Background: The mechanism of immune suppression caused by Francisella tularensis SchuS4 strain, a category A agent, are yet unknown.
Results: FTL_0325/FTT0831c genes of F. tularensis suppress proinflammatory cytokines by preventing activation of NF-κB signaling.
Conclusion: FTL_0325/FTT0831c of Francisella is a key virulence factor and functions as an immunosuppressant.
Significance: Understanding of such pathogenic mechanisms will define vaccine candidates to prevent tularemia acquired naturally or through an act of bioterrorism.
Keywords: Bacterial Pathogenesis, Cytokine, Host-Pathogen Interactions, Immunosuppression, Immunosupressor, Infectious Diseases, Innate Immunity, Macrophages, NF-κB (NF-KB), Francisella tularensis
Abstract
Francisella tularensis, the causative agent of tularemia, is one of the deadliest agents of biological warfare and bioterrorism. Extremely high virulence of this bacterium is associated with its ability to dampen or subvert host innate immune response. The objectives of this study were to identify factors and understand the mechanisms of host innate immune evasion by F. tularensis. We identified and explored the pathogenic role of a mutant interrupted at gene locus FTL_0325, which encodes an OmpA-like protein. Our results establish a pathogenic role of FTL_0325 and its ortholog FTT0831c in the virulent F. tularensis SchuS4 strain in intramacrophage survival and suppression of proinflammatory cytokine responses. This study provides mechanistic evidence that the suppressive effects on innate immune responses are due specifically to these proteins and that FTL_0325 and FTT0831c mediate immune subversion by interfering with NF-κB signaling. Furthermore, FTT0831c inhibits NF-κB activity primarily by preventing the nuclear translocation of p65 subunit. Collectively, this study reports a novel F. tularensis factor that is required for innate immune subversion caused by this deadly bacterium.
Introduction
Francisella tularensis is a Gram-negative facultative intracellular bacterium capable of causing lethal disease called tularemia in various species including humans. F. tularensis exists as two clinically relevant strains; the highly virulent Type A (F. tularensis ssp. tularensis (SchuS4)) strains that are often associated with severe clinical course and pneumonic tularemia in North America (1) and Type B (F. tularensis ssp. holarctica) strains that cause acute but mild self-limiting infections in the Eurasian and American continents (2). The live vaccine strain (LVS)5 is a derivative of F. tularensis ssp. holarctica. The other two subspecies, Francisella novicida and Francisella mediasiatica, are not associated with human disease. In the recent past, bioterror threats have renewed interest in understanding the pathogenesis of Francisella. The Centers for Disease Control has classified F. tularensis as a category A agent based on its high virulence and potential use in a terrorist attack. Use of F. tularensis as a bioterrorism agent arises from its high infectivity, ease of aerosolization, and dissemination to cause severe pulmonary disease (1, 3, 4). The control of pneumonic tularemia in a large population is difficult due to a lack of a licensed vaccine and ineffective therapies against antibiotic-resistant strains (5). Another critical characteristic of F. tularensis is its ability to actively suppress host innate immune responses (6). However, the factors and mechanisms that F. tularensis utilizes to interfere with innate immune development are yet unknown.
Activation of NF-κB and MAPK signaling play a central role in immune-dependent bacterial clearance. To dampen the host innate immune response, it is not surprising that several bacterial pathogens have evolved mechanisms to circumvent these signaling events. To inhibit NF-κB, bacterial pathogens adopt mechanisms that either involve secretion of effectors with inhibitory TLR-like domains or use type III or IV secretion systems to inject their effectors directly into the host cells (7, 8). F. tularensis lacks type III and IV secretion systems; however, it does contain a type IV pilus biogenesis system that secretes soluble proteins via type II-like secretion machinery (9–12). Additionally, a type VI secretion system encoded by Francisella pathogenicity island has recently been identified in F. tularensis, which is required for intracellular survival and modulation of host cell signaling (13, 14). F. tularensis also encodes a functional type I secretion system that is required for pathogenesis (11). A recent study has shown that an unknown Francisella factor suppresses proinflammatory cytokine production from infected as well as uninfected bystander cells (6). Another report has speculated that this factor may be secreted in a TolC-dependent fashion to cause immune suppression (15). To date, a limited number of F. tularensis factors including intracellular growth locus C, RipA, and antioxidant enzyme catalase (KatG) of F. tularensis LVS have been shown to cause innate immune subversion through inhibition of MAPK and NF-κB signaling (16–19). In contrast, F. tularensis SchuS4-mediated cytokine suppression is independent of intracellular growth locus C (20), and the roles of RipA or KatG in the immune subversion have not been fully established.
The objectives of this study were to identify factors and understand the mechanisms of host innate immune evasion by F. tularensis LVS (Type B) and the highly virulent F. tularensis SchuS4 (Type A) strain. We characterized mutants in the FTL_0325 gene encoding OmpA-like protein, which was identified in a transposon screen of F. tularensis LVS and its ortholog FTT0831c in the virulent F. tularensis SchuS4 for their role in innate immune subversion. We report that OmpA-like proteins encoded by FTL_0325/FTT0831c genes of F. tularensis LVS and SchuS4 strains, respectively, are required for intramacrophage survival and suppression of proinflammatory cytokines. We further demonstrate that FTL_0325/FTT0831c proteins of LVS and SchuS4 interfere with NF-κB signaling to restrict proinflammatory cytokines. This study provides an understanding of the innate immune subversion mechanisms of F. tularensis especially with reference to the highly virulent SchuS4 strain.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Culture
F. tularensis LVS (ATCC 29684; American Type Culture Collection) was provided by Dr. K. Elkins (U. S. Food and Drug Administration, Bethesda, MD). F. tularensis SchuS4, originally isolated from a human case of tularemia (21), was obtained from the U. S. Army Medical Research Institute for Infectious Diseases (Frederick, MD). All experiments using SchuS4 were conducted within the Centers for Disease Control-certified Biosafety level-3 (BSL-3) facility at Albany Medical College. F. novicida was obtained from the Microbiology Core Facility at Albany Medical College. All the Francisella strains were cultured in brain heart infusion, Mueller Hinton broth, or Mueller Hinton chocolate agar plates. Brain heart infusion was supplemented with 10% heat inactivated fetal bovine serum (FBS), whereas Mueller Hinton broth was supplemented with calcium chloride, magnesium chloride, ferric pyrophosphate, glucose, and isovitaleX (BD Biosciences). For the selection of transposon and gene deletion mutants and transcomplemented strains, kanamycin was included at a concentration of 10–20 μg/ml in Mueller Hinton broth culture or chocolate agar medium (in the case of F. tularensis) or 35 μg/ml (in the case of Escherichia coli). The FTL_0325 mutant was identified by screening a transposon mutant library in murine alveolar macrophage cell line, MH-S.6 An in-frame gene deletion mutant of FTL_0325 ortholog, FTT0831c, in SchuS4 and its transcomplemented strain were generated in this study (supplemental Table S1). Wild type (WT) F. tularensis LVS and FTL_0325 mutant were killed by exposing to UV light as described earlier (22).
Macrophages and Cell Lines
MH-S (murine alveolar macrophage cell line of BALB/c origin) (23) and human THP-1 cells were maintained and cultured in RPMI 1640 medium containing 25 mm l-glutamine, 25 mm HEPES, 10% heat inactivated FBS, 1% sodium pyruvate, and penicillin-streptomycin. The THP-1 cells were differentiated by treating with 100 nm phorbol 12-myristate 13-acetate overnight. The bone marrow-derived macrophages (BMDMs) from WT C57BL/6 and TLR2−/− mice and human embryonic kidney 293 (HEK293) T cells were cultured in Dulbecco's modified Eagle's medium containing 4.5 g/liter glucose, 10% FCS, 1% HEPES, 1% sodium pyruvate, and 1% l-glutamine.
Site-directed Mutagenesis and Transcomplementation
An allelic replacement method was used to construct an in-frame FTT0831c gene deletion mutant (ΔFTT0831c) of F. tularensis SchuS4 (24). For construction of the ΔFTT0831c mutant, the entire coding region of the FTT0831c gene was deleted by employing an approach described earlier (25, 26). For transcomplementation, the full-length FTT0831c gene was amplified by PCR and cloned in pKK214 vector downstream of GroEL promoter of F. tularensis following our recently published protocol (26, 27). The plasmid constructs, bacterial strains, and the primer sequences used in this study are shown in supplemental Table S1.
Macrophage Invasion and Replication Assays
Gentamicin protection assays were performed as described earlier (26). In macrophage assays involving live or UV-killed WT F. tularensis LVS, SchuS4, FTL_0325, or ΔFTT0831c mutant and its transcomplemented strain, a multiplicity of infection (m.o.i.) of 100 was used. After infection, the culture supernatants were collected at various time points for quantification of cytokines. The macrophages were lysed at various time points, and lysates were diluted 10-fold and plated on Mueller Hinton chocolate agar plates to enumerate the intracellular bacterial replication. The colonies were counted after 48 h, and the results were expressed as cfu/ml. The cell lysates were also used for Western blot analysis and quantification of NF-κB luciferase activity. To quantitate the intramacrophage lysis, F. tularensis LVS, F. novicida, and FTL_0325 mutant were transformed with pFNLTP6:luciferase construct (kindly provided by Dr. Denise Monack, Stanford University) and used in an assay as recently published (28).
Cytokine Measurement
A mouse inflammation cytometric bead array kit (BD Biosciences) was used for the measurement of TNF-α, IL-6, and IL-1β. Data were acquired on a FACSArray instrument (BD Biosciences) and analyzed using cytometric bead array software Version 1.1 (BD Biosciences). The cytokine levels were expressed as pg/ml.
Protein Expression, Cell Culture, DNA Transfections, and Luciferase Assay
FTT0831c open reading frame was cloned into the eukaryotic expression vector pcDNA3.1 (under the control of the CMV promoter). HEK293T cells were transfected with pcDNA3.1-expressing FTT0831c using empty vector as a control. The expression of FTT0831c in transfected cells was validated by Western blot analysis using anti-FTT0831c primary and goat anti-rat HRP-conjugated secondary antibodies.
For luciferase gene reporter assays, HEK293T cells (2 × 105 cells/well) were transfected with 100 ng of plasmid encoding 3xNF-κB luciferase reporter (29) (a generous gift from Dr. Albert S. Baldwin, Lineberger Comprehensive Cancer Center Chapel Hill, NC) in FTT0831c-expressing or control cells transfected with 1000 ng of pcDNA:FTT0831c using FuGENE 6 transfection kit (Roche Applied Science) as per the manufacturer's instructions. The amount of transfected DNA was kept constant by using pcDNA3.1 empty vector. The cells were activated either by treating with TNF-α (20 ng/ml × 3 h) or co-transfecting with a vector containing NF-κB p65 (50 ng). For ISRE-luciferase reporter assays, Cignal ISRE reporter ((Luc) kit (Qiagen)) was used, and the transfections were performed as per the manufacturer's instructions. After 24 h of transfection, the cells were treated with 1000 units/ml human IFN-β (PBL Biosciences) for 18 h. The cells were lysed using a passive reporter lysis buffer (Promega), and luciferase activity was monitored using a Dual-Luciferase assay kit (Promega) according to the manufacturer's protocol and read in a Victor Luminometer (Wallac). The luciferase activity was normalized to Renilla levels.
Western Blot Analysis
HEK293T cells were transfected with empty or pcDNA3.1 expressing FTT0831c (1000 ng) as described above. The cells were lysed in 1% Nonidet P-40 lysis buffer (50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1% Nonidet P-40 (v/v), 2 mm EDTA, 2 mm DTT supplemented with protease inhibitors (Roche Applied Science)). The protein concentrations were measured by BCA assay. Equal amounts of proteins (∼30 μg) were resolved on a SDS-PAGE gel (4–20%; Bio-Rad), transferred to nitrocellulose membrane (0.2 μm), and probed with anti-FTT0831c or anti-β actin antibodies (Santa Cruz Biotechnology). For IL-1β blots, BMDMs were infected with WT F. tularensis LVS or the FTL_0325 mutant at 100 m.o.i., and the cells were lysed, resolved on SDS-PAGE gels, and blotted using anti IL-1β antibodies (Santa Cruz). The blots were developed using a chemiluminescent substrate and visualized either by autoradiography or on a chemiluminescent imager.
Immunofluorescence
HEK293T cells were transfected with pcDNA3.1:FTT0831c or empty vector as described above. After TNF-α stimulation (20 ng/ml × 30 min), the cells were washed with PBS and fixed with a 3:2 ratio (v/v) of acetone:PBS for 5 min. After 3 washes with 1% bovine serum albumin fraction V (BSA), PBS, the cells were blocked with 10% normal goat-serum in 1% (BSA), PBS for 30 min. The cells were stained for endogenous NF-κB p65 using rabbit anti-p65 IgG (1:300) in blocking solution for 1.5 h followed by Alexa 594-conjugated goat anti-rabbit IgG (1: 500) for 1 h. The cells were washed three times with 1% BSA, followed by PBS and distilled H2O. The cells were stained with DAPI to visualize nucleus, mounted with a coverslip, and visualized using an Axio Observer Z1 fluorescence microscope (Zeiss). Images were taken at 20× magnification. Fields were chosen randomly to count the number of cells showing nuclear, cytoplasmic, or diffused staining. More than 200 cells were counted for each transfection condition for quantification of p65 localization.
Binding of anti-FTT0831c antibodies to the cell surface of F. tularensis was detected by immunofluorescent staining. Bacterial cells (1 × 109) untreated or treated with proteinase-K were suspended in 500 μl of PBS containing 5 % BSA and then incubated for 2 h with anti-FTT0831c polyclonal monospecific antibodies, anti-F. tularensis LPS monoclonal, or polyclonal anti-KatG antibodies at a dilution of 1:50 in PBS containing 5 % BSA. After washing twice with PBS, the pellets were incubated for 2 h with 50 μl of Alexa Fluor 488-conjugated goat anti-rat IgG antibodies for FTT0831c or goat anti-mouse antibodies (Invitrogen) at a 1:100 dilution. Cell pellets were washed 3 times with PBS and resuspended in 50 μl of PBS. One drop of the cell suspension was smeared onto a microscope slide, mounted using slowfade gold antifade reagent (Invitrogen), covered with a coverslip, and viewed under a fluorescence microscope. Bacterial cells stained with anti-F. tularensis LPS and another outer membrane protein, anti-KatG, antibodies were used as positive and negative controls, respectively.
Localization of FTT0831c
Spheroplasting and sucrose density gradient centrifugation for localization of FTT0831c was essentially performed as previously reported (30). Sequential fractions were collected from gradients, and densities (g/ml) were calculated based upon refractive indices. The proteins were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted to detect outer and inner membrane proteins in fractionated sucrose density gradients. Anti-FTT0831c polyclonal monospecific antibodies were used to determine FTT0831c localization in the various membrane fractions. Polyclonal monospecific antibodies against FopA and Mip were used as localization controls for outer membrane, whereas SecY antibodies were used as inner membrane controls.
RNA Isolation and Quantitative RT-PCR
HEK293T cells were transfected with pcDNA3.1:FTT0831c or empty vector as described above. Total RNA was isolated using RNeasy RNA purification columns (Qiagen) following the manufacturer's protocol and treated with DNase I to remove any residual genomic DNA. Quantitative real-time PCR was performed using the SuperScript III Platinum SyBR Green One-Step qRT-PCR kit (Invitrogen) and a CFX96 Real-time PCR Detection System instrument (Bio-Rad). Intron spanning primer sequences were used to amplify: IL-8 (forward, 5′-GCT CTG TGT GAA GGT GCA GT-3′; reverse, 5′-CCA GAC AGA GCT CTC TTC CA-3′) and β-actin (forward, 5′-CCC CCA TGC CAT CCT GCG TCT G-3′; reverse, 5′-CTC GGC CGT GGT GGT GAA GC-3′). All reactions were run in triplicate, and the specificity of PCR amplification was analyzed by melting curve analysis. Ct values were normalized to β-actin, and relative copy number was calculated by the standard 2−ΔΔCT method. Results are represented as -fold change over untreated.
Statistical Analysis
All results were expressed as means ± S.E. or S.D. Statistical comparisons between the groups were made using one-way ANOVA followed by Bonferroni's correction, non-parametric Mann-Whitney test, or unpaired Student's t test. Differences between the experimental groups were considered statistically significant at a p < 0.05 level.
RESULTS
FTL_0325 Mutant of F. tularensis Induces Significantly Increased Proinflammatory Cytokines in Macrophages
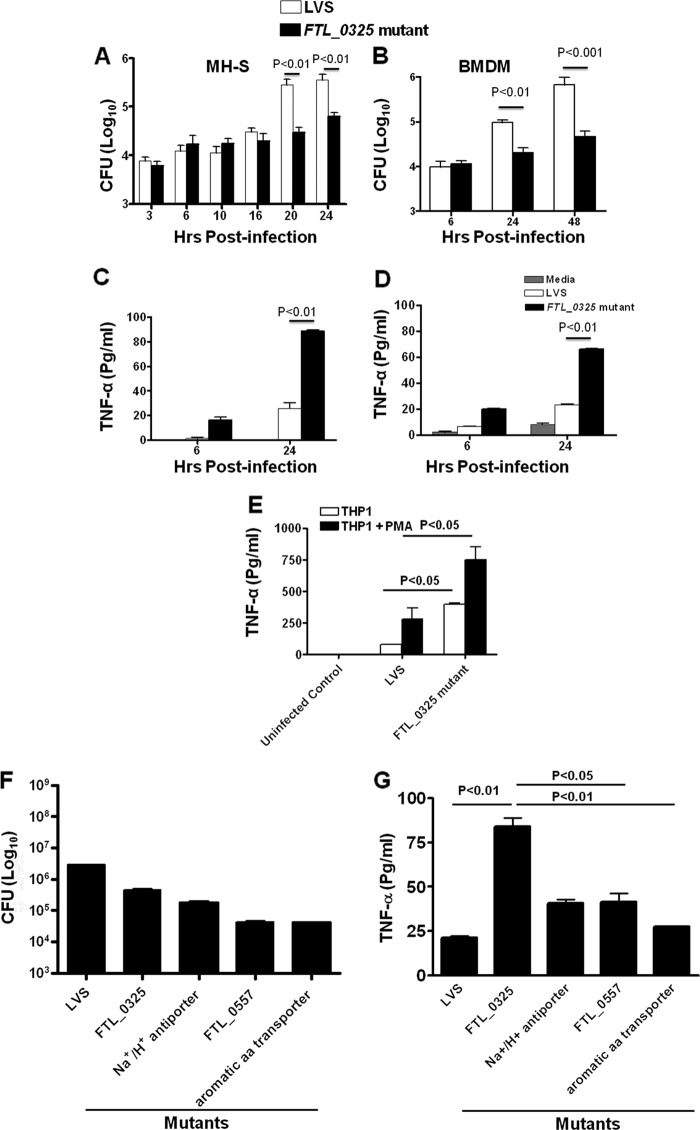
In our transposon mutant screen, a mutant in FTL_0325 gene of F. tularensis was identified that exhibited a 7-fold reduction in the number of bacteria recovered at 24 h post-infection (PI) as compared with the WT F. tularensis. We further investigated the role of FTL_0325 in intramacrophage survival by performing in vitro assays. Infection of MH-S cells or primary BMDMs with FTL_0325 mutant resulted in attenuated growth of the mutant (Fig. 1, A and B). Analysis of proinflammatory cytokines revealed significantly elevated TNF-α levels in culture supernatants of FTL_0325 mutant as compared with those observed for WT F. tularensis LVS-infected macrophages (Fig. 1, C and D). Similar results were obtained when human undifferentiated or phorbol 12-myristate 13-acetate-differentiated THP1 cells were used (Fig. 1E). These results suggested that FTL_0325 is required for intramacrophage survival and participates in restricting TNF-α production.
FIGURE 1.
FTL_0325 mutant induces significantly increased proinflammatory cytokines in macrophages. Macrophage cell culture invasion assay was performed in MH-S cells (A) and BMDMs (B) derived from C57BL/6 mice. The macrophages were infected with 100 m.o.i. of FTL_0325 mutant or WT F. tularensis LVS. The intracellular replication was quantitated at the indicated times and expressed as log10 cfu. TNF-α levels were measured in culture supernatants from infected MH-S cells (C), BMDMs (D), and untreated or phorbol 12-myristate 13-acetate differentiated THP1 cells (E). F, the macrophages were infected with 100 m.o.i. of indicated mutants or WT F. tularensis LVS. The intracellular replication was quantitated 24 h PI and expressed as log10 cfu. G, TNF-α levels were measured in culture supernatants from infected MHS cells 24 h PI. The values represent the mean ± S.D. (A and C) of quadruplicate samples from one of the three experiments conducted or mean ± S.E. (B, D, E, F, and G) and cumulative of three independent experiments conducted. The p values were determined using one-way ANOVA.
Our observation that the FTL_0325 mutant of F. tularensis LVS induced proinflammatory cytokine production prompted us to investigate if this feature was specific to the FTL_0325 mutant or is common to other attenuated mutants of F. tularensis LVS. It was found that infection of macrophages with FTL_0557 (ΔpmrA) or other gene mutants that are as attenuated for intramacrophage growth as FTL_0325 mutant (Fig. 1F) did not induce significant amounts of TNF-α and that the quantitated cytokine levels in the culture supernatants were similar to those observed after infection with WT F. tularensis LVS. By comparison, as observed earlier, significantly elevated levels of TNF-α were observed in macrophages infected with FTL_0325 mutant (Fig. 1G). Collectively, these results demonstrate that FTL_0325 is specifically required for the suppression of cytokine responses, and its loss results in significantly increased induction of proinflammatory cytokine TNF-α.
Subcellular Localization of FTL_0325/FTT0831c of F. tularensis
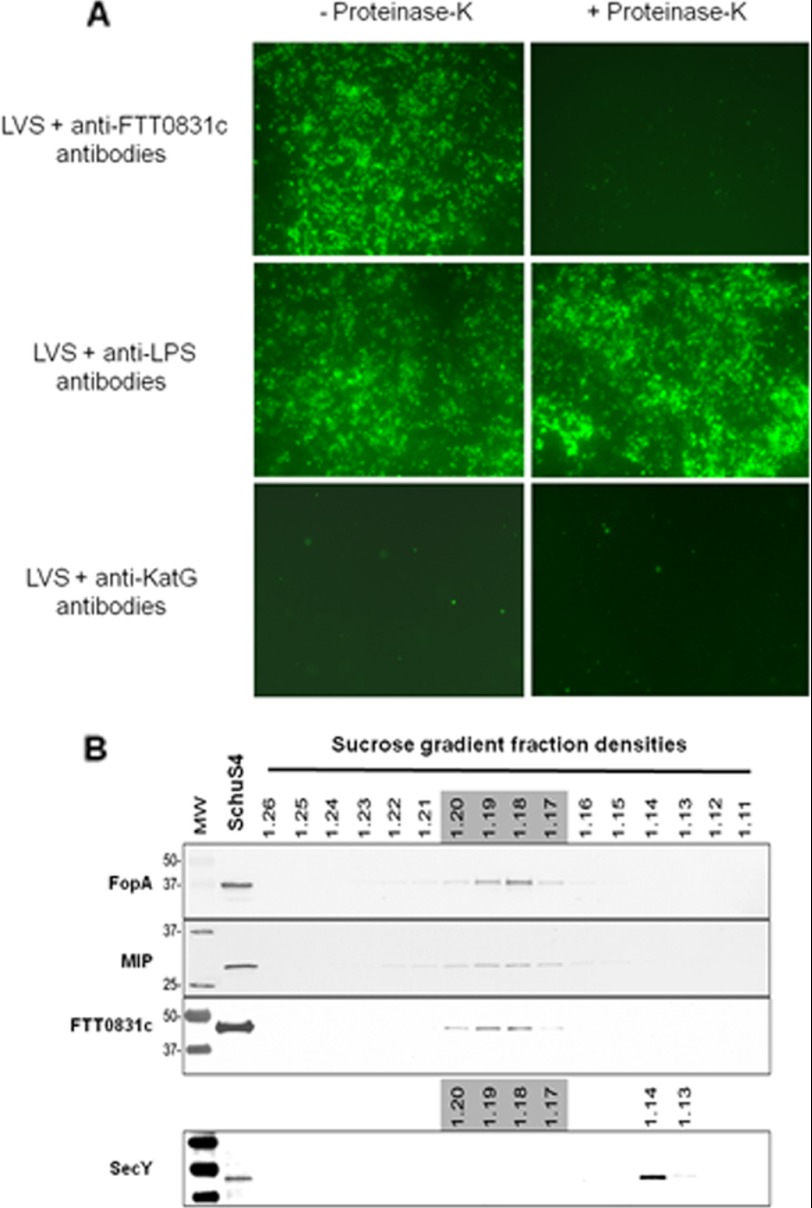
Immunofluorescence staining was performed using polyclonal anti-FTT0831c antibodies to determine if FTL_0325 is exposed on the bacterial surface. The anti-FTT0831c antibodies positively stained the bacterial surface, and the staining was lost after treatment with proteinase-K, indicating that the protein is surface-exposed. No staining was observed when the bacteria were stained with antibodies against KatG, indicating the specificity of the surface staining with anti-FTT0831c antibodies. The surface staining of bacterial cells with anti-F. tularensis LPS antibodies either untreated or treated with proteinase-K indicated that the loss of anti-FTT0831c staining after proteinase-K treatment is not due to the removal of antibodies by residual protease activity (Fig. 2A). Furthermore, sucrose density gradient fractionation and immunoblotting localized FTT0831c to the outer membrane fractions in F. tularensis SchuS4 strain (Fig. 2B), indicating that FTL_0325/FTT0831c proteins localize to the bacterial outer membrane with surface-exposed regions.
FIGURE 2.
Localization of OmpA-like protein of F. tularensis. A, surface localization of FTL_0325 by immunofluorescent staining with polyclonal anti-FTT0831c antibodies and Alexa Fluor 488-conjugated goat anti-rat IgG secondary antibodies. Anti-F. tularensis LPS monoclonal and polyclonal anti-KatG antibodies were used as positive and negative controls, respectively (left panel). Bacteria were treated with proteinase-K, washed, and stained (right panel) (magnification ×63). B, shown is subcellular localization of FTT0831c in F. tularensis SchuS4 by immunoblotting of sucrose density gradient fractions. Sequential fractions were collected from gradients, and immunoblotting was performed using anti-FTT0831c antibodies. Known outer membrane proteins FopA and Mip and inner membrane protein SecY were used as controls for localization.
Induction of Proinflammatory Cytokines TNF-α and IL-6 by FTL_0325 Mutant Is not Due to Increased Release of TLR Ligands
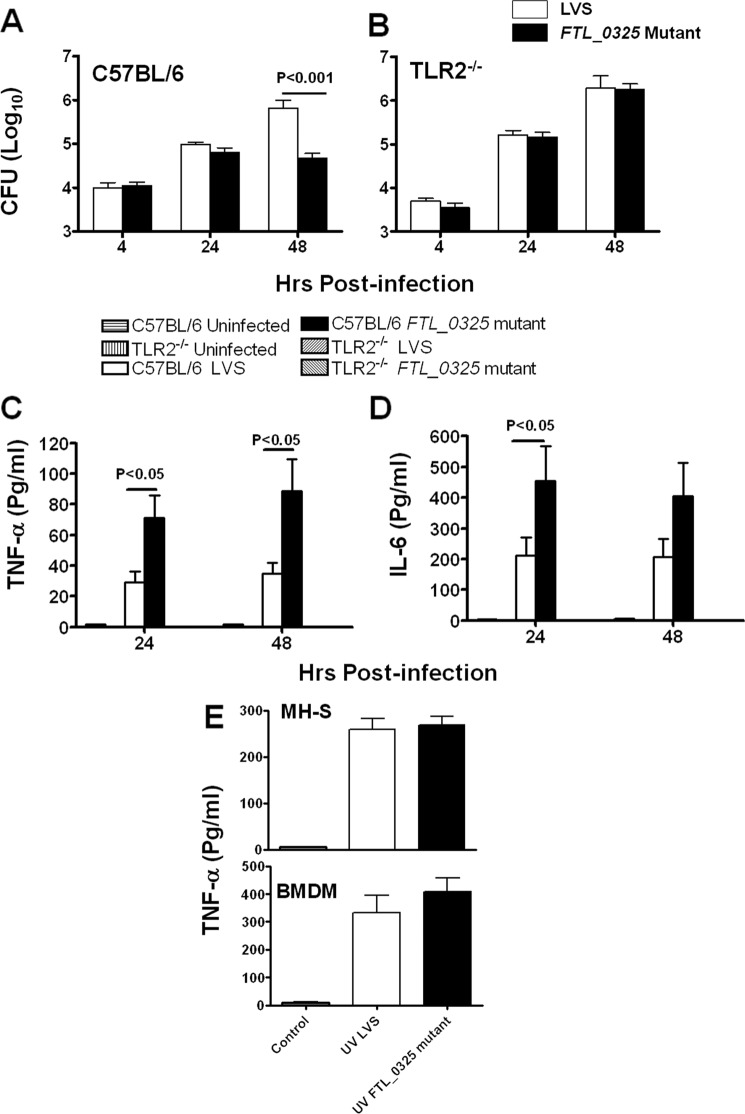
Toll-like receptor-2 (TLR2) plays a major role in innate immunity and induction of proinflammatory cytokines after Francisella infection (31). Because FTL_0325 is localized to the outer membrane with surface-exposed structures, we investigated whether its loss results in induction of proinflammatory cytokines due to enhanced TLR2 signaling. The BMDMs derived from WT or TLR2−/− C57BL/6 mice were infected with FTL_0325 mutant or WT F. tularensis LVS at an m.o.i. of 100. The results showed that significantly lower numbers of FTL_0325 mutant bacteria were recovered from WT BMDMs at 48 h PI as compared with the WT LVS (Fig. 3A). Contrary to this, FTL_0325 mutant replicated similarly as WT F. tularensis LVS in TLR2−/− BMDMs (Fig. 3B). As observed earlier, FTL_0325 mutant induced higher TNF-α and IL-6 levels than WT F. tularensis LVS in BMDMs derived from WT C57BL/6 mice. However, both TNF-α and IL-6 were not observed in TLR2−/− BMDMs infected with either F. tularensis LVS or FTL_0325 mutant (Fig. 3, C and D). These results indicate that TLR2 signaling is required to control the growth of the FTL_0325 mutant and to drive TNF-α and IL-6 production. These results also led us to hypothesize that loss of FTL_0325 may affect the outer membrane structure, thereby exposing a higher proportion of TLR2 ligands, which may result in an enhanced TLR2-dependent proinflammatory cytokine response. To address this, MH-S cells or the BMDMs derived from C57BL/6 mice were infected with ultraviolet (UV) killed WT LVS (UV LVS) or FTL_0325 mutant (UV FTL_0325), and TNF-α levels were measured in culture supernatants 24 h later. No differences in the levels of TNF-α were observed between UV FTL_0325 and UV LVS-infected macrophages (Fig. 3E). These results indicate that elevated levels of proinflammatory cytokines are not due to an increase in TLR2-dependent recognition of bacteria because of an alteration in the outer membrane structures caused by the loss of FTL_0325. Furthermore, these results also suggest that live Francisella and phagosome escape is required for suppression of proinflammatory cytokines and that FTL_0325-mediated suppression of proinflammatory cytokines may not be due to the inhibition of TLR2 per se, but rather, the likely targets are TLR2-dependent inflammatory pathways.
FIGURE 3.
Elevated proinflammatory cytokines by FTL_0325 mutant is not due to increased release of TLR ligands. A macrophage cell culture invasion assay was performed in BMDMs derived from WT (A) and TLR2−/− C57BL/6 (B) mice. The macrophages were infected with 100 m.o.i. of FTL_0325 mutant or WT F. tularensis LVS. The intracellular replication was quantitated at the indicated times and expressed as log10 cfu. TNF-α levels were measured in culture supernatants from WT BMDMs (C) and TLR2−/− (D) macrophages. E, the macrophages were infected with 100 m.o.i. of UV-killed FTL_0325 mutant or UV-killed WT F. tularensis LVS, and TNF-α levels were measured in culture supernatants 24 h later. The results are expressed as the mean ± S.E. and are representative of at least two-three independent experiments. The p values were determined using one-way ANOVA.
FTL_0325 Mutant Escapes from the Phagosomes
Escape of Francisella from the phagosomes and its subsequent replication in the host cell cytosol is associated with activation and secretion of IL-1β as a result of inflammasome activation (28, 32). To investigate if FTL_0325 mutant of F. tularensis escapes the phagosomes similar to its WT counterpart, we performed Western blot analysis to detect proteolytic cleavage and activation of IL-1β. It was observed that higher levels of bioactive IL-1β were observed in FTL_0325 mutant-infected macrophages as compared with the WT F. tularensis LVS-infected cells 24 h PI (Fig. 4A). Because the cleavage of the pro-form of IL-1β results in active IL-1β, which is secreted in the culture supernatants, levels of secreted IL-1β were also measured in the culture supernatants of infected BMDMs at 12 and 24 h PI. Similar to the Western blot analysis results, significantly higher levels of secreted IL-1β levels were detected in culture supernatants from FTL_0325-infected macrophages 24 h PI (Fig. 4B). To further investigate if the activation of IL-1β was due to an enhanced intramacrophage lysis of FTL_0325 mutant, a recently described luminescence assay was performed (28). No differences in the levels of intramacrophage lysis were observed between WT F. tularensis and FTL_0325 mutant (Fig. 4C). These results indicated that similar to the WT F. tularensis LVS, FTL_0325 mutant escapes from the phagosomes into the cytosol and activates the inflammasome that results in the activation and secretion of proinflammatory cytokine IL-1β. Furthermore, the IL-1β activation/secretion is not in response to the excessive release of ligands due to intracellular bacterial lysis in FTL_0325-infected macrophages.
FIGURE 4.
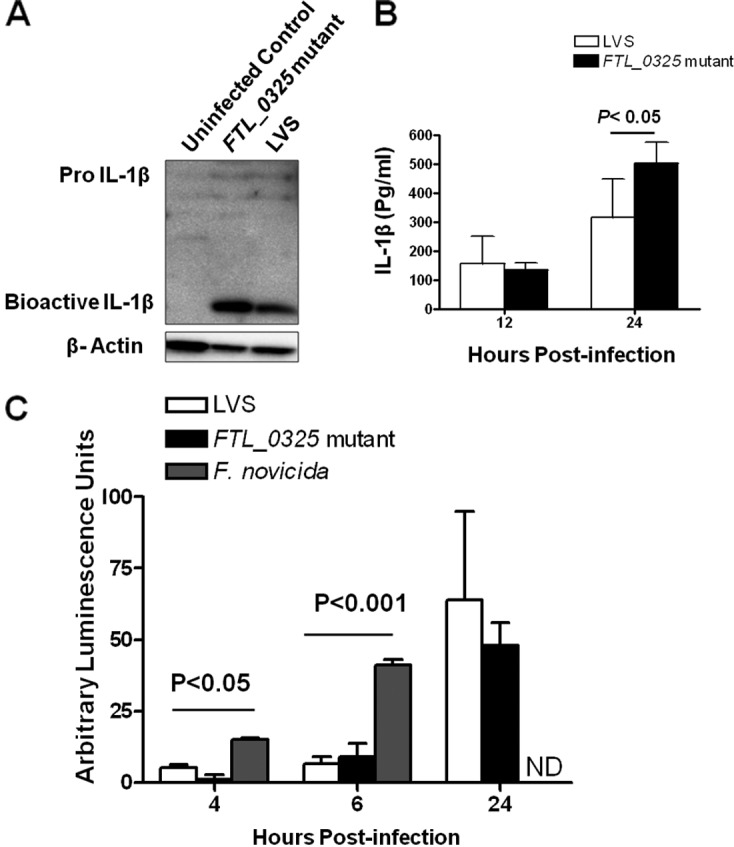
FTL_0325 mutant escapes from the phagosomes. A, shown is detection of IL-1β activation. Lysates from BMDMs infected with 100 m.o.i. of LVS or FTL_0325 mutant at 24 h PI were run on a SDS-gel, transferred, and blotted with anti-IL-1β antibodies. Blotting with anti-β actin antibodies was used as the loading control. B, levels of secreted IL-1β were measured in culture supernatants from infected BMDMs at the indicated times. C, quantification of intramacrophage bacterial lysis. Macrophages were infected with an m.o.i. of 100 with the indicated strains carrying pFNLTP6:luciferase construct. The macrophages were lysed at the indicated times, and luminescence was measured in a luminometer (PerkinElmer Life Sciences). F. novicida was used as a positive control. The results are representative of two to three independent experiments, expressed as the mean ± S.D., and the p values were determined using one-way ANOVA. In C, comparisons are shown between F. novicida and LVS or FTL_0325 mutant strain. ND, not determined.
Hyper-induced Proinflammatory Cytokine Response in FTL_0325 Mutant-infected Macrophages Is Due to Enhanced Activation of NF-κB Signaling
Our observation that FTL_0325 likely inhibits signaling pathways downstream of TLR2 led us to investigate downstream signaling events. Because activation of TLR2 signaling leads to activation of NF-κB, we monitored the effects of FTL_0325 on NF-κB activity by using a NF-κB-responsive firefly luciferase reporter system in HEK293T cells. We first established that both the F. tularensis LVS and FTL_0325 mutant were equally capable of infecting HEK293T cells (not shown). Infection of HEK293T cells transfected with NF-κB-luciferase reporter construct with WT F. tularensis LVS suppressed NF-κB-dependent luciferase activity. However, infection with FTL_0325 mutant induced significantly higher NF-κB-dependent luciferase activity (Fig. 5). These results indicated that FTL_0325 likely suppresses the induction of cytokines by impairing NF-κB activation.
FIGURE 5.
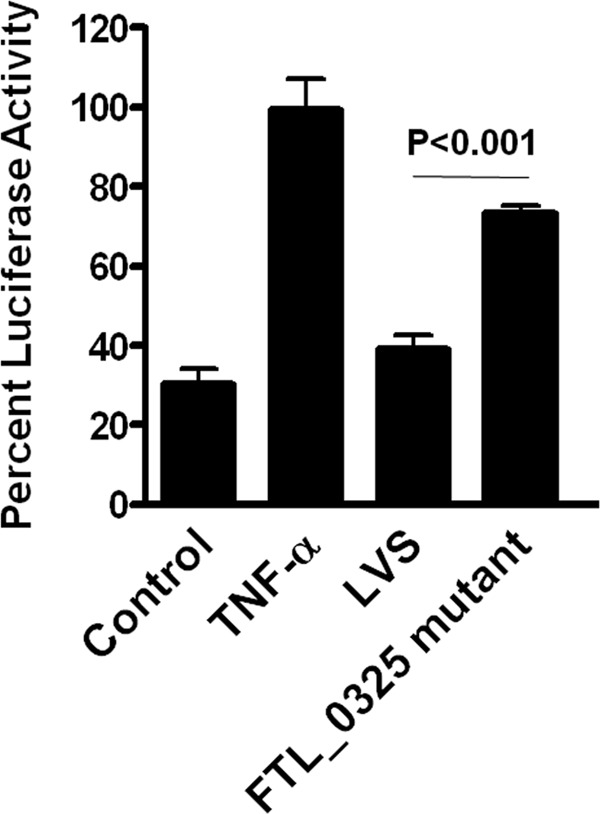
Hyper-induced proinflammatory cytokine response in FTL_0325 mutant-infected macrophages is due to enhanced activation of NF-κB. MH-S cells were transfected with NF-κB-luciferase construct and infected with FTL_0325 mutant and F. tularensis LVS at 100 m.o.i. for 4 h. The cells were lysed, and luciferase activity was monitored using the Dual Luciferase Assay kit (Promega). The data are expressed as percent luciferase activity and represent the mean ± S.D. from one of the three independent experiments. The statistical significance of the results was examined with Student's t test, and p values were recorded.
FTT0831c Protein Inhibits NF-κB Activity by Preventing Nuclear Translocation of p65 Subunit
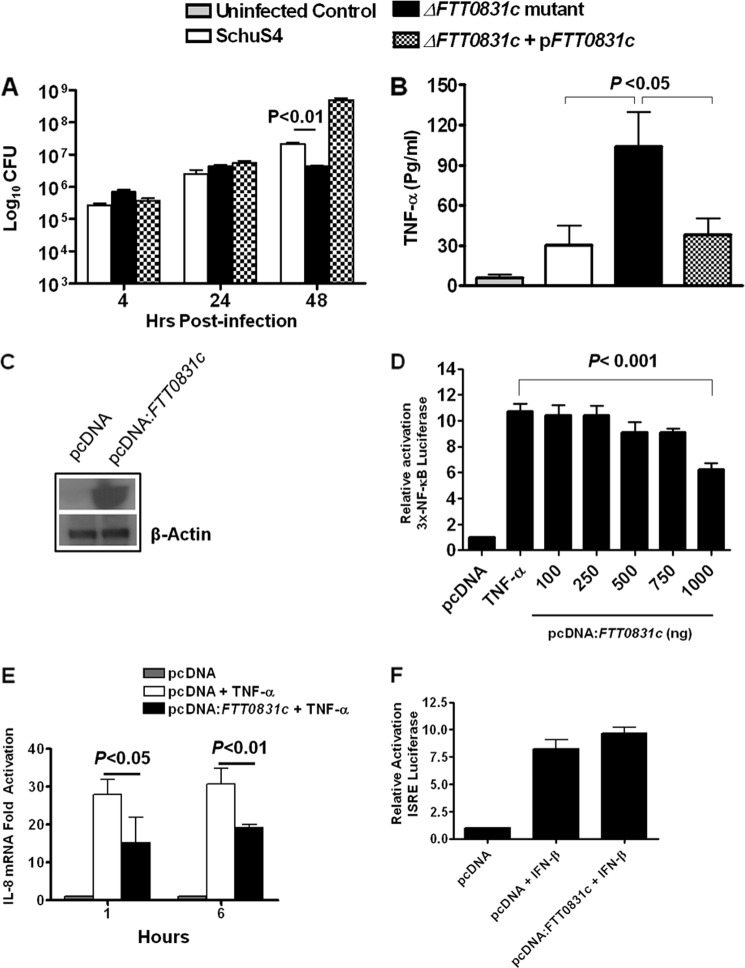
To further establish the role of FTL_0325 in mediating the suppression of cytokines, we generated an in-frame deletion (ΔFTT0831c) and transcomplemented strain (ΔFTT0831c+pFTT0831c) of its ortholog FTT0831c in the highly virulent F. tularensis SchuS4 strain. These bacterial strains were analyzed in a macrophage cell culture assay using MH-S cells. The cell lysates were quantified for bacterial replication, whereas the cell culture supernatants were quantified for TNF-α levels. As observed for the FTL_0325 mutant, ΔFTT0831c was attenuated for intramacrophage survival at 48 h PI (Fig. 6A). Furthermore, a significantly decreased TNF-α production was observed in MH-S cells infected with the WT SchuS4 strain as compared with ΔFTT0831c-infected macrophages (Fig. 6B). The phenotype of the ΔFTT0831c was restored to its WT F. tularensis SchuS4 counterpart by transcomplementing the FTT0831c gene, confirming that the suppression of cytokine response is mediated specifically by FTT0831c.
FIGURE 6.
FTT0831c protein inhibits NF-κB activity. A, a macrophage cell culture invasion assay was performed in MH-S cells. The macrophages were infected with 100 m.o.i. of ΔFTT0831c mutant, WT F. tularensis SchuS4, and the transcomplemented ΔFTT0831c+pFTT0831c strain. The intracellular replication was quantitated and is expressed as Log10 cfu. B, the TNF-α levels were measured in culture supernatants from infected MH-S cells 24 h PI. The values represent the mean ± S.D. of six simples each and are representative of two experiments conducted. The p values were determined using one-way ANOVA. C, shown is confirmation of FTT0831c expression by Western blot analysis of HEK293T cells transfected with 1000 ng of pcDNA:FTT0831c vector. D, shown is a dose-dependent reduction in NF-κB activity in HEK293T cells transfected with indicated concentration of pcDNA expressing FTT0831c. The luciferase activity was determined as a measure of NF-κB activity in HEK293T cells treated with TNF-α. E, IL-8 mRNA expression in HEK293T cells transfected with pcDNA:FTT0831c (1000 ng) by quantitative real-time PCR is shown. F, shown is determination of ISRE-dependent luciferase activity in HEK293T cells transfected with Cignal ISRE-Luciferase reporter and empty or FTT0831c expressing pcDNA (1000 ng) 18 h post-treatment with IFNβ. Data presented in D, E, and F are derived from quadruplicate samples, are cumulative of two independent experiments, and are represented as the mean ± S.E. The p values were determined by one-way ANOVA.
We observed that FTL_0325 or FTT0831c mutants were able to induce significantly elevated levels of proinflammatory cytokines in infected macrophages. To rule out pleiotropic affects associated with gene deletion in these mutants, the role of FTT0831c was examined in the absence of any other Francisella protein. To investigate the direct effects of FTT0831c protein on NF-κB inhibition, HEK293T cells were co-transfected with plasmids expressing the NF-κB-luciferase and FTT0831c-expressing pcDNA3.1 vector. The expression of FTT0831c in the transfected cells was confirmed by Western blot analysis (Fig. 6C) and immunofluorescence staining. It was observed that FTT0831c was expressed in the cytoplasm of the transfected cells (not shown). The transfected cells were stimulated with TNF-α to induce NF-κB-dependent luciferase gene expression. The luciferase activity increased after TNF-α stimulation in cells transfected with the empty pcDNA3.1 vector, but this level was reduced significantly in the cells co-transfected with the FTT0831c-expressing pcDNA3.1 vector. It was observed that transfection with graded amounts of pcDNA expressing FTT0831c reduced luciferase activity in a dose-dependent fashion (Fig. 6D). These results corroborated the findings observed with FTL_0325 mutant of LVS (Fig. 5) and demonstrated that FTT0831c can restrict NF-κB-regulated gene expression in the absence of any other Francisella gene products.
To validate the specificity of FTT0831c-induced attenuation of NF-κB activity and to avoid any bias linked to the luciferase assay, effects on IL-8 mRNA expression were also examined by quantitative real-time PCR. Expression of FTT0831c in HEK293T cells suppressed TNF-α-mediated IL-8 expression by nearly 40–50% at 1 and 6 h post-TNF-α treatment (Fig. 6E), demonstrating that FTT0831c acts specifically on NF-κB to attenuate its activity and also affects IL-8 expression. Specificity of attenuation of NF-κB activity by FTT0831c was validated further by performing an interferon-stimulated response element (ISRE)-luciferase assay. The HEK293T cells were transfected with ISRE-luciferase reporter plasmid, FTT0831c-expressing, or an empty pcDNA plasmid, the transfected cells were treated with IFN-β for 18 h, and luciferase activity was measured. In this assay, FTT0831c did not inhibit IFN-β-mediated activation of ISRE-luciferase activity, suggesting that FTT0831c has no effect on ISRE-dependent transcription factors (Fig. 6F). Taken together, these data suggest that FTT0831c selectively inhibits NF-κB activation.
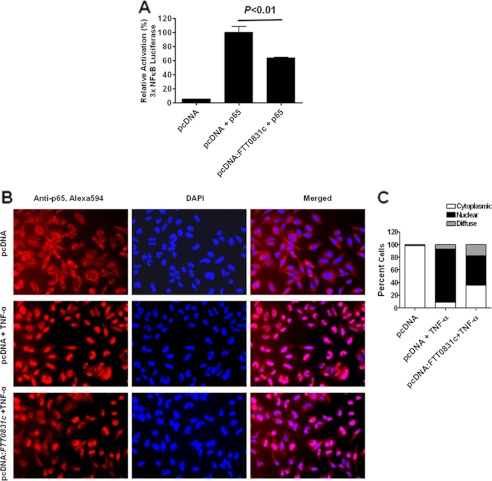
We next tested whether the expressed FTT0831c protein acts at the level of p65/RelA subunit of NF-κB and prevents its nuclear translocation. HEK293T cells were transiently transfected with plasmids expressing NF-κB-dependent luciferase reporter and p65 subunit in the presence or absence of FTT0831c-expressing pcDNA3.1. The luciferase activity was quantified as a measure of NF-κB activity. In the cells overexpressing p65 subunit, the FTT0831c protein expression blocked p65-mediated luciferase activity (Fig. 7A), whereas a robust luciferase activity was observed in the control cells. Consistently, HEK293T cells transfected with empty or FTT0831c-expressing pcDNA3.1 plasmid showed a marked reduction in the nuclear translocation of endogenous p65 after TNF-α treatment as detected by immunofluorescence staining with anti-p65 antibodies (Fig. 7, B and C). These results demonstrate that FTT0831c protein exerts its inhibitory effect on NF-κB primarily by preventing the translocation of p65 subunit to the nucleus.
FIGURE 7.
FTT0831c protein inhibits NF-κB activity by preventing nuclear translocation of p65 subunit. A, HEK293T cells expressing FTT0831c were co-transfected with vector expressing p65 subunit. p65-induced NF-κB luciferase reporter activity was measured. B, HEK293T cells were transfected as in A and treated with TNF-α for 30 min. Immunofluorescence staining was performed to detect the cellular localization of p65subunit of NF-κB (magnification 20× and 100×; red, p65; blue, nucleus). C, quantification of nuclear localization of p65 subunit of NF-κB is shown. At least 200 cells were counted in randomly chosen fields for each condition and expressed as percent cells with p65 staining. Data presented in A are derived from quadruplicate samples and are cumulative of five independent experiments and are represented as the mean ± S.E. The p values were determined by one-way ANOVA.
DISCUSSION
OmpA, a major protein component of the E. coli outer membrane, has been very well characterized. OmpA in E. coli serves a multitude of functions including being a structural protein, a phage, and colicin receptor, mediating serum resistance, playing a role in F-factor-dependent conjugation, and contributing to immune evasion (33). OmpA is expressed at a very high level in E. coli but regulated tightly at posttranscriptional level (34). OmpA-expressing E. coli prevents macrophage apoptosis by inducing the anti-apoptotic factor, Bclxl (35), and suppresses cytokine and chemokine induction by the infected monocytes through inactivation of both NF-κB and MAP kinases (36). Furthermore, OmpA from a meningitis-causing strain of E. coli subverts dendritic cell maturation and expression of co-stimulatory molecules CD40, HLA-DR, and CD86. This inhibitory effect on dendritic cell maturation is also associated with an increase in TGF-β and a decrease in TNF-α, IL-6, and IL-12 (37). Based on the homology to the OmpA domain of E. coli OmpA protein, several proteins have been classified as OmpA-like proteins, and their role in virulence has been very well established in several bacterial pathogens (38–42). In the virulent E. coli strain K1 and other bacterial pathogens such as Pasteurella, Klebsiella, and Neisseria, OmpA-like protein is required for entry and survival within macrophages (43–45).
The OmpA-like protein of F. tularensis is encoded by FTL_0325 gene in LVS and by FTT0831c in the virulent SchuS4 strain. This protein is a 417-amino acid sequence that encodes a 45.75-kDa protein. The annotation of FTL_0325 and FTT0831c as an OmpA-like protein is based on the homologies of their conserved OmpA domains to the C-terminal domain of E. coli OmpA protein (46–48). The OmpA domain of F. tularensis LVS and SchuS4, similar to E. coli OmpA domain, possesses a periplasmic peptidoglycan-associating motif. However, the remainder of OmpA-like protein of Francisella appears to be structurally unique as it does not bear any sequence homology to E. coli OmpA or OmpA-like proteins from other Gram-negative bacteria. The surface localization studies confirmed that the OmpA-like protein of Francisella is localized to the outer membrane, suggesting that surface exposed regions exist. Similar features have been reported for OmpA-like protein of Leptospira interrogans (41). In vivo, the FTL_0325 mutant of F. tularensis LVS was found to be highly attenuated for virulence in mice and induced an early inflammatory response.6 We report here that FTL_0325 and FTT0831c of F. tularensis LVS and SchuS4, respectively, mediate their immune subversive effects by interfering with NF-κB signaling. Overall, this study describes a novel virulence factor of F. tularensis that is required for intramacrophage survival and innate immune subversion.
The innate immune response represents the first line of defense during infection and uses a number of pathogen-associated molecular pattern recognizing TLRs that converge in the activation of downstream signaling pathways. NF-κB is a key factor downstream of TLRs, and its activation is responsible for controlling the expression of multiple genes involved in the inflammatory response such as TNF-α, IL-1, IL-6, IL-8, and IL-12. Several bacterial pathogens have evolved sophisticated mechanisms to subvert the host immune response by suppressing NF-κB activity either at the level of IκB phosphorylation or nuclear translocation. Mycobacterium, E. coli, and Brucella secrete effector proteins homologous to Toll/IL-1R domains (7, 8, 50, 51), whereas Salmonella (52, 53) and Chlamydia (54) utilize their secretion systems to inject effector proteins that interfere with the host ubiquitin signaling pathways to suppress IκB ubiquitination and NF-κB activity. In addition to NF-κB, some bacterial pathogens have evolved mechanisms to suppress MAPKs, another key antimicrobial signaling pathway involved in bacterial clearance. Yersinia, Shigella, and Bacillus anthracis inhibit MAPK signaling to suppress inflammatory response that permits bacterial growth and dissemination (55–58). It has been reported that F. tularensis LVS suppresses MAPK signaling via intracellular growth locus C and RipA (16, 19). This study reports that the FTL_0325/FTT0831c proteins of both the F. tularensis LVS and SchuS4 suppress proinflammatory cytokine response by inhibiting NF-κB signaling. The beneficial effects of NF-κB activation appeared to be related to increased production of TNF-α as observed in the culture supernatants of macrophages infected with either FTL_0325 or FTT0831c mutants as compared with WT F. tularensis LVS or SchuS4. It was also observed that infection of FTL_0325 mutant induced significantly elevated levels of IL-1β. It is expected that this may be due to higher pro-IL-1β levels induced in response to enhanced NF-κB signaling observed in FTL_0325 mutant-infected macrophages. Alternatively, it is also possible that FTL_0325 interferes with activation of inflammasome and is a subject of ongoing investigation.
Several previous studies have reported the requirement of F. tularensis structural components for virulence and immune modulation. FopC protein of F. novicida and F. tularensis SchuS4 is an important virulence factor (59, 60). Moreover, it has been demonstrated that FopC suppresses IFN-γ signaling and thus interferes with generation of an effective innate immune response (59). Several other structural components of both the LVS and F. novicida such as MviN, a lipid II filppase (61), RipA, a cytoplasmic membrane protein (19), or the LPS O-antigen (62) and capsular components have been shown to be required for virulence, suppression of proinflammatory cytokines, and cell death in infected macrophages. Results from the present study indicate that FTL_0325 serves to play a role different than these structural proteins. First, unlike FopC, FTL_0325 is not required for maintaining the structural integrity of the bacteria; second, its loss does not result in as severe attenuation of intramacrophage growth as observed for mviN, ripA, or the LPS O-antigen-deficient mutants. A recent report by Peng et al. (28) has challenged the existing paradigm by demonstrating that the hyperinflammatory nature of F. tularensis LVS and F. novicida mutants deficient in membrane components is due to compromised structural integrity and excessive release of bacterial ligands resulting from bacterial lysis in macrophages and not due to the loss of gene in question. In this study we took three different approaches to rule out that the hyperinflammatory nature of FTL_0325 or FTT0831c mutants of LVS and SchuS4 is not due to 1) compromised membrane integrity, 2) release/exposure of surface TLR ligands, and 3) excessive intramacrophage lysis that releases TLR ligands in the macrophage cytosol to cause NF-κB activation. Our data demonstrate that FTL_0325 mutant does not exhibit any in vitro growth defect and increased sensitivity to antibiotics, detergents, or serum, indicating that the membrane integrity is not compromised in this mutant (supplemental Figs. S1 and S2). Furthermore, an identical cytokine response of the MH-S cells and BMDMs to UV-killed LVS and FTL_0325 mutant also suggests that loss of OmpA-like protein does not lead to increased surface-exposed TLR ligands. The activation and secretion of IL-1β in FTL_0325-infected macrophages suggests that the mutant escapes the phagosomes, indicating that the mutant is similar to WT F. tularensis in every other respect. Furthermore, intramacrophage lysis studies revealed that both the WT LVS and FTL_0325 mutant exhibit an increased intramacrophage lysis at 24 h PI; however, the extent of lysis did not differ between the two strains and, thus, does not account for the differences in the proinflammatory cytokine profiles. When comparisons were made with other mutants deficient in membrane components or a hypothetical membrane protein, it was observed that all mutants including FTL_0325 were attenuated for intramacrophage growth (Fig. 1F). However, enhanced innate immune response was observed only in macrophages infected with FTL_0325 mutant, indicating that loss of structural components does not necessarily result in hyper cytokine response (Fig. 1G). Several recent studies have highlighted the differences between the pathogenesis of F. novicida, LVS, and SchuS4 strain (63–65) and have shown that infection of mice or macrophages with the former strain results in rapid induction of proinflammatory cytokines, whereas these responses are extremely muted after infection with the latter two strains. F. novicida, which was used as a positive control in intramacrophage lysis studies, underwent significantly higher lysis as compared with F. tularensis LVS or FTL_0325 mutant. This observation strongly supports the notion that extensive lysis of F. novicida may be a major contributor to its hyperinflammatory nature. Collectively, our data demonstrate that the enhanced inflammatory response after infection with mutants of F. tularensis deficient in FTL_0325/FTT0831c is not due to altered membrane integrity, surface exposed ligands, or enhanced intracellular lysis but more specifically to the loss of these proteins.
Our data support the notion that in the absence of OmpA-like protein, the mutant bacteria escape the phagosomes and replicate for a while without undergoing extensive lysis. However, these data do not exclude the possibility that this may enhance blebbing and shedding of outer membrane material in the host cell cytosol resulting in a hypercytokine response. We, therefore, performed functional expression studies with FTT0831c protein of SchuS4 (Figs. 6, D, E, and F, and 7) to eliminate any bias associated with the excessive release of bacterial ligands in macrophages infected with mutants and resulting in a hyperinflammatory phenotype. Our cell-based assays showed that overexpression of FTT0831c suppressed the canonical NF-κB pathway by inhibiting nuclear translocation of the p65 protein, resulting in blockade of NF-κB-dependent activation of inflammatory cytokines. Reduced IL-8 expression and no effect on IFN-β-dependent activation of ISRE further established that FTT0831c-mediated inhibition is relatively specific for NF-κB. Our data suggest that the FTL_0325/FTT0831c proteins of F. tularensis LVS and SchuS4 are targeted into the host cells to specifically inactivate NF-κB to dampen the innate immune response. We speculate that FTL_0325/FTT0831c protein mediates this effect either by direct physical interaction with p65 subunit or by preventing its phosphorylation and subsequent nuclear translocation. Therefore, targeting p65 subunit in the host cell cytoplasm and preventing its nuclear translocation may represent an efficient strategy that F. tularensis adopts to inhibit NF-κB transactivating activity. A similar role has recently been reported for OmpA protein of Klebsiella pneumoniae (66, 67). Collectively, these results demonstrate that FTT0831c of F. tularensis SchuS4 offers this pathogen a selective advantage by allowing immune evasion that is independent of any other Francisella protein. Understanding the molecular mechanism of action of FTT0831c of F. tularensis requires additional studies.
Our initial software predictions indicate that OmpA-like protein of F. tularensis may not span the membrane several times as has been shown for E. coli (47), and thus, it is likely that majority of the protein is surface-exposed. Results of our surface localization and immunofluorescence staining experiments support these predictions and demonstrate that FTT0831c/FTL_0325 localizes to the outer membrane and is exposed on the surface of Francisella. However, in the context of the Francisella infection, the mechanism through which OmpA-like protein on the bacterial surface comes in contact with the NF-κB signaling components remained an unanswered question in this study. Our hypothesis is that Francisella sheds OmpA-like protein during its residence in the mildly acidic environment of the phagosomes and during its rapid replication in the macrophage cytosol. The support for our hypothesis comes from a recent report demonstrating that Francisella produces outer membrane vesicles that may serve as a “vesicle-mediated secretion” system to deliver bacterial proteins in the extracellular milieu (68). Outer membrane vesicles are spherical fragments of bacterial outer membrane that are produced continuously without the concomitant bacterial lysis by several Gram-negative bacteria to mediate inflammatory response and virulence in vivo (49, 69). Interestingly, proteome characterization of outer membrane vesicles from F. novicida revealed the presence of FTN_0346, an ortholog of FTT0831c/FTL_0325 proteins of F. tularensis LVS and SchuS4, respectively (68). Based on these observations, we speculate that outer membrane vesicles containing surface-exposed OmpA-like protein shed abundantly by Francisella in the macrophage cytosol may interact with cytosolic NF-κB-signaling components to mediate its suppressive effect. Additionally, OmpA-like protein may also be released from the dying bacterial cells. A comprehensive understanding of how OmpA-like protein interacts with p65 to subvert NF-κB signaling and induction of proinflammatory cytokines remains a matter of continued investigation in our laboratory.
In conclusion, this study establishes FTL_0325/FTT0831c protein of Francisella as a key virulence factor that functions as an immunosuppressant to facilitate bacterial persistence. Stimulation of the inflammatory response at mucosal surfaces is an essential antimicrobial defense mechanism, and NF-κB plays a central role in the transcriptional regulation of proinflammatory cytokine genes. Suppression of this key signaling pathway via FTT0831c protein by virulent F. tularensis SchuS4 explains one of the mechanisms adopted by this pathogen to down-modulate the host innate immune responses. This study has identified one of the structural proteins of F. tularensis playing a role in the modulation of innate immune response. However, keeping in view the extreme virulence of F. tularensis SchuS4, there might be several other unknown factors that may have a similar role. A better understanding of such pathogenic mechanisms will lead to the identification of defined subunit vaccine candidates for the prevention of tularemia acquired naturally or through an act of bioterrorism.
Supplementary Material
Acknowledgment
We thank Dr. Zhongtao Zhang, Assistant Professor, New York Medical College, Valhalla for bioinformatics analysis.
This work was supported, in whole or in part, by National Institutes of Health Grants 2P01AI056320, 1R21AI075250-01A2 (to M. Malik), and 7R56AI090072-02 (to C. S. B.).

This article contains supplemental Table S1 and Figs. S1 and S2.
M. Mahawar, M. K. Atianand, R. J. Dotson, V. Mora, S. M. Rabadi, D. W. Metzger, J. F. Huntley, J. A. Harton, M. Malik, and C. S. Bakshi, unpublished data.
- LVS
- live vaccine strain
- ISRE
- interferon-stimulated response element
- PI
- post-infection
- BMDM
- bone marrow-derived macrophage
- m.o.i.
- multiplicity of infection
- ANOVA
- analysis of variance
- TLR
- Toll-like receptor.
REFERENCES
- 1. Oyston P. C., Sjostedt A., Titball R. W. (2004) Tularemia. Bioterrorism defense renews interest in Francisella tularensis. Nat. Rev. Microbiol. 2, 967–978 [DOI] [PubMed] [Google Scholar]
- 2. Tärnvik A., Priebe H. S., Grunow R. (2004) Tularemia in Europe. An epidemiological overview. Scand. J. Infect. Dis. 36, 350–355 [DOI] [PubMed] [Google Scholar]
- 3. Dennis D. T., Inglesby T. V., Henderson D. A., Bartlett J. G., Ascher M. S., Eitzen E., Fine A. D., Friedlander A. M., Hauer J., Layton M., Lillibridge S. R., McDade J. E., Osterholm M. T., O'Toole T., Parker G., Perl T. M., Russell P. K., Tonat K. (2001) Tularemia as a biological weapon. Medical and public health management. JAMA 285, 2763–2773 [DOI] [PubMed] [Google Scholar]
- 4. Gallagher-Smith M., Kim J., Al-Bawardy R., Josko D. (2004) Francisella tularensis. Possible agent in bioterrorism. Clin. Lab. Sci. 17, 35–39 [PubMed] [Google Scholar]
- 5. Saslaw S., Eigelsbach H. T., Prior J. A., Wilson H. E., Carhart S. (1961) Tularemia vaccine study. II. Respiratory challenge. Arch. Intern. Med. 107, 702–714 [DOI] [PubMed] [Google Scholar]
- 6. Bosio C. M., Bielefeldt-Ohmann H., Belisle J. T. (2007) Active suppression of the pulmonary immune response by Francisella tularensis Schu4. J. Immunol. 178, 4538–4547 [DOI] [PubMed] [Google Scholar]
- 7. Alvarez J. I. (2005) Inhibition of Toll-like receptor immune responses by microbial pathogens. Front. Biosci. 10, 582–587 [DOI] [PubMed] [Google Scholar]
- 8. Noss E. H., Pai R. K., Sellati T. J., Radolf J. D., Belisle J., Golenbock D. T., Boom W. H., Harding C. V. (2001) Toll-like receptor 2-dependent inhibition of macrophage class II MHC expression and antigen processing by 19-kDa lipoprotein of Mycobacterium tuberculosis. J. Immunol. 167, 910–918 [DOI] [PubMed] [Google Scholar]
- 9. Chakraborty S., Monfett M., Maier T. M., Benach J. L., Frank D. W., Thanassi D. G. (2008) Type IV pili in Francisella tularensis. Roles of pilF and pilT in fiber assembly, host cell adherence, and virulence. Infect. Immun. 76, 2852–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Forslund A. L., Kuoppa K., Svensson K., Salomonsson E., Johansson A., Byström M., Oyston P. C., Michell S. L., Titball R. W., Noppa L., Frithz-Lindsten E., Forsman M., Forsberg A. (2006) Direct repeat-mediated deletion of a type IV pilin gene results in major virulence attenuation of Francisella tularensis. Mol. Microbiol. 59, 1818–1830 [DOI] [PubMed] [Google Scholar]
- 11. Gil H., Benach J. L., Thanassi D. G. (2004) Presence of pili on the surface of Francisella tularensis. Infect. Immun. 72, 3042–3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hager A. J., Bolton D. L., Pelletier M. R., Brittnacher M. J., Gallagher L. A., Kaul R., Skerrett S. J., Miller S. I., Guina T. (2006) Type IV pili-mediated secretion modulates Francisella virulence. Mol. Microbiol. 62, 227–237 [DOI] [PubMed] [Google Scholar]
- 13. Bröms J. E., Sjöstedt A., Lavander M. (2010) The Role of the Francisella tularensis pathogenicity island in type VI secretion, intracellular survival, and modulation of host cell signaling. Front. Microbiol. 1, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bröms J. E., Lavander M., Sjöstedt A. (2009) A conserved α-helix essential for a type VI secretion-like system of Francisella tularensis. J. Bacteriol. 191, 2431–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Platz G. J., Bublitz D. C., Mena P., Benach J. L., Furie M. B., Thanassi D. G. (2010) A tolC mutant of Francisella tularensis is hypercytotoxic compared to the wild type and elicits increased proinflammatory responses from host cells. Infect. Immun. 78, 1022–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Telepnev M., Golovliov I., Grundström T., Tärnvik A., Sjöstedt A. (2003) Francisella tularensis inhibits Toll-like receptor-mediated activation of intracellular signaling and secretion of TNF-α and IL-1 from murine macrophages. Cell Microbiol. 5, 41–51 [DOI] [PubMed] [Google Scholar]
- 17. Telepnev M., Golovliov I., Sjöstedt A. (2005) Francisella tularensis LVS initially activates but subsequently down-regulates intracellular signaling and cytokine secretion in mouse monocytic and human peripheral blood mononuclear cells. Microb. Pathog. 38, 239–247 [DOI] [PubMed] [Google Scholar]
- 18. Melillo A. A., Bakshi C. S., Melendez J. A. (2010) Francisella tularensis antioxidants harness reactive oxygen species to restrict macrophage signaling and cytokine production. J. Biol. Chem. 285, 27553–27560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang M. T., Mortensen B. L., Taxman D. J., Craven R. R., Taft-Benz S., Kijek T. M., Fuller J. R., Davis B. K., Allen I. C., Brickey W. J., Gris D., Wen H., Kawula T. H., Ting J. P. (2010) Deletion of ripA alleviates suppression of the inflammasome and MAPK by Francisella tularensis. J. Immunol. 185, 5476–5485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chase J. C., Celli J., Bosio C. M. (2009) Direct and indirect impairment of human dendritic cell function by virulent Francisella tularensis Schu S4. Infect. Immun. 77, 180–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Eigelsbach H. T., Braun W., Herring R. D. (1951) Studies on the variation of Bacterium tularense. J. Bacteriol. 61, 557–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baron S. D., Singh R., Metzger D. W. (2007) Inactivated Francisella tularensis live vaccine strain protects against respiratory tularemia by intranasal vaccination in an immunoglobulin A-dependent fashion. Infect. Immun. 75, 2152–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mbawuike I. N., Herscowitz H. B. (1989) MH-S, a murine alveolar macrophage cell line. Morphological, cytochemical, and functional characteristics. J. Leukoc Biol. 46, 119–127 [DOI] [PubMed] [Google Scholar]
- 24. Golovliov I., Sjöstedt A., Mokrievich A., Pavlov V. (2003) A method for allelic replacement in Francisella tularensis. FEMS Microbiol. Lett. 222, 273–280 [DOI] [PubMed] [Google Scholar]
- 25. Bakshi C. S., Malik M., Regan K., Melendez J. A., Metzger D. W., Pavlov V. M., Sellati T. J. (2006) Superoxide dismutase B gene (sodB)-deficient mutants of Francisella tularensis demonstrate hypersensitivity to oxidative stress and attenuated virulence. J. Bacteriol. 188, 6443–6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mahawar M., Kirimanjeswara G. S., Metzger D. W., Bakshi C. S. (2009) Contribution of citrulline ureidase to Francisella tularensis strain Schu S4 pathogenesis. J. Bacteriol. 191, 4798–4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Melillo A. A., Mahawar M., Sellati T. J., Malik M., Metzger D. W., Melendez J. A., Bakshi C. S. (2009) Identification of Francisella tularensis live vaccine strain CuZn superoxide dismutase as critical for resistance to extracellularly generated reactive oxygen species. J. Bacteriol. 191, 6447–6456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Peng K., Broz P., Jones J., Joubert L. M., Monack D. (2011) Elevated AIM2-mediated pyroptosis triggered by hypercytotoxic Francisella mutant strains is attributed to increased intracellular bacteriolysis. Cellular Microbiology 13, 1586–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bedoya F., Sandler L. L., Harton J. A. (2007) Pyrin-only protein 2 modulates NF-κB and disrupts ASC:CLR interactions. J. Immunol. 178, 3837–3845 [DOI] [PubMed] [Google Scholar]
- 30. Huntley J. F., Conley P. G., Hagman K. E., Norgard M. V. (2007) Characterization of Francisella tularensis outer membrane proteins. J. Bacteriol. 189, 561–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Malik M., Bakshi C. S., Sahay B., Shah A., Lotz S. A., Sellati T. J. (2006) Toll-like receptor 2 is required for control of pulmonary infection with Francisella tularensis. Infect. Immun. 74, 3657–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sanapala S., Yu J. J., Murthy A. K., Li W., Guentzel M. N., Chambers J. P., Klose K. E., Arulanandam B. P. (2012) Perforin- and granzyme-mediated cytotoxic effector functions are essential for protection against francisella tularensis following vaccination by the defined F. tularensis subsp. novicida fopC vaccine strain. Infect. Immun. 80, 2177–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Prasadarao N. V., Wass C. A., Weiser J. N., Stins M. F., Huang S. H., Kim K. S. (1996) Outer membrane protein A of Escherichia coli contributes to invasion of brain microvascular endothelial cells. Infect. Immun. 64, 146–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Koebnik R., Locher K. P., Van Gelder P. (2000) Structure and function of bacterial outer membrane proteins. Barrels in a nutshell. Mol. Microbiol. 37, 239–253 [DOI] [PubMed] [Google Scholar]
- 35. Sukumaran S. K., Selvaraj S. K., Prasadarao N. V. (2004) Inhibition of apoptosis by Escherichia coli K1 is accompanied by increased expression of BclXL and blockade of mitochondrial cytochrome c release in macrophages. Infect. Immun. 72, 6012–6022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Selvaraj S. K., Prasadarao N. V. (2005) Escherichia coli K1 inhibits proinflammatory cytokine induction in monocytes by preventing NF-κB activation. J. Leukoc. Biol. 78, 544–554 [DOI] [PubMed] [Google Scholar]
- 37. Mittal R., Prasadarao N. V. (2008) Outer membrane protein A expression in Escherichia coli K1 is required to prevent the maturation of myeloid dendritic cells and the induction of IL-10 and TGF-β. J. Immunol. 181, 2672–2682 [DOI] [PubMed] [Google Scholar]
- 38. Hu Q., Han X., Zhou X., Ding C., Zhu Y., Yu S. (2011) OmpA is a virulence factor of Riemerella anatipestifer. Vet. Microbiol. 150, 278–283 [DOI] [PubMed] [Google Scholar]
- 39. Nair M. K., Venkitanarayanan K., Silbart L. K., Kim K. S. (2009) Outer membrane protein A (OmpA) of Cronobacter sakazakii binds fibronectin and contributes to invasion of human brain microvascular endothelial cells. Foodborne Pathog. Dis. 6, 495–501 [DOI] [PubMed] [Google Scholar]
- 40. Nicholson T. F., Watts K. M., Hunstad D. A. (2009) OmpA of uropathogenic Escherichia coli promotes postinvasion pathogenesis of cystitis. Infect. Immun. 77, 5245–5251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ristow P., Bourhy P., da Cruz McBride F. W., Figueira C. P., Huerre M., Ave P., Girons I. S., Ko A. I., Picardeau M. (2007) The OmpA-like protein Loa22 is essential for leptospiral virulence. PLoS Pathog. 3, e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Starnino S., Leuzzi R., Ghisetti V., De Francesco M. A., Cusini M., Impara G., Galluppi E., Pizza M., Stefanelli P. (2010) Molecular analysis of two novel Neisseria gonorrhoeae virulent components. The macrophage infectivity potentiator and the outer membrane protein A. New Microbiol. 33, 167–170 [PubMed] [Google Scholar]
- 43. Sukumaran S. K., Shimada H., Prasadarao N. V. (2003) Entry and intracellular replication of Escherichia coli K1 in macrophages require expression of outer membrane protein A. Infect. Immun. 71, 5951–5961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dabo S. M., Confer A. W., Quijano-Blas R. A. (2003) Molecular and immunological characterization of Pasteurella multocida serotype A:3 OmpA. Evidence of its role in P. multocida interaction with extracellular matrix molecules. Microbial Pathogenesis 35, 147–157 [DOI] [PubMed] [Google Scholar]
- 45. Serino L., Nesta B., Leuzzi R., Fontana M. R., Monaci E., Mocca B. T., Cartocci E., Masignani V., Jerse A. E., Rappuoli R., Pizza M. (2007) Identification of a new OmpA-like protein in Neisseria gonorrhoeae involved in the binding to human epithelial cells and in vivo colonization. Mol. Microbiol. 64, 1391–1403 [DOI] [PubMed] [Google Scholar]
- 46. Marsh D. (2000) Infrared dichroism of twisted β-sheet barrels. The structure of E. coli outer membrane proteins. J. Mol. Biol. 297, 803–808 [DOI] [PubMed] [Google Scholar]
- 47. Smith S. G., Mahon V., Lambert M. A., Fagan R. P. (2007) A molecular Swiss army knife. OmpA structure, function, and expression. FEMS Microbiol. Lett. 273, 1–11 [DOI] [PubMed] [Google Scholar]
- 48. Wang Y. (2002) The function of OmpA in Escherichia coli. Biochem. Biophys. Res. Commun. 292, 396–401 [DOI] [PubMed] [Google Scholar]
- 49. Bomberger J. M., Maceachran D. P., Coutermarsh B. A., Ye S., O'Toole G. A., Stanton B. A. (2009) Long distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog. 5, e1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yadav M., Zhang J., Fischer H., Huang W., Lutay N., Cirl C., Lum J., Miethke T., Svanborg C. (2010) Inhibition of TIR domain signaling by TcpC. MyD88-dependent and -independent effects on Escherichia coli virulence. PLoS Pathog. 6, e1001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cirl C., Wieser A., Yadav M., Duerr S., Schubert S., Fischer H., Stappert D., Wantia N., Rodriguez N., Wagner H., Svanborg C., Miethke T. (2008) Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain-containing proteins. Nat. Med. 14, 399–406 [DOI] [PubMed] [Google Scholar]
- 52. Pavlova B., Volf J., Ondrackova P., Matiasovic J., Stepanova H., Crhanova M., Karasova D., Faldyna M., Rychlik I. (2011) SPI-1-encoded type III secretion system of Salmonella enterica is required for the suppression of porcine alveolar macrophage cytokine expression. Vet. Res. 42, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Haraga A., Miller S. I. (2003) A Salmonella enterica serovar typhimurium translocated leucine-rich repeat effector protein inhibits NF-κB-dependent gene expression. Infect. Immun. 71, 4052–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Betts H. J., Wolf K., Fields K. A. (2009) Effector protein modulation of host cells. Examples in the Chlamydia spp. arsenal. Curr. Opin. Microbiol. 12, 81–87 [DOI] [PubMed] [Google Scholar]
- 55. Mukherjee S., Orth K. (2008) In vitro signaling by MAPK and NFκB pathways inhibited by Yersinia YopJ. Methods Enzymol. 438, 343–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Reiterer V., Grossniklaus L., Tschon T., Kasper C. A., Sorg I., Arrieumerlou C. (2011) Shigella flexneri type III secreted effector OspF reveals new cross-talks of proinflammatory signaling pathways during bacterial infection. Cell. Signal. 23, 1188–1196 [DOI] [PubMed] [Google Scholar]
- 57. Chopra A. P., Boone S. A., Liang X., Duesbery N. S. (2003) Anthrax lethal factor proteolysis and inactivation of MAPK kinase. J. Biol. Chem. 278, 9402–9406 [DOI] [PubMed] [Google Scholar]
- 58. Soundararajan V., Patel N., Subramanian V., Sasisekharan V., Sasisekharan R. (2011) The many faces of the YopM effector from plague causative bacterium Yersinia pestis and its implications for host immune modulation. Innate Immun. 17, 548–557 [DOI] [PubMed] [Google Scholar]
- 59. Nallaparaju K. C., Yu J. J., Rodriguez S. A., Zogaj X., Manam S., Guentzel M. N., Seshu J., Murthy A. K., Chambers J. P., Klose K. E., Arulanandam B. P. (2011) Evasion of IFN-γ signaling by Francisella novicida is dependent upon Francisella outer membrane protein C. PLoS ONE. 6, e18201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Twine S., Byström M., Chen W., Forsman M., Golovliov I., Johansson A., Kelly J., Lindgren H., Svensson K., Zingmark C., Conlan W., Sjöstedt A. (2005) A mutant of Francisella tularensis strain SCHU S4 lacking the ability to express a 58-kDa protein is attenuated for virulence and is an effective live vaccine. Infect. Immun. 73, 8345–8352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ulland T. K., Buchan B. W., Ketterer M. R., Fernandes-Alnemri T., Meyerholz D. K., Apicella M. A., Alnemri E. S., Jones B. D., Nauseef W. M., Sutterwala F. S. (2010) Cutting edge. Mutation of Francisella tularensis mviN leads to increased macrophage absent in melanoma 2 inflammasome activation and a loss of virulence. J. Immunol. 185, 2670–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lindemann S. R., Peng K., Long M. E., Hunt J. R., Apicella M. A., Monack D. M., Allen L. A., Jones B. D. (2011) Francisella tularensis Schu S4 O-antigen and capsule biosynthesis gene mutants induce early cell death in human macrophages. Infect. Immun. 79, 581–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hall J. D., Woolard M. D., Gunn B. M., Craven R. R., Taft-Benz S., Frelinger J. A., Kawula T. H. (2008) Infected host-cell repertoire and cellular response in the lung following inhalation of Francisella tularensis Schu S4, LVS, or U112. Infect. Immun. 76, 5843–5852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mortensen B. L., Fuller J. R., Taft-Benz S., Kijek T. M., Miller C. N., Huang M. T., Kawula T. H. (2010) Effects of the putative transcriptional regulator IclR on Francisella tularensis pathogenesis. Infect. Immun. 78, 5022–5032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kieffer T. L., Cowley S., Nano F. E., Elkins K. L. (2003) Francisella novicida LPS has greater immunobiological activity in mice than F. tularensis LPS and contributes to F. novicida murine pathogenesis. Microbes Infect. 5, 397–403 [DOI] [PubMed] [Google Scholar]
- 66. March C., Moranta D., Regueiro V., Llobet E., Tomás A., Garmendia J., Bengoechea J. A. (2011) Klebsiella pneumoniae outer membrane protein A is required to prevent the activation of airway epithelial cells. J. Biol. Chem. 286, 9956–9967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Regueiro V., Moranta D., Frank C. G., Larrarte E., Margareto J., March C., Garmendia J., Bengoechea J. A. (2011) Klebsiella pneumoniae subverts the activation of inflammatory responses in a NOD1-dependent manner. Cell. Microbiol. 13, 135–153 [DOI] [PubMed] [Google Scholar]
- 68. Pierson T., Matrakas D., Taylor Y. U., Manyam G., Morozov V. N., Zhou W., van Hoek M. L. (2011) Proteomic characterization and functional analysis of outer membrane vesicles of Francisella novicida suggests possible role in virulence and use as a vaccine. J. Proteome Res. 10, 954–967 [DOI] [PubMed] [Google Scholar]
- 69. McBroom A. J., Johnson A. P., Vemulapalli S., Kuehn M. J. (2006) Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J. Bacteriol. 188, 5385–5392 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.