Abstract
Purpose
The catalytic selectivity of human UGT1A9, an important membrane-bound enzyme catalyzing glucuronidation of xenobiotics were determined experimentally using 145 phenolics, and analyzed by 3D-QSAR methods.
Methods
The catalytic efficiency of UGT1A9 was determined by kinetic profiling. Quantitative structure activity relationships were analyzed using the CoMFA and CoMSIA techniques. Molecular alignment of the substrate structures was made by superimposing the glucuronidation site and its adjacent aromatic ring to achieve maximal steric overlap. For a substrate with multiple active glucuronidation sites, each site was considered as a separate substrate.
Results
The 3D-QSAR analyses produced statistically reliable models with good predictive power (CoMFA: q2 = 0.548, r2= 0.949, r2pred = 0.775; CoMSIA: q2 = 0.579, r2= 0.876, r2pred = 0.700). The contour coefficient maps were applied to elucidate structural features among substrates that are responsible for the selectivity differences. Furthermore, the contour coefficient maps were overlaid in the catalytic pocket of a homology model of UGT1A9; this enabled us to identify the UGT1A9 catalytic pocket with a high degree of confidence.
Conclusion
The CoMFA/CoMSIA models can predict the substrate selectivity and in vitro clearance of UGT1A9. Our findings also provide a possible molecular basis for understanding UGT1A9 functions and its substrate selectivity.
Keywords: UGT1A9, Glucuronidation, CoMFA, CoMSIA, Homology modeling
Introduction
The high rate of attrition in drug development has become a conundrum in pharmaceutical industry. One root cause for attrition is the unfavorable absorption, distribution, metabolism, and elimination (ADME) characteristics (1). Accordingly, there are considerable interests in developing either computational (in silico) or in vitro ADME methods to aid the lead compound selection (2,3).A main advantage of a computational model is that it allows the ADME properties predicted for a new structure without experimental determination. This merit is rather tempting when thousands of (even more) drug candidates need to be screened. In fact, the use of ADME modeling/prediction has become an effective approach for industry to reduce late-stage attrition in drug discovery (2).
Predicting the metabolic fate of a drug candidate is an indispensible component of ADME evaluation. Extensive metabolism may result in poor bioavailability and/or drug inefficacy, whereas poor metabolism can be associated with drug toxicity. Significant advances have been made to predict cytochrome p450 (CYPs)-mediated metabolism using molecular modeling techniques such as two dimensional/three dimensional (2D/3D) quantitative structure-activity relationships (QSAR), pharmacophore, and homology modeling (4,5). And a number of software packages to predict CYP metabolism have been commercialized (e.g., MetaSite, Simcyp) (6,7). However, relatively fewer efforts are directed to develop such models and to characterize structural features of substrates for other important drug metabolizing enzymes such as UDP-glucuronosyltransferases (UGTs) (8).
UGTs catalyze the glucuronidation reaction which has been recognized as a prevailing metabolic and detoxification pathway for many drugs, sometimes targeting the products (hydroxylated phenols) of CYP-mediated metabolism (3). Human UGTs constitute a large family of enzymes, and are systematically classified into four subfamilies, UGT1, UGT2, UGT3, and UGT8.9 One unique feature about human UGTs is that these enzymes show remarkably broad substrate specificity. A UGT substrate usually contains one nucleophilic group (i.e., hydroxyl (-OH) group, carboxylic acid (-COOH), and amines) to which the glucuronic acid derived from the cofactor UDP-glucuronic acid (UDPGA) is transferred. Although it is rare, the acidic carbons and thiol group can also be glucuronidated (10). Another important feature of human UGTs is that they exhibit vast overlapping substrate specificity; this has challenged the identification of specific probe substrates (and possibly inhibitors) for a particular UGT enzyme (11). Lacking of an in vivo selective UGT probe is a significant barrier to in vivo glucuronidation studies with respect to evaluation of the role of a UGT enzyme (12)
A complete three dimensional structure of human UGTs is not yet available. Only a partial crystal structure of UGT2B7 (C-terminal domain) was resolved in 2007 (13). This structure, combined with molecular modeling studies, provides substantial insights into the UDPGAbinding and possible catalytic mechanism (13,14). Due to the lack of structural information of the N-terminus (for substrate binding), relatively little is known about the specific molecular interactions that govern UGT selectivity for its substrates. Nonetheless, Miners and colleagues demonstrate that substrate hydrophobicity and the spatial arrangement of two hydrophobic regions (close to the glucuronidation site) are important for substrate recognition by several human UGT isoforms based on (2D/3D) regression models and a pharmacophore model (8,15,16).
UGT1A9 is a major UGT1A isoform in human liver (17). Its role in clearance of both chemotherapeutic and non-chemotherapeutic drugs (e.g., SN-38, tamoxifen and acetaminophen) and in detoxification of carcenogens (e.g., NNAL and benzo[a]pyrene) has been widely recognized (18–20). Moreover, UGT1A9 polymorphisms (e.g., M33T, C183G and V167A) are being identified; those genetic variants have impaired glucuronidation activity that might have clinical implications (19,21). Given its importance in clearance of many xenobiotics/drugs, UGT1A9 has received considerable studies in recent years (22–26). The aim of this work is to enhance our understanding of molecular interactions of UGT1A9 with its substrates, and to develop a more generalized model that can be used to predict UGT1A9-mediated glucuronidation of novel drug candidates. To this end, ligand-based three-dimensional quantitative structure-activity relationship (3D-QSAR) methods (i.e., Comparative Molecular Field Analysis (CoMFA) and Comparative Molecular Similarity Indices Analysis (CoMSIA)) were applied to yield statistically reliable models with good predictive power. The correlation results obtained by CoMFA/CoMSIA were graphically interpreted in terms of field contribution maps. The catalytic pocket (or binding pocket) geometry and its physiochemical properties indicated from the CoMFA/CoMSIA analyses were compared with that from a homology model of UGT1A9.
Materials and Methods
Materials
Expressed human UGT1A9 isoform (Supersomes™) was purchased from BD Biosciences (Woburn, MA). 4-Methylumbelliferone-glucuronide, baicalin (baicalein-7-O-glucuronide), uridine diphosphoglucuronic acid (UDPGA), alamethicin, D-saccharic-1,4-lactone monohydrate, and magnesium chloride were purchased from Sigma-Aldrich (St Louis, MO). Ammonium acetate was purchased from J.T. Baker (Phillipsburg, NT). SN-38-glucuronide, and propofol-glucuronide were obtained from Toronto Research Chemicals (North York, Ontario, Canada). Wogonoside (wogonin-7-O-glucuronide) was purchased from Chengdu Mansite Pharmaceutical Co. Ltd. (Chengdu, China). All (145) UGT1A9 substrates (Table 1) were obtained from commercial sources. The chemical structures of these UGT1A9 substrates are shown in Fig. S1 (Supporting Information).
Table 1.
Experimentally determined kinetic parameters for UGT1A9-mediated glucuronidation of 145 compounds, including 2 catecins (No.1–2), 5 chalcones (No.3–7), 1 chromone (No.8), 6 courmarins (No.9–14), 3 curcumins (No.15–17), 6 aromatic hydrocarbons (No.18–23), 4 flavanones (No.24–27), 31 flavones (No.28–58), 36 flavonols (No.59–94), 3 hydroxycinnamic acids (No.95–97), 11 isoflavones (No.98–108), 27 phenols (No.109–135), and 7 other compounds (No.136–145). The chemical structures (including the SMILES codes) of all compounds are shown in Fig.S1 (Supporting Information). The site of glucuronidation is indicated in the parenthesis. For a compound with multiple glucuronides generated, a lower-case letter is appended for a distinction.
| No. | Name | Km | Vmax | CLint (Vmax/Km) | Log (CLint) | Log (CLint) | Log (CLint) |
|---|---|---|---|---|---|---|---|
| μM | pmol/mg/min | μl/mg/min | (Actual) | (CoMFA) | (CoMSIA) | ||
| 1.a | (−)-Epigallocatechingallate (3′-OH) | 18.7 | 2955 | 158 | 2.20 | 2.32 | 2.42 |
| 1.b | (−)-Epigallocatechingallate (4″-OH) | 15.3 | 9984 | 646 | 2.81 | 2.72 | 3.36 |
| 2 | (−)-Epigallocatechin(3′-OH) | 135 | 5130 | 38 | 1.58 | 1.40 | 1.90 |
| 3 | 2-Hydroxychalcone$ | 0.23 | 552 | 2400 | 3.38 | 3.83 | 3.36 |
| 4 | 4-Hydroxychalcone$ | 1.62 | 54.8 | 34 | 1.53 | 1.68 | 1.32 |
| 5 | 2′-Hydroxychalcone$ | 0.77 | 1008 | 1309 | 3.12 | 2.80 | 2.40 |
| 6 | 4′-Hydroxychalcone$ | 1.72 | 339 | 197 | 2.29 | 1.95 | 1.97 |
| 7.a | Phloretin (2′-OH)$ | 0.63 | 467 | 741 | 2.87 | 2.88 | 2.64 |
| 7.b | Phloretin (4′-OH)$ | 0.43 | 636 | 1479 | 3.17 | 3.21 | 3.14 |
| 8 | 7-Hydroxychromone | 82.8 | 668 | 8 | 0.91 | 1.37 | 1.62 |
| 9 | 3,4-Diphenyl-7-hydroxycoumarin | 3.37 | 137 | 41 | 1.61 | 1.53 | 1.46 |
| 10 | 4-Methylumbelliferone | 12 | 3653 | 304 | 2.48 | 2.43 | 2.06 |
| 11 | 4-Hydroxy-6-methylcoumarin | 57.8 | 225 | 4 | 0.59 | 1.06 | 0.84 |
| 12 | 6-Hydroxy-7-methoxyl-4-phenylcoumarin | 312 | 3744 | 12 | 1.07 | 1.04 | 1.69 |
| 13 | 8-Hydroxywarfarin | 369 | 9229 | 25 | 1.40 | 1.69 | 1.67 |
| 14 | Scopoletin | 5.53 | 1064 | 192 | 2.28 | 2.60 | 2.56 |
| 15 | Curcumin | 35.4 | 1958 | 55 | 1.74 | 1.59 | 1.60 |
| 16 | Demethoxycurcumin | 18.6 | 258 | 14 | 1.14 | 1.37 | 1.16 |
| 17 | Bisdemethoxycurcumin * | 257 | 2070 | 8 | 0.90 | 1.04 | 0.68 |
| 18 | Emodin | 2.87 | 2928 | 1020 | 3.01 | 3.10 | 3.05 |
| 19 | Endoxifen * | 316 | 2086 | 6.6 | 0.82 | 1.97 | 1.01 |
| 20 | Enterolactone (3-OH) | 57.4 | 5292 | 92 | 1.96 | 1.95 | 2.04 |
| 21 | Naphthol$ | 0.25 | 72.1 | 288 | 2.46 | 1.89 | 1.55 |
| 22 | Raloxifene (6-OH) | 1.93 | 39.3 | 20 | 1.31 | 1.15 | 1.06 |
| 23.a | Tyrphostin B42 (3-OH) | 3.60 | 3677 | 1020 | 3.01 | 3.02 | 3.16 |
| 23.b | Tyrphostin B42 (4-OH) * | 14.5 | 2008 | 138 | 2.14 | 2.27 | 2.63 |
| 24 | 7-Hydroxyflavanone * | 3.34 | 5001 | 1497 | 3.18 | 2.88 | 2.58 |
| 25 | 4′-Hydroxy-3-methoxyflavanone | 83.4 | 1687 | 20 | 1.31 | 1.29 | 1.39 |
| 26.a | Hesperetin (3′-OH) | 4.29 | 4192 | 977 | 2.99 | 2.99 | 3.31 |
| 26.b | Hesperetin (7-OH) | 6.00 | 2444 | 407 | 2.61 | 2.45 | 2.49 |
| 27 | Narigenin (7-OH) | 3.34 | 5001 | 1497 | 3.18 | 3.35 | 2.98 |
| 28 | 2′-Hydroxyflavone | 0.40 | 74.4 | 186 | 2.27 | 2.44 | 2.26 |
| 29 | 3,4′-Dimethoxy-5,7,3′-trihydroxyflavone (7-OH) | 1.64 | 7494 | 4570 | 3.66 | 3.43 | 3.43 |
| 30 | 3′-Benzyloxy-5,7-dihydroxy-3,4′-dimethoxyflavone (7-OH) | 1.25 | 2079 | 1663 | 3.22 | 3.25 | 3.22 |
| 31 | 3′-Hydroxyflavone | 2.02 | 926 | 458 | 2.66 | 2.66 | 2.63 |
| 32 | 4′-Hydroxyflavone | 1.46 | 98.7 | 67.6 | 1.83 | 1.83 | 2.11 |
| 33 | 5,7-Dihydroxy-3′,4′,5′-trimethoxyflavone (7-OH) | 0.45 | 382 | 849 | 2.93 | 3.25 | 2.84 |
| 34 | 5-Hydroxyflavone | 0.96 | 118 | 123 | 2.09 | 2.09 | 2.77 |
| 35 | 6,3′,4′-Trihydroxyflavone (3′-OH)# | 1.87 | 6470 | 3450 | 3.54 | 2.93 | 2.86 |
| 36 | 6,7,3′-Trihydroxflavone (7-OH) | 3.01 | 2333 | 775 | 2.89 | 3.05 | 3.14 |
| 37 | 6,7-Dihydroxyflavone (7-OH) | 4.49 | 4090 | 910 | 2.96 | 3.07 | 3.18 |
| 38 | 6-Hydroxyflavone | 2.27 | 76.1 | 34 | 1.53 | 1.30 | 1.64 |
| 39 | 6-Methoxyluteolin (7-OH) | 1.15 | 1902 | 1654 | 3.22 | 3.28 | 3.18 |
| 40 | 5,7,2′-Trihydroxyflavone (7-OH) * | 0.23 | 862 | 3748 | 3.57 | 3.44 | 3.39 |
| 41.a | 7,3′,4′-Trihydroxyflavone (3′-OH) | 2.45 | 2921 | 1192 | 3.08 | 3.23 | 2.92 |
| 41.b | 7,3′,4′-Trihydroxyflavone (4′-OH) | 2.44 | 744 | 305 | 2.48 | 2.38 | 2.74 |
| 41.c | 7,3′,4′-Trihydroxyflavone (7-OH) | 6.38 | 820 | 128 | 2.11 | 2.70 | 2.75 |
| 42 | 7,2′-Dihydroxyflavone (7-OH)* | 0.65 | 381 | 586 | 2.77 | 2.74 | 2.94 |
| 43 | 7,3′-Dihydroxyflavone (7-OH) | 29.6 | 16166 | 546 | 2.74 | 2.85 | 2.86 |
| 44 | 7-Hydroxy-2′-methoxyflavone | 2.21 | 2228 | 1008 | 3.00 | 3.04 | 2.82 |
| 45 | 7-Hydroxy-3-methylflavone | 1.46 | 2929 | 2006 | 3.30 | 3.19 | 2.83 |
| 46 | 7-Hydroxy-3′-methoxyflavone* | 25.3 | 132 | 5 | 1.02 | 2.81 | 2.80 |
| 47 | 7-Hydroxy-4′-methoxyflavone | 3.37 | 764 | 227 | 2.36 | 3.00 | 3.24 |
| 48 | 7-Hydoxy-5-methylflavone | 0.94 | 5369 | 5712 | 3.76 | 3.34 | 3.03 |
| 49 | 7-Hydroxyflavone | 3.59 | 4895 | 1364 | 3.13 | 2.88 | 2.90 |
| 50 | Apigenin (7-OH) | 1.93 | 3289 | 1704 | 3.23 | 2.96 | 3.24 |
| 51 | Baicalein (OH)# | 0.70 | 5310 | 7586 | 3.88 | 3.37 | 3.64 |
| 52 | Chrysin (7-OH) | 0.25 | 4537 | 18148 | 3.96 | 3.56 | 3.34 |
| 53 | Chrysoeriol (7-OH) | 0.91 | 2765 | 3038 | 3.48 | 3.16 | 3.09 |
| 54 | Diosmetin (7-OH) | 0.56 | 5361 | 9573 | 3.41 | 2.63 | 2.99 |
| 55 | Flavopiridol (7-OH) | 37.5 | 219 | 5.8 | 0.77 | 0.57 | 1.02 |
| 56.a | Luteolin (3′-OH) | 0.17 | 660 | 3870 | 3.58 | 3.56 | 3.08 |
| 56.b | Luteolin (7-OH) | 0.33 | 700 | 2100 | 3.32 | 3.38 | 3.20 |
| 57 | OroxylinA (7-OH) | 2.38 | 2060 | 866 | 2.94 | 3.46 | 3.32 |
| 58 | Wogonin (7-OH) | 1.27 | 5070 | 3992 | 3.60 | 3.98 | 3.63 |
| 59 | 3,2′-Dihydroxyflavone (3-OH) | 0.67 | 52 | 78 | 1.89 | 2.68 | 3.34 |
| 60 | 3,3′-Dihydroxyflavone (3-OH)* | 0.11 | 3100 | 29000 | 4.44 | 4.31 | 4.30 |
| 61 | 3,5-Dihydroxyflavone (3-OH)* | 0.25 | 1900 | 7400 | 3.87 | 4.05 | 3.88 |
| 62 | 3,6,4′-Trihydroxyflavone (3-OH)* | 0.31 | 6800 | 22000 | 4.34 | 4.31 | 3.97 |
| 63 | 3,3′,4′-Trihydroxyflavone (3-OH) | 0.10 | 2200 | 22000 | 4.34 | 4.21 | 3.92 |
| 64 | 3,4′-Dihydroxyfavone (3-OH) | 0.13 | 1800 | 14000 | 4.14 | 3.89 | 3.77 |
| 65 | 3,6-Dihydroxyfavone (3-OH) | 0.62 | 13000 | 21000 | 4.32 | 4.42 | 4.34 |
| 66 | 3-Hydroxy-6-methylflavone * | 0.062 | 6750 | 110000 | 5.04 | 4.34 | 4.29 |
| 67.a | 3,7-Dihydroxy-3′,4′,5′-trimethoxyflavone (3-OH) | 0.63 | 2872 | 4551 | 3.66 | 3.67 | 3.70 |
| 67.b | 3,7-Dihydroxy-3′,4′,5′-trimethoxyflavone (7-OH) * | 1.61 | 289 | 180 | 1.00 | 1.15 | 2.17 |
| 68.a | 3,7-Dihydroxyflavone (3-OH)# | 0.22 | 4600 | 21000 | 4.32 | 3.69 | 4.18 |
| 68.b | 3,7-Dihydroxyflavone (7-OH) | 1.50 | 2040 | 1360 | 3.13 | 3.09 | 3.05 |
| 69.a | 3,7-Dihydroxy-3′,4′-dimethoxyflavone (3-OH) | 1.03 | 753 | 731 | 3.16 | 3.48 | 3.79 |
| 69.b | 3,7-Dihydroxy-3′,4′-dimethoxyflavone (7-OH) | 1.16 | 11.6 | 10 | 1.00 | 1.22 | 2.37 |
| 70 | 3,7,3′-Trihydroxyflavone (3-OH) * | 3.37 | 764 | 227 | 2.86 | 4.00 | 4.33 |
| 71 | 3-Hydroxy-2′,3′-dimethoxyflavone | 0.52 | 4400 | 16000 | 3.92 | 3.60 | 3.91 |
| 72 | 3-Hydroxy-2′-methoxyflavone | 0.21 | 3800 | 18000 | 4.25 | 4.10 | 3.47 |
| 73 | 3-Hydroxy-3′-methoxyflavone | 0.063 | 2900 | 46000 | 4.66 | 4.56 | 4.58 |
| 74 | 3-Hydroxy-4′-methoxyflavone | 0.059 | 1900 | 32000 | 4.51 | 4.24 | 4.47 |
| 75 | 3-Hydroxy-5,7-dimethoxyflavone | 0.13 | 3300 | 26000 | 4.40 | 4.35 | 4.59 |
| 76 | 3-Hydroxy-5-methoxyflavone | 0.082 | 1900 | 24000 | 4.36 | 4.56 | 4.46 |
| 77 | 3-Hydroxy-6,4′-dimethoxyflavone | 0.043 | 2200 | 51000 | 4.71 | 4.97 | 5.05 |
| 78 | 3-Hydroxy-6-methoxyflavone | 0.074 | 5400 | 73000 | 4.86 | 4.97 | 4.79 |
| 79 | 3-Hydroxy-7,4′-dimethoxyflavone | 0.19 | 8700 | 46000 | 4.66 | 4.47 | 4.60 |
| 80 | 3-Hydroxy-7-methoxyflavone | 0.27 | 10000 | 37000 | 4.57 | 4.36 | 4.28 |
| 81 | 3-Hydroxyflavone | 0.30 | 2100 | 7000 | 3.84 | 4.01 | 4.15 |
| 82 | 3-Hydroxy-6-methyl-3′,4′-methylenedioxyflavone | 0.066 | 6000 | 91000 | 4.96 | 4.82 | 4.63 |
| 83 | Datiscetin (3-OH) * | 2.46 | 246 | 100 | 2.00 | 2.42 | 3.10 |
| 84.a | Fisetin (3-OH) | 0.63 | 1500 | 2300 | 3.38 | 3.90 | 3.95 |
| 84.b | Fisetin (3′-OH) | 0.74 | 2620 | 3520 | 3.55 | 3.37 | 3.33 |
| 84.c | Fisetin (4′-OH) | 0.52 | 1620 | 3090 | 3.49 | 3.63 | 3.54 |
| 85.a | Galangin (3-OH)# | 0.68 | 7400 | 11000 | 4.04 | 3.71 | 3.91 |
| 85.b | Galangin (7-OH) | 0.54 | 2560 | 4740 | 3.68 | 3.77 | 3.50 |
| 86 | Geraldol (3-OH) | 1.47 | 5885 | 4003 | 3.60 | 3.89 | 3.76 |
| 87 | Isorhamnetin (3-OH) | 0.32 | 12000 | 38000 | 4.59 | 4.13 | 3.49 |
| 88.a | Kaempferol (3-OH) | 0.32 | 1900 | 6000 | 3.78 | 3.60 | 3.54 |
| 88.b | Kaempferol (7-OH) * | 3.87 | 870 | 220 | 2.34 | 3.36 | 3.41 |
| 89 | Morin (3-OH) | 0.68 | 40 | 60 | 1.77 | 2.33 | 2.73 |
| 90.a | Myricetin (3-OH) | 0.61 | 490 | 800 | 2.90 | 3.33 | 3.39 |
| 90.b | Myricetin (3′-OH)# | 0.67 | 1530 | 2290 | 3.36 | 3.25 | 3.73 |
| 90.c | Myricetin (4′-OH) | 0.64 | 3170 | 4930 | 3.69 | 3.69 | 3.30 |
| 91.a | Quercetin (3-OH) * | 0.36 | 3300 | 9200 | 3.97 | 3.91 | 3.68 |
| 91.b | Quercetin (3′-OH) | 0.90 | 1880 | 2100 | 3.32 | 3.49 | 3.44 |
| 91.c | Quercetin (7-OH) | 0.85 | 3820 | 4490 | 3.65 | 3.52 | 3.24 |
| 92.a | Resokaempferol (3-OH) | 0.36 | 2600 | 7300 | 3.86 | 3.59 | 3.80 |
| 92.b | Resokaempferol (7-OH) * | 2.67 | 1150 | 430 | 2.63 | 2.67 | 2.94 |
| 93 | Rhamnetin (3-OH) | 0.23 | 10000 | 45000 | 4.65 | 4.34 | 3.84 |
| 94.a | Syringetin (3OH) | 0.98 | 7810 | 7333 | 3.87 | 4.11 | 3.46 |
| 94.b | Syringetin (7OH) | 1.61 | 289 | 178 | 2.26 | 2.05 | 2.88 |
| 95 | Ferulic Acid * | 884 | 372 | 0.4 | 0.30 | 1.50 | 0.09 |
| 96 | Isoferulic Acid | 237 | 15640 | 66 | 1.82 | 1.62 | 1.16 |
| 97.a | Caffeic Acid (3-OH) | 564 | 6204 | 11 | 1.04 | 1.50 | 0.86 |
| 97.b | Caffeic Acid (4-OH) | 1012 | 2024 | 2 | 0.30 | 0.51 | 0.50 |
| 98 | 7-Hydroxy-6-methoxyisoflavone | 3.08 | 1143 | 371 | 2.57 | 2.44 | 2.21 |
| 99 | 8-Hydroxy-7-methoxyisoflavone | 1.42 | 5299 | 3732 | 3.57 | 3.76 | 3.34 |
| 100 | Biochanin A (7-OH) * | 1.13 | 379 | 335 | 2.53 | 2.17 | 1.93 |
| 101 | Daidzein (7-OH) | 14.3 | 1107 | 77 | 1.89 | 2.17 | 1.78 |
| 102 | Dihydrodaidzein (7-OH) | 77.7 | 628 | 8.1 | 0.91 | 0.89 | 0.74 |
| 103 | Equol (7-OH) | 213 | 794 | 3.7 | 0.57 | 0.44 | 0.68 |
| 104 | Formononetin (7-OH) | 4.59 | 139 | 30 | 1.48 | 1.48 | 1.48 |
| 105 | Genistein (7-OH) | 2.09 | 1290 | 617 | 2.79 | 2.96 | 2.23 |
| 106 | Glycitein (7-OH) | 1.47 | 403 | 274 | 2.44 | 2.56 | 2.33 |
| 107 | Maackiain | 3.12 | 234 | 75 | 1.87 | 1.57 | 1.29 |
| 108 | Prunetin (5-OH) | 1.29 | 230 | 178 | 2.25 | 2.20 | 2.42 |
| 109 | 4-Bromophenol *# | 30.9 | 3057 | 99 | 2.00 | 1.53 | 1.49 |
| 110 | 4-n-Butylphenol | 43.1 | 647 | 15 | 1.18 | 1.45 | 1.53 |
| 111 | 4-Chlorophenol | 34.0 | 3230 | 95 | 1.98 | 1.44 | 1.60 |
| 112 | 4-Cyclopentylphenol# | 18.2 | 1083 | 60 | 1.78 | 1.40 | 1.11 |
| 113 | 4-Ethoxyphenol | 454 | 3761 | 8.3 | 0.92 | 1.25 | 1.57 |
| 114 | 4-Ethylphenol *# | 15.8 | 1179 | 75 | 1.87 | 1.45 | 1.39 |
| 115 | 4-Fluorophenol * | 112 | 1680 | 15 | 1.18 | 1.56 | 1.79 |
| 116 | 4-Iodophenol | 20.5 | 3034 | 148 | 1.87 | 1.40 | 1.35 |
| 117 | 4-Isopropylphenol | 36.4 | 2184 | 60 | 1.78 | 1.62 | 1.17 |
| 118 | 4-Methoxyphenol | 154 | 2310 | 15 | 1.18 | 1.01 | 1.41 |
| 119 | 4-Methylphenol | 57.3 | 1433 | 25 | 1.39 | 1.30 | 1.41 |
| 120 | 4-Nitrophenol# | 80.6 | 2902 | 36 | 1.56 | 1.50 | 1.92 |
| 121 | 4-Hydroxyacetophenone | 317 | 4438 | 14 | 1.15 | 1.40 | 1.61 |
| 122 | 4-Hydroxybenzophenone | 35.9 | 2441 | 68 | 1.83 | 1.90 | 1.38 |
| 123 | 4-Phenylphenol | 56.2 | 1236 | 22 | 1.34 | 1.50 | 1.33 |
| 124 | 4-Phenylazophenol | 82.5 | 1155 | 14 | 1.15 | 1.29 | 1.48 |
| 125 | 4-Propoxyphenol | 156 | 1560 | 10 | 1.00 | 1.06 | 1.55 |
| 126 | 4-n-Propylphenol | 47.6 | 1999 | 42 | 1.62 | 1.49 | 1.39 |
| 127 | 4-sec-Butylphenol * | 87.5 | 3237 | 37 | 1.57 | 1.66 | 1.19 |
| 128 | 4-tert-Butylphenol * | 78.4 | 5253 | 67 | 1.83 | 1.90 | 1.47 |
| 129 | Butyl-4-hydroxybenzoate | 84.1 | 2691 | 32 | 1.51 | 1.51 | 1.91 |
| 130 | Ethyl-4-hydroxybenzoate | 74.5 | 3055 | 41 | 1.61 | 1.43 | 1.97 |
| 131 | Eugenol | 25 | 1004 | 40 | 1.60 | 1.80 | 1.95 |
| 132 | Methyl-4-hydroxybenzoate | 101 | 5454 | 54 | 1.73 | 1.61 | 1.98 |
| 133 | Propofol | 26 | 148 | 6 | 0.76 | 0.86 | 1.32 |
| 134 | Propyl-4-hydroxybenzoate | 152 | 4560 | 30 | 1.48 | 1.54 | 2.08 |
| 135 | Bisphenol A | 91.8 | 4420 | 48 | 1.68 | 1.85 | 1.02 |
| 136 | Combretastatin A4# | 4.18 | 9009 | 2155 | 3.33 | 3.34 | 2.88 |
| 137 | Pterostilbene | 35.9 | 40.4 | 1.1 | 0.05 | −0.09 | 0.52 |
| 138 | Resveratrol (3-OH) | 3.09 | 196 | 63 | 1.8 | 2.23 | 2.14 |
| 139 | A-769662 (6-OH) | 0.78 | 10.8 | 14 | 1.14 | 0.89 | 0.62 |
| 140 | Entacapone (3-OH) | 8.64 | 88400 | 10233 | 4.01 | 4.10 | 4.43 |
| 141 | Ezetimibe | 268 | 73.6 | 0.30 | −0.56 | −0.68 | −0.50 |
| 142 | Mycophenolic Acid | 71.5 | 6538 | 91 | 1.96 | 1.99 | 1.92 |
| 143 | Psilocin | 748 | 5984 | 8 | 0.90 | 0.90 | 0.66 |
| 144 | SN-38 | 26.4 | 73.1 | 3 | 0.44 | 0.68 | 0.41 |
| 145 | Tolcapone (3-OH) * | 43.4 | 18100 | 417 | 2.62 | 2.79 | 2.08 |
These 25 compounds were used as a test set and not included in the derivation of model equations.
Described using the substrate inhibition equation (eq.2). Ksi value is not shown because it is unimportant in calculation of CLint (please see Materials and Methods).
Described using a biphasic kinetics model (eq.3). Km1and Vmax1 values are shown in the columns Km and Vmax, respectively. CLint2 value is not shown because it is unimportant in calculation of CLint (please see Materials and Methods).
Enzyme Assays
Enzyme assays using expressed UGT1A9 were conducted following a standard protocol as described in our earlier publications (27,28). Briefly, the incubation procedures were as follows: [1] UGT1A9 (13–53 μg/ml as optimum for the reaction), magnesium chloride (0.88 mM), saccharolactone (4.4 mM), alamethicin (0.022 mg/ml), different concentrations of substrates in a 50 mM potassium phosphate buffer (pH 7.4), and UDPGA (3.5 mM, added last) were mixed; [2] the mixture (final volume, 200 μl) was incubated at 37°C for a predetermined period of time (15–120 min); and [3] the reaction was stopped by the addition of 50 μl of 94% acetonitrile/6% glacial acetic acid. Great effort was made to ensure that the rates of metabolite formation were linear with respect to time (15–120 min) and protein concentration (13–53 μg/ml), so we can obtain accurate and reliable initial metabolism rates. Apparent glucuronidation rates were calculated as the amount of glucuronide(s) formed per protein concentration per reaction time (or pmol/mg/min). All experiments were performed in triplicates.
UPLC analysis
The Waters ACQUITY UPLC (Ultra performance liquid chromatography) system was used to analyze the UGT1A9 substrates and their glucuronides (27,28). Except for the elution gradient, all other UPLC conditions (such as the mobile phase and column) were kept the same. The gradient was adjusted carefully to ensure a good separation of a substrate from its glucuronide(s) and of a glucuronide isomer from the other one(s). Quantitation of propofol glucuronide, SN-38 glucuronide, 4-methylumbelliferone-glucuronide, baicalein-7-O-glucuronide, and wogonin-7-O-glucuronide was made with the authentic reference standards. For all other glucuronides without commercial availability, the conversion factor of glucuronide vs. aglycone was determined using our published method (29). Quantitation of these glucuronides was based on the standard curve of the parent compound and further calibrated with the conversion factors.
Identification of glucuronide and glucuronidation site
Glucuronide formation by UGT1A9 was confirmed via the hydrolysis (by β-D-glucuronidase) experiment and the molecular weight detection by UPLC/MS/MS, a standard procedure in our lab (27,28). The site (-OH group) of glucuronidation is an important information that was incorporated into CoMFA/CoMSIA analyses (see later section). However, regular MS/MS is unable to probe the site of glucuronidation (or deduce the exact structure of a glucuronide); because the glucuronic acid moiety is readily detached from a glucuronide once collision energy is applied. For a substrate with a single -OH group, the site of glucuronidation has to be this -OH group and no addition effort is needed. By contrast, for a substrate containing multiple -OH groups, three methods were used to elucidate a glucuronidation site. That is (1) for flavones and flavonols, the site of glucuronidation was assigned by the “UV spectrum maxima (λmax) shift method” (30). This method is based on the characteristic UV shifts caused by glucuronic acid substitution on a particular -OH group; (2) the information regarding the site of glucuronidation was collated from the literature, the references are provide in Table S1 (Supporting Information); (3) the preferred site of glucuronidation was assigned as 2′-OH for 7 phloretin, and 3-OH for 23 tyrphostin B42. These assignments are uncertain, even though they demonstrate a good consistency in later 3D-QSAR analyses.
Kinetics analysis
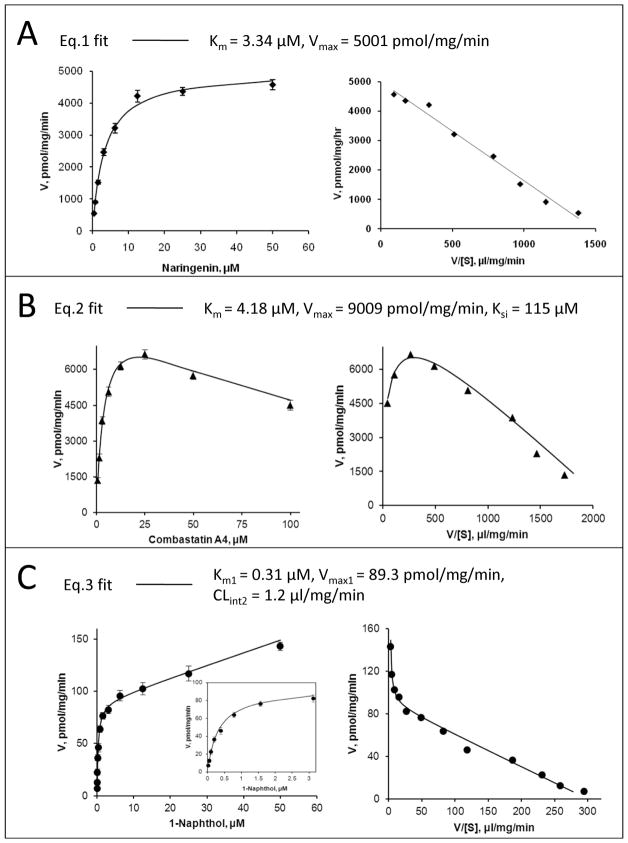
Kinetic data points were model-fitted using a nonlinear least-squares regression method performed by GraphPad Prism V5 for Windows (GraphPad Software, San Diego, CA). The model used to fit a kinetic profile was carefully selected based on a diagnostic plot (i.e., Eadie-hofstee plot). Overall, Michaelis-Menten equation (eq.1), the substrate inhibition equation (eq.2), and a biphasic kinetic model (eq.3) were used. The intrinsic clearance CLint representing the catalytic efficiency was calculated as Vmax/Km for eq.1 and eq.2 fitting and Vmax1/Km1 for eq.3 fitting. Representative fitting of the equations to kinetic data was demonstrated in Fig. 1.
Figure 1. Representative fitting of the model equations (eqs.1-3) to kinetic data of UGT1A9 with its substrates, both rate plot (left) and Eadie-Hofstee plot (right) are given.
Panel A: Eq.1 (Michaelis-Menten model) is used to describe glucuronidation of 27 naringenin by UGT1A9. Panel B: Eq.2 (a substrate inhibition model) is used to describe glucuronidation of 136 combrestatin A4 by UGT1A9. Panel C: Eq.3 (a biphasic model) is used to describe glucuronidation of 21 1-naphthol by UGT1A9. Points are experimentally determined values, while the solid lines show the computer-derived curves of best fit.
| Eq.1 |
| Eq.2 |
| Eq.3 |
Molecular Alignment
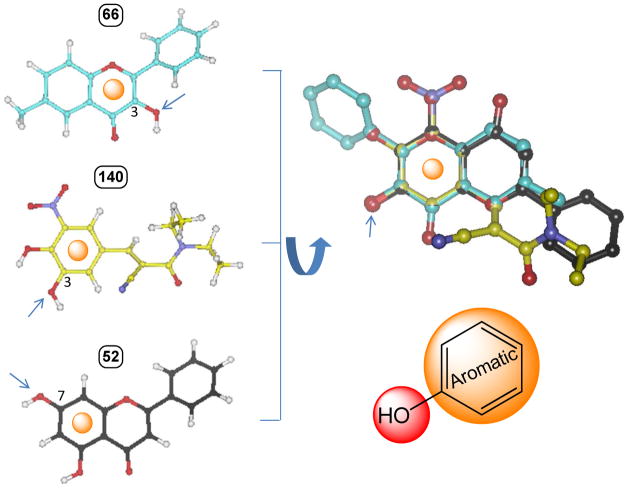
All substrate structures were prepared using SYBYL 8.0 (Tripos, US). Carboxylate groups were considered to be deprotonated. Energy minimizations were performed using the Tripos force field with partial atomic charges (assigned by Gasteiger-Hückel method). One of the most important factors affecting the quality of a model is the alignment of the individual molecules. The frequently used alignment methods (i.e., substructure overlap, pharmacophore overlap, and docking) are not suitable for this study, because a common core of atoms or a common feature pharmacophore cannot be defined for the 145 structurally diverse substrates (Table 1) and the protein structure is not available. To achieve our goals, we performed a flexible alignment of three most active substrates (66: 3-hydroxy-6-methylflavone, 140: entacapone; 52: chrysin) with a constraint that the glucuronidation site must be overlaid. The important common features of the most active substrates were found to be the glucuronidation site and its adjacent aromatic ring (Fig. 2). All other substrates were then aligned to superimpose these two features. Further, a maximal steric overlap was used to determine the best alignment when more than one alignment was possible. In the case of a substrate with multiple active glucuronidation sites, more than one structural pose was aligned corresponding to the glucuronidation at each site. This treatment rendered a final total 166 aligned structure conformations (Table 1). The structural diversity of the aligned ligands is shown in Fig. 3A. (The atomic coordinates of all molecules of the dataset are available from the authors upon request, bwu3@uh.edu).
Figure 2. A flexible alignment of three most active substrates (66: 3-hydroxy-6-methylflavone, 140: entacapone; 52: chrysin) to identify their common structural features.
The flexible alignment was performed with a constraint that the glucuronidation site must be overlaid. The important commonalities of the most active substrates are found to be the glucuronidation site and its adjacent aromatic ring.Arrows indicate the site of glucuronidation.
Figure 3. A wide diversity in both chemical structure (A) and activity (B) for UGT1A9 substrates in this work.
Panel A: Overview of all aligned structures in training set and test set (n=166).Panel B: UGT1A9 activity distribution (expressed as log(CLint)) of the training and test set.
CoMFAand CoMSIA analyses
The whole data set was divided into two parts, the training set (n= 141) and the test set (n = 25) (Table 1). The training set and test set molecules was classified to ensure that both sets could cover the whole range of glucuronidation activity and structural diversity studied. Other than these two criteria, the compounds were randomly assigned. The training set was used for model building and the test set for an external validation of the model. All comparative molecular field evaluations were performed using SYBL 8.0 (Tripos, US). The CoMFA steric energy (Lennard-Jones) and electrostatic (Coulomb) energy were calculated with SYBYL standard parameters (TRIPOS standard field, 2 Å grid spacing, dielectric distance 1/r2, cutoff 30 kcal/mol) using a sp3 carbon probe atom with a charge of +1. We also performed CoMSIA analysis (an extension of the CoMFA methodology) because CoMSIA may produce better or complementary results (31). In CoMSIA, three different similarity fields (steric, electrostatic, and hydrophobic) were evaluated with SYBYL standard parameters (2 Å grid spacing, attenuation factor α = 0.3) using a probe atom with 1 Å radius, charge +1, and hydrophobicity +1. Hydrogen bond donor and acceptor fields were not considered because CoMSIA analyses with these two extra features did not result in significant improvement of model quality. Partial least squares (PLS) analyses were performed following the CoMFA standard implementation in SYBYL. To check statistical significance of the models, cross-validations were performed by means of the “leave-one-out” procedure using enhanced version of PLS, the SAMPLS method. The optimal number of components was determined by selecting the smallest Spress (corresponds to the highest q2 value).The same number of components was subsequently used to derive the final QSAR models with no validation (column filtering was set to 2.0 kcal/mol); these models were used for prediction of activity. The statistical results are summarized in Table 2. The q2 (cross-validated r2), Spress (cross-validated standard error of prediction), r2 (non-cross-validated r2), and standard error of estimate (SEE) values were computed as defined in SYBYL. Predictive power of the obtained CoMFA and CoMSIA models was further validated with the test set which was not included in the model derivation. The predictive correlation coefficient, r2pred, of the CoMFA and CoMSIA models were calculated according to the definition of Cramer et al and are also shown in Table 2 (32).
Table 2.
Summary of modeling parameters from CoMFA and CoMSIA analyses
| Statistics | CoMFA | CoMSIA |
|---|---|---|
| q2a | 0.548 | 0.579 |
| Spressb | 0.885 | 0.799 |
| r2c | 0.949 | 0.876 |
| rpredd | 0.775 | 0.700 |
| SEEe | 0.282 | 0.435 |
| Componentsf | 8 | 5 |
| Fg | 271.8 | 190.0 |
| Pr2=0h | 0.000 | 0.000 |
| Fraction | ||
| Steric | 0.465 | 0.185 |
| Electrostatic | 0.535 | 0.435 |
| Hydrophobic | / | 0.380 |
Cross-validated correlation coefficient after the leave-one-out procedure.
Cross-validated standard error of prediction.
Non-cross-validated correlation coefficient.
Correlation coefficient for test set predictions.
Standard error of estimate
Optimum number of components.
F-test value.
Probability of obtaining the observed F ratio value by chance alone.
Homology modeling and molecular docking
A homology model for human UGT1A9 was constructed using Modeler 9v6 with a standard protocol (33). The best model with the lowest objective function values (DOPE) was selected for loop refinement and followed by energy minimization in GROMACS 3.3 program package (34). Energy minimization was done by steepest gradient descent, with an initial step size of 0.01 nm and a maximum of 1000 step. The VvGT1 (PDB code: 2c1z) from red grape was used as the template for two reasons (35): First, both UGT1A9 and VvGT1 belong to the glycosyltransferase 1 family (GT1) according to the CAZY database (36). Although sharing a low sequence identity, GT1 members adopt a similar α/β/α folding (so called “GT-B” fold) and their 3D structures are predicted to be highly conserved (17). Second, UGT1A9 and VvGT1 surprisingly share an overlapping substrate specificity (including regioselectivity), for example, they both preferentially metabolize flavonols at the 3-OH position (28,35). Due to a low sequence identity between the target and template proteins (~15 %), sequence alignment (shown in Supporting Information Fig. S2) was aided with secondary structure predictions, a strategy used earlier (37). The co-crystalized cofactor (UDP-2-fluoro glucose) was copied to the homology model as a block residue. Molecular docking of kaempferol (88.a) for 3-O-glucuronidation to the UGT1A9 model with the program GOLD (CCDC, Cambridge, UK) was performed using a distant constraint, a procedure similar to our earlier publications(38,39). A distance constraint (2.5~4 Å) was set between 3-OH and His37 (the catalytic residue). GOLDscore was used to identify the lowest energy docking results. The UGT1A9 model (with kaempferol docked) coordinates are available upon request.
Results
Experimental dataset
A large database of kinetic parameters was experimentally determined by kinetic profiling for UGT1A9-mediated glucuronidation of 145 phenolics (structures in Supporting Information Fig. S1), which are from 12 different classes (see Table 1 for compound names, their classification and kinetic parameters). The log(CLint) values for glucuronidation of selected substrates range from −0.56 (141) to 5.04 (66), demonstrating a wide diversity in the catalytic activity (Fig. 3B). To our knowledge, this is the largest dataset of kinetic data obtained using expressed UGT1A9 in current literature. Among all UGT substrates here, 17 compounds form more than one glucuronide; in particular, there are 4 compounds (i.e., 41, 84, 90, 91) from which three glucuronides at different positions (-OH groups) are generated. Differentiated kinetic properties for glucuronidation at each position suggests that distinct (productive) binding modes within the catalytic domain are possible (27,28). Based on this “expert” knowledge (i.e., the existence of multiple binding modes for a same substrate), multiple active poses (for each of the aforementioned 17 substrates) were incorporated to the molecular alignment for QSAR analyses (please also see Discussion section).
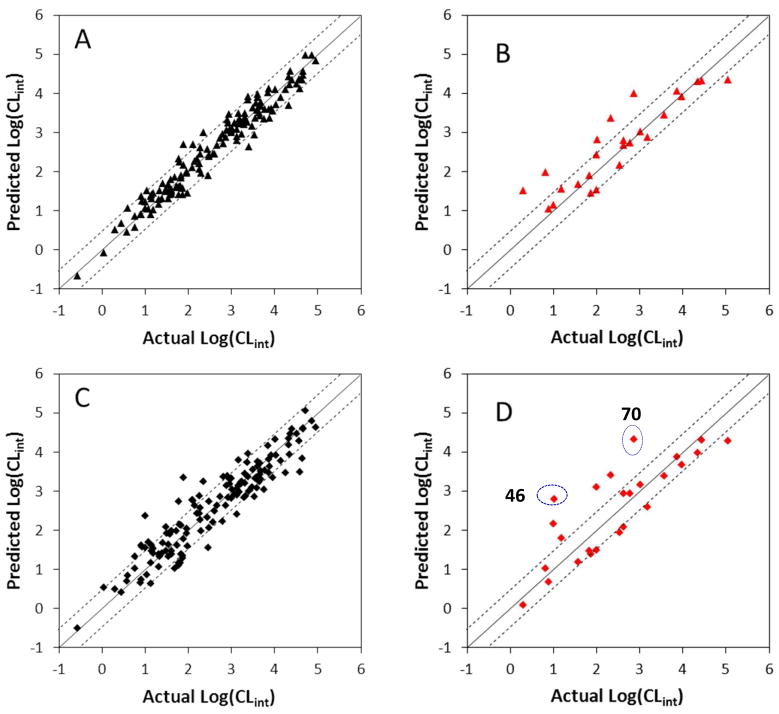
Predictive power of the analyses
A training dataset of UGT1A9 substrates allows the derivation of two separate QSAR models with statistical significance (Table 2). The predictive power of the two models was validated by predicting the catalytic efficiency of 25 additional substrates not included in the training set (Fig. 4). For almost all substrates, the predicted values fall close to the observed log(CLint) values, deviating by no more than 1 logarithmic unit (Table 1 & Fig. 4). However, in CoMSIA prediction, the activities of 46 (7-hydroxy-3′-methoxyflavone) and 70 (3,7,3′-trihydroxyflavone) are significantly over-estimated more than 1 logarithmic unit, even though CoMSIA analysis reveals significantly better correlation in terms of a higher q2.
Figure 4. Correlations between the experimental glucuronidation parameters and the predicted ones from the 3D-QSAR models.
Panel A: Fitted predictions versus actual catalytic efficiencies for the training set. The predicted values were obtained using the CoMFA method. Panel B: Predicted versus actual catalytic efficiencies for the test set not included in model derivation. The predicted values were obtained using the CoMFA method. Panel C: Fitted predictions versus actual catalytic efficiencies for the training set. The predicted values were obtained using the CoMSIA method. Panel D: Predicted versus actual catalytic efficiencies for the test set not included in model derivation. The predicted values were obtained using the CoMSIA method. Dashed lines represent the observed prediction bias of 3.0-fold deviation from unity. Solid lines represent the observed prediction bias of 10.0-fold deviation from unity.
CoMFA model
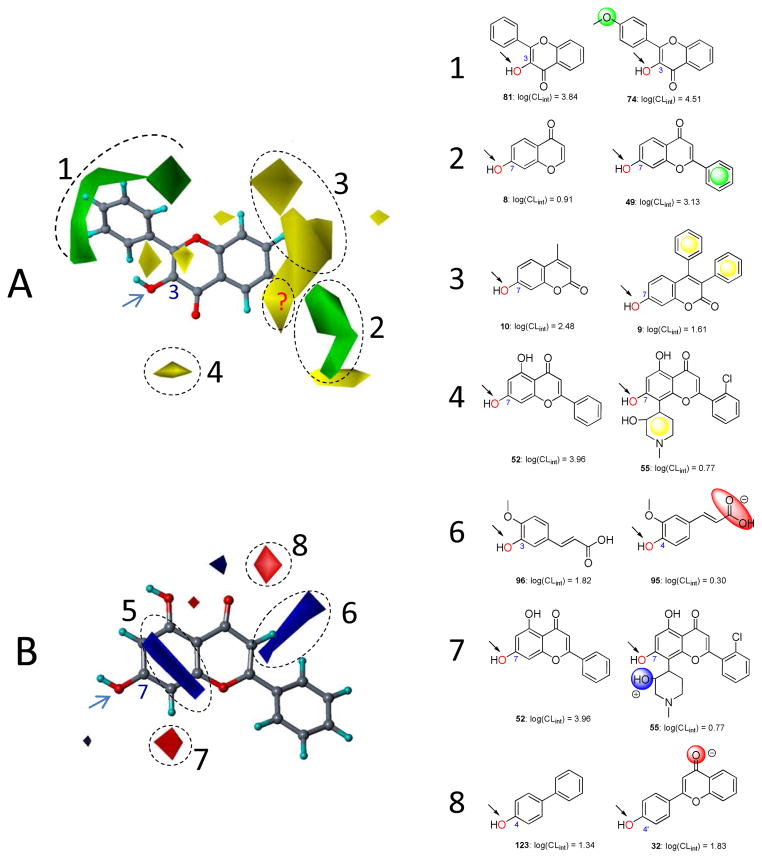
The usual way of understanding CoMFA is by graphing the associated fields. In Fig. 5A, the steric maps derived from CoMFA are displayed. Areas indicated by green contours (numbered 1 and 2) correspond to regions where steric occupancy with bulky groups will increase catalytic activity. The yellow contours (numbered 3 and 4) mean bulky groups should be avoided; otherwise reduced activity can be expected. 74 is more active than 81, which is possibly explained by the fact that the former orients its 4′-methoxy group into the favored region (contour 1) (Fig. 5). Similarly, compared to the less active 8, the more active 49 fills the favorable region (contour 2) by the 2-benzene moiety (Fig. 5). 9 is less active than 10, this is most likely because it orients the 3,4-diphenyl group into the disfavored region (contour 3) (Fig. 5). Likewise, due to the occupancy of the disfavored region (contour 4) by a piperidinyl group, 55 has a lower activity than 52 (Fig. 5).
Figure 5. Field contribution maps from the CoMFA analysis.
Panel A presents the steric maps. 3-Hydroxyflavone (81) is shown inside the field for reference. Green: Areas in which bulky groups are sterically favorable for glucuronidation; Yellow: Areas in which bulky groups are unfavorable for glucuronidation. Panel B presents the electrostatic maps. Chrysin (52) is shown inside the field for reference. Blue: Areas in which electropositive atoms/groups are favorable for glucuronidation; Red: Areas in which electronegative atoms/groups are favorable for glucuronidation. Examples are given to on the right side matching the CoMFA results to experimental data. Favored and disfavored contour levels for CoMFA were fixed at 85% and 15%, respectively.
The maps of electrostatic properties are shown in Fig. 5B. The areas contoured in blue (numbered 5 and 6) correspond to regions where electropositive groups will enhance the catalytic activity, as will electronegative groups placed into areas indicated in red (number 7 and 8). Contour 5 is close to the glucuronidation site and in parallel with the bicyclic ring of 52 chrysin (shown in Fig. 5B). The contribution of contour 5 to the activity-structure correlation is uncertain since we could not find a pair to demonstrate its significance. Although being isomers, 95 and 96 possess distinct catalytic activity. Due to a substitution difference of the acrylic acid group on the benzene skeleton, the less active 95 orients its carboxylate group into an area (contour 6) indicated to be unfavorable for negatively charged groups (Fig. 5). 55 occupies the contour 7 (unfavorable for electropositive groups) by the 3-hydroxyl group of the piperidinyl moiety; this is consistent with the fact that 55 is less active compared to 52 (Fig. 5). 32 orients a carbonyl group with an electronegative oxygen into a site highlighted to be favorable for negatively charged residues, thus it possesses a higher activity than 123 (Fig. 5).
CoMSIA model
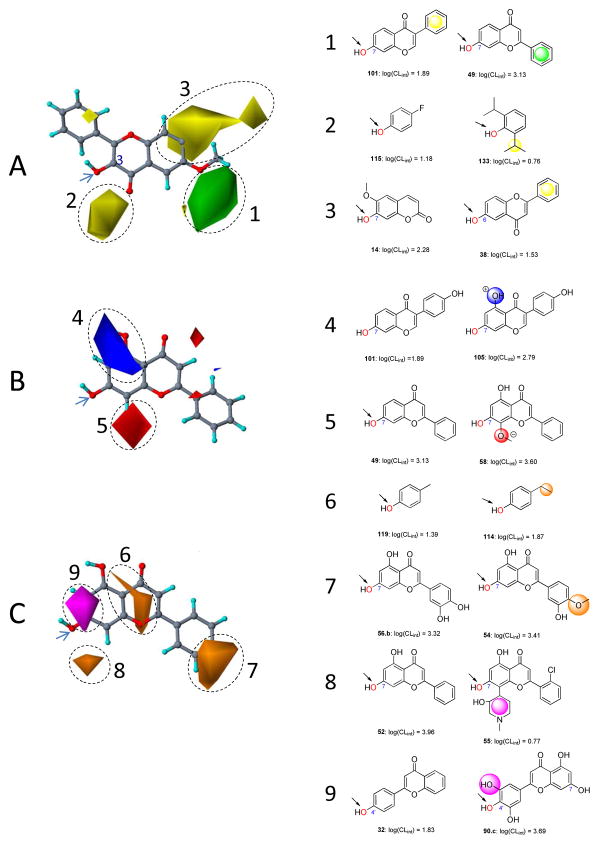
The CoMSIA method also provides field contribution contours that allow the correlation results to be mapped back onto the molecular structures. These contours are given in Fig. 6 together with some exemplary substrates. For consistency, coloring scheme of the contoured areas (to indicate a property contribution) for steric and electrostatic fields is the same as used in CoMFAmaps. In Fig. 6A, the steric property is displayed. Interestingly, the contours (numbered 1, 2, and 3) are largely consistent with those derived from CoMFA. The more active 49 orients its 2-benzene ring into the favorable region (contour 1), whereas the less active 101 orients its 3-benzene ring into the unfavorable region (contour 3) (Fig. 6). For a similar reason, 133 partially occupies the unfavorable region (contour 2) by a isopropyl group and shows a lower activity compared to 115. As an additional example pair (molecules 14 and 38) is given, 14 avoids the unfavorable steric groups and are more active than 38, whose 2-benzene ring is positioned into contour 3 highlighted to be unfavorable for bulky groups.
Figure 6. Field contribution maps from the CoMSIA analysis.
Panel A presents the steric maps. 3-Hydroxy-6-methxylflavone (66) is shown inside the field for reference. Green: Areas in which bulky groups are sterically favorable for glucuronidation; Yellow: Areas in which bulky groups are unfavorable for glucuronidation. Panel B presents the electrostatic maps. Chrysin (52) is shown inside the field for reference. Blue: Areas in which electropositive atoms/groups are favorable for glucuronidation; Red: Areas in which electronegative atoms/groups are favorable for glucuronidation. Panel C presents the hydrophobic maps. Orange: Areas where hydrophobic groups enhance glucuronidation; Magenta: Areas where hydrophilic groups decrease glucuronidation. Examples are given on the right side matching the CoMFA results to experimental data. Favored and disfavored contour levels for CoMSIA were fixed at 85% and 15%, respectively.
The maps of electrostatic properties (numbered 4 and 5) show fewer features in space (Fig. 6B), compared to those from CoMFA. 105 is more active than 101, largely because it has a hydroxyl group (with an electropositive hydrogen) overlaid with the favorable region (contour 4). By contrast, 58 is more active than 49 for the reason that an ether group (with an electronegative oxygen) occupied in contour 5 indicated to be favorable for electronegative groups.
The maps for hydrophobic properties are shown in Fig. 6C. Substrates orienting groups with increasing hydrophobicity into areas contoured in orange (numbered 6, 7, and 8) will enhance activity, as will groups with increasingly hydrophilicity placed in areas indicated in purple (numbered 9). 114 is more active than 119; this is probably because both ethyl and methyl groups substituted on the phenol backbone are adjacent to the hydrophobic favorable region (contour 6), but the former is more hydrophobic than the latter (Fig. 6). An increased hydrophobicity (from a hydroxyl group to a methoxy group) in contour 7 (hydrophobicity favorable) leads to an enhanced activity, as is seen from a comparison between 54 and 56.b (Fig. 6). 55 possesses an activity significantly lower than 32, which is in complete agreement with the indication that the presence of a hydrophilic piperidinyl group in the unfavorable region (contour 8) would result in a diminished activity compared to its absence (Fig. 6). Contour 9 highlights an area where occupancy of a hydrophilic (polar) group would enhance the activity, which exemplified by a comparison of 32 and 90.c. 90.c orients a 3′-hydroxyl group into the favorable region contour 9, thus is more active than 32.
Exploring UGT1A9 catalytic pocket using a homology model and CoMFA/CoMSIA maps
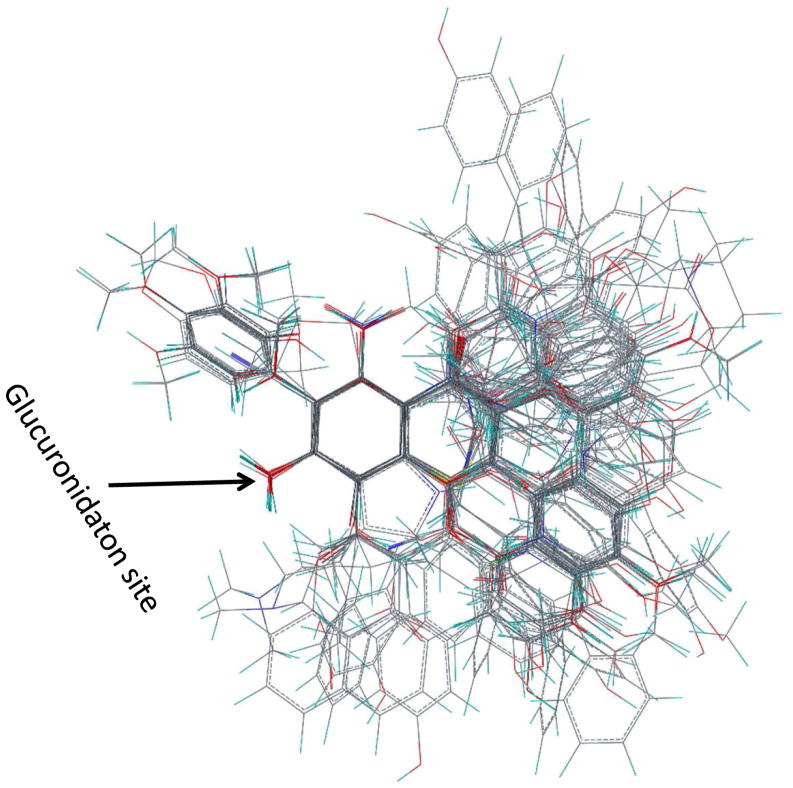
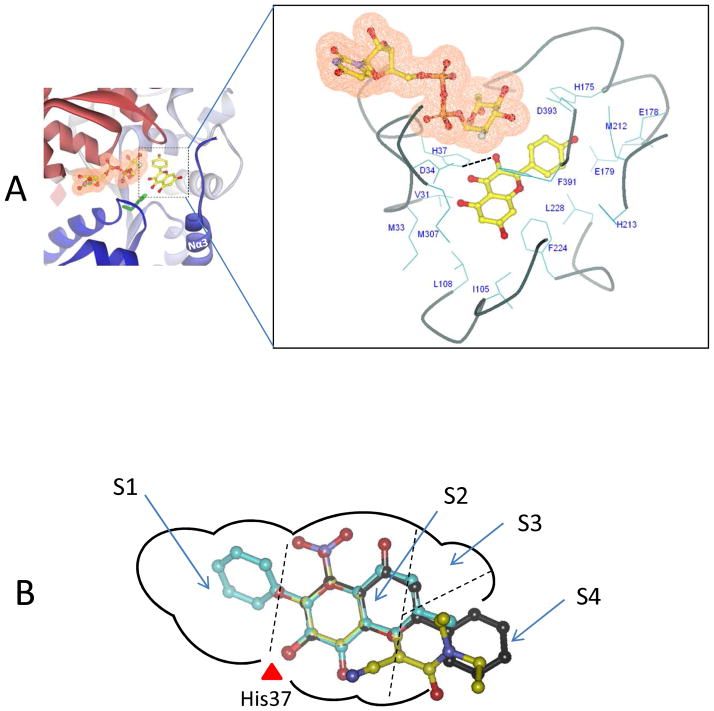
To explore the molecular mechanisms of UGT1A9-substrate interaction, a homology model of UGT1A9 was constructed. This structural model incorporates a simulated binding of 88.a kaempferol (in an active 3-O-glucuronidation mode) where 3-OH group of kaempferol is reasonably hydrogen-bonded with the catalytic residue His37. The protein residues forming the pocket wall were identified and presented in Fig. 7 (more detailed description of the binding pocket is provided in Supporting Information Fig. S3). The binding pocket is divided into four sub-pockets designated as S1, S2, S3 and S4 (Fig. 7B), among which pocket S3 is relatively small in size due to the steric hindrance by residues from helix Nα3 and its preceding loop. Interestingly, pocket S4 appears to be open to solvent, and potentially contributes to accommodation of a long-chain substrate such as 15 curcumin. The three most active substrates (66: 3-hydroxy-6-methylflavone, 140: entacapone; 52: chrysin) were mapped into the pocket by an alignment with kaempferol. The B-ring of 66 is fitted to S1, and the common aromatic ring to S2. S4 accommodates the B-ring of 52 or the N,N-dimethylamide group of 140.
Figure 7. Three-dimensional model of the UGT1A9-kaempferol (88.a) complex.
Panel A , a side view of the three-dimensional model of UGT1A9-kaempferol complex. Kaempferol is indicated in a ball-and-stick model. The cofactor is indicated in a ball-and-stick model with a molecular surface. An expanded view of residues potentially involved in the interactions with kaempferol (3-O-glucuronidation) in the model is presented. Dashed black line indicates the potential hydrogen bond. Panel B, A two-dimensional schematic representation of UGT1A9 catalytic pocket. The three most active substrates (66: 3-hydroxy-6-methylflavone, 140: entacapone; 52: chrysin) are mapped into the pocket. The binding pocket is divided into four sub-pockets designated as S1, S2, S3 and S4.
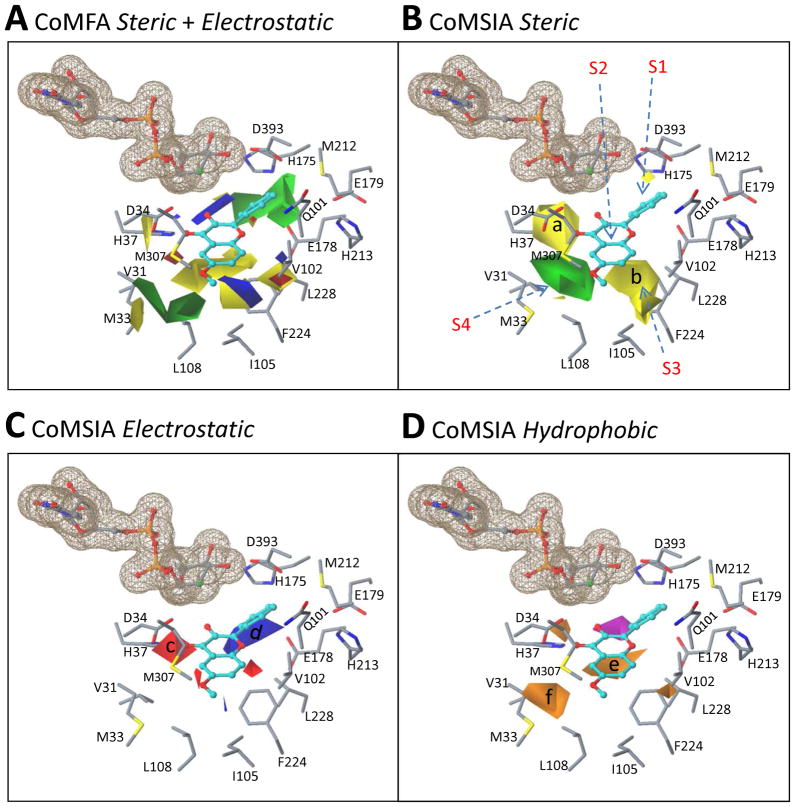
The CoMFA/CoMSIA maps were superimposed on the binding site of this structural model based on the binding mode of kaempferol. The contour maps are largely consistent with the 3D shape of the UGT1A9 catalytic site (Fig. 8). The green regions where bulky groups favor activity correspond to the regions of the active site where unfilled spaces exist. This indicates that a bulky group in these regions increases the van de Waals interaction between a substrate and UGT1A9, thus increasing the activity (Fig. 8A/B).Also, the yellow regions (e.g., a and b in Fig. 8B) where bulky groups disfavor activity correspond to the regions of the active site where steric hindrance is noted.
Figure 8. Superposition of the CoMFA/CoMSIA contour maps over the binding site of a homology-modeled UGT1A9 structure based on a simulated binding model of kaempferol (3-OH).
The UGT1A9 protein is shown in a stick model. Kaempferol is indicated in a ball-and-stick model and the cofactor is shown in a ball-and-stick model with a molecular surface. Panel A: Overlay of the CoMFA steric and electrostatic maps with the UGT1A9 binding site. Panel B–D: Overlay of the CoMSIA steric (B), electrostatic (C), and hydrophobic (D) fields with the UGT1A9 binding site.
Unexpectedly, electrostatic maps between CoMFA and CoMSIA are not consistent with each other; this might indicate an uncertainty in correlating the electrostatic property of substrates with their activity (Fig. 8A/C). In contrast to the fact that CoMFA electrostatic maps appear in the regions where no polar residues can be identified, CoMSIA electrostatic maps show a good compatibility with the surrounding residues. An electronegative group in its favorable region (contour c or previously named contour 5 in Figure 6) presumably interacts with UGT1A9 by forming a hydrogen bond or through electrostatic interactions. As only polar residuesAsp34 (with an electronegative side chain) and His37 (with an electropositive side chain) appear in the neighborhood of contour c, His37 is proposed to contribute to the interaction of UGT1A9 with an electronegative group (of a substrate) positioned in this region (Fig. 8). The residues that are around region d (Fig. 8C) favoring an electropositive group on a substrate are Glu178, Glu179 and Asp393 (all are electronegative). In addition, the hydrophobic regions (e and f) are lined with the hydrophobic residues Val31, Met33, Leu108, Phe224, Leu228, Met307, and Phe391 (Fig. 8D).
Discussion
In this study, we report two 3D-QSAR models for UGT1A9-mediated glucuronidation. These models are more generalized thus more useful in comparison to our earlier ones (27), which are somewhat limited to predicting UGT1A9-mediated 3-O-glucuronidation of flavonols, a subset of phenolics. Our model is challenged by the fact that many substrates form more than one glucuronide at different sites (so called “glucuronide isomers”). Recent studies indicate that the glucuronide isomers are resulted from distinct binding modes of the same substrate in the catalytic domain of UGT protein (27,28). Each binding mode orients a glucuronidation site (-OH) towards the catalytic residue (usually a histidine) for reaction, and to generate a corresponding glucuronide isomer. Accordingly, we treat such a UGT substrate as multiple “substrates” that adopt distinct spatial conformations; each of which corresponds to a binding mode to form a regiospecific glucuronide isomer. Our derived models with statistical significance and high predictive capability suggest that this treatment is reasonable and useful in terms of in silico modeling of UGT substrates. It is also highlighted that our model can be used to predict regiospecific glucuronidation mediated by UGT1A9, which was not accomplished before (27).
Another challenge to our model is that it incorporates a large diversity of (n=145) substrate structures. As a result, difficulty is raised with respect to the molecular alignment of those structures. We proposed a unique alignment method that is to superimpose the glucuronidation site and its adjacent aromatic ring, the two important features identified by a flexible alignment of three most active substrates (Fig. 2). Glucuronidation sites should be aligned to improve model quality as suggested from previous modeling experiences (8,15).The importance of positioning of a glucuronidation site into an area close to both cofactor and the catalytic histidine residue for glucuronidation is also demonstrated in a recent study (40). Early studies identify a common feature pharmacophore for several UGT isoforms, which unanimously includes two separate hydrophobic regions (adjacent to the glucuronidation site) (8,15,41). However, we believe two hydrophobic regions spatially isolated might not be universal features for all UGT1A9 substrates. For example, such features cannot be found in 109 (a simple phenol) structure. Token together, the alignment rule here conforms to the catalytic mechanism and is applicable to a simple phenol. More importantly the rule is demonstrated to be satisfactory when measured by the quality of resulting models.
The use of log (CLint), a measure of catalytic efficiency, as the parameter for correlation analyses is more reliable and meaningful, compared to other kinetic parameters such as Km and Vmax. As stated by Sharma and Duffel (42,43), in the case of enzymes that may exhibit nonproductive binding interactions with some substrates, a highly relevant kinetic parameter for a CoMFA correlation of the structure to its ability to serve as a substrate for such enzymes is the CLint value, or log (CLint). This is because CLint is independent of nonproductive binding contributions. Although there is no direct evidence (e.g., crystal structure) of nonproductive binding of a substrate to a UGT protein, kinetic characterization has indicated that nonproductive binding of substrates to a UGT isoform can be a major reason why the enzymes frequently exhibit substrate inhibition kinetics (44). Therefore, the use of CLint value for our CoMFA/CoMSIA analyses of UGT1A9 is justified. Moreover, in vitro intrinsic clearance (CLint) is frequently used to predict in vivo clearance such as hepatic clearance with a reasonable success rate (45,46). Hence, this parameter is a more appropriate indicator as the susceptibility of a substrate to glucuronidation in vivo.
The recognition of glucuronidation as an important metabolic pathway has lent increasing efforts towards better understanding of the molecular mechanisms of UGT functions and of the substrate structural features associated with UGT selectivity. Inevitably, a molecular-level structural elucidation of the protein is necessary for such pursuits. Here, a homology model of UGT1A9 was constructed aiming to enhance our understanding of interactions between UGT1A9 and substrates, in addition to the CoMFA/CoMSIA results. The structural information of our UGT1A9 model was imported from the template protein VvGT1 (a plant UGT). At present, the use of a plant UGT for homology modeling of human UGTs is preferred and justifiable, because (1) plant and human UGTs are classified into the same superfamily GT1; GT1 members adopt a GT-B fold and their tertiary structures are predicted to be highly conserved; (2) plant and human UGTs share a similar catalytic mechanism (i.e., serine hydrolase-like mechanism); (3) a determined partial crystal structure (for C-terminal or UDPGA binding domain portion only) of human UGT2B7 agrees well with its counterparts in plant UGT crystal structures (13,14). Such modeling effort appears to be useful here and has also been utilized to elucidate the amino acids that are responsible for the large activity differences between UGT1A9 and 1A10(25).
A good consistency between the CoMFA/CoMSIA maps and a homology model of UGT1A9 is highlighted in terms of the steric and hydrophobic interactions (Fig. 8). The results provide a highly possible 3D structure of UGT1A9 binding pocket as well as substantial insights into the molecular mechanisms regarding the recognition of a substrate by UGT1A9. The models can be used to guide de novo design of compounds with desired UGT1A9 activity. For example, a more active compound should have its -OH group towards the catalytic residue, and the rest of its structure occupies those green regions (cavities in the binding site), and avoids those yellow regions where steric hindrance exists. We anticipate that this approach of CoMFA/CoMSIA coupled with a protein homology model may be applicable to other UGT isoforms. A more exhaustive elucidation of molecular interactions of other UGT isoforms and a more complete comparison of substrate selectivity across UGT isoforms might be necessary in order to ultimately predict overall glucuronidation and uncover the fine substrate selectivity difference. This is important as the knowledge can be used to accelerate drug development and to promote human health.
Although model construction is based on the active poses of UGT1A9 substrates, it is of interest to see if the model can be used to distinguish a non-substrate from a substrate. We experimentally identified three UGT1A9 non-substrates, namely, estradiol (a selective probe for UGT1A1), salicylic acid, and aminosalicylic acid (no metabolite can be detected when incubating these compounds with UGT1A9). The predicted log (CLint) values from the CoMFA model are 1.41, 1.11, and 1.42, respectively, indicating these compounds are very poor substrates of UGT1A9. Therefore, the model is fairly accurate; though its capability to predict an absolute non-substrate is somewhat limited.
In conclusion, we have performed 3D-QSAR analyses using the powerful techniques Comparative Molecular Field Analysis (CoMFA) and Comparative Molecular Similarity Indices Analysis (CoMSIA) based on a large training dataset with a 106-fold range of relative catalytic activity. The derived models show statistical significance and substantive predictability (CoMFA: q2 = 0.548, r2= 0.949, r2pred = 0.775; CoMSIA: q2 = 0.579, r2= 0.876, r2pred = 0.700). The real-world use of these models is fully expected to predict the catalytic activity of structural diverse chemicals (including drug candidates) towards UGT1A9. Moreover, the field contribution maps from CoMFA/CoMSIA were applied to elucidate the catalytic pocket of UGT1A9 with the aid of a homology model of UGT1A9. The results consistently depict a plausible catalytic pocket with a set of geometry configuration and a hydrophobic interacting environment, even though the electrostatic interactions are less defined. Our findings for the first time provide a possible molecular basis for understanding UGT1A9 functions and its substrate selectivity.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (GM070737) to MH.
Abbreviations used
- UGTs
UDP-glucuronosyltransferases
- UDPGA
uridine diphosphoglucuronic acid
- CoMFA
Comparative Molecular Field Analysis
- CoMSIA
Comparative Molecular Similarity Indices Analysis
- CYPs
Cytochrome P450
- ADME
absorption, distribution, metabolism, and elimination
- UPLC
ultra performance liquid chromatography
- MS
mass spectroscopy
- Km
Michaelis-Menten constant
- Vmax
maximal velocity
- CLint
intrinsic clearance
- 2D/3D
2-dimensional/3-dimensional
- QSAR
Quantitative Structure-Activity Relationship
- PLS
Partial Least Squares
References
- 1.Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov. 2004;3(8):711–5. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- 2.Wishart DS. Improving early drug discovery through ADME modelling: an overview. Drugs R D. 2007;8(6):349–62. doi: 10.2165/00126839-200708060-00003. [DOI] [PubMed] [Google Scholar]
- 3.Emoto C, Murayama N, Rostami-Hodjegan A, Yamazaki H. Methodologies for investigating drug metabolism at the early drug discovery stage: prediction of hepatic drug clearance and P450 contribution. Curr Drug Metab. 2010;11(8):678–85. doi: 10.2174/138920010794233503. [DOI] [PubMed] [Google Scholar]
- 4.Wang JF, Chou KC. Molecular modeling of cytochrome P450 and drug metabolism. Curr Drug Metab. 2010;11(4):342–6. doi: 10.2174/138920010791514180. [DOI] [PubMed] [Google Scholar]
- 5.Mendieta-Wejebe JE, Correa-Basurto J, García-Segovia EM, Ceballos-Cancino G, Rosales-Hernández MC. Molecular Modeling Used to Evaluate CYP2C9-Dependent Metabolism: Homology Modeling, Molecular Dynamics and Docking Simulations. Curr Drug Metab. 2011 doi: 10.2174/138920011795713670. (in press) [DOI] [PubMed] [Google Scholar]
- 6.Cruciani G, Carosati E, De Boeck B, Ethirajulu K, Mackie C, Howe T, Vianello R. MetaSite: understanding metabolism in human cytochromes from the perspective of the chemist. J Med Chem. 2005;48(22):6970–9. doi: 10.1021/jm050529c. [DOI] [PubMed] [Google Scholar]
- 7.Jamei M, Marciniak S, Feng K, Barnett A, Tucker G, Rostami-Hodjegan A. The Simcyp population-based ADME simulator. Expert Opin Drug Metab Toxicol. 2009;5(2):211–23. doi: 10.1517/17425250802691074. [DOI] [PubMed] [Google Scholar]
- 8.Smith PA, Sorich MJ, McKinnon RA, Miners JO. Pharmacophore and quantitative structure-activity relationship modeling: complementary approaches for the rationalization and prediction of UDP-glucuronosyltransferase 1A4 substrate selectivity. J Med Chem. 2003;46(9):1617–26. doi: 10.1021/jm020397c. [DOI] [PubMed] [Google Scholar]
- 9.Mackenzie PI, Bock KW, Burchell B, Guillemette C, Ikushiro S, Iyanagi T, Miners JO, Owens IS, Nebert DW. Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenetics Genomics. 2005;15:677–685. doi: 10.1097/01.fpc.0000173483.13689.56. [DOI] [PubMed] [Google Scholar]
- 10.Radominska-Pandya A, Czernik PJ, Little JM, Battaglia E, Mackenzie PI. Structural and functional studies of UDP-glucuronosyltransferases. Drug Metab Rev. 1999;31(4):817–99. doi: 10.1081/dmr-100101944. [DOI] [PubMed] [Google Scholar]
- 11.Court MH. Isoform-selective probe substrates for in vitro studies of human UDP-glucuronosyltransferases. Methods Enzymol. 2005;400:104–16. doi: 10.1016/S0076-6879(05)00007-8. [DOI] [PubMed] [Google Scholar]
- 12.Miners JO, Mackenzie PI, Knights KM. The prediction of drug-glucuronidation parameters in humans: UDP-glucuronosyltransferase enzyme-selective substrate and inhibitor probes for reaction phenotyping and in vitro-in vivo extrapolation of drug clearance and drug-drug interaction potential. Drug Metab Rev. 2010;42(1):196–208. doi: 10.3109/03602530903210716. [DOI] [PubMed] [Google Scholar]
- 13.Miley MJ, Zielinska AK, Keenan JE, Bratton SM, Radominska-Pandya A, Redinbo MR. Crystal structure of the cofactor-binding domain of the human phase II drug-metabolism enzyme UDP-glucuronosyltransferase 2B7. J Mol Biol. 2007;369(2):498–511. doi: 10.1016/j.jmb.2007.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radominska-Pandya A, Bratton SM, Redinbo MR, Miley MJ. The crystal structure of human UDP-glucuronosyltransferase 2B7 C-terminal end is the first mammalian UGT target to be revealed: the significance for human UGTs from both the 1A and 2B families. Drug Metab Rev. 2010;42(1):133–44. doi: 10.3109/03602530903209049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sorich MJ, Smith PA, McKinnon RA, Miners JO. Pharmacophore and quantitative structure activity relationship modelling of UDP-glucuronosyltransferase 1A1 (UGT1A1) substrates. Pharmacogenetics. 2002;12(8):635–45. doi: 10.1097/00008571-200211000-00008. [DOI] [PubMed] [Google Scholar]
- 16.Sorich MJ, Miners JO, McKinnon RA, Smith PA. Multiple pharmacophores for the investigation of human UDP-glucuronosyltransferase isoform substrate selectivity. Mol Pharmacol. 2004;65(2):301–8. doi: 10.1124/mol.65.2.301. [DOI] [PubMed] [Google Scholar]
- 17.Wu B, Kulkarni K, Basu S, Zhang S, Hu M. First-pass metabolism via UDP-glucuronosyltransferase: a barrier to oral bioavailability of phenolics. J Pharm Sci. 2011;100(9):3655–81. doi: 10.1002/jps.22568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang JL, Beland FA, Doerge DR, Wiener D, Guillemette C, Marques MM, Lazarus P. Characterization of benzo(a)pyrene-trans-7,8-dihydrodiol glucuronidation by human tissue microsomes and overexpressed UDP-glucuronosyltransferase enzymes. Cancer Res. 2002;62(7):1978–86. [PubMed] [Google Scholar]
- 19.Olson KC, Dellinger RW, Zhong Q, Sun D, Amin S, Spratt TE, Lazarus P. Functional characterization of low-prevalence missense polymorphisms in the UDP-glucuronosyltransferase 1A9 gene. Drug Metab Dispos. 2009;37(10):1999–2007. doi: 10.1124/dmd.108.024596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lazarus P, Sun D. Potential role of UGT pharmacogenetics in cancer treatment and prevention: focus on tamoxifen and aromatase inhibitors. Drug Metab Rev. 2010;42(1):182–94. doi: 10.3109/03602530903208652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villeneuve L, Girard H, Fortier LC, Gagné JF, Guillemette C. Novel functional polymorphisms in the UGT1A7 and UGT1A9 glucuronidating enzymes in Caucasian and African-American subjects and their impact on the metabolism of 7-ethyl-10-hydroxycamptothecin and flavopiridol anticancer drugs. J Pharmacol Exp Ther. 2003;307(1):117–28. doi: 10.1124/jpet.103.054072. [DOI] [PubMed] [Google Scholar]
- 22.Kurkela M, García-Horsman JA, Luukkanen L, Mörsky S, Taskinen J, Baumann M, Kostiainen R, Hirvonen J, Finel M. Expression and characterization of recombinant human UDP-glucuronosyltransferases (UGTs). UGT1A9 is more resistant to detergent inhibition than other UGTs and was purified as an active dimeric enzyme. J Biol Chem. 2003;278(6):3536–44. doi: 10.1074/jbc.M206136200. [DOI] [PubMed] [Google Scholar]
- 23.Patana AS, Kurkela M, Finel M, Goldman A. Mutation analysis in UGT1A9 suggests a relationship between substrate and catalytic residues in UDP-glucuronosyltransferases. Protein Eng Des Sel. 2008;21(9):537–43. doi: 10.1093/protein/gzn030. [DOI] [PubMed] [Google Scholar]
- 24.Fujiwara R, Nakajima M, Yamamoto T, Nagao H, Yokoi T. In silico and in vitro approaches to elucidate the thermal stability of human UDP-glucuronosyltransferase (UGT) 1A9. Drug Metab Pharmacokinet. 2009;24(3):235–44. doi: 10.2133/dmpk.24.235. [DOI] [PubMed] [Google Scholar]
- 25.Itaaho K, Laakkonen L, Finel M. How many and which amino acids are responsible for the large activity differences between the highly homologous UDP-glucuronosyltransferases (UGT) 1A9 and UGT1A10? Drug Metab Dispos. 2010;38(4):687–96. doi: 10.1124/dmd.109.031229. [DOI] [PubMed] [Google Scholar]
- 26.Miners JO, Bowalgaha K, Elliot DJ, Baranczewski P, Knights KM. Characterization of niflumic acid as a selective inhibitor of human liver microsomal UDP-glucuronosyltransferase 1A9: application to the reaction phenotyping of acetaminophen glucuronidation. Drug Metab Dispos. 2011;39(4):644–52. doi: 10.1124/dmd.110.037036. [DOI] [PubMed] [Google Scholar]
- 27.Wu B, Morrow JK, Singh R, Zhang S, Hu M. Three-dimensional quantitative structure-activity relationship studies on UGT1A9-mediated 3-O-glucuronidation of natural flavonols using a pharmacophore-based comparative molecular field analysis model. J Pharmacol Exp Ther. 2011;336(2):403–13. doi: 10.1124/jpet.110.175356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu B, Xu B, Hu M. Regioselective Glucuronidation of Flavonols by Six Human UGT1A Isoforms. Pharm Res. 2011;28(8):1905–18. doi: 10.1007/s11095-011-0418-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang L, Singh R, Liu Z, Hu M. Structure and Concentration Changes Affect Characterization of UGT Isoform-Specific Metabolism of Isoflavones. Mol. Pharm. 2009;6(5):1466–82. doi: 10.1021/mp8002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh R, Wu B, Tang L, Liu Z, Hu M. Identification of the position of mono-O-glucuronide of flavones and flavonols by analyzing shift in online UV spectrum (lambdamax) generated from an online diode array detector. J Agric Food Chem. 2010;58(17):9384–95. doi: 10.1021/jf904561e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Böhm M, St rzebecher J, Klebe G. Three-dimensional quantitative structure-activity relationship analyses using comparative molecular field analysis and comparative molecular similarity indices analysis to elucidate selectivity differences of inhibitors binding to trypsin, thrombin, and factor Xa. J Med Chem. 1999;42(3):458–77. doi: 10.1021/jm981062r. [DOI] [PubMed] [Google Scholar]
- 32.Cramer RD, Patterson DE, Bunce JD. Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J Am Chem Soc. 1988;110:5959–358. 5967. doi: 10.1021/ja00226a005. [DOI] [PubMed] [Google Scholar]
- 33.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 34.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 35.Offen W, Martinez-Fleites C, Yang M, Kiat-Lim E, Davis BG, Tarling CA, Ford CM, Bowles DJ, Davies GJ. Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 2006;25(6):1396–405. doi: 10.1038/sj.emboj.7600970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.http://www.cazy.org/
- 37.Laakkonen L, Finel M. A molecular model of the human UGT1A1, its membrane orientation and the interactions between different parts of the enzyme. Mol Pharmacol. 2010;77(6):931–939. doi: 10.1124/mol.109.063289. [DOI] [PubMed] [Google Scholar]
- 38.Shao H, He X, Achnine L, Blount JW, Dixon RA, Wang X. Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant Cell. 2005;17:3141–3154. doi: 10.1105/tpc.105.035055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li L, Modolo LV, Escamilla-Trevino LL, Achnine L, Dixon RA, Wang X. Crystal Structure of Medicago truncatula UGT85H2 - Insights into the Structural Basis of a Multifunctional (Iso)flavonoid Glycosyltransferase. J Mol Biol. 2007;370:951–963. doi: 10.1016/j.jmb.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 40.Takaoka Y, Ohta M, Takeuchi A, Miura K, Matsuo M, Sakaeda T, Sugano A, Nishio H. Ligand orientation governs conjugation capacity of UDP-glucuronosyltransferase 1A1. J Biochem. 2010;148(1):25–8. doi: 10.1093/jb/mvq048. [DOI] [PubMed] [Google Scholar]
- 41.Smith PA, Sorich MJ, Low LS, McKinnon RA, Miners JO. Towards integrated ADME prediction: past, present and future directions for modelling metabolism by UDP-glucuronosyltransferases. J Mol Graph Model. 2004;22(6):507–17. doi: 10.1016/j.jmgm.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 42.Sharma V, Duffel MW. Comparative molecular field analysis of substrates for an aryl sulfotransferase based on catalytic mechanism and protein homology modeling. J Med Chem. 2002;45(25):5514–22. doi: 10.1021/jm010481c. [DOI] [PubMed] [Google Scholar]
- 43.Sharma V, Duffel MW. A comparative molecular field analysis-based approach to prediction of sulfotransferase catalytic specificity. Methods Enzymol. 2005;400:249–63. doi: 10.1016/S0076-6879(05)00014-5. [DOI] [PubMed] [Google Scholar]
- 44.Luukkanen L, Taskinen J, Kurkela M, Kostiainen R, Hirvonen J, Finel M. Kinetic characterization of the 1A subfamily of recombinant human UDP-glucuronosyltransferases. Drug Metab Dispos. 2005;33(7):1017–26. doi: 10.1124/dmd.105.004093. [DOI] [PubMed] [Google Scholar]
- 45.Kilford PJ, Stringer R, Sohal B, Houston JB, Galetin A. Prediction of drug clearance by glucuronidation from in vitro data: use of combined cytochrome P450 and UDP-glucuronosyltransferase cofactors in alamethicin-activated human liver microsomes. Drug Metab Dispos. 2009;37(1):82–9. doi: 10.1124/dmd.108.023853. [DOI] [PubMed] [Google Scholar]
- 46.Cubitt HE, Houston JB, Galetin A. Relative importance of intestinal and hepatic glucuronidation-impact on the prediction of drug clearance. Pharm Res. 2009;26(5):1073–83. doi: 10.1007/s11095-008-9823-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.