Abstract
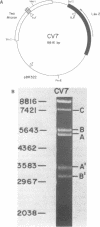
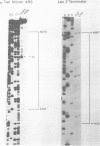
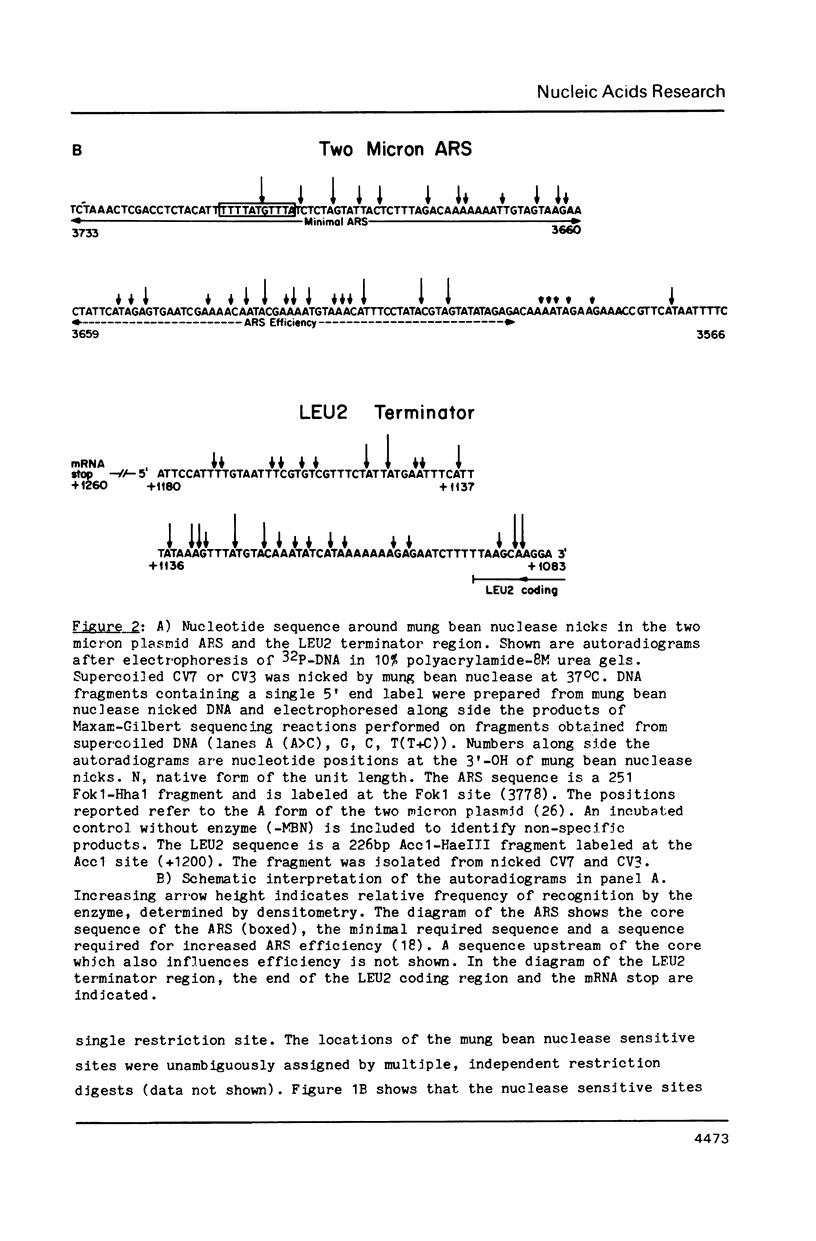
Mung bean nuclease was used to probe for DNA unwinding in torsionally-stressed chimeric plasmids containing two micron plasmid sequences and the yeast LEU2 gene in a pBR322 vector. The yeast sequences are cleaved at only two sites, both of which map to regulatory regions: (1) the autonomously replicating sequence (ARS), an origin of DNA replication, of the two micron plasmid and (2) the transcription terminator region of the LEU2 gene. Nucleotide level analysis of the nuclease cleavage pattern shows that an A + T-rich structure, distinct from other non-B DNA conformations, is recognized. A computer analysis reveals that A + T content alone is not sufficient to explain the preferential occurrence of the A + T-rich structure in the ARS over other sequences of equal A + T content. The A + T-rich structure detected in the ARS maps to sequences required for DNA replication. Our findings demonstrate the DNA conformational flexibility of certain yeast regulatory regions and provide support for the hypothesis that the A + T-rich sequence in the ARS plays a role in DNA unwinding during the initiation of DNA replication.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andreadis A., Hsu Y. P., Hermodson M., Kohlhaw G., Schimmel P. Yeast LEU2. Repression of mRNA levels by leucine and primary structure of the gene product. J Biol Chem. 1984 Jul 10;259(13):8059–8062. [PubMed] [Google Scholar]
- Benham C. J. Theoretical analysis of competitive conformational transitions in torsionally stressed DNA. J Mol Biol. 1981 Jul 25;150(1):43–68. doi: 10.1016/0022-2836(81)90324-7. [DOI] [PubMed] [Google Scholar]
- Brennan C. A., Dombroski A. J., Platt T. Transcription termination factor rho is an RNA-DNA helicase. Cell. 1987 Mar 27;48(6):945–952. doi: 10.1016/0092-8674(87)90703-3. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Hicks J. B. Replication and recombination functions associated with the yeast plasmid, 2 mu circle. Cell. 1980 Sep;21(2):501–508. doi: 10.1016/0092-8674(80)90487-0. [DOI] [PubMed] [Google Scholar]
- Campbell J. L. Eukaryotic DNA replication. Annu Rev Biochem. 1986;55:733–771. doi: 10.1146/annurev.bi.55.070186.003505. [DOI] [PubMed] [Google Scholar]
- Cantor C. R., Efstratiadis A. Possible structures of homopurine-homopyrimidine S1-hypersensitive sites. Nucleic Acids Res. 1984 Nov 12;12(21):8059–8072. doi: 10.1093/nar/12.21.8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker S. E., Sweder K., Srienc F., Bailey J. E., Campbell J. L. Deletion mutations affecting autonomously replicating sequence ARS1 of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Nov;4(11):2455–2466. doi: 10.1128/mcb.4.11.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson M., Echols H., Wickner S., Alfano C., Mensa-Wilmot K., Gomes B., LeBowitz J., Roberts J. D., McMacken R. Specialized nucleoprotein structures at the origin of replication of bacteriophage lambda: localized unwinding of duplex DNA by a six-protein reaction. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7638–7642. doi: 10.1073/pnas.83.20.7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristensky B., Lis J., Wu R. Portable microcomputer software for nucleotide sequence analysis. Nucleic Acids Res. 1982 Oct 25;10(20):6451–6463. doi: 10.1093/nar/10.20.6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froman B. E., Tait R. C., Rodriguez R. L. Nucleotide sequence of the 3' terminal region of the LEU2 gene from Saccharomyces cerevisiae. Gene. 1984 Nov;31(1-3):257–261. doi: 10.1016/0378-1119(84)90218-x. [DOI] [PubMed] [Google Scholar]
- Funnell B. E., Inman R. B. Comparison of partial denaturation maps with the known sequence of simian virus 40 and phi X174 replicative form DNA. J Mol Biol. 1979 Jun 25;131(2):331–340. doi: 10.1016/0022-2836(79)90079-2. [DOI] [PubMed] [Google Scholar]
- Gellert M. DNA topoisomerases. Annu Rev Biochem. 1981;50:879–910. doi: 10.1146/annurev.bi.50.070181.004311. [DOI] [PubMed] [Google Scholar]
- Hartley J. L., Donelson J. E. Nucleotide sequence of the yeast plasmid. Nature. 1980 Aug 28;286(5776):860–865. doi: 10.1038/286860a0. [DOI] [PubMed] [Google Scholar]
- Hentschel C. C. Homocopolymer sequences in the spacer of a sea urchin histone gene repeat are sensitive to S1 nuclease. Nature. 1982 Feb 25;295(5851):714–716. doi: 10.1038/295714a0. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Kearsey S. Structural requirements for the function of a yeast chromosomal replicator. Cell. 1984 May;37(1):299–307. doi: 10.1016/0092-8674(84)90326-x. [DOI] [PubMed] [Google Scholar]
- Komaromy M., Govan H. An inexpensive semi-automated sequence reader for the Apple II computer. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):675–678. doi: 10.1093/nar/12.1part2.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski D. Changes in site specificity of single-strand-specific endonucleases on supercoiled PM2 DNA with temperature and ionic environment. Nucleic Acids Res. 1984 Sep 25;12(18):7071–7086. doi: 10.1093/nar/12.18.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski D., Kroeker W. D., Laskowski M., Sr Mung bean nuclease I. Physical, chemical, and catalytic properties. Biochemistry. 1976 Oct 5;15(20):4457–4463. doi: 10.1021/bi00665a019. [DOI] [PubMed] [Google Scholar]
- Kowalski D., Sanford J. P. Action of mung bean nuclease on supercoiled PM2 DNA. J Biol Chem. 1982 Jul 10;257(13):7820–7825. [PubMed] [Google Scholar]
- Laskowski M., Sr Purification and properties of the mung bean nuclease. Methods Enzymol. 1980;65(1):263–276. doi: 10.1016/s0076-6879(80)65036-8. [DOI] [PubMed] [Google Scholar]
- Lilley D. M. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickol J. M., Felsenfeld G. DNA conformation at the 5' end of the chicken adult beta-globin gene. Cell. 1983 Dec;35(2 Pt 1):467–477. doi: 10.1016/0092-8674(83)90180-0. [DOI] [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cruciform structures in supercoiled DNA. Nature. 1981 Feb 5;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- Pruitt S. C., Reeder R. H. Effect of topological constraint on transcription of ribosomal DNA in Xenopus oocytes. Comparison of plasmid and endogenous genes. J Mol Biol. 1984 Mar 25;174(1):121–139. doi: 10.1016/0022-2836(84)90368-1. [DOI] [PubMed] [Google Scholar]
- Radloff R., Bauer W., Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich A., Nordheim A., Wang A. H. The chemistry and biology of left-handed Z-DNA. Annu Rev Biochem. 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- Ryoji M., Worcel A. Chromatin assembly in Xenopus oocytes: in vivo studies. Cell. 1984 May;37(1):21–32. doi: 10.1016/0092-8674(84)90297-6. [DOI] [PubMed] [Google Scholar]
- Sawadaishi K., Miura K., Ohtsuka E., Ueda T., Ishizaki K., Shinriki N. Ozonolysis of supercoiled pBR322 DNA resulting in strand scission to open circular DNA. Nucleic Acids Res. 1985 Oct 25;13(20):7183–7194. doi: 10.1093/nar/13.20.7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawadaishi K., Miura K., Ohtsuka E., Ueda T., Shinriki N., Ishizaki K. Structure- and sequence-specificity of ozone degradation of supercoiled plasmid DNA. Nucleic Acids Res. 1986 Feb 11;14(3):1159–1169. doi: 10.1093/nar/14.3.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon E., Evans T., Welsh J., Efstratiadis A. Conformation of promoter DNA: fine mapping of S1-hypersensitive sites. Cell. 1983 Dec;35(3 Pt 2):837–848. doi: 10.1016/0092-8674(83)90116-2. [DOI] [PubMed] [Google Scholar]
- Sheflin L. G., Kowalski D. Altered DNA conformations detected by mung bean nuclease occur in promoter and terminator regions of supercoiled pBR322 DNA. Nucleic Acids Res. 1985 Sep 11;13(17):6137–6154. doi: 10.1093/nar/13.17.6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheflin L. G., Kowalski D. Mung bean nuclease cleavage of a dA + dT-rich sequence or an inverted repeat sequence in supercoiled PM2 DNA depends on ionic environment. Nucleic Acids Res. 1984 Sep 25;12(18):7087–7104. doi: 10.1093/nar/12.18.7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton C. K., Kilpatrick M. W., Wells R. D. S1 nuclease recognizes DNA conformational junctions between left-handed helical (dT-dG n. dC-dA)n and contiguous right-handed sequences. J Biol Chem. 1984 Feb 10;259(3):1963–1967. [PubMed] [Google Scholar]
- Wang J. C. DNA topoisomerases. Annu Rev Biochem. 1985;54:665–697. doi: 10.1146/annurev.bi.54.070185.003313. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Peck L. J., Becherer K. DNA supercoiling and its effects on DNA structure and function. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):85–91. doi: 10.1101/sqb.1983.047.01.011. [DOI] [PubMed] [Google Scholar]