Abstract
Selenium (Se) is an essential micronutrient. Its biological functions are associated with selenoproteins, which contain this trace element in the form of the 21st amino acid, selenocysteine. Genetic defects in selenocysteine insertion into proteins are associated with severe health issues. The consequences of selenoprotein deficiency are more variable, with several selenoproteins being essential, and several showing no clear phenotypes. Much of these functional studies benefited from the use of rodent models and diets employing variable levels of Se. This review summarizes the data obtained with these models, focusing on mouse models with targeted expression of individual selenoproteins and removal of individual, subsets or all selenoproteins in a systemic or organ-specific manner.
Keywords: selenium, selenocysteine, selenoproteins, mouse models
1. Selenoproteins: identification and oxidoreductase functions
Glutathione peroxidase 1 (GPx1) [1] was the first identified selenoprotein. Initially isolated from human erythrocytes, it was shown to protect hemoglobin from oxidative damage. Later, it was found to be dependent on selenium (Se), [2–5]. Se is incorporated into GPx1 in the form of the 21st amino acid, selenocysteine (Sec). In comparison to cysteine (Cys), Sec has a lower pKa and is a stronger nucleophile [6]. Almost all known selenoproteins are oxidoreductases with Sec in the active center. Sec insertion requires the presence of an in-frame UGA codon and the Sec insertion sequence (SECIS) element, a kink-turn RNA structure. In eukaryotes, the SECIS element is located in the 3’ UTRs of selenoprotein mRNAs. Biosynthesis of Sec occurs on its own tRNA, tRNA[Ser]Sec, which is initially charged with Ser. SECIS-binding protein 2 (SBP2 or Secisbp2) binds the SECIS element and recruits Sec-tRNA[Ser]Sec along with other factors involved in Sec insertion [7, 8]. Characterization of the structure and conserved sequences of the SECIS element allowed development of computational programs for identification of selenoprotein genes in sequence databases [9–11]. The SECISsearch program was designed to recognize sequence, structural and thermodynamic parameters of SECIS elements [12]. By searching for SECIS elements, in-frame UGA codons in the ORFs and the presence of Cys-containing orthologs of selenoproteins, selenoprotein genes could be identified in genomic sequences. Accordingly, the human genome was found to contain 25 selenoprotein genes (Table 1). Most of these proteins participate in maintaining cellular redox homeostasis, including three thioredoxin reductases (TRs), five glutathione peroxidases (GPx1), methionine sulfoxide reductase (MsrB1), and three thyroid hormone deiodinases (DIs).
Table 1.
Mammalian selenoproteins: localization and functions
| Selenoprotein | Localization | Function | References |
|---|---|---|---|
| 15 kDa selenoprotein (Sep15) | ER |
|
[21, 117–119] |
| Thyroid hormone deiodinase 1 (DI1, Dio1) | Plasma membrane |
|
[120, 121] |
| Thyroid hormone deiodinase 2 (DI2, Dio2) | ER |
|
[48] |
| Thyroid hormone deiodinase 3 (DI3, Dio3) | Plasma membrane |
|
[121, 122] |
| Glutathione peroxidase 1 (GPx1) | Cytosol |
|
[13] |
| Glutathione peroxidase 2 (GPx2) | Cytosol |
|
[123, 124] |
| Glutathione peroxidase 3 (GPx3) | Plasma |
|
[125] |
| Glutathione peroxidase 4 (GPx4, PHGPx) | Cytosol Mitochondria nucleus (testis-specific) |
|
[36] |
| Glutathione peroxidase 6 (GPx6) | Cytosol |
|
[124] |
| Selenoprotein H (SelH) | Nucleus |
|
[126–128] |
| Selenoprotein I (SelI) | Membrane | unknown function | [12] |
| Selenoprotein K (SelK) | ER membrane |
|
[66] |
| Selenoprotein M (SelM) | ER |
|
[129] |
| Selenoprotein N (SelN, SEPN1, SelN1) | ER membrane |
|
[24, 130] |
| Selenoprotein O (SelO) | Mitochondria |
|
[12] |
| Selenoprotein P (SelP) | Plasma |
|
[62, 131, 132] |
| Selenoprotein R (SelR, MsrB1, Selx1) | Cytosol |
|
[18] |
| Selenoprotein S (SelS, SEPS1, Tanis, VIMP, and SELENOS) | ER membrane |
|
[23, 24] |
| SPS2 | Cytosol |
|
[82, 133] |
| Selenoprotein T (SelT) | ER and Golgi |
|
[134] |
| Thioredoxin reductase 1 (TR1, Txnrd1) | Cytosol |
|
[15, 135] |
| Thioredoxin/glutathi one reductase (TGR, TR2, Txnrd3) | Cytosol |
|
[136] |
| Thioredoxin reductase 3 (Txnrd2, TR3) | Mitochondria |
|
[137] |
| Selenoprotein V (SelV) | Cytosol |
|
[126] |
| Selenoprotein W (SelW) | Cytosol |
|
[138] |
As shown in Table 1, the functions of several selenoproteins have been established, but the majority of selenoproteins have no known functions. Indeed, besides TRs, GPxs, MsrB1, DIs, and SPS2, the specific reactions catalyzed by selenoproteins are not known. However, conservation of selenoproteins among species and preservation of the complex biosynthetic pathway for their production indicate the importance of this class of proteins. So far, the common feature of all selenoproteins with the identified functions is their participation in oxidoreductase reactions. This type of reaction is important in intracellular redox homeostasis and antioxidant defense. GPxs (and possibly the N-terminal domain of SelP) are capable of reducing various peroxides [13, 14]. TRs and MsrB1 participate in the reduction of disulfides and methionine (Met) sulfoxide residues in proteins, respectively [15–18]. DIs catalyze reductive removal of iodine (I) from the outer ring of the prohormone thyroxine (T4) yielding various forms of thyroid hormones [19, 20]. Sep15, SelM, SelH, SelS, SelK, SelN, SelT, SelW are less characterized, whereas almost no studies have been done on SelV, SelO, and SelI. Most likely, many these proteins are also oxidoreductases with Sec in the active site. More than half of mammalian selenoproteins are characterized by the thioredoxin-like fold. This fold is a two-layer α/β/α sandwich structure that includes a conserved CxxC motif (i.e., two Cys separated by two other residues). In some cases, one of the Cys residues can be substituted with Ser or Thr. This fold is especially common for enzymes that catalyze formation or isomerization of disulfide bonds or perform other functions that change the redox state of cysteine residues. In addition, at least 6 out of 25 selenoproteins (Sep15, SelK, SelM, SelN, SelS, and SelT) reside in the ER lumen, an additional selenoprotein (D2 or Dio2) is associated with ER membranes (its catalytic site faces the cytosol), and several secreted selenoproteins pass through this compartment. The enrichment of the ER with selenoproteins suggests the roles of selenoproteins in ER-associated pathways, such as protein secretion/modification (Sep15, SelM [21]) and ER-associated protein degradation ERAD (SelS, SelK [22, 23, 24]).
2. Mouse models for studying selenoproteins
Knockout (KO) and transgenic models can be used for evaluating protein functions as well as for their impact on physiology and pathology. To examine selenoprotein functions, a number of mouse models have been developed and characterized. Generally, these models can be divided into two groups. The first group includes animals lacking (or overexpressing) one or two selenoproteins. The second group includes various mouse models characterized by the altered selenoprotein biosynthesis pathway. These animals develop systemic selenoprotein deficiency. The use of these animal groups is discussed in the following sections.
2.1 Targeted removal of individual selenoproteins
Several mouse models with targeted inactivation of one or two selenoproteins have been developed and characterized thus far [25, 26]. Their overview is given in Table 2. Three selenoproteins were found to be essential for embryogenesis: TR1, TR3 and GPx4. Knockout of cytosolic TR1 leads to embryonic death between days E8.5 and E10.5 [27, 28]. While the cardiomyocyte-specific TR1 KO mice developed normally, the neuronal system (NS)-specific TR1 KO caused severe neurological symptoms, such as ataxia and tremor [29]. These symptoms were the result of cerebellar hypoplasia, abnormal foliation, perturbed lamination and reduced proliferation of granule cell precursors in the cerebellum [29]. Mitochondrial TR3 KO induced embryonic lethality between days E13.5 and E15.5. Compared to controls, embryos were smaller, developed anemia and showed high levels of liver apoptosis. NS-specific TR3 KO mice developed normally without signs of neurodegeneration; however, the cardiomyocyte-specific TR3 KO mice died from the heart failure within a few hours of birth [30]. Disruption of mitochondrial TR3 in B and T cells did not affect viability and functions of immune cells [31]. Similar to mice, TR3 polymorphism was found to be associated with dilated cardiomyopathy in humans. Both nucleotide substitutions were in the open reading frame and were part of the FAD-binding domain [32].
Table 2.
Knockout of individual selenoprotein genes in mice
| Gene | Approach | Phenotype | References |
|---|---|---|---|
| GPx1 | Whole body |
|
[13, 139] |
| GPx2 | Whole body |
|
[42] |
| GPx1+GPx2 | Whole body |
|
[43] |
| GPx3 | Whole body |
|
[44] |
| GPx4 | Whole body |
|
[33] |
| GPx4 | Neuron specific |
|
[34, 36] |
| GPx4 | Spermatid-specific |
|
[38] |
| nGPx4 | Whole body |
|
|
| mGPx4 | Whole body |
|
|
| TR1 | Whole body |
|
[27] |
| TR1 | Cardiomyocyte-specific |
|
[27] |
| TR1 | Neuron-specific |
|
[29] |
| TR3 | Whole body |
|
[30] |
| TR3 | Cardiomyocyte-specific |
|
[30] |
| TR3 | Neuron-specific, T-and B cells specific |
|
[30, 31] |
| DI1 | Whole body |
|
[46] |
| DI2 | Whole body |
|
[48, 49] |
| DI3 | Whole body |
|
[52, 53] |
| DI1+DI2 | Whole body |
|
[51] |
| SelP |
|
[54–56] | |
| MsrB1 | Whole body |
|
[65] |
| Sep15 | Whole body |
|
[67] |
| SelK | Whole body |
|
[66] |
| SEPN1 | Whole body |
|
[69, 70] |
GPx4 is another essential selenoenzyme: its homozygous genetic inactivation was found to be lethal by E7.5 [33–35]. GPx4 is represented by cytosolic (cGPx4), nuclear (nGPx4) and mitochondrial (mGPx4) isoforms. These isoforms are synthesized from the same gene by alternative initiation of transcription and differ by their N-terminal sequences. nGPx4 expression is driven by its own testes-specific promoter, which lies inside the first intron of the cytosolic GPx4 transcript [36]. The role of GPx4 in sperm maturation was supported by the finding that this protein was a structural component of the mitochondrial capsule of male germ cells [37]. In addition, spermatid-specific knockout of GPx4 led to infertility in mice [38]. Moreover, inducible inactivation of GPx4 in mice and primary cells led to an increased 12/15-lipoxygenase-derived lipid peroxidation followed by apoptosis triggered by activation of apoptosis-inducing factor. To further access the function of each isoform, several KO/transgenic mouse models were prepared. nGPx4 KO mice developed normally; neither testicular structure nor fertility were affected in these mice; however, the delayed sperm chromatin condensation was observed [39]. The sequence between the two alternative translation initiation codons corresponding to mitochondrial and cytosolic forms encodes a mitochondrial signal peptide. Thus, introduction of the in-frame stop codon between these start codons resulted in the specific disruption of the mGPx4 form without affecting the expression of cGPx4. mGPx4 KO male mice were infertile [40]. These experiments revealed an essential role of mGPx4 in male reproduction.
KO of other GPxs did not affect viability and fertility. The major findings with GPx KO mice are summarized in Table 2. GPx1 KO mice did not show significant phenotypes; however, they were more susceptible to oxidative stress and viral myocarditis, as well as to reoxygenation damage due to ischemia-reperfusion injury [41]. GPx2 is mainly expressed in the epithelial tissues, and its disruption affects intestinal cells [42]. GPx1 and GPx2 double KO mice are characterized by severe colitis when maintained on an atherogenic diet [43]. Recently, GPx3 KO mice were developed [44]. Even though no significant phenotype was observed, this model revealed the specific binding of GPx3 to the basement membranes of renal cortical proximal and distal convoluted tubules.
Experiments designed to understand the function of selenoproteins in the thyroid gave ambiguous results. Both general and liver-specific knockout of DI1did not lead to significant changes in the thyroid hormone axis. [45, 46]. DI2 is expressed in the pituitary and is thought to be a T4 sensor, which is known to be a part of the negative feedback loop for thyroid hormone production. DI2 KO mice showed pituitary resistance to T4 [47]. In addition, DI2 was found to be important in the conversion of T4 to T3 in peripheral tissues (T3 stimulation is critical for the development of the auditory functions) [48, 49]. DI2 activity was increased in the WT mouse cochlea at postnatal day 7, and then declined by day 10. This DI2 activity correlated with the onset of hearing. This observation suggests that DI2 plays an important role in producing local T3 for the proper cochlear development [50]. At the same time, DI2 deficiency resulted in delayed cochlear differentiation that was the reason for irreversible deafness of DI2 KO mice [49]. DI1/DI2 double KO mice did not augment the phenotype of D1 or D2 KO mice; it was rather the sum of each single KO [51]. DI3 is responsible for inactivation of T3 and T4. DI3 KO mice showed signs of central hypothyroidism, suggesting the importance of T3 degradation for maintaining the thyroid hormone axis [52]. D3 KO mice, like DI2 KO mice, were characterized by impaired auditory function, but with different pathogenesis. Unlike DI2 KO, DI3 KO mice displayed accelerated cochlear differentiation, which also resulted in deafness. This might suggest a critical role of DI3 in local protection of the developing tissues from premature T3 exposure and differentiation [53].
Experiments with SelP KO mice revealed that the major function of SelP is the transport of Se from liver to peripheral tissues [54, 55]. SelP KO mice developed symptoms of general Se deficiency, such as ataxia, seizures and male infertility [56, 57]. All these symptoms (except male infertility) could be rescued by an increase in dietary Se. Some of these phenotypes could be rescued by an increase in dietary Se. Apparently, SelP KO mice are a particularly well suited model to study Se deficiency. Analysis of the liver-specific Sec tRNA[Ser]Sec KO (liver Trsp KO) mice (these mice lack expression of all selenoproteins in hepatocytes and will be discussed later in this review) showed decreased expression and activity of selenoproteins in peripheral tissues, which confirmed the transport function for the hepatic SelP [58]. The levels of Se in the brain remained unaffected in liver-specific Trsp KO mice; also, these mice did not show neurological phenotypes. These findings suggested another essential SelP function in the brain. Restoration of liver SelP expression in SelP KO mice restored Se transport and removed symptoms associated with Se deficiency [59]. Thus, hepatocyte-derived SelP provides the major Se supply for kidney, testis and brain. However, under Se deficiency, overexpression of SelP in the liver was unable to rescue the phenotypes of SelP KO mice, which indicates the importance of local SelP production to support selenoprotein biosynthesis under limiting Se conditions [59]. SelP was found to be recognized by two receptors: ApoER2 (mostly in the testes) and megalin. Mice, lacking these receptors demonstrated Se deficiency in testes and kidney proximal tubule epithelial cells, respectively [60, 61]. SelP consists of two parts. The N-terminal region contains a conserved UxxC motif, which is part of the domain characterized by the thioredoxin-like fold [62]. The C-terminal part of SelP contains multiple Sec residues and is involved in providing Se for the synthesis of other selenoproteins. Deletion of the C-terminal region of SelP resulted in a milder phenotype compared to the KO of the entire protein. Overall, the C-terminus plays a critical role in Se transport [63]. Infection of mice lacking the C-terminal domain of SelP with the African tripanosomiasis resulted in lower tissue injury in comparison with SelP KO mice. These mice also showed decreased production of reactive oxygen species and decreased apoptosis in the liver immune cells, increased parasite clearance capacity of myeloid cells, and increased survival. All these observations indicate that the N-terminal part of SelP plays an important role in these processes [64].
Recently, three additional KO models were described [65]. A KO of MsrB1 did not lead to strong phenotypes: the KO mice were viable and fertile. However, various tissues of MsrB1 KO mice were characterized by a decreased level of MsrA (methionine sulfoxide reductase specific for the S-diastereomer of Met sulfoxide) and increased levels of malondialdehyde, protein carbonyls, protein Met sulfoxide, as well as higher levels of oxidized glutathione and reduced levels of free and protein thiols; all this indicates the persistent oxidative stress in MsrB1 KO mice.
Systemic inactivation of SelK in mice also did not affect viability and reproduction [66]. However, as a result of the receptor mediated Ca2+ flux, SelK KO mice showed compromised functions of the immune cells, including T cell proliferation, T cell and neutrophil migration, and Fcγ receptor-mediated oxidative burst in macrophages; they also showed higher susceptibility to viral infection.
Unexpected results were obtained from the analysis of the mouse model characterized by targeted inactivation of the Sep15 gene. These Sep15 KO mice developed congenital nuclear cataracts. Sep15 mRNA was enriched during lens development, which suggested Sep15 function in lens formation. These cataracts did not appear to be due to severe oxidative stress or glucose dysregulation and presumably are associated with improper folding status of lens proteins caused by Sep15 deficiency [67].
Genetic defects in SEPN1 gene are associated with a human disorder called SEPN1 related myopathy, which includes early-onset muscle atrophy, myotendinous contractures and muscle weakness [68]. These symptoms lead to respiratory insufficiency, spine rigidity and severe scoliosis. Recently, a mouse model for SelN (Sepn1) deficiency was developed and characterized. Although SEPN1 KO mice showed normal embryogenesis and growth, they demonstrated limited motility and body rigidity during physical exercise [69]. By 4 months of age, these animals displayed a reduced pool of muscle satellite cells (SC), which are essential for adult muscle growth and repair. SelN expression was drastically increased during muscle regeneration followed by cardiotoxin-induced injury. Under these conditions, SelN KO mice showed poorer recovery, characterized by lower injured-to-colateral muscle mass ratio and excessive SC loss. The essential role of SelN in SC homeostasis is consistent with the observation that biopsies from patients with SEPN1 related myopathies showed a significant SC loss [70].
There are several selenoproteins, which are still poorly characterized, and which would benefit from the development and characterization of KO models. These proteins include SPS2, SelI, SelO, SelS, SelT, SelV, and SelW.
2.2 Overexpression of selenoproteins in mice
Besides selenoprotein gene KO mice, several studies described animals with overexpression of individual selenoproteins. One of the best such models is the GPx-overexpressing mice (GPx1oe). These animals were shown to develop hyperglycemia and hyperinsulinemia, and they also developed high levels of blood insulin and increased islet β-cell mass [71–73]. It should be noted that similar phenotypes were observed in Type 2 diabetes models. When maintained on a high fat diet, these mice developed obesity and insulin resistance, unlike GPx1 KO mice, which showed reduced insulin levels and decreased islet β-cell mass [72]. This phenotype of GPx1oe mice might be explained by insufficient ROS-mediated signaling in islet β-cells. In a different model of diabetes, expression of GPx1 had a beneficial effect. Here, overexpression of GPx1 in the islet β-cell of the db/db mice alleviated hyperglycemia at an early age and completely reversed it by 20 weeks of age [74]. Since redox signaling plays a critical role in β-cell signal transduction, both deficiency and excess of GPx1 are capable of deregulating signaling pathways. These results suggest the importance of controlled GPx1 expression for prevention of Type 2 diabetes.
Another research group developed mice with transgenic overexpression of mitochondrial GPx4 (mGPx4) [75]. Compared to littermate controls, these mice developed attenuated cardiac dysfunction in response to ischemia/reperfusion injury. Overexpression of mGPx4 reduced the levels of lipid peroxidation and slightly increased the activity of the electron transport chain (ETC) complexes I, III, and IV.
Another example of overexpression of a selenoprotein in an animal model is the overexpression of SelM in rats [76]. These animals showed a better response to oxidant treatment. When fed with high Se diet, transgenic rats showed altered ERK signal transduction in the brain, which was characterized by inhibition of the alpha/gamma-secretase activity and Tau protein phosphorylation. These observations suggest a possible protective role of SelM in the Alzheimer's disease [77].
3. Mouse models targeting the Sec biosynthesis pathway
Inactivating a single selenoprotein in the mouse can provide information about its function and reveal phenotypes associated with its deficiency. However, targeting the Sec incorporation machinery allows modulation of the expression of subsets or even all selenoproteins. Many such models have been developed.
3.1 Sec incorporating machinery
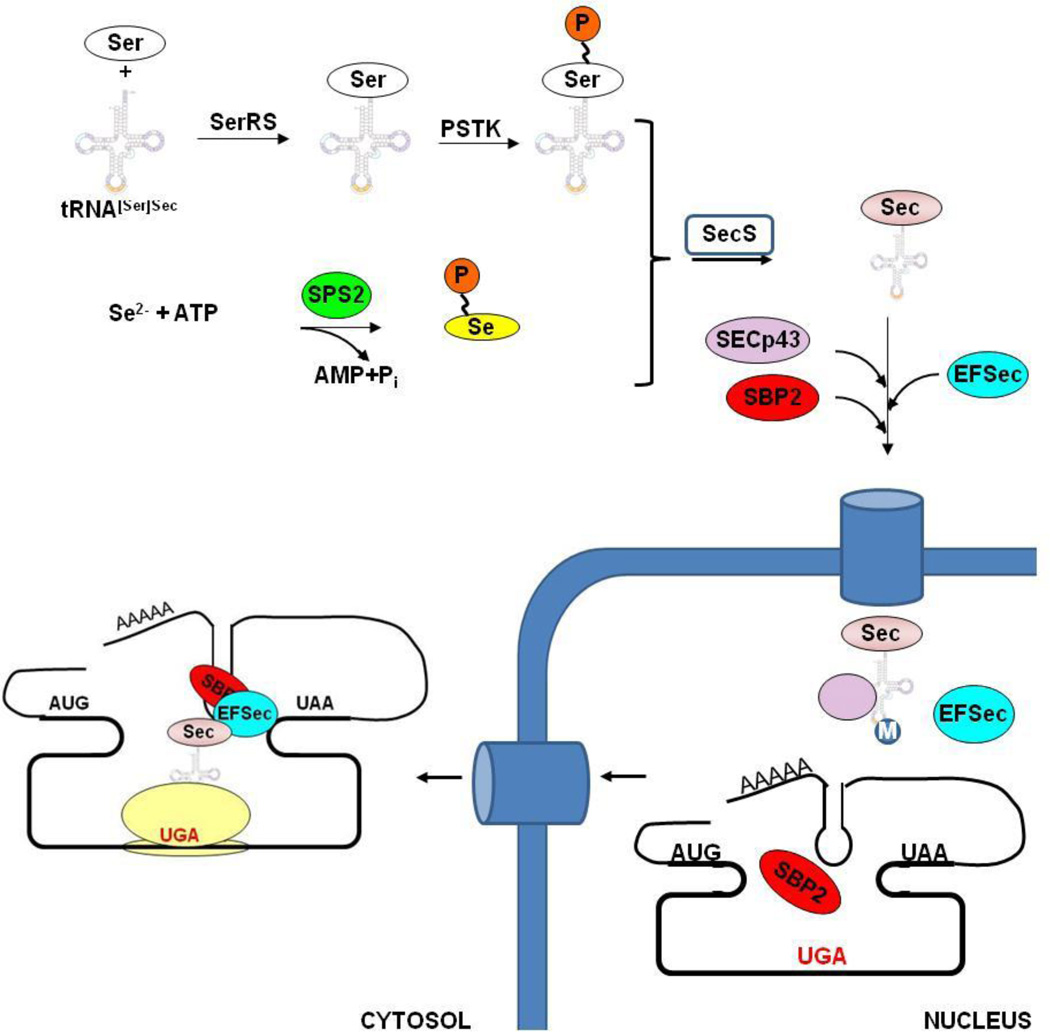
In eukaryotic cells, Sec biosynthesis and incorporation is a complex multi-stage process [7, 8, 78, 79]. The overall pathway of Sec incorporation is illustrated in Fig 1. Sec is synthesized on its own tRNA, tRNA[Ser]Sec, which is the product of the Trsp gene. Initially, this tRNA is charged with Ser, forming Ser-tRNA[Ser]Sec. This reaction is catalyzed by seryl-tRNA synthetase (SerRS). Ser-tRNA[Ser]Sec is further phosphorylated by phosphoseryl-tRNA[Ser]Sec kinase (PSTK). The Se donor compound for the Sec biosynthesis, selenophosphate, is synthesized by selenophosphate synthetase 2 (SPS2). Sec synthase (SecS or SepSecS) catalyzes the pyridoxal phosphate-dependent reaction which results in Sec-tRNA[Ser]Sec formation. Once formed, Sec-tRNA[Ser]Sec associates with EFSec and SBP2, and this supramolecular complex is translocated to the nucleus [80]. SBP2 recognizes the SECIS element, which is located in the 3’-UTR of selenoprotein mRNAs. This complex then supports the incorporation of Sec in response to the in-frame-UGA codon. There are several features which are critical for proper function of the pathway: 1) as shown in Fig 1, SBP2 and EFSec shuttle between the nucleus and cytosol. This allows binding selenoprotein mRNAs in the nucleus and inhibition of the nonsense mediated decay induced by the in-frame stop codon [81]; 2) SPS2 is itself a selenoprotein, forming a positive feedback loop [82]; 3) recently, it was found that SPS2 can also synthesize thiophosphate, promoting incorporation of Cys in place of Sec; in mice maintained on the Se-deficient diet, insertion of Cys at UGA codon of TR1 equaled that of Sec [83]; and 4) Sec tRNA[Ser]Sec is a unique tRNA, which undergoes multiple modifications, further regulating Sec incorporation. These modifications include isopentenyladenosine modification at position 37 and methylcarboxymethyl-5’-uridine (mcm5U) at position 34. The last step in Sec-tRNA[Ser]Sec maturation is the methylation of mcm5U, which may be assisted by Secp43 and results in the formation of methylcarboxymethyl-5’-uridine-2’-O-hydroxymethylribose (mcm5Um) [84]. This process is highly sensitive to the primary, secondary and tertiary structure of the tRNA as well as to overall Se status. mcm5U supports the synthesis of “housekeeping” selenoproteins, such as GPx4, TR1 and TR3, whereas the methylated tRNA is needed for expression of “stress-related” selenoproteins, such as GPx1, GPx3, and MsrB1. This change in selenoprotein expression pattern is commonly observed during Se deficiency, but the precise molecular mechanism is unknown.
Figure 1. Mechanisms of eukaryotic Sec biosynthesis and incorporation.
Sec tRNA[Ser]Sec is initially charged with Ser, which is further phosphorylated by PSTK. SPS2 facilitates the synthesis of selenophosphate, the selenium donor compound. SecS then catalyzes Sec formation. SECp43 may be involved in the methylation of Sec tRNA[Ser]Sec at the A34 position. Protein factors, including SBP2 and EFSec, bind the SECIS element, located in the 3’-UTRs of selenoprotein mRNAs. After translocation to the cytosol, protein factors support interaction with the ribosome and Sec incorporation.
There are several ways to regulate efficiency of Sec incorporation. In order to modulate expression of selenoproteins, the easiest way is to change the levels of dietary Se. To examine the effects of dietary Se on various health parameters, one can adjust Se concentration in rodent chow. For example, 0.1 ppm Se in the diet corresponds approximately to the human Recommended Dietary Allowance for adults, whereas 0.4 ppm Se may correspond to the diet supplemented with 200 µg Se/day, which is the dose most often used in clinical trials involving Se [85–87]. This approach was successfully applied to examine Se function in diabetes [88], cancer [89], the immune response [90, 91], etc. The disadvantage of this approach, however, is that with the change in dietary Se in order to regulate selenoprotein expression, the levels of low molecular weight Se compounds are also changed, which might itself influence certain pathways.
3.2 Trsp transgenic mouse models
Stable expression of mutant Trsp was shown to severely affect selenoprotein biosynthesis by interfering with the Sec incorporation pathway by a dominant-negative mechanism. According to this hypothesis, two mouse models were generated. In the first model, A37 was substituted with G37 [92], and in the second, T34 was replaced with A34 [93]. Both models lacked mcm5Um34; thus, expression of stress-related selenoproteins was severely reduced, whereas expression of housekeeping selenoprotein genes was little affected. The effect of G37 transgene was tissue-specific: it was significant in the liver and kidney, but not in testes [92]. The G37 transgenic mice were studied for various health parameters. These mice were found to be more susceptible to viral infection [94], colon cancer [95] and X-ray damage [96]. Crossing the G37 and C3/Tag mice provided a good model for studying the function of selenoproteins in prostate cancer. Such mice were found to accelerate the development of prostatic epithelial neoplasia (PIN), suggesting a protecting role of selenoproteins during prostate cancer development [97]. The G37 mice demonstrated enhanced muscle growth in the setting that modeled exercise overload. These data correlated with the initial activation of the insulin signaling pathway, which included increased Akt and p70 phosphorylation [98]. Abnormal insulin signaling might be, in part, the reason for glucose intolerance and lead to a diabetes-like phenotype, that was recently observed in the G37 mice [88].
3.3 Trsp knockout mouse models
Another approach of inactivating selenoprotein function in mice is to target the Trsp gene. The complete KO of Trsp leads to embryonic lethality [99], but a conditional removal of Trsp is possible [100]. Development of the tissue-specific KO models helped examining important functions of selenoproteins in the heart and skeletal muscle, endothelial cells [101], skin [102], bone [103], neurons [104] and the immune cells (macrophages, T cells and hematopoietic tissues) [105–107], and also studying more dispensable selenoprotein functions in the liver [58, 108], mammary gland [100] and podocytes [109]. KO of Trsp in the endothelial cells led to embryonic death at day E14.5 due to necrosis of the central nervous system, erythrocyte immaturity and subcutaneous hemorrhage. Mice with the myocyte-specific Trsp KO died 12 days after birth from acute myocardial failure [101]. Deletion of Trsp in the skin resulted in runt phenotype, epidermal neoplasia, and abnormal development of the hair follicles. Altogether, these abnormalities induced weight loss and early death. Thus, selenoproteins have a role in maintaining skin integrity [110]. Osteo-chondroprogenitor-specific Trsp KO mice showed multiple skeletal abnormalities, including growth retardation, abnormal epiphyseal plates, delayed ossification, and chondronecrosis of cartilage [103]. The neuron-specific Trsp KO induced severe neurodegeneration in the hippocampus and led to the absence of certain interneurons [110] (similar to what was observed in the neuron-specific GPx4 KO model). Besides, these mice showed degeneration of the Purkinje and granule cells that led to cerebral hyperplasia. In several studies, Se modulated the immune response. To understand the function of selenoproteins in immune cells, T and B cell-specific Trsp KO mice were developed. The T-cell-specific Trsp KO decreased the pool of mature T-cells and impaired T-cell dependent antibody response. Lack of antioxidant enzymes caused extensive oxidative stress and weak proliferation in response to T-cell receptor stimulation [106]. Macrophage specific-Trsp KO mice showed impaired invasiveness, which might be explained by hyperproduction of ROS and altered expression of extracellular matrix proteins [105]. Ablation of the Trsp gene in hematopoietic tissues resulted in anemia, which led to an increased production of erythroid progenitors in the bone marrow as well as to thymus atrophy [107]. The liver-specific Trsp KO induced expression of phase II enzymes, including various GSTs [111]. By preventing SelP synthesis and secretion, the liver-specific Trsp KO dramatically decreased plasma SelP. Thus, these mice showed symptoms of Se deficiency, which could be rescued by increased Se intake [93]. The mammary gland-specific Trsp KO mice showed increased levels of p53 and decreased expression of BRCA1 tumor suppressor [100]. In addition, mice carrying a knockout of Trsp in the liver were found to have an increased apolipoprotein E (ApoE) level and elevated cholesterol levels in plasma that was accompanied by enhanced expressed of the genes involved in cholesterol biosynthesis, metabolism and transport [112]. Interestingly, transgenic mouse models that express housekeeping, but not stress-related selenoproteins restored the expression of these genes (made them close to the corresponding levels observed in wild type controls). These studies showed that housekeeping selenoproteins have a role in regulating lipoprotein biosynthesis and metabolism and were consistent with the earlier studies showing that selenium deficiency increased ApoE expression. Overall, mouse models with conditional Trsp KO turned out to be a powerful tool for understanding functions of selenoproteins in various tissues.
3.4 Knockout/transgenic mouse models
An additional strategy to investigate the effect of transgene overexpression is to develop KO/transgenic animal models. In the case of selenoproteins, liver Trsp KO mice were crossed with the G37 or A34 mice. In both cases, similar expression patterns of housekeeping selenoproteins were observed. As discussed above, restoration of housekeeping selenoprotein genes partially decreased elevated levels of ApoE and serum cholesterol that had been observed in the liver-specific Trsp KO. Another useful knockout/transgene mouse model was also described [113]. STAF (Sec tRNA gene transcription activating factor) is a transcription factor for several RNA PolII and RNA PolIII-dependent genes. In this study, the authors overexpressed Trsp lacking the STAF binding promoter region and afterwards removed the WT Trsp. Interestingly, removal of the STAF binding site did not affect Trsp levels in the heart and testis, but showed severe reduction of the transgene in the liver, kidney, lung, spleen, and brain. Moreover, methylation of Trsp at A34 was significantly decreased, and expression of stress-related selenoproteins was reduced. These mice demonstrated the neurological phenotype similar to that of SelP KO mice. These findings indicated the importance of the STAF binding region in regulation of Sec tRNA[Ser]Sec expression and its proper modification status.
4. Concluding remarks
Development of appropriate animal models is a critical step in the characterization of biological functions of genes. A great deal of research in the area of Se biology was devoted to the understanding of functions of this micronutrient and selenoproteins in health and disease. It is clear from the discussion above that the functions of several selenoproteins and their forms could not have been determined without the use of appropriate KO models. However, there are also several obstacles resulting from the analysis of human diseases associated with the genetic defects in selenoprotein biosynthesis. For example, the presence of hypomorphic alleles of SBP2 gene was associated with retarded growth due to thyroid axis imbalance in children. At the same time, these patients experienced myopathy, waddling gait and mental retardation [114, 115] and were characterized by bilateral hearing loss and infertility. Recent research demonstrated that mutations in SecS gene are associated with the development of autosomal-recessive progressive cerebellocerebral atrophy [116] and that the observed phenotypes could be partially reproduced in the corresponding KO animal models. Indeed, analysis of the SelP KO mice could explain all symptoms, except for abnormalities in the thyroid function. While hypothyroidism is one of the first complaints in patients with impaired Sec incorporation pathway, in mice this effect is less pronounced. There are also several open questions. For example, mice maintained on the Se-deficient diet survive for more than one year with no visible abnormalities. The lifespan of the G37 transgenic mice was not affected: these mice were fully fertile, and did not develop symptoms similar to those in the patients with defects in SBP2. Understanding the reasons for the differences between human and mouse phenotypes could provide important new insights into the role of Se, Sec and selenoproteins in human health, and also into the molecular mechanisms of Sec incorporation and selenoprotein function. This research may also reveal novel regulatory mechanisms.
Highlights.
-
➢
Se regulates pathways through incorporation into selenoproteins in the form of Sec
-
➢
Recent findings in selenoprotein biosynthesis and functions are summarized
-
➢
Overview of available knockout mouse models relevant to Se biology is provided
-
➢
Mouse models with targeted expression of selenoproteins are described
-
➢
Limitations of using animal models and insights into human health are discussed
Abbreviations
- Cys
cysteine
- DI1
thyroid hormone deiodinase type 1
- DI2
thyroid hormone deiodinase type 2
- DI3
thyroid hormone deiodinase type 3
- ER
endoplasmic reticulum
- ERAD
ER associated degradation
- ICP-MS
inductively coupled plasma mass spectrometry
- GF
germ-free
- GPx1
glutathione peroxidase 1
- GPx2
glutathione peroxidase 2
- GPx3
glutathione peroxidase 3
- GPx4
glutathione peroxidase 4
- GSH
glutathione
- I
iodine
- MsrA
methionine-S-sulfoxide reductase
- MsrB
methionine-R-sulfoxide reductase
- NS
neuronal system
- SC
satellite cells
- Se
selenium
- Sec
selenocysteine
- SECIS
selenocysteine insertion sequence
- SelH
selenoprotein H
- SelI
selenoprotein I
- SelK
selenoprotein K
- SelM
selenoprotein M
- SelN
selenoprotein N
- SelO
selenoprotein O
- SelP
selenoprotein P
- SelS
selenoprotein S
- SelT
selenoprotein T
- SelV
selenoprotein V
- SelW
selenoprotein W
- Sep15
the 15 kDa selenoprotein
- SPS1
selenophosphate synthetase 1
- SPS2
selenophosphate synthetase 2
- TR1
thioredoxin reductase 1
- TR3
thioredoxin reductase 3
- Trsp
Sec tRNA[Ser]Sec gene
- UGT
UDP-glucose:glycoprotein glucosyltransferase
- UPR
unfolded protein response
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mills GC. Hemoglobin catabolism. I. Glutathione peroxidase, an erythrocyte enzyme which protects hemoglobin from oxidative breakdown. J Biol Chem. 1957;229:189–197. [PubMed] [Google Scholar]
- 2.Rotruck JT, Pope AL, Ganther HE, Swanson AB, Hafeman DG, Hoekstra WG. Selenium: biochemical role as a component of glutathione peroxidase. Science. 1973;179:588–590. doi: 10.1126/science.179.4073.588. [DOI] [PubMed] [Google Scholar]
- 3.Flohe L, Gunzler WA, Schock HH. Glutathione peroxidase: a selenoenzyme. FEBS Lett. 1973;32:132–134. doi: 10.1016/0014-5793(73)80755-0. [DOI] [PubMed] [Google Scholar]
- 4.Kraus RJ, Foster SJ, Ganther HE. Identification of selenocysteine in glutathione peroxidase by mass spectroscopy. Biochemistry. 1983;22:5853–5858. doi: 10.1021/bi00294a026. [DOI] [PubMed] [Google Scholar]
- 5.Flohe L. The glutathione peroxidase reaction: molecular basis of the antioxidant function of selenium in mammals. Curr Top Cell Regul. 1985;27:473–478. doi: 10.1016/b978-0-12-152827-0.50047-5. [DOI] [PubMed] [Google Scholar]
- 6.Arner ES. Selenoproteins-What unique properties can arise with selenocysteine in place of cysteine? Exp Cell Res. 2010;316:1296–1303. doi: 10.1016/j.yexcr.2010.02.032. [DOI] [PubMed] [Google Scholar]
- 7.Donovan J, Copeland PR. Threading the needle: getting selenocysteine into proteins. Antioxid Redox Signaling. 2010;12:881–892. doi: 10.1089/ars.2009.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hatfield DL, Carlson BA, Xu XM, Mix H, Gladyshev VN. Selenocysteine incorporation machinery and the role of selenoproteins in development and health. Prog Nucleic Acid Res Mol Biol. 2006;81:97–142. doi: 10.1016/S0079-6603(06)81003-2. [DOI] [PubMed] [Google Scholar]
- 9.Kryukov GV, Kryukov VM, Gladyshev VN. New mammalian selenocysteine-containing proteins identified with an algorithm that searches for selenocysteine insertion sequence elements. J Biol Chem. 1999;274:33888–33897. doi: 10.1074/jbc.274.48.33888. [DOI] [PubMed] [Google Scholar]
- 10.Gladyshev VN, Kryukov GV. Evolution of selenocysteine-containing proteins: significance of identification and functional characterization of selenoproteins. Biofactors. 2001;14:87–92. doi: 10.1002/biof.5520140112. [DOI] [PubMed] [Google Scholar]
- 11.Kryukov GV, Gladyshev VN. Mammalian selenoprotein gene signature: identification and functional analysis of selenoprotein genes using bioinformatics methods. Methods Enzymol. 2002;347:84–100. doi: 10.1016/s0076-6879(02)47010-3. [DOI] [PubMed] [Google Scholar]
- 12.Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 13.Lubos E, Loscalzo J, Handy DE. Glutathione Peroxidase-1 in Health and Disease: From Molecular Mechanisms to Therapeutic Opportunities. Antioxid Redox Signaling. 2011;15:1957–1997. doi: 10.1089/ars.2010.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takebe G, Yarimizu J, Saito Y, Hayashi T, Nakamura H, Yodoi J, Nagasawa S, Takahashi K. A comparative study on the hydroperoxide and thiol specificity of the glutathione peroxidase family and selenoprotein P. J Biol Chem. 2002;277:41254–41258. doi: 10.1074/jbc.M202773200. [DOI] [PubMed] [Google Scholar]
- 15.Holmgren A, Lu J. Thioredoxin and thioredoxin reductase: current research with special reference to human disease. Biochem Biophys Res Commun. 2010;396:120–124. doi: 10.1016/j.bbrc.2010.03.083. [DOI] [PubMed] [Google Scholar]
- 16.Lu J, Berndt C, Holmgren A. Metabolism of selenium compounds catalyzed by the mammalian selenoprotein thioredoxin reductase. Biochim Biophys Acta. 2009;1790:1513–1519. doi: 10.1016/j.bbagen.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 17.Oien DB, Moskovitz J. Selenium and the methionine sulfoxide reductase system. Molecules. 2009;14:2337–2344. doi: 10.3390/molecules14072337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee BC, Dikiy A, Kim HY, Gladyshev VN. Functions and evolution of selenoprotein methionine sulfoxide reductases. Biochim Biophys Acta. 2009;1790:1471–1477. doi: 10.1016/j.bbagen.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marsili A, Zavacki AM, Harney JW, Larsen PR. Physiological role and regulation of iodothyronine deiodinases: a 2011 update. J Endocrinol Invest. 2011;34:395–407. doi: 10.3275/7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohrle J, Gartner R. Selenium and thyroid. Best Pract Res Clin Endocrinol Metab. 2009;23:815–827. doi: 10.1016/j.beem.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 21.Labunskyy VM, Hatfield DL, Gladyshev VN. The Sep15 protein family: roles in disulfide bond formation and quality control in the endoplasmic reticulum. IUBMB Life. 2007;59:1–5. doi: 10.1080/15216540601126694. [DOI] [PubMed] [Google Scholar]
- 22.Shchedrina VA, Everley RA, Zhang Y, Gygi SP, Hatfield DL, Gladyshev VN. Selenoprotein K binds multiprotein complexes and is involved in the regulation of endoplasmic reticulum homeostasis. J Biol Chem. 2011;286:42937–42948. doi: 10.1074/jbc.M111.310920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- 24.Shchedrina VA, Zhang Y, Labunskyy VM, Hatfield DL, Gladyshev VN. Structure-function relations, physiological roles, and evolution of mammalian ER-resident selenoproteins. Antioxid Redox Signaling. 2010;12:839–849. doi: 10.1089/ars.2009.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conrad M, Schweizer U. Unveiling the molecular mechanisms behind selenium-related diseases through knockout mouse studies. Antioxid Redox Signaling. 2010;12:851–865. doi: 10.1089/ars.2009.2912. [DOI] [PubMed] [Google Scholar]
- 26.Conrad M. Transgenic mouse models for the vital selenoenzymes cytosolic thioredoxin reductase, mitochondrial thioredoxin reductase and glutathione peroxidase 4. Biochim Biophys Acta. 2009;1790:1575–1585. doi: 10.1016/j.bbagen.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 27.Jakupoglu C, Przemeck GK, Schneider M, Moreno SG, Mayr N, Hatzopoulos AK, de Angelis MH, Wurst W, Bornkamm GW, Brielmeier M, Conrad M. Cytoplasmic thioredoxin reductase is essential for embryogenesis but dispensable for cardiac development. Mol Cell Biol. 2005;25:1980–1988. doi: 10.1128/MCB.25.5.1980-1988.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bondareva AA, Capecchi MR, Iverson SV, Li Y, Lopez NI, Lucas O, Merrill GF, Prigge JR, Siders AM, Wakamiya M, Wallin SL, Schmidt EE. Effects of thioredoxin reductase-1 deletion on embryogenesis and transcriptome. Free Radic Biol Med. 2007;43:911–923. doi: 10.1016/j.freeradbiomed.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soerensen J, Jakupoglu C, Beck H, Forster H, Schmidt J, Schmahl W, Schweizer U, Conrad M, Brielmeier M. The role of thioredoxin reductases in brain development. PloS One. 2008;3:e1813. doi: 10.1371/journal.pone.0001813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conrad M, Jakupoglu C, Moreno SG, Lippl S, Banjac A, Schneider M, Beck H, Hatzopoulos AK, Just U, Sinowatz F, Schmahl W, Chien KR, Wurst W, Bornkamm GW, Brielmeier M. Essential role for mitochondrial thioredoxin reductase in hematopoiesis, heart development, and heart function. Mol Cell Biol. 2004;24:9414–9423. doi: 10.1128/MCB.24.21.9414-9423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geisberger R, Kiermayer C, Homig C, Conrad M, Schmidt J, Zimber-Strobl U, Brielmeier M. B- and T-cell-specific inactivation of thioredoxin reductase 2 does not impair lymphocyte development and maintenance. Biol Chem. 2007;388:1083–1090. doi: 10.1515/BC.2007.131. [DOI] [PubMed] [Google Scholar]
- 32.Sibbing D, Pfeufer A, Perisic T, Mannes AM, Fritz-Wolf K, Unwin S, Sinner MF, Gieger C, Gloeckner CJ, Wichmann HE, Kremmer E, Schafer Z, Walch A, Hinterseer M, Nabauer M, Kaab S, Kastrati A, Schomig A, Meitinger T, Bornkamm GW, Conrad M, von Beckerath N. Mutations in the mitochondrial thioredoxin reductase gene TXNRD2 cause dilated cardiomyopathy. Eur Heart J. 2011;32:1121–1133. doi: 10.1093/eurheartj/ehq507. [DOI] [PubMed] [Google Scholar]
- 33.Imai H, Hirao F, Sakamoto T, Sekine K, Mizukura Y, Saito M, Kitamoto T, Hayasaka M, Hanaoka K, Nakagawa Y. Early embryonic lethality caused by targeted disruption of the mouse PHGPx gene. Biochem Biophys Res Commun. 2003;305:278–286. doi: 10.1016/s0006-291x(03)00734-4. [DOI] [PubMed] [Google Scholar]
- 34.Seiler A, Schneider M, Forster H, Roth S, Wirth EK, Culmsee C, Plesnila N, Kremmer E, Radmark O, Wurst W, Bornkamm GW, Schweizer U, Conrad M. Glutathione peroxidase 4 senses and translates oxidative stress into 12/15-lipoxygenase dependent- and AIF-mediated cell death. Cell Metab. 2008;8:237–248. doi: 10.1016/j.cmet.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 35.Yant LJ, Ran Q, Rao L, Van Remmen H, Shibatani T, Belter JG, Motta L, Richardson A, Prolla TA. The selenoprotein GPX4 is essential for mouse development and protects from radiation and oxidative damage insults. Free Rad Biol Med. 2003;34:496–502. doi: 10.1016/s0891-5849(02)01360-6. [DOI] [PubMed] [Google Scholar]
- 36.Conrad M, Schneider M, Seiler A, Bornkamm GW. Physiological role of phospholipid hydroperoxide glutathione peroxidase in mammals. Biol Chem. 2007;388:1019–1025. doi: 10.1515/BC.2007.130. [DOI] [PubMed] [Google Scholar]
- 37.Ursini F, Heim S, Kiess M, Maiorino M, Roveri A, Wissing J, Flohe L. Dual function of the selenoprotein PHGPx during sperm maturation. Science. 1999;285:1393–1396. doi: 10.1126/science.285.5432.1393. [DOI] [PubMed] [Google Scholar]
- 38.Imai H, Hakkaku N, Iwamoto R, Suzuki J, Suzuki T, Tajima Y, Konishi K, Minami S, Ichinose S, Ishizaka K, Shioda S, Arata S, Nishimura M, Naito S, Nakagawa Y. Depletion of selenoprotein GPx4 in spermatocytes causes male infertility in mice. J Biol Chem. 2009;284:32522–32532. doi: 10.1074/jbc.M109.016139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conrad M, Moreno SG, Sinowatz F, Ursini F, Kolle S, Roveri A, Brielmeier M, Wurst W, Maiorino M, Bornkamm GW. The nuclear form of phospholipid hydroperoxide glutathione peroxidase is a protein thiol peroxidase contributing to sperm chromatin stability. Mol Cell Biol. 2005;25:7637–7644. doi: 10.1128/MCB.25.17.7637-7644.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider M, Forster H, Boersma A, Seiler A, Wehnes H, Sinowatz F, Neumuller C, Deutsch MJ, Walch A, Hrabe de Angelis M, Wurst W, Ursini F, Roveri A, Maleszewski M, Maiorino M, Conrad M. Mitochondrial glutathione peroxidase 4 disruption causes male infertility. FASEB J. 2009;23:3233–3242. doi: 10.1096/fj.09-132795. [DOI] [PubMed] [Google Scholar]
- 41.Thu VT, Kim HK, Ha SH, Yoo JY, Park WS, Kim N, Oh GT, Han J. Glutathione peroxidase 1 protects mitochondria against hypoxia/reoxygenation damage in mouse hearts. Pflugers Arch. 2010;460:55–68. doi: 10.1007/s00424-010-0811-7. [DOI] [PubMed] [Google Scholar]
- 42.Florian S, Krehl S, Loewinger M, Kipp A, Banning A, Esworthy S, Chu FF, Brigelius-Flohe R. Loss of GPx2 increases apoptosis, mitosis, and GPx1 expression in the intestine of mice. Free Rad Biol Med. 2010;49:1694–1702. doi: 10.1016/j.freeradbiomed.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Esworthy RS, Aranda R, Martin MG, Doroshow JH, Binder SW, Chu FF. Mice with combined disruption of Gpx1 and Gpx2 genes have colitis. Am J Physiol. 2001;281:G848–G855. doi: 10.1152/ajpgi.2001.281.3.G848. [DOI] [PubMed] [Google Scholar]
- 44.Olson GE, Whitin JC, Hill KE, Winfrey VP, Motley AK, Austin LM, Deal J, Cohen HJ, Burk RF. Extracellular glutathione peroxidase (GPx3) binds specifically to basement membranes of mouse renal cortex tubule cells. Am J Physiol Renal Physiol. 2010;298:F1244–F1253. doi: 10.1152/ajprenal.00662.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Streckfuss F, Hamann I, Schomburg L, Michaelis M, Sapin R, Klein MO, Kohrle J, Schweizer U. Hepatic deiodinase activity is dispensable for the maintenance of normal circulating thyroid hormone levels in mice. Biochem Biophys Res Commun. 2005;337:739–745. doi: 10.1016/j.bbrc.2005.09.102. [DOI] [PubMed] [Google Scholar]
- 46.Schneider MJ, Fiering SN, Thai B, Wu SY, St Germain E, Parlow AF, St Germain DL, Galton VA. Targeted disruption of the type 1 selenodeiodinase gene (Dio1) results in marked changes in thyroid hormone economy in mice. Endocrinology. 2006;147:580–589. doi: 10.1210/en.2005-0739. [DOI] [PubMed] [Google Scholar]
- 47.Rosene ML, Wittmann G, Arrojo e Drigo R, Singru PS, Lechan RM, Bianco AC. Inhibition of the type 2 iodothyronine deiodinase underlies the elevated plasma TSH associated with amiodarone treatment. Endocrinology. 2010;151:5961–5970. doi: 10.1210/en.2010-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Williams GR, Bassett JH. Deiodinases: the balance of thyroid hormone: local control of thyroid hormone action: role of type 2 deiodinase. J Endocrinol. 2011;209:261–272. doi: 10.1530/JOE-10-0448. [DOI] [PubMed] [Google Scholar]
- 49.Ng L, Goodyear RJ, Woods CA, Schneider MJ, Diamond E, Richardson GP, Kelley MW, Germain DL, Galton VA, Forrest D. Hearing loss and retarded cochlear development in mice lacking type 2 iodothyronine deiodinase. Proc Nat Acad Sci USA. 2004;101:3474–3479. doi: 10.1073/pnas.0307402101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Campos-Barros A, Amma LL, Faris JS, Shailam R, Kelley MW, Forrest D. Type 2 iodothyronine deiodinase expression in the cochlea before the onset of hearing. Proc Nat Acad Sci USA. 2000;97:1287–1292. doi: 10.1073/pnas.97.3.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Galton VA, Schneider MJ, Clark AS, St Germain DL. Life without thyroxine to 3,5,3'-triiodothyronine conversion: studies in mice devoid of the 5'-deiodinases. Endocrinology. 2009;150:2957–2963. doi: 10.1210/en.2008-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hernandez A, Martinez ME, Liao XH, Van Sande J, Refetoff S, Galton VA, St Germain DL. Type 3 deiodinase deficiency results in functional abnormalities at multiple levels of the thyroid axis. Endocrinology. 2007;148:5680–5687. doi: 10.1210/en.2007-0652. [DOI] [PubMed] [Google Scholar]
- 53.Ng L, Hernandez A, He W, Ren T, Srinivas M, Ma M, Galton VA, St Germain DL, Forrest D. A protective role for type 3 deiodinase, a thyroid hormone-inactivating enzyme, in cochlear development and auditory function. Endocrinology. 2009;150:1952–1960. doi: 10.1210/en.2008-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schomburg L, Schweizer U, Holtmann B, Flohe L, Sendtner M, Kohrle J. Gene disruption discloses role of selenoprotein P in selenium delivery to target tissues. Biochem J. 2003;370:397–402. doi: 10.1042/BJ20021853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hill KE, Zhou J, McMahan WJ, Motley AK, Atkins JF, Gesteland RF, Burk RF. Deletion of selenoprotein P alters distribution of selenium in the mouse. J Biol chem. 2003;278:13640–13646. doi: 10.1074/jbc.M300755200. [DOI] [PubMed] [Google Scholar]
- 56.Hill KE, Zhou J, McMahan WJ, Motley AK, Burk RF. Neurological dysfunction occurs in mice with targeted deletion of the selenoprotein P gene. J Nutr. 2004;134:157–161. doi: 10.1093/jn/134.1.157. [DOI] [PubMed] [Google Scholar]
- 57.Olson GE, Winfrey VP, Nagdas SK, Hill KE, Burk RF. Selenoprotein P is required for mouse sperm development. Biol Reprod. 2005;73:201–211. doi: 10.1095/biolreprod.105.040360. [DOI] [PubMed] [Google Scholar]
- 58.Schweizer U, Streckfuss F, Pelt P, Carlson BA, Hatfield DL, Kohrle J, Schomburg L. Hepatically derived selenoprotein P is a key factor for kidney but not for brain selenium supply. Biochem. J. 2005;386:221–226. doi: 10.1042/BJ20041973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Renko K, Werner M, Renner-Muller I, Cooper TG, Yeung CH, Hollenbach B, Scharpf M, Kohrle J, Schomburg L, Schweizer U. Hepatic selenoprotein P (SePP) expression restores selenium transport and prevents infertility and motor-incoordination in Sepp-knockout mice. Biochem J. 2008;409:741–749. doi: 10.1042/BJ20071172. [DOI] [PubMed] [Google Scholar]
- 60.Burk RF, Hill KE, Olson GE, Weeber EJ, Motley AK, Winfrey VP, Austin LM. Deletion of apolipoprotein E receptor-2 in mice lowers brain selenium and causes severe neurological dysfunction and death when a low-selenium diet is fed. J Neurosci. 2007;27:6207–6211. doi: 10.1523/JNEUROSCI.1153-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Olson GE, Winfrey VP, Hill KE, Burk RF. Megalin mediates selenoprotein P uptake by kidney proximal tubule epithelial cells. J Biol Chem. 2008;283:6854–6860. doi: 10.1074/jbc.M709945200. [DOI] [PubMed] [Google Scholar]
- 62.Burk RF, Hill KE. Selenoprotein P-expression, functions, and roles in mammals. Biochim Biophys Acta. 2009;1790:1441–1447. doi: 10.1016/j.bbagen.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hill KE, Zhou J, Austin LM, Motley AK, Ham AJ, Olson GE, Atkins JF, Gesteland RF, Burk RF. The selenium-rich C-terminal domain of mouse selenoprotein P is necessary for the supply of selenium to brain and testis but not for the maintenance of whole body selenium. J Biol Chem. 2007;282:10972–10980. doi: 10.1074/jbc.M700436200. [DOI] [PubMed] [Google Scholar]
- 64.Bosschaerts T, Guilliams M, Noel W, Herin M, Burk RF, Hill KE, Brys L, Raes G, Ghassabeh GH, De Baetselier P, Beschin A. Alternatively activated myeloid cells limit pathogenicity associated with African trypanosomiasis through the IL-10 inducible gene selenoprotein P. J Immunol. 2008;180:6168–6175. doi: 10.4049/jimmunol.180.9.6168. [DOI] [PubMed] [Google Scholar]
- 65.Fomenko DE, Novoselov SV, Natarajan SK, Lee BC, Koc A, Carlson BA, Lee TH, Kim HY, Hatfield DL, Gladyshev VN. MsrB1 (methionine-R-sulfoxide reductase 1) knock-out mice: roles of MsrB1 in redox regulation and identification of a novel selenoprotein form. J Biol Chem. 2009;284:5986–5993. doi: 10.1074/jbc.M805770200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Verma S, Hoffmann FW, Kumar M, Huang Z, Roe K, Nguyen-Wu E, Hashimoto AS, Hoffmann PR. Selenoprotein K knockout mice exhibit deficient calcium flux in immune cells and impaired immune responses. J Immunol. 2011;186:2127–2137. doi: 10.4049/jimmunol.1002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kasaikina MV, Fomenko DE, Labunskyy VM, Lachke SA, Qiu W, Moncaster JA, Zhang J, Wojnarowicz MW, Jr, Natarajan SK, Malinouski M, Schweizer U, Tsuji PA, Carlson BA, Maas RL, Lou MF, Goldstein LE, Hatfield DL, Gladyshev VN. Roles of the 15-kDa Selenoprotein (Sep15) in Redox Homeostasis and Cataract Development Revealed by the Analysis of Sep 15 Knockout Mice. J Biol Chem. 2011;286:33203–33212. doi: 10.1074/jbc.M111.259218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moghadaszadeh B, Petit N, Jaillard C, Brockington M, Roy SQ, Merlini L, Romero N, Estournet B, Desguerre I, Chaigne D, Muntoni F, Topaloglu H, Guicheney P. Mutations in SEPN1 cause congenital muscular dystrophy with spinal rigidity and restrictive respiratory syndrome. Nat Genet. 2001;29:17–18. doi: 10.1038/ng713. [DOI] [PubMed] [Google Scholar]
- 69.Rederstorff M, Castets P, Arbogast S, Laine J, Vassilopoulos S, Beuvin M, Dubourg O, Vignaud A, Ferry A, Krol A, Allamand V, Guicheney P, Ferreiro A, Lescure A. Increased muscle stress-sensitivity induced by selenoprotein N inactivation in mouse: a mammalian model for SEPN1-related myopathy. PLoS One. 2011;6:e23094. doi: 10.1371/journal.pone.0023094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Castets P, Bertrand AT, Beuvin M, Ferry A, Le Grand F, Castets M, Chazot G, Rederstorff M, Krol A, Lescure A, Romero NB, Guicheney P, Allamand V. Satellite cell loss and impaired muscle regeneration in selenoprotein N deficiency. Hum Mol Genet. 2011;20:694–704. doi: 10.1093/hmg/ddq515. [DOI] [PubMed] [Google Scholar]
- 71.Mysore TB, Shinkel TA, Collins J, Salvaris EJ, Fisicaro N, Murray-Segal LJ, Johnson LE, Lepore DA, Walters SN, Stokes R, Chandra AP, O'Connell PJ, d'Apice AJ, Cowan PJ. Overexpression of glutathione peroxidase with two isoforms of superoxide dismutase protects mouse islets from oxidative injury and improves islet graft function. Diabetes. 2005;54:2109–2116. doi: 10.2337/diabetes.54.7.2109. [DOI] [PubMed] [Google Scholar]
- 72.Lei XG, Cheng WH. New roles for an old selenoenzyme: evidence from glutathione peroxidase-1 null and overexpressing mice. J Nutr. 2005;135:2295–2298. doi: 10.1093/jn/135.10.2295. [DOI] [PubMed] [Google Scholar]
- 73.Wang XD, Vatamaniuk MZ, Wang SK, Roneker CA, Simmons RA, Lei XG. Molecular mechanisms for hyperinsulinaemia induced by overproduction of selenium-dependent glutathione peroxidase-1 in mice. Diabetologia. 2008;51:1515–1524. doi: 10.1007/s00125-008-1055-3. [DOI] [PubMed] [Google Scholar]
- 74.Harmon JS, Bogdani M, Parazzoli SD, Mak SS, Oseid EA, Berghmans M, Leboeuf RC, Robertson RP. beta-Cell-specific overexpression of glutathione peroxidase preserves intranuclear MafA and reverses diabetes in db/db mice. Endocrinology. 2009;150:4855–4862. doi: 10.1210/en.2009-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dabkowski ER, Williamson CL, Hollander JM. Mitochondria-specific transgenic overexpression of phospholipid hydroperoxide glutathione peroxidase (GPx4) attenuates ischemia/reperfusion-associated cardiac dysfunction. Free Rad Biol Med. 2008;45:855–865. doi: 10.1016/j.freeradbiomed.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 76.Hwang DY, Sin JS, Kim MS, Yim SY, Kim YK, Kim CK, Kim BG, Shim SB, Jee SW, Lee SH, Bae CJ, Lee BC, Jang MK, Cho JS, Chae KR. Overexpression of human selenoprotein M differentially regulates the concentrations of antioxidants and H2O2, the activity of antioxidant enzymes, and the composition of white blood cells in a transgenic rat. In J Mol Med. 2008;21:169–179. [PubMed] [Google Scholar]
- 77.Yim SY, Chae KR, Shim SB, Hong JT, Park JY, Lee CY, Son HJ, Sheen YY, Hwang DY. ERK activation induced by selenium treatment significantly downregulates beta/gamma-secretase activity and Tau phosphorylation in the transgenic rat overexpressing human selenoprotein M. In J Mol Med. 2009;24:91–96. doi: 10.3892/ijmm_00000211. [DOI] [PubMed] [Google Scholar]
- 78.Stoytcheva ZR, Berry MJ. Transcriptional regulation of mammalian selenoprotein expression. Biochim Biophys Acta. 2009;1790:1429–1440. doi: 10.1016/j.bbagen.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reeves MA, Hoffmann PR. The human selenoproteome: recent insights into functions and regulation. Cell Mol Life Sci. 2009;66:2457–2478. doi: 10.1007/s00018-009-0032-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Palioura S, Sherrer RL, Steitz TA, Soll D, Simonovic M. The human SepSecS-tRNASec complex reveals the mechanism of selenocysteine formation. Science. 2009;325:321–325. doi: 10.1126/science.1173755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Papp LV, Lu J, Striebel F, Kennedy D, Holmgren A, Khanna KK. The redox state of SECIS binding protein 2 controls its localization and selenocysteine incorporation function. Mol Cell Biol. 2006;26:4895–4910. doi: 10.1128/MCB.02284-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xu XM, Carlson BA, Irons R, Mix H, Zhong N, Gladyshev VN, Hatfield DL. Selenophosphate synthetase 2 is essential for selenoprotein biosynthesis. Biochem J. 2007;404:115–120. doi: 10.1042/BJ20070165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu XM, Turanov AA, Carlson BA, Yoo MH, Everley RA, Nandakumar R, Sorokina I, Gygi SP, Gladyshev VN, Hatfield DL. Targeted insertion of cysteine by decoding UGA codons with mammalian selenocysteine machinery. Proc Nat Acad Sci USA. 2010;107:21430–21434. doi: 10.1073/pnas.1009947107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu XM, Mix H, Carlson BA, Grabowski PJ, Gladyshev VN, Berry MJ, Hatfield DL. Hatfield, Evidence for direct roles of two additional factors, SECp43 and soluble liver antigen, in the selenoprotein synthesis machinery. J Biol Chem. 2005;280:41568–41575. doi: 10.1074/jbc.M506696200. [DOI] [PubMed] [Google Scholar]
- 85.McLachlan SK, Thomson CD, Ferguson EL, McKenzie JE. Dietary and biochemical selenium status of urban 6- to 24-month-old South Island New Zealand children and their postpartum mothers. J Nutr. 2004;134:3290–3295. doi: 10.1093/jn/134.12.3290. [DOI] [PubMed] [Google Scholar]
- 86.Thomson CD. Selenium and iodine intakes and status in New Zealand and Australia. The Br J Nutr. 2004;91:661–672. doi: 10.1079/BJN20041110. [DOI] [PubMed] [Google Scholar]
- 87.Thomson CD. Assessment of requirements for selenium and adequacy of selenium status: a review. Eur Journal Clin Nutr. 2004;58:391–402. doi: 10.1038/sj.ejcn.1601800. [DOI] [PubMed] [Google Scholar]
- 88.Labunskyy VM, Lee BC, Handy DE, Loscalzo J, Hatfield DL, Gladyshev VN. Both Maximal Expression of Selenoproteins and Selenoprotein Deficiency Can Promote Development of Type 2 Diabetes-Like Phenotype in Mice. Antioxid Redox Signaling. 2011;14:2327–2336. doi: 10.1089/ars.2010.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Novoselov SV, Calvisi DF, Labunskyy VM, Factor VM, Carlson BA, Fomenko DE, Moustafa ME, Hatfield DL, Gladyshev VN. Selenoprotein deficiency and high levels of selenium compounds can effectively inhibit hepatocarcinogenesis in transgenic mice. Oncogene. 2005;24:8003–8011. doi: 10.1038/sj.onc.1208940. [DOI] [PubMed] [Google Scholar]
- 90.Hoffmann FW, Hashimoto AC, Shafer LA, Dow S, Berry MJ, Hoffmann PR. Dietary selenium modulates activation and differentiation of CD4+ T cells in mice through a mechanism involving cellular free thiols. J Nutr. 2010;140:1155–1161. doi: 10.3945/jn.109.120725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hoffmann PR, Jourdan-Le Saux C, Hoffmann FW, Chang PS, Bollt O, He Q, Tam EK, Berry MJ. A role for dietary selenium and selenoproteins in allergic airway inflammation. J Immunol. 2007;179:3258–3267. doi: 10.4049/jimmunol.179.5.3258. [DOI] [PubMed] [Google Scholar]
- 92.Moustafa ME, Carlson BA, El-Saadani MA, Kryukov GV, Sun QA, Harney JW, Hill KE, Combs GF, Feigenbaum L, Mansur DB, Burk RF, Berry MJ, Diamond AM, Lee BJ, Gladyshev VN, Hatfield DL. Selective inhibition of selenocysteine tRNA maturation and selenoprotein synthesis in transgenic mice expressing isopentenyladenosine-deficient selenocysteine tRNA. Mol Cell Biol. 2001;21:3840–3852. doi: 10.1128/MCB.21.11.3840-3852.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Carlson BA, Moustafa ME, Sengupta A, Schweizer U, Shrimali R, Rao M, Zhong N, Wang S, Feigenbaum L, Lee BJ, Gladyshev VN, Hatfield DL. Selective restoration of the selenoprotein population in a mouse hepatocyte selenoproteinless background with different mutant selenocysteine tRNAs lacking Um34. J Biol Chem. 2007;282:32591–32602. doi: 10.1074/jbc.M707036200. [DOI] [PubMed] [Google Scholar]
- 94.Sheridan PA, Zhong N, Carlson BA, Perella CM, Hatfield DL, Beck MA. Decreased selenoprotein expression alters the immune response during influenza virus infection in mice. J Nutr. 2007;137:1466–1471. doi: 10.1093/jn/137.6.1466. [DOI] [PubMed] [Google Scholar]
- 95.Irons R, Carlson BA, Hatfield DL, Davis CD. Both selenoproteins and low molecular weight selenocompounds reduce colon cancer risk in mice with genetically impaired selenoprotein expression. J Nutr. 2006;136:1311–1317. doi: 10.1093/jn/136.5.1311. [DOI] [PubMed] [Google Scholar]
- 96.Baliga MS, Diwadkar-Navsariwala V, Koh T, Fayad R, Fantuzzi G, Diamond AM. Selenoprotein deficiency enhances radiation-induced micronuclei formation. Mol Nutr Food Res. 2008;52:1300–1304. doi: 10.1002/mnfr.200800020. [DOI] [PubMed] [Google Scholar]
- 97.Diwadkar-Navsariwala V, Prins GS, Swanson SM, Birch LA, Ray VH, Hedayat S, Lantvit DL, Diamond AM. Selenoprotein deficiency accelerates prostate carcinogenesis in a transgenic model. Proc Nat Acad Sci USA. 2006;103:8179–8184. doi: 10.1073/pnas.0508218103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hornberger TA, Sukhija KB, Wang XR, Chien S. mTOR is the rapamycin-sensitive kinase that confers mechanically-induced phosphorylation of the hydrophobic motif site Thr(389) in p70(S6k) FEBS Lett. 2007;581:4562–4566. doi: 10.1016/j.febslet.2007.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bosl MR, Takaku K, Oshima M, Nishimura S, Taketo MM. Early embryonic lethality caused by targeted disruption of the mouse selenocysteine tRNA gene (Trsp) Proc Nat Acad Sci USA. 1997;94:5531–5534. doi: 10.1073/pnas.94.11.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kumaraswamy E, Carlson BA, Morgan F, Miyoshi K, Robinson GW, Su D, Wang S, Southon E, Tessarollo L, Lee BJ, Gladyshev VN, Hennighausen L, Hatfield DL. Selective removal of the selenocysteine tRNA [Ser]Sec gene (Trsp) in mouse mammary epithelium. Mol Cell Biol. 2003;23:1477–1488. doi: 10.1128/MCB.23.5.1477-1488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shrimali RK, Weaver JA, Miller GF, Starost MF, Carlson BA, Novoselov SV, Kumaraswamy E, Gladyshev VN, Hatfield DL. Selenoprotein expression is essential in endothelial cell development and cardiac muscle function. Neuromuscul Disord. 2007;17:135–142. doi: 10.1016/j.nmd.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sengupta A, Lichti UF, Carlson BA, Ryscavage AO, Gladyshev VN, Yuspa SH, Hatfield DL. Selenoproteins are essential for proper keratinocyte function and skin development. PloS One. 2010;5:e12249. doi: 10.1371/journal.pone.0012249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Downey CM, Horton CR, Carlson BA, Parsons TE, Hatfield DL, Hallgrimsson B, Jirik FR. Osteo-chondroprogenitor-specific deletion of the selenocysteine tRNA gene, Trsp, leads to chondronecrosis and abnormal skeletal development: a putative model for Kashin-Beck disease. PLoS Genetics. 2009;5:e1000616. doi: 10.1371/journal.pgen.1000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wirth EK, Conrad M, Winterer J, Wozny C, Carlson BA, Roth S, Schmitz D, Bornkamm GW, Coppola V, Tessarollo L, Schomburg L, Kohrle J, Hatfield DL, Schweizer U. Neuronal selenoprotein expression is required for interneuron development and prevents seizures and neurodegeneration. FASEB J. 2010;24:844–852. doi: 10.1096/fj.09-143974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Suzuki T, Kelly VP, Motohashi H, Nakajima O, Takahashi S, Nishimura S, Yamamoto M. Deletion of the selenocysteine tRNA gene in macrophages and liver results in compensatory gene induction of cytoprotective enzymes by Nrf2. J Biol Chem. 2008;283:2021–2030. doi: 10.1074/jbc.M708352200. [DOI] [PubMed] [Google Scholar]
- 106.Shrimali RK, Irons RD, Carlson BA, Sano Y, Gladyshev VN, Park JM, Hatfield DL. Selenoproteins mediate T cell immunity through an antioxidant mechanism. The J Biol Chem. 2008;283:20181–20185. doi: 10.1074/jbc.M802559200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kawatani Y, Suzuki T, Shimizu R, Kelly VP, Yamamoto M. Nrf2 and selenoproteins are essential for maintaining oxidative homeostasis in erythrocytes and protecting against hemolytic anemia. Blood. 2011;117:986–996. doi: 10.1182/blood-2010-05-285817. [DOI] [PubMed] [Google Scholar]
- 108.Carlson BA, Novoselov SV, Kumaraswamy E, Lee BJ, Anver MR, Gladyshev VN, Hatfield DL. Specific excision of the selenocysteine tRNA[Ser]Sec (Trsp) gene in mouse liver demonstrates an essential role of selenoproteins in liver function. The J Biol Chem. 2004;279:8011–8017. doi: 10.1074/jbc.M310470200. [DOI] [PubMed] [Google Scholar]
- 109.Blauwkamp MN, Yu J, Schin MA, Burke KA, Berry MJ, Carlson BA, Brosius FC, 3rd, Koenig RJ. Podocyte specific knock out of selenoproteins does not enhance nephropathy in streptozotocin diabetic C57BL/6 mice. BMC Nephrology. 2008;9:7. doi: 10.1186/1471-2369-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Carlson BA, Yoo MH, Tsuji PA, Gladyshev VN, Hatfield DL. Mouse models targeting selenocysteine tRNA expression for elucidating the role of selenoproteins in health and development. Molecules. 2009;14:3509–3527. doi: 10.3390/molecules14093509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sengupta A, Carlson BA, Weaver JA, Novoselov SV, Fomenko DE, Gladyshev VN, Hatfield DL. A functional link between housekeeping selenoproteins and phase II enzymes. Biochem J. 2008;413:151–161. doi: 10.1042/BJ20080277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sengupta A, Carlson BA, Hoffmann VJ, Gladyshev VN, Hatfield DL. Loss of housekeeping selenoprotein expression in mouse liver modulates lipoprotein metabolism. Biochem Biophys Res Commun. 2008;365:446–452. doi: 10.1016/j.bbrc.2007.10.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Carlson BA, Schweizer U, Perella C, Shrimali RK, Feigenbaum L, Shen L, Speransky S, Floss T, Jeong SJ, Watts J, Hoffmann V, Combs GF, Gladyshev VN, Hatfield DL. The selenocysteine tRNA STAF-binding region is essential for adequate selenocysteine tRNA status, selenoprotein expression and early age survival of mice. Biochem J. 2009;418:61–71. doi: 10.1042/BJ20081304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schweizer U, Dehina N, Schomburg L. Disorders of selenium metabolism and selenoprotein function. Curr Opin Pediatr. 2011;23:429–435. doi: 10.1097/MOP.0b013e32834877da. [DOI] [PubMed] [Google Scholar]
- 115.Schoenmakers E, Agostini M, Mitchell C, Schoenmakers N, Papp L, Rajanayagam O, Padidela R, Ceron-Gutierrez L, Doffinger R, Prevosto C, Luan J, Montano S, Lu J, Castanet M, Clemons N, Groeneveld M, Castets P, Karbaschi M, Aitken S, Dixon A, Williams J, Campi I, Blount M, Burton H, Muntoni F, O'Donovan D, Dean A, Warren A, Brierley C, Baguley D, Guicheney P, Fitzgerald R, Coles A, Gaston H, Todd P, Holmgren A, Khanna KK, Cooke M, Semple R, Halsall D, Wareham N, Schwabe J, Grasso L, Beck-Peccoz P, Ogunko A, Dattani M, Gurnell M, Chatterjee K. Mutations in the selenocysteine insertion sequence-binding protein 2 gene lead to a multisystem selenoprotein deficiency disorder in humans. J Clin Invest. 2010;120:4220–4235. doi: 10.1172/JCI43653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Agamy O, Ben Zeev B, Lev D, Marcus B, Fine D, Su D, Narkis G, Ofir R, Hoffmann C, Leshinsky-Silver E, Flusser H, Sivan S, Soll D, Lerman-Sagie T, Birk OS. Mutations disrupting selenocysteine formation cause progressive cerebello-cerebral atrophy. Am J Hum Genet. 2010;87:538–544. doi: 10.1016/j.ajhg.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ferguson AD, Labunskyy VM, Fomenko DE, Arac D, Chelliah Y, Amezcua CA, Rizo J, Gladyshev VN, Deisenhofer J. NMR structures of the selenoproteins Sep15 and SelM reveal redox activity of a new thioredoxin-like family. J Biol Chem. 2006;281:3536–3543. doi: 10.1074/jbc.M511386200. [DOI] [PubMed] [Google Scholar]
- 118.Labunskyy VM, Ferguson AD, Fomenko DE, Chelliah Y, Hatfield DL, Gladyshev VN. A novel cysteine-rich domain of Sep15 mediates the interaction with UDP-glucose: glycoprotein glucosyltransferase. J Biol Chem. 2005;280:37839–37845. doi: 10.1074/jbc.M508685200. [DOI] [PubMed] [Google Scholar]
- 119.Labunskyy VM, Yoo MH, Hatfield DL, Gladyshev VN. Sep15, a thioredoxin-like selenoprotein, is involved in the unfolded protein response and differentially regulated by adaptive and acute ER stresses. Biochemistry. 2009;48:8458–8465. doi: 10.1021/bi900717p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Maia AL, Goemann IM, Meyer EL, Wajner SM. Deiodinases: the balance of thyroid hormone: type 1 iodothyronine deiodinase in human physiology and disease. J Endocrinol. 2011;209:283–297. doi: 10.1530/JOE-10-0481. [DOI] [PubMed] [Google Scholar]
- 121.Schomburg L, Kohrle J. On the importance of selenium and iodine metabolism for thyroid hormone biosynthesis and human health. Mol Nutr Food Res. 2008;52:1235–1246. doi: 10.1002/mnfr.200700465. [DOI] [PubMed] [Google Scholar]
- 122.Dentice M, Salvatore D. Deiodinases: the balance of thyroid hormone: local impact of thyroid hormone inactivation. J Endocrinol. 2011;209:273–282. doi: 10.1530/JOE-11-0002. [DOI] [PubMed] [Google Scholar]
- 123.Brigelius-Flohe R, Kipp A. Glutathione peroxidases in different stages of carcinogenesis. Biochim Biophys Acta. 2009;1790:1555–1568. doi: 10.1016/j.bbagen.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 124.Brigelius-Flohe R. Glutathione peroxidases and redox-regulated transcription factors. Biol Chem. 2006;387:1329–1335. doi: 10.1515/BC.2006.166. [DOI] [PubMed] [Google Scholar]
- 125.Bouayed J, Bohn T. Exogenous antioxidants - Double-edged swords in cellular redox state: Health beneficial effects at physiologic doses versus deleterious effects at high doses. Oxid Med Cell Longev. 2010;3:228–237. doi: 10.4161/oxim.3.4.12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dikiy A, Novoselov SV, Fomenko DE, Sengupta A, Carlson BA, Cerny RL, Ginalski K, Grishin NV, Hatfield DL, Gladyshev VN. SelT, SelW, SelH, and Rdx12: genomics and molecular insights into the functions of selenoproteins of a novel thioredoxin-like family. Biochemistry. 2007;46:6871–6882. doi: 10.1021/bi602462q. [DOI] [PubMed] [Google Scholar]
- 127.Mendelev N, Mehta SL, Witherspoon S, He Q, Sexton JZ, Li PA. Upregulation of human selenoprotein H in murine hippocampal neuronal cells promotes mitochondrial biogenesis and functional performance. Mitochondrion. 2011;11:76–82. doi: 10.1016/j.mito.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Panee J, Stoytcheva ZR, Liu W, Berry MJ. Selenoprotein H is a redox-sensing high mobility group family DNA-binding protein that up-regulates genes involved in glutathione synthesis and phase II detoxification. J Biol Chem. 2007;282:23759–23765. doi: 10.1074/jbc.M702267200. [DOI] [PubMed] [Google Scholar]
- 129.Reeves MA, Bellinger FP, Berry MJ. The neuroprotective functions of selenoprotein M and its role in cytosolic calcium regulation. Antioxid Redox Signaling. 2010;12:809–818. doi: 10.1089/ars.2009.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jurynec MJ, Xia R, Mackrill JJ, Gunther D, Crawford T, Flanigan KM, Abramson JJ, Howard MT, Grunwald DJ. Selenoprotein N is required for ryanodine receptor calcium release channel activity in human and zebrafish muscle. Proc Nat Acad Sci USA. 2008;105:12485–12490. doi: 10.1073/pnas.0806015105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Richardson DR. More roles for selenoprotein P: local selenium storage and recycling protein in the brain. Biochem J. 2005;386:e5–e7. doi: 10.1042/BJ20050149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Schweizer U, Brauer AU, Kohrle J, Nitsch R, Savaskan NE. Selenium and brain function: a poorly recognized liaison. Brain Res. 2004;45:164–178. doi: 10.1016/j.brainresrev.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 133.Small-Howard A, Morozova N, Stoytcheva Z, Forry EP, Mansell JB, Harney JW, Carlson BA, Xu XM, Hatfield DL, Berry MJ. Supramolecular complexes mediate selenocysteine incorporation in vivo. Mol Cell Biol. 2006;26:2337–2346. doi: 10.1128/MCB.26.6.2337-2346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sengupta A, Carlson BA, Labunskyy VM, Gladyshev VN, Hatfield DL. Selenoprotein T deficiency alters cell adhesion and elevates selenoprotein W expression in murine fibroblast cells. Biochem Cell Biol. 2009;87:953–961. doi: 10.1139/o09-064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Turanov AA, Kehr S, Marino SM, Yoo MH, Carlson BA, Hatfield DL, Gladyshev VN. Mammalian thioredoxin reductase 1: roles in redox homoeostasis and characterization of cellular targets. Biochem J. 2010;430:285–293. doi: 10.1042/BJ20091378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Su D, Novoselov SV, Sun QA, Moustafa ME, Zhou Y, Oko R, Hatfield DL, Gladyshev VN. Mammalian selenoprotein thioredoxin-glutathione reductase. Roles in disulfide bond formation and sperm maturation. J Biol Chem. 2005;280:26491–26498. doi: 10.1074/jbc.M503638200. [DOI] [PubMed] [Google Scholar]
- 137.Turanov AA, Su D, Gladyshev VN. Characterization of alternative cytosolic forms and cellular targets of mouse mitochondrial thioredoxin reductase. J Biol Chem. 2006;281:22953–22963. doi: 10.1074/jbc.M604326200. [DOI] [PubMed] [Google Scholar]
- 138.Noh OJ, Park YH, Chung YW, Kim IY. Transcriptional regulation of selenoprotein W by MyoD during early skeletal muscle differentiation. J Biol Chem. 2010;285:40496–40507. doi: 10.1074/jbc.M110.152934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.de Haan JB, Bladier C, Griffiths P, Kelner M, O'Shea RD, Cheung NS, Bronson RT, Silvestro MJ, Wild S, Zheng SS, Beart PM, Hertzog PJ, Kola I. Mice with a homozygous null mutation for the most abundant glutathione peroxidase, Gpx1, show increased susceptibility to the oxidative stress-inducing agents paraquat and hydrogen peroxide. J Biol Chem. 1998;273:22528–22536. doi: 10.1074/jbc.273.35.22528. [DOI] [PubMed] [Google Scholar]