Background: NLRP1 mediates the release of the inflammatory cytokine IL-1β and is linked to several human inflammatory diseases.
Results: Autolytic proteolysis occurs within the C terminus of NLRP1 and is modulated by polymorphisms and alternative mRNA splicing.
Conclusion: Autolytic cleavage is a key regulator of the NLRP1 inflammasome and downstream IL-1β production.
Significance: Understanding the mechanisms underlying NLRP1 activation is required to develop effective therapeutics.
Keywords: Cell Biology, Cell Signaling, Inflammation, Inflammatory Bowel Disease, Molecular Cell Biology, Inflammasome, NLRP1
Abstract
Nucleotide-binding domain leucine-rich repeat proteins (NLRs) play a key role in immunity and disease through their ability to modulate inflammation in response to pathogen-derived and endogenous danger signals. Here, we identify the requirements for activation of NLRP1, an NLR protein associated with a number of human pathologies, including vitiligo, rheumatoid arthritis, and Crohn disease. We demonstrate that NLRP1 activity is dependent upon ASC, which associates with the C-terminal CARD domain of NLRP1. In addition, we show that NLRP1 activity is dependent upon autolytic cleavage at Ser1213 within the FIIND. Importantly, this post translational event is dependent upon the highly conserved distal residue His1186. A disease-associated single nucleotide polymorphism near His1186 and a naturally occurring mRNA splice variant lacking exon 14 differentially affect this autolytic processing and subsequent NLRP1 activity. These results describe key molecular pathways that regulate NLRP1 activity and offer insight on how small sequence variations in NLR genes may influence human disease pathogenesis.
Introduction
NLR proteins belong to a larger family of pattern recognition receptors that sense conserved molecular signatures derived from pathogens and host tissues. Upon activation, pattern recognition receptors trigger diverse signaling pathways that coordinate acute inflammatory responses and seed the development of long term adaptive immunity. Select members of the pattern recognition receptor family, including several NLR proteins, participate in these responses by triggering the release of IL-1β, a potent proinflammatory cytokine. This is accomplished through the assembly of a large macromolecular complex called the inflammasome (1–4). Caspase-1 is recruited to these NLR containing complexes generally via the adaptor protein ASC. This leads to caspase-1 activation followed by IL-1β maturation and cellular release.
Given the inflammatory potential of IL-1β, strict regulation of NLR-driven inflammasome activation is critical to human health. This has been highlighted by genomic analyses that have identified links between polymorphisms in certain NLR genes and human diseases that are marked by high levels of serum IL-1β. Rare gain of function mutations in NLRP3, for example, are linked to cryopyrinopathies, a class of related diseases that manifest as recurring flares of fever, arthritis, and skin urticaria but can also lead to bone malformation and mental retardation (5–7). Polymorphisms in NLRP1, on the other hand, are linked to a more diverse group of diseases, including vitiligo, rheumatoid arthritis, systemic sclerosis, Crohn disease, and melanoma (8–14). The fact that IL-1β-blocking therapies, such as anakinra, can alleviate symptoms of some of these diseases suggests that pathology is in part a result of uncontrolled NLR-driven inflammasome activity (15, 16).
Most NLR proteins share a similar domain architecture consisting of an N-terminal homotypic interaction domain, such as a pyrin (PYD), CARD (caspase activation and recruitment domain), or baculoviral inhibitor repeat domain, a central nucleotide binding domain, and a series of C-terminal leucine-rich repeats (LRR).3 In addition, homologs of human NLRs can be found in mice where their function is largely conserved (17). NLRP1, however, is distinct among the members of this family. First, in addition to encoding PYD, the nucleotide binding domain, and LRR domains, NLRP1 encodes two additional domains at its C terminus, a FIIND (function to find domain) and a CARD (18, 19). The presence of two homotypic interaction domains (PYD, CARD) has led to questions regarding the role of ASC in caspase-1 recruitment to the inflammasome, given that the C-terminal CARD could directly interact with caspase-1 as has been shown for NLRC4 (20). Second, distinct from most NLRs, NLRP1 protein structure is not conserved across species. In mice, the Nlrp1 gene does not encode an N-terminal PYD and has multiple alleles that differ in response to anthrax lethal toxin (21). Strains expressing Nlrp-1b A1 mount a protective immune response when challenged with Bacillus anthracis, whereas strains expressing other alleles that are not activated by anthrax lethal toxin succumb to infection. This function is not conserved in human cells where there is little evidence of a role for NLRP1 in anthrax lethal toxin-mediated responses.
In addition to NLRP1, CARD8 is the only other protein known to possess a FIIND (22). Recently, this domain was shown to contain adjacent ZU5-UPA domains that undergo autolytic cleavage in a manner analogous to other proteins harboring these domains (23). These authors went on to show a cleavage occurs in a C-terminal-truncated form of NLRP1. However, the functional consequences of NLRP1 autolytic cleavage remain unknown.
Here, we confirm that NLRP1 undergoes autolytic proteolysis within the FIIND, and we demonstrate that NLRP1 inflammasome activity is dependent strictly upon this cleavage. Following cleavage, the two fragments remained associated, and ASC is recruited to the C-terminal CARD domain of the processed NLRP1 molecule. Single nucleotide polymorphisms or alternative mRNA splicing modulate this post-translational event and downstream IL-1β processing. These results provide insight into the intramolecular mechanisms that govern NLRP1 activity and may provide the first step in understanding how this unique NLR protein is linked to diverse human pathologies.
EXPERIMENTAL PROCEDURES
Expression Vectors, Mutagenesis, and Antibodies
All expression vectors were constructed in pFNcmv (N-terminal HA tag), pcDNA3.1, or pcDNA3.1/V5-His (C-terminal V5-His tag). The pFNcmv vector was used for generation of BacMam viruses containing ASC (pFNcmv-ASC), procaspase-1 (pFNcmv-ProC1-FLAG), and pro-IL-1β (pFNcmv-pro-IL-1β). The pcDNA3.1 vector was modified to contain an N-terminal HA tag and C-terminal FLAG-His tag, and the pcDNA3.1/V5-His vector was modified to contain an N-terminal HA tag using standard PCR techniques employing custom-designed primers containing multiple restriction sites. Mutagenesis was performed using the QuikChange XL site-directed mutagenesis kit from Stratagene. Sequences of all plasmid inserts were confirmed by sequence analysis. Anti-HA antibody was purchased from Cell Signaling. Anti-HA-agarose beads were purchased from Roche Applied Science. Anti-FLAG M2 antibody was purchased from Sigma. Anti-His, anti-V5, and secondary detection antibodies were purchased from Invitrogen. Two rabbit anti-NLRP1 polyclonal and mouse monoclonal antibodies were raised against immunogens using genomic antibody technology corresponding to NLRP1(137–236), and NLRP1(1275–1374).
NLRP1 and Truncation Mutants
All NLRP1 sequences aligned with GenBankTM accession no. NM_033004, with the exception of ΔEx14 (GenBankTM accession no. NM_014922). Domain boundaries used for NLRP1 were as follows: PYD(1–88), FIIND(985–1378), and CARD(1379–1473). The ΔEx14 isoform is missing amino acids 1262–1305 of full-length NLRP1.
Cell Culture, Viral Transduction, and Transient Transfection
HEK293T and U2OS cells (ATCC) were cultured in DMEM+Glutamax (Invitrogen), supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). THP1 and U937cells were grown in RPMI (Invitrogen) supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. BacMam viruses were transduced into human U2OS cells using standard methods. All plasmids were transiently transfected into HEK293T cells using FuGENE 6 reagent (Roche Molecular Biochemicals).
Reconstituted Inflammasome Assays
For transduction assays, human U2OS cells (100,000 cells/cm2) were seeded in 24-well plates in the presence of BacMam virus at the indicated multiplicity of infection (MOI). IL-1β was assayed from cell culture supernatants at 24 h. For transfection assays, HEK293T cells (50,000 cells/cm2) were seeded in 24-well plates and incubated overnight at 37 °C in a humidified air incubator. The following day, 1 ng of NLRP1 or NLRP1 mutants, 1 ng of pFNcmv-ASC, 1 ng pFNcmv-ProC1-FLAG, and 50 ng pFNcmv-proIL-1β were transfected as described. Cell culture supernatants were collected ∼48 h after transfection and assayed for IL-1β (Mesoscale Discovery).
Western Blot Analysis and Immunoprecipitation
Transfected cells or other cell lines were lysed in lysis buffer containing 50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 20 mm EDTA, 0.6% Non-Idet P-40, 1 mm dithiothreitol, 50 mm Na3VO4, and CompleteTM protease inhibitor (Roche Molecular Biochemicals) for 1 h on ice. In some experiments, cells were lysed in 1% Triton X-100, 0.05% DOC, 0.05% SDS, 50 mm Tris, pH 7.5, 150 mm NaCl supplemented with protease inhibitor mixture (Pierce). Cellular debris was removed by centrifugation at 21,000 × g for 20 min at 4 °C. For Western analysis, lysates were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and immunoblotted using standard protocols.
In Vitro Cleavage Assay
HEK293T cells were transfected with NLRP1-expressing plasmids. Cells were lysed in 0.5% Triton X-100, 150 mm NaCl, 50 mm HEPES, pH 7.4, and NLRP1 was immunoprecipitated using anti-HA-agarose beads. The beads were washed three times and suspended in lysis buffer. Hydroxylamine was added to a final concentration of 400 mm, and samples were incubated at 30 °C for 18 h. The reactions were stopped by the addition of 4× sample buffer (Invitrogen) and 10× reducing buffer (Invitrogen). The samples were analyzed by Western blot using antibodies to the C terminus of NLRP1.
Fluorescence Microscopy
U2OS cells were plated in glass bottomed 96-well plates (PerkinElmer Life Science) in the presence of BacMam virus expressing ASC-GFP, NLRP1, or NLRP3. The cells were incubated for 18 h, washed in PBS, and fixed in 3.7% paraformaldehyde. Five images (10×) were captured for each sample, and ASC specks were counted.
RESULTS
NLRP1 Inflammasome Activity Requires C-terminal CARD Domain and Adaptor Protein, ASC
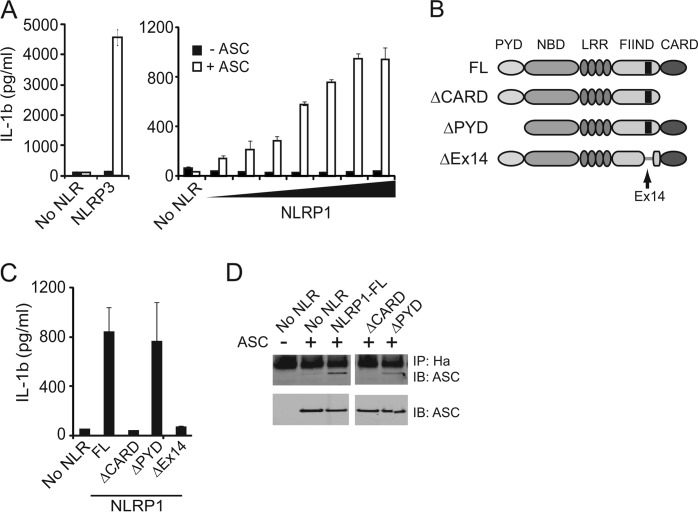
To determine the molecular requirements for human NLRP1 inflammasome activity in cells, we developed an in vitro assay in which components of the NLRP1 inflammasome were reconstituted in U2OS cells through BacMam transduction (24). This assay was validated using NLRP3, the best characterized member of inflammasome-forming NLRs. In agreement with published results, NLRP3 mediated IL-1β release required co-expression of ASC, together with pro-IL-1β and caspase-1 (Fig. 1A). To determine whether ASC was required for NLRP1 mediated IL-1β release, increasing titers of NLRP1 BacMam virus were transduced into cells in the presence or absence of ASC (Fig. 1A). Similar to NLRP3, ASC was required for NLRP1-mediated caspase-1 activation as mature IL-1β was not detected in samples expressing NLRP1 in the absence of this adaptor protein. In addition to IL-1β release, we also determined the ability of NLRP1 to drive the redistribution of ASC from diffuse cytoplasmic staining into a single speck (25). In these assays, GFP-tagged ASC was co-expressed with NLRP3 or NLRP1, and ASC distribution was assessed by fluorescence microscopy. Similar to NLRP3, NLRP1 expression induced the redistribution of cytoplasmic GFP-tagged ASC into a single speck within the cells (supplemental Fig. 1A).
FIGURE 1.
NLRP1 inflammasome activity requires the adaptor protein, ASC, elements encoded with exon 14 and the C-terminal CARD. A, IL-1β ELISA of supernatants from U2OS cells transduced with increasing MOI of BacMam virus expressing NLRP3 or increasing amounts of NLRP1, procaspase-1 (MOI of 1.25), and pro-IL-1β (MOI of 25) in the presence or absence of ASC (MOI of 1.25). B, domain architecture of NLRP1. PYD (pryin domain), nucleotide binding domain (NBD), LRR, FIIND (function to find), CARD (caspase-recruitment domain). Exon 14 is located within FIIND domain (amino acids 1262–1305). C, IL-1β ELISA from supernatants of HEK293T cells transfected with NLRP1 truncation mutants, ASC, procaspase-1, and pro-IL-1β. D, immunoprecipitation of NLRP1 truncation mutants from transfected HEK293T cells. Immunoblots (IB) were probed for ASC.
NLRP3 expresses an N-terminal PYD that recruits ASC during inflammasome formation. In contrast, NLRP1 expresses an N-terminal PYD as well as a C-terminal CARD, both of which have the potential to recruit ASC. Therefore, to determine which of these domains was required for ASC recruitment, NLRP1 mutants lacking these domains were generated. In addition, we also included a naturally occurring mRNA splice isoform, which lacks exon 14 and is present in approximately equal abundance to full-length NLRP1 in THP-1 cells (Fig. 1B and data not shown). This isoform, NLRP1ΔEx14, lacks 44 amino acids within the FIIND and was used in previous cell-free NLRP1 inflammasome studies (26). Cells expressing NLRP1ΔPYD promoted IL-1β release to levels comparable with full-length NLRP1, indicating that this N-terminal domain is dispensable for inflammasome function (Fig. 1C). In contrast, CARD deletion completely abrogated IL-1β release, demonstrating a strict requirement for this C-terminal domain. In line with these results, deletion of the CARD also resulted in loss of ASC speck formation (supplemental Fig. 1B). Interestingly, deletion of exon 14 also ablated NLRP1 induced IL-1β release and ASC speck formation (Fig. 1C and supplemental Fig. 1B), suggesting that elements encoded within this exon are also required for NLRP1 inflammasome activation. Finally, to directly assess the role of the CARD in ASC binding, immunoprecipitation experiments were performed. The NLRP1 mutant lacking the CARD domain failed to co-precipitate ASC, whereas loss of the PYD had little effect on ASC binding (Fig. 1D). Thus, NLRP1 inflammasome activity is dependent upon interactions with ASC that are mediated through its C-terminal CARD as well as sequences encoded within the FIIND.
NLRP1 Undergoes Proteolytic Cleavage within C-terminal FIIND
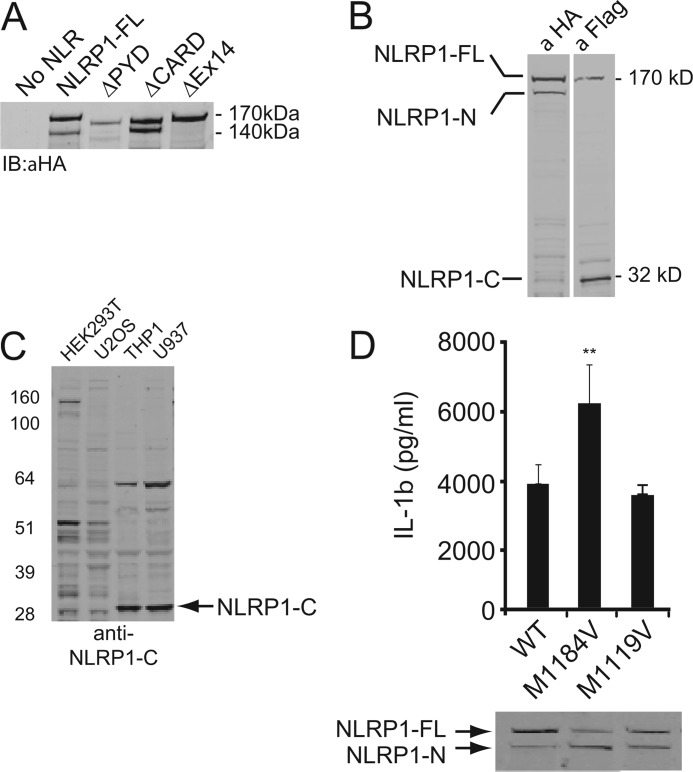
During the course of these studies, we observed that NLRP1 migrated as a high molecular weight doublet except in the absence of exon 14 (Fig. 2A). This migration pattern was likely due to proteolysis of NLRP1 within the FIIND (23). To confirm processing of NLRP1, the full-length protein was modified to express an N-terminal HA tag and a C-terminal FLAG tag. HEK293T cells were transiently transfected, and cell extracts were evaluated using anti-tag-specific antibodies (Fig. 2B). Immunoblot analysis of the N-terminal HA tag confirmed the presence of the 170-kDa band, which represented full-length NLRP1 (NLRP1-FL), and a 140 kDa band, which represented an N-terminal fragment of the processed protein (NLRP1-N). Antibodies directed to the C-terminal FLAG tag detected full-length NLRP1 and a ∼32 kDa C-terminal cleavage fragment of NLRP1 (NLRP1-C). The 140-kDa NLRP1-N band did not appear in immunoblots probed with anti-FLAG antibodies to detect the C terminus (Fig. 2B). To examine whether endogenously expressed NLRP1 also undergoes cleavage, we generated antibodies to the C terminus of NLRP1. In THP1 and U937 monocytic cells, a prominent band corresponding to NLRP1-C was observed (Fig. 2C). This band was absent in cells that do not express NLRP1 mRNA (HEK293T cells), and it was not detected in the same samples probed with control antibodies (data not shown). Endogenous full-length NLRP1 was difficult to consistently detect, suggesting that the endogenous protein may be fully processed under basal conditions.
FIGURE 2.
NLRP1 is cleaved within the FIIND. A, immunoblot (IB) analysis of HEK293T cells transfected with NLRP1 truncation mutants. B, immunoblot analysis of HEK293T cells transfected with full-length NLRP1 expressing an N-terminal HA tag and a C-terminal FLAG tag. C, immunoblot analysis of endogenous NLRP1 in different cell lines with antibodies directed to the C terminus. D, upper panel, IL-1β ELISA of supernatants from HEK293T cells expressing wild type NLRP1 or the indicated SNP. The lower panel shows the NLRP1 immunoblot probed with anti-HA antibodies.
Similar to other NLRs linked to human autoimmune diseases; single nucleotide polymorphisms of NLRP1 have been identified (8, 9, 11, 14). We tested two commonly occurring SNPs for their ability to undergo processing and mediate IL-1β maturation. Interestingly, M1184V showed increased processing as demonstrated by the increased amount of the N-terminal NLRP1 cleavage fragment in Western blot analysis (Fig. 2D). Furthermore, this SNP induced significantly more IL-1β release in reconstituted inflammasome assays. A separate SNP, M1119V, underwent proteolytic processing and induced IL-1β production at levels comparable with WT NLRP1. Thus, these results support the notion that NLRP1 inflammasome activity is proportional to the level of proteolytic processing. Furthermore, these results demonstrate that single amino acid substitutions can have profound effects on NLRP1 processing and inflammasome activity. This may underlie, at least in part, the role of certain SNPs in the development of human disease.
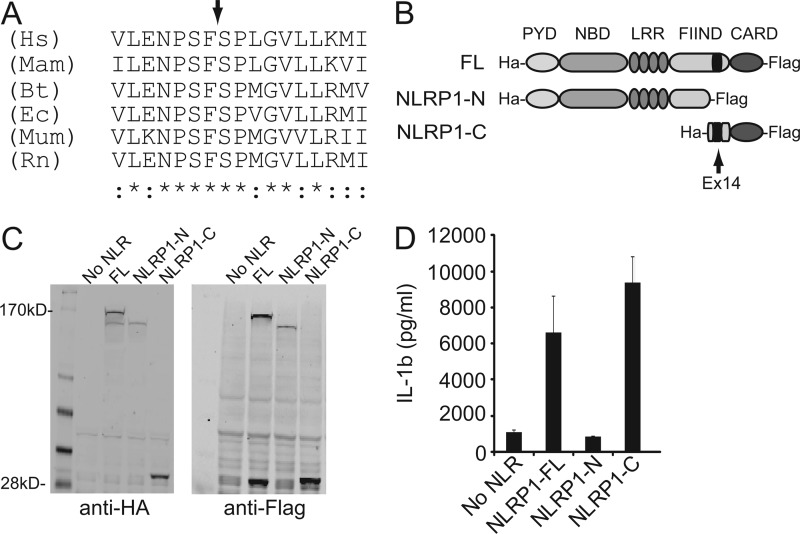
The apparent molecular weight of the NLRP1-N and NLRP1-C bands coupled with the loss of these fragments in the NLRP1ΔEx14 isoform provided strong evidence that sequences within the FIIND may play a critical role in NLRP1 processing. To precisely map the cleavage site, recombinant His-tagged NLRP1 was purified on a nickel-chelate column and transferred to a PVDF membrane. The NLRP1-C fragment was isolated and subjected to Edman degradation (supplemental Fig. 2). The N-terminal residues of this fragment were identified as 1213SPLGVLLKMIHN. This allowed us to map the cleavage site within a well conserved region of the FIIND domain between Phe1212/Ser1213 (Fig. 3A). This is consistent with what has been seen previously when only the FIIND-CARD is expressed (23).
FIGURE 3.
The C-terminal cleavage fragment of NLRP1 is necessary and sufficient for NLRP1 inflammasome activity. A, multiple species sequence alignment of the NLRP1 cleavage site. Asterisks denote completely conserved residues; colon denotes highly conserved residues; arrow denotes site of peptide cleavage. B, diagram depicting the N- and C-terminal cleavage fragments. C, Western blot showing expression of N- and C- terminal NLRP1 cleavage fragments. D, IL-1β ELISA of supernatants from HEK293 cells expressing full-length NLRP1 or the indicated cleavage fragment. Hs, Homo sapiens; Mam, Macaca mulatta; Bt, Bos taurus; Ec, Equus caballus; Mum, Mus musculus; Rn, Rattus norvegicus.
With the sequence identity of the two NLRP cleavage fragments confirmed, we prepared plasmids expressing each fragment and tested their ability to induced IL-1β release in reconstituted inflammasome assays (Fig. 3, B and C). The C-terminal fragment of NLRP1 (NLRP1-C) promoted IL-1β release at levels comparable with full-length NLRP1 (NLRP1-FL). However, no IL-1β was detected in samples expressing the N-terminal fragment (NLRP1-N) (Fig. 3D). These results demonstrated that the C-terminal fragment of NLRP1 is necessary and sufficient to drive NLRP1 inflammasome function.
Proteolysis within FIIND Is Required for NLRP1 Inflammasome Activity
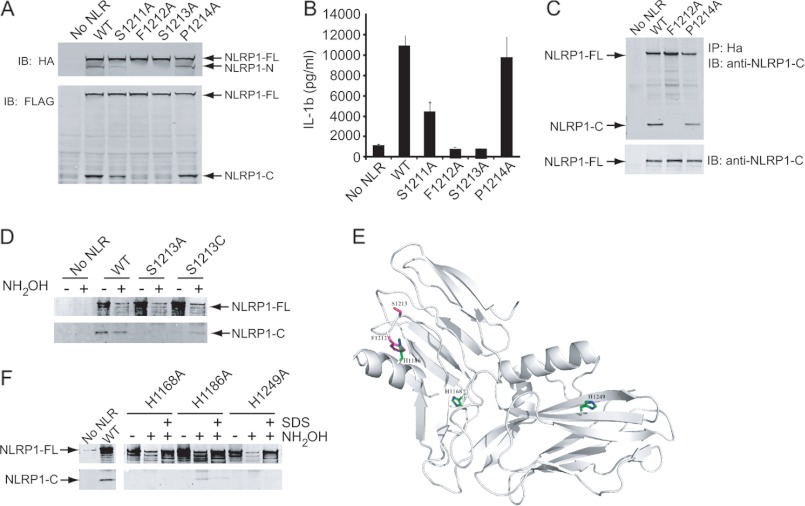
We next sought to provide a more thorough demonstration that NLRP1 undergoes autolytic cleavage via a mechanism similar to other ZU5-UPA domain containing proteins and confirm its functional role in inflammasome activity. Substitution at the P1 residue (F1212A) or P1′ residue (S1213A) completely blocked the appearance of the NLRP1-N and NLRP1-C fragments, suggesting these residues are critical for NLRP1 cleavage (Fig. 4A). In contrast, substitution at the P2 position (S1211A) and P2′ position (P1214A) had less of an effect on NLRP1 cleavage. In functional assays, mutations that blocked cleavage (F1212A and S1213A) also blocked NLRP1 mediated IL-1β release (Fig. 4B). Notably, the S1211A mutation, which exhibited an ∼50% reduction in processing resulted in an ∼50% decrease in IL-1β release, suggesting that cleavage of NLRP1 and inflammasome activity are proportional. These results demonstrate that autolytic processing is linked directly to NLRP1 inflammasome activity.
FIGURE 4.
Cleavage within the FIIND is critical for NLRP1 inflammasome activity. A, immunoblot (IB) analysis of HEK293T cells transfected with NLRP1 containing point mutations within the cleavage site. NLRP1 was identified with antibodies directed to HA or FLAG tags located at the N or C termini, respectively. B, IL-1β ELISA of supernatants from HEK293 cells expressing the indicated point mutation. C, NLRP1 was immunoprecipitated with N-terminal HA antibodies, and immunoblots were probed with antibodies specific for the C terminus of NLRP1. D and F, wild type NLRP1 or the indicated point mutant was immunoprecipitated from cell extracts via an N-terminal HA tag. Captured proteins were treated with NH2OH and analyzed by immunoblot using antibodies specific for the C terminus of NLRP1. Some samples were treated with SDS to disrupt tertiary structure. E, predicted ribbon structure of the NLRP1 FIIND domain showing the position of the three conserved His residues and the cleavage site.
The PIDD (p53-inducible protein with a death domain) is another ZU5-UPA-containing protein that undergoes autoproteolysis (27). Following this post-translational event, the two cleavage fragments of PIDD remain associated. Next, we asked whether the two cleavage fragments of NLRP1 similarly remain associated following proteolytic processing within the cell. In these studies, NLRP1 was immunoprecipitated from cellular extracts with antibodies directed to the N-terminal HA tag. The samples were washed extensively and fractionated by SDS-PAGE, and immunoblots were probed with antibodies specific for the C terminus of NLRP1. In cells expressing cleavable forms of NLRP1, the C-terminal cleavage fragment, NLRP1-C, was detected, suggesting that the cleavage products remain associated after processing (Fig. 4C).
Autolytic Processing of NLRP1 Requires Histidine Residue Distal to Cleavage Site
Cleavage of PIDD occurs through an autolytic pathway triggered when the His residue at the P2 position of the cleavage site deprotonates the hydroxyl group side chain of Ser at P1. The reactive oxygen of Ser then performs a nucleophilic attack in cis on the α-carbonyl of the Phe residue at the P1 position leading to cleavage of the peptide bond. Substitution of Cys at P1′ blocks processing because the His residue is unable to deprotonate the sulfhydryl group of Cys (27). The reaction can be rescued by the addition of hydroxylamine (NH2OH), which deprotonates the Cys sulfhydryl group, allowing the reactive sulfur to perform nucleophilic attack at the α-carbonyl of Phe.
To determine whether NLRP1 activation follows an autolytic cleavage mechanism similar to PIDD, the P1′ position of NLRP1 was mutated to either an Ala or a Cys residue. WT and mutant NLRP1 were expressed, and cell extracts were immunoprecipitated with antibodies to the N-terminal HA tag. Some samples were then treated with hydroxylamine prior to immunoblot analysis. In agreement with results presented above, the C-terminal cleavage product was observed in samples expressing WT NLRP1 (Fig. 4D). The addition of hydroxylamine had little effect on cleavage and likely represented the constitutive nature of this mechanism. In contrast to WT NLRP1, the C-terminal cleavage fragment was not detected in samples expressing NLRP1, in which the P1′ Ser residue was mutated to an Ala or Cys. However, treatment with hydroxylamine induced proteolysis in samples expressing NLRP1 S1213C but not those expressing NLRP1 S1213A. These results suggested that, similar to PIDD, NLRP1 activity requires autolytic peptide bond cleavage for activity.
Autolytic cleavage of PIDD is dependent upon a His residue at the P2 position flanking the cleavage site (27). However, unlike PIDD, NLRP1 possesses a Ser residue at this site, suggesting that a His “trigger” was likely to exist elsewhere but in close proximity to the cleavage site through three-dimensional folding. To identify this critical His residue, we compared the orthologous NLRP1 FIIND sequences across multiple species (supplemental Fig. 3). Three invariant His residues, His1168, His1186, and His1249, were identified (Fig. 4E). Separate Ala mutations were introduced for each conserved His and evaluated in reconstituted inflammasome assays and by Western blot analysis. In all three cases, Ala substitution resulted in loss of NLRP1 cleavage and loss of NLRP1 inflammasome activity (supplemental Fig. 4). This was not entirely surprising given the invariant nature of these His residues across species. Thus, it is likely that one or more of these amino acids acts to maintain a functional conformation and this function is separate from the autolytic activity.
To delineate between the His residues required for functional conformation and the His residue responsible for initiating autolytic cleavage, these NLRP1 point mutants were immunoprecipitated from cell extracts and treated with hydroxylamine. The addition of this base should compensate for the absence of a His trigger by deprotonating the hydroxyl group of the P1′ Ser residue, thereby initiating the autolytic cleavage event. Only the H1186A mutation displayed inducible cleavage, suggesting that this residue serves as the trigger for NLRP1 autolytic processing (Fig. 4F). NLRP1-C was not detected in samples expressing His1168 or His1249, suggesting that these residues likely function in maintaining an appropriately folded conformation. Exposure of samples to SDS prior to the addition of hydroxylamine greatly reduced autolytic cleavage, highlighting the importance of tertiary structure to the autoproteolytic function of NLRP1. These findings were further supported through in silico structural alignment of the ZU5-UPA domain of NLRP1 with that of UNC5b. This analysis indicated that His1186 will be located within a loop that exists in close proximity to the cleavage site (Fig. 4E). This demonstrated the important role of the ZU5-UPA domain fold in NLRP1 processing and inflammasome activity.
DISCUSSION
NLRP1 was one of the first members of the NLR family to be identified, yet little is known regarding the mechanism of NLRP1 activation (18, 19). In this report, we demonstrate that NLRP1 inflammasome activity requires the adaptor protein, ASC, and autolytic cleavage within the C-terminal FIIND domain. Previous reports have demonstrated that the adaptor protein ASC enhances but is not required for NLRP1-mediated caspase-1 activation (16). Similar results have been shown for the murine homolog (28). Our studies set out to reconstitute the human NLRP1 inflammasome as a means to develop a cell-based small molecule screening assay. During the development of these assays, we discovered that NLRP1 was incapable of inducing caspase-1 activation and IL-1β release in the absence of ASC, indicating that this adaptor protein is an integral part of an active NLRP1 inflammasome complex.
Different models of NLRP1 inflammasome assembly have been hypothesized, although they have not been proven formally (29, 30). One model suggests that a PYD-PYD interaction between ASC and the N terminus of NLRP1 facilitates recruitment of procaspase-1 via CARD-CARD interactions with ASC (28). However, our data demonstrate that the PYD of NLRP1 is not required for inflammasome activity. Given the dependence upon ASC and the requirement of the C-terminal CARD domain of NLRP1, our results suggest an alternative model, whereby ASC dimers form via PYD-PYD associations. In this model, the two “free” CARD domains of an ASC dimer facilitate the interaction with the CARD of NLRP1 and the CARD of procaspase-1 to assemble the inflammasome. Confirmation of this model will require more rigorous biochemical analyses.
Autolytic processing has been reported for other eukaryotic proteins (23, 27, 31). Perhaps the best described mammalian protein known to undergo autoprocessing within its ZU5-UPA domain is PIDD, where autolytic cleavage regulates its ability to trigger either NF-kB activation or apoptosis (27). The C terminus of NLRP1 is structurally similar to PIDD, in that an LRR and a CARD flank the site of proteolysis. A key difference, however, is the location of the His residue that initiates this intramolecular cleavage. In PIDD, this trigger residue is located at the P2 position, adjacent to the cleavage site (27). In contrast, NLRP1 possesses a Ser residue at the P2 position and this residue is incapable of forming the requisite ester intermediate, which is formed during the NO-acyl rearrangement. This led us to hypothesize that the catalytic His residue required to initiate NLRP1 proteolysis was held in proximity to the cleavage site via the tertiary structure of the FIIND. Sequence comparison of this domain across multiple species revealed three conserved His residues: His1168, His1186, and His1249. When proteins containing single nucleotide mutations were analyzed, only NLRP1 H1186A was able to undergo autolytic cleavage upon the addition of hydroxylamine. Interestingly, three-dimensional modeling of the FIIND, using the crystal structure of Unc5b, indicated that this residue is located in a loop that is held in close proximity to the cleavage site. This strongly suggests that His1186 is the critical trigger residue required for NLRP1 cleavage and subsequent activity.
Many mRNA splice isoforms have been identified for individual NLR proteins (32). Most of these isoforms result from excision of exons encoding the LRRs of which the functional consequences remain unclear. In this report, we demonstrate that a naturally occurring mRNA splice variant of NLRP1, in which nucleotides encoding exon 14 have been excised, failed to undergo autolytic processing and did not mediate IL-1β maturation. One hypothesis is that loss of exon 14 alters the three-dimensional structure of the FIIND and increases the distance between the distal His trigger residue and the cleavage site. However, our results indicate that the role of exon 14 is more complex. If sequences corresponding to exon 14 are removed from the active NLRP1-C cleavage fragment, IL-1β release is blocked (data not shown). This suggests that additional elements within exon 14 function in NLRP1 inflammasome activity following the autolytic processing event.
Polymorphisms of NLRP1 have recently been reported to confer risk of a variety of human diseases (8, 9, 11, 14). Our analysis of two NLRP1 polymorphisms revealed an interesting correlation with NLRP1 cleavage and inflammasome activity. It has been suggested that the M1184V polymorphism is associated with Crohn disease (14). When Met1184 was mutated to Val1184, autoproteolytic cleavage and IL-1β processing were increased markedly, whereas another coding polymorphism (M1119V) resulted in no significant effects. Notably, M1184V resides only two amino acids removed from the catalytic histidine trigger that induces cleavage. It is likely that the M1184V mutation results in a slight structural change, allowing for an increased reaction rate, possibly by bringing the catalytic histidine into closer proximity to the NLRP1 cleavage site. Interestingly, among the NLRP1 sequences analyzed in supplemental Fig. 3, most harbor a Val residue at the position analogous to human NLRP1 Met1184. This list includes mouse Nlrp1, and we did observe an increase in the basal autolytic proteolysis in overexpression studies examining the function of murine Nlrp1b (data not shown) (33). Future three-dimensional structural analysis of the NLRP1 FIIND domain and more specifically the NLRP1 cleavage site would shed additional light on this hypothesis.
In conclusion, we have shown that the adaptor protein ASC and the CARD domain of NLRP1 are absolutely required for NLRP1 inflammasome function. NLRP1 inflammasome function depends upon a recently identified cleavage event that occurs within the FIIND domain of NLRP1 (23, 31). This cleavage-dependent inflammasome activity is altered with at least one known disease-associated coding polymorphism. Future studies will determine what effects other polymorphisms have on NLRP1 autolytic cleavage and activity. Our finding that NLRP1 must undergo autolytic cleavage for inflammasome activity opens the door to developing new classes of compounds that inhibit this process to treat diseases such as vitiligo and other associated autoimmune diseases.
Supplementary Material
Acknowledgments
We thank Molecular Discovery Research for providing BacMam and plasmid reagents with special thanks to J. Fornwald and R. Lehr; Analytical Biochemistry and Biophysics for providing protein sequencing services with special thanks to G. Scott; the GlaxoSmithKline Core Sequencing facility for nucleic acid sequencing; and Jong Yu for discussions and providing technical assistance.
This work was supported by GlaxoSmithKline.

This article contains supplemental Figs. 1–5.
- LRR
- leucine-rich repeat
- MOI
- multiplicity of infection
- ASC
- apoptosis-associated speck-like protein containing a caspase recruit domain.
REFERENCES
- 1. Martinon F., Burns K., Tschopp J. (2002) The inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of pro-IL-β. Mol. Cell 10, 417–426 [DOI] [PubMed] [Google Scholar]
- 2. Srinivasula S. M., Poyet J. L., Razmara M., Datta P., Zhang Z., Alnemri E. S. (2002) The PYRIN-CARD protein ASC is an activating adaptor for caspase-1. J. Biol. Chem. 277, 21119–21122 [DOI] [PubMed] [Google Scholar]
- 3. Wang L., Manji G. A., Grenier J. M., Al-Garawi A., Merriam S., Lora J. M., Geddes B. J., Briskin M., DiStefano P. S., Bertin J. (2002) PYPAF7, a novel PYRIN-containing Apaf1-like protein that regulates activation of NF-κB and caspase-1-dependent cytokine processing. J. Biol. Chem. 277, 29874–29880 [DOI] [PubMed] [Google Scholar]
- 4. Geddes B. J., Wang L., Huang W. J., Lavellee M., Manji G. A., Brown M., Jurman M., Cao J., Morgenstern J., Merriam S., Glucksmann M. A., DiStefano P. S., Bertin J. (2001) Human CARD12 is a novel CED4/Apaf-1 family member that induces apoptosis. Biochem. Biophys. Res. Commun. 284, 77–82 [DOI] [PubMed] [Google Scholar]
- 5. Aganna E., Martinon F., Hawkins P. N., Ross J. B., Swan D. C., Booth D. R., Lachmann H. J., Bybee A., Gaudet R., Woo P., Feighery C., Cotter F. E., Thome M., Hitman G. A., Tschopp J., McDermott M. F. (2002) Association of mutations in the NALP3/CIAS1/PYPAF1 gene with a broad phenotype including recurrent fever, cold sensitivity, sensorineural deafness, and AA amyloidosis. Arthritis Rheum. 46, 2445–2452 [DOI] [PubMed] [Google Scholar]
- 6. Agostini L., Martinon F., Burns K., McDermott M. F., Hawkins P. N., Tschopp J. (2004) NALP3 forms an IL-1β-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity 20, 319–325 [DOI] [PubMed] [Google Scholar]
- 7. Aksentijevich I., Nowak M., Mallah M., Chae J. J., Watford W. T., Hofmann S. R., Stein L., Russo R., Goldsmith D., Dent P., Rosenberg H. F., Austin F., Remmers E. F., Balow J. E., Jr., Rosenzweig S., Komarow H., Shoham N. G., Wood G., Jones J., Mangra N., Carrero H., Adams B. S., Moore T. L., Schikler K., Hoffman H., Lovell D. J., Lipnick R., Barron K., O'Shea J. J., Kastner D. L., Goldbach-Mansky R. (2002) De novo CIAS1 mutations, cytokine activation, and evidence for genetic heterogeneity in patients with neonatal-onset multisystem inflammatory disease (NOMID): A new member of the expanding family of pyrin-associated autoinflammatory diseases. Arthritis Rheum. 46, 3340–3348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jin Y., Riccardi S. L., Gowan K., Fain P. R., Spritz R. A. (2010) Fine-mapping of vitiligo susceptibility loci on chromosomes 7 and 9 and interactions with NLRP1 (NALP1). J. Invest. Dermatol. 130, 774–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zurawek M., Fichna M., Januszkiewicz-Lewandowska D., Gryczyńska M., Fichna P., Nowak J. (2010) Hum. Immunol. 71, 530–534 [DOI] [PubMed] [Google Scholar]
- 10. Sui J., Li H., Fang Y., Liu Y., Li M., Zhong B., Yang F., Zou Q., Wu Y. (2012) NLRP1 gene polymorphism influences gene transcription and is a risk factor for rheumatoid arthritis in han chinese. Arthritis Rheum. 64, 647–654 [DOI] [PubMed] [Google Scholar]
- 11. Dieudé P., Guedj M., Wipff J., Ruiz B., Riemekasten G., Airo P., Melchers I., Hachulla E., Cerinic M. M., Diot E., Hunzelmann N., Caramaschi P., Sibilia J., Tiev K., Mouthon L., Riccieri V., Cracowski J. L., Carpentier P. H., Distler J., Amoura Z., Tarner I., Avouac J., Meyer O., Kahan A., Boileau C., Allanore Y. (2011) NLRP1 influences the systemic sclerosis phenotype: A new clue for the contribution of innate immunity in systemic sclerosis-related fibrosing alveolitis pathogenesis. Ann. Rheum. Dis. 70, 668–674 [DOI] [PubMed] [Google Scholar]
- 12. Verma D., Bivik C., Farahani E., Synnerstad I., Fredrikson M., Enerback C., Rosdahl I., Soderkvist P. (2012) Inflammasome polymorphisms confer susceptibility to sporadic malignant melanoma. Pigment Cell Melanoma Res., in press [DOI] [PubMed] [Google Scholar]
- 13. Pontillo A., Girardelli M., Kamada A. J., Pancotto J. A., Donadi E. A., Crovella S., Sandrin-Garcia P. (2012) Polimorphisms in inflammasome genes are involved in the predisposition to systemic lupus erythematosus. Autoimmunity 45, 271–278 [DOI] [PubMed] [Google Scholar]
- 14. Cummings J. R., Cooney R. M., Clarke G., Beckly J., Geremia A., Pathan S., Hancock L., Guo C., Cardon L. R., Jewell D. P. (2010) The genetics of NOD-like receptors in Crohn's disease. Tissue Antigens 76, 48–56 [DOI] [PubMed] [Google Scholar]
- 15. Hedrich C. M., Fiebig B., Sallmann S., Bruck N., Hahn G., Roesler J., Roesen-Wolff A., Heubner G., Gahr M. (2008) Good response to IL-1β blockade by anakinra in a 23-year-old CINCA/NOMID patient without mutations in the CIAS1 gene. Cytokine profiles and functional studies. Scand. J. Rheumatol. 37, 385–389 [DOI] [PubMed] [Google Scholar]
- 16. Mertens M., Singh J. A. (2009) Anakinra for rheumatoid arthritis: A systematic review. J. Rheumatol. 36, 1118–1125 [DOI] [PubMed] [Google Scholar]
- 17. Mariathasan S. (2007) ASC, Ipaf and Cryopyrin/Nalp3: Bona fide intracellular adapters of the caspase-1 inflammasome. Microbes Infect. 9, 664–671 [DOI] [PubMed] [Google Scholar]
- 18. Hlaing T., Guo R. F., Dilley K. A., Loussia J. M., Morrish T. A., Shi M. M., Vincenz C., Ward P. A. (2001) Molecular cloning and characterization of DEFCAP-L and -S, two isoforms of a novel member of the mammalian Ced-4 family of apoptosis proteins. J. Biol. Chem. 276, 9230–9238 [DOI] [PubMed] [Google Scholar]
- 19. Bertin J., DiStefano P. S. (2000) The PYRIN domain: A novel motif found in apoptosis and inflammation proteins. Cell Death Differ. 7, 1273–1274 [DOI] [PubMed] [Google Scholar]
- 20. Poyet J. L., Srinivasula S. M., Tnani M., Razmara M., Fernandes-Alnemri T., Alnemri E. S. (2001) Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J. Biol. Chem. 276, 28309–28313 [DOI] [PubMed] [Google Scholar]
- 21. Boyden E. D., Dietrich W. F. (2006) Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat. Genet. 38, 240–244 [DOI] [PubMed] [Google Scholar]
- 22. Pathan N., Marusawa H., Krajewska M., Matsuzawa S., Kim H., Okada K., Torii S., Kitada S., Krajewski S., Welsh K., Pio F., Godzik A., Reed J. C. (2001) TUCAN, an antiapoptotic caspase-associated recruitment domain family protein overexpressed in cancer. J. Biol. Chem. 276, 32220–32229 [DOI] [PubMed] [Google Scholar]
- 23. D'Osualdo A., Weichenberger C. X., Wagner R. N., Godzik A., Wooley J., Reed J. C. (2011) CARD8 and NLRP1 undergo autoproteolytic processing through a ZU5-like domain. PLoS One 6, e27396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Davenport E. A., Nuthulaganti P., Ames R. S. (2009) BacMam: Versatile gene delivery technology for GPCR assays. Methods Mol. Biol. 552, 199–211 [DOI] [PubMed] [Google Scholar]
- 25. Richards N., Schaner P., Diaz A., Stuckey J., Shelden E., Wadhwa A., Gumucio D. L. (2001) Interaction between pyrin and the apoptotic speck protein (ASC) modulates ASC-induced apoptosis. J. Biol. Chem. 276, 39320–39329 [DOI] [PubMed] [Google Scholar]
- 26. Faustin B., Lartigue L., Bruey J. M., Luciano F., Sergienko E., Bailly-Maitre B., Volkmann N., Hanein D., Rouiller I., Reed J. C. (2007) Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol. Cell 25, 713–724 [DOI] [PubMed] [Google Scholar]
- 27. Tinel A., Janssens S., Lippens S., Cuenin S., Logette E., Jaccard B., Quadroni M., Tschopp J. (2007) Autoproteolysis of PIDD marks the bifurcation between prodeath caspase-2 and prosurvival NF-κB pathway. EMBO J. 26, 197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nour A. M., Yeung Y. G., Santambrogio L., Boyden E. D., Stanley E. R., Brojatsch J. (2009) Anthrax lethal toxin triggers the formation of a membrane-associated inflammasome complex in murine macrophages. Infect. Immun. 77, 1262–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tschopp J., Martinon F., Burns K. (2003) NALPs: A novel protein family involved in inflammation. Nat. Rev. Mol. Cell Biol. 4, 95–104 [DOI] [PubMed] [Google Scholar]
- 30. Stutz A., Golenbock D. T., Latz E. (2009) Inflammasomes: too big to miss. J. Clin. Invest. 119, 3502–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rosenblum J. S., Blobel G. (1999) Autoproteolysis in nucleoporin biogenesis. Proc. Natl. Acad. Sci. U.S.A. 96, 11370–11375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ting J. P., Davis B. K. (2005) CATERPILLER: A novel gene family important in immunity, cell death, and diseases. Annu. Rev. Immunol. 23, 387–414 [DOI] [PubMed] [Google Scholar]
- 33. Frew B. C., Joag V. R., Mogridge J. (2012) Proteolytic processing of Nlrp1b is required for inflammasome activity. PLoS Pathog. 8, e1002659. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.