Abstract
Adenosine-to-inosine (A-to-I) RNA editing targets double-stranded RNA stem–loop structures in the mammalian brain. It has previously been shown that miRNAs are substrates for A-to-I editing. For the first time, we show that for several definitions of edited miRNA, the level of editing increases with development, thereby indicating a regulatory role for editing during brain maturation. We use high-throughput RNA sequencing to determine editing levels in mature miRNA, from the mouse transcriptome, and compare these with the levels of editing in pri-miRNA. We show that increased editing during development gradually changes the proportions of the two miR-376a isoforms, which previously have been shown to have different targets. Several other miRNAs that also are edited in the seed sequence show an increased level of editing through development. By comparing editing of pri-miRNA with editing and expression of the corresponding mature miRNA, we also show an editing-induced developmental regulation of miRNA expression. Taken together, our results imply that RNA editing influences the miRNA repertoire during brain maturation.
Adenosine-to-inosine (A-to-I) RNA editing is a co-transcriptional or post-transcriptional processing event that converts adenosine to inosine within double-stranded RNA. The reaction is catalyzed by the ADAR (adenosine deaminase that acts on RNA) enzymes (Bass et al. 1997). Due to structural similarities, inosine is read as guanosine (G) by the cellular machineries. Thus, an inosine in the RNA sequence will appear as an A-to-G change. Editing of the RNA sequence will yield a product different from the one encoded by the genome and thereby enhance product diversity. In mammals the ADAR gene family consists of three members: ADAR, ADARB1, and ADARB2 (also known as ADAR1, 2, and 3). However, only ADAR and ADARB1 have been shown to be active. They modify both messenger RNA (mRNA) and non-coding RNA sequences, particularly in the brain (Melcher et al. 1996; Higuchi et al. 2000; Hartner et al. 2004; Wang et al. 2004). Several genes involved in neurotransmission are edited in the coding sequence, changing the readout of the protein and thereby increasing the number of protein isoforms (Sommer et al. 1991; Burns et al. 1997; Hoopengardner et al. 2003; Ohlson et al. 2007). Furthermore, small non-coding RNAs such as microRNAs (miRNAs) have been shown to undergo A-to-I editing (Luciano et al. 2004; Blow et al. 2006; Yang et al. 2006; Kawahara et al. 2007a,b). Mature miRNAs are small single-stranded (ss) RNA molecules of ∼21–23 nt in length, whose main function is to down-regulate gene expression. The miRNA matures from a primary RNA transcript (pri-miRNA), which contains short inverted repeats in the sequence and, therefore, forms a stem–loop structure (Kim 2005). The stem–loop is recognized and cleaved by the nuclear RNase DROSHA, together with an essential RNA-binding protein, the DiGeorge syndrome critical region 8 (DGCR8) protein. The pri-miRNA is processed into a 70-nt-long stem–loop precursor miRNA (pre-miRNA) (Lee et al. 2003, 2006; Denli et al. 2004; Han et al. 2006). If processed correctly, exportin-5 and RanGTP transport the pre-miRNA to the cytoplasm. The length of the stem–loop and the presence of 3′ overhangs are critical for correct recognition of the pre-miRNA by exportin-5. In addition, exportin-5 will protect the pre-miRNA from digestion (Yi et al. 2003; Lund et al. 2004). In the cytoplasm, another RNase, DICER1 (also known as Dicer), in concert with the dsRNA binding protein TRBP, processes the pre-miRNA into a mature miRNA*–miRNA duplex (Hutvagner et al. 2001; Chendrimada et al. 2005). The strands are then separated and incorporated into the RNA-induced silencing complex (RISC). This configuration has the capacity to down-regulate protein production by targeting sequences complementary to the mature miRNA, in 3′ UTRs of mRNAs (Gregory et al. 2005; Filipowicz et al. 2007). One or both strands of the duplex may serve as the mature miRNA, although the 5′-strand (5p) has been shown to be dominant (Khvorova et al. 2003; Schwarz et al. 2003). In complex with RISC, the miRNA prevents expression either by mRNA cleavage or translational repression (Bass 2000; Sempere et al. 2004; Kim et al. 2005; Zamore and Haley 2005; Filipowicz et al. 2007). Interestingly, recent studies indicate that up to 84% of the protein down-regulation, shown upon miRNA repression, is due to mRNA destabilization rather than inhibition of translation initiation (Guo et al. 2010). To be active, the miRNA has to bind with full complementarity in the seed sequence that encompasses nucleotides 2–8 in the 5′ end of the miRNA. Thus, a change in this region may change the target recognition and thereby biogenesis.
We have previously shown that site-selective A-to-I editing of mRNA substrates in general increases during brain development from embryogenesis to adulthood (Wahlstedt et al. 2009). In the present investigation, we have analyzed whether editing of miRNAs can change target selection during brain development. Using high-throughput (HTP) RNA sequencing (RNA-seq), we performed an unbiased search for edited, mature miRNAs in mouse brain tissue from several developmental stages. This resulted in novel sites of editing as well as evidence of a dramatic increase in editing of miRNAs from the embryonic to the postnatal brain. These results indicate that A-to-I editing diversifies target recognition by miRNAs during development. Furthermore, our data suggest that specific genes, targeted by non-edited and edited miRNAs, are regulated in a manner that agrees with differences in editing frequencies.
Results
We wanted to investigate whether A-to-I editing of miRNA is regulated through development, as is the case for mRNA. The ADAR enzymes have been shown to recognize pri-miRNAs for editing, but in principle also pre-miRNAs can be edited. We were particularly interested in (1) editing leading to processing inefficiency or deficiency of miRNAs, and (2) editing of seed sequences altering target recognition, i.e., acting as co-translational or post-translational modifications that increase target variety during brain development.
To determine the frequency of editing in mature miRNAs in the brain, we applied HTP RNA-seq to small RNAs (10–50 nt in length) from three different developmental stages of the mouse brain: embryonic day 15 (E15), postnatal day 2 (P2), and postnatal day 21 (P21). After RNA extraction from total brain, the samples were fractionated; this was followed by adapter hybridization, reverse transcription, and cDNA library amplification using standard emulsion PCR. We primarily used the SOLiD system for the HTP RNA-seq analysis, but, in order to increase specificity, we also took advantage of Illumina sequenced miRNA from Chi et al. (2009). Since inosine is read as guanosine during reverse transcription, A-to-I editing can be detected as A:G mismatches between the RNA-seq reads and genomic DNA.
Initial translation, trimming, and filtering
Obtaining a good specificity, i.e., excluding non-genuine editing sites, was the main challenge in analyzing the SOLiD reads. We took advantage of the special properties of the SOLiD system's di-base coding in order to remove as many reads with errors as possible. As explained in Methods, the reads were translated from color space into nucleotide sequences, and only the reads for which the 3′-end adapter was properly translated were used in the subsequent analysis. In this way, we could exclude many reads containing errors, and the error rate in the analyzed sequences became significantly lower than in merely translated SOLiD sequences.
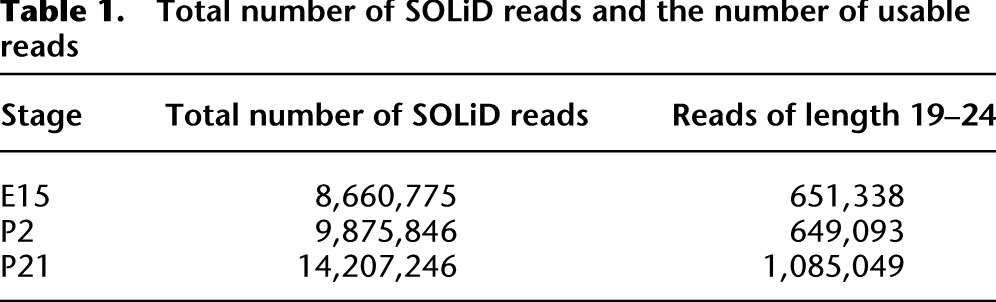
In the SOLiD system, the adapter at the 3′ end of the sequence can be attached to a fragmented RNA sequence of any length. Before applying our analysis pipeline, we removed all reads with a length shorter than 19 nt or longer than 24 nt, i.e., not resembling a mature miRNA. Supplemental Figures 1–3 show the distribution of read lengths after removal of adapter sequences in the three developmental stages. As shown, in each stage, more than 200,000 reads (read length 0) were solely adapter sequences. Table 1 shows the total number of SOLiD reads and the number of reads with length 19–24 after removing the adapter sequences. After this initial translation and filtering, the remaining reads were aligned against miRBase (Griffiths-Jones et al. 2006) using SHRiMP (Rumble et al. 2009) in order to identify A-to-G mismatches between miRNAs in the database and the read, i.e., preliminary indications of A-to-I editing.
Table 1.
Total number of SOLiD reads and the number of usable reads
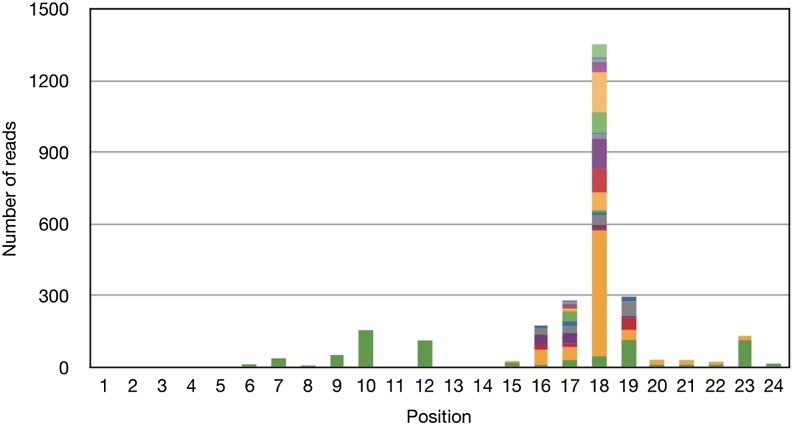
The resulting alignments showed a clear bias toward mismatches at the 3′ end of miRNAs, e.g., a substantial number of A:G mismatches at positions 16–24 (Fig. 1) as well as mismatches in general (Supplemental Fig. 4). These mismatches could a priori be due to a real biological phenomenon, but turned out to be SOLiD sequencing errors. A key observation underlying this conclusion was that exonic SOLiD reads, of various lengths, showed the same type of mismatch peak at the 3′ end (Supplemental Figs. 5–7), but Illumina miRNA reads from Chi et al. (2009) did not (Supplemental Fig. 8). For a more detailed explanation, see Supplemental Section S2.
Figure 1.
Bar chart for number of A:G mismatches in miRNA reads at developmental day E15 from SOLiD. The chart shows alignment of the reads against miRBase. Each color band means a species of miRNA. Note that the same color in different bars can represent different species of miRNA.
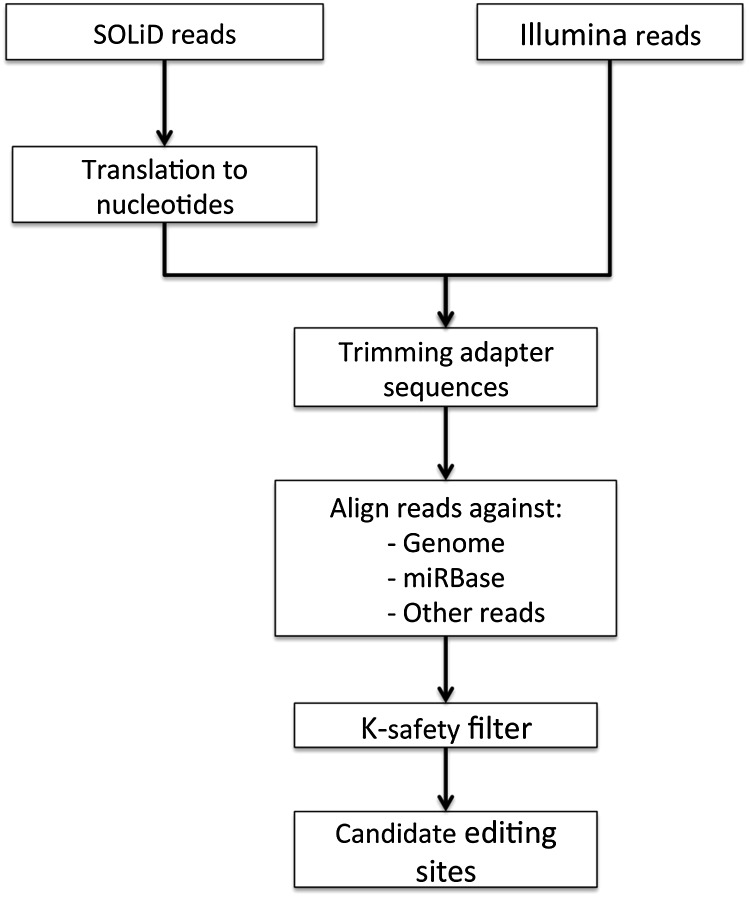
In our bioinformatics analysis pipeline (Fig. 2), the reads were aligned to precursor miRNA sequences in miRBase and also to the mouse genome mm9. The reason that we use precursor miRNA sequences from the miRBase instead of mature miRNAs is because there is a considerable variation of start and end positions for mature sequences corresponding to the very same miRBase entry. Each read with an alignment to precursor miRNA sequences starting on or before the initial position of the miRBase annotated mature sequence, and ending on or after the annotated end, were kept. One of the aims of the bioinformatics pipeline was to distinguish between A:G mismatches, which arose from genuine editing events, and other changes such as single nucleotide polymorphisms (SNPs). However, none of the A:G mismatches between our reads (translated SOLiD as well as Illumina) and precursor miRNAs in miRBase were known SNPs (Sherry et al. 2001).
Figure 2.
The analysis pipeline. Initially, all of the SOLiD reads are translated to nucleotides from color space. Thereafter all adapters were removed, and the reads with length 19–24 were aligned against miRBase, the mouse genome, and other reads. The alignments were further analyzed with the hairpin test and K-safety filter in the analysis pipeline.
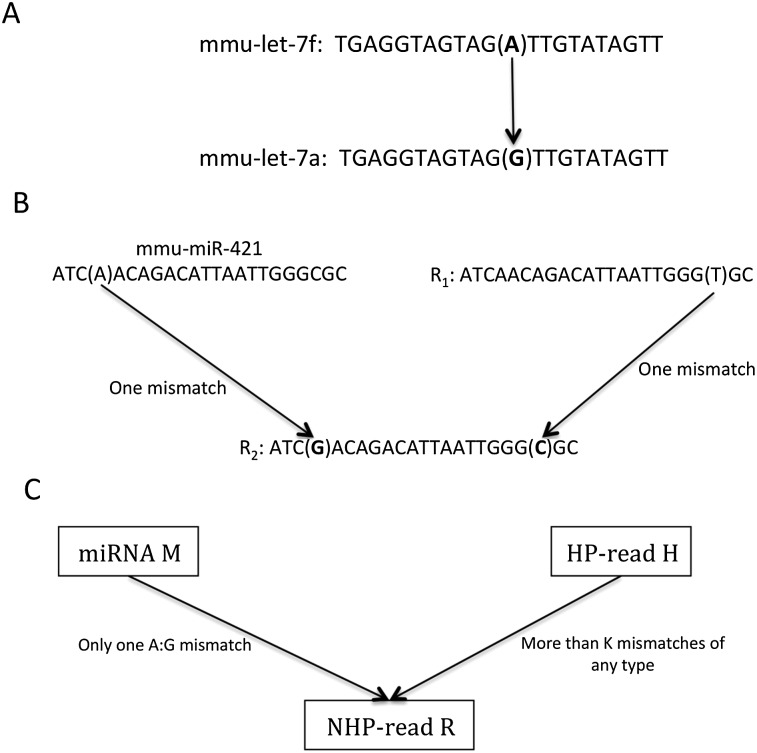
To further increase specificity, we developed the K-safety filter. Families of miRNA consist of miRNAs with similar sequences. In some cases, two family members differ by an A:G mismatch only (Fig. 3A). A read with an A:G mismatch to one miRNA and with a perfect alignment to a second miRNA, or a genomic locus with a hairpin-forming potential, was not considered to provide sufficient evidence of editing. With no perfectly matching miRNA or genomic loci with hairpin-forming potential, a read passed the 0-safety filter and was counted as the result of an editing event. Higher values of K were used to obtain more stringent filters. In general, a read passed the K-safety filter if it had (1) an A:G mismatch to a known miRNA; and (2) more than K mismatches to any other miRNA as well as a genomic locus with hairpin-forming potential.
Figure 3.
Illustration of the K-safety filter. (HP-read) Read with hairpin-forming potential. (NHP-read) Read without hairpin-making potential. (A) A read can simply be another known miRNA with an A:G mismatch to the source. (B) Read R1 is an HP-read that has a C:T mismatch with read R2. Read R2 looks like the edited version of miR-421 with an A:G mismatch in position 4. (C) Shows a read, R, with a negative hairpin test (NHP-read), comprising only one mismatch with any known miRNA, M. The closest read with a positive hairpin to this read has more than K mismatches with it, thus the read R passes the K-safety filter.
Given the high computational cost to search for all loci with a hairpin-forming potential that a given read can be aligned to with at most K mismatches, we performed a slightly different search that in practice gave the same result. Reasoning that if a locus with hairpin-forming potential actually hosted an miRNA, there should be reads originating from it that align perfectly with the genomic locus, allowed us to search instead for hairpin reads, i.e., reads aligning perfectly to a locus with hairpin-forming potential, with at most K mismatches to the currently filtered read.
For example, in Figure 3B, read R2 appears to be an edited version of mmu-miR-421. However, there is also an assumed hairpin read, R1, that has one mismatch to read R2. Thus, R2 does not pass the 1-safety filter. Furthermore, in our HTP screen, we also detect other nucleotide changes than A to G (see Supplemental Figs. 9–16).
Sets of putatively edited miRNAs of varying stringency
Editing candidates were identified, at various stringency levels, and compared with candidates from previous analyses (Kawahara et al. 2007a,b, 2008; Chiang et al. 2010).
Table 2 shows, for various values of K, the number of miRNAs with A:G mismatches for SOLiD (Applied Biosystems/Life Technologies), Illumina, and both SOLiD and Illumina data that pass the K-safety filter. The first row of the table contains the number of potential edited sites obtained when no K-safety filter is applied. As is seen in the table, increasing stringency by applying the 4-safety filter narrows down the number of candidates from 180 to 75 for SOLiD, from 35 to 17 for Illumina, and from 19 to eight for the intersection of the Illumina and the SOLiD data. Using K equal to 1 and 4 gives the same result for the Illumina data as well as for the intersection.
Table 2.
Number of A:G mismatches with different values of K for SOLiD, Illumina, and intersection of SOLiD and Illumina data
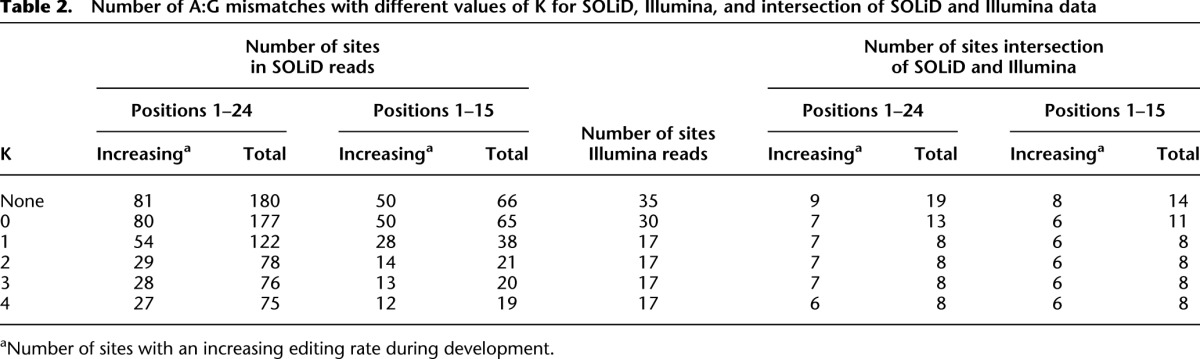
Table 3 contains all candidate A-to-I editing sites that pass the 4-safety filter and appear in both the SOLiD and the Illumina data, i.e., our most reliable identified edited miRNAs. More information about these candidates and other candidates with a lower value of K can be found in Supplemental Table 3. Six of the eight candidates in Table 3 have an increasing ratio of editing through development. Of the eight miRNAs in Table 3, miR-376b, miR-376c, miR-376a*, and miR-379 have previously been shown to be edited in samples from adult brain (Kawahara et al. 2007b, 2008). In all but one (miR-130a), the editing frequency is higher in the adult brain than in the developmental stages.
Table 3.
A:G mismatches common between SOLiD and Illumina reads with 4-safety filter
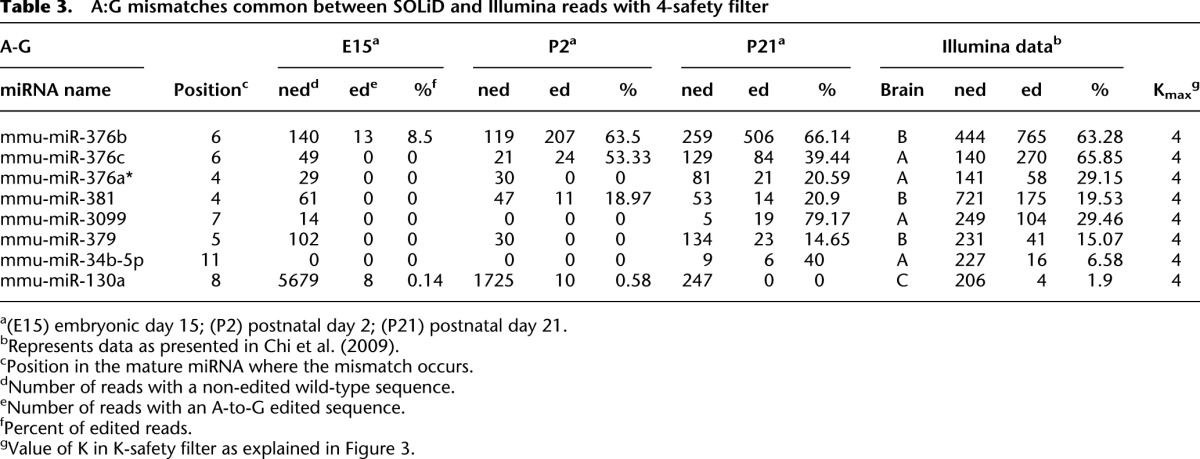
By lowering the stringency level one step, we get the mismatch sites that pass the 4-safety filter and (1) appear in positions 1–15 and only in the SOLiD data; or (2) appear only in the Illumina data. Supplemental Tables 1 and 2 contain the list of our candidates for SOLiD and Illumina, respectively, for each value of K. Twelve of the 19 SOLiD sites are increasing. Two of these previously not reported candidates, miR-24-2* and miR-337-3p, are edited in positions 7 and 11, respectively. In the Illumina data, there are five candidates on this stringency level. Among these five candidates, the most interesting one is miR-1196, which is edited at position 7, i.e., in the seed region. This site is highly edited; the frequencies are 66.57% and 63.64% in brains A and B, respectively.
When relaxing stringency to the 0-safety filter for the intersection of SOLiD and Illumina data, we found miR-301a, let-7c, miR-669a-3p, miR-669o-3p, and miR-101a to be edited at positions 10, 17, 10, 10, and 16, respectively (Supplemental Table 3). These sites are novel and pass the Chiang et al. (2010) test (see Methods) but were not reported in Chiang et al. (2010) or Kawahara et al. (2007a,b, 2008). There is an interesting observation regarding miR-669a-3p and miR-669o-3p; these two miRNAs have identical mature sequences, so they merely pass the 0-safety filter. A closer inspection showed that the nearest hairpin read to the edited sequence of these two miRNAs is the miRNA miR-669a-3-3p, which has two mismatches to the edited sequence. Taking this fact into account, miR-669a-3p and miR-669o-3p can be considered to be more reliable editing candidates. Editing of these two miRNAs is also increasing during development. However, we have not determined whether both miRNAs are edited or just one of them. Finally, as can be seen, for each K value between 0 and 4, the majority of the miRNAs in the SOLiD data that are edited in positions 1–15 and pass the K-safety filter have an increasing ratio of edited-to-unedited reads during development (Table 2).
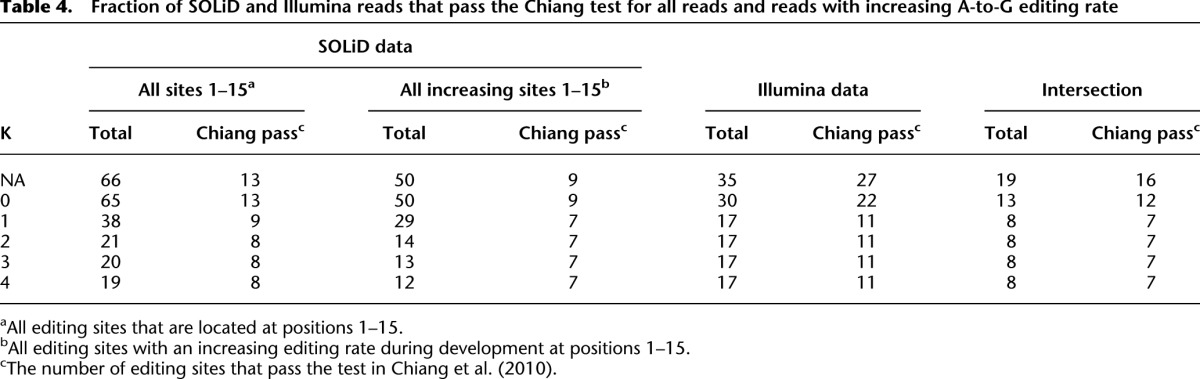
Table 4 contains a summary of the number of SOLiD and Illumina reads that passed the test in Chiang et al. (2010) and the K-safety filter at various values of K. As Table 4 shows for both SOLiD and Illumina data, the 4-safety filter gives a few more candidates than the Chiang test. In particular, some candidates that passed the 4-safety filter did not satisfy the Chiang test requirement that each candidate should be supported by at least 10 reads. For example, miR-34b-5p, which also appears in the intersection of the SOLiD and the Illumina data and is edited at 40% at stage P21, is rejected by the Chiang test due to this requirement.
Table 4.
Fraction of SOLiD and Illumina reads that pass the Chiang test for all reads and reads with increasing A-to-G editing rate
Evidence for C-to-T editing from HTP data
By comparing our SOLiD HTP data with the Illumina data, we also detected candidates for C-to-U editing. Several novel substrates for C-to-U editing, possibly by the APOBEC1 enzyme, have recently been identified by HTP RNA-seq, but none of these reside in transcripts coding for miRNAs (Rosenberg et al. 2011). Supplemental Tables 4 and 5 contain lists of C:T mismatch sites for SOLiD and Illumina data, respectively. Supplemental Table 6 contains the list of C:T mismatched sites that are both in SOLiD and Illumina data and shows C-to-U editing candidates for all values of K. We consider those mismatch sites that passed the 4-safety filter and the Chiang test to be our most reliable C-to-U editing candidates. Editing of miR-379, miR-140*, and miR376a was analyzed by Sanger sequencing of RT-PCR products at the pri-miRNA level in an attempt to verify C-to-U editing. However, C:T mismatches, indicative of C-to-U editing, could not be found (data not shown) at the pri-miRNA level. One possible explanation is that this particular C-to-U editing occurs only in pre- or mature miRNA.
Dramatic increase in A-to-I editing of miRNAs after birth
To confirm the findings in the SOLiD data of increased editing in the seed sequences of the mature miRNAs during development, we analyzed editing of the candidate miRNAs at the pri-miRNA level in mouse brain during embryogenesis as well as several postnatal stages. cDNAs as well as genomic DNA from the same mouse strain were amplified by PCR using primers specific for each pri-miRNA. Editing was considered to be verified whenever the putatively edited position had a visible G peak in a chromatogram obtained by direct Sanger sequencing; editing frequency was in this step estimated based on the relative heights of A and G peaks in the chromatogram. However, due to technical limitations, Sanger sequencing cannot reliably detect editing levels below ∼10%. An increase in editing of the adenosines in the seed sequence was verified for six different miRNAs. Since this method is applied to pri-miRNA, it cannot exclude editing occurring on the pre-miRNA level.
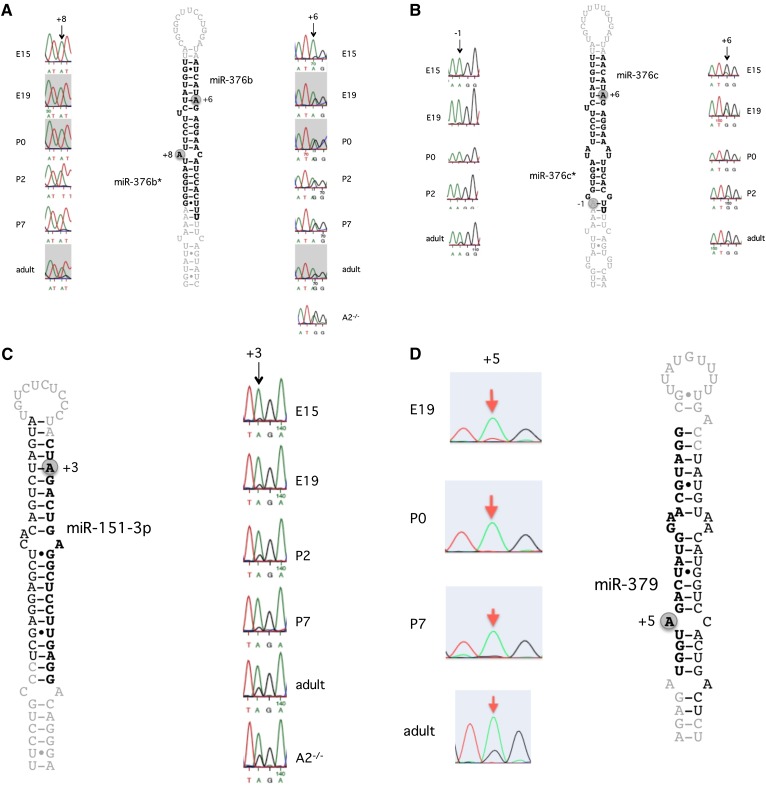
It has previously been shown that mmu-miR-376b is edited in the adult brain (Kawahara et al. 2007b). miR-376b is edited at position +6, which resides in the seed sequence (Fig. 4A). In our validation, the following observations were made. At E15 only a low level of editing was detected (Fig. 4A). At birth the A and G peaks in the chromatogram were equal, indicating that at least 50% of the pri-miRNAs are edited, but no further increase in editing was detected in the adult brain, rather a slight decrease. When editing of pri-miRNA-376b was analyzed in the adult ADARB1−/− knockout mouse brain, a higher level of editing could be detected than in the wild-type mouse. This result corroborates earlier reports that editing of miR-376b is performed by ADAR. The higher level in the knockout also suggests that ADARB1 interferes with ADAR editing, possibly by binding competition. Despite differences in editing levels between pri-miRNA-376b and the mature form, the result of the Sanger sequencing agrees well with the HTP RNA-seq data. As can be seen in Table 3, the latter shows an increase in editing at the +6 site of the mature miR-376b, with 8,5%, 63%, and 66% of the transcripts being edited at E15, P2, and P21, respectively. Thus the editing level is low during embryogenesis, but increases dramatically after birth and is then kept at a constant high level.
Figure 4.
Editing of pri-miRNAs at different developmental stages, detected by direct Sanger sequencing. RNA from whole mouse brain was amplified by RT-PCR followed by Sanger sequencing. Typical samples of biological triplicates are shown. The different developmental stages used for sequencing are indicated in each figure. The edited position is indicated by an arrow. (A) Editing of pri-miR-376b. (B) Editing of pri-miR-376c. (C) Editing of pri-miR-151-3p. (D) Editing of pri-miR-379-5p.
In miR-376b*, editing has previously been detected at position +8 of the mature miRNA (Kawahara et al. 2007b). For the +8 site, in our RNA-seq data we can only see editing in the adult brain, where 38% of the miRNAs are edited (Supplemental Table 1). However, miR-376b* is not commonly incorporated into the RISC complex. The mature frequency of this miRNA is very low in our data, possibly due to degradation. For this site, Sanger sequencing verification of editing levels at the pri-miRNA level shows absent or very low editing levels (no visible G peak) at the earlier developmental stages and low, but visible, editing at the adult stage (Fig. 4A). This result confirms the RNA-seq data on mature miRNAs and shows that editing in this case is not interfering with the maturation.
It has been shown that mmu-miR-376c is edited at position +6 of the mature miRNA (Kawahara et al. 2007b). Our HTP sequencing data detect no editing of the mature miRNA before birth but ∼40%–50% editing at the postnatal stages (Table 3). This level of editing is different from that at the pri-miRNA level. Already at E15, the +6 site is highly edited, and it stays highly edited throughout development (Fig. 4B). This result indicates that editing interferes with pri-miR-376c maturation, and it is therefore detected at a higher level in the pri-miRNA.
Editing of the mature mmu-miR-151 was not detected in our HTP screen (data not shown), which is supported by previous analysis showing that editing of mmu-miR-151 leads to a DICER1 processing deficiency (Kawahara et al. 2007a). Nevertheless, it has been shown that pri-miR-151 is edited at positions −1 and +3. We therefore wanted to investigate whether we could find an increase in editing during development at the pri-miRNA level, before DICER1 processing. Indeed, we found editing to increase at the +3 site during development (Fig. 4C). One obvious explanation for the absence of miR-151 at the adult stage is the very low frequency of mature product. However, why mature miR-151 is absent at the embryonic stage is less obvious. One reasonable explanation is that it is not the editing event per se that leads to the inability of further processing, but rather that ADAR binding affects the accessibility of the miRNA to be processed by DICER1.
In the adult human brain, miR-379 has been shown to be edited at the +5 site to 60% at the pri-miRNA level and to 15% in the mature miR-379 RNA (Kawahara et al. 2008). We can confirm editing at the adult stage of both mature and pri-miR-379 (Fig. 4D; Table 3). However, up to P7, no editing could be detected in this miRNA. This result indicates that it is only at the adult stage that editing has the potential to influence target recognition by changing the seed sequence of the miRNA.
In summary, we can conclude that most of the miRNAs, shown by us and others to be edited, have low levels of editing before birth that increase until adulthood. These sequence differences are post-transcriptional, not detected in the genomic DNA, as shown in Supplemental Figure 17.
Editing of miRNAs can lead to a change in gene regulation during development
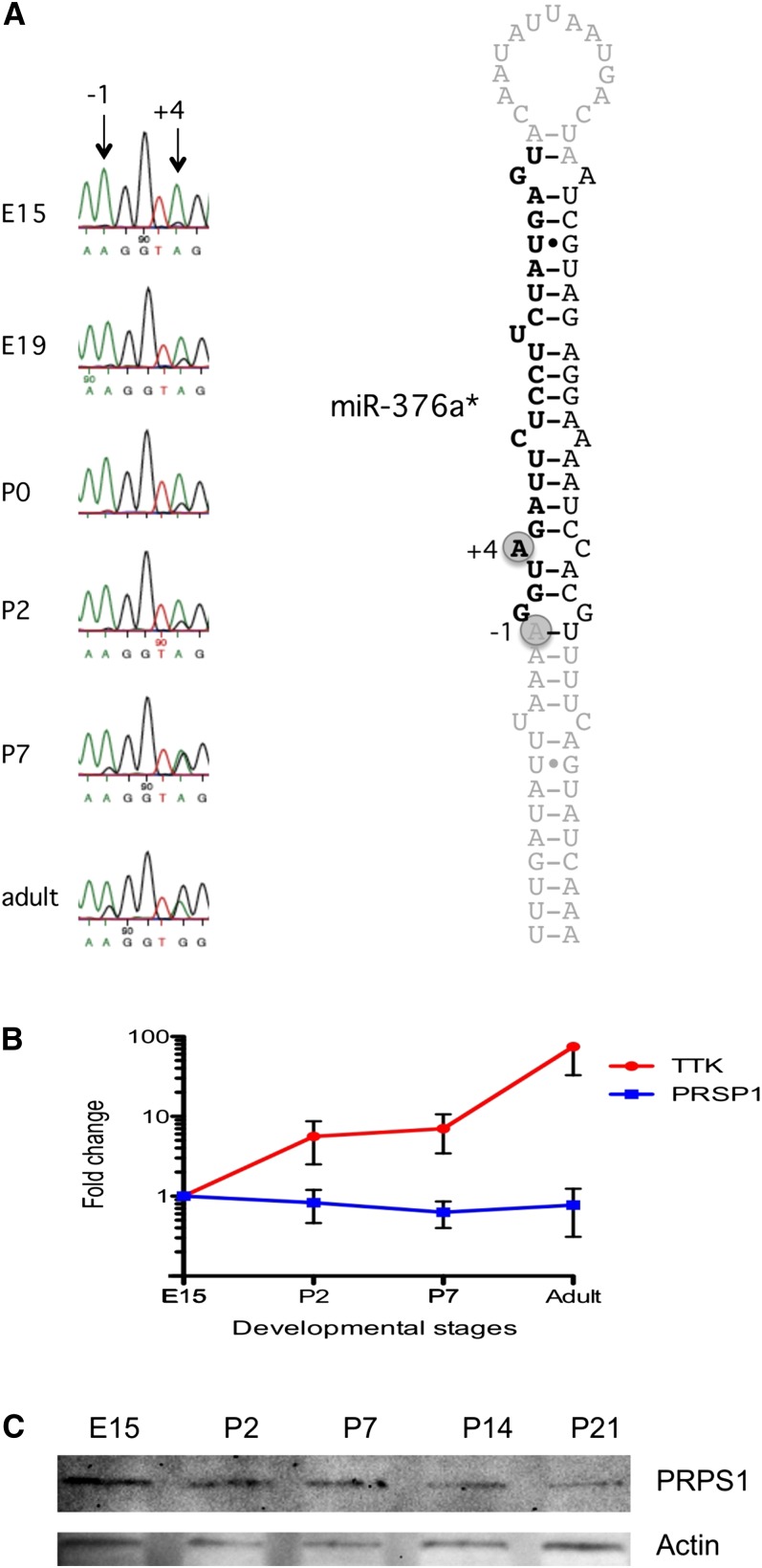
miR-376a* has previously been shown to be edited at two sites, −1 and +4 (Kawahara et al. 2007b). In our HTP sequencing screen, editing at the +4 site is estimated to be 21% in the adult mouse brain, but it is not detectable in the earlier developmental stages (Table 3). For pri-miR-376a*, the frequency of editing increased throughout development, with a very low level at E15 that gradually increases until the adult stage, where the majority of all miRNAs are edited (Fig. 5A). Thus, the editing frequency is significantly higher at the pri-miRNA stage than at the mature miRNA stage, indicating a possible processing deficiency due to the editing event.
Figure 5.
Editing of miR-376a redirects targeting during development. (A) Editing of pri-miR-376a at six different developmental stages. RNA from whole mouse brain was amplified by RT-PCR and sequenced. The different developmental stages used for sequencing are as indicated. (Arrows) Edited positions. (B) RNA from whole mouse brain at four different developmental stages (E15, P2, P7, and adult) was amplified by RT-PCR, and mRNA levels of TTK and PRPS1 were analyzed by qPCR. All error bars represent the SEM. (C) Protein levels of PRPS1 during development of the mouse brain. Immunoblot analysis showing PRPS1 protein expression during developmental day E19, P2, P7, P14, and adult. An anti-actin antibody was used as loading control.
Two targets for miR-376a* have been verified, TTK (threonine and tyrosine kinase), targeted by the non-edited miR-376a*; and PRPS1 (phosphoribosyl pyrophosphate synthetase 1), a target of the miRNA when it is edited in the seed sequence at position +4 (Kawahara et al. 2007b). To investigate the functional consequences of increased editing of this miRNA, we assayed the expression of these targets during brain development. Total RNA from E15, P2, P7, and adult was extracted from whole mouse brain. RT-PCR with primers specific for either TTK (target for the non-edited miRNA) or PRPS1 (target for the edited miRNA) was performed followed by quantitative PCR (q-PCR) (Fig. 5B). Interestingly, the expression of the two targets changes concurrent with the editing event changing target recognition of the miRNA. The PRPS1 transcript is highly expressed in the embryo and decreases gradually until adulthood, while the TTK mRNA increases after birth. These results agree with the recent results showing that changes in the mRNA levels closely mirror the influence that miRNAs have on gene expression, which point toward mRNA destabilization being the major reason for reduced protein levels (Guo et al. 2010).
To further verify the developmental regulation of PRPS1, we analyzed the protein expression by immunoblotting. Protein was extracted from five different developmental stages (E15, P2, P7, P14, and adult). Also, at the protein level, a slight decrease in the amount of PRPS1 throughout development can be identified (Fig. 5C).
Editing may promote dendrite outgrowth during early development
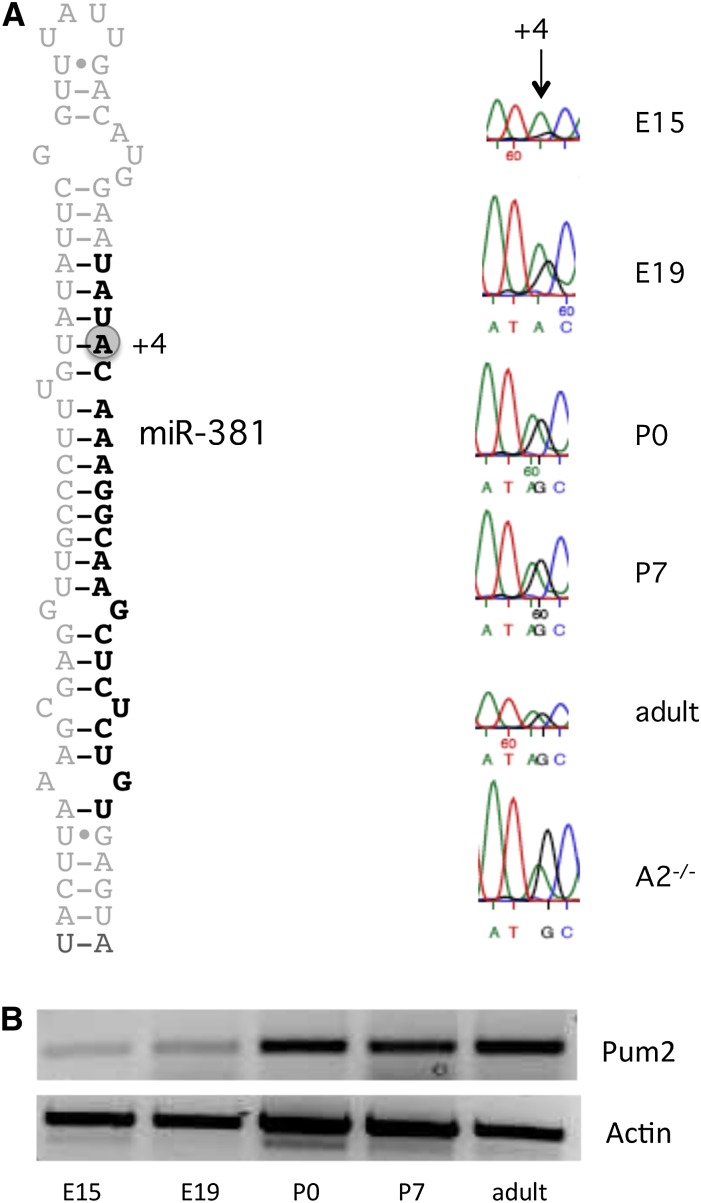
In our HTP screen for miRNAs targeted for A-to-I editing, we found a novel candidate, miR-381, to have increased editing at the +4 site of the seed sequence during development (Fig. 6A). Editing of the adult miR-381 was also recently suggested by the Bartel laboratory (Chiang et al. 2010). Interestingly, this miRNA is situated in the same cluster (miR379-410) as most of the other known edited miRNAs. The mature miR-381 shows no sign of editing at E15, but a clear presence of edited transcripts is seen in the two postnatal stages, P2 and P21, where the proportions of edited reads are 19% and 21%, respectively (Table 3). We also observe a steady increase in editing of pri-miR-381, with low editing levels at E15 that increase to at least as many edited as non-edited pri-miRNAs in the adult brain (Fig. 6A). Although both of these results, from the mature miR-381 and the pri-miRNA, support an increasing trend of editing, the level of editing at the pri-level is clearly higher, supporting the idea that editing leads to processing deficiency.
Figure 6.
Consequence of editing at the +4 site in miR-381 throughout development. (A) Editing of pri-miR-381 at five different developmental stages. RNA from whole mouse brain was amplified by RT-PCR and sequenced. The different developmental stages used for sequencing are indicated in the figure. (Arrow) The edited position. Editing of miR-381 in RNA extracted from an ADARB1-null mouse (A2−/−) indicates that ADAR is the major enzyme to edit this site. (B) Detection of Pum2 mRNA levels using semiquantitative RT-PCR. RNA from whole mouse brain at five different developmental stages (E15, E19, P0, P7, and adult) was amplified by RT-PCR with primers specific for Pum2.
Hitherto, no target for miR-381 has been confirmed, but, in a recent publication by Schratt and co-workers, miR-381 has been suggested to be an important player in activity-induced denditrogenesis (Fiore et al. 2009). As previously mentioned, miR-381 is expressed together with other miRNAs in a cluster identified as miR379-410. It has been shown that, during activity-dependent dendritogenesis, Mef2 induces the expression of this cluster. Pumilio 2 (Pum2) is a translational repressor that negatively regulates dendritic outgrowth (Vessey et al. 2006, 2010). The non-edited miR-381 has at least two potential target sites in the 3′ UTR of the Pum2 mRNA that are evolutionarily conserved between mouse, rat, and human. miR-376b and miR-134 are two other miRNAs from the miR379-410 cluster that have conserved target sites in Pum2. Interestingly, as shown above, both of these are also edited. Editing of mature miR-376b increases drastically during development and reaches a level of 66% in the adult brain, while the mature miR-134 could only be detected at the embryonic stage, where 40% of the sequences showed A-to-I editing at position 18 (Table 3; Supplemental Table 1). However, editing of miR-134 could not be confirmed at the pri-miRNA level (data not shown). Although editing at position 18 will not affect its targeting, it is likely that ADAR binding to position 18 will interfere with the maturation process.
To determine whether the level of Pum2 transcript is related to editing of miR-381, miR-376b, and possibly miR-134, we extracted RNA from five different developmental stages and performed semiquantitative RT-PCR using primers specific for Pum2. At the embryonic stages, when mature miR-381 and miR-376b are edited to a low extent or not at all, only low levels of Pum2 transcripts could be detected. As editing of these two miRNAs increases during development, the expression of Pum2 increases (Fig. 6B; Table 3). We cannot exclude that Pum2 expression is regulated also in other ways, but our results point to a developmental regulation of miR-381 and miR-376b editing, which prevents them from targeting the Pum2 mRNA for degradation.
Discussion
In this study, we have analyzed whether A-to-I editing of miRNAs is regulated during brain development. Editing of miRNA has been shown to either lead to new targets, if it occurs within the crucial seed sequence, or maturation defects due to insufficient processing of the miRNA by either DROSHA or DICER1 (Yang et al. 2006; Kawahara et al. 2007a,b). It is also possible that editing affects strand preference when loaded into the RISC complex. All of these steps can consequently affect biogenesis. With current HTP RNA-seq methods, it is possible to detect low levels of editing in mature miRNAs to an unprecedented precision. We have primarily used SOLiD HTP RNA-seq to evaluate A-to-I editing levels of mature miRNAs in mouse brain throughout development. We used total mouse brain RNA from three developmental stages—E15, P2, and P21—to search for known and novel editing sites within short RNA structures resembling the length of mature miRNAs with altered levels of editing during development. For five out of our eight most stringent editing candidates, we verified increased editing on the pri-miRNA level during brain maturation (Table 3). Out of the three that were not verified experimentally in the pri-miRNA, we are particularly confident about editing at position 11 of miR-34b-5p because it also passed the test in Chiang et al. (2010).
There are editing sites reported in Chiang et al. (2010) that cannot be found in our SOLiD data. The reason may be that the miRNAs are not extracted from the same tissue types. In Chiang et al. (2010), three mouse tissues are used: brain, ovary, and testes, from embryonic E7.5, E9.5, E12.5, and newborn mice. We used only brain tissue from E15, P2, and P21 mice. It is clearly possible that these miRNAs are expressed and edited differently in ovary and testis.
We can conclude that the gradual increase in editing efficiency seen for most selectively edited sites involved in neurotransmission also applies to miRNAs during development of the mammalian brain. The most striking editing events all occur in the crucial seed sequence, essential for target recognition. This will render the miRNAs to change targets during the course of development. Two clear examples of editing events within the seed sequence are miR-381 (novel) and miR-376b. One potential target for these miRNAs is Pum2, which increases in expression concurrent with the increase in miRNA editing throughout development (Fig. 6B). Pum2 is an important player as a negative regulator of dendrite outgrowth that has also been suggested to modulate the activity of RISC in neurons (Schratt 2009).
The edited forms of miR-381 and miR376b are less likely to target Pum2. Thus, editing could be a way to indirectly regulate the growth of dendrites during development. We show that both miR-381 and miR-376b are predominantly, if not exclusively, edited by ADAR (Figs. 4A, 6A). The long form of ADAR, p150, has been shown to shuttle between the nucleus and cytoplasm (Patterson and Samuel 1995). It is therefore possible that these miRNAs are edited also at the pre-miRNA level directly, at their site of action, in the dendrites. However, we cannot exclude that other regulatory determinants are involved in this process. For example, Schratt and co-workers have shown that down-regulation of Pum2 by miR-134 is important for activity-dependent dendritogenesis, but miR-134 had no effect under basal conditions (Fiore et al. 2009). Although we show that miR-134 is edited in our HTP sequencing screen at E15, the edited nucleotide should not change its targets. However, editing at position 18 could interfere with its maturation and, thereby, regulate miR-134 levels in uninduced neurons. It is also noteworthy that the edited form of mmu-miR-381 shares the same seed sequence as mmu-miR-300, although the rest of the sequence differs between the two miRNAs. However, similarly to the case of miR-381, the targets of miR-300 are yet to be determined.
Interestingly, our HTP screen on mature miRNAs combined with direct Sanger sequencing indicates that editing not only affects miRNA targeting, but it also affects the maturation and processing of several miRNAs. These processes are also increasingly affected during development. For example, in a previous study, it was shown that editing of miR-151-3p at the −1 and +3 sites leads to inhibition of DICER1 processing (Kawahara et al. 2007a). In line with this, our data show no editing of miR-151-3p at the mature level (Table 3). However, we can verify editing at the pri-level showing an increased frequency throughout development (Fig. 4C). This result indicates that, due to editing-induced inhibition of maturation, there may be more edited miRNAs than those detected in our analysis of mature miRNAs.
Another miRNA with processing deficiencies is miR-142, which has been shown to be edited at nine different sites in both the 5p and the 3p strand of the pri-miRNA in hematopoietic tissue (Yang et al. 2006). The functional consequence of these editing events is an inability to be further processed by DROSHA; that is, editing regulates the maturation from pri-miRNA to pre-miRNA. The unprocessed edited miRNA is degraded by the ribonuclease Tudor-SN previously shown to degrade inosine-containing transcripts (Scadden 2005). Direct Sanger sequencing of pri-miR-142 uncovered no editing in mouse brain from embryogenesis to adulthood (data not shown). This result may indicate that editing of this miRNA is tissue-specific.
In the case of both miR-376a* and miR-381, the editing event leads to a change or prevention of target recognition during development. The two genes shown to be targets of miR-376a* are TTK (target of the non-edited) and PRPS1 (target of the edited) (Kawahara et al. 2007b). We can show that the expression levels of these two targets change concurrently and in agreement with the increased editing, suggesting change of targets of the miRNAs as the brain matures. However, the apparent effect of editing on TTK, the former target, is much larger than that on PRPS1, the new target. In addition to the change of targets, miR-376a* and miR-381 also seem to be subjected to processing deficiencies. The most likely explanation of this is that the binding of ADARs to the miRNA not only leads to the editing event, but it also competes with the processing by DROSHA and DICER1, even in cases in which no editing takes place. We therefore propose that increased ADAR activity during development most often leads to a down-regulation of the miRNA with which it is interacting.
Some of the mature miRNAs in our HTP screen that show high levels of editing at all developmental stages could not be verified by direct Sanger sequencing on the pri-miRNA level. One explanation for this is that editing occurs on the pre-miRNA level. An editing event that occurs on the pre-miRNA level has the potential to be edited both in the nucleus and in the cytoplasm. Noteworthy, editing of miR-93 cannot be verified at the pri-mRNA by Sanger sequencing, although it is confirmed at the mature miRNA level in all three developmental stages (data not shown). Conversely to editing at the pri-miRNA level, the efficiency of editing in miR-93 is decreasing as the brain matures. Unpublished data from our laboratory indicate that both ADAR and ADARB1 are located in the cytoplasm during early development, and as development proceeds, they translocate to the nucleus. If miR-93 gets edited at the pre-miRNA level in the cytoplasm, a decrease in editing could be explained by the change in localization of the ADAR enzymes during development. We cannot, however, exclude that the reason we do not observe editing at the pri-miRNA level is the limitations of direct Sanger sequencing.
We here show that RNA editing influences miRNA expression extensively, not only on the level of expression, but also by redirecting targeting. Since this regulation increases as the brain matures, it suggests that editing of miRNAs is important for brain development. Furthermore, detecting editing by the ADAR enzymes in miRNAs by HTP RNA-seq is powerful, but functional studies are required to identify downstream effects of the editing events, i.e., verification of miRNA target loss and the impact it may have. We here show the power of combining HTP sequencing, bioinformatics, and specific experimental verifications in order to shed light on the impact of A-to-I editing on the process of RNA interference.
Methods
RNA extraction
Total brains from NMRI mice were collected at different developmental stages: E15, E19, P0, P2, P7, and P21. Total brain RNA was extracted from adult ADARB1−/− mice. The RNA was isolated using TRIzol (Invitrogen) according to the manufacturer's instructions. Before cDNA synthesis, the RNA was DNase (Fermentas) treated.
Determination of mature miRNA editing sites
For SOLiD high-throughput sequencing, 10 μg of total mouse brain RNA from three developmental stages—E15, P2, and P21—was isolated as described above. Small RNAs (10–40 nt) were purified form total RNA using flashPAGETM fractionation followed by adapter hybridization, ligation, reverse transcription, RNase H digest, and library amplification. This was followed by a small RNA library cleanup and size selection (105–150 bp) by PAGE. Before sequencing, the DNA was amplified by emulsion PCR. Emulsion PCR isolates individual DNA molecules along with primer-coated beads in aqueous droplets within an oil phase. The resulting beads, each containing only copies of the same DNA molecule, are deposited on a glass slide. The template on the selected beads undergoes a 3′ modification to allow covalent attachment to the slide. Samples were then analyzed on the SOLiD Analyzer. For the E15 stage, 8,660,775 reads were obtained; for P2, 9,875,846 reads; and for P21, 14,207,246 reads.
Verification of edited sites in the pri-miRNA sequence
To verify edited sites detected by HTP sequencing, cDNA synthesis was made from 3 μg of total RNA (see above) using random hexamer deoxyoligonucleotides (Invitrogen) and SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer's instructions. The cDNA was amplified by PCR using specific primers for each pri-miRNA, Pum2, and actin. Upon request, we can provide the sequence of the primers. PCR samples were gel-purified using NucleoSpin Extract II (Macherey-Nagel), and editing of each miRNA was verified by direct sequencing as a ratio between the A and the G peaks in the chromatogram (Eurofins MWG Operon). All cDNA sequences were compared with genomic DNA sequences from the same individual, and A-to-I editing sites were identified as an A in the genomic DNA sequence. Biological triplicates were used for PCR amplification and sequencing of both cDNA and DNA samples.
Q-PCR
The RNA levels for TTK and PRPS1 were determined by quantitative PCR (Q-PCR) at four different developmental stages—E15, P2, P7, and adult. Real-time PCR amplifications were performed in a reaction volume of 20 μL on the ABI PRISM 7000 sequencing detection system (Applied Biosystems). The thermal cycling started for 2 min at 50°C followed by 10 min at 95°C. This was followed by 40 cycles of 15 sec at 95°C and 1 min at 60°C. Each sample was analyzed in triplicate and repeated at least once.
Western blot
Mouse brain crude extracts from five different developmental stages were obtained by homogenizing the brain with 3 volumes of Lysis-M buffer (Roche) according to the manufacturer's instructions. Lysates containing 30 mg of protein and 2× sample buffer (Bio-Rad) were boiled for 5 min, separated on a 10% polyacrylamide gel, and transferred to a polyvinylidine fluoride membrane (Bio-Rad). Following blocking in 10% nonfat-milk in TBS-Tween, membranes were incubated with antibodies against PRPS1 (NUVUS) and Actin (Sigma-Aldrich) as an internal control. A swine anti-rabbit secondary antibody coupled to horseradish peroxidase (DakoCytomation) was used, and the blot was visualized by the ECL-plus Western Blot Detection Kit (GE Healthcare).
The bioinformatics pipeline
Figure 2 shows a flowchart of the bioinformatics pipeline. For SOLiD reads, we take advantage of two properties of the reads in color space in order to obtain nucleotide sequences with very low error rates. The first property is that a single incorrect color in a color-space sequence gives rise to many incorrect nucleotides after the position corresponding to the incorrect color. The second property is that all reads in color space contain a known adapter sequence in their 3′ ends. The way we take advantage of these properties is by removing all reads in which the 3′ end does not translate into a proper 3′ adapter. We also remove the 3′ adapter from each read. Finally, we remove all reads of length <19 or >24. After translation to the letter space, this base is removed. This means that each remaining sequence is correct or originates from a read with at least two incorrect colors, which is very unlikely. We refer to the remaining reads as the “translated reads.”
For Illumina reads, we remove the adapter sequence and barcode. Then we separate reads of length 19–24 nt for further analysis in the pipeline.
We align each translated SOLiD read and trimmed Illumina read against the mouse reference genome (Mus musculus NCBI37), miRBase 17, as well as the entire set of translated reads. To facilitate a highly sensitive identification of de novo miRNAs among the translated SOLiD reads and Illumina reads, we apply a simple and liberal hairpin test. We call the reads that pass the hairpin tests “HP-reads.” For details of the hairpin test, see the next section, “Hairpin Test.” The HP-reads are used in the K-safety filter. The K-safety filter is a method we developed to identify reads that may be edited versions of a known miRNA, but also can have originated from another miRNA, known or unknown, through K read errors. For details, see the section, “The K-Safety Filter.”
Hairpin test
We search for the HP reads by mapping the reads of length 19–24 nt to the mouse genome using SHRiMP (Rumble et al. 2009), and then we search for any read that matched perfectly to a locus in the genome for which the reverse complement (with at most three mismatches) can be located within 100 bp.
The K-safety filter
If an miRBase member has an A:G mismatch to a read, we call the miRBase member the “source.” We use the K-safety filter to remove any candidate editing site that may originate from a known miRNA or an unknown miRNA (a genomic locus with hairpin-making potential) different from its source. That is, in the K-safety filter, for each read R with an A:G mismatch to a source, we search for other known miRNAs or HP-reads with at most K mismatches to the read R, and if one is found, the read is removed. In Figure 3C, which illustrates the K-safety filter, the NHP-read R has an A:G mismatch to a known miRNA, M. Assume that the read H has more than K mismatches to the read R and that this is the minimum among all HP-reads, then R passes the K-safety filter. For example, if there are three mismatches between H and R, then R passes the 2-safety filter.
The Chiang et al. test
In Chiang et al. (2010), three simple criteria were used to gain specificity; an miRNA position was considered to be edited if (1) the relative level of editing was at least 5%; (2) it was supported by at least 10 reads; and (3) it was not in the two last positions at the 3′ end of an miRNA.
Data access
Data are archived at the NCBI Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra) under accession number SRP008143.
Acknowledgments
We thank the Science for Life Laboratory, Uppsala University for the SOLiD sequencing. The Center for Metagenomic Sequence analysis (CMS) and the Swedish Research Council funded this work.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.131912.111.
References
- Bass BL 2000. Double-stranded RNA as a template for gene silencing. Cell 101: 235–238 [DOI] [PubMed] [Google Scholar]
- Bass BL, Nishikura K, Keller W, Seeburg PH, Emeson RB, O'Connell MA, Samuel CE, Herbert A 1997. A standardized nomenclature for adenosine deaminases that act on RNA. RNA 3: 947–949 [PMC free article] [PubMed] [Google Scholar]
- Blow MJ, Grocock RJ, van Dongen S, Enright AJ, Dicks E, Futreal PA, Wooster R, Stratton MR 2006. RNA editing of human microRNAs. Genome Biol 7: R27 doi: 10.1186/gb-2006-7-4-r27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB 1997. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387: 303–308 [DOI] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436: 740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB 2009. Argonaute HITS-CLIP decodes microRNA–mRNA interaction maps. Nature 460: 479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. 2010. Mammalian microRNAs: Experimental evaluation of novel and previously annotated genes. Genes Dev 24: 992–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ 2004. Processing of primary microRNAs by the Microprocessor complex. Nature 432: 231–235 [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya S, Artus C, Habermacher R 2007. Mechanisms and regulation of the miRNA-mediated repression in mammalian cells. FEBS J 274: 43 [Google Scholar]
- Fiore R, Khudayberdiev S, Christensen M, Siegel G, Flavell SW, Kim TK, Greenberg ME, Schratt G 2009. Mef2-mediated transcription of the miR379-410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J 28: 697–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R 2005. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123: 631–640 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34: D140–D144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JJ, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho YJ, Zhang BT, Kim VN 2006. Molecular basis for the recognition of primary microRNAs by the Drosha–DGCR8 complex. Cell 125: 887–901 [DOI] [PubMed] [Google Scholar]
- Hartner JC, Schmittwolf C, Kispert A, Muller AM, Higuchi M, Seeburg PH 2004. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J Biol Chem 279: 4894–4902 [DOI] [PubMed] [Google Scholar]
- Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH 2000. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406: 78–81 [DOI] [PubMed] [Google Scholar]
- Hoopengardner B, Bhalla T, Staber C, Reenan R 2003. Nervous system targets of RNA editing identified by comparative genomics. Science 301: 832–836 [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838 [DOI] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K 2007a. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer–TRBP complex. EMBO Rep 8: 763–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K 2007b. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science 315: 1137–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Megraw M, Kreider E, Iizasa H, Valente L, Hatzigeorgiou AG, Nishikura K 2008. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res 36: 5270–5280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell 115: 209–216 [DOI] [PubMed] [Google Scholar]
- Kim VN 2005. MicroRNA biogenesis: Coordinated cropping and dicing. Nat Rev Mol Cell Biol 6: 376–385 [DOI] [PubMed] [Google Scholar]
- Kim J, Jung JH, Reyes JL, Kim YS, Kim SY, Chung KS, Kim JA, Lee M, Lee Y, Kim VN, et al. 2005. microRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J 42: 84–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419 [DOI] [PubMed] [Google Scholar]
- Lee Y, Han J, Yeom KH, Jin H, Kim VN 2006. Drosha in primary microRNA processing. Cold Spring Harb Symp Quant Biol 71: 51–57 [DOI] [PubMed] [Google Scholar]
- Luciano DJ, Mirsky H, Vendetti NJ, Maas S 2004. RNA editing of a miRNA precursor. RNA 10: 1174–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U 2004. Nuclear export of microRNA precursors. Science 303: 95–98 [DOI] [PubMed] [Google Scholar]
- Melcher T, Maas S, Herb A, Sprengel R, Seeburg PH, Higuchi M 1996. A mammalian RNA editing enzyme. Nature 379: 460–464 [DOI] [PubMed] [Google Scholar]
- Ohlson J, Pedersen JS, Haussler D, Öhman M 2007. Editing modifies the GABAA receptor subunit α3. RNA 13: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson JB, Samuel CE 1995. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: Evidence for two forms of the deaminase. Mol Cell Biol 15: 5376–5388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg BR, Hamilton CE, Mwangi MM, Dewell S, Papavasiliou FN 2011. Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3′ UTRs. Nat Struct Mol Biol 18: 230–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumble SM, Lacroute P, Dalca AV, Fiume M, Sidow A, Brudno M 2009. SHRiMP: Accurate mapping of short color-space reads. PLoS Comput Biol 5: e1000386 doi: 10.1371/journal.pcbi.1000386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scadden AD 2005. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat Struct Mol Biol 12: 489–496 [DOI] [PubMed] [Google Scholar]
- Schratt G 2009. microRNAs at the synapse. Nat Rev Neurosci 10: 842–849 [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208 [DOI] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V 2004. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol 5: R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K 2001. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res 29: 308–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B, Kohler M, Sprengel R, Seeburg PH 1991. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 67: 11–19 [DOI] [PubMed] [Google Scholar]
- Vessey JP, Vaccani A, Xie Y, Dahm R, Karra D, Kiebler MA, Macchi P 2006. Dendritic localization of the translational repressor Pumilio 2 and its contribution to dendritic stress granules. J Neurosci 26: 6496–6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vessey JP, Schoderboeck L, Gingl E, Luzi E, Riefler J, Di Leva F, Karra D, Thomas S, Kiebler MA, Macchi P 2010. Mammalian Pumilio 2 regulates dendrite morphogenesis and synaptic function. Proc Natl Acad Sci 107: 3222–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlstedt H, Daniel C, Ensterö M, Öhman M 2009. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res 19: 978–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Miyakoda M, Yang W, Khillan J, Stachura DL, Weiss MJ, Nishikura K 2004. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J Biol Chem 279: 4952–4961 [DOI] [PubMed] [Google Scholar]
- Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K 2006. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol 13: 13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17: 3011–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Haley B 2005. Ribo-gnome: The big world of small RNAs. Science 309: 1519–1524 [DOI] [PubMed] [Google Scholar]