Abstract
Pollen development is disturbed in the early tetrad stage of the YX-1 male sterile mutant of wolfberry (Lycium barbarum L.). The present study aimed to identify differentially expressed anther proteins and to reveal their possible roles in pollen development and male sterility. To address this question, the proteomes of the wild-type (WT) and YX-1 mutant were compared. Approximately 1760 protein spots on two-dimensional differential gel electrophoresis (2D-DIGE) gels were detected. A number of proteins whose accumulation levels were altered in YX-1 compared with WT were identified by mass spectrometry and the NCBInr and Viridiplantae EST databases. Proteins down-regulated in YX-1 anthers include ascorbate peroxidase (APX), putative glutamine synthetase (GS), ATP synthase subunits, chalcone synthase (CHS), CHS-like, putative callose synthase catalytic subunit, cysteine protease, 5B protein, enoyl-ACP reductase, 14-3-3 protein and basic transcription factor 3 (BTF3). Meanwhile, activities of APX and GS, RNA expression levels of apx and atp synthase beta subunit were low in YX-1 anthers which correlated with the expression of male sterility. In addition, several carbohydrate metabolism-related and photosynthesis-related enzymes were also present at lower levels in the mutant anthers. In contrast, 26S proteasome regulatory subunits, cysteine protease inhibitor, putative S-phase Kinase association Protein 1(SKP1), and aspartic protease, were expressed at higher levels in YX-1 anthers relative to WT anthers. Regulation of wolfberry pollen development involves a complex network of differentially expressed genes. The present study lays the foundation for future investigations of gene function linked with wolfberry pollen development and male sterility.
Introduction
In flowering plants, male reproductive processes take place in the stamen, a part of the anther, which contains diploid sporogenous cells that experience meiosis to form haploid microspores and finally develop into pollen grains or the male gametophyte [1]–[5]. Detailed analysis of anther development has shown that cell differentiation occurs in a precise chronological order, with distinct stages, which can be related to bud size [6]. Once this process becomes disordered, pollen production might be aborted, resulting in male sterility. Many genes controlling stamen and pollen development have been identified, and their specific roles characterized. These functions appear to be conserved in higher plants [7]–[11].
In recent years there has been an increasing application of proteomic approaches to study anther development and pollen reproduction, such as in Arabidopsis [12]–[15], rice [16]–[19], tomato [2], [20], and Brassica napus [21]–[23]. By applying proteomic analyses, many proteins specifically expressed in anthers with roles in pollen development [24], tapetum degradation [25], programmed cell death (PCD) [26], and callose hydrolyzation [27] were identified. Proteins involved in metabolic process, stress resistance [16], [28], and several transcription and translational regulating factors [29]–[31] were also characterized. It is difficult to find male sterile related genes affecting pollen development directly; therefore, methods for globally detecting different expression patterns between the male sterile mutant and the WT must be applied. The exploitation of plant male sterility is enhanced by combining transcriptome with proteome analysis of developing pollen [32]. In tomato, proteomic analysis of male-sterile 7B-1mutant anthers at the stage when tetrads formed revealed that proteasome and 5B protein, with potential roles in tapetum degeneration, were down-regulated. Cystatin, regulator of endogenous proteolytic activities during seed maturation and germination and in PCD, were up-regulated and correlated with the male sterility [2]. In addition, proteins associated with carbohydrate and energy metabolism, photosynthesis and flavonoid synthesis were also down-regulated in CMS anthers of Brassica napus, all of which might have roles in pollen development [23]. Moreover, several proteins correlated with male sterility were identified in rice, with roles in protein synthesis, signal transduction, cell death and carbohydrate metabolism. The occurrence of male sterile mutants is usually explained by the lack of certain proteins/enzymes involved in pollen development. In the gametophytic male-sterile mutant gaMS-2 of maize, reduced Zea m1 level is associated with the sterility [33]. GS was inactivated in tobacco anthers and microspores by a dominant-negative mutant approach, resulting in male sterility [34]. The conditional male sterile mutant of Arabidopsis was associated with the FLP1 protein, likely playing a role in the synthesis of the components of tryphine, sporopollenin of exine and the wax of stems and siliques. [35]. In addition, several housekeeping proteins with potential roles in microspore development also showed altered abundance. All of these identified proteins have important functions in pollen development in higher plants.
Lycium barbarum L, a woody bush, is a famous traditional Chinese herbal medicine that nourishes the kidneys and liver, brightens eyes, reduces blood glucose and serum lipids, and has anti-aging, immunomodulating, anticancer, anti-fatigue, and male fertility-facilitating properties [36], [37]. It has been widely used as a health food for 2300 years, with its fruit being used to produce various types of health products and foods, such as medicinal beverages and dietary soups [38], [39]. However, it is difficult to meet the need for substantial improvement of existing varieties of wolfberry by natural selection.
Plant male sterility is important for developmental and molecular studies and in hybrid seed programs [40]; therefore, many male sterile mutants in higher plants have been characterized. To the best of our knowledge, few studies on male sterility of wolfberry have been reported. The spontaneous male sterile mutant YX-1 produces flowers with shrunken stamens, in which pollen production is aborted in early tetrad stage. YX-1 mutants also show reduced stamen filament length relative to the WT, and appear beneath the receptive stigma at flower opening [41]. As the male sterile varieties are valuable resources that greatly facilitate the production of hybrids via cross-pollination, in-depth study of anther and pollen development, YX-1 male sterile mutant has important application value in hybrid breeding programs and important theoretical significance.
In this present study, we report the proteomes of the anthers of the WT and YX-1 wolfberry and identify differentially expressed proteins (in terms of protein spot volume). We discuss their possible biological roles and their potential effects on anther development and pollen fertility, with the aim of understanding the molecular mechanism of the biological process at the proteomic level. The protein profiles of anthers at early tetrad stage, when tetrad and tapetum show normal development in WT anthers, but tapetum and tetrads degeneration are observed in YX-1, were compared by 2D-DIGE, and interesting proteins were identified by mass spectrometry. More than 1760 spots were observed on DIGE gels. Compared with the WT, in YX-1 mutant anthers, proteins related to energy, carbohydrate and amino acid metabolism, pollen development, stress response, and signaling were down-regulated. In contrast, proteins involved in proteolysis were up-regulated. The possible functions of these proteins in pollen development and in male sterility are discussed. The results will provide important information for further studies on the molecular mechanism of male sterility in wolfberry.
Materials and Methods
Plant Growth and Anther Collection
The YX-1 male sterile mutant and the WT were grown in the wolfberry orchard of Yinchuan Yuxin Wolfberry seed industry Co., Ltd., located in Yinchuan, Ningxia Municipality, China. Plants were watered regularly and fertilized weekly with a commercial fertilizer (40% Compound Fertilizer, Nantong Hailing Fertilizer Co., Ltd., Nantong, China).
Flower buds of about 4.8–5.0 mm(WT)and 5.0–5.8 mm(YX-1)length (Fig. 1A, corresponding the early stage of tetrad) were picked using a combination of the bud size and anther cytological investigation, as previously reported [42], dissected under a microscope. Stamens from buds at the same stage,when tetrads and tapetum cell were degrading in YX-1 anthers but showed normal development in WT anthers(confirmed by anther squashes, Fig. 1D)were isolated for 2D-DIGE analysis. Stamen samples were either used immediately or frozen in liquid nitrogen and stored at −80°C until further analysis.
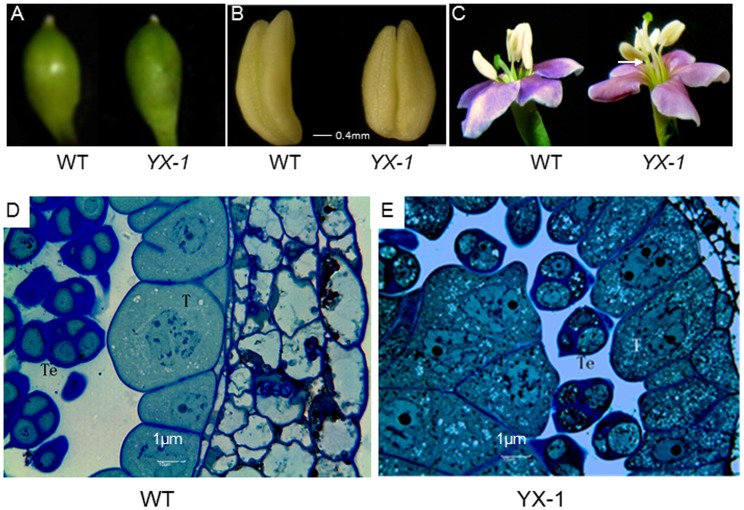
Figure 1. Morphological changes of YX-1 mutant compared with WT about flower organs. A.
Flower buds of WT and YX-1 mutant of wolfberry. B. Stamens from WT and YX-1 buds. C. Flowers of WT and YX-1, note the reduced stamen filament relative to the WT, appearing beneath the receptive stigma. D. Cross-section of WT anther from the same stage as in B, note the normal developed tapetum (T) and tetrads. E. Cross-section of YX-1 anther from the same stage as in B, note the premature tapetum (T) and degraded tetrads (Te), both with masses of small bright vacuoles.
Protein Sample Preparation
Anther protein extractions were performed using a trichloroacetic acid (TCA)-acetone protocol [23] with some modifications. The samples of fresh or frozen wolfberry anthers were finely powdered in liquid nitrogen and homogenized with chilled acetone/10% TCA for 30 min, and then precipitated overnight at −20°C. Precipitated proteins were centrifuged at 12000×g for 45 min at 4°C. After three washes with acetone, the pellets were vacuum dried and proteins extracted with buffer containing 7 mol·L–1 urea, 2 mol·L–1 thiourea, 150 mmol·L–1 Tris-HCl (pH8.5), 4% CHAPS, and 1 mmol·L–1 PMSF, by vortexing for 1 h at room temperature, and centrifugation at 12 000 g for 45 min at 4°C. The supernatants were collected and the protein content was determined according to the Bradford method, using the Bio-Rad protein assay reagent (Bio-Rad, Hercules, CA, USA), and samples were stored at −80°C until 2-DE.
Fluorescence Labeling Protein with CyDyes
Protein labeling with fluorescent cyanine dyes was performed according to the manufacturer’s instructions (GE Amersham, Fairfield, CT, USA). Individual samples from three groups (pooled internal standard, YX-1, and the WT) were labeled with Cy2, Cy3, and Cy5, respectively. The three dyes were designed to ensure that proteins common to each sample have the same relative mobility regardless of the dye used to tag them. CyDyes were reconstituted in anhydrous DMF and combined with samples at a ratio of 400 pmol of CyDye to 50 µg of protein. Labeling was performed on ice and in the dark for 30 min. The reaction was then quenched by incubating with 1.5 µL of 10 mM lysine on ice in the dark for 10 min.
2-DE and Image Acquisition
Proteins were focused on 13 cm Immobiline Drystrips at pH3-10, with non-linear pH gradients (GE Amersham), using an Ettan IPGphor Isoelectric Focusing System (GE Amersham) for a total of 70 000 volt hours. After isoeletric focusing, IPG strips were equilibrated in buffer (6 M urea, 50 mM Tris-HCl, 30% glycerol, 2% SDS) supplemented with 1% DTT to maintain the proteins in a fully reduced state, followed by 3% iodoacetamide to prevent reoxidation of thiol during electrophoresis. Proteins were then separated on 12.5% SDS polyacrylamide gels using a Hofer SE 600 (GE Amersham). Samples were run in three biological replicates. The gels were scanned using a Typhoon FLA9000 (GE Amersham). Excitation/emission wavelengths for Cy2, Cy3, and Cy5 are 488/520, 532/580, and 633/670 nm, respectively.
DIGE Analysis
Relative protein quantification across male sterile mutant YX-1 and WT samples was performed using DeCyder 2-D Differential Analysis Software (v 6.05.11, GE Amersham). The Cy2-labeled pooled internal standard on every gel allowed accurate relative quantitation of protein spot features across different gels. The spots that were present on at least two gels based on the image analysis were considered as expressed protein spots. Student’s t-test (p<0.05) and one-way ANOVA were used to calculate significant differences in relative abundances of protein spot-features in the male sterile anther compared with the WT anthers. Spots with reproducible and significant variations, at least 1.5-fold up- or down-regulated, were considered differentially expressed proteins.
In-gel Digestion
Protein spots were cut from gels, destained for 20 min in 30 mM potassium ferricyanide/100 mM sodium thiosulfate (1∶1 v/v), washed in Milli-Q water until the gels were colorless, and then lyophilized. Each spot was digested in 5 µL 10 ng/µL trypsin (sequencing-grade reagent, Promega, Fitchburg, WI, USA) at 37°C overnight. The peptides were extracted three times with 60% ACN/0.1% TFA. The extracts were dried completely by centrifugal lyophilization. The resulting tryptic digests were concentrated and desalted using C18 ZipTips (Millipore Corporation, Bedford, MA, USA) according to the manufacturer’s protocol.
Mass Spectrometry and Data Analysis
Samples were mixed (1∶1 v/v) with 5 mg/ml HCCA matrix and analyzed by a 4800 Plus MALDI-TOF/TOF™ Analyzer (Applied Biosystems, Carlsbad, CA, USA). Calibration for MS/MS mode was carried out using fragment ion masses from Glufibrinopeptide. Initial analysis of protein digests on the 4800 instrument was carried out by MS in positive ion reflectron mode, using trypsin autolysis products (m/z 842.510, 1045.564 and/or 2211.105) for internal mass calibration where possible, or else the default calibration. Parent mass peaks with a mass range of 800–4000 Da and minimum S/N 50 were picked out for tandem TOF/TOF-MS/MS analysis. The 10 most abundant precursor ions in each spectrum (excluding trypsin autolysis fragments) were subsequently selected by Protein Pilot™ v4.0 software for MS/MS analysis, with collision induced dissociation (CID) closed, increased laser fluence, and 2500 laser shots collected per sample. The UV laser was operated at a 25 Hz repetition rate with a wavelength of 355 nm and the accelerated voltage was 2 kV.
Protein identifications were conducted by combining search (MS plus MS/MS) to the entries of a non-redundant protein and/or EST-viridiplantae database downloaded from the National Center for Biotechnology Information using MASCOT open source (http://www.matrixscience.com) in the NCBI non-redundant database. Search parameters were the enzyme trypsin, taxonomy restrictions to Viridiplantae, ±50 ppm peptide mass tolerance in MS, ±0.2 Da for MS/MS data, one missed cleavage, carbamidomethyl (C) as a fixed modification and methionine oxidation as a variable modification. The confidence in the peptide mass fingerprinting matches (p<0.05) was based on the MOWSE score and confirmed by the accurate overlapping of the matched peptides with the major peaks of the mass spectrum. Only significant hits, as defined by the MASCOT probability analysis (p<0.05), were accepted. The functional classification of the identified ESTs was performed following a BLAST search, and the protein sequences of matching ESTs were then searched against the NCBInr protein database. Only BLAST matches with E values ≤10−30 were selected [19].
Quantitative Real-time RT-PCR Analysis
Quantitative real-time RT-PCR was used to assay gene expression levels for apx and atp synthase beta subunit. Primers for quantitative real-time RT-PCR analysis are shown in Table 1. Wolfberry actin (Accession number HQ415754.1) was used for RNA normalization. All wolfberry RNA samples were diluted to 200 ng µl–1. The SuperScript™ III platinum® two-step qRT-PCR kit with SYBR® Green (Invitrogen, Carlsbad, CA, USA) was used for detecting the expression levels of the genes. Quantitative real-time RT-PCR was carried out in a final volume of 20 µl containing 10 µl Platinum® SYBR® Green qPCR SuperMix-UDG, 10 µM forward and reverse primers in a 7500 Real time PCR System (Applied Biosystems). The relative expression levels of all the samples were analyzed according to recommendations in the User Bulletin for the 7500 Real time PCR System. All reactions were performed in three biological replicates. The threshold cycles (Ct value) of the target genes and actin in different samples were obtained by quantitative real-time RT-PCR.
Table 1. Specific primers used for quantitative real-time RT-PCR analysis.
| Gene | Forward primer | Reverse primer |
| apx | 5′-AACCTGAGCAATGCCCAGAA-3′ | 5′-TCATTTAGCCCCATCCTGTAGAA-3′ |
| atp synthase beta subunit | 5′-GGATCCGAAGTATCGGCCTTA-3′ | 5′-TGCGGGTACATAAACTGCTTGA-3′ |
| actin | 5′-GACCTTCAATGTTCCCGCTATG-3′ | 5′- GCCATCACCAGAGTCCAACAC-3′ |
Enzyme Activity Assay
To monitor the activity of APX and GS, protein was extracted with 100 mmol L−1 sodium phosphate buffer (pH 7.0) containing 5 mmol L−1 ascorbate and 1 mmol L−1 EDTA, and 10 mmol L−1 Tris-HCl buffer (pH 7.6) containing 1 mmolL−1 MgCl2, 1 mmol L−1 EDTA and 1 mmol L−1 β-mercaptoethanol ), respectively from WT and mutant anthers. APX activity was determined by the decrease of absorbance at 290 nm (extinction coefficient 2.8 mM cm−1) as described by Chen and Asada [43].The reaction mixture was composed of 50 mM potassium phosphate buffer (pH 7.0), 0.5 mM ascorbate, 0.2 mM H2O2 and the appropriate volume of protein extract.
Determination of GS activity was performed by the method of Oaks et al [44]. The reaction mixture contained 80 mmol L−1 glutamate, 1 mmol L−1 hydroxylamine, 8 mmol L−1 ATP, 0.2 mol L−1 N-tris- (hydroxymethyl)methyl glycine (Tricine) (pH 7.8), 4 mmol L−1 MgSO4 and 0.2 mmol L−1 EDTA and the appropriate volume of protein extract, and the absorbance of the hydroxamate derivative of glutamic acid measured at 540 nm. One unit of GS activity was defined as 1 µmol L-glutamate γ-monohydroxamate formed per min. Protein in enzyme extract was estimated using Bradford method [45]. Samples were performed in three biological replicates.
Results
Anther Development
Under the growth conditions as mentioned in “Plant growth and anther collection”, buds at the early tetrad stage from YX-1 mutant were longer (approximate 5.0–5.8 mm length) than that from the WT (approximate 4.8–5.0 mm length, Fig. 1A); and the stamens of the YX-1 appeared less slender compared with those of the WT (Fig. 1B ). Also, YX-1 mutants showed reduced stamen filament length relative to the WT, and appear beneath the receptive stigma at flower opening (Fig. 1C). Corresponding to the stage and bud lengths outlined in Fig. 1A the tapetal cells were intact, with a dense cytoplasm containing a few small vacuoles, both tetrads and tapetum showed normal development in WT anthers (Fig. 1D). In contrast, in YX-1 anthers (Fig. 1E), tetrads and tapetal cells had started to degenerate, and numerous small vacuoles were present in the tapetum,also small vacuoles appeared in tetraspores. Anther squashes at an earlier stage (buds of 3.0–5.0 length mm) in both WT and YX-1 showed tetrads enveloped within a thick callose wall, a rich cytoplasm in the tetraspores and tapetal cells radially enlarged with a dense cytoplasm. At later stages (buds of 6.0–7.0 mm length), microspores were observed in WT anthers, but in YX-1 anthers, tetrads and the tapetal cells had completely disintegrated and disappeared, resulting in an empty anther chamber (not shown). This confirmed our earlier observation that in the YX-1 mutant, pollen development breaks down at the early tetrad stage [42].
2D-DIGE Analysis of Anther Proteomes
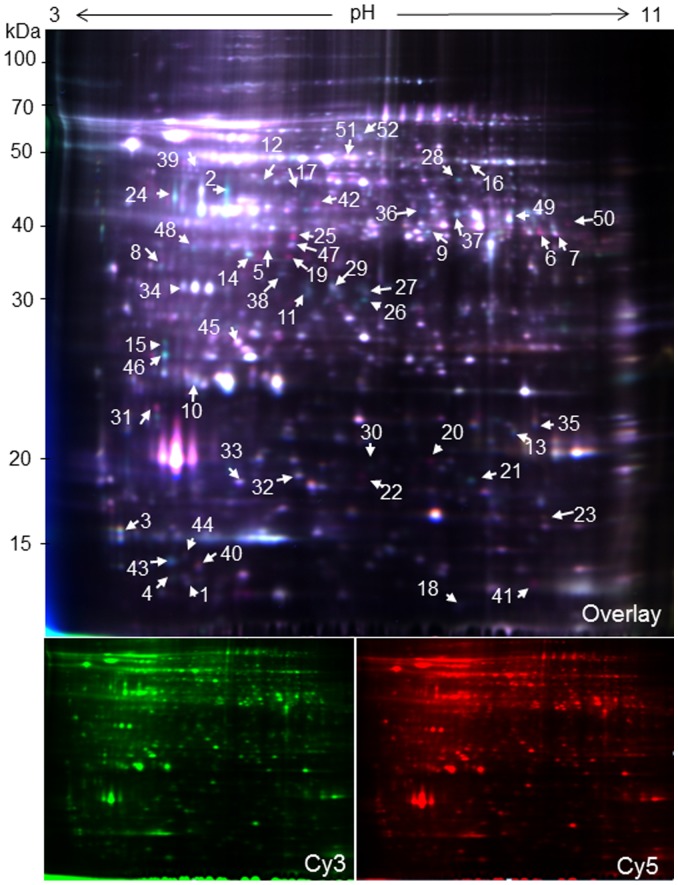
Soluble proteins of wolfberry anthers from YX-1 (approximate 5.0–5.8 mm length) and the WT buds (approximate 4.8–5.0 mm length) were extracted. After preparative experiments, we selected pH 3–11 strips for proteome analysis. Due to the limitations of conventional 2-DE for reproducibility and sensitivity, we used the DIGE technology to study differentially expressed proteins in WT and YX-1 anthers. Equal amounts (50 µg) of protein samples of WT and YX-1 anthers were labeled with Cy2 (internal standard), Cy3, or Cy5 dyes. An overlay of the Cy3 and Cy5 images from the 2D-DIGE gels is shown in Fig. 2. The protein expression patterns of YX-1 mutant were generally similar to those of WT anthers, and more than 1760 spots were observed by DIGE methodology.
Figure 2. 2D-DIGE images of anther proteins from the WT and male sterile mutant YX-1 in wolfberry.
Extracts from the WT and YX-1 anther of three independent biological repeat experiments were differentially labeled with the spectrally resolvable CyDye fluors Cy3 and Cy5 and separated by two-dimensional electrophoresis (2-DE) on 13-cm (pH 3–11) IPG strips and 12.5% polyacrylamide gels. A merged image of Cy5-labeled YX-1 (red) and Cy3-labeled WT (green) is shown. Arrowed and numbered spots in the image are differentially expressed protein spots. Molecular markers (in kDa) are shown on the left.
From the result of 2-DE image analysis, we found a number of spots with lower or higher protein abundances (measured as the relative spot volume) in YX-1 anthers compared with those in the WT. A threshold limit of 1.5-fold was set in this study as previously reported[2] and three replicates were performed to reduce the number of potential false positives because of the sensitivity and reproducibility of DIGE technology. Fig. 2 shows a representative DIGE image of WT and YX-1 anther protein extracts labeled with Cy3 and Cy5 and separated with IPG 3–11 strips and the numbered spots used for mass spectrometry analysis. Some of these differential spots are shown in the enlarged portion of gels in Fig. 3. Fifty-two spots showed at least a 1.5-fold change in protein abundance (p<0.05), among which, 13 showed an increase in YX-1 mutant anthers and 39 showed decreased abundance compared with their levels in WT anthers.
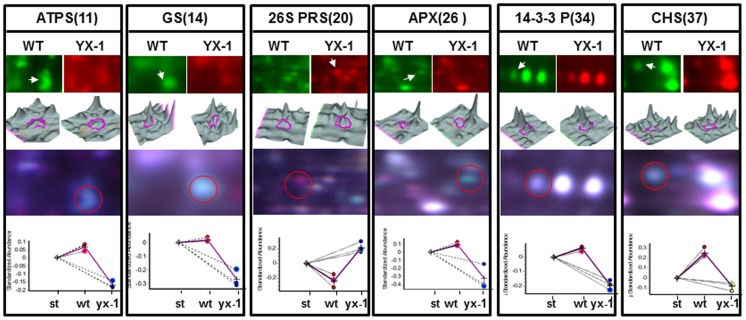
Figure 3. Analysis of several identified proteins.
The readout of the DeCyder Biological Variation Analysis (BVA) module is shown for ATP synthase subunit E (ATPS, spot 11), putive glutamine synthetase (GS, spot No. 14), 26S proteasome regulatory subunit (26S PRS, spot No. 20), ascorbate peroxidase (APX, spot No. 26), 14-3-3 protein (14-3-3, spot No. 34) and chalcone synthase family protein (CHS, spot No. 37). Enlarged regions of 2D-DIGE gels for Cy3-labeled WT (green) and Cy5-labeled YX-1 (red), and the corresponding 3D views, are represented. The bottom panel shows a graphic representation of the differences in abundance of these proteins across three independent experiments. For normalization purposes, a Cy2-labeled internal standard was included, corresponding to a pool of protein from all extracts used in the analysis (st, standard).
Protein Identification
For MALDI-TOF/TOF MS and MALDI-TOF/TOF MS/MS analysis, a total of 52 differently expressed spots with greater than 1.5-fold change in both genotypes (as marked in Fig. 2) were excised. Ultimately, 45 spots (86%) were successfully identified as 41 individual proteins by searching against NCBInr and Viridiplantae EST databases, proteins associated with pollen development and tapetal activities in other plants were found to be differentially expressed in YX-1, as shown in table 2. Four proteins were observed as multiple spots, e.g., three spots corresponding to ascorbate peroxidase (APX, with low levels in YX-1 anthers), two spots corresponding to ribulose-1,5-bisphosphate carboxylase, two spots corresponding to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and two spots corresponding to copper chaperone. In WT anthers, proteins with higher expression, relative to the mutant, included putative glutamine synthetase (GS), ATP synthase subunits, malate dehydrogenase (MDH), plastid aldolase homolog, GAPDH, fructokinase-like protein, ribulose-1,5-bisphosphate carboxylase, APX, chalcone synthase (CHS), CHS-like protein, 5B protein, cysteine protease, protein disulfide isomerase, basic transcription factor 3(BTF3), calmodulin-like protein 1, 14-3-3 protein, putative callose synthase catalytic subunit, and enoyl-ACP reductase. In YX-1 anthers, the up-regulated proteins included cysteine protease inhibitor 5, putative S-phase Kinase association Protein 1(SKP1), disulfide isomerase, 26S proteasome subunits, ubiquitin-protein ligase, and an aspartic protease.
Table 2. Proteins from WT and YX-1 anthers of wolfberry analyzed by DIGE and MALDI-TOF/TOF, and identified by searching against NCBInr and Viridiplantae EST databases.
| aSpot No. | bAcc. No. | cProtein name (Species) | dMr/pI | eScore | fS.C. (%) | gS.V.R. (YX-1/WT) | h p-value |
| Photosynthesis-related | |||||||
| 1 | gi|170320 | ribulose-1,5-bisphosphate carboxylase (NS) | 10.3/5.37 | 100 | 80 | −1.57 | 0.0083 |
| 2 | gi|445628 | RuBisCO activase(NT) | 42.9/5.52 | 100 | 85 | −2.76 | 0.0013 |
| 3 | gi|230922 | unactivated form Of ribulose-1,5- bisphosphatecarboxylase(NT) | 14.7/5.19 | 100 | 80 | −2.20 | 0.0048 |
| 4 | gi|170320 | ribulose-1,5-bisphosphate carboxylase (NS) | 10.3/5.37 | 100 | 77 | −1.70 | 0.00015 |
| Carbohydrate and energy metabolism | |||||||
| 5 | gi|1781348 | homologous to plastidic aldolases (ST) | 38.6/5.89 | 100 | 17 | −1.65 | 0.0011 |
| 6 | gi|4539543 | glyceraldehyde-3-phosphate dehydrogenase (NT) | 36.8/7.7 | 100 | 42 | −2.52 | 0.00024 |
| 7 | gi|4539543 | glyceraldehyde-3-phosphate dehydrogenase (NT) | 36.8/7.7 | 100 | 42 | −2.38 | 0.0066 |
| 8 | gi|21592495 | fructokinase-like protein (AT) | 35.2/5.12 | 100 | 27 | −2.65 | 0.0027 |
| 9 | gi|21388550 | malate dehydrogenase(ST) | 36.4/8.48 | 100 | 22 | −1.88 | 0.0076 |
| 10 | gi|48209968 | ATP synthase D chain, mitochondrial (ST) | 29.8/5.34 | 100 | 60 | −2.61 | 0.0011 |
| 11 | gi|9652289 | putative ATP synthase subunit E (SL) | 27.4/6.63 | 100 | 47 | −2.73 | 0.00024 |
| 12 | gi|56784991 | putative ATP synthase beta subunit (OS) | 45.9/5.33 | 100 | 22 | −1.72 | 0.00078 |
| Nucleic acid metabolism | |||||||
| 13 | gi|8272416 | nucleoside diphosphate kinase 3(BR) | 21.5/7.98 | 100 | 36 | −1.83 | 0.0012 |
| Amino acid metabolism | |||||||
| 14 | gi|28393681 | putative glutamine synthetase(AT) | 38.9/5.59 | 97.2 | 14 | −2.97 | 0.00023 |
| 15 | gi|18414289 | ARD(AT) | 23.5/4.99 | 99.9 | 14 | −2.48 | 0.0033 |
| 16 | gi|4049354 | glycine hydroxymethyltransferase-like protein (AT) | 50.9/8.13 | 100 | 22 | −2.67 | 0.0052 |
| Protein metabolism | |||||||
| 17 | gi|19851 | cysteine protease (NT) | 40.8/6.0 | 96.2 | 26 | −2.42 | 0.0017 |
| 18 | gi|415833 | 5B protein (SL) | 11.6/8.16 | 100 | 29 | −1.95 | 0.00005 |
| 19 | gi|37805883 | putative S-phase Kinase association Protein 1(SKP1) (OS) | 35.5/5.57 | 98.6 | 9 | 2.42 | 0.0039 |
| 20 | gi|18424049 | 26S proteasome regulatory subunit (AT) | 24.4/5.08 | 99.5 | 27 | 2.59 | 0.0054 |
| 21 | gi|30693656 | ubiquitin-protein ligase (AT) | 16.8/6.2 | 100 | 46 | −2.89 | 0.0024 |
| *22 | gi|309372357 | NT2C-EST-0809 (NT) similar to aspartic protease gi|226503984(ZM) | 18.7/6.64 | 99.9 | 10 | 2.23 | 0.011 |
| 23 | gi|20137686 | cysteine protease inhibitor 5(ST) | 17.1/8.63 | 98 | 16 | 2.10 | 0.0052 |
| 24 | gi|30692346 | ribosomal protein S1(AT) | 45.3/5.13 | 100 | 19 | −2.88 | 0.00026 |
| 25 | gi|1848212 | protein disulfide-isomerase (NT) | 40.1/5.99 | 100 | 14 | 2.74 | 0.0089 |
| Stress related | |||||||
| 26 | gi|34809902 | ascorbate peroxidase(NT) | 32.3/5.96 | 98.7 | 15 | −2.76 | 0.0094 |
| 27 | gi|34809902 | ascorbate peroxidase(NT) | 32.3/5.96 | 100 | 26 | −1.53 | 0.0048 |
| 28 | gi|21039134 | ascorbate peroxidase (SL) | 42.4/8.65 | 100 | 26 | −2.44 | 0.0021 |
| 29 | gi|55296784 | putative peroxidase (OS) | 35.4/6.43 | 95.1 | 25 | −2.34 | 0.002 |
| Transcription factor | |||||||
| 30 | gi|15220876 | putative transcription factor BTF3 (AT) | 17.9/6.62 | 100 | 30 | −2.17 | 0.00017 |
| Signaling-related | |||||||
| 31 | gi|9979177 | translationally-controlled tumor protein(NT) | 18.9/4.54 | 98.9 | 20 | 2.65 | 0.000038 |
| 32 | gi|75319566 | calmodulin-like protein 1(OS) | 21.1/4.75 | 100 | 31 | −1.77 | 0.006 |
| 33 | gi|15233402 | putative calcium-binding protein (AT) | 21.2/4.59 | 100 | 40 | −1.79 | 0.000012 |
| 34 | gi|3766535 | 14-3-3 protein (ST) | 29.4/4.78 | 100 | 58 | −1.99 | 0.00035 |
| Anther development | |||||||
| 35 | gi|4588012 | putative callose synthase catalytic subunit (GH) | 21.9/8.42 | 100 | 27 | −2.43 | 0.00067 |
| 36 | gi|2326772 | chalcone synthase -like protein(NS) | 40.7/5.59 | 100 | 30 | −2.59 | 0.0011 |
| Flavonoid synthesis | |||||||
| 37 | gi|15217605 | chalcone synthase family protein (AT) | 43.9/6.01 | 100 | 20 | −2.78 | 0.0047 |
| Fatty acid synthesis | |||||||
| 38 | gi|2204236 | enoyl-ACP reductase(NT) | 33.9/6.41 | 99.9 | 8 | −2.32 | 0.0003 |
| Unknown proteins | |||||||
| 39 | gi|115474835 | Os08g0154300 (OS) | 43.1/5.18 | 100 | 18 | −1.87 | 0.0016 |
| 40 | gi|2673909 | hypothetical protein (AT) | 13.1/4.59 | 99 | 18 | 2.24 | 0.00073 |
| 41 | gi|54291158 | hypothetical protein (OS) | 14.0/7.7 | 99.8 | 21 | 2.43 | 0.0015 |
| Others | |||||||
| 42 | gi|56562181 | formate dehydrogenase (SL) | 42.4/6.87 | 100 | 30 | 2.54 | 0.00022 |
| 43 | gi|15228869 | copper chaperone (AT) | 13.1/4.91 | 99.8 | 30 | −2.96 | 0.0061 |
| 44 | gi|15228869 | copper chaperone (AT) | 13.1/4.91 | 99.9 | 26 | −1.50 | 0.0032 |
| 45 | gi|30580343 | caffeoyl-CoA O-methyltransferase 6(NT) | 27.9/5.3 | 100 | 32 | 2.33 | 0.00022 |
Spot number in 2-DE gel, as shown in Fig. 2.
Accession number in NCBInr/EST database.
Protein names and species from the NCBInr/EST database. NT, Nicotiana tabacum; NS, Nicotiana sylvestris; AT, Arabidopsis thaliana; SL, Solanum lycopersicum; ST, Solanum tuberosum; BR, Brassica rapa; ZM, Zea mays; OS, Oryza sativa.GH, Gossypium hirsutum.
Theoretical molecular weight and pI of the identified proteins.
Mascot protein score for ions complemented by the percentage of the confidence index (C.I.).
Sequence Coverage.
Spot volume ratios are the average for each spot from three replicate gels.
Protein spots with a significant change in abundance (1.5-fold or above) between the WT and YX-1, and a P-value of≤0.05 were considered statistically significant.
Spots from EST database.
Unidentified differently spots not listed.
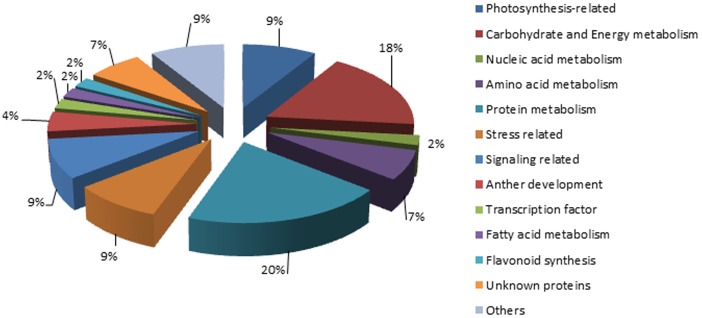
The 41 identified proteins could be classified into various functional groups (as shown in Fig. 4) based on the known functions from NCBI gene annotations and the literature. The largest numbers of protein spots with 1.5-fold or higher changes in abundance were those related to protein metabolism (20%), i.e., protein synthesis and folding, proteases and protease inhibitor(4 were up-regulated in WT and 5 in YX-1 anthers), and carbohydrate and energy metabolism (18%, 8 highly expressed in WT anthers). Other groups of differentially expressed proteins were related to signaling (9%, 3 were up-regulated in WT and 1 in YX-1 anthers), photosynthesis (9%, 4 highly expressed in WT anthers), stress response (9%, 1 were up-regulated in WT and 3 in YX-1 anthers) and other proteins (9%). Relatively few proteins were classified into amino acid metabolism (7%), unknown proteins (7%), anther development (4%), transcription factor (2%), fatty acid metabolism (2%), nucleic acid metabolism (2%) and flavonoid synthesis (2%) groups (Fig. 4).
Figure 4. Function classifications of identified proteins in WT and YX-1 anthers of wolfberry.
As can be seen from Table 2, between the WT and YX-1 anthers, three differently expressed proteins were identified as mitochondrial ATP synthase related subunits (spots No. 10, 11 and 12), which are involved in energy metabolism. Three proteins correspond to APX (spots No. 26, 27, and 28), which is involved in stress response processes. GS (spot No. 15, Table 2) was one of the proteins that showed a major expression difference (Fig. 2 and 3) between the WT and YX-1 anthers in DIGE gel images. Therefore, we selected these three enzymes to further confirm changes using enzyme activities or mRNA expression.
Expression of apx and atp Synthase Beta Subunit mRNA
To further understand the differential expressions of APX and component of the mitochondrial ATP synthase complex at the level of gene expression between the WT and YX-1 anthers, quantitative real-time RT-PCR was used to analyze their expression levels at the stage of the early tetrad development (the same stage as the proteome analysis) between the male sterile anthers and the WT. It should be noted that these two genes had been isolated by 5′- and 3′-race methods in our previous experiments [46]. Fig. 5 shows the apx and atp synthase beta subunit mRNA expression levels in the wolfberry anthers. At the early tetrad stage of anther development, the expression level of apx and atp synthase beta subunit mRNA in YX-1 anthers was only 11% and 31% to that in WT anthers, respectively. The results clearly indicated that expression of apx and atp synthase beta subunit mRNA is significantly reduced during the process of anthers abortion.
Figure 5. Quantitative real-time RT-PCR using SYBR Green assays for quantitative analysis of apx and atp synthase beta subunit mRNA expression levels in wolfberry anthers.
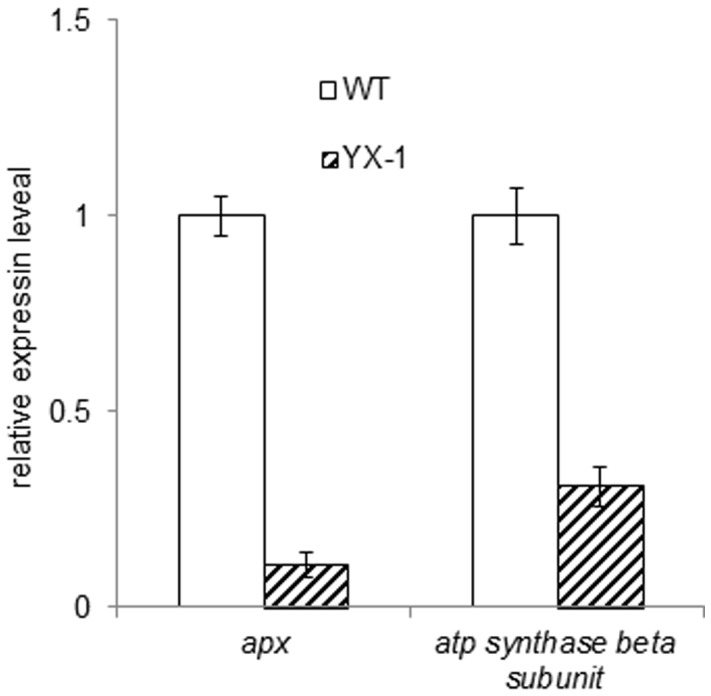
Data are the mean ± SD from three replications.
Enzyme Activity
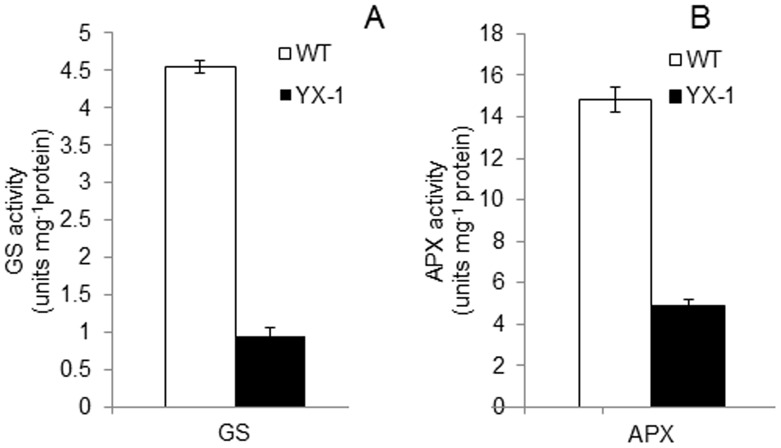
GS (spot No. 14) showed maximum difference in expression between the WT and the mutant in DIGE gel images (Fig. 2); in addition, three spots (spot No. 26, 27 and 28) were identified to be APX, which displayed low abundance in mutant anthers. Therefore, we analyzed the activity of these two enzymes in WT and YX-1 anthers. The activity of GS was nearly 5 times, and that of APX approximately 3 times, higher in WT compared with that in YX-1 anthers (Fig. 6A and 6B), which correlated with the 2-D DIGE results.
Figure 6. The activities of glutamine synthetase (GS) (A) and ascorbate peroxidase (APX) (B) in WT and YX-1 anthers (of the same stage as shown in Fig. 1B).
Error bars indicate standard deviation.
Discussion
There has been little detailed proteomic characterization of the male sterile wolfberry. Therefore, we conducted a comprehensive proteomics analysis between mutant YX-1 and WT anthers to gain an understanding of the mechanisms of wolfberry male sterility. The potential roles of some differentially expressed proteins in anther development and pollen fertility are discussed below.
The tapetum is universally present in higher plant anthers. In addition to its main function of supplying nutrition for meiocytes/spores, the tapetum has other roles, such as the production of the locular fluid, callase, pollenkitt/tryphine, sporophytic proteins and enzymes, and the formation of exine precursors, all of which are required for the normal development of microspores to pollen grains [4]. The tapetum undergoes cellular degradation during the late stage of anther development, which is considered a programmed cell death (PCD) event. Furthermore, tapetal cell disintegration accords well with the post-meiotic anther development processes. A premature or delayed degradation of the tapetum can result in male sterility [47]. In normal anthers of wolfberry, both tetrads and tapetum show normal development, with a dense cytoplasm and only a few small vacuoles present at the early tetrad stage (the stage used in the present study, Fig. 1D). In contrast, in the YX-1 mutant anthers at the same stage, the tapetum begins to degenerate, coupled to numerous small vacuoles which also appear in tetraspore (Fig. 1E), which suggests that premature tapetum degeneration in YX-1 could contribute to male sterility.
A number of enzymes, belonging to the carbohydrate and energy metabolism group, are reduced in YX-1 anthers, relative to the WT. These down-regulated enzymes include mitochondrial ATP synthase subunits, fructokinase-like protein, MDH, aldolase, and GAPDH (Fig. 2 and Table 2). In CMS lines of rice and B. napus, some of the enzymes involved in energy and carbohydrate metabolism are also down-regulated [16], [23]. In particular, three protein spots were identified to be mitochondrial ATP synthase D chain (spot No.10, Fig. 2 and Table 2), putative ATP synthase subunit E (spot No.11, Fig. 2, 3 and Table 2), and mitochondrial ATP synthase beta subunit (spot No.12, Fig. 2 and Table 2). Cytoplasmic male sterility in plants is generally the consequence of dysfunction of mitochondria in the pollen, and several mitochondrion DNA regions encoding F0F1-ATPase (i.e. ATP synthase) subunits have been identified associated with CMS [48]. Plant mitochondrial genomes contain approximately 60 open reading frames (ORFs), among which, orf25 is implicated playing a role in the CMS of T cytoplasm maize [49], and orfB is involved in CMS in several plant species [50]–[52]. An accepted hypothesis on the mechanism of CMS is that the increased demand for respiratory function and cellular energy in the form of ATP during anther development may be compromised by expression of the aberrant mitochondria genes. ATP synthase β-subunit was observed in pollen mitochondria and was found to be generally important for male gametophytic development [53]. If the β-subunit is defective, it will cause the dysfunction of F0F1-ATPase, which may impact the energy output of mitochondria, resulting in abnormal anther development with non-functional pollens [48], [54], [55]. In the present study, protein spot No.12 was identified as beta subunit of ATP synthase F1 sector and it is inferred to be a defective protein, which may lead to the dysfunction of F0F1- ATPase by incorporation into the ATP synthase complex. Quantitative real-time RT-PCR showed that the expression of atp synthase beta subunit RNA was reduced by approximately 70% in YX-1 anthers, relative to the WT (Fig. 5), which is consistent with the difference in protein abundance. In fact, it has been suggested that during microspore development, demand for energy is especially high. The reduced level of ATP synthase beta subunit suggests that the male sterile mutant plants are in an energy starved state. This is in accordance with down-regulation of genes controlling enzymes associated with energy in a CMS line of B. napus [23], [56].
Four down-regulated proteins are involved in carbohydrate metabolism: fructokinase-like protein, MDH, aldolase, and GAPDH. Starch is synthesized in anthers before meiosis and subsequently hydrolyzed to provide energy for lipid synthesis in both tapetum and microspores [57]. Decreased abundance of these enzymes in YX-1 anthers could alter levels of sugar and starch, two molecules key to biosynthesis and energy balance.
Several other proteins/enzymes, including GS, APX, putative callose synthase catalytic subunit, CHS, CHS-like and enoyl-ACP reductase, were down-regulated in YX-1 anthers, and these proteins/enzymes may have a role in tapetum and pollen development. One of the most notable difference in WT and YX-1 anther gels was the location of the protein putative GS (spot No.14, Fig. 2, 3, and Table 2), and may be involved in pollen development. GS is found as two isoforms, cytoplasmic GS1 and chloroplastic GS2, catalyzing the ATP-dependent conversion of glutamine to glutamate. Isolated and in vitro-cultured microspores were unable to develop into functional pollen grains in a medium lacking glutamine [58], which indicated that glutamine plays a key role in plant amino acid metabolism and pollen development. The importance of GS1 in pollen reproduction has been shown in rice [59] and maize [60]. In tobacco, GS1 was inhibited by introducing mutated tobacco GS genes fused to the tapetum-specific TA29 and microspore-specific NTM19 promoters, and pollen aborted close to the first pollen mitosis in the transgenic plants, resulting in male sterility [61]. YX-1 anthers also showed lower GS activity relative to WT (Fig. 6A). The decreased GS activity in YX-1 anthers could cause a reduction in glutamine, which is required for pollen development thereby resulting male sterility.
It is notable that three spots (spot No.26, 27 and 28; Fig. 2 and Table 2) were identified as APXs and all of them showed lower amounts in YX-1 relative to WT anthers. Just as in cotton and rice [62], [63], lower activity/amounts of oxidative stress enzymes in cytoplasmic male-sterile anthers was detected compared with fertile anthers. Moreover, the expression of apx RNA and the APX activity were lower in YX-1 anthers, respectively, relative to the WT (Fig. 5 and Fig. 6B), during the process of anther abortion, when a great deal of ROS might be generated in the anther cell.
Callose synthase is responsible for the synthesis of callose deposited at the primary cell wall of meiocytes, tetrads and microspores in Arabidopsis, and T-DNA insertion mutations of the CalS5 gene resulted in degeneration of microspores, thereby, male sterility [64]. Several studies have also described mutations in callose wall formation and dissolution in petunia [65] and tobacco [66] that disrupt fertility. Putative callose synthase catalytic subunit (spot No.35; Fig. 2 and Table 2) showed low spot volume in YX-1 anthers. Collectively, the evidence indicates that the timing of callose formation and dissolution are critical for normal fertility.
Most plant phenolics, including flavonoids, are products of phenylpropanoid metabolism. CHS is one of the main enzymes in the flavonoid biosynthesis pathway, and an alteration in CHS abundance would be expected to affect the accumulation of all classes of phenolic compounds. Generally, tapetal cells produce proteins and lipids, as well as flavonoids, which are secreted into the pollen sac and form part of the exine [4]. Several enzymes involved in secondary metabolism, including CHS, are specifically or predominantly expressed in the tapetum [67]. It is reported that CHS is essential for pollen development and fertility in several plant species, and disruptions to CHS activity in the anthers resulted in the production of sterile pollen [68]–[72]. In addition, in recent research in Arabidopsis anthers, LAP5 and LAP6, encoding anther-specific proteins with similarity to CHS, were suggested to play a role in pollen development and exine formation [73]. All of the above results suggest that flavonoids play an important role in the development of functional pollen. This study showed that two proteins, CHS (spot No.37, Fig. 2, 3, and Table 2) and CHS-like protein (spot 36, Fig. 2 and Table 2) are down-regulated in the mutant, indicating the premature degradation of the tapetum in YX-1 mutant is concomitant with the reduction of anther specific CHS abundance. Thus, the level of flavonoids might decrease to below the level required to generate the pollen exine, leading to male sterility.
As a catalytic component of the fatty acid synthetase system in plants, enoyl-ACP reductase is prominently expressed in the tapetum, developing pollen grains, and vascular tissue of anthers. In the Arabidopsis mod1 mutant, reduced activity of enoyl-ACP reductase led to abnormal development of various organs and reduced fertility [74]. It is also reported that the DPW gene, encoding a fatty-ACP reductase, is expressed in both tapetal cells and microspores during anther development in rice, and in a dpw mutant, defective anther development and degenerated pollen grains with an irregular exine appeared [75]. In YX-1, a protein identified as enoyl-ACP reductase (spot No. 38, Fig. 2, and Table 2) showed reduced abundance, which might affect fatty acid synthesis and anther development.
Besides metabolic pathways, proper anther development requires diverse regulatory processes. 14-3-3 proteins, being conserved phosphopeptide binding proteins in eukaryotic organisms [76], [77], regulate diverse biological processes in plants, such as metabolism, transcription, organellar protein trafficking, and stress responses [78], [79]. There have been some reports that 14-3-3 proteins are associated with ATP synthases in a phosphorylation-dependent style, playing a regulatory role in starch accumulation [80], regulation of PCD as a MAPKKKa-interacting protein in pollen development [81], In maize, reduced abundance of the14-3-3 protein led to temporal gene expression changes and, ultimately, pollen sterility [31]. In YX-1 anthers, a 14-3-3 protein (spot No.34, Fig. 2, 3, and Table 2) abundance was down-regulated compared with the WT. The aberrant abundances of such 14-3-3 factors could contribute directly to YX-1 defects. Another protein, BTF3 (spot No.30, Fig. 2 and Table 2) was also detected as having a reduced abundance level in YX-1 anthers relative to the WT. BTF3 is the β-subunit of the nascent-polypeptide-associated complex, with a conserved role in regulating protein localization during translation in plants [82]. In a photoperiod-sensitive male-sterile mutant of rice, defects in pollen development were related to abnormal protein localization in anther tissue layers, including the tapetum [83]. The reduced abundance of BTF3 in male sterile anthers was similar to results obtained in tomato [2], which was considered to affect protein localization in the anther and hence affect pollen development.
In YX-1 anthers at the early tetrad stage, some of proteolytic enzymes, including aspartic protease, 26S proteasome regulatory subunit and SKP1,as well as cysteine protease inhibitor, were up-regulated and these proteins may have a role in tapetum degeneration. Aspartic protease acts as an anti-cell-death factor participating in PCD, and overexpression of the gene encoding aspartic protease resulted in male sterility in Arabidopsis [84]. In common with the observation in the 7B-1 male sterile mutant of tomato [2], spot No.22 (Fig. 2 and Table 2) was identified as aspartic protease with increased amounts in mutant anther. The higher abundance of aspartic protease in the YX-1 anther could disturb PCD of the tapetum and pollen development, causing male sterility.
Another protein with higher spot volume in YX-1, relative to WT anthers, was 26S proteasome regulatory subunit (spot No.20, Fig. 2, 3 and Table 2). Proteasomes are regulators of many processes such as the cell cycle, embryogenesis, metabolism, gametophyte survival, hormone signaling, senescence and defense [85], [86], and have been identified in plant reproductive organs, such as anthers [2], [86]. During PCD, proteasomes are released into the extracellular space and have the potential to damage nearby cells. The higher level of this protein in YX-1 compared with WT anthers might disturb the degradation of regulatory proteins in anther tissues, hence leading to premature degradation of the tapetum and male sterility. Selective proteolysis of proteins mediated by the ubiquitin pathway is an important pathway for controlling many biological events. The SCF class of E3 ubiquitin ligases controls the ubiquitination of a wide variety of substrates, thereby mediating their degradation by the 26S proteasome. In Arabidopsis, it was reported that the Skp1 homologue ASK1 involved in the regulation of pollen development, and the ask1-1 mutant produces polyads containing microspores of variable number and size, leading to non-viable pollen grains and male sterility [87]. In this study, the reduced level of protein spot putative SKP1 (spot No.19; Fig. 2 and Table 2) in YX-1 anthers may affect pollen development.
In plants, cysteine protease inhibitor act as regulators of endogenous proteolytic activities. In rice, the TDR gene controls tapetum degeneration by targeting anther specific cysteine protease and protease inhibitor genes [88]. The male sterile mutant showed a reduced activity of cysteine protease (spot No.17; Fig. 2 and Table 2) but higher activity of cysteine inhibitor (spot No.23; Fig. 2 and Table 2) and this would inhibit cysteine protease activity and disturb tapetum development thereby effecting male sterility.
Other proteins were also reduced in YX-1 anthers, such as 5B protein, considered to be related to tapetum degradation by inhibiting proteasome activity, are cysteine-rich and are specifically expressed in the tapetum and stamen in plants [25], [89]. In tomato, the 5B protein showed lower abundance at the tetrad stage in male sterile mutant 7B-1 [2]. In our study, the abundance of the 5B protein (spot No.18, Fig. 2 and Table 2) was reduced in the YX-1 anther relative to the WT, which indicated that tapetum development was disturbed by abnormal abundance of the 5B protein, leading to pollen abortion. In addition, a number of other proteins showed altered abundance, including calmodulin-like protein 1 (spot No.32; Fig. 2 and Table 2) and putative calcium-binding protein(spot No.33; Fig. 2 and Table 2), which translates a signal of cytosolic Ca2+ elevation to downstream protein targets in numerous signal transduction cascades [90]. The altered abundance level of these enzymes/proteins in YX-1 anthers might affect the abundance of regulatory proteins in anther tissues and, ultimately, pollen development.
Conclusions
This study applied a proteomic approach to identify regulating proteins in the anthers of a male sterile mutant of wolfberry. We conclude that the breakdown of pollen development at the early tetrad stage of YX-1 mutant anthers is associated with the differential expression of several proteins, including energy conversion related (e. g., ATP synthase subunits), amino acid metabolism related (e.g., GS), stress response related (e.g., APX), proteins with roles in signaling (e.g., 14-3-3 protein), anther development (e.g., putative callose synthase catalytic subunit), as well as proteases and protease inhibitor (e.g., 5B protein, 26S proteasome regulatory subunits, aspartic protease, cysteine protease, cysteine protease inhibitor and putative SKP1). Significantly, the abnormal protein complex of ATP synthase may cause the dysfunction of mitochondrion, and alterations of some protein abundances (such as APX, GS) could be the consequences of mitochondrial dysfunction in the pollen as indicated by changes in abundance of subunits of ATP synthase. These data indicate regulating patterns of wolfberry pollen development is a complex network. The significant impact on pollen fertility in YX-1 appears to be the result of regulation of multiple metabolic pathways with differentially expressed genes potentially involved, which is supported by the results in male-sterile mutants of Arabidopsis, rice, tomato and maize [2], [32], [91], [92].
Acknowledgments
We thank the Shanghai Institutes of Biological Sciences, Research Center for Proteome Analysis of the 2D in-gel electrophoresis and MS analysis.
Funding Statement
This research was supported by the National Natural Science Foundation of China (30960208, 31060162, 30800692, 31000718 and 31171573). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Liu J, Qu LJ (2008) Meiotic and mitotic cell cycle mutants involved in gametophyte development in Arabidopsis. Mol Plant 1: 564–574. [DOI] [PubMed] [Google Scholar]
- 2. Sheoran IS, Ross AR, Olson DJ, Sawhney VK (2009) Differential expression of proteins in the wild type and 7B-1 male-sterile mutant anthers of tomato (Solanum lycopersicum): a proteomic analysis. J Proteomics 71: 624–636. [DOI] [PubMed] [Google Scholar]
- 3. Scott RJ, Spielman M, Dickinson HG (2004) Stamen structure and function. Plant Cell 16 Suppl: S46–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goldberg RB, Beals TP, Sanders PM (1993) Anther development: basic principles and practical applications. Plant Cell 5: 1217–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McCormick S (2004) Control of male gametophyte development. Plant Cell 16 Suppl: S142–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Scott R, Hodge R, Paul W, Draper J (1991) The molecular biology of anther differentiation. Plant Sci 80: 167–191. [Google Scholar]
- 7. Sorensen AM, Krober S, Unte US, Huijser P, Dekker K, et al. (2003) The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J 33: 413–423. [DOI] [PubMed] [Google Scholar]
- 8. Endo M, Tsuchiya T, Saito H, Matsubara H, Hakozaki H, et al. (2004) Identification and molecular characterization of novel anther-specific genes in Oryza sativa L. by using cDNA microarray. Genes Genet Syst 79: 213–226. [DOI] [PubMed] [Google Scholar]
- 9. Lou P, Kang J, Zhang G, Bonnema G, Fang Z, et al. (2007) Transcript profiling of a dominant male sterile mutant (Ms-cd1) in cabbage during flower bud development. Plant Sci 172: 111–119. [Google Scholar]
- 10. Hu SW, Fan YF, Zhao HX, Guo XL, Yu CY, et al. (2006) Analysis of MS2Bnap genomic DNA homologous to MS2 gene from Arabidopsis thaliana in two dominant digenic male sterile accessions of oilseed rape (Brassica napus L.). Theor Appl Genet 113: 397–406. [DOI] [PubMed] [Google Scholar]
- 11. Chen W, Yu XH, Zhang K, Shi J, De Oliveira S, et al. (2011) Male Sterile 2 encodes a plastid-localized fatty acyl carrier protein reductase required for pollen exine development in Arabidopsis. Plant Physiol 157: 842–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Holmes-Davis R, Tanaka CK, Vensel WH, Hurkman WJ, McCormick S (2005) Proteome mapping of mature pollen of Arabidopsis thaliana . Proteomics 5: 4864–4884. [DOI] [PubMed] [Google Scholar]
- 13. Sheoran IS, Sproule KA, Olson DJH, Ross ARS, Sawhney VK (2006) Proteome profile and functional classification of proteins in Arabidopsis thaliana (Landsberg erecta) mature pollen. Sex Plant Reprod 19: 185–196. [Google Scholar]
- 14. Grobei MA, Qeli E, Brunner E, Rehrauer H, Zhang R, et al. (2009) Deterministic protein inference for shotgun proteomics data provides new insights into Arabidopsis pollen development and function. Genome Res 19: 1786–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Noir S, Brautigam A, Colby T, Schmidt J, Panstruga R (2005) A reference map of the Arabidopsis thaliana mature pollen proteome. Biochem Biophys Res Commun 337: 1257–1266. [DOI] [PubMed] [Google Scholar]
- 16. Kerim T, Imin N, Weinman JJ, Rolfe BG (2003) Proteome analysis of male gametophyte development in rice anthers. Proteomics 3: 738–751. [DOI] [PubMed] [Google Scholar]
- 17. Sun Q, Hu C, Hu J, Li S, Zhu Y (2009) Quantitative proteomic analysis of CMS-related changes in Honglian CMS rice anther. Protein J 28: 341–348. [DOI] [PubMed] [Google Scholar]
- 18. Dai S, Chen T, Chong K, Xue Y, Liu S, et al. (2007) Proteomics identification of differentially expressed proteins associated with pollen germination and tube growth reveals characteristics of germinated Oryza sativa pollen. Mol Cell Proteomics 6: 207–230. [DOI] [PubMed] [Google Scholar]
- 19. Imin N, Kerim T, Weinman JJ, Rolfe BG (2001) Characterisation of rice anther proteins expressed at the young microspore stage. Proteomics 1: 1149–1161. [DOI] [PubMed] [Google Scholar]
- 20. Sheoran IS, Ross AR, Olson DJ, Sawhney VK (2007) Proteomic analysis of tomato (Lycopersicon esculentum) pollen. J Exp Bot 58: 3525–3535. [DOI] [PubMed] [Google Scholar]
- 21. Mihr C, Baumgärtner M, Dieterich J-H, Schmitz UK, Braun H-P (2001) Proteomic approach for investigation of cytoplasmic male sterility (CMS) in Brassica . J Plant Physiol 158: 787–794. [Google Scholar]
- 22. Sheoran IS, Pedersen EJ, Ross AR, Sawhney VK (2009) Dynamics of protein expression during pollen germination in canola (Brassica napus). Planta 230: 779–793. [DOI] [PubMed] [Google Scholar]
- 23. Sheoran IS, Sawhney VK (2010) Proteome analysis of the normal and Ogura(ogu) CMS anthers of Brassica napus to identify proteins associated with male sterility. Botany 88: 217–230. [Google Scholar]
- 24. Wu S, O’Leary SJ, Gleddie S, Eudes F, Laroche A, et al. (2008) A chalcone synthase-like gene is highly expressed in the tapetum of both wheat (Triticum aestivum L.) and triticale (xTriticosecale Wittmack). Plant Cell Rep 27: 1441–1449. [DOI] [PubMed] [Google Scholar]
- 25. McNeil KJ, Smith AG (2005) An anther-specific cysteine-rich protein of tomato localized to the tapetum and microspores. J Plant Physiol 162: 457–464. [DOI] [PubMed] [Google Scholar]
- 26. Wu HM, Cheun AY (2000) Programmed cell death in plant reproduction. Plant Mol Biol 44: 267–281. [DOI] [PubMed] [Google Scholar]
- 27. Wan L, Zha W, Cheng X, Liu C, Lv L, et al. (2011) A rice beta-1,3-glucanase gene Osg1 is required for callose degradation in pollen development. Planta 233: 309–323. [DOI] [PubMed] [Google Scholar]
- 28. Imin N, Kerim T, Rolfe BG, Weinman JJ (2004) Effect of early cold stress on the maturation of rice anthers. Proteomics 4: 1873–1882. [DOI] [PubMed] [Google Scholar]
- 29. McNeil KJ, Smith AG (2010) A glycine-rich protein that facilitates exine formation during tomato pollen development. Planta 231: 793–808. [DOI] [PubMed] [Google Scholar]
- 30. Kim OK, Jung JH, Park CM (2010) An Arabidopsis F-box protein regulates tapetum degeneration and pollen maturation during anther development. Planta 232: 353–366. [DOI] [PubMed] [Google Scholar]
- 31. Datta R, Chamusco KC, Chourey PS (2002) Starch biosynthesis during pollen maturation is associated with altered patterns of gene expression in maize. Plant Physiol 130: 1645–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang D, Oses-Prieto JA, Li KH, Fernandes JF, Burlingame AL, et al. (2010) The male sterile 8 mutation of maize disrupts the temporal progression of the transcriptome and results in the mis-regulation of metabolic functions. Plant J 63: 939–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang W, Scali M, Vignani R, Milanesi C, Petersen A, et al. (2004) Male-sterile mutation alters Zea m 1 (β-expansin 1) accumulation in a maize mutant. Sex Plant Reprod 17: 41–47. [Google Scholar]
- 34. Ribarits A, Mamun A, Li S, Resch T, Fiers M, et al. (2009) A novel and reversible male sterility system using targeted inactivation of glutamine synthetase and doubled haploidy. Advances in Haploid Production in Higher Plants. 285–294. [Google Scholar]
- 35. Ariizumi T, Hatakeyama K, Hinata K, Sato S, Kato T, et al. (2003) A novel male-sterile mutant of Arabidopsis thaliana, faceless pollen-1, produces pollen with a smooth surface and an acetolysis-sensitive exine. Plant Mol Biol 53: 107–116. [DOI] [PubMed] [Google Scholar]
- 36. Chang RC, So KF (2008) Use of anti-aging herbal medicine, Lycium barbarum, against aging-associated diseases. What do we know so far? Cell Mol Neurobiol 28: 643–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang Y, Zhao H, Sheng X, Gambino PE, Costello B, et al. (2002) Protective effect of Fructus Lycii polysaccharides against time and hyperthermia-induced damage in cultured seminiferous epithelium. J Ethnopharmacol 82: 169–175. [DOI] [PubMed] [Google Scholar]
- 38.Li Q (2001) Healthy functions and medicinal prescriptions of Lycium barbarum (Gou Ji Zi). Jindun Press, 26p. [Google Scholar]
- 39. Li XL, Zhou AG (2007) Evaluation of the antioxidant effects of polysaccharides extracted from Lycium barbarum . Med Chem Res 15: 471–482. [Google Scholar]
- 40. Kaul MLH (1987) Male sterility in higher plants. Springer Verlag: Berlin 1005: 116–117. [Google Scholar]
- 41. Qin K, Tian Y, Li YX, Hong FY, Zhong SY, et al. (2006) Discovery and identification of male sterility Lycium barbarum germplasm YX-1. Acta Botanica Boreali-Occidentalia Sinica 9: 1838–1841. [Google Scholar]
- 42. Xu Q, Qin K, Feng AL, Qi JZ, Han L, et al. (2009) Cytological investigation on anther development of a male sterile and fertile Line in Lycium barbarum L. Journal of Ningxia University (Natural Science Edition). 30: 263–267. [Google Scholar]
- 43. Chen GX, Asada K (1989) Ascorbate peroxidase in tea leaves: occurrence of two isozymes and the differences in their enzymatic and molecular properties. Plant Cell Physiol 30: 987–998. [Google Scholar]
- 44. Oaks A, Stulen I, Jones K, Winspear MJ, Misra S, et al. (1980) Enzymes of nitrogen assimilation in maize roots. Planta 148: 477–484. [DOI] [PubMed] [Google Scholar]
- 45. Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254. [DOI] [PubMed] [Google Scholar]
- 46.Han L (2012) Differential analysis of fertility-related protein in male-sterile Lycium barbarum L. A Master Thesis of Ningxia University.40–45. [Google Scholar]
- 47. Pacini E FG, Hesse M (1985) The tapetum: its form, function and possible phylogeny in embryophyta. Plant Syst Evol 149: 155–185. [Google Scholar]
- 48. Hanson MR, Bentolila S (2004) Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell 16: S154–S169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Levings CS (1993) Thoughts on cytoplasmic male sterility in cms-T maize. Plant Cell 5: 1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Laver H, Reynolds S, Moneger F, Leaver C (1991) Mitochondrial genome organization and expression associated with cytoplasmic male sterility in sunflower (Helianthus annuus). Plant J 1: 185–193. [DOI] [PubMed] [Google Scholar]
- 51. Schnable PS, Wise RP (1998) The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci 3: 175–180. [Google Scholar]
- 52. Nakajima Y, Yamamoto T, Muranaka T, Oeda K (2001) A novel orfB-related gene of carrot mitochondrial genomes that is associated with homeotic cytoplasmic male sterility (CMS). Plant Mol Biol 46: 99–107. [DOI] [PubMed] [Google Scholar]
- 53. De Paepe R, Forchioni A, Chetrit P, Vedel F (1993) Specific mitochondrial proteins in pollen: presence of an additional ATP synthase beta subunit. P Natl A Sci 90: 5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wei L, Fei Z, Wu X, Dong H, Zhou P, et al. (2010) Mitochondrial comparative proteomic analysis of sterile line and its maintain line of purple cytoplasmic rice (Oryza sativa). Adv Biosci Biotech 1: 145–151. [Google Scholar]
- 55. Linke B, Börner T (2005) Mitochondrial effects on flower and pollen development. Mitochondrion 5: 389. [DOI] [PubMed] [Google Scholar]
- 56. Carlsson J, Lagercrantz U, Sundstrom J, Teixeira R, Wellmer F, et al. (2007) Microarray analysis reveals altered expression of a large number of nuclear genes in developing cytoplasmic male sterile Brassica napus flowers. Plant J 49: 452–462. [DOI] [PubMed] [Google Scholar]
- 57. Vizcay-Barrena G, Wilson ZA (2006) Altered tapetal PCD and pollen wall development in the Arabidopsis ms1 mutant. J Exp Bot 57: 2709–2717. [DOI] [PubMed] [Google Scholar]
- 58. Kyo M, Harada H (1986) Control of the developmental pathway of tobacco pollen in vitro. Planta 168: 427–432. [DOI] [PubMed] [Google Scholar]
- 59. Tabuchi M, Sugiyama K, Ishiyama K, Inoue E, Sato T, et al. (2005) Severe reduction in growth rate and grain filling of rice mutants lacking OsGS1;1, a cytosolic glutamine synthetase1;1. Plant J 42: 641–651. [DOI] [PubMed] [Google Scholar]
- 60. Martin A, Lee J, Kichey T, Gerentes D, Zivy M, et al. (2006) Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. Plant Cell 18: 3252–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ribarits A, Mamun A, Li S, Resch T, Fiers M, et al. (2007) Combination of reversible male sterility and doubled haploid production by targeted inactivation of cytoplasmic glutamine synthetase in developing anthers and pollen. Plant Biotechnol J 5: 483–494. [DOI] [PubMed] [Google Scholar]
- 62. Jiang P, Zhang X, Zhu Y, Zhu W, Xie H, et al. (2007) Metabolism of reactive oxygen species in cotton cytoplasmic male sterility and its restoration. Plant Cell Rep 26: 1627–1634. [DOI] [PubMed] [Google Scholar]
- 63. Wan C, Li S, Wen L, Kong J, Wang K, et al. (2007) Damage of oxidative stress on mitochondria during microspores development in Honglian CMS line of rice. Plant Cell Rep 26: 373–382. [DOI] [PubMed] [Google Scholar]
- 64. Dong X, Hong Z, Sivaramakrishnan M, Mahfouz M, Verma DPS (2005) Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant J 42: 315–328. [DOI] [PubMed] [Google Scholar]
- 65. Warmke H, Overman M Ann (1972) Cytoplasmic male sterility in Sorghum I. Gallose behavior in fertile and sterile anthers. J Hered 63: 103–108. [Google Scholar]
- 66. Worrall D, Hird DL, Hodge R, Paul W, Draper J, et al. (1992) Premature dissolution of the microsporocyte callose wall causes male sterility in transgenic tobacco. Plant Cell 4: 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shen JB, Hsu FC (1992) Brassica anther-specific genes: characterization and in situ localization of expression. Mol Gen Genet 234: 379–389. [DOI] [PubMed] [Google Scholar]
- 68. van der Meer IM, Stam ME, van Tunen AJ, Mol JN, Stuitje AR (1992) Antisense inhibition of flavonoid biosynthesis in petunia anthers results in male sterility. Plant Cell 4: 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Napoli CA, Fahy D, Wang HY, Taylor LP (1999) white anther: A Petunia mutant that abolishes pollen flavonol accumulation, induces male sterility, and is complemented by a chalcone synthase transgene. Plant Physiol 120: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mo Y, Nagel C, Taylor LP (1992) Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proc Natl Acad Sci U S A 89: 7213–7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yang S, Terachi T, Yamagishi H (2008) Inhibition of chalcone synthase expression in anthers of Raphanus sativus with Ogura male sterile cytoplasm. Ann Bot 102: 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Atanassov I RE, Antonov L, Atanassov A (1998) Expression of an anther-specific chalcone synthase-like gene is correlated with uninucleate microspore development in Nicotiana sylvestris . Plant Mol Biol 38: 10. [DOI] [PubMed] [Google Scholar]
- 73. Dobritsa AA, Lei Z, Nishikawa S, Urbanczyk-Wochniak E, Huhman DV, et al. (2010) LAP5 and LAP6 encode anther-specific proteins with similarity to chalcone synthase essential for pollen exine development in Arabidopsis. Plant Physiol 153: 937–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mou Z, He Y, Dai Y, Liu X, Li J (2000) Deficiency in fatty acid synthase leads to premature cell death and dramatic alterations in plant morphology. Plant Cell 12: 405–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Shi J, Tan H, Yu XH, Liu Y, Liang W, et al. (2011) Defective pollen wall is required for anther and microspore development in rice and encodes a Fatty acyl carrier protein reductase. Plant Cell 23: 2225–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ferl RJ, Manak MS, Reyes MF (2002) The 14-3-3s. Genome Biol 3: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bridges D, Moorhead GB (2005) 14-3-3 proteins: a number of functions for a numbered protein. Sci STKE 2005: re10. [DOI] [PubMed] [Google Scholar]
- 78. Sehnke PC, Ferl RJ (2000) Plant 14-3-3s: omnipotent metabolic phosphopartners? Sci STKE 2000: pe1. [DOI] [PubMed] [Google Scholar]
- 79. Kaiser M, Ottmann C (2010) The first small-molecule inhibitor of 14-3-3s: modulating the master regulator. Chembiochem 11: 2085–2087. [DOI] [PubMed] [Google Scholar]
- 80. Sehnke PC, Chung HJ, Wu K, Ferl RJ (2001) Regulation of starch accumulation by granule-associated plant 14-3-3 proteins. Proc Natl Acad Sci U S A 98: 765–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Oh CS, Pedley KF, Martin GB (2010) Tomato 14-3-3 protein 7 positively regulates immunity-associated programmed cell death by enhancing protein abundance and signaling ability of MAPKKKα. Plant Cell 22: 260–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10: 79–87. [DOI] [PubMed] [Google Scholar]
- 83. Jiang SY, Cai M, Ramachandran S (2007) ORYZA SATIVA MYOSIN XI B controls pollen development by photoperiod-sensitive protein localizations. Dev Biol 304: 579–592. [DOI] [PubMed] [Google Scholar]
- 84. Ge X, Dietrich C, Matsuno M, Li G, Berg H, et al. (2005) An Arabidopsis aspartic protease functions as an anti-cell-death component in reproduction and embryogenesis. EMBO Rep 6: 282–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kurepa J, Smalle JA (2008) Structure, function and regulation of plant proteasomes. Biochimie 90: 324–335. [DOI] [PubMed] [Google Scholar]
- 86. van der Hoorn RA (2008) Plant proteases: from phenotypes to molecular mechanisms. Annu Rev Plant Biol 59: 191–223. [DOI] [PubMed] [Google Scholar]
- 87. Zhao D, Han T, Risseeuw E, Crosby WL, Ma H (2003) Conservation and divergence of ASK1 and ASK2 gene functions during male meiosis in Arabidopsis thaliana . Plant Mol Biol 53: 163–173. [DOI] [PubMed] [Google Scholar]
- 88. Li N, Zhang DS, Liu HS, Yin CS, Li X, et al. (2006) The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Aguirre PJ, Smith AG (1993) Molecular characterization of a gene encoding a cysteine-rich protein preferentially expressed in anthers of Lycopersicon esculentum . Plant Mol Biol 23: 477–487. [DOI] [PubMed] [Google Scholar]
- 90. McCormack E, Braam J (2003) Calmodulins and related potential calcium sensors of Arabidopsis. New Phytol 159: 585–598. [DOI] [PubMed] [Google Scholar]
- 91. Jung KH, Han MJ, Lee YS, Kim YW, Hwang I, et al. (2005) Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell 17: 2705–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Benedetti CE, Costa CL, Turcinelli SR, Arruda P (1998) Differential expression of a novel gene in response to coronatine, methyl jasmonate, and wounding in the Coi1 mutant of Arabidopsis. Plant Physiol 116: 1037–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]