SUMMARY
Olfactory stimuli are detected by over 1,000 odorant receptor genes in mice, with each receptor being mapped to specific glomeruli in the olfactory bulb. The Trace Amine-Associated Receptors (TAARs) are a small family of evolutionarily conserved olfactory receptors whose contribution to olfaction remains enigmatic. Here, we show that a majority of the TAARs are mapped to a discrete subset of glomeruli in the dorsal olfactory bulb of the mouse. This TAAR projection is distinct from the previously described class I and class II domains, and is formed by a sensory neuron population that is restricted to express TAAR genes prior to choice. We further show that the dorsal TAAR glomeruli are selectively activated by low concentrations of amines. Our data uncover a hard-wired, parallel input stream in the main olfactory pathway that is specialized for the detection of volatile amines.
INTRODUCTION
Information about volatile chemicals (odorants) in the environment is transduced by odorant receptors (ORs) expressed by olfactory sensory neurons (OSNs) in the olfactory epithelium (Buck and Axel, 1991). Each OSN expresses one allele of a single OR gene (Chess et al., 1994; Malnic et al., 1999; Serizawa et al., 2004). The expressed receptor dictates odorant responsiveness (Bozza et al., 2002; Zhao et al., 1998) and influences axon guidance (Feinstein and Mombaerts, 2004; Mombaerts et al., 1996; Serizawa et al., 2006; Wang et al., 1998). OSNs expressing a given OR send convergent axonal projections to defined glomeruli in the main olfactory bulb (Mombaerts et al., 1996; Ressler et al., 1994; Vassar et al., 1994), thereby mapping the OR repertoire onto the glomerular array.
In order to detect structurally diverse odorants, mammalian OR repertoires are typically large (Niimura, 2009; Zhang and Firestein, 2009). Mice have over 1,000 ORs divided into two phylogenetically distinct groups: ~150 class I ORs, and >900 class II ORs (Freitag et al., 1995; Glusman et al., 2001; Zhang and Firestein, 2002). OSNs in the dorsal epithelium express most of the class I ORs and a subset of class II ORs (Zhang et al., 2004; Tsuboi et al., 2006). The axons of these intermingled populations segregate and form glomeruli in two discrete domains (DI and DII) in the dorsal bulb (Bozza et al., 2009; Kobayakawa et al., 2007; Tsuboi et al., 2006; Zhang et al., 2004). The DI and DII domains can be observed functionally and may play a role in representing odor quality (Bozza et al., 2009; Kobayakawa et al., 2007; Matsumoto et al., 2010).
The trace amine-associated receptors (TAARs) were identified as a second class of chemosensory receptors (Liberles and Buck, 2006) that are evolutionarily conserved in vertebrates, including humans (Lindemann et al., 2005). While this conservation suggests a critical role in olfaction, the contribution of this small receptor repertoire to olfactory function is unknown. The mouse has 15 TAAR genes, 14 of which are expressed in the main olfactory epithelium (Liberles and Buck, 2006). TAARs are phylogenetically related to biogenic amine receptors (Borowsky et al., 2001; Bunzow et al., 2001; Grandy, 2007) and respond selectively to amines in heterologous systems (Ferrero et al., 2011; Ferrero et al., 2012; Liberles and Buck, 2006; Staubert et al., 2010; Zucchi et al., 2006). Some TAARs respond to odorants that could serve as social cues and/or predator-derived signals (Ferrero et al., 2011; Liberles and Buck, 2006). However, nothing is known about the wiring and functional properties of TAAR-expressing OSNs in vivo.
We used gene targeting and optical imaging to visualize and functionally characterize TAAR projections in the mouse olfactory system. We find that a majority of the TAARs are mapped to a cluster of glomeruli located between the previously characterized DI and DII domains in the dorsal olfactory bulb. The TAAR projection is associated with a distinct subset of OSNs that is biased to choose TAARs over other ORs, similar to the bias observed for OSNs expressing class I and class II ORs. We also provide evidence that this bias in gene expression is already established prior to initial receptor gene choice. Using in vivo optical imaging, we show that the dorsal TAAR glomeruli are selectively activated by low concentrations of volatile amines. Our data reveal that the TAARs feed into a genetically and anatomically distinct olfactory input stream that is selectively activated by volatile amines.
RESULTS
We characterized the expression of all of the individual TAAR genes in the olfactory epithelium by in situ hybridization. Ten of the fourteen TAARs (Taar2, 3, 4, 5, 7e, 7f, 8a, 8b, 8c and 9) were expressed specifically in the dorsal epithelium (Figure S1) and are referred to here as “dorsal TAARs”. In contrast, Taar 7a and 7b were expressed predominantly in the ventral epithelium, named here as “ventral” TAARs. Taar6 and 7d were expressed in both zones, described here as “broad TAARs”. Thus, a majority of TAARs are expressed in the dorsal epithelium, which suggested to us that they might project to glomeruli in the dorsal olfactory bulb.
TAAR-expressing OSNs project to glomeruli in the dorsal olfactory bulb
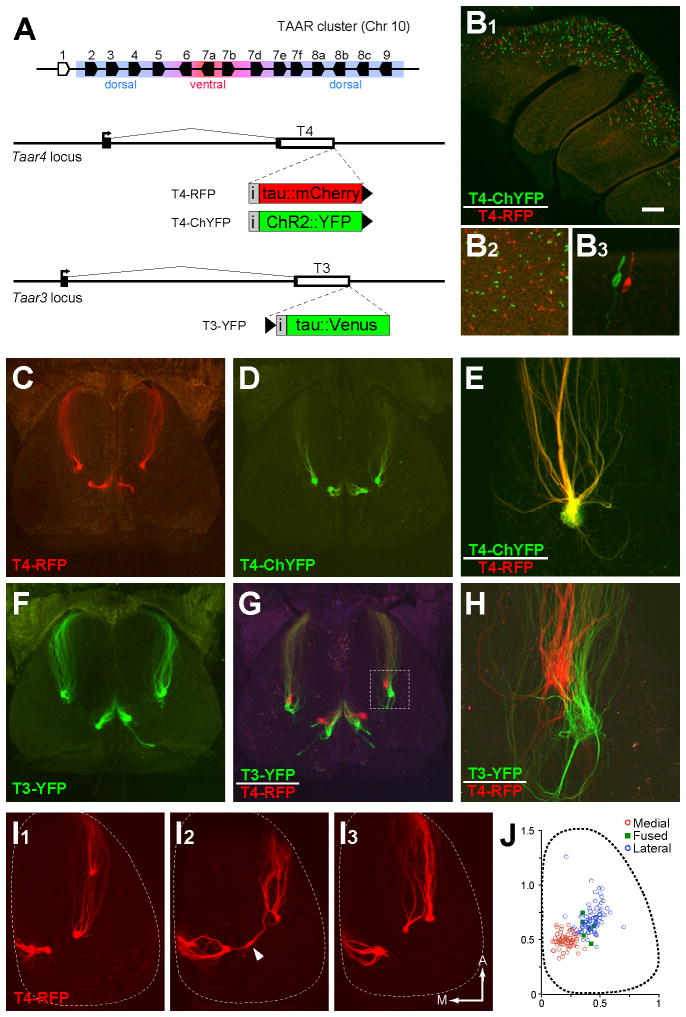
To directly visualize TAAR-expressing OSNs and their projections, we modified two dorsal TAAR genes, Taar3 (T3) and Taar4 (T4), by gene targeting to drive co-expression of fluorescent markers, tauVenus (YFP), tauCherry (RFP) or channelrhodopsin2-YFP (ChYFP). We generated three strains of gene-targeted mice: T3-YFP, T4-RFP, and T4-ChYFP (Figure 1A). Each strain exhibited spatially distributed subsets of labeled OSNs in the dorsal epithelium (Figure 1B). In mice harboring alternately tagged T4 alleles, we found no double-labeled cells (n=2,588 OSNs; Figure 1B) indicating that TAARs are subject to monoallelic expression, like classical ORs.
Figure 1. Gene targeting reveals TAAR projections to the olfactory bulb.
(A) (Top) Diagram of the TAAR gene cluster. Polygons indicate gene orientation; olfactory TAARs are shown in black, non-olfactory Taar1 in white. Expression in the dorsal or ventral epithelium is indicated. (Bottom) Structure of Taar3 and Taar4 loci showing coding sequences (white boxes), non-translated regions (black boxes) and transcription start sites (arrows). Targeted insertions contain an internal ribosome entry site (grey box marked, “i”) and coding sequences for fluorescent markers (colored boxes). Black triangles indicate the location and orientation of loxP sites.
(B1) Medial view of olfactory epithelium in a T4-RFP/T4-ChYFP mouse. (B2) Higher magnification shows that OSNs are either green or red. (B3) Detailed view of an epithelial section showing two neighboring OSNs, each expressing one of the two Taar4 alleles.
(C) Dorsal view of the olfactory bulbs of a heterozygous T4-RFP mouse showing axonal projections and glomeruli formed by tagged OSNs.
(D) Dorsal view of the bulbs of a heterozygous T4-ChYFP mouse.
(E) High magnification image of a lateral glomerulus in a compound heterozygous T4-RFP/T4-ChYFP mouse. Axons coalesce into the same glomerulus.
(F) Dorsal view of the bulbs of a heterozygous T3-YFP mouse.
(G) Dorsal view of the bulbs of a double heterozygous T3-YFP/T4-RFP mouse. Boxed area is shown in (H).
(H) Higher magnification image of lateral glomeruli in T3-YFP/T4-RFP mice.
(I) Right olfactory bulbs from three T4-RFP mice illustrating different patterns of glomerular convergence by T4 axons. Arrowhead points to a “fused” glomerulus (receiving both lateral and medial innervation). Anterior and medial are indicated.
(J) Positions of medial (red circles), lateral (blue circles), and “fused” glomeruli (green squares) taken from the left and right olfactory bulb across animals (n=62 bulbs, ages P15 to P75). Glomerular positions were measured with respect to the length and width of the bulbs and projected onto a bulb of average size. Scale bar = 200 μm in B1, 65 μm in B2, 20 μm in B3, 350 μm in C, D, F and G, 90 μm in E, 80 μm in H, and 250 μm in I.
See also Figures S1 and S2.
OSNs expressing T3 and T4 project to glomeruli in the dorsal aspect of the main olfactory bulb (Figure 1C–1H). T3 and T4 axons form neighboring but distinct glomeruli (Figure 1G and 1H). Axons from OSNs expressing the genetically tagged alleles intermingle with unlabeled axons in heterozygous mice, but not in homozygous mice (Figure S2), suggesting that the genetically tagged axons exhibit normal targeting with corresponding wild-type axons (Bozza et al., 2002; Vassalli et al., 2002). In addition, T4 axons expressing RFP or ChYFP co-innervated the same glomeruli (Figure 1E), indicating that the markers do not differentially affect axon guidance.
As with canonical ORs (Mombaerts et al., 1996), T3 and T4 axons have medial and lateral projections to each bulb (Figures 1C and 1F). However, we note several characteristic features of TAAR projections. First, the medial and lateral glomeruli are closer to each other than is typically observed for dorsal ORs: T4 glomeruli were separated by 491 μm on average (range= 306–666 μm, n=23 bulbs). By comparison, glomeruli for class I projections are typically separated by roughly 700–1000 μm (data not shown). Second, in T4-RFP mice, 6 out of 62 bulbs had a glomerulus that received input from both medial and lateral axons. These “fused” glomeruli were usually located between the medial and lateral convergence sites (Figure 1I). Third, the incidence of supernumerary glomeruli (>2 per bulb) was higher for T4 than other ORs; the average number of glomeruli per bulb was 2.5 ± 0.5 (1.1 ± 0.3 medial; 1.3 ± 0.5 lateral; 0.1 ± 0.2 fused; mean ± SD; n=66 bulbs) in mice ranging from P15 to P75. No difference was observed at P15, P30, P45, P60 and P75 (data not shown). Despite this overall variability, T4 glomeruli were restricted to a circumscribed region of the caudal bulb (Figure 1J). Similar projection patterns were seen for Taar5 and Taar9 in gene-targeted mice (data not shown), suggesting that the dorsal TAARs might map to a discrete caudal region on the dorsal bulb.
TAAR-expressing OSNs form a third dorsal projection (DIII)
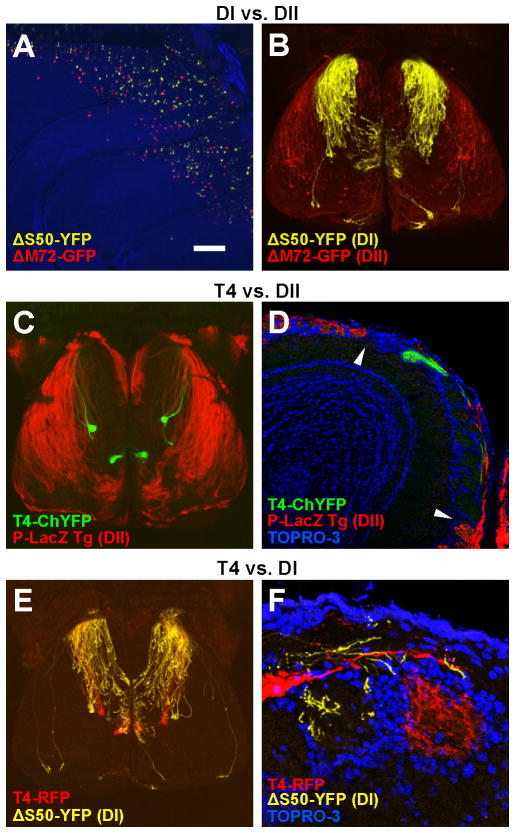
The dorsal bulb has been previously divided into two known projections or domains, DI and DII (Figure 2A and 2B). To examine whether TAAR-expressing OSNs project to the DI or DII domains, we crossed T4-ChYFP mice with the transgenic marker P-LacZ (Figure 2C), which drives β-galactosidase expression in a large proportion of class II-expressing OSNs and labels DII (Bozza et al., 2009; Vassalli et al., 2011). In these mice, T4 glomeruli were located in the LacZ-negative region, medial to DII (Figure 2C and 2D). We also crossed T4-RFP mice to ΔS50-YFP animals, in which the DI domain is labeled. In these mice, T4 glomeruli were located near the caudal margin of DI (Figure 2E and 2F), often partially intermingled with YFP-labeled, class I glomeruli. Despite being closely associated with DI, significantly fewer T4 glomeruli were innervated by ΔS50 axons than glomeruli for a model class I OR, MOL2.3 (data not shown). This suggested that T4 glomeruli have a different axon guidance identity than DI glomeruli.
Figure 2. T4 glomeruli lie outside DII and are closely associated with DI.
(A) Medial view of the olfactory epithelium showing overlapping distribution of ΔS50 (class I) and ΔM72 (class II) OSNs in the dorsal epithelium.
(B) Dorsal view of the olfactory bulbs in which ΔS50 and ΔM72 axons innervate DI (yellow) and DII (red).
(C) Dorsal view of the bulbs of a P-LacZ Tg+; T4-ChYFP heterozygous mouse showing DII axons (red) and T4 glomeruli (green). T4 glomeruli fall outside of DII.
(D) Coronal section through the bulb of a P-LacZ Tg+; T4-ChYFP heterozygous mouse showing a T4 glomerulus (green) in the LacZ-negative region. The boundary between this region and DII (red) is indicated by arrowheads. Nuclei are stained with TOPRO-3 (blue).
(E) Dorsal view of the bulbs of a ΔS50-YFP; T4-RFP double heterozygous mouse showing the DI axons (yellow) and T4 glomeruli (red).
(F) Coronal section showing absence of innervation of T4 glomerulus (red) by ΔS50-YFP axons (yellow). Counterstained with TOPRO-3 (blue).
Scale bar = 260 μm in A; 500 μm in B, C, E; 130 μm in D; 30 μm in F.
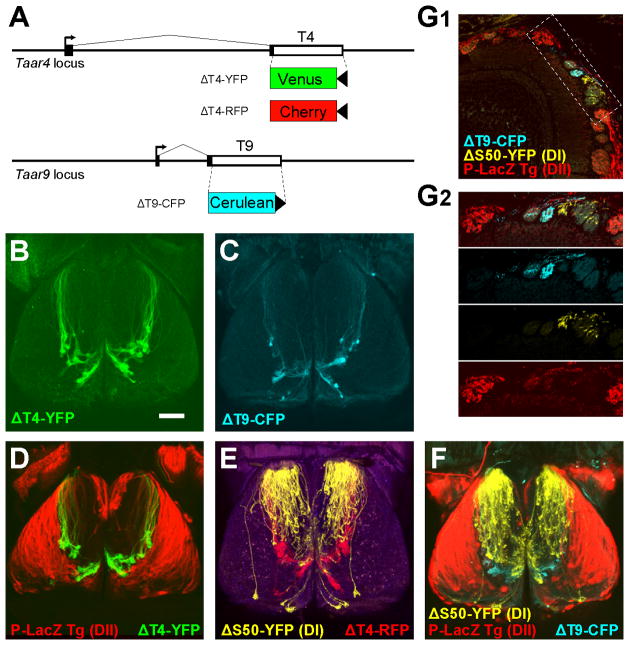
If TAAR glomeruli are part of the DI projection, then TAAR-deletion alleles should diffusely label the entire DI domain, as is seen for class I OR deletions (Bozza et al., 2009). To test this, we generated three gene-targeted mouse strains with TAAR coding sequence deletions: ΔT4-YFP, in which the Taar4 coding sequence is replaced with Venus YFP; ΔT4-RFP in which the Taar4 coding sequence is replaced with gap-cherry; ΔT9-CFP, in which the Taar9 coding sequence is replaced with Cerulean CFP (Figure 3A). Strikingly, OSNs expressing these TAAR-deletion alleles densely innervate a small cluster of glomeruli in the dorsal bulb (Figure 3B and 3C). The cluster comprised 19 ± 3 glomeruli (mean ± SD; n=8 bulbs) in heterozygous ΔT4-YFP mice. This pattern is in stark contrast to class I and class II OR-deletions, which exhibit sparse labeling of a large number of glomeruli (Figure 2B; Bozza et al., 2009). When crossed to the class I and class II markers, the ΔT4- and ΔT9-labeled glomeruli were distinct from DI and DII glomeruli (Figure 3D–3G). Thus, the TAAR-deletion alleles reveal a new dorsal projection, named here DIII, that appears distinct from DI and DII.
Figure 3. TAAR coding sequence deletions reveal a third domain.
(A) Gene targeting to generate three TAAR-deletion alleles. Non-coding regions are shown as black boxes. Transcription start sites shown as arrows. The coding sequence (white box) of Taar4 is replaced with either Venus YFP (yellow) or gap-Cherry (red); Taar9 is replaced with Cerulean CFP. LoxP sites are shown as black triangles.
(B) Dorsal view of the olfactory bulbs of a heterozygous ΔT4-YFP mouse showing a cluster of glomeruli (green) located in the caudal bulb.
(C) Dorsal view of the bulbs of a homozygous ΔT9-CFP mouse.
(D) Dorsal view of the bulbs of a P-LacZ Tg+; ΔT4-YFP heterozygous mouse. The axons and glomeruli labeled by ΔT4-YFP axons (green) are located outside of the DII domain (red).
(E)Dorsal view of the bulbs of a ΔS50-YFP; ΔT4-RFP double heterozygous mouse showing that the glomeruli labeled by the ΔT4-RFPallele (red) are distinct from DI (yellow).
(F) Dorsal view of the bulbs of a P-LacZ Tg+; ΔS50-YFP; ΔT9-CFPtriple-mutant mouse showing that the region labeled by the T9-CFPallele (cyan) is largely separate from DI (yellow) and DII (red).
(G) Coronal section through the bulb of a P-LacZ Tg+; ΔS50-YFP; ΔT9-CFPtriple-mutant showing that ΔT9 axons (cyan) are outside of DI (yellow) and DII (red). DII glomeruli are densely innervated by LacZ-positive axons (red). (G2) Higher magnification view of the area indicated in (G1).
Scale bar = 500 μm in B–F; 120 μm in G1, 80 μm in G2.
TAAR-expressing OSNs are biased to co-express TAAR genes
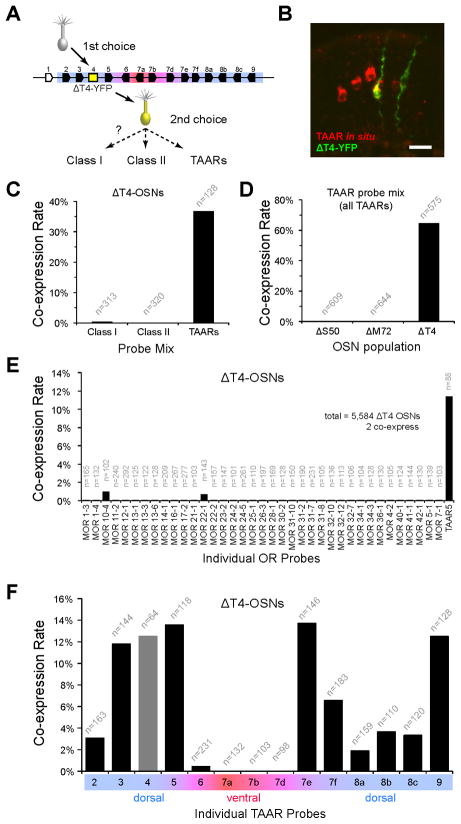
The DI and DII projections are generated by two types of OSNs (d1 and d2) which selectively expresses class I or class II genes, respectively (Bozza et al., 2009). The dense labeling of relatively few glomeruli by ΔT4-OSNs is consistent with the idea that they also exhibit biased co-expression from a limited repertoire of OR genes—perhaps the TAARs. To test this, we quantified co-expression in ΔT4-YFP OSNs by combined immunohistochemistry/in situ hybridization using equally sized class-specific probe mixes (Figure 4A and 4B). Indeed, ΔT4-YFP OSNs were much more likely to be labeled by the TAAR mix than the class I or class II mixes (TAAR=37%, 47/128 cells; class I=0.3%, 1/313 cells; class II: 0/320 cells; Fisher’s Exact test, p<0.001; Figure 4C). Because we observed low levels of class I OR expression, we tested 37 individual class I OR probes. The data show that class I co-expression occurs rarely (Figure 4E).
Figure 4. ΔT4-YFP OSNs selectively co-express TAARs.
(A) Schematic of gene choice in ΔT4-OSNs. A nascent OSN (grey) chooses the ΔT4 allele (yellow box) and is labeled (yellow cell), does not express a functional receptor (grey cilia), and makes an alternate choice (dashed arrows).
(B) Epithelial section from a ΔT4-YFP mouse. OSNs expressing the ΔT4-YFP allele are labeled (green) by immunohistochemistry for YFP; TAAR-expressing OSNs are labeled (red) by in situ hybridization using a TAAR probe mix. Double-labeled OSNs (yellow) co-express ΔT4-YFP and a dorsal TAAR gene. Scale bar = 20 μm.
(C) Co-expression rates for class I ORs, class II ORs, and TAARs in ΔT4-OSNs. Each class is represented by a mixture of 5 dorsally expressed probes (see Experimental Procedures).
(D) Co-expression rates for TAARs in class I OR-, class II OR-, and TAAR-deletion OSNs (ΔS50, ΔM72 and ΔT4) using the probe mix for all TAARs. The higher co-expression rate for ΔT4 compared with (D) is expected given that this probe mix includes all 14 TAAR genes.
(E) Co-expression rates for 37 individual OR probes and a Taar5 probe in ΔT4-OSNs.
(F) Co-expression rates for individual TAAR genes inΔT4-OSNs. Co-expression was observed for all dorsal TAARs. Co-expression of ventral/broad TAAR genes was rare. The rate for Taar4 (grey bar) was measured in heterozygous (ΔT4-YFP/wt) mice, which retain one intact copy of Taar4. All data are pooled from counts using at least two animals. See also Figures S1 and S3.
If TAARs are selectively expressed by a third OSN-type, then d1- and d2-OSNs should not co-express the TAAR genes. To test this prediction, we quantified TAAR co-expression in ΔS50-YFP (class I) and ΔM72-GFP (class II) OSNs using a probe mix that hybridizes to all 14 olfactory TAARs (Figure 4D). As predicted, we observed very low levels of co-expression (or no co-expression) of TAARs in class I and class II OR-deletion neurons (1/609 ΔS50-OSNs; 0/644 ΔM72-OSNs). In contrast, a large proportion of ΔT4-YFP OSNs was labeled by the same TAAR probe mix (64.5%, 371/575 ΔT4 OSNs; Fisher’s exact test, p<0.001). We note that this is less than 100% that one might predict (see Discussion). Overall, these data indicate that the d1- and d2-OSNs are strongly biased against expressing TAAR genes, consistent with the idea that TAARs are chosen selectively by a specific OSN-type (d3).
Using gene-specific probes, we observe that ΔT4-YFP OSNs selectively co-express the dorsal TAARs (Taar2, 3, 4, 5, 7e, 7f, 8a, 8b, 8c and 9).Co -expression rates ranged from 1.9 to 13.7% (Figure 4F). This is up to 20-fold higher than rates observed for OR co-expression in class I and class II OR-deletions (Bozza et al., 2009), further suggesting that ΔT4-OSNs choose from a smaller repertoire of potential alleles. Co-expression of the ventral and broad TAARs, however, was either infrequent or not observed. This likely reflects the normal paucity (or absence) of expression of these genes in the dorsal epithelium (Figure S1), where ΔT4-OSNs are located. Thus, as a population, ΔT4-OSNs are biased to co-express the 10 dorsal TAAR genes.
OSNs expressing deletion alleles project to glomeruli corresponding to the co-expressed OR (Feinstein et al., 2004; Lewcock and Reed, 2004). Similarly, we observe that ΔT4-YFP OSNs that co-express the T4-RFP allele innervate T4-RFP glomeruli (Figure S3). Thus, it is likely that the glomeruli revealed in ΔT4-YFP mice correspond to all 10 of the dorsal TAAR genes.
Biased co-expression of TAARs is not a cluster effect
All of the TAAR genes are located in a single cluster on chromosome 10. Thus, biased co-expression could be caused by preferential choice of genes in cis to the deletion allele (Lewcock and Reed, 2004) rather than the presence of a biased OSN-type. We therefore investigated the contribution of cis co-expression to the TAAR co-expression bias. To do this, we generated a TAAR cluster deletion allele (ΔT2-9) in which a ~250kb genomic interval encompassing Taar2 through Taar9 was excised (Figure 5A; see also Extended Experimental Procedures). We then crossed ΔT2–9 to the ΔT4-YFP strain, thereby removing all of the olfactory TAAR genes that are in trans to the ΔT4-YFP allele.
Figure 5. Biased TAAR co-expression is not a cluster effect.
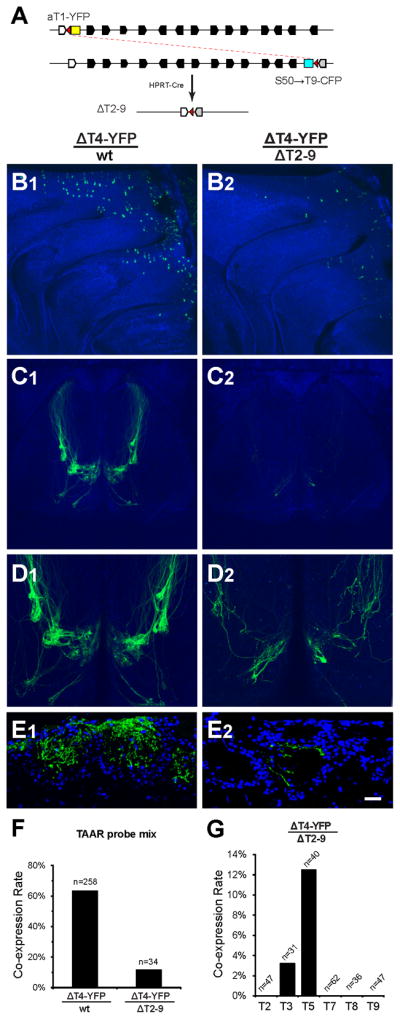
(A) In vivo recombination strategy to generate the TAAR cluster deletion allele. Black polygons show location and orientation of olfactory TAAR genes. Non-olfactory Taar1 is shown in white. Two targeted mutations, aT1-YFP and S50→T9-CFP, introduce loxP sites (red triangles) into both ends of the cluster. When crossed in the presence of HPRT-Cre, F1 mice harboring the cluster deletion, ΔT2-9, can be recovered. S50 coding sequence shown as grey polygon, CFP as cyan box, YFP as yellow box. Both fluorescent markers and all olfactory TAAR genes are absent from the ΔT2-9 allele.
(B) Medial views of the olfactory epithelium from a control ΔT4-YFP/wt (B1) mouse and a ΔT4-YFP/ΔT2–9 littermate (B2). OSNs expressing the ΔT4-YFP allele can be seen in the dorsal epithelium (green). There is a decrease in the number of OSNs expressing the ΔT4-YFP allele in the absence of transco -expression. Both images were scanned using the same gain settings. Blue is background autofluorescence.
(C) Dorsal views of olfactory bulbs from control ΔT4-YFP/wt (C1) and ΔT4-YFP/ΔT2-9 littermate (C2) mice. The pattern of intensely labeled glomeruli (green) normally seen (C1) is severely reduced in the absence of transco -expression (C2). Both images were scanned using the same settings.
(D) High magnification image of the same bulbs in (C). The image (D2) was scanned at higher gain.
(E) Coronal sections of glomeruli in ΔT4-YFP/wt (E1) and a ΔT4-YFP/ΔT2–9 (E2).Glomeruli in DIII are densely innervated by ΔT4-YFP axons in ΔT4-YFP/wt mice. Only sparse innervation is seen in the absence of the trans cluster. Sections are counterstained with the nuclear label TOPRO-3 (blue).
(F) Co-expression rates for all TAARs in ΔT4 -OSNs, in the presence (ΔT4/wt) or absence (ΔT4-YFP/ΔT2-9) of the wild-type TAAR cluster in trans. Co-expression is drastically reduced when the choice of TAAR alleles in trans is no longer available.
(G) Co-expression rates for TAAR genes in ΔT4-OSNs when the trans cluster is absent (only alleles in cis are available). Co-expression is biased towards the genes immediately flanking the ΔT4-YFP allele. T7 and T8 indicate coding-sequence probes for Taar7a and Taar8b, which hybridize to all members of their respective subfamilies.
Scale bar = 200 μm in B and D, 400 μm in C, and 30 μm in E.
The absence of the trans TAAR cluster had a dramatic effect on the population of ΔT4-YFP OSNs. First, the number of labeled ΔT4-OSNs was reduced and the dense labeling of spatially circumscribed TAAR glomeruli was eliminated (Figure 5B–5E). Second, the rate of TAAR co-expression was decreased (ΔT4-YFP/wt: 64 %; ΔT4-YFP/ΔT2-9: 12%; Fisher’s exact test, p<0.0001; Figure 5F), suggesting that co-expression in cis does not readily occur, or is not readily detectable. Finally, the rare co-expression that did occur in cis was observed only for the genes immediately flanking the ΔT4-YFP locus—Taar3 and Taar5 (Figure 5G). This positional bias in co-expression is not observed in the presence of the trans cluster (Figure 4F). Taken together, the data indicate that co-expression in ΔT4-OSNs occurs preferentially in trans, and that a cluster effect cannot account for the gene expression bias that we observe. Rather, we propose that the bias in gene choice is an inherent property of the OSNs that choose from the TAAR repertoire.
TAAR-expressing OSNs show biased “first” choice
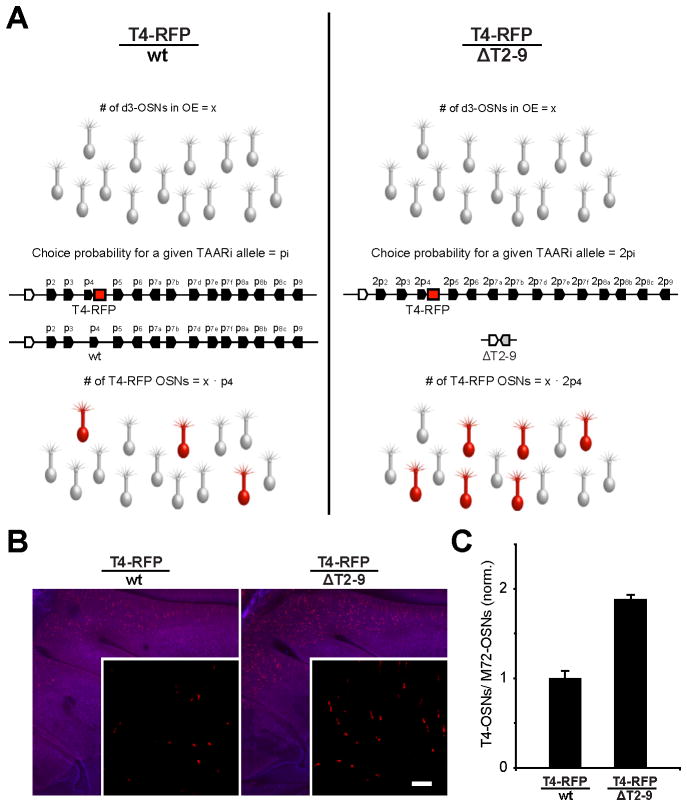
A hallmark of our model is that different OSN-types exhibit biased gene choice. However, we have only observed biases in alternate choice (i.e. co-expression in OSNs that have first chosen an OR-deletion locus). It has not been possible to observe biases in first choice. Taking advantage of the small TAAR gene family, we looked for evidence of biased first choice by examining the effect of gene dosage on the probability of TAAR gene expression (Figure 6A). Aside from TAARs, it is estimated that 400 OR genes are expressed in the dorsal epithelium (Zhang et al., 2004). If each dorsal OSN initially chooses from all dorsal genes (i.e. there are no OSN-types), then removing one copy of the TAAR cluster (14 out of ~800 available alleles) would have a negligible effect on the number of OSNs expressing any of the remaining TAAR alleles. On the other hand, if there were a fixed population of OSNs restricted to express TAARs (i.e. a d3-OSN type), removing one copy of the TAAR cluster (14 out of 28 available alleles) should double the number of OSNs that express any of the remaining TAAR alleles (Figure 6A).
Figure 6. Reducing the number of TAAR alleles increases the probability of T4-RFP expression.
(A) Diagram of the hypothesis that there is a population of fixed size, d3-OSNs (grey neurons), that is restricted to choose among the TAAR alleles. (Left) In the presence of both copies of the TAAR cluster, each individual allele, i, has a specific probability of expression, pi. In heterozygous T4-RFP animals, a subset of d3-OSNs express the tagged allele as dictated by its probability, pT4. (Right) In the absence of the trans cluster (i.e. half the number of available TAAR alleles), the probability of choosing the remaining Taar4 allele (T4-RFP) and the number of RFP-labeled OSNs in the epithelium would double. If OSNs can choose among all ~800 dorsal OR alleles (~400 genes), the loss of 14 alleles should have a negligible effect on choice probability.
(B) Wholemount views of the olfactory epithelia in T4-RFP/wt and T4-RFP/ΔT2-9 mice. Insets show higher magnification. Scale bar = 400 μm, 50 μm in insets.
(C) Normalized counts of RFP-labeled OSNs in epithelial sections of T4-RFP/wt, and T4-RFP/ΔT2-9 mice. The number of T4-RFP OSNs was normalized to a dorsal class II population, M72-GFP, in the same sections. The number of OSNs expressing the T4-RFP allele nearly doubles in the absence of the trans TAAR cluster (error bars show standard deviations).
To distinguish between these possibilities, we compared the number of OSNs expressing the T4-RFP allele in the presence or absence of the trans TAAR cluster. Indeed, the number of T4-RFP OSNs nearly doubled when the repertoire of available TAAR alleles was reduced by half (1.9 ± 0.05-fold increase; mean ± SD; n=4 mice per genotype; Figure 6B and 6C). Similar data were obtained for T3 (data not shown). Our interpretation is that there is a fixed population of neurons (an OSN-type) that is restricted to choose from the TAAR repertoire prior to first choice.
Taken together, our anatomical and genetic data demonstrate that the ΔT4 mutation marks a third OSN-type, d3, which creates a third dorsal projection, DIII, comprising glomeruli corresponding to all of the dorsally expressed TAAR genes.
The DIII glomeruli form an amine-selective domain
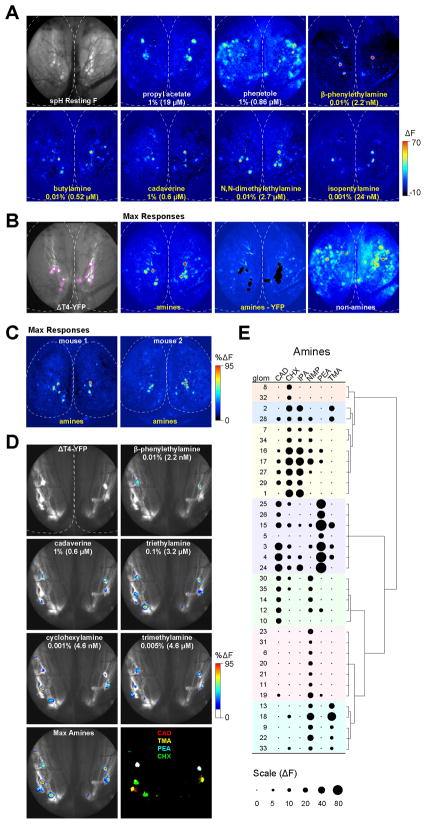
The TAARs respond preferentially to amines in heterologous expression systems (Liberles and Buck, 2006). However, nothing is known about the function of the olfactory TAARs in vivo. To examine their functional properties, we imaged odor-evoked activity from the olfactory bulbs of mice expressing the genetically encoded activity reporter synaptop Hluorin in all mature OSNs (Bozza et al., 2004). By recording from mice that were also heterozygous for the ΔT4-YFP allele, we could directly visualize the DIII glomeruli (including T4). We asked whether amines preferentially activate the DIII glomeruli by using the lowest concentration of amines that elicited robust responses across animals (determined experimentally).
Surprisingly, we found that low concentrations of amines selectively activated a few highly sensitive glomeruli clustered in the dorsal, caudal bulb. (Figure 7A). The cluster of amine-selective glomeruli was observed across animals (Figure 7C; n=11 wild-type or ΔT4-YFP mice) and corresponded to the genetically tagged DIII projection (Figures 7A and 7B). DIII glomeruli were the most sensitive amine-responsive glomeruli in the dorsal bulb. While some amines activated glomeruli outside of DIII, these responses were always much smaller in amplitude (Figure 7B). Within DIII, the amines were represented in a combinatorial fashion, with each eliciting responses in one or a few glomeruli (Figure 7D). Across animals, we were able to identify up to 12 amine-responsive glomeruli per bulb (n=6 mice) suggesting that our odorant set activates a subset of the ~20 dorsal TAAR glomeruli. The response profiles of DIII glomeruli could be classified into 7 groups with variable breadth of tuning (Figure 7E; n=3 mice). The most effective stimulus was β-phenylethylamine, which elicited the largest amplitude responses despite being presented at the lowest vapor concentration. Imaging experiments in T4-RFP mice show that these β-phenylethylamine-responsive glomeruli correspond to T4 (R.P., A.D., J. Zhang and T.B., unpublished data).
Figure 7. The dorsal TAAR glomeruli are selectively activated by amines.
(A) In vivo glomerular imaging in a ΔT4-YFP heterozygous mouse. (Top left) Resting spH fluorescence imaged through thinned bone. Pseudocolored panels show odor-evoked changes in fluorescence (ΔF) in response to the indicated odorants. All panels are scaled to the same maximum ΔF. Dotted line approximates the edges of the left and right olfactory bulbs. Anterior is up.
(B) (Left panel) Same mouse as in (A) showing YFP-labeled glomeruli (outlined in magenta). Pseudocolored images show maximum response projections for amine and non-amine odorants. Panel marked “amines-YFP” shows max amines responses with the outlined YFP-labeled region subtracted. Only small responses lateral to DIII remain. (Blood vessel-derived intrinsic signals can be seen in left anterior bulb). Amine responses include the amines in panel (A) plus 0.005% trimethylamine, 0.01% N-methylpiperidine, 0.01% triethylamine, 0.01% cyclohexylamine, 0.01% octylamine, 1% ethylenediamine. Non-amine odors include 1% propyl acetate, phenetole, isopropyl tiglate, 2-heptanone, and propionic acid.
(C) Maximum response projections in two other mice showing clustering of amine-responsive glomeruli. Responses are scaled from 0–95% of the maximum ΔF within a given animal. Amine odors: 0.01% β-phenylethylamine, 0.001% isopentylamine, 0.005% trimethylamine, 0.01% N-methylpiperidine (mouse 1), plus 0.01% cyclohexylamine (mouse 2).
(D) Odorant responses in another ΔT4-YFP heterozygous mouse overlaid onto the YFP signal. Greyscale image shows YFP-labeled projections in the left and right bulbs. Outline of midline and caudal bulb is indicated (dotted line). Response maps were thresholded below 16% maximum ΔF, pseudocolored and overlaid on top of the greyscale image. Responses to different amines are confined to YFP-labeled glomeruli. Lower right panel shows thresholded maximum projections indicating the locations of glomeruli responding to four amines cadaverine, trimethylamine, β-phenylethylamine and cyclohexylamine. A broadly tuned, β-phenylethylamine-responsive glomerulus appears white in overlay. Odor responses are scaled from 0–95% of the maximum ΔF for a given odorant presentation.
(E) Response profiles of DIII (YFP-positive) glomeruli to a set of 6 odorants: CAD= 1% cadaverine, CHX= 0.001% cyclohexylamine, IPA= 0.001% isopentylamine, NMP= 0.01% N-methylpiperidine, PEA= 0.01% β-phenylethylamine, TMA= 0.005% trimethylamine. The area of each dot is proportional to response amplitude (ΔF). Response profiles are ordered by hierarchical clustering (Ward’s method) with groups indicated (colored shading).
Taken together, the data demonstrate that the DIII projection represents glomeruli for the dorsally expressed TAARs and that these glomeruli are the most sensitive amine-responsive glomeruli in the dorsal olfactory bulb.
DISCUSSION
Our data reveal for the first time the anatomy and functional properties of TAAR projections to the olfactory bulb. Our data argue strongly that 10 out of 14 olfactory TAARs are mapped to a cluster of typical glomeruli in the dorsal olfactory bulb of the mouse. We show this directly for T3 and T4, and indirectly for other TAARs by analyzing TAAR-deletion alleles. We provide genetic evidence that the OSNs that form this projection are restricted to express TAAR genes prior to initial choice and are different from those underlying the previously described DI and DII domains. We thus refer to the TAAR projection as “DIII”. We go on to show that the DIII projection comprises highly sensitive, amine-responsive glomeruli, establishing an amine-selective, functional domain in the dorsal bulb. Thus, the TAARs feed into a genetically and anatomically distinct olfactory subsystem that is selectively activated by volatile amines.
A TAAR-specific OSN-type
Our data place the TAARs firmly into an existing framework (Figure S4) describing the mapping of OR inputs to the olfactory bulb by distinct types of OSNs (Bozza et al., 2009). Several lines of evidence support the view that there is an OSN-type corresponding to the dorsal TAARs. First, we observe a strong bias in gene choice—OSNs expressing a TAAR-deletion allele preferentially co-express TAARs over class I and class II ORs, and OSNs expressing class I and class II OR-deletion alleles rarely co-express the TAAR genes. Second, the gene choice bias is not a cluster effect (i.e. a byproduct of choosing neighboring TAAR genes in cis). Under our experimental conditions, co-expression in cis appears to be disfavored and cannot account for the high frequency of TAAR co-expression we observe in ΔT4-OSNs. Third, this bias is not a consequence of having already chosen a TAAR gene. Removing half of the available TAAR alleles causes an approximate doubling of the number of OSNs expressing one of the remaining alleles, indicating a bias in “first choice”. This provides the strongest evidence to date for the existence of intermingled OSN-types, one of which is dedicated to expressing TAARs.
One might predict that 100% of ΔT4-OSNs should co-express TAARs. Our in situ hybridization experiments show that 65% of ΔT4-OSNs hybridize with a mix of probes of all the TAARs. Similarly, the sum of co-expression rates from individual TAAR probes is 72%. We note that it is unknown what proportion of OSNs goes on to successfully choose an intact gene after choosing a nonfunctional OR or TAAR—some may fail to successfully express another receptor altogether. Consistent with this idea, the number of labeled OSNs in ΔT4-YFP mice is only 43% of the number of T4-RFP OSNs, and 30% of ΔT4-OSNs are immature (OMP-negative) as compared with 9% of T4-RFP OSNs (data not shown)—this suggests that expression of the ΔT4 allele has an effect on OR maturation.
Alternatively, ΔT4-OSNs could choose (non-TAAR) OR genes, in one of the two following scenarios. In the first, 35% of ΔT4-OSNs could choose randomly from among the ~400 dorsal OR genes. In fact, we detect some class I OR co-expression, but this is at relatively low rates. The doubling of T4-OSNs in the cluster deletion argues that TAAR-expressing OSNs choose from a small gene repertoire, suggesting that non-TAAR expression is generally a rare event. We note also that if ΔT4-OSNs co-express many OR genes, they must not form corresponding glomeruli as there are only ~20 labeled glomeruli in ΔT4-YFP mice. Thus, such hypothetical OR expression would appear to be functionally irrelevant.
In the second scenario, the 35% of ΔT4-OSNs could choose from a few specific OR genes. Our results also argue against this. If TAAR-expressing OSNs can choose from a few, specific OR genes, these genes should still be available for co-expression in ΔT4-YFP/ΔT2-9 mice. In this case, we would not likely see the dramatic reduction in ΔT4 cell number, and we would expect to see a few densely innervated glomeruli in these animals. We do not observe this. Taken together, the data suggest that the lower-than-expected co-expression rate of TAARs may be partly explained by ΔT4-OSNs that have not yet initiated transcription of another TAAR (at least in detectable amounts) or that have failed to choose another receptor.
The underlying cause of the reduction in cell number and axon labeling in ΔT4-YFP/ΔT2-9 mice is not known. Co-expression in cis might be disfavored leading to cell death. Alternately, ΔT4-OSNs might switch in cis to a neighboring TAAR gene, silencing the ΔT4-YFP allele. The rare labeled OSNs we observe in ΔT4-YFP/ΔT2-9 mice could represent those that have switched to express a TAAR allele in cis, have terminated transcription of the deletion allele, but still retained enough YFP protein to be labeled. Similar experiments using a vomeronasal gene cluster deletion demonstrate a complete absence of cisco -expression, a so-called “cluster lock” (Roppolo et al., 2007). We see a similar bias, though not a strict lock since cisco -expression was detected, albeit at low frequency. Thus, the two systems likely share common mechanisms. More pertinent to our present argument, our experiments demonstrate that the TAAR co-expression bias in ΔT4-OSNs is not the byproduct of a cluster effect.
The DIII domain
The previously described DI and DII domains are formed by afferent projections of OSN-types to the olfactory bulb. Our data suggest that the dorsal TAAR projection, DIII, is similarly generated. We note that DIII glomeruli are often partially intermingled with DI glomeruli. Thus, there is not as distinct a “boundary” between DI and DIII glomeruli as there is between DI and DII. This likely reflects that fact that the DIII projection comprises far fewer glomeruli than the other projections. However, like the other two OSN-types, the d3-OSN type appears to possess a specific axon guidance identity. Preliminary data indicate that TAAR-expressing OSNs project to the same region of the bulb even if they are forced to express a class I OR (our unpublished observations). Thus, the DIII domain likely arises from the same mechanisms as DI and DII. Due to their close association, DI and DIII glomeruli may share a similar function, or information from the two projections may be combined for downstream processing.
Four of the 14 TAAR genes have ventral or broad expression patterns that are not confined to the dorsal epithelium. Given what is known about epithelium-to-bulb topography (Schoenfeld and Cleland, 2005), Taar7a and Taar7b may map to ventral glomeruli while Taar6 and Taar7d may map to both ventral and dorsal glomeruli. Alternately, they could represent an OSN-type with an atypical projection pattern, analogous to the OR37 family (Strotmann et al., 2000). Interestingly, the ΔT4 and ΔT9 strains reported here, and tagged ΔT5 and ΔT6 strains (data not shown), exhibit spurious projections to glomeruli that are more caudal and ventral than DIII. This suggests that Taar6, 7a, 7b, and/or 7d may project to more ventral and caudal glomeruli. In any case, our data indicate that all three known classes of mammalian ORs are systematically mapped on the dorsal surface of the bulb.
Amine responses in the dorsal olfactory bulb
Few studies have characterized responses to amines in the mammalian olfactory bulb, perhaps due to their generally offensive odor. Previous data show that the amines activate broad regions of the rodent olfactory bulb (Ma et al., 2012; Takahashi et al., 2004). One previous study in the rat found that amines activate a set of anterior, dorsal glomeruli that also responds to carboxylic acids and aldehydes, as well as a cluster of caudal amine-selective glomeruli (Takahashi et al., 2004). Our data suggest that the observed amine-selective cluster may correspond to the TAARs. In our experiments, amine responses were restricted to the DIII region, except at high concentrations, which could elicit activation in the DI region as well (data not shown). Therefore, the broader amine responses in previous studies were likely due to the higher stimulus concentrations used (100–1000 fold higher than those used here). The data also suggest that the organization of dorsal TAAR projections may be similar in rats and mice.
The DIII glomeruli were by far the most sensitive in the dorsal bulb to the tested amines, though glomeruli outside the DIII cluster were also amine-responsive. One obvious limitation of our experiments is that the imaging is limited to the dorsal bulb. It is therefore unclear whether there are also ventral glomeruli with comparable sensitivity to amines. Future studies mapping odorant responses in the ventral bulb and characterizing sensory thresholds in the TAAR cluster deletion mice may shed light on this issue.
In addition, it is unclear whether the DIII glomeruli are activated exclusively by amines or whether they respond to other classes of compounds. Our imaging data support the idea that TAARs respond preferentially to amines (Liberles and Buck, 2006). However, responses to non-amines might be seen if enough odorants are tested, consistent with a lack of chemotopy in the olfactory bulb (Bozza et al., 2004; Ma et al., 2012; Soucy et al., 2009). Regardless, from our perspective, the important question is not whether the DIII domain is activated solely by amines, but whether selective activation of the DIII glomeruli relays specific information that is relevant for certain olfactory behaviors.
Function of the TAARs
The functional significance of the dorsal domains, including DIII, is unclear. Odorants associated with decay and fermentation such as short chain carboxylic acids preferentially activate DI (Bozza et al., 2009; Kobayakawa et al., 2007; Matsumoto et al., 2010; Takahashi et al., 2004). Similarly, we find that amines, which are byproducts of tissue decomposition (Silla Santos, 1996) and that have a repulsive odor to humans, preferentially activate DIII. It has been suggested that the dorsal bulb may mediate innate aversion in rodents (Kobayakawa et al., 2007; Matsumoto et al., 2010), and both acids and amines have been reported as aversive stimuli in mice, albeit at extremely high concentrations (Kobayakawa et al., 2007). Similarly, the T4 ligand β-phenylethylamine, which is enriched in predator cat urine, has been reported to elicit innate aversion in rats and mice (Ferrero et al., 2011). It is tempting to speculate that selective activation of the DIII domain may represent the aversive quality of amines.
The TAAR repertoire is evolutionarily retained in mammals, suggesting that it serves a critical function in olfaction that is not met by the much larger repertoire of canonical ORs. One view is that TAARs selectively encode information about pheromones (Liberles, 2009; Munger et al., 2009), socially relevant odors, or predator-derived cues (Ferrero et al., 2011; Liberles and Buck, 2006). Our data suggest an alternate (though not mutually exclusive) view—that this phylogenetically distinct class of aminergic receptors is simply required for high sensitivity detection of amines. In some species, such as rodents, the TAAR inputs may feed into circuits that elicit innate avoidance or context specific behaviors. But, more generally, the small TAAR family may contribute significantly to setting the behavioral thresholds of animals to a subset of olfactory stimuli.
EXPERIMENTAL PROCEDURES
Gene targeting
Gene targeting to modify TAAR alleles via homologous recombination was performed as described (Bozza et al., 2009) with some modifications. Genomic fragments containing TAAR loci originated from 129S7 BAC clones (Adams et al., 2005; bMQ-215O19 for Taar3 and Taar4, bMQ-329H19 for Taar9). BAC recombineering (Warming et al., 2005) was used to insert AscI-flanked selection cassettes (galK or kan) into the targeted loci on the BACs just downstream of the TAAR coding sequences. Targeting vectors were then isolated by gap-repair, and the selection cassettes replaced with marker cassettes by standard cloning. See Extended Experimental Procedures for detailed cloning information for the 6 gene targeted alleles described here.
Targeting vectors were linearized and electroporated into 129 or 129B6F1 ES cells and G418-resistant clones were screened by long range PCR and/or Southern blotting. Chimeras were generated by aggregation of ES cells with CD1 morulae (Wood et al., 1993) or by injection into C57BL/6 blastocysts. Chimeras were crossed to C57BL/6 females and all strains are on a mixed 129/B6 background.
The TAAR cluster deletion (ΔT2-9) was generated by Cre-mediated trans-allelic recombination in vivo (Wu et al., 2007).
In situ hybridization and immunohistochemistry
The combination of in situ hybridization/immunohistochemistry was performed as described (Bozza et al., 2009; Ishii et al., 2004) with minor modifications (see also Extended Experimental Procedures). TAAR coding-sequence probes were transcribed directly from T7-tagged PCR products which were amplified from the genomic BAC clones. Gene-specific probes for individual members of the Taar7 and Taar8subfamilies were generated to 3 UTR sequence obtained by PCR from a cDNA library of olfactory epithelium. These 3 UTR probes (<1kb) were hybridized at 55°C and washed at 59°C. The standard coding-sequence probes (~1kb) were hybridized at 62°C and washed at 65°C. The single probes and probe mixes for class I and class II dorsal ORs were as previously described (Bozza et al., 2009). The probe mix for TAARs comprised Taar2, Taar3, Taar4, Taar5 and Taar9. The all-TAARs probe mix comprised all nine coding-sequence probes. See Table S1 for probe target sequences.
In vivo imaging
Imaging was performed as described (Bozza et al., 2004) with the following modifications. Mice were 5–12 weeks old and were heterozygous for both the OMP-spH and ΔT4-YFP alleles. Mice were anesthetized with sodium pentobarbital (65 mg/kg; Sigma) and given atropine sulfate (5.4 mg/kg; Med-Pharmex); chlorprothixene hydrochloride (13 mg/kg; Sigma) as a sedative; pyridoxalphosphate-6-azophenyl-2′, 4′-disulfonic acid (PPADS) and suramin (51 mg/kg and 130 mg/kg; Tocris) to counteract effects of nasal inflammation (Hegg and Lucero, 2006). A double tracheotomy was performed so that animals could breathe freely while odor delivery via artificial sniffing was controlled through the forward tube (Wachowiak and Cohen, 2001).
The bone overlying the olfactory bulbs was thinned using a dental drill. Light excitation was provided using a 200 W metal-halide lamp (Prior Scientific) filtered through standard filters sets for YFP (86001 JP3, Chroma) and GFP (96343, Nikon). Optical signals for synaptopHluorin were recorded using a CCD camera (NeuroCCD SM256; RedShirtImaging) at 7 Hz for 10 s, which included a 2 s pre-stimulus interval and a 2 s odor pulse. Inter-stimulus interval was at least 120 s. Artificial sniffing during the stimulus period was 150 mL/min at 3 Hz. Odorants were delivered using a computer-controlled, flow dilution olfactometer that delivered nitrogen and purified air to a Teflon mixing chamber/odor port. Disposable odor cartridges were attached directly this port. This design was required to prevent contamination of the olfactometer with even low concentrations of amines. Blank trials between odor stimuli were compared throughout the experiment to monitor potential odorant contamination in the nasal passages. In order to obtain low enough concentrations, amines were typically diluted 1:100 or 1:1000 in water, then further by flow dilution. Vapor concentrations were calculated using published vapor pressures (US EPA, Estimation Programs Interface Suite, v 4.0).
To control for photobleaching, blank trials were subtracted from odor trials prior to analysis. Response maps were obtained by subtracting a 1.4 s temporal average preceding the stimulus from a 1.4 s temporal average at the end of the trial. Responses are expressed as ΔF to account for the fact that the background spH fluorescence is not correlated with the pool of indicator that reports neuronal activity (Bozza et al., 2004; McGann et al., 2005). Max response images were produced by projecting maximum pixel values across individual responses onto a single image. Stimuli were presented at least twice, but data were often taken from the first trial as run down of the amine responses was prevalent. Images were processed and analyzed in Neuroplex (RedShirtImaging) and Image J (NIH) software.
Individual glomerular responses for hierarchical cluster analysis were obtained by drawing regions-of-interest (ROIs) around YFP-positive, amine-responsive glomeruli. Responses were thresholded at two standard deviations from background noise (measured from multiple non-responsive regions) and scaled to the maximum response of each glomerulus for clustering analysis.
Supplementary Material
HIGHLIGHTS.
A majority of TAARs are mapped to a spatially distinct subset of dorsal glomeruli
TAARs are expressed by a specific neuron type that exhibits biased gene choice
The gene choice bias is established prior to first choice
The dorsal TAAR glomeruli are specifically activated by low concentrations of amines
Acknowledgments
This work was supported by grants from the NIH/NIDCD and by the Visiting Scientist Program at Janelia Farm Research Campus. We thank Paul Feinstein for scientific discussions, for assistance in generating mouse strains, and for critical comments on the manuscript; Ivan Rodriguez for comments on the manuscript; Dima Rinberg for help with olfactometry and scientific discussions; Matt Wachowiak for advice on bulb imaging and for comments on the manuscript; Chingwen Yang and Rada Norinsky and the Gene Targeting and Transgenic Resource Centers at Rockefeller University, and Lynn Doglio and the Transgenic and Targeted Mutagenesis Lab at Northwestern University for assistance with gene targeting; Jim Cox and the staff at Janelia Farm for mouse colony maintenance. Thanks to Brian Weiland and Stephanie Leung for technical assistance. We also thank Ann Palmenberg for providing IRES constructs, and Walter Tsark (City of Hope) for HPRT-Cre mice.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES AND CITATIONS
- Adams DJ, Quail MA, Cox T, van der Weyden L, Gorick BD, Su Q, Chan WI, Davies R, Bonfield JK, Law F, et al. A genome-wide, end-sequenced 129Sv BAC library resource for targeting vector construction. Genomics. 2005;86:753–758. doi: 10.1016/j.ygeno.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Borowsky B, Adham N, Jones KA, Raddatz R, Artymyshyn R, Ogozalek KL, Durkin MM, Lakhlani PP, Bonini JA, Pathirana S, et al. Trace amines: identification of a family of mammalian G protein-coupled receptors. Proc Natl Acad Sci U S A. 2001;98:8966–8971. doi: 10.1073/pnas.151105198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozza T, Feinstein P, Zheng C, Mombaerts P. Odorant receptor expression defines functional units in the mouse olfactory system. J Neurosci. 2002;22:3033–3043. doi: 10.1523/JNEUROSCI.22-08-03033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozza T, McGann JP, Mombaerts P, Wachowiak M. In vivo imaging of neuronal activity by targeted expression of a genetically encoded probe in the mouse. Neuron. 2004;42:9–21. doi: 10.1016/s0896-6273(04)00144-8. [DOI] [PubMed] [Google Scholar]
- Bozza T, Vassalli A, Fuss S, Zhang JJ, Weiland B, Pacifico R, Feinstein P, Mombaerts P. Mapping of class I and class II odorant receptors to glomerular domains by two distinct types of olfactory sensory neurons in the mouse. Neuron. 2009;61:220–233. doi: 10.1016/j.neuron.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Bunzow JR, Sonders MS, Arttamangkul S, Harrison LM, Zhang G, Quigley DI, Darland T, Suchland KL, Pasumamula S, Kennedy JL, et al. Amphetamine, 3,4-methylenedioxymethamphetamine, lysergic acid diethylamide, and metabolites of the catecholamine neurotransmitters are agonists of a rat trace amine receptor. Mol Pharmacol. 2001;60:1181–1188. doi: 10.1124/mol.60.6.1181. [DOI] [PubMed] [Google Scholar]
- Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- Feinstein P, Bozza T, Rodriguez I, Vassalli A, Mombaerts P. Axon guidance of mouse olfactory sensory neurons by odorant receptors and the beta2 adrenergic receptor. Cell. 2004;117:833–846. doi: 10.1016/j.cell.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Feinstein P, Mombaerts P. A contextual model for axonal sorting into glomeruli in the mouse olfactory system. Cell. 2004;117:817–831. doi: 10.1016/j.cell.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Ferrero DM, Lemon JK, Fluegge D, Pashkovski SL, Korzan WJ, Datta SR, Spehr M, Fendt M, Liberles SD. Detection and avoidance of a carnivore odor by prey. Proc Natl Acad Sci U S A. 2011;108:11235–11240. doi: 10.1073/pnas.1103317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrero DM, Wacker D, Roque MA, Baldwin MW, Stevens RC, Liberles SD. Agonists for 13 Trace Amine-Associated Receptors Provide Insight into the Molecular Basis of Odor Selectivity. ACS Chem Biol. 2012 doi: 10.1021/cb300111e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag J, Krieger J, Strotmann J, Breer H. Two classes of olfactory receptors in Xenopus laevis. Neuron. 1995;15:1383–1392. doi: 10.1016/0896-6273(95)90016-0. [DOI] [PubMed] [Google Scholar]
- Glusman G, Yanai I, Rubin I, Lancet D. The complete human olfactory subgenome. Genome Res. 2001;11:685–702. doi: 10.1101/gr.171001. [DOI] [PubMed] [Google Scholar]
- Grandy DK. Trace amine-associated receptor 1-Family archetype or iconoclast? Pharmacol Ther. 2007;116:355–390. doi: 10.1016/j.pharmthera.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegg CC, Lucero MT. Purinergic receptor antagonists inhibit odorant-induced heat shock protein 25 induction in mouse olfactory epithelium. Glia. 2006;53:182–190. doi: 10.1002/glia.20258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T, Omura M, Mombaerts P. Protocols for two- and three-color fluorescent RNA in situ hybridization of the main and accessory olfactory epithelia in mouse. J Neurocytol. 2004;33:657–669. doi: 10.1007/s11068-005-3334-y. [DOI] [PubMed] [Google Scholar]
- Kobayakawa K, Kobayakawa R, Matsumoto H, Oka Y, Imai T, Ikawa M, Okabe M, Ikeda T, Itohara S, Kikusui T, et al. Innate versus learned odour processing in the mouse olfactory bulb. Nature. 2007;450:503–508. doi: 10.1038/nature06281. [DOI] [PubMed] [Google Scholar]
- Lewcock JW, Reed RR. A feedback mechanism regulates monoallelic odorant receptor expression. Proc Natl Acad Sci U S A. 2004;101:1069–1074. doi: 10.1073/pnas.0307986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberles SD. Trace amine-associated receptors are olfactory receptors in vertebrates. Ann N Y Acad Sci. 2009;1170:168–172. doi: 10.1111/j.1749-6632.2009.04014.x. [DOI] [PubMed] [Google Scholar]
- Liberles SD, Buck LB. A second class of chemosensory receptors in the olfactory epithelium. Nature. 2006;442:645–650. doi: 10.1038/nature05066. [DOI] [PubMed] [Google Scholar]
- Lindemann L, Ebeling M, Kratochwil NA, Bunzow JR, Grandy DK, Hoener MC. Trace amine-associated receptors form structurally and functionally distinct subfamilies of novel G protein-coupled receptors. Genomics. 2005;85:372–385. doi: 10.1016/j.ygeno.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Ma L, Qiu Q, Gradwohl S, Scott A, Yu EQ, Alexander R, Wiegraebe W, Yu CR. Distributed representation of chemical features and tunotopic organization of glomeruli in the mouse olfactory bulb. Proc Natl Acad Sci U S A. 2012;109:5481–5486. doi: 10.1073/pnas.1117491109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96:713–723. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- Matsumoto H, Kobayakawa K, Kobayakawa R, Tashiro T, Mori K, Sakano H. Spatial arrangement of glomerular molecular-feature clusters in the odorant-receptor class domains of the mouse olfactory bulb. J Neurophysiol. 2010;103:3490–3500. doi: 10.1152/jn.00035.2010. [DOI] [PubMed] [Google Scholar]
- McGann JP, Pirez N, Gainey MA, Muratore C, Elias AS, Wachowiak M. Odorant representations are modulated by intra- but not interglomerular presynaptic inhibition of olfactory sensory neurons. Neuron. 2005;48:1039–1053. doi: 10.1016/j.neuron.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Mombaerts P, Wang F, Dulac C, Chao SK, Nemes A, Mendelsohn M, Edmondson J, Axel R. Visualizing an olfactory sensory map. Cell. 1996;87:675–686. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- Munger SD, Leinders-Zufall T, Zufall F. Subsystem organization of the mammalian sense of smell. Ann Rev Physiol. 2009;71:115–140. doi: 10.1146/annurev.physiol.70.113006.100608. [DOI] [PubMed] [Google Scholar]
- Niimura Y. Evolutionary dynamics of olfactory receptor genes in chordates: interaction between environments and genomic contents. Human Genomics. 2009;4:107–118. doi: 10.1186/1479-7364-4-2-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressler KJ, Sullivan SL, Buck LB. Information coding in the olfactory system: evidence for a stereotyped and highly organized epitope map in the olfactory bulb. Cell. 1994;79:1245–1255. doi: 10.1016/0092-8674(94)90015-9. [DOI] [PubMed] [Google Scholar]
- Roppolo D, Vollery S, Kan CD, Luscher C, Broillet MC, Rodriguez I. Gene cluster lock after pheromone receptor gene choice. EMBO J. 2007;26:3423–3430. doi: 10.1038/sj.emboj.7601782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfeld TA, Cleland TA. The anatomical logic of smell. Trends Neurosci. 2005;28:620–627. doi: 10.1016/j.tins.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Sakano H. One neuron-one receptor rule in the mouse olfactory system. Trends Genet. 2004;20:648–653. doi: 10.1016/j.tig.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Takeuchi H, Yamagishi Y, Suzuki M, Sakano H. A neuronal identity code for the odorant receptor-specific and activity-dependent axon sorting. Cell. 2006;127:1057–1069. doi: 10.1016/j.cell.2006.10.031. [DOI] [PubMed] [Google Scholar]
- Silla Santos MH. Biogenic amines: their importance in foods. Int J Food Microbiol. 1996;29:213–231. doi: 10.1016/0168-1605(95)00032-1. [DOI] [PubMed] [Google Scholar]
- Soucy ER, Albeanu DF, Fantana AL, Murthy VN, Meister M. Precision and diversity in an odor map on the olfactory bulb. Nat Neurosci. 2009;12:210–220. doi: 10.1038/nn.2262. [DOI] [PubMed] [Google Scholar]
- Staubert C, Boselt I, Bohnekamp J, Rompler H, Enard W, Schoneberg T. Structural and functional evolution of the trace amine-associated receptors TAAR3, TAAR4 and TAAR5 in primates. PLoS One. 2010;5:e11133. doi: 10.1371/journal.pone.0011133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strotmann J, Conzelmann S, Beck A, Feinstein P, Breer H, Mombaerts P. Local permutations in the glomerular array of the mouse olfactory bulb. J Neurosci. 2000;20:6927–6938. doi: 10.1523/JNEUROSCI.20-18-06927.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi YK, Nagayama S, Mori K. Detection and masking of spoiled food smells by odor maps in the olfactory bulb. J Neurosci. 2004;24:8690–8694. doi: 10.1523/JNEUROSCI.2510-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi A, Miyazaki T, Imai T, Sakano H. Olfactory sensory neurons expressing class I odorant receptors converge their axons on an antero-dorsal domain of the olfactory bulb in the mouse. Eur J Neurosci. 2006;23:1436–1444. doi: 10.1111/j.1460-9568.2006.04675.x. [DOI] [PubMed] [Google Scholar]
- Vassalli A, Feinstein P, Mombaerts P. Homeodomain binding motifs modulate the probability of odorant receptor gene choice in transgenic mice. Mol Cell Neurosci. 2011;46:381–396. doi: 10.1016/j.mcn.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassalli A, Rothman A, Feinstein P, Zapotocky M, Mombaerts P. Minigenes impart odorant receptor-specific axon guidance in the olfactory bulb. Neuron. 2002;35:681–696. doi: 10.1016/s0896-6273(02)00793-6. [DOI] [PubMed] [Google Scholar]
- Vassar R, Chao SK, Sitcheran R, Nunez JM, Vosshall LB, Axel R. Topographic organization of sensory projections to the olfactory bulb. Cell. 1994;79:981–991. doi: 10.1016/0092-8674(94)90029-9. [DOI] [PubMed] [Google Scholar]
- Wachowiak M, Cohen LB. Representation of odorants by receptor neuron input to the mouse olfactory bulb. Neuron. 2001;32:723–735. doi: 10.1016/s0896-6273(01)00506-2. [DOI] [PubMed] [Google Scholar]
- Wang F, Nemes A, Mendelsohn M, Axel R. Odorant receptors govern the formation of a precise topographic map. Cell. 1998;93:47–60. doi: 10.1016/s0092-8674(00)81145-9. [DOI] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood SA, Allen ND, Rossant J, Auerbach A, Nagy A. Non-injection methods for the production of embryonic stem cell-embryo chimaeras. Nature. 1993;365:87–89. doi: 10.1038/365087a0. [DOI] [PubMed] [Google Scholar]
- Wu S, Ying G, Wu Q, Capecchi MR. Toward simpler and faster genome-wide mutagenesis in mice. Nat Genet. 2007;39:922–930. doi: 10.1038/ng2060. [DOI] [PubMed] [Google Scholar]
- Zhang X, Firestein S. The olfactory receptor gene superfamily of the mouse. Nat Neurosci. 2002;5:124–133. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]
- Zhang X, Firestein S. Genomics of olfactory receptors. Results Probl Cell Differ. 2009;47:25–36. doi: 10.1007/400_2008_28. [DOI] [PubMed] [Google Scholar]
- Zhang X, Rogers M, Tian H, Zou DJ, Liu J, Ma M, Shepherd GM, Firestein SJ. High-throughput microarray detection of olfactory receptor gene expression in the mouse. Proc Natl Acad Sci U S A. 2004;101:14168–14173. doi: 10.1073/pnas.0405350101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Ivic L, Otaki JM, Hashimoto M, Mikoshiba K, Firestein S. Functional expression of a mammalian odorant receptor. Science. 1998;279:237–242. doi: 10.1126/science.279.5348.237. [DOI] [PubMed] [Google Scholar]
- Zucchi R, Chiellini G, Scanlan TS, Grandy DK. Trace amine-associated receptors and their ligands. Br J Pharmacol. 2006;149:967–978. doi: 10.1038/sj.bjp.0706948. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.