Abstract
The 17q21.31 inversion polymorphism exists either as direct (H1) or inverted (H2) haplotypes with differential predispositions to disease and selection. We investigated its genetic diversity in 2700 individuals with an emphasis on African populations. We characterize eight structural haplotypes that vary in size from 1.08 to 1.49 Mbp as a result of complex rearrangements and provide evidence for a 30 kbp H1/H2 double recombination event. We show that recurrent partial duplications of the KANSL1 (previously known as KIAA1267) gene have occurred on both H1 and H2 haplotypes and risen to high frequency in European populations. We identify a likely ancestral H2 haplotype (H2′) lacking these duplications, enriched among African hunter-gatherer groups yet essentially absent from West Africans populations. While H1 and H2 segmental duplications arose independently and prior to the human migration out of Africa, they have reached high frequencies recently among Europeans either due to extraordinary genetic drift or selective sweeps.
INTRODUCTION
Chromosomal rearrangements occur in many species and can contribute to phenotypic variability and genomic evolution 1–5. Compared to other structural variants, inversions may be under different selective pressures because recombination is suppressed between heterokaryotypes6–9. The 17q21.31 inversion locus represents one of the most dynamic and complex regions of the human genome. Two haplotypes exist, in direct (H1) and inverted (H2) orientation, which previous studies demonstrate do not recombine over nearly 2 Mbp, resulting in extended linkage disequilibrium (LD) 10. The H2 haplotype is enriched in Europeans, and carriers are predisposed to the 17q21.31 microdeletion syndrome as a result of NAHR between directly oriented segmental duplications mapping on the inverted chromosome 11–14. A recent study of copy number variation in the 1000 Genomes Project demonstrated that a 205 kbp duplication is associated with 30% of European H1 haplotypes while a smaller 155 kbp duplication in the same region is fixed in European H2 haplotypes 15. The latter predisposes to NAHR and subsequent the 17q21.31 microdeletion syndrome.
Using short tandem repeats, Donnelly et al. 16 estimated the time to the most recent common ancestor (TMRCA) of the H2 haplotype between approximately 16–108 thousand years ago and that the H2 haplotype originated in Africa; however, sequence divergence between H1 and H2 indicate a more ancient coalescence of 2.3 million years ago (mya). The discovery of an H2 haplotype without duplication from the genome sequence of a Khoisan Bushman 17 suggested recent structural changes in the evolution of the H2 lineage. Given the importance of the H2-specific duplication in disease and its significant population stratification, we explored the architecture of this region in more detail using a combination of next-generation sequencing (NGS), array comparative genomic hybridization (CGH), and fluorescence in situ hybridization (FISH) in a total of 2700 individuals from diverse geographic populations. We specifically surveyed the distribution of the H2 haplotype in African ethnic groups with variable modes of subsistence focusing on hunter-gatherer populations to capture potentially ancient structural and nucleotide diversity.
RESULTS
Duplication architecture of 17q21.31 locus
Using NGS from 620 individuals from three major continental groups (Africans, Asians, and Europeans; 1000 Genomes Project) and 185 admixed individuals (total n = 805), we estimate the copy number of this locus using sequence read-depth as described in Sudmant et al. 15 (Supplementary Tables 1,2). The region consists of three large copy number polymorphic (CNP) segmental duplications (Figure 1), which include short (155 kbp) and long (205 kbp) duplications corresponding to the promoter and first exon of KANSL1 associated with the H2 and H1 haplotypes, respectively. For simplicity, we refer to these as CNP155 and CNP205. We find that almost 60% of Europeans carry at least one of these duplications (Figure 1a,b); however, they are virtually nonexistent in the African and Asian populations (Figure 1c). The third polymorphism is 210 kbp in length and spans most of NSF upstream of KANSL1 (CNP210) 15. Asian populations show higher copy number of CNP210 when compared to European and African populations. In fact, individuals with four haploid copies of this duplication—an estimated 800 kbp of tandem repeat—are exclusively of Asian descent (Figure 1c).
Figure 1. Duplication architecture of 17q21.31.

(a) Frequency of haplotypes (H2D, H1D) carrying duplications (CNP155 and CNP205) and those not carrying duplications (ND) are shown for three major continental groups (Africans, Asians, and Europeans) based on analysis of 620 individuals. (b) Read-depth-based copy number estimates of the 17q21.31 region from 46 representative European genomes show different patterns of duplications for the KANSL1 and NSF regions. Colors indicate the absolute copy number genome-wide for each given segment 15. The heatmap is aligned to the H1 haplotype structure at the bottom (reference genome) where colored boxes indicate segmental duplications as described in Zody et al. 13, and the black line represents single-copy regions. The heatmap distinguishes genotypes for a 205 kbp copy-number polymorphism (CNP205) associated with H2D, a 155 kbp polymorphism (CNP155) associated with H1D, and copy-number variation of a 210 kbp segment of NSF, which ranges from 2–8 copies (CNP210). Note that CNP205 and CNP155 have a diploid copy number of two in the reference genome assembly (shown at the bottom) and CNP210 has instead a diploid copy number of four. (c) Population stratification of duplicated alleles. CNP155 and CNP205 show increased allele frequency (23.1% and 19.6%, respectively) among Europeans; CNP210 shows a significant increase in copy number among Asians, compared to Europeans and Africans.
Alternative structural configurations of 17q21.31
In order to further investigate the genomic organization of the region, we performed FISH experiments on 30 ethnically diverse individuals (Figure 2a, Supplementary Table 3). 35/60 chromosomes tested were inverted and all 35 were concordant with the PCR assay diagnostic for the H2 haplotype inversion (Supplementary Figure 1) 18. We did not observe any non-inverted H2 chromosomes as recently reported by Rao et al. 19. FISH and array CGH experiments confirm three haploid copy number states for the KANSL1 locus (CN = 1, 2 and 3) and three haploid copy number states for the NSF locus (CN = 1, 2 and 3). We analyzed 21 samples of African descent (Supplementary Figure 2 and Supplementary Table 4) and found that the H2-specific duplication (CNP205) was highly polymorphic among Africans.
Figure 2. Alternative structural haplotypes of 17q21.31.
(a) FISH cohybridization experiments using probes mapping to CNP155/CNP205 (WIBR2-2342H02 in green), NSF CNP210 duplication (WIBR2-1321L07 in red), and at the single-copy region (WIBR2-3237D21 in blue) are shown. (b) Shown are eight distinct structural haplotypes (five H1 and three H2) ranging in size from 1.08 to 1.49 Mbp. Colored boxes indicate segmental duplications as determined by complete sequencing of large-insert BAC clones by Zody et al. 13. Hashed boxes correspond to regions present in single copy in that specific haplotype but duplicated in others. The locations of three core duplicons mapping in close proximity to the inversion breakpoints are shown. These represent some of the most abundant and rapidly evolving duplicated sequences in the human genome 49. The duplication content for each haplotype is indicated in parentheses. Four main haplotypes are defined based on KANSL1 copy number and on the length of the duplication (Boettger et al. 20 nomenclature in parentheses): H1′ (direct haplotype) and H2′ (inverted haplotype) with one copy each of KANSL1, H1D (H1.β2.γ1) with a long duplication of the gene, and H2D (H2.α2.γ2) with a short duplication. H1′ configurations with one copy of NSF are defined as H1.1 (H1.β1.γ1), with two copies as H1.2 (H1.β1.γ2), and with three copies as H1.3 (H1.β1.γ3). H1D configurations with three copies of the long duplication are defined as H1D.3 (H1.β3.γ1). Similarly, H2′ configurations with one copy of NSF are defined as H2.1 and with two copies as H2.2 (H2.α1.γ2).
Using two constructed BAC-based assemblies corresponding to one direct and one inverted haplotype 13, together with read-depth-based copy number estimates, BAC pool sequencing (Antonacci et al., unpublished), and FISH, we characterize at least eight alternate structural configurations of the 17q21.31 region, which differ dramatically in their organization and duplication content (Figure 2b). Four main structural haplotypes may be defined based on the inversion status and the copy number status (CNP205 and CNP155) of KANSL1 duplications: H1′ (direct) and H2′ (inverted) carry no duplications of KANSL1, H1D (direct) has two copies of CNP205, and H2D (inverted) has two copies of CNP155. We further identify configurations with three copies of CNP205 as H1D.3 while H1′ configurations with no NSF duplications are defined as H1.1, two copies of CNP210 as H1.2, and three copies as H1.3. Similarly, H2′ configurations with one copy of NSF are defined as H2.1 and two copies as H2.2, etc. Interestingly, all H1D haplotypes analyzed showed a fixed copy number of 1 for NSF and all H2D haplotypes have a fixed copy number of 2 (CNP210). The H2.1 haplotype is among the simplest carrying single copies of all CNPs including CNP210. The NSF CNP is highly variable among H1′ ranging from 2–8 diploid copies. FISH experiments using a probe tagging both CNP155 and CNP205 and a probe tagging uniquely CNP205 reveal that the proximal breakpoints of the duplications are different and the duplications map in different locations (tandem duplication on H1D and interspersed on H2D), strongly suggesting that the two duplications are independent events (Supplementary Figure 3). An independent study published in this issue finds multiple distinct haplotypes also defined based on the duplication content and organization 20 (Supplementary Table 5).
Inversion and duplication frequency in Africa
Previous studies of this locus in African populations have suggested that the inverted haplotype was rare or nonexistent in most of Africa. Diversity sample surveys, however, have been biased to populations primarily of Western or South African descent. We sought to explore the diversity more systematically by analyzing genetic data from a larger collection of African samples including the HapMap Project, 1000 Genomes Project, Human Genome Diversity Project (HGDP), the African Diversity Panel (unpublished data), and the Hunter-Gatherer/Bushman Panels 17,21 (Supplementary Table 6,7; Supplementary Figure 4). We utilized previously published inversion tagging SNPs 10,16 and our copy number estimates in combination with publicly available phased SNP data to identify additional inversion- and haplotype-specific duplication tagging SNPs (Supplementary Table 8).
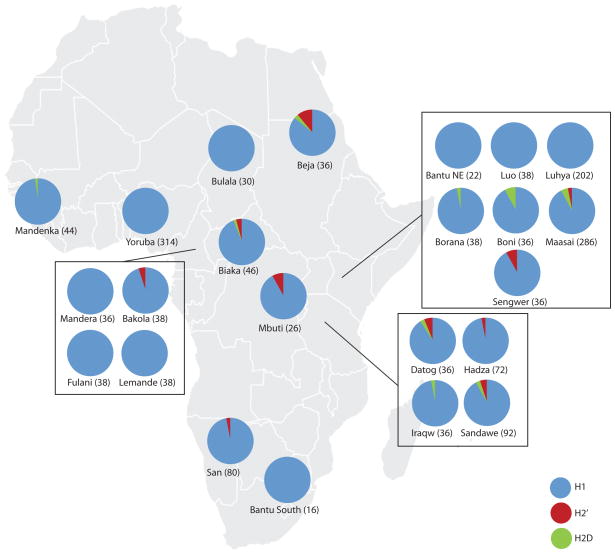
We were able to accurately type 818 African individuals from 23 diverse ethnic groups for the H1, H2′, and H2D haplotypes. We tested the H2 orientation of nine samples for which cell lines were available and confirmed the inversion orientation status in all individuals (Supplementary Table 3). We were unable to identify common SNPs that could distinguish H1′ from H1D haplotypes. We find that the H2 haplotypes are almost absent in Western African populations but are much more prevalent in the Eastern African populations (Figure 3) than originally estimated 10,16. For example we examined 286 Maasai individuals, we found the H2 haplotype frequency to be approximately 7%, which was thought to be absent from this population 16 (Table 1). The highest inversion frequency is 13% found in the Beja from Sudan—a population that has experienced significant gene flow from Middle Eastern populations16 where the frequency of H2 haplotypes is enriched. The inversion is nonexistent from populations speaking Niger-Kordofanian languages with the major exception of the Biaka and Bakola Pygmies where the inversion is found at roughly 5% frequency. The H2′ is primarily found in the hunter-gatherer populations—the San, Hadza, Bakola, Biaka, Mbuti, and Sengwer. The highest frequency of H2D is found in the Boni, Maasai, and Sandawe.
Figure 3. Haplotype frequency of 17q21.31 inversion in Africa.
Frequency of direct (H1), inverted (H2′), and inverted with duplication (H2D) haplotypes in 818 individuals (1636 chromosomes) from 23 African populations. The H2′ haplotype is absent from virtually all Western African individuals except for the Pygmy populations (Bakola, Biaka, and Mbuti). The H2′ haplotype frequency is highest in the Beja from Sudan likely due to admixture from neighboring Middle Eastern countries. The inversion is also at appreciable frequencies in the other hunter-gatherer populations (San, Hadza, Sandawe, Boni, and Sengwer).
Table 1.
Frequencies of H1′, H2′, and H2D in 23 diverse African ethnic groups
| Population | Country | Number of individuals | Frequency of H1′ | Frequency of H2 | Frequency of H2D | Subsistence Pattern | Language Family | Language Major Subgrouping | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Bakola | Cameroon | 19 | 94.74% | 5.26% | 0.00% | Hunter-gatherer | Niger-Kordofanian | Bantoid | African Diversity Panel |
| Bantu_N.E. | Kenya | 11 | 100.00% | 0.00% | 0.00% | Farmer | Niger-Kordofanian | Bantoid | HGDP |
| Bantu_South | South Africa | 8 | 100.00% | 0.00% | 0.00% | Farmer | Niger-Kordofanian | Bantoid | HGDP |
| Beja | Sudan | 18 | 86.11% | 11.11% | 2.78% | Herder | Afroasiatic | Cushitic | African |
| Biaka | Central African Republic | 23 | 93.48% | 4.35% | 2.17% | Hunter-gatherer | Niger-Kordofanian | Adamawa-Ubangi | HGDP, H2 Diversity Panel |
| Boni | Kenya | 18 | 92.11% | 0.00% | 7.89% | Hunter-gatherer | Afroasiatic | Cushitic | African Diversity Panel |
| Borana | Kenya | 19 | 97.37% | 0.00% | 2.63% | Herder | Afroasiatic | Cushitic | African Diversity Panel |
| Bulala | Chad | 15 | 100.00% | 0.00% | 0.00% | Farmer | Nilo-Saharan | Central Sudanic | African Diversity Panel |
| Datog | Tanzania | 18 | 91.67% | 5.56% | 2.78% | Herder | Nilo-Saharan | Eastern Sudanic | African Diversity Panel |
| Fulani | Cameroon | 19 | 100.00% | 0.00% | 0.00% | Herder | Niger-Kordofanian | Senegambian | African Diversity Panel |
| Hadza | Tanzania | 36 | 97.22% | 2.78% | 0.00% | Hunter-gatherer | Khoesan | Hadza | African Diversity Panel, Hunter-Gatherer |
| Iraqw | Tanzania | 18 | 97.22% | 0.00% | 2.78% | Mixed farmer | Afroasiatic | Cushitic | African Diversity Panel |
| Lemande | Cameroon | 19 | 100.00% | 0.00% | 0.00% | Farmer | Niger-Kordofanian | Bantoid | African Diversity Panel |
| Luhya | Kenya | 101 | 100.00% | 0.00% | 0.00% | Farmer | Niger-Kordofanian | Bantoid | HapMap, 1000 Genomes Project |
| Luo | Kenya | 19 | 100.00% | 0.00% | 0.00% | Herder | Niger-Kordofanian | Bantoid | African Diversity Panel |
| Maasai | Kenya | 143 | 92.66% | 2.45% | 4.90% | Farmer | Nilo-Saharan | Eastern Sudanic | HapMap |
| Mandenka | Senegal | 22 | 97.73% | 0.00% | 2.27% | Herder | Niger-Kordofanian | Mande | HGDP |
| Mandera | Cameroon | 18 | 100.00% | 0.00% | 0.00% | Farmer | Niger-Kordofanian | Mande | African Diversity Panel |
| Mbuti | Democratic Republic of Congo | 13 | 92.31% | 7.69% | 0.00% | Hunter-gatherer | Nilo-Saharan | Central Sudanic | African Diversity Panel, HGDP |
| San | Namibia, South Africa | 40 | 97.50% | 2.50% | 0.00% | Hunter-gatherer | Khoesan | Southern | HGDP, Bushman Collection, Hunter-Gatherer |
| Sandawe | Tanzania | 46 | 91.30% | 5.43% | 3.26% | Hunter-gatherer | Khoesan | Sandawe | African, Hunter-Gatherer Panel |
| Sengwer | Kenya | 18 | 91.67% | 8.33% | 0.00% | Hunter-gatherer | Nilo-Saharan | Eastern Sudanic | African Diversity Panel |
| Yoruba | Nigeria | 157 | 100.00% | 0.00% | 0.00% | Farmer | Niger-Kordofanian | Defoid | African Diversity Panel, HGDP, HapMap, 1000 Genomes Project |
Evolutionary age of duplication events
To understand the evolutionary history without the complication of recombination, we used the alignment of phased SNPs from a 136 kbp LD block within the inversion interval to build a maximum likelihood tree for each haplotype and ethnic group in the HapMap Project (Figure 4a). There were four important observations. First, there is strong bootstrap support indicating that the H1 and H2 haplotype clades are completely distinct. Second, there is strong support for the hypothesis that the H1′ haplotype is ancestral to the H1D haplotype. Third, the H2 and H2D show little to no variation. Finally, the analysis strongly suggests that the H1- and H2-specific duplications of the KANSL1 locus were separate, derived events.
Figure 4. Phylogenetic relationship between H1 and H2 haplotypes.
(a) Alignment of 43 SNPs from a 136 kbp LD block within the inversion region (chr17:41466118-41602794, NCBI build36) from HapMap individuals (total N = 728 individuals; 1456 chromosomes) were used to build a maximum likelihood tree with 1000 bootstrap replicates (all branches with 100% bootstrap support). (b) An unrooted neighbor-joining tree was constructed using MEGA4 50 complete deletion option based on 204,447 aligned base pairs from unique sequence within the inversion. The number of mutations for each branch is indicated above the branch. African individuals are highlighted in red and Europeans are highlighted in blue.
To overcome SNP ascertainment biases we obtained complete genomic sequence from the representative haplotypes including all possible single nucleotide variants (SNVs). We sequenced H2D haplotype-resolved fosmids derived from a European individual (NA12156) carrying this duplication 22; a European H2′ homozygote (NA20589) and a Maasai H2D homozygote (NA21599) using NGS; and used publicly available sequence from the San Bushman, KB1 17 carrying the H2′ haplotype and a H1D homozygous individual (NA12878) from the 1000 Genomes Project. We also included previously published assemblies of the H1′ and H2D haplotypes from the RP11 BAC assemblies 13 and used these references to assist in sequence alignment from these additional genomes.
We constructed an unrooted neighbor-joining tree (Figure 4b) using Kimura 2-parameter distance estimates based on sequence alignments to the unique 204,447 bp portion of this region. Consistent with previous analyses, we estimate that the H1 and H2 haplotypes coalesced approximately 2.3 mya. We note, however, a striking dearth of genetic diversity on the H2 lineage (Table 2). We expect nucleotide diversity (π) between any two chromosomes from a constant population size evolving neutrally to be approximately 0.001 23. For the H1 lineage, π is equal to 0.00047 but is nearly four times lower for the H2 lineage (π = 0.00012). Although our sample size is small, we note that π is lowest for the H2D haplotype when compared to H2′ (0.00004 vs. 0.00025). We observe virtually no sequence differences between the genomic sequences for H2D individuals. This is unlikely to represent cryptic ancestry as whole-genome comparison of RP11 and NA12156 suggests an average heterozygosity of 0.000943 ± 0.000597. The topology of the tree as well as the lack of diversity on the H2 haplotypes is suggestive of a recent bottleneck followed by population expansion or selective sweep.
Table 2.
Nucleotide diversity between haplotype groups
| Population | N | Nucleotide diversity (pi) |
|---|---|---|
| All H1 | 2 | 0.00047 |
| All H2 | 5 | 0.00012 |
| H2D | 3 | 0.00004 |
| H2′ | 2 | 0.00025 |
| Human (all haplotypes) | 7 | 0.00207 |
| Nonhuman Primate* | 2 | 0.03281 |
Nonhuman primate = chimpanzee and orangutan
We estimate the coalescence time of the H2 and H2D haplotype in African populations to be approximately 136,000 ± 19,000 years old and the coalescence time between African and European H2 haplotypes at 48,000 ± 11,000 years old (Supplementary Table 9). The European H2′ and H2D are more similar than the African and European H2D suggesting that the European H2′ has possibly undergone homogenization with the more predominant H2D haplotypes. The H1′ and H1D haplotypes have a much older coalescence of approximately 250,000 ± 26,000 years ago consistent with Stefansson and colleagues′ data 10. We present these dates with the caveat that they represent an average coalescence time over the entire interval given that H1′ and H1D haplotypes and H2′ and H2D haplotypes can freely recombine, and these segments may represent sequence from multiple common ancestors.
We compared the sequence in the duplicated regions for each haplotype clade to obtain a more accurate evolutionary age for the duplication events that is not biased by recombination events across the inversion interval. We aligned the NA12878 H1D sequence to the RP11 H1′ sequence at CNP205 to estimate the age of the H1 duplication, which equals 247,000 ± 20,000 years. We repeated this analysis for the H2D sequence (CNP155) from NA21599 and the reference H2D sequence from RP11 and estimated the age of the duplication at 1.3 million ± 106,000 years. This is more consistent with the range of values estimated for the duplication from the Stefansson et al. 10 and Zody et al. 13 analyses. We note that the age of the H2 duplication is much deeper than the coalescent times estimated from the unique sequence of the sampled H2 haplotypes which range from ~48,000 to ~136,000 years depending on the pairs of haplotypes chosen to analyze as well as the segment of DNA analyzed. These younger coalescent times for H2 haplotypes are consistent with the recent TMRCA of H2 haplotypes observed in Donnelly et al. 16 as these authors used short tandem repeat polymorphisms within the inversion interval. These discrepancies are striking and are suggestive of selection on the H2 haplotype resulting in the recent coalescence of extant H2 haplotypes.
H1 and H2 haplotype exchange
In general, inversions are predicted to result in complete suppression of recombination; therefore, sequence divergence is expected to be higher than in freely recombining chromosomal segments. We examined the sequence divergence between the H1′ and H2D haplotype sequences from the RP11 BAC assembly. The average value of π between the two haplotypes is 0.00416; however, we discovered a 30 kbp stretch of sequence over which the average value of π is equal to 0.0005. This level of divergence is significantly different from the distribution of nucleotide diversity over the entire inversion interval (Kolmogorov-Smirnov D = 0.9345; p = 0) (Figure 5a). The region of relatively high sequence identity overlaps the 5′ region of the corticotropin-releasing hormone receptor 1 (CRHR1) gene including the promoter and first two exons. CRHR1 is involved in anxiety-related behavior and stress adaptation 24–28.
Figure 5. Historical exchange between H1 and H2 haplotypes.
(a) Divergence plotted in 5 kbp sliding windows. A 30 kbp region (chr17:41213364-41248960, middle red bar) of reduced divergence over the 5′ end of CRHR1 is revealed. The spike in divergence at 41.35 Mbp corresponds to a simple TATA repeat tract. Median-joining haplotype networks based on the HapMap collection for (b) the region proximal to the 5′ end of CRHR1 (chr17:41011056-41091056, far left red bar), (c) the region of reduced divergence, and (d) the region distal to the region of reduced divergence (chr17:41410073-41425073, far right red bar). The proportion of H1′ (blue), H1D (orange), H2′ (red), and H2D (green) haplotypes are shown. The haplotypes form distinct clades proximal and distal to the CRHR1 region while over the region of reduced divergence the haplotypes are mixed creating a large haplogroup where H1 chromosomes have sequence similar to the H2 chromosomes. Red tick marks represent the number of mutations separating each haplogroup.
To study the history of this region in greater detail, we constructed a series of median-joining haplotype networks (http://www.fluxus-engineering.com) using HapMap phase 3 SNPs for 728 unrelated individuals in the region of reduced diversity as well as proximal and distal loci for comparison (Supplementary Note). Over the proximal and distal intervals, the H1 and H2 chromosomes are cleanly divided into distinct haplotype clades (Figure 5b,c). In contrast, over the homogenized CRHR1 region we found that 15 H2′, 123 H2D chromosomes as well as 197 H1′ and 27 H1D chromosomes grouped together in a single haplogroup (Figure 5d). Thus, over the 5′ segment of CRHR1, some H1 haplotypes have a sequence unusually similar to that found on H2 haplotypes. As this region is too large for a gene conversion event, this likely represents a historical double recombination event between H1 and H2 haplotypes. This haplogroup configuration is found in all major continental groupings of HapMap suggesting that the double recombination event predates the dispersal of modern humans out of Africa.
DISCUSSION
Based on our survey of structural genetic diversity from 2700 diverse population samples, we conclude that the H1- and H2-specific duplications evolved independently and the absence of duplication was ancestral in both the H1 and H2 lineages. We have resolved eight distinct structural haplotypes that vary in size from 1.08 to 1.49 Mbp. Five of these haplotypes belong to the H1 lineage while three belong to the H2 lineage. The least complex haplotype with regards to duplication architecture is the H2.1 haplotype, consistent with H2 haplotype reported for the San Bushman. European and Mediterranean populations show a dramatic enrichment of duplicated haplotypes (60% frequency) when compared to any other worldwide population group. When compared to the inversion, these duplications show greater population stratification.
These population differences have important disease ramifications with respect to the 17q21.31 microdeletion syndrome 11,14,29. Since the H2D haplotype is the only one out of eight possible configurations with homologous segmental duplications in direct orientation flanking the disease-critical region, only carriers of the H2D haplotype are predisposed to the 17q21.31 microdeletion through NAHR. Thus, European populations are much more at risk for the 17q21.31 microdeletion syndrome than Asians and Africans. The H2′ inversion haplotype (enriched among Africans and Southern Europeans) does not carry the predisposing duplication and therefore is not at risk for this recurrent deletion. We find 97% of the cases reported in the literature occur among individuals of European descent 30. A screen of 1084 African American samples with developmental delay using a TaqMan assay found no occurrences of the 17q21.31 microdeletion 31. The only known African American 17q21.31 microdeletion reported 30 had breakpoints mapping outside of the segmental duplications and thus occurred by a mechanism other than NAHR.
Our analyses demonstrate that either the H1′ or H2′ haplotype is ancestral; however, combined with previous analyses, the results presented here favor H2′ as the ancestral haplotype of the genus Homo. First, SNPs monomorphic among H2 haplotypes but polymorphic among the H1 haplotypes matched the chimpanzee allele 90% of the time, but SNPs monomorphic in H1 haplotypes and polymorphic in H2 haplotypes only matched the chimpanzee allele 60% of the time 13. Second, the inverted configuration is the ancestral state based on an analysis of Old World and New World monkeys 13. Finally, our phylogenetic and coalescent analyses provide strong evidence for an African origin of the H2 haplotype. We find the H2′ haplotype among populations thought to map near the root of the human phylogeny such as the San Bushman and other hunter-gatherers such as the click-speaking populations of Tanzania and Pygmies. Previous studies suggest an ancient genetic affinity and shared ancestry among these groups 21,32. Additionally our analyses indicate the San H2′ haplotype has an older evolutionary age when compared to the African H2D and European H2′ haplotypes.
Despite its high frequency among European populations, we observe extraordinary homogeneity among the H2 haplotypes. The ancient coalescence of H1 and H2 and the excess of rare polymorphisms among H2 haplotypes indicate a recent bottleneck or selective sweep particularly in the European H2D lineage where nucleotide diversity is the lowest. Recent analyses support the original observation that the H2 haplotype is associated with increased mean rates of recombination in females 33. It is also known that females with increased mean rates of recombination have more offspring 34,35 strengthening the evidence for a selective advantage for H2D carriers. This observation remains intriguing since the duplication architecture associated with H2D carriers clearly predisposes to microdeletion 14, and, therefore, must be subjected to weak purifying selection.
Among the 887 informative SNPs between the H1 and H2 haplotypes; there are 9 missense, 9 synonymous, and 22 UTR mutations on the H2 haplotype (Supplementary Table 10). Seven of the nine missense mutations are in IMP5 (intramembrane protease 5), and four of these are predicted to alter the protein structure by PolyPhen 36. The H1 alleles of two of these four amino acid altering mutations have been previously associated with Parkinson’s disease 37 and therefore the H2 missense mutations may represent substitutions under positive selection. Interestingly, the H2 haplotypes carry the derived allele for these four SNPs whereas the other SNPs that are not predicted to alter the protein retain the ancestral allele when compared to the chimpanzee. The H2 has potentially changed functionally while the H1 haplotype has kept a structure more similar to the chimpanzee, lending some support to the hypothesis of selection on the H2 haplotype. The IMP5 gene is predicted to be under purifying selection using maximum likelihood analyses of eight mammalian species (data not shown), although non-synonymous changes are not uncommon during evolution. The two remaining missense mutations are in KANSL1—a gene when mutated results in a phenotype similar to the 17q21.31 microdeletion syndrome 38. Both variants are not predicted to alter the protein structure as predicted by PolyPhen, but are highly conserved (GERP scores between 2 and 4).
It is also not clear why the H1D haplotypes have risen to such high frequencies in European populations. Given that certain H1 haplotypes are associated with neurological disorders such as Parkinson’s disease (PD) 39,40, Alzheimer’s disease (AD)41, and progressive supranuclear palsy (PSP) 18,42,43, we examined association between the duplication and these disease predisposing haplotypes in the 1000 Genomes Project individuals (Supplementary Table 11). We found that the duplication was present less often than predicted by linkage equilibrium for the H1c haplotype (p<0.0001), which is a risk haplotype for AD and PSP 41,42. The duplication was extremely rare on PD risk haplotypes 39,42,44; however, given the sample size and low frequencies of the PD risk haplotypes in the population, we were unable to assess whether these associations were significant. Nevertheless, these observations suggest that the disease risk haplotypes likely arose on H1′ haplotypes, warranting further investigation into the possible protective role of the duplication in these diseases. The H2 haplotype, which almost always bears the duplication in Europeans, is protective against many of these diseases 39,44 but predisposes to microdeletion associated with intellectual disability.
If there is a selective advantage to both the H1D and H2D haplotypes, one possibility may be the recurrent duplications involving both the promoter and first exon of KANSL1. KANSL1 is a chromatin modifier gene thought to have a role in complex brain function and which has been associated with the 17q21.31 microdeletion syndrome 11,38,45. We note that KANSL1 gene expression is increased in the brains of PD cases when compared to controls 39 suggesting that dysregulation of KANSL1 expression may have phenotypic consequences of disease relevance.
Another interesting observation is the striking absence of genetic diversity within a 30 kbp stretch of CRHR1 between H1 and H2 haplotypes despite the deep evolutionary divergence of the haplotypes 2.3 mya. The observed decrease in divergence at the CRHR1 locus is reminiscent of patterns observed among some highly divergent HLA haplotypes—a group of haplotypes that, like the 17q21.31 interval, are otherwise characterized by high sequence diversity and reduced recombination between divergent clades 46. It seems implausible that this 30 kbp stretch has been maintained at such a degree of sequence identity relative to the rest of the region since the coalescence of the two haplotypes. We propose that the observed pattern results from a classical double crossover event via an inverted loop structure during meiosis resulting in the transfer of sequence between these two haplotypes sometime after their initial separation. We have no evidence of the reciprocal event from sequence data suggesting that it may have been lost from the human population.
In conclusion, we propose that the ancestral H2′ haplotype arose in Eastern or Central Africa and spread to Southern Africa before the emergence of anatomically modern humans (Figure 6). Approximately 2.3 mya, the inversion rearranged to what we now refer as the “direct” orientation haplotype (H1′). This haplotype spread throughout the Homo ancestral populations in the African continent virtually replacing the H2′ haplotype and becoming the predominant haplotype. We note that both Denisova and Neandertal sister groups are predicted to be H1′ 47,48. These early haplotypes were much simpler in their duplication architecture similar to great apes. We find that the more complex duplication architectures are particularly enriched among out-of-Africa populations. Based on sequence at the duplication loci, we estimate that the H2-specific duplication event occurred approximately 1.3 mya. Independent of the H2 duplication, the H1-specific duplication event occurred much more recently, approximately 250,000 years ago. Interestingly, we do not observe this haplotype in any of the African or Asian populations suggesting that it may have been lost in these populations as a result of genetic drift. The H2D haplotype has risen to frequencies of 10–25% in European populations with virtually no genetic variation, suggesting an extremely recent and rapid expansion of this haplotype. High-coverage sequencing of more individuals along with fecundity data will likely shed further light on whether the high frequency of the haplotype-specific duplication in Europeans is due to selection or the effects of demographic history particular to this locus.
Figure 6. Evolutionary history of 17q21.31 haplotypes.
We propose a model where the H2′ haplotype represents the ancestral configuration of the 17q21.31 region in humans. Approximately 2.3 mya, the inversion toggled back to the direct orientation and spread to South Africa prior to the emergence of modern humans. The H2D duplication arose in Africa 1.3 mya, and the H1D duplication independently arose much more recently, approximately 250,000 years ago. The H1′ haplotype spread throughout Western Africa, and all haplotypes spread to the Middle East and Europe as part of the out-of-Africa migration.
METHODS
Samples
Genomic DNA and lymphoblast cell lines from HapMap individuals were obtained from Coriell Cell Repository (Camden, NJ). Genomic DNA and lymphoblast cell lines from African individuals were obtained as described in Donnelly et al. 16 and Tishkoff et al. 32. See Supplementary Tables 1 and 2 for samples used. Publicly available SNP and sequence data for the 17q21.31 region were downloaded from HapMap Phase III, HGDP, 1000 Genomes Project, and Stanford University.
Array CGH
We designed custom-targeted microarray (Roche NimbleGen) for the 17q21.31 region (12,468 probes tiled across 1.9 Mbp). DNA was labeled using the NimbleGen Dual-Color DNA Labeling Kit (Roche NimbleGen, Inc., Madison, WI) using NA19240 as a reference sample. Hybridizations were performed at 42°C for 60 hr using the NimbleGen Wash Buffer Kit as described previously 51. Scanned images (aGenePix 4000B Scanner) were analyzed using NimbleScan v2.5 and copy number variants were called using the segMNT algorithm.
Fluorescence in situ hybridization
Metaphase spreads and interphase nuclei were obtained from human lymphoblast cell lines. FISH experiments were performed using fosmid clones (Supplementary Table 8) directly labeled by nick-translation with Cy3-dUTP (Perkin-Elmer), Cy5-dUTP (Perkin-Elmer), and fluorescein-dUTP (Enzo) as described previously. 52. Digital images were obtained using a Leica DMRXA2 epifluorescence microscope equipped with a cooled CCD camera (Princeton Instruments). DAPI, Cy3, Cy5, and fluorescein fluorescence signals, detected with specific filters, were recorded separately as gray-scale images. Pseudocoloring and merging of images were performed using Adobe Photoshop software. A minimum of 50 interphase nuclei were scored for each inversion to statistically determine the orientation of the examined region.
Illumina sequencing of H2D haplosorted fosmid clones
Fosmid clones from NA12156 were assigned to haplotypes using previously described methods 22 and DNA was isolated by a modified alkaline lysis miniprep procedure as follows: cell pellet was resuspended in 250 μL Qiagen buffer P1 with RNAse and lysed with 250 μL of 0.2M NaOH/1%SDS solution for five minutes. Lysis was neutralized with 250 μL 3M NaOAc, pH 4.8. Neutralized lysate was incubated on ice for 40 minutes, collected by centrifugation for 15 min at 13000 rpm and 4°C, concentrated by standard ethanol precipitation, and resuspended in 50 μL 10 mM Tris-Cl pH 8.5. Libraries were prepared from fosmid clone DNA using Illumina-compatible Nextera DNA sample prep kits (Epicentre, Cat. No. GA09115). The manufacturer’s protocol was followed with modifications including a set of barcoded oligos as described 53. Barcoded libraries were combined for size selection using E-Gel SizeSelect 2% (Invitrogen, Cat. No.G6610-02). The band spanning 600–700 bp was amplified via limited-cycle PCR with iProof High-Fidelity polymerase (Bio-Rad) with the following program: initial denaturation at 98°C for 30 sec, followed by a 6–12 cycles of denaturation at 98°C for 10 sec, annealing at 64°C for 30 sec, and extension at 72°C for 40 sec. Amplified, size-selected libraries were then purified with QIAquick PCR Purification Kit (Cat. No. 28104), quantified on an Invitrogen Qubit Fluorometer, and paired-end sequenced (101 bp reads) on an Illumina HiSeq 2000. Sequence reads were aligned to the chr17_ctg5_hap1 reference sequence (GRCh37) using BWA (version 0.5.9), and variants were called using SAMtools mpileup (version 0.1.16).
Illumina sequencing of H2 and H2D homozygous genomes
3 μg of genomic DNA from NA20589 (H2′/H2′) and NA21599 (H2D/H2D) were sheared, end-repaired, an A-tail was added, and adaptors were ligated to the fragments as described54. After ligation samples were run on a 6% pre-cast polyacrylamide gel (Invitrogen, Cat. No. EC6265BOX). The band at 400 bp was excised, diced, and incubated as described above. Size selected fragments were amplified with 0.5 μL of primers, 25 μL of 2X iProof, 0.25 μL of SYBR Green, and 8.25 μL of dH2O under the following conditions: 98°C for 30 sec, 30 cycles of 98°C for 10 sec, 60°C for 30 sec, 72 °C for 30 sec, 72 °C for 15 sec followed by 72 °C for 2 min. Fluorescence was assessed between the 30 and 15 sec 72 °C step. Amplified, size-selected libraries were quantified using an Agilent 2100 Bioanalyzer and paired-end sequenced (101 bp reads) on an Illumina HiSeq 2000. We generated a total of 13–14 fold sequence coverage.
Haplotype assignment, coalescent and phylogenetic analyses
We assigned haplotypes to 728 phased HapMap samples (1456 chromosomes) using previously ascertained inversion marking SNPs 10,16 and H2-specific duplication tagging SNPs identified in the present study in combination with array CGH 55 copy number estimates. We used a 3-SNP haplotype (rs1800547, rs2957297, rs199451) to assign phased haplotypes to H1, H2, and H2D haplogroups. We required that all three SNPs match the expected haplotype. Phase switch errors were manually corrected as described13. We confirmed H1/H2 status with PCR genotyping results and were able to resolve all but one haplotype; this individual was eliminated from further analysis. We used array CGH 55 copy number estimates to assign genotypes for these 728 individuals. We finally combined these sources of information to assign the final haplotype. Six out of 728 individuals were discordant between the SNP genotype and array CGH-based copy number estimate. Three of these were resolved by FISH experiments while three were resolved by PCR genotyping. Fifty-seven individuals were H1/H1D and therefore the haplotypes could not be confidently assigned; these individuals are included in haplotype frequency data but not in phylogenetic analyses.
We combined the read-depth-based copy number estimates with the phased SNP data from the 1000 Genomes Project using a 4-SNP haplotype (rs1396862, rs17650901, rs1052553, rs199448) to assign phased haplotypes to H1, H2, and H2D haplogroups. We required that all four SNPs match the expected haplotype; 0.5% of haplotypes had one discordant SNP and these haplotypes were flagged for manual inspection for phase switch and genotyping errors. Phase switch errors were manually corrected as described in Zody et al. 13. We then used read-depth-based copy number estimates to assign phased haplotypes to H1, H1D, H2, and H2D haplogroups. Finally, we combined these sources of information to assign the final haplotype. Sixteen out of 805 individuals were discordant between the SNP genotype and read-depth-based copy number estimate. For five of these individuals we also had array CGH and/or FISH data; for 4/5 of these discordances the SNP haplotype was concordant with the array CGH/FISH data while for 1/5 instances the read-depth-based haplotype was consistent with the array CGH/FISH data. For the remaining 11 discordant individuals we eliminated them from the rest of the analysis since we could not confidently assign the haplotype. Sixty-seven individuals were H1/H1D and therefore the haplotypes could not be confidently assigned; these individuals are included in haplotype frequency data but not in phylogenetic analyses.
We also used the Illumina 650Y SNP genotyping data from 936 unrelated individuals from the HGDP collection. We phased the 650Y data using BEAGLE 56,57 and assigned haplotypes (H1, H2′, and H2D) based on a 4 SNP haplotype (rs175635986, rs19871997, rs2668692, rs199533). We required that all four SNPs match the expected haplotype; 0.5% of haplotypes had one discordant SNP or missing data and these were eliminated from further analysis. Illumina 1M SNP data from the African Diversity Panel (unpublished data) were used to assign haplotypes (H1, H2′, and H2D) based on a 3 SNP haplotype (rs1800547, rs2957297, rs199451). Four individuals from the African Diversity Panel were discordant between SNP haplotype and PCR genotyping results and were removed from further analysis. Illumina 550K SNP data from the Hunter-Gatherer Panel 21 were used to assign haplotypes (H1, H2′, and H2D) based on a 4 SNP haplotype (rs17563986, rs1800547, rs1981997, rs2668692, rs199533). We required that all four SNPs match the expected haplotype; two haplotypes had one SNP that did not match the expected haplotype and were excluded from further analysis. We were able to type a total of 351 African samples from the African Diversity and the Hunter-Gatherer Panels. We were unable to find any tag SNPs for the H1D haplotype so the HGDP, African Diversity Panel, and Hunter-Gatherer analyses do not contain any chromosomes assigned to that haplotype as we did not have independent copy number information on these individuals.
We used PHYLIP 58 to build a maximum likelihood tree based on the alignment of 43 SNPs from the 136 kbp LD block identified in the HapMap Project populations. BAC-based assemblies of the RP11 H1′ and H2D haplotypes and chimpanzee and orangutan haplotypes were aligned to KB1, NA12878, NA20589, NA21599 whole-genome sequence and NA12156 haploresolved fosmid sequence with CLUSTALW 59. We constructed a neighbor-joining phylogeny using Kimura-2 parameter distance (complete deletion option) using MEGA4 50. Nucleotide diversity and other population genetic analyses were performed using DnaSp v5 60.
Supplementary Material
Acknowledgments
The authors wish to thank J. Akey, M. Dennis, and B. Dumont for helpful discussions and C. Alkan for computational assistance. We thank Z. Jiang for his initial work on the H1/H2 alignments. We are grateful to T. Brown for assistance with manuscript preparation, C. Lee for technical assistance and to the anonymous reviewers of this paper who provided insightful comments. We thank the 1000 Genomes Project Consortium for access to unpublished sequence data for the 17q21.31 locus. K.M.S. was supported by a Ruth L. Kirschstein National Research Service Award (NRSA) training grant to the University of Washington (T32HG00035) and an individual NRSA Fellowship (F32GM097807). C.D.C. was supported by an individual NRSA Fellowship (F32HG006070). P.H.S. was supported by a Natural Sciences and Engineering Research Council of Canada Fellowship. JMK was supported by a Ruth L. Kirschstein National Research Service Award (NRSA) training grant to Stanford University (T32HG000044). This work was supported by National Institutes of Health Grants HG002385 and HG004120 to E.E.E. E.E.E. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
ACCESSION NUMBERS
All sequence data has been submitted to the Short Read Archive (SRA) under the ID: SRA046964.
AUTHOR CONTRIBUTIONS
This study was designed by K.M.S., F.A. and E.E.E. K.M.S. performed array CGH, genotyping and sequence analysis. F.A. performed FISH experiments and fosmid shotgun sequencing library construction. P.H.S. performed read-depth-based copy number analysis. J.M.K. performed sequence analysis on double recombination region. C.D.C performed array CGH analysis. L.V. and M.M. performed whole-genome shotgun sequencing library construction and PCR genotyping. L.S. and W.B. performed PCR genotyping and SNP array genotyping. M.I., G.L. T.B.N., S.A.O., J-M.B., and A.F. contributed to African sample collection. M.P.D. and K.K.K. contributed to H2 Diversity Panel sample collection and genotyping. S.A.T. contributed to African sample collection and SNP array data. K.M.S., F.A., J.M.K., S.A.T., and E.E.E. contributed to data interpretation. K.M.S., F.A., and E.E.E. wrote the manuscript.
URL
HapMap Phase III (http://hapmap.ncbi.nlm.nih.gov/)
HGDP (http://www.cephb.fr/en/hgdp/diversity.php/)
1000 Genomes Project (http://www.1000genomes.org/)
Stanford University (http://www-evo.stanford.edu/repository/paper0002/)
San Bushman, KB1 (ftp://ftp.bx.psu.edu/data/bushman/)
References
- 1.Dobzhansky T. The genetics of natural populations. Genetics. 1950;35:288–302. doi: 10.1093/genetics/35.3.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dobzhansky T, Sturtevant AH. Inversions in the chromosomes of Drosophila pseudoobscura. Genetics. 1938;23:28–64. doi: 10.1093/genetics/23.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lowry DB, Willis JH. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tuzun E, et al. Fine-scale structural variation of the human genome. Nat Genet. 2005;37:727–32. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 5.Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailey JA, et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–7. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 7.Sharp AJ. Emerging themes and new challenges in defining the role of structural variation in human disease. Hum Mutat. 2009;30:135–44. doi: 10.1002/humu.20843. [DOI] [PubMed] [Google Scholar]
- 8.Lupski JR. Genome structural variation and sporadic disease traits. Nat Genet. 2006;38:974–6. doi: 10.1038/ng0906-974. [DOI] [PubMed] [Google Scholar]
- 9.Antonacci F, et al. Characterization of six human disease-associated inversion polymorphisms. Hum Mol Genet. 2009;18:2555–66. doi: 10.1093/hmg/ddp187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stefansson H, et al. A common inversion under selection in Europeans. Nat Genet. 2005;37:129–37. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 11.Sharp AJ, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet. 2006;38:1038–42. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- 12.Koolen DA, et al. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat Genet. 2006;38:999–1001. doi: 10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- 13.Zody MC, et al. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat Genet. 2008;40:1076–83. doi: 10.1038/ng.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koolen DA, et al. Clinical and molecular delineation of the 17q21.31 microdeletion syndrome. J Med Genet. 2008;45:710–20. doi: 10.1136/jmg.2008.058701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sudmant PH, et al. Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–6. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donnelly MP, et al. The distribution and most recent common ancestor of the 17q21 inversion in humans. Am J Hum Genet. 2010;86:161–71. doi: 10.1016/j.ajhg.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuster SC, et al. Complete Khoisan and Bantu genomes from southern Africa. Nature. 2010;463:943–7. doi: 10.1038/nature08795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baker M, et al. Association of an extended haplotype in the tau gene with progressive supranuclear palsy. Hum Mol Genet. 1999;8:711–5. doi: 10.1093/hmg/8.4.711. [DOI] [PubMed] [Google Scholar]
- 19.Rao PN, Li W, Vissers LE, Veltman JA, Ophoff RA. Recurrent inversion events at 17q21.31 microdeletion locus are linked to the MAPT H2 haplotype. Cytogenet Genome Res. 2010;129:275–9. doi: 10.1159/000315901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boettger LM, Handsaker RE, Zody MC, McCarroll SA. Structural haplotypes and recent evolution of the human 17q21.31 region. Nat Genet. 2012 doi: 10.1038/ng.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henn BM, et al. Hunter-gatherer genomic diversity suggests a southern African origin for modern humans. Proc Natl Acad Sci U S A. 2011;108:5154–62. doi: 10.1073/pnas.1017511108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kidd JM, et al. Haplotype sorting using human fosmid clone end-sequence pairs. Genome Res. 2008;18:2016–23. doi: 10.1101/gr.081786.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cargill M, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22:231–8. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- 24.Heim C, et al. Effect of Childhood Trauma on Adult Depression and Neuroendocrine Function: Sex-Specific Moderation by CRH Receptor 1 Gene. Front Behav Neurosci. 2009;3:41. doi: 10.3389/neuro.08.041.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Z, et al. Association of corticotropin-releasing hormone receptor1 gene SNP and haplotype with major depression. Neurosci Lett. 2006;404:358–62. doi: 10.1016/j.neulet.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 26.Liu Z, et al. Association study of corticotropin-releasing hormone receptor1 gene polymorphisms and antidepressant response in major depressive disorders. Neurosci Lett. 2007;414:155–8. doi: 10.1016/j.neulet.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 27.Bradley RG, et al. Influence of child abuse on adult depression: moderation by the corticotropin-releasing hormone receptor gene. Arch Gen Psychiatry. 2008;65:190–200. doi: 10.1001/archgenpsychiatry.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polanczyk G, et al. Protective effect of CRHR1 gene variants on the development of adult depression following childhood maltreatment: replication and extension. Arch Gen Psychiatry. 2009;66:978–85. doi: 10.1001/archgenpsychiatry.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaw-Smith C, et al. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nat Genet. 2006;38:1032–7. doi: 10.1038/ng1858. [DOI] [PubMed] [Google Scholar]
- 30.Cooper GM, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–46. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mefford HC, et al. A method for rapid, targeted CNV genotyping identifies rare variants associated with neurocognitive disease. Genome Res. 2009;19:1579–85. doi: 10.1101/gr.094987.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tishkoff SA, et al. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–44. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fledel-Alon A, et al. Variation in human recombination rates and its genetic determinants. PLoS One. 2011;6:e20321. doi: 10.1371/journal.pone.0020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kong A, et al. Recombination rate and reproductive success in humans. Nat Genet. 2004;36:1203–6. doi: 10.1038/ng1445. [DOI] [PubMed] [Google Scholar]
- 35.Coop G, Wen X, Ober C, Pritchard JK, Przeworski M. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science. 2008;319:1395–8. doi: 10.1126/science.1151851. [DOI] [PubMed] [Google Scholar]
- 36.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simon-Sanchez J, et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet. 2009;41:1308–12. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koolen DA, et al. Mutations in the chromatin modifier gene KANSL1 cause the 17q21.31 microdeletion syndrome. Nat Genet. doi: 10.1038/ng.2262. In press. [DOI] [PubMed] [Google Scholar]
- 39.Tobin JE, et al. Haplotypes and gene expression implicate the MAPT region for Parkinson disease: the GenePD Study. Neurology. 2008;71:28–34. doi: 10.1212/01.wnl.0000304051.01650.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skipper L, et al. Linkage disequilibrium and association of MAPT H1 in Parkinson disease. Am J Hum Genet. 2004;75:669–77. doi: 10.1086/424492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Myers AJ, et al. The MAPT H1c risk haplotype is associated with increased expression of tau and especially of 4 repeat containing transcripts. Neurobiol Dis. 2007;25:561–70. doi: 10.1016/j.nbd.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 42.Pittman AM, et al. The structure of the tau haplotype in controls and in progressive supranuclear palsy. Hum Mol Genet. 2004;13:1267–74. doi: 10.1093/hmg/ddh138. [DOI] [PubMed] [Google Scholar]
- 43.Hoglinger GU, et al. Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nat Genet. 2011;43:699–705. doi: 10.1038/ng.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seto-Salvia N, et al. Dementia risk in Parkinson disease: disentangling the role of MAPT haplotypes. Arch Neurol. 2011;68:359–64. doi: 10.1001/archneurol.2011.17. [DOI] [PubMed] [Google Scholar]
- 45.Dubourg C, et al. Clinical and molecular characterization of 17q21.31 microdeletion syndrome in 14 French patients with mental retardation. Eur J Med Genet. 2011;54:144–51. doi: 10.1016/j.ejmg.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 46.Dawkins R, et al. Genomics of the major histocompatibility complex: haplotypes, duplication, retroviruses and disease. Immunol Rev. 1999;167:275–304. doi: 10.1111/j.1600-065x.1999.tb01399.x. [DOI] [PubMed] [Google Scholar]
- 47.Reich D, et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–60. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–22. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang Z, et al. Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat Genet. 2007;39:1361–8. doi: 10.1038/ng.2007.9. [DOI] [PubMed] [Google Scholar]
- 50.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 51.Antonacci F, et al. A large and complex structural polymorphism at 16p12.1 underlies microdeletion disease risk. Nature genetics. 2010;42:745–50. doi: 10.1038/ng.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Antonacci F, et al. A large and complex structural polymorphism at 16p12.1 underlies microdeletion disease risk. Nat Genet. 2010;42:745–50. doi: 10.1038/ng.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adey A, et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010;11:R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Igartua C, et al. Targeted enrichment of specific regions in the human genome by array hybridization. Curr Protoc Hum Genet. 2010;Chapter 18(Unit 18):3. doi: 10.1002/0471142905.hg1803s66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Campbell CD, et al. Population-genetic properties of differentiated human copy-number polymorphisms. Am J Hum Genet. 2011;88:317–32. doi: 10.1016/j.ajhg.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–23. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–97. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Felsenstein J. PHYLIP -- Phylogeny Inference Package (Version 3.2) Cladistics. 1989:164–166. [Google Scholar]
- 59.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 60.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–2. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.