provided a negative control for brain staining (fig. 6A, Siglec-6-Fc is positive for staining the spleen). Since human Siglec-11-Fc also showed binding to chimpanzee brain tissue sections (data not shown, the binding is less prominent), potential Siglec-11 ligands might have already been present in ancestral hominid brains, prior to the recruitment of human Siglec-11 expression to brain microglia. The presence of Siglec-11 ligands in the brain is also consistent with the previous reports that α2-8–linked Neu5Ac (the best natural ligand for human Siglec-11) is enriched in the brain (Sato et al. 2000).
FIG. 6.
Detection of Siglec-11 ligands in human brain. (A) Sections of human cerebral cortex or spleen were probed with human Siglec-11-Fc or Siglec-6-Fc (negative control) to detect Siglec binding sites. Brownish-pink staining is positive. Magnification is 400×, scale bar shows 50 μ. (B) Western blot analysis of the biotinylated human brain membrane extract precipitated with human Siglec-11 Fc (Lane 1) or Siglec-6 Fc (Lane 3). Lane 2 is a marker lane. The blot was stripped and reprobed with anti-FLAG antibody to confirm the presence of Siglec-11 Fc and Siglec-6 Fc, both of which have the FLAG tag.
Precipitation of Siglec-11-Fc binding proteins from human brain lysate showed multiple bands in a western blot, thus indicating the presence of more than one type of Siglec-11 ligand in the human brain (fig. 6B, as a control, Siglec-6 does not bind any ligands from the human brain lysate). Some of these bands were observed even after treatment of the brain lysate with sialidase, suggesting sialidase-resistant sialic acids and/or protein–protein interactions (data not shown). Regardless, it appears likely that human Siglec-11 developed new functions following the SIGLEC16P → SIGLEC11 gene conversion event, and that the evolutionary gain of Siglec-11 brain expression in the hominin lineage has potential meaning to the biology of human brain microglia.
High Frequency of SIGLEC16P Allele in Human Populations
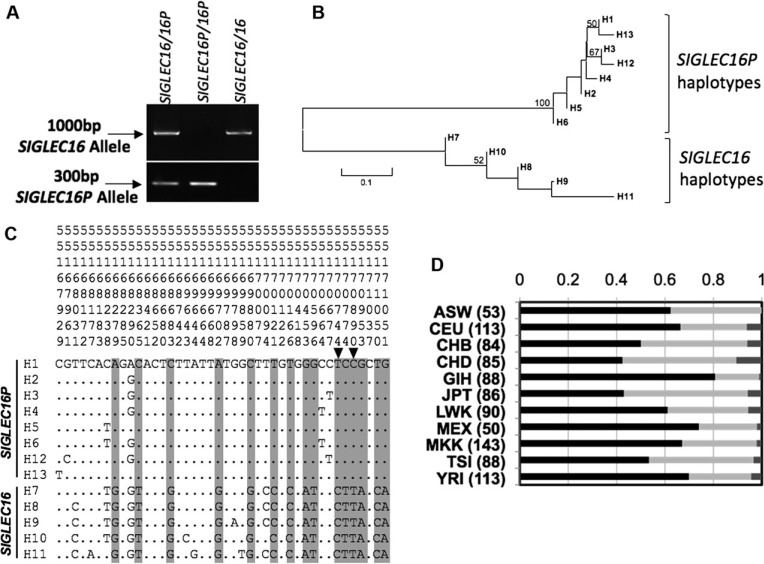
We performed genomic PCR on a number of individuals from diverse geographic origins to determine the allele frequency of the functional SIGLEC16 and the null SIGLEC16P (fig. 7A). The sample set consisted of 12 sub-Saharan African samples and 27 samples obtained from the HapMap sample set (table 1). The samples showed a combined SIGLEC16 allele frequency of 0.17.
FIG. 7.
SIGLEC16/P haplotype analysis in the human population. (A). Genomic PCR showing the presence of SIGLEC16 or SIGLEC16P alleles. (B). Neighbor-Joining tree of 13 identified haplotypes of human SIGLEC16/P from 27 human individuals. PHASE2.1.1 was used to reconstruct all of the haplotypes. Bootstrap values of more than 50% from 1,000 replications were shown on the tree branches. (C). Sequence alignment of SIGLEC16 and SIGLEC16P haplotypes. SNPs strongly linked to the SIGLEC16 or SIGLEC16P genotype are highlighted in gray. Two SNPs (site 55170744 and 55170890) present in HapMap database were indicated by the arrowheads. The location of each SNP on chromosome 19 is shown above using a sequence of numbers. All of the positions are based on human hg18 assembly. (D). SIGLEC16 genotyping using HapMap SNPs information from 993 individuals. The numbers of individuals are shown in parentheses. The bars represent the frequency of homozygous SIGLEC16P (−/−), black; heterozygous (+/−), light gray; and homozygous SIGLEC16 (+/+), dark gray. The geographic origins of the genotyped individuals are—ASW: African ancestry in Southwest USA; CEU: Utah residents with Northern and Western European ancestry from the CEPH collection; CHB: Han Chinese in Beijing, China; CHD: Chinese in Metropolitan Denver, Colorado; GIH: Gujarati Indians in Houston, Texas; JPT: Japanese in Tokyo, Japan; LWK: Luhya in Webuye, Kenya; MEX: Mexican ancestry in Los Angeles, California; MKK: Maasai in Kinyawa, Kenya; TSI: Tuscan in Italy; YRI: Yoruban in Ibadan, Nigeria.
The 3′ end of SIGLEC16/P (3,122 bp) was further sequenced by the NHGRI sequencing center in the 27 HapMap samples (table 1). This part of SIGLEC16/P was not involved in SIGLEC16P → SIGLEC11 conversion and is thus free from any PCR and sequencing ambiguity caused by the converted SIGLEC11. In total, 42 SNPs were found among the sequenced samples. PHASE2.1.1 (Stephens et al. 2001; Stephens and Scheet 2005) was used to reconstruct the haplotypes for SIGLEC16 locus. In total, 13 haplotypes were predicted. Interestingly, all of the SIGLEC16P alleles were clustered together and all of the SIGLEC16 alleles were separately clustered together on the phylogenetic tree (fig. 7B) showing that the two groups shared a distant common ancestor.
We took advantage of the observation above and correlated the SIGLEC16 locus genotype (SIGLEC16 or SIGLEC16P) to several SNPs identified from the 27 HapMap samples (highlighted in gray, fig. 7C). Two SNPs identified were also included in the HapMap database (rs12611411 and rs12984584) and in perfect linkage disequilibrium with the SIGLEC16/P polymorphism. This allowed us to estimate SIGLEC16/P allele frequencies from 993 individuals of diverse geographic origins available in HapMap (fig. 7D). Thus, we calculated the worldwide frequency of SIGLEC16 allele as 0.22, which is close to that acquired from PCR analysis above. Interestingly, several HapMap SNPs located next to these two sites also showed a similar pattern of genotype distributions, including rs12462972, rs4802644, and rs3786671 (data not shown). Particularly, rs3786671 is almost 10 kb away from rs12611411.
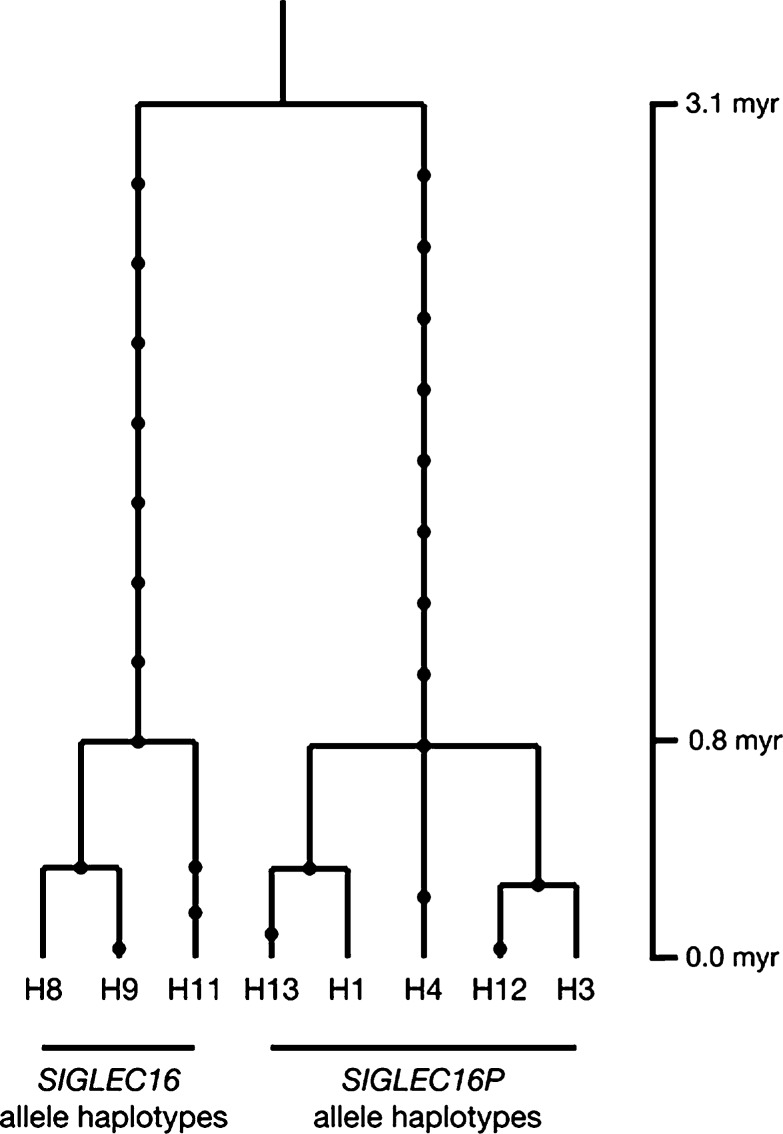
As shown in figure 8, human haplotypes are divided into two lineages: SIGLEC16P and SIGLEC16 allele lineages. The TMRCA of deduced haplotypes was estimated as 3.1 myr. This supports the notion that two alleles have coexisted for a very long time in hominins. We also estimated the divergence time between SIGLEC16 and SIGLEC16P allele lineages by using the haplotypes in our TMRCA estimation. The genetic distance between two alleles is 0.007 ± 0.001 and that between human haplotypes and chimpanzee sequence is 0.011 ± 0.002. Assuming 6 myr as human–chimpanzee divergence time, the divergence time between SIGLEC16 and SIGLEC16P allele lineages becomes 3.8 ± 0.5 myr. This timing is very close to the estimated time from the TMRCA analysis. These data also supported the notion that the original functional loss of human SIGLEC16 must have predated the SIGLEC16P → SIGLEC11 gene conversion events. Consistent with this, the gene conversions of SIGLEC16P → SIGLEC11 have been dated to ∼1 Ma.
FIG. 8.
Gene tree of human SIGLEC16 and SIGLEC16P haplotypes. The chimpanzee SIGLEC16 sequence was used to infer the most recent common ancestral sequence. Each dot represents a nucleotide substitution.
Evidence for a More Recent Second-Round Gene Conversion
In addition to the SIGLEC16P → SIGLEC11 gene conversions in A/A′1 and A/A′2, the sequence comparison of hSIGLEC16 and hSIGLEC11 also raises the possibility that another gene conversion event may have occurred between two loci. It was found that like hSIGLEC16P, hSIGLEC16 showed a high sequence identity (99.7%) to hSIGLEC11 at the A/A′1 region. hSIGLEC11 is more closely related to hSIGLEC16 than hSIGLEC16P in the A/A′1 (fig. 3A) and 170-bp regions (supplementary fig. S3, Supplementary Material online). This suggested that there might have been another gene conversion between hSIGLEC11 and hSIGLEC16, including both A/A′1 and 170-bp regions. The phylogenetic tree comparison between the A/A′1 and A/A′2 regions (fig. 3A and B) suggested that the direction of this gene conversion is from hSIGLEC11 to hSIGLEC16, instead of vice versa, due to the close relationship of hSIGLEC16 to hSIGLEC16P on the A/A′1 tree (fig. 3A). One may also argue that this second-round gene conversion is actually one of the two tandem gene conversion events we claimed earlier, with hSIGLEC16 instead of hSIGLEC16P as the donor gene sequence at the A/A′1 region. However, if hSIGLEC16 converted hSIGLEC11 in the first-round A/A′1 gene conversion, the divergence time of hSIGLEC16P and hSIGLEC11 A/A′1 regions should be identical to the TMRCA of hSIGLEC16P and hSIGLEC16, which is more than 3 Ma. In fact, our estimated divergence time of the A/A′1 region between hSIGLEC16P and hSIGLEC11 is around 1 Ma, which is much lesser than that expected. Thus, it is more likely that a gene conversion hSIGLEC11 → hSIGLEC16 at the A/A′1 and 170-bp regions occurred after the hSIGLEC16P → hSIGLEC11 gene conversions (fig. 9). Further analysis is required to verify this scenario and date this more recent event. Regardless, the fact that the ORFs of hSIGLEC11 and hSIGLEC16 have been maintained after an unusual series of very complex gene conversion events suggest that these events might have been subject to some selection forces. Notably, the long persistence of the human SIGLEC16P allele and its high frequency in human populations might have been driven by the hitchhiking of this allele along with the fixation of the converted SIGLEC11, which would be part of a broader evolutionary picture including two loci. Figure 10 attempts to summarize all of these issues into the most likely scenario.
FIG. 9.
Proposed scenario of gene conversions between human SIGLEC11 and SIGLEC16 loci. Human SIGLEC16P converted human SIGLEC11 in both of the A/A′1 and A/A′2 regions ∼1.0–1.2 Ma. After these gene conversions, human SIGLEC11 converted human SIGLEC16 in the A/A′1 and 170-bp divergent regions. SIGLEC11 and SIGLEC16 loci are shown in a head-to-head orientation. Arrowheads indicate the 4-bp deletion in human SIGLEC16P.
FIG. 10.
Proposed Scenario for the evolution of SIGLEC11 and SIGLEC16 loci over the last 20 myr. See text for discussion.
Discussion
Our analysis extends our original suggestion that the SIGLEC16 locus converted SIGLEC11 in the hominin lineage. It is well known that gene conversions by pseudogenes cause deleterious mutations resulting in human diseases, such as congenital adrenal hyperplasia, Gaucher disease, and Shwachman–Diamond syndrome (Tusie-Luna and White 1995; Cormand et al. 2000; Boocock et al. 2003; Forest 2004; Chen et al. 2007). On the other hand, the generation of antibody diversity in the chicken IgY locus, both at the somatic and the germ-line level, is known as a classic case of pseudogenes productively converting genes (Benatar and Ratcliffe 1993; Flajnik 2002). The chicken has single functional Ig V and J genes, and Ig diversity is generated mainly by gene conversion between a functional V gene and multiple upstream V pseudogenes (reviewed in Flajnik 2002).
To our knowledge, the only other vertebrate case of a pseudogene donor conversion that results in an intact and novel gene is the SIGLEC16P → SIGLEC11 conversion event we studied here. Moreover, this appears to be the first instance of human-specific productive gene conversion event involving a pseudogene donor. Although the study of pseudogenes in humans has been quite thorough, the possible paucity of conversion events between pseudogenes and genes might be explained by their sequence divergence: gene conversion normally involves pairs of donor/acceptor sequences sharing at least 92% identity (Chen et al. 2007), but most pseudogenes diverge more than 8% from their parent genes. Remarkably, despite the fact that hSIGLEC16P accumulated mutations at a neutral rate, two tandem gene conversions delivered relatively large sequence changes into human SIGLEC11. The low probability that these complex events were able to maintain the open reading frame of the recipient genes suggests that they might have been subject to some selection forces.
Ongoing gene conversion between SIGLEC5 and SIGLEC14 has been reported in multiple primates, including humans (Angata et al. 2006). Interestingly, a (TG)n tract, a genomic signature specifically favoring gene conversion, is located at the 3′ conversion boundaries in SIGLEC5 and SIGLEC14 (Angata et al. 2006). However, there is no such distinctive sequence showing around the conversion boundaries of A/A′1 and A/A′2 regions in SIGLEC11 and SIGLEC16/P. Moreover, a previous study has found many other motifs related to gene conversions (Chuzhanova et al. 2009). We searched the presence of all of the potential motifs in human and chimpanzee SIGLEC11 and SIGLEC16 loci and identified several relevant motifs around the conversion boundaries. However, they are either found in hSIGLEC11 only, or shared between human and chimpanzee sequences. Thus, we did not see a clear association between the SIGLEC16/P and SIGLEC11 gene conversion with any of these motifs.
Gene conversion of SIGLEC11 in humans resulted in microglial specific expression in the human brain. Microglial cells have been shown to be highly active, continually surveying their microenvironment with extremely motile processes and protrusions (Nimmerjahn et al. 2005; Ransohoff and Perry 2009). Microglia can also support neuronal survival by secreting cytokines and growth factors (Lu et al. 2005). The importance of Siglec-11 in the brain was recently demonstrated in the murine microglial cells transduced with a Siglec-11 vector (Wang and Neumann 2010). Siglec-11 expression reduced microglial phagocytosis of apoptotic neuronal material. It also reduced expression of Lipopolysaccharide (LPS) induced proinflammatory substances such as Nitric oxide synthase-2 and Interleukin-1β (IL-1β). Moreover, Siglec-11 was shown to have a neuroprotective effect in microglial-neuronal mixed cultures, when the neurons had polysialic acid (PSA) on their surface. Since PSA has α2-8–linked sialic acids, it is a potential ligand for Siglec-11.
In this regard, our results indicate that Siglec-11 ligands are present in the human brain and that human-specific recruitment of Siglec-11 in the brain could have had a significant impact on intracellular signaling pathways. The cytosolic tail of Siglec-11 is known to have an inhibitory ITIM that recruits the protein-tyrosine phosphatase SHP-1 (Src homology domain 2-containing phosphatase 1), an intracellular regulator of many signaling pathways (Angata et al. 2002). Notably, mice with hypomorphic mutations of SHP-1 (Shultz et al. 1993) have shown a marked decrease in the number of their microglial cells (Wishcamper et al. 2001), suggesting that SHP-1 is essential for the maintenance of microglia in the central nervous system. It has also been shown that in the microglia of SHP-1 hypomorphic mice, there is an elevated production of neurotoxic substances such as nitric oxide, Tumor Necrosis Factor-α (TNF-α), and IL-1β when challenged with LPS (Zhao et al. 2006). Of note, SHP-1 hypomorphic mice also have a slightly smaller brain size than littermate controls (Wishcamper et al. 2001). Thus, the human-specific gain of Siglec-11 expression likely affects SHP-1 biology and this in turn may have influenced microglial numbers and/or physiology and contributed to human brain evolution.
There is also evidence that microglia play a prominent role in several common human brain diseases such as Alzheimer disease and HIV-1-associated dementia (Minagar et al. 2002; Monsonego and Weiner 2003; Streit 2004). Interestingly, while the pathology of such diseases is common among humans, they have not been reported to date in the closely related great apes (Varki et al. 2011). Further studies are underway to address this issue at the histological and genotypic level. In this regard, previous study (Cao et al. 2008) has shown that SIGLEC16 message is also found in some human brain mRNA preparations. Thus, the Siglec-16 protein is also likely expressed in the microglia of individuals who have at least one SIGLEC16 allele. Regardless, since our Siglec-11 antibody also recognizes Siglec-16, we can say with some confidence that any brain Siglec-16 expression will be limited to the microglia. Consistent with this, our sequence comparison also showed that hSIGLEC16 contains a weak GATA-1 binding site same as that in hSIGLEC11 (fig. 5).
Siglec-16 does not have a cytosolic signaling motif but instead interacts with the transmembrane adapter molecule DAP12 that has a cytosolic activating ITAM. Since human Siglec-16 and Siglec-11 have identical ligand binding domains, they are expected to bind to the same set of ligands in the brain. But once activated, these Siglecs would be expected to have opposing effects, Siglec-11 would activate phosphatases such as SHP-1 and SHP-2 (via its ITIM), where Siglec-16 would recruit activating kinases such as Syk and ZAP-70 (via the ITAM). In ∼60% of humans homozygous for the SIGLEC16P (table 1), this ITAM-induced signaling is absent. This genetic difference between individuals could possibly affect microglial biology in healthy, as well as diseased brains such as Alzheimer's and HIV-induced dementia where microglia have been shown to be involved in disease process. In this regard, microglia have been found surrounding the Alzheimer's plaques (Perlmutter et al. 1992) and have been implicated in both neurotoxic functions (neuron loss in live Alzheimer'’s model brain) (Fuhrmann et al. 2010) and neuroprotective functions (clearance of β-amyloid fibrils) (El Khoury et al. 2007). These and other in vivo studies were performed on Alzheimer's mouse models that lack Siglec-11, whose ligands are present in the human brain. Thus, microglia in mouse Alzheimer disease models and microglia in humans with Alzheimer'’s disease might behave differently.
It has been noted that Alzheimer's plaques have aggregates of glycolipids (gangliosides) and glycoproteins (apolipoprotein E and Clusterin) that are sialylated and thus are potential ligands for Siglec-11 (Salminen and Kaarniranta 2009). It has also been proposed that this ligand binding leads to Siglec-11-mediated immunosupression of microglia. Although we have shown in an earlier study that Siglec-11 does not bind to gangliosides with α2-8–linked sialic acid (Angata et al. 2002), it remains to be seen whether the plaque associated glycoproteins bind Siglec-11.
The SIGLEC11 gene in humans clearly survived an unusual series of complex gene conversion events to maintain an active reading frame. This suggests a beneficial effect of the converted gene in human evolution. It is unknown what presumed selection force has driven the spread and fixation of the converted SIGLEC11 gene in ancestral human populations. Benefits on neuronal development conferred by its microglial expression could be a potential force. In this regard, the gene conversion-induced changes in a GATA-1 binding site in the SIGLEC11 promoter region require further analysis.
Due to their highly similar extracellular domains, there is at present no reliable way of distinguishing human Siglec-16 from Siglec-11 using immunohistochemical methods. In the future, we would like to produce specific antibodies to probe and differentiate the expression of Sigec-11 and Siglec-16 in normal and diseased brains. In addition, we would like to know the identity of the ligands these proteins associate with and how they might affect brain physiology in humans. These studies are likely relevant to the evolution of the hominin brain during the last few million years.
Supplementary Material
Supplementary figures S1–S4 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This research was supported by National Institutes of Health grants P01HL107150 and R01GM32373 to A.V., a Japan Society for the Promotion of Science Postdoctoral Fellowship for Research Abroad and the Ministry of Education, Culture, Sports, Science and Technology grant 23570271 to T.H., and by the G. Harold and Leila Y. Mathers Charitable Foundation. All anonymized human frozen and paraffin samples for immunohistochemistry assays were provided by the National Cancer Institute's Cooperative Human Tissue Network.
References
- Angata T, Hayakawa T, Yamanaka M, Varki A, Nakamura M. Discovery of Siglec-14, a novel sialic acid receptor undergoing concerted evolution with Siglec-5 in primates. FASEB J. 2006;20:1964–1973. doi: 10.1096/fj.06-5800com. [DOI] [PubMed] [Google Scholar]
- Angata T, Kerr SC, Greaves DR, Varki NM, Crocker PR, Varki A. Cloning and characterization of human Siglec-11. A recently evolved signaling molecule that can interact with SHP-1 and SHP-2 and is expressed by tissue macrophages, including brain microglia. J Biol Chem. 2002;277:24466–24474. doi: 10.1074/jbc.M202833200. [DOI] [PubMed] [Google Scholar]
- Angata T, Varki A. Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem Rev. 2002;102:439–469. doi: 10.1021/cr000407m. [DOI] [PubMed] [Google Scholar]
- Benatar T, Ratcliffe MJ. Polymorphism of the functional immunoglobulin variable region genes in the chicken by exchange of sequence with donor pseudogenes. Eur J Immunol. 1993;23:2448–2453. doi: 10.1002/eji.1830231011. [DOI] [PubMed] [Google Scholar]
- Boocock GR, Morrison JA, Popovic M, Richards N, Ellis L, Durie PR, Rommens JM. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat Genet. 2003;33:97–101. doi: 10.1038/ng1062. [DOI] [PubMed] [Google Scholar]
- Cao H, Crocker PR. Evolution of CD33-related siglecs: regulating host immune functions and escaping pathogen exploitation? Immunology. 2011;132:18–26. doi: 10.1111/j.1365-2567.2010.03368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Lakner U, de Bono B, Traherne JA, Trowsdale J, Barrow AD. SIGLEC16 encodes a DAP12-associated receptor expressed in macrophages that evolved from its inhibitory counterpart SIGLEC11 and has functional and non-functional alleles in humans. Eur J Immunol. 2008;38:2303–2315. doi: 10.1002/eji.200738078. [DOI] [PubMed] [Google Scholar]
- Carlin AF, Chang YC, Areschoug T, Lindahl G, Hurtado-Ziola N, King CC, Varki A, Nizet V. Group B Streptococcus suppression of phagocyte functions by protein-mediated engagement of human Siglec-5. J Exp Med. 2009;206:1691–1699. doi: 10.1084/jem.20090691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JM, Cooper DN, Chuzhanova N, Ferec C, Patrinos GP. Gene conversion: mechanisms, evolution and human disease. Nat Rev Genet. 2007;8:762–775. doi: 10.1038/nrg2193. [DOI] [PubMed] [Google Scholar]
- Chen X, Varki A. Advances in the biology and chemistry of sialic acids. ACS Chem Biol. 2010;5:163–176. doi: 10.1021/cb900266r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuzhanova N, Chen JM, Bacolla A, Patrinos GP, Ferec C, Wells RD, Cooper DN. Gene conversion causing human inherited disease: evidence for involvement of non-B-DNA-forming sequences and recombination-promoting motifs in DNA breakage and repair. Hum Mutat. 2009;30:1189–1198. doi: 10.1002/humu.21020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormand B, Diaz A, Grinberg D, Chabas A, Vilageliu L. A new gene-pseudogene fusion allele due to a recombination in intron 2 of the glucocerebrosidase gene causes Gaucher disease. Blood Cells Mol Dis. 2000;26:409–416. doi: 10.1006/bcmd.2000.0317. [DOI] [PubMed] [Google Scholar]
- Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7:255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- El Khoury J, Toft M, Hickman SE, Means TK, Terada K, Geula C, Luster AD. Ccr2 deficiency impairs microglial accumulation and accelerates progression of Alzheimer-like disease. Nat Med. 2007;13:432–438. doi: 10.1038/nm1555. [DOI] [PubMed] [Google Scholar]
- Flajnik MF. Comparative analyses of immunoglobulin genes: surprises and portents. Nat Rev Immunol. 2002;2:688–698. doi: 10.1038/nri889. [DOI] [PubMed] [Google Scholar]
- Forest MG. Recent advances in the diagnosis and management of congenital adrenal hyperplasia due to 21-hydroxylase deficiency. Hum Reprod Update. 2004;10:469–485. doi: 10.1093/humupd/dmh047. [DOI] [PubMed] [Google Scholar]
- Fuhrmann M, Bittner T, Jung CK, Burgold S, Page RM, Mitteregger G, Haass C, LaFerla FM, Kretzschmar H, Herms J. Microglial Cx3cr1 knockout prevents neuron loss in a mouse model of Alzheimer's disease. Nat Neurosci. 2010;13:411–413. doi: 10.1038/nn.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Greter M, Leboeuf M, et al. (12 co-authors) Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, et al. (56 co-authors) A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths RC. Ancestral inference from gene trees. In: Slatkin M, Venuille M, editors. Modern developments in theoretical population genetics. New York: Oxford University Press; 2002. pp. 94–117. [Google Scholar]
- Griffiths RC, Tavare S. Simulating probability distribution in the coalescent. Theor Popul Biol. 1994;46:131–159. [Google Scholar]
- Guillemin GJ, Brew BJ. Microglia, macrophages, perivascular macrophages, and pericytes: a review of function and identification. J Leukoc Biol. 2004;75:388–397. doi: 10.1189/jlb.0303114. [DOI] [PubMed] [Google Scholar]
- Hanisch UK, Kettenmann H. Microglia: active sensor and versatile effector cells in the normal and pathologic brain. Nat Neurosci. 2007;10:1387–1394. doi: 10.1038/nn1997. [DOI] [PubMed] [Google Scholar]
- Hayakawa T, Angata T, Lewis AL, Mikkelsen TS, Varki NM, Varki A. A human-specific gene in microglia. Science. 2005;309:1693. doi: 10.1126/science.1114321. [DOI] [PubMed] [Google Scholar]
- Inoko E, Nishiura Y, Tanaka H, Takahashi T, Furukawa K, Kitajima K, Sato C. Developmental stage-dependent expression of an alpha2,8-trisialic acid unit on glycoproteins in mouse brain. Glycobiology. 2010;20:916–928. doi: 10.1093/glycob/cwq049. [DOI] [PubMed] [Google Scholar]
- Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HN, editor. Mammalian protein metabolism III. New York: Academic Press; 1969. pp. 21–132. [Google Scholar]
- Lange-Dohna C, Zeitschel U, Gaunitz F, Perez-Polo JR, Bigl V, Rossner S. Cloning and expression of the rat BACE1 promoter. J Neurosci Res. 2003;73:73–80. doi: 10.1002/jnr.10639. [DOI] [PubMed] [Google Scholar]
- Lu YZ, Lin CH, Cheng FC, Hsueh CM. Molecular mechanisms responsible for microglia-derived protection of Sprague-Dawley rat brain cells during in vitro ischemia. Neurosci Lett. 2005;373:159–164. doi: 10.1016/j.neulet.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, et al. (16 co-authors) TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minagar A, Shapshak P, Fujimura R, Ownby R, Heyes M, Eisdorfer C. The role of macrophage/microglia and astrocytes in the pathogenesis of three neurologic disorders: hIV-associated dementia, Alzheimer disease, and multiple sclerosis. J Neurol Sci. 2002;202:13–23. doi: 10.1016/s0022-510x(02)00207-1. [DOI] [PubMed] [Google Scholar]
- Monsonego A, Weiner HL. Immunotherapeutic approaches to Alzheimer's disease. Science. 2003;302:834–838. doi: 10.1126/science.1088469. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–1318. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- Patel N, Brinkman-Van der Linden ECM, Altmann SW, et al. (11 co-authors) OB-BP1/Siglec-6-A leptin- and sialic acid-binding protein of the immunoglobulin superfamily. J Biol Chem. 1999;274:22729–22738. doi: 10.1074/jbc.274.32.22729. [DOI] [PubMed] [Google Scholar]
- Perlmutter LS, Scott SA, Barron E, Chui HC. MHC class II-positive microglia in human brain: association with Alzheimer lesions. J Neurosci Res. 1992;33:549–558. doi: 10.1002/jnr.490330407. [DOI] [PubMed] [Google Scholar]
- Ransohoff RM, Perry VH. Microglial physiology: unique stimuli, specialized responses. Annu Rev Immunol. 2009;27:119–145. doi: 10.1146/annurev.immunol.021908.132528. [DOI] [PubMed] [Google Scholar]
- Reich D, Green RE, Kircher M, et al. (28 co-authors) Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Rozas R. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Salminen A, Kaarniranta K. Siglec receptors and hiding plaques in Alzheimer's disease. J Mol Med. 2009;87:697–701. doi: 10.1007/s00109-009-0472-1. [DOI] [PubMed] [Google Scholar]
- Sato C, Fukuoka H, Ohta K, Matsuda T, Koshino R, Kobayashi K, Troy FAII, Kitajima K. Frequent occurrence of pre-existing alpha2-->8-linked disialic and oligosialic acids with chain lengths up to 7 Sia residues in mammalian brain glycoproteins—prevalence revealed by highly sensitive chemical methods and anti-di-, oligo-, and poly-Sia antibodies specific for defined chain lengths. J Biol Chem. 2000;275:15422–15431. doi: 10.1074/jbc.275.20.15422. [DOI] [PubMed] [Google Scholar]
- Shultz LD, Schweitzer PA, Rajan TV, Yi T, Ihle JN, Matthews RJ, Thomas ML, Beier DR. Mutations at the murine motheaten locus are within the hematopoietic cell protein-tyrosine phosphatase (Hcph) gene. Cell. 1993;73:1445–1454. doi: 10.1016/0092-8674(93)90369-2. [DOI] [PubMed] [Google Scholar]
- Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 2005;76:449–462. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit WJ. Microglia and Alzheimer's disease pathogenesis. J Neurosci Res. 2004;77:1–8. doi: 10.1002/jnr.20093. [DOI] [PubMed] [Google Scholar]
- Takezaki N, Rzhetsky A, Nei M. Phylogenetic test of the molecular clock and linearized trees. Mol Biol Evol. 1995;12:823–833. doi: 10.1093/oxfordjournals.molbev.a040259. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tusie-Luna MT, White PC. Gene conversions and unequal crossovers between CYP21 (steroid 21-hydroxylase gene) and CYP21P involve different mechanisms. Proc Natl Acad Sci U S A. 1995;92:10796–10800. doi: 10.1073/pnas.92.23.10796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A. Since there are PAMPs and DAMPs, there must be SAMPs? Glycan “self-associated molecular patterns” dampen innate immunity, but pathogens can mimic them. Glycobiology. 2011;21:1121–1124. doi: 10.1093/glycob/cwr087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Angata T. Siglecs—the major subfamily of I-type lectins. Glycobiology. 2006;16:1R–27R. doi: 10.1093/glycob/cwj008. [DOI] [PubMed] [Google Scholar]
- Varki A, Schauer R. Sialic acids. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of glycobiology. 2nd ed. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2009. pp. 199–218. [PubMed] [Google Scholar]
- Varki NM, Strobert E, Dick EJ, Benirschke K, Varki A. Biomedical differences between human and nonhuman hominids: potential roles for uniquely human aspects of sialic acid biology. Annu Rev Pathol. 2011;6:365–393. doi: 10.1146/annurev-pathol-011110-130315. [DOI] [PubMed] [Google Scholar]
- von Gunten S, Bochner BS. Basic and clinical immunology of Siglecs. Ann N Y Acad Sci. 2008;1143:61–82. doi: 10.1196/annals.1443.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chow R, Deng L, et al. (11 co-authors) Expression of Siglec-11 by human and chimpanzee ovarian stromal cells, with uniquely human ligands: implications for human ovarian physiology and pathology. Glycobiology. 2011;21:1038–1048. doi: 10.1093/glycob/cwr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Neumann H. Alleviation of neurotoxicity by microglial human Siglec-11. J Neurosci. 2010;30:3482–3488. doi: 10.1523/JNEUROSCI.3940-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney G, Wang S, Chang H, Cheng KY, Lu P, Zhou XD, Yang WP, McKinnon M, Longphre M. A new siglec family member, siglec-10, is expressed in cells of the immune system and has signaling properties similar to CD33. Eur J Biochem. 2001;268:6083–6096. doi: 10.1046/j.0014-2956.2001.02543.x. [DOI] [PubMed] [Google Scholar]
- Wishcamper CA, Coffin JD, Lurie DI. Lack of the protein tyrosine phosphatase SHP-1 results in decreased numbers of glia within the motheaten (me/me) mouse brain. J Comp Neurol. 2001;441:118–133. [PubMed] [Google Scholar]
- Zhang M, Angata T, Cho JY, Miller M, Broide DH, Varki A. Defining the in vivo function of Siglec-F, a CD33-related Siglec expressed on mouse eosinophils. Blood. 2007;109:4280–4287. doi: 10.1182/blood-2006-08-039255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Brooks DM, Lurie DI. Lipopolysaccharide-activated SHP-1-deficient motheaten microglia release increased nitric oxide, TNF-alpha, and IL-1beta. Glia. 2006;53:304–312. doi: 10.1002/glia.20283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.