Abstract
HIV-1 envelope glycoprotein is the primary target for HIV-1–specific antibodies. The native HIV-1 envelope spike on the virion surface is a trimer, but trimeric gp140 and monomeric gp120 currently are believed to induce comparable immune responses. Indeed, most studies on the immunogenicity of HIV-1 envelope oligomers have revealed only marginal improvement over monomers. We report here that suitably prepared envelope trimers have nearly all the antigenic properties expected for native viral spikes. These stable, rigorously homogenous trimers have antigenic properties markedly different from those of monomeric gp120s derived from the same sequences, and they induce potent neutralizing antibody responses for a cross-clade set of tier 1 and tier 2 viruses with titers substantially higher than those elicited by the corresponding gp120 monomers. These results, which demonstrate that there are relevant immunologic differences between monomers and high-quality envelope trimers, have important implications for HIV-1 vaccine development.
HIV-1 envelope glycoprotein mediates the fusion of viral and target cell membranes, the first critical step leading to infection (1). The precursor of the envelope protein, gp160, forms a trimer and is then cleaved by a furin-like protease into two noncovalently associated fragments: gp120 for receptor binding and gp41 for membrane fusion. Three copies of each fragment make up the mature envelope spike (gp120/gp41)3. This trimeric complex undergoes large, irreversible structural rearrangements after binding to the primary receptor CD4 and coreceptor (e.g., CCR5 and CXCR4) and drives the membrane fusion process. Monomeric gp120 can dissociate from the complex either spontaneously or upon CD4 binding in certain viral strains (2).
The envelope glycoprotein also is the primary target of humoral responses in HIV-1–infected patients. Strong evidence for the potential of antibody protection comes from passive transfer and mucosal simian-human immunodeficiency virus challenge studies in macaques and from a vectored immunoprophylaxis study in humanized mice (3–6). Although most antibodies induced during infection are nonneutralizing or are strain specific, recent studies indicate that 10–25% of patients produce broadly neutralizing antibodies (bNAbs) during the course of HIV-1 infection (7), raising the hope that immunogens capable of inducing such responses may lead to an effective vaccine. A number of broadly reactive neutralizing antibodies (NAbs) have been isolated that recognize conserved regions of the envelope glycoprotein. mAbs VRC01-03, CH31, 3BNC60, HJ16, and b12 target the CD4 binding site (CD4 BS) on gp120 with strong, broadly neutralizing activity (reviewed in ref. 8); PG9 and PG16 appear to recognize a quaternary epitope, which is trimer specific and glycan dependent, in the relatively conserved regions of the variable V2 and V3 loops of gp120 (9); 2G12 is another bNAb that recognizes an epitope on the outer surface of gp120 in a glycan- and conformational-dependent manner (10). Very recently, another group of bNAbs, designated “PGT antibodies,” has been identified; these antibodies react with glycan-dependent epitopes near the base of the V3 loop (11). Additional bNAbs, including 2F5 and 4E10, interact with a region on gp41 adjacent to the viral membrane called the “membrane-proximal external region” (MPER) (12, 13). Among these bBAbs, those against gp120 are believed to target directly the native, functional envelope trimer on the surface of virion, whereas the gp41-directed antibodies have been shown to block viral infection by attacking the prehairpin intermediate conformation of gp41 (14, 15).
Anti-gp41 NAbs are rare both in natural infection and after immunization with envelope-based vaccine candidates, and gp120, in principle, contains most of the neutralizing epitopes. Monomeric gp120 is relatively easy to manufacture and has been used as a subunit vaccine in three large efficacy trials. In the two early trials, gp120 vaccines failed to show any protection against HIV-1 infection or delay in disease progression (16, 17). The recent RV144 trial using a regimen involving priming with an ALVAC vector and boosting with a gp120 protein afforded 31.2% efficacy (18). A key question thus concerns the optimal form of the envelope glycoprotein for inducing HIV-1–specific NAbs. Monomeric gp120 is safe and easy to manufacture, but there are several reservations concerning its use as an immunogen. First, gp120 vaccines alone provided little or no protection in human efficacy trials (16–18). Second, antibodies elicited by monomeric gp120 react mainly with epitopes that are poor neutralization targets and presumably are occluded on primary HIV-1 isolates (19). Third, the strongly immunogenic and ineffective epitopes on monomeric gp120 could distract the immune system from targeting the more relevant, broadly neutralizing epitopes.
Is the envelope trimer a better immunogen than the gp120 monomer? Cleaved and functional (gp120/gp41)3 complexes are unstable and are difficult to produce as recombinant products. Thus, gp140, the ectodomain of trimeric gp160, has been used to mimic the native state of the envelope spikes (20–23). Because convincing evidence has been lacking to show that envelope trimers or oligomers induce stronger NAb responses than monomeric gp120, there is a general belief that both forms of the envelope glycoprotein have comparable immunogenicity. The envelope trimers or oligomers used in previous immunogenicity studies often had “gp120-like” characteristics, however, such as binding to CD4-induced (CD4i) antibodies in the absence of CD4 and exhibiting high affinity for nonneutralizing CD4 BS antibodies (24–28). Moreover, they often either aggregate or dissociate into dimers and monomers during expression or purification and probably fail to imitate the native envelope spikes.
We demonstrate here the feasibility of producing high-quality HIV-1 envelope trimers in human cells with a yield suitable for large-scale immunogenicity studies. We compare antigenic properties of two biochemically stable and homogenous gp140 trimers with the corresponding gp120 monomers derived from the same precursor sequences. We find that the gp140 trimers react strongly only with anti-CD4 BS bNAbs but not with nonneutralizing CD4 BS antibodies. We further show that the gp120 portions of the gp140 trimer maintain a conformation distinct from the CD4-bound state and do not undergo conformational changes upon binding to antibody VRC01. The trimers also show detectable binding to the bNAbs PG9 and PG16, which fail to react with the gp120 monomers. These stable trimers have different stoichiometries when binding soluble CD4 (1:1) and VRC01 Fab (1:3). Moreover, the gp140 trimers induce potent, cross-clade NAb responses with titers substantially higher than those elicited by the corresponding gp120 monomers for a diverse set of both tier 1 and tier 2 viruses. These results support the concept that HIV-1 envelope trimers more accurately mimic the antigenic properties of the native envelope spike on the surface of virions than do gp120 monomers and thus are better immunogens.
Results
Production of Stable gp140 Trimers and gp120 Monomers.
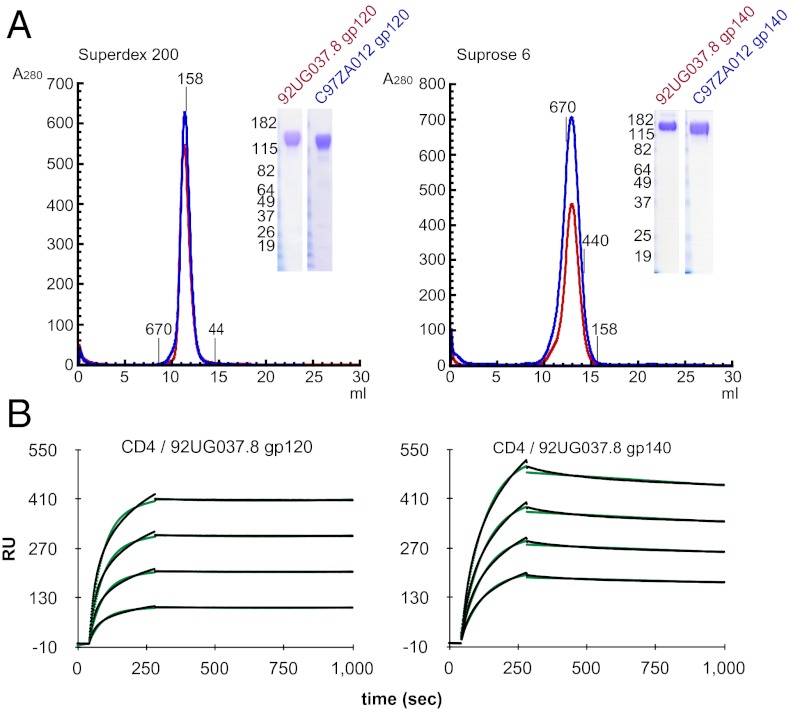
We previously identified two sequences (92UG037.8 from clade A and CZA97012 from clade C) for which the gp140 trimers are stable and homogeneous when expressed in insect cells (6, 15). There has been increasing recognition that N-linked glycans are important components of broadly neutralizing epitopes (9, 29, 30). To produce these trimers with authentic human glycosylation, we have generated stable 293T cell lines transfected with the expression constructs of 92UG037.8 and C97ZA012 gp140 trimers. As shown in Fig. 1A, stable and homogenous gp140 proteins can be purified from the supernatants of these cell lines, and the yield for each of them is >5 mg/L. To compare antigenic and immunogenic properties of the monomeric gp120 and trimeric gp140 made from the same precursor sequences, we also generated stable 293T cell lines that express gp120 from the same isolates. The yield for the two monodisperse gp120 proteins is ∼8 mg/L (Fig. 1A). All purified proteins elute as a single sharp peak from a sizing column and show a high level of purity, as judged by SDS/PAGE stained by Coomassie (Fig. 1A, Insets). Analytical ultracentrifugation and multiangle light-scattering analyses confirm that gp140 is trimeric and gp120 is monomeric (Table S1). The gp140 trimers do not dissociate into dimers or monomers during or after purification, and they remain stable at 4 °C for at least 4 wk. As expected, all the gp140 trimers and gp120 monomers bind tightly to soluble CD4, as detected by surface plasmon resonance (SPR) binding experiments (Fig. 1B, Fig. S1, and Table S2).
Fig. 1.
Production of stable and homogenous HIV-1 gp120 and gp140 proteins in 293T cells. (A) His-tagged gp140 proteins derived from the HIV-1 92UG037.8 (clade A) and CZA97012 (clade C) isolates and their corresponding full-length gp120s were purified from supernatants of 293T cells stably transfected with the expression constructs of these proteins. The purified gp120 (Left) and gp140 (Right) proteins were resolved by gel-filtration chromatography on a Superdex 200 and a Superose 6 column, respectively. The molecular weight standards include thyoglobulin (670 kDa), ferritin (440 kDa), γ-globulin (158 kDa), and ovalbumin (44 kDa). Peak fractions were pooled and analyzed by Coomassie-stained SDS/PAGE (Insets). (B) Soluble 4D CD4 was immobilized on a CM-5 chip, and various concentrations (50–1,000 nM) of 92UG037.8 gp120 or 92UG037.8 gp140 were passed over the chip surface. Binding kinetics was evaluated using a 1:1 Langmuir binding model; binding constants are summarized in Table S2. The sensorgrams are shown in black and the fits in green. All injections were carried out in duplicate and gave essentially identical results. Only one of the duplicates is shown.
Antigenic Properties of Trimeric gp140 and Monomeric gp120.
CD4 BS epitopes.
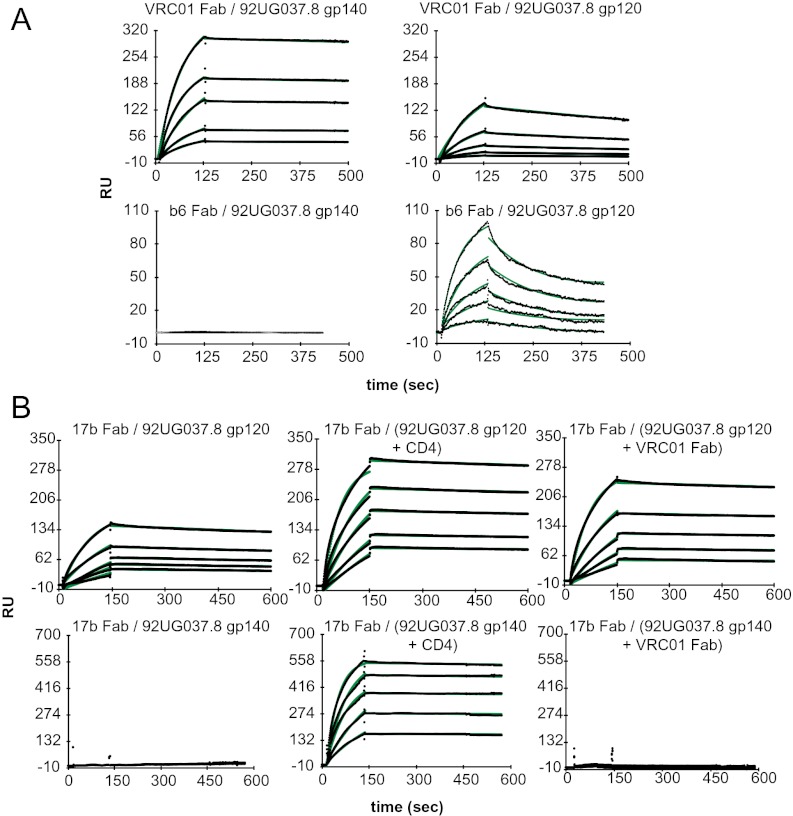
The CD4 BS is one of the most vulnerable regions of the envelope protein. It is targeted by a number of bNAbs. On the native, functional envelope trimer, the site probably is accessible only to CD4 and bNAbs and inaccessible to nonneutralizing mAbs, but on monomeric gp120 it is well exposed and open to all CD4 BS ligands (31). We used SPR to assess reactivity of VRC01, one of the most potent bNAbs targeting the CD4 BS, to both gp140 and gp120. To avoid potential artifacts associated with chemical immobilization, we used the human antibody binder, a mixture of monoclonal antibodies recognizing the Fab region of human antibodies, to capture and orient the VRC01 Fab fragments. As shown in Fig. 2A and Fig. S2, all the gp140 trimers and gp120 monomers bind tightly to the VRC01 Fab, with affinities ranging from 2.1 to 96 nM. The slower off-rate, and hence higher affinity, for the VRC01–gp140 interactions, compared with those of VRC01–gp120s interactions, may result from avidity effects when the monomeric Fab is immobilized on the sensor chip. Both the gp140 and gp120 proteins derived from the clade A isolate also react strongly with another CD4 BS bNAb, CH31, but the proteins from the clade C isolate bind CH31 less tightly, consistent with the lower sensitivity of this isolate to CH31 neutralization (Fig. S3 and Table S3). In contrast, a nonneutralizing CD4 BS antibody, b6, reacts with the 92UG037.8 gp120 with an affinity of 50 nM but does not bind at all to the 92UG037.8 gp140 trimer (Fig. 2A), indicating that the b6 epitope is effectively occluded by the trimer configuration. Thus, the conformation of the CD4 BS in our stable gp140 trimers is consistent with the neutralization profile of a native, functional envelope trimer.
Fig. 2.
Interactions of gp140 and gp120 with CD4 BS antibodies and CD4i antibody. (A) Fab of the CD4 BS antibody VRC01 or b6 was captured on a surface coated with human Fab binder, and varous concentratons(50–1,000 nM) of 92UG037.8 gp120 or 92UG037.8 gp140 were passed over the chip surface. (B) Fab of the CD4i antibody 17b was immobilized on a chip surface using human Fab binder. Then 92UG037.8 gp120 and 92UG037.8 gp140, purified 92UG037.8 gp120-CD4 and 92UG037.8 gp140-CD4 complexes, or purified 92UG037.8 gp120-VRC01 Fab and 92UG037.8 gp140-VRC01 Fab complexes at various concentrations (50–1,000 nM) were passed over the 17b surface. Binding kinetics was evaluated using a 1:1 Langmuir binding model; binding constants are summarized in Table S2. The sensorgrams are shown in black and the fits in green. RU, response units.
CD4i epitopes.
CD4 binding induces the formation of the bridging sheet in gp120, creating the coreceptor binding site as well as CD4i epitopes (32–34). CD4i antibodies, such as 17b, often are used to detect conformational changes in gp120. In Fig. 2B, immobilized 17b Fab can capture the full-length gp120 even in the absence of CD4, suggesting that the free monomeric gp120 is not fully rigid and that it samples the CD4-bound conformation in solution. As expected, CD4 binding greatly enhances the strength of the interaction between gp120 and 17b (Fig. 2B, and Fig. S4, and Table S2). In contrast, the gp140 trimer does not bind the 17b Fab at all in the absence of CD4, even at high Fab concentrations, but binds tightly in the presence of soluble CD4 (Fig. 2B, Fig. S4, and Table S2). Thus, the gp120 portions in the unliganded envelope trimer are tightly confined in a conformation distinct from the CD4-bound state.
VRC01 induces conformational changes in gp120 similar to those triggered by CD4, but it does not induce those changes in the functional envelope trimers expressed on the cell surfaces (35). We asked whether VRC01 can induce conformational changes in our recombinant gp120 and gp140. As shown in Fig. 2B and Fig. S4, 17b indeed binds the preformed gp120-VRC01 Fab complex more tightly than does gp120 alone, but it does not bind the gp140-VRC01 Fab complex at all (also see Table S2). These results also are confirmed by ELISA. We conclude that our stable gp140 trimers, like the cell-surface–expressed envelope trimers, resist conformational changes induced by VRC01 binding (35).
Quaternary epitopes.
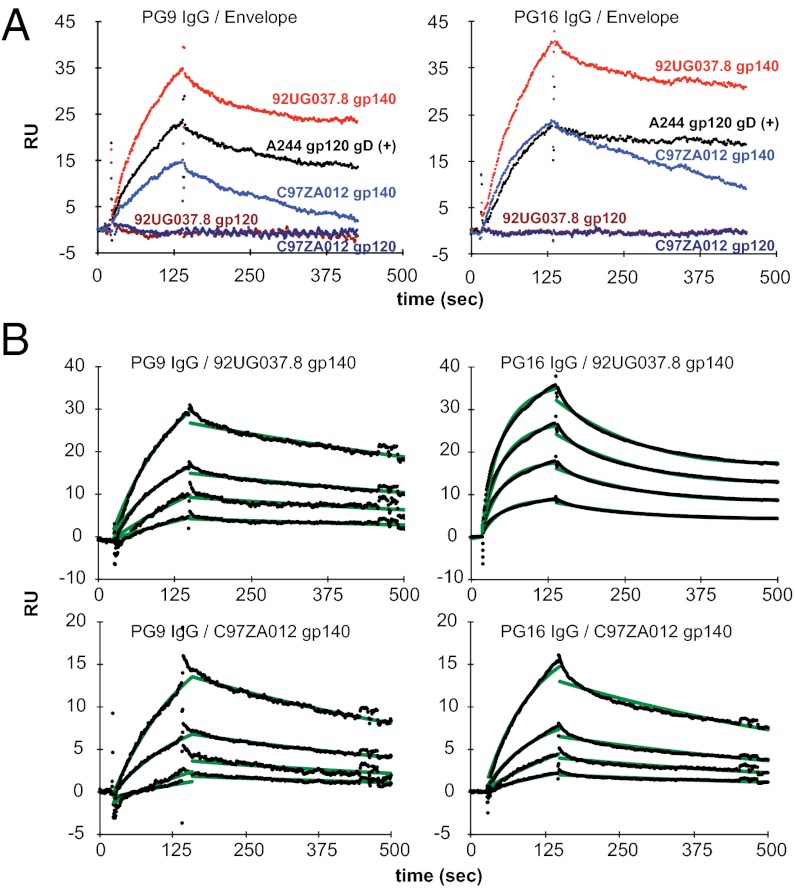
The intact quaternary epitope, first defined by antibodies 2909, PG9, and PG16, is believed to form only on the surface of the functional trimer (9, 36). Binding of PG9/16 to soluble envelope monomers as well as to trimers has been reported, and a crystal structure of PG9 in complex with an isolated V1V2 loop presented on an unrelated scaffold also has been determined (9, 37–39). However, the reported PG9/16 binding is not strictly trimer specific, nor does the binding fully correlate with neutralization. In our experiments, both the 92UG037.8 and C97ZA012 gp140 trimers show detectable binding to PG9 and PG16 (Fig. 3A), whereas monomeric gp120s derived from the two isolates fail to bind either of these antibodies. Thus, the trimer organization of all gp140 species restores, at least partially, the epitopes targeted by PG9 and PG16. Interactions of the PG 9/16 antibodies to our trimers are still weaker than their neutralization potency (Table S3 and Fig. 3B) but are comparable to the affinity of these antibodies for the monomeric A244 gp120 gD(+), used in the RV144 trial, which has been reported to bind these antibodies tightly (38). Moreover, the binding pattern of the trimers to PG9 and P16 agrees with the neutralization profile of the two antibodies for these isolates (Table S3). For example, the 92UG037.8 gp140 trimer binds more tightly to PG9 and PG16 than does the C97ZA012 gp140, consistent with the greater sensitivity of the 92UG037.8 isolate to neutralization by these antibodies. Antibody binding of our gp140 trimers thus correlates directly with neutralization of the purified antibodies. As shown in Fig. 3B, the affinities of the two gp140 trimers for the two antibodies range from 43 to 310 nM.
Fig. 3.
Binding of gp140 and gp120 to bNAbs against the quaternary epitopes. (A) The bNAb PG9 IgG (Left) or PG16 IgG (Right) was captured on a chip surface coated with protein A. Sensorgrams were recorded by passing 92UG037.8 gp140 trimer (1 μM; in red); C97ZA012 gp140 trimer (1 μM; in blue); 92UG037.8 gp120 monomer (1 μM; in dark red); C97ZA012 gp120 monomer (1 μM; in dark blue); or A244 gp120 gD(+) monomer (1 μM; in black) over the antibody surfaces. The apparent differences in response units are caused partly by differences in molecular masses of the flowing analytes. (B) PG9 (Left) or PG16 IgG (Right) was immobilized on the chip surface using human Fab binder. Then various concentrations (50–1,000 nM) of 92UG037.8 gp140 or C97ZA012 gp140 were passed over the antibody surfaces. Binding kinetics was evaluated using a 1:1 Langmuir binding model; binding constants are summarized in Table S2. The sensorgrams are shown in black and the fits in green.
Glycan-dependent neutralizing epitopes.
Additional broadly neutralizing epitopes on gp120 require N-linked glycans, in particular, at the residue Asn332 on the outer domain of gp120, which is covered by densely packed carbohydrates. These antibodies either must bind directly to the carbohydrates or penetrate the glycan shield (30). As shown in Fig. S5, 2G12 binds gp120 and gp140 almost equally well, although gp140 shows a slower off-rate, perhaps because of avidity effects when the Fab is immobilized. Likewise, another bNAb, PGT123, targeting the V3 loop stem, also reacts well with both gp120 and gp140 (Figs. S5 and S6), indicating that these epitopes are fully accessible in both monomeric and trimeric forms of the envelope protein.
Finally, as expected, the mammalian cell–expressed 92UG037.8 trimer does not bind MPER-directed bNAbs, which target the fusion intermediate state of gp41 (Fig. S7), and binds only weakly to anti-gp41 cluster II antibodies (40). Overall, our stable and homogeneous gp140 trimers exhibit a number of characteristics expected for a native, functional envelope trimer.
Stoichiometry of the interactions of gp140 trimers with the CD4 BS ligands.
To characterize further the binding of the gp140 trimers with the CD4 BS ligands, we determined the stoichiometry of these interactions by analytical ultracentrifugation and multiangle light scattering. Values of the molecular mass of the same protein or protein complex determined by the two independent approaches generally were in excellent agreement with each other (Table S1 and Figs. S8 and S9). The molecular masses determined for the two gp140s were ∼412 kDa, as expected for a trimer, and were ∼127 kDa for the two monomeric gp120s. The molecular masses determined for the complexes of four-domain CD4 with gp140 trimers were ∼456 kDa, showing there is only one bound CD4 molecule per trimer in the complex. In contrast, the mass determined for the VRC01 Fab-gp140 complex was ∼567 kDa, corresponding to a stoichiometry of 3:1 for the VRC01–gp140 interaction. In addition, the measured molecular mass of the gp140-CD4-17b complexes was ∼508 kDa. Thus, it appears that one CD4-triggered gp140 trimer can bind just one 17b Fab, suggesting that the other two gp120 subunits preserve their unliganded conformations.
Immunogenicity in Guinea Pigs.
Binding antibody responses.
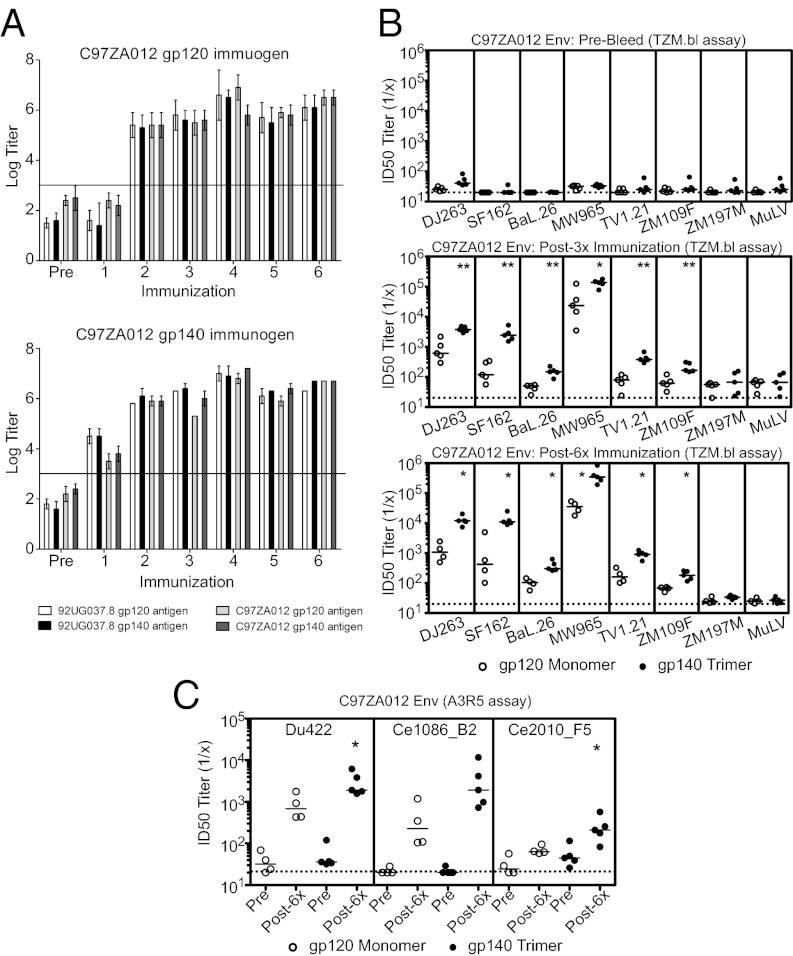
To evaluate the immunogenicity of these gp120s and gp140s, we immunized outbred female Hartley guinea pigs (n = 5 per group) at weeks 0, 4, 8, 20, 24, and 28 by i.m. injection with 100 μg protein per animal, using immunostimulatory di-nucleotide CpG DNA and 15% (vol/vol) oil-in-water emulsion Emulsigen/PBS as adjuvant. This adjuvant combination has been shown to give a more balanced immune response with higher levels of NAbs than each adjuvant on its own (41). Because the molar ratio of gp120 in the monomer immunogen and the gp120 moiety in the trimer immunogen was 1.2:1, the effective dosage of gp120 was slightly higher in the monomer than in the trimer. Envelope-specific antibodies to all four immunogens were assessed by ELISA of sera obtained 4 wk after each immunization. High-titer binding antibody responses were observed in all immunized animals, but responses after a single immunization were higher in the animals that received the gp140 trimers (Fig. 4A and Fig. S10A). Binding antibody titers induced by the monomer and trimer were comparable after the third immunization.
Fig. 4.
Immunogenicity of the C97ZA012 trimeric gp140 and monomeric gp120 in guinea pigs. (A) Sera of guinea pigs, vaccinated with either C97ZA012 gp120 or gp140 obtained prevaccination (Pre) and 4 wk after each immunization, were tested in endpoint ELISAs against all 92UG037.8 and C97ZA012 antigens as indicated. Binding antibody responses against 92UG037.8 gp120 monomer (white bars), 92UG037.8 gp140 trimer (black bars), C97ZA012 gp120 monomer (light gray bars), and C97ZA012 gp140 trimer (dark gray bars) are presented as geometric mean titers at each time point ± SD. Horizontal lines indicate background threshold. (B) Guinea pig sera obtained prevaccination (Pre-Bleed), 4 wk after the third vaccination (Post-3×), and 4 wk after the sixth vaccination (Post-6×) were tested against a multiclade panel of tier 1 neutralization-sensitive isolates, including clade A (DJ263.8), clade B (SF162.LS and Bal.26), and cladeC (MW965.26, TV1.21, ZM109F, and ZM197M) HIV-1 Env pseudoviruses and MuLV (negative control) in TZM.bl neutralization assays. NAb titers induced by C97ZA012 gp120 and C97ZA012 gp140 are represented graphically. *P < 0.05; **P < 0.01; unpaired two-tailed t test. Horizontal bars indicate median titers. (C) Sera obtained prevaccination (Pre) and 4 wk after the sixth vaccination (Post-6×) were tested against three tier 2 clade C isolates (Du422, Ce1086_B2, and Ce2010_F5 IMC viruses) in A3R5 neutralization assays. NAb titers induced by C97ZA012 gp120 and C97ZA012 gp140 are represented y graphically. *P < 0.05; unpaired two-tailed t test.
NAb responses.
We compared neutralizing antibody responses elicited by our stable gp140 trimers and corresponding gp120 monomers using a standardized multiclade panel of tier 1 pseudoviruses from clades A, B, and C in the TZM.bl assay (42) as well as tier 2 clade C infectious molecular clone (IMC) viruses in the A3R5 assay. NAb responses were assessed at baseline and after three and six immunizations. We defined the criteria for NAb positivity as ID50 titers that were (i) more than threefold above preimmune background, (ii) more than twofold above a concurrent murine leukemia virus (MuLV) control, and (iii) an absolute titer >60.
Guinea pigs immunized with either the 92UG037.8 or C97ZA012 gp140 trimer developed robust, cross-clade neutralizing activity against several tier 1 clade A (DJ263.8), clade B (SF162.LS and BaL.26), and clade C (MW965.26, TV1.21, ZM109F.PB4, and ZM197M.PB7) viruses with ID50 titers against the most sensitive virus in the panel, MW965.26, ranging from 57,247–196,648 after the third immunization and from 113,956–848,672 after the sixth immunization (Fig. 4B and Figs. S10B and S11). Tier 1 NAb titers also were elicited by gp120 monomers, but the overall magnitudes of these titers were substantially lower than those observed for their trimeric counterparts (Fig. 4B and Fig. S10B). Specifically, mean ID50 titers induced by the C97ZA012 gp140 trimer were 3.3-, 4.1-, 15.1-, and 3.4-fold greater than median titers induced by the gp120 monomer after the third immunization against MW965.26, DJ263.8, SF162.LS, and BaL.26, respectively, and were 15.1-, 9.9-, 9.0-, and 3.4-fold greater than mean titers induced by the gp120 monomer after the sixth immunization against these same viruses (Fig. 4B). Statistical analyses confirmed that C97ZA012 trimers gave significantly higher tier 1 NAb titers than did their monomeric counterparts [P < 0.01 (DJ263.8, SF162.LS, BaL.26, TV1.21, and ZM109F.PB4) and P < 0.05 (MW965.26)]. Similar results were observed with the 92UG037.8 gp140 trimer [P < 0.01 (DJ263.8, SF162.LS, MW965.26, TV1.21, and ZM109F.PB4)] (Fig. S10B).
We could not detect NAb activity in the TZM.bl assay against two neutralization-resistant tier 2 clade C Env pseudoviruses (ZM233M.PB6 and Du422.1) with sera from animals immunized with either gp120 monomer or gp140 trimer (Fig. S11). We then tested sera against three tier 2 clade C IMC viruses using the more sensitive A3R5 neutralization assay (Fig. 4C and Figs. S10C and S12). We observed that the C97ZA012 gp140 trimer induced higher frequencies of NAb responses as well as higher ID50 titers than did the gp120 monomer against the tier 2 viruses Du422.1 (P < 0.05), Ce1086_B2 (P = NS), and Ce2010_F5 (P < 0.05) (Fig. 4C). As expected, tier 2 NAb titers were lower than tier 1 NAb titers but were clearly detectable in this assay. Taken together, these data demonstrate that in guinea pigs the gp140 trimers elicited higher-magnitude tier 1 and tier 2 NAbs as well as more rapid kinetics of antibody responses than did the corresponding monomeric gp120s.
Discussion
The recent RV144 trial has provided evidence that an HIV-1 vaccine containing an envelope protein boost might afford some degree of protection against HIV-1 infection in clinical settings (18). It remains uncertain, however, which form of the envelope glycoprotein is better at inducing NAbs. Our current immunogenicity studies in guinea pigs clearly show that the gp140 trimers induce potent, cross-clade NAb responses with titers that are substantially higher than those induced by the gp120 monomers for a diverse set of both tier 1 and tier 2 viruses. We conclude that the HIV-1 envelope trimer is indeed a better immunogen than the monomer and suggest that the well-characterized gp140 trimers should be considered for future clinical trials.
Env-based immunogens, including gp120 and gp140, have been tested extensively in animal models and induce high titers of binding antibodies, typically with limited potency and breadth of neutralizing activity (19). Several studies comparing the immunogenicity of monomeric gp120 and trimeric or oligomeric gp140 showed only marginal improvement when the latter was used (24–27, 43), leading to the widely held belief that these immunogens are comparable. Most gp140s in the previous immunogenicity studies had suboptimal stability (26, 44, 45) and also exhibited gp120-like characteristics, such as binding to CD4i antibodies in the absence of CD4 and interacting with nonneutralizing CD4 BS antibodies. Some also reacted with MPER-directed NAbs (46). Binding CD4i antibodies in the absence of CD4 or interacting with nonneutralizing CD4 BS antibodies is not expected for the native envelope spikes, because such binding will compete directly with receptor or coreceptor and confer high neutralization potency. Sophisticated molecular design was required to produce a “resurfaced” gp120 protein or a gp140 variant that can discriminate the neutralizing and nonneutralizing CD4 BS antibodies (28, 47), but our stable trimers distinguish between these two types of antibodies (Fig. 2). The MPER-directed antibodies neutralize by targeting the prehairpin intermediate conformation of gp41, and their epitopes are not present on the native envelope spikes (14, 15). Thus, reactivity with MPER antibodies by a gp140 preparation, which supposedly mimics the untriggered state of the envelope, would indicate that parts of the gp41 moiety in the prepared protein are very flexible or that they have undergone unintended conformational changes. Some gp140 trimers reported previously probably contain three gp120s with few mutual structural constraints, held together only by a common gp41 “stem.” Therefore it is not surprising that these gp140 proteins show gp120-like characteristics and exhibit immunogenicity similar to that of the corresponding gp120 monomers. From the data presented here, we suggest that a gp140 trimer in the prefusion conformation should have the following characteristics: (i) high stability and conformational homogeneity; (ii) no binding of CD4i antibodies in the absence of CD4; (iii) discrimination between neutralizing and nonneutralizing CD4 BS antibodies; (iv) no conformational changes upon binding to VRC01; (v) reactivity with bNAbs, such as PG9 and PG16, that target the trimer-dependent, quaternary epitopes; and (vi) no interaction with MPER-directed antibodies. Of course there may be other distinct antigenic properties associated with the fully cleaved, functional envelope trimer.
The stoichiometry (3:1) of VRC01 binding to the stable gp140 trimers confirms that the three CD4 BS in the trimer are well exposed and that they can accommodate three molecules with the size of an Fab. Why, then, can only one CD4 molecule bind to an envelope trimer? Reinherz and colleagues first reported the stoichiometry of CD4 binding to a stable SIV gp140 trimer as 1:1 (48) but later concluded that one HIV-1 ADA strain gp140 trimer can bind more than one CD4 molecule (49). Those investigators postulated that the HIV-1 envelope may have evolved to be more open, closer to a “fusion-ready” transition than the simian immunodeficiency virus (SIV) envelope, implying that HIV-1 envelope may be more sensitive to antibody neutralization. We now have demonstrated that the stable HIV-1 gp140 trimers have the same stoichiometry for CD4 binding as the SIV trimer, suggesting that this phenomenon may be physiologically relevant. VRC01 binding, with a stoichiometry of 3:1, does not induce conformational changes in gp140 trimers, whereas CD4, which reacts with only one gp120 and breaks the symmetry of the trimer, triggers large structural rearrangements in gp120. This observation indicates that the asymmetry introduced by CD4 is caused by the conformational changes induced by CD4 binding, not by steric interference. Crystallographic analysis of gp120 core proteins revealed that CD4 binding induces formation of the bridging sheet as well as reorganization of the inner domain of gp120 and possibly large movements of the V1V2 loop (34). Conceivably, these movements can block the other two CD4 BS, thereby creating an asymmetric structure. Moreover, the asymmetry of CD4-liganded gp140 prevents the other subunits from undergoing similar structural rearrangements, because only one 17b Fab binds a CD4-triggered trimer (Table S1). Srivastava et al. (45) reported that all three CD4 BS are available to CD4 on certain gp140 trimers that have a deletion in the V2 loop. Thus, modifications in the variable loop can change the valency of the gp140 trimers for CD4 association.
If our observations on the CD4-induced asymmetry are indeed relevant to HIV-1 entry, what are the advantages of such negative cooperativity of CD4 binding for the functional trimer? First, requiring only one CD4 and, presumably, one coreceptor to trigger the envelope trimer is an efficient way to initiate membrane fusion, given the scarcity of the envelope spikes on the surface of virion (50). Second, blocking the other two CD4 BS and preventing them from further conformational changes would effectively reduce the number of vulnerable sites during the fusion-intermediate steps that can be attacked by the host immune responses. Thus, the unusual stoichiometry by the receptor binding may be still another mechanism by which HIV-1 evades the host immune system. Third, three CD4-induced rearrangements might destabilize the trimer and lead to premature gp120 shedding, thereby inactivating it.
The bNAbs isolated from infected patients show remarkable potency and breadth. Induction of such responses is a critical goal for vaccine development, but none of the envelope-based immunogens, including our stable gp140 trimers presented here, has been able to raise high levels of bNAbs in small animal models. Detailed analysis of these monoclonal antibodies suggests that somatic hypermutation, as well as breaking host tolerance to carbohydrates or lipids, may be required for developing potent broadly neutralizing activities over an extended period (9, 11, 14, 51). We note that the broadly neutralizing epitopes involving N-linked glycans on the rigid surface of the gp120 outer domain appear to be well exposed on both monomeric gp120 and trimeric gp140 but failed to induce high titers of 2G12- or PGT123-like antibodies in the current study. Moreover, significant protection against stringent virus challenges can be achieved in nonhuman primates even without high titers of NAbs that neutralize resistant isolates (52). Thus, the full potential of our trimer immunogens may need to be evaluated further in more relevant clinical settings. Overall, we believe that generating improved trimer immunogens that closely mimic the native functional spikes on the HIV-1 virion is an important step toward an effective HIV-1 vaccine.
Methods and Materials
Production of HIV-1 Envelope Proteins, CD4, and Antibodies.
Expression constructs and protein production of HIV-1 envelope protein, CD4, and antibodies are described in SI Materials and Methods. All envelope proteins used were produced in 293T cells.
SPR Binding Assays, Multiangle Light Scattering, and Analytical Ultracentrifugation.
Details of all binding experiments, multiangle light scattering, and analytical ultracentrifugation are described in SI Materials and Methods.
Animals, Immunizations and ELISA.
Details of animals, immunizations, and ELISA are described in SI Materials and Methods.
NAb Assays.
NAb responses against HIV-1 Env pseudoviruses were measured using luciferase-based neutralization assays in TZM.bl cells as described (6, 42). Details are given in SI Materials and Methods. For neutralization assays in A3R5 cells, serial dilutions of serum samples were performed in 10% RPMI growth medium (100 μL per well) in 96-well flat-bottomed plates. IMC HIV-1 expressing Renilla luciferase (53) was added to each well in a volume of 50 μL, and plates were incubated for 1 h at 37 °C. Then A3R5 cells were added (9 × 104 cells per well in a volume of 100 μL) in 10% RPMI growth medium containing diethylaminoethyl-dextran (11 μg/mL). Assay controls included replicate wells of A3R5 cells alone (cell control) and A3R5 cells with virus (virus control). After incubation for 4 d at 37 °C, 90 μL of medium was removed from each assay well, and 75 μL of cell suspension was transferred to a 96-well white, solid plate. Diluted ViviRen Renilla luciferase substrate (Promega) was added to each well (30 μL), and after 4 min the plates were read on a Victor 3 luminometer. The A3R5 cell line was generously provided by R. McLinden and J. Kim (US Military HIV Research Program, Rockville, MD). Tier 2 clade C IMC Renilla luciferase viruses Du422.1.LucR.T2A.ecto, Ce2010_F5.LucR.T2A.ecto, and Ce1086_B2.LucR.T2A.ecto were generously provided by C. Ochsenbauer (University of Alabama at Birmingham, Birmingham, AL), and stocks were prepared in 293T/17 cells as previously described (53).
Supplementary Material
Acknowledgments
We thank Stephen Harrison, Michael Freeman, Sophia Rits-Volloch, Joanna Mangar, and Gary Frey for generous advice and assistance; and Larry Liao, Barton Haynes, Dennis Burton, John Mascola, and James Robinson for reagents. This work was supported by National Institutes of Health Grants AI084794, AI078526, and GM083680; the Ragon Institute of MGH, MIT, and Harvard; and the Center for HIV/AIDS Vaccine Immunology.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1204533109/-/DCSupplemental.
References
- 1.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore JP, McKeating JA, Weiss RA, Sattentau QJ. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science. 1990;250:1139–1142. doi: 10.1126/science.2251501. [DOI] [PubMed] [Google Scholar]
- 3.Mascola JR, et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med. 2000;6:207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 4.Hessell AJ, et al. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat Med. 2009;15:951–954. doi: 10.1038/nm.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balazs AB, et al. Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature. 2012;481:81–84. doi: 10.1038/nature10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nkolola JP, et al. Breadth of neutralizing antibodies elicited by stable, homogeneous clade A and clade C HIV-1 gp140 envelope trimers in guinea pigs. J Virol. 2010;84:3270–3279. doi: 10.1128/JVI.02252-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stamatatos L, Morris L, Burton DR, Mascola JR. Neutralizing antibodies generated during natural HIV-1 infection: Good news for an HIV-1 vaccine? Nat Med. 2009;15:866–870. doi: 10.1038/nm.1949. [DOI] [PubMed] [Google Scholar]
- 8.Clapham PR, Lu S. Vaccinology: Precisely tuned antibodies nab HIV. Nature. 2011;477:416–417. doi: 10.1038/477416a. [DOI] [PubMed] [Google Scholar]
- 9.Walker LM, et al. Protocol G Principal Investigators Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trkola A, et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walker LM, et al. Protocol G Principal Investigators Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muster T, et al. Cross-neutralizing activity against divergent human immunodeficiency virus type 1 isolates induced by the gp41 sequence ELDKWAS. J Virol. 1994;68:4031–4034. doi: 10.1128/jvi.68.6.4031-4034.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stiegler G, et al. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 2001;17:1757–1765. doi: 10.1089/08892220152741450. [DOI] [PubMed] [Google Scholar]
- 14.Alam SM, et al. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc Natl Acad Sci USA. 2009;106:20234–20239. doi: 10.1073/pnas.0908713106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frey G, et al. A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. Proc Natl Acad Sci USA. 2008;105:3739–3744. doi: 10.1073/pnas.0800255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flynn NM, et al. rgp120 HIV Vaccine Study Group Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J Infect Dis. 2005;191:654–665. doi: 10.1086/428404. [DOI] [PubMed] [Google Scholar]
- 17.Pitisuttithum P, et al. Bangkok Vaccine Evaluation Group Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J Infect Dis. 2006;194:1661–1671. doi: 10.1086/508748. [DOI] [PubMed] [Google Scholar]
- 18.Rerks-Ngarm S, et al. MOPH-TAVEG Investigators Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 19.Burton DR, et al. HIV vaccine design and the neutralizing antibody problem. Nat Immunol. 2004;5:233–236. doi: 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- 20.Chen B, et al. Expression, purification, and characterization of gp160e, the soluble, trimeric ectodomain of the simian immunodeficiency virus envelope glycoprotein, gp160. J Biol Chem. 2000;275:34946–34953. doi: 10.1074/jbc.M004905200. [DOI] [PubMed] [Google Scholar]
- 21.Earl PL, et al. Immunogenicity and protective efficacy of oligomeric human immunodeficiency virus type 1 gp140. J Virol. 2001;75:645–653. doi: 10.1128/JVI.75.2.645-653.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X, et al. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J Virol. 2002;76:4634–4642. doi: 10.1128/JVI.76.9.4634-4642.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Binley JM, et al. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol. 2000;74:627–643. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim M, et al. Comparison of HIV Type 1 ADA gp120 monomers versus gp140 trimers as immunogens for the induction of neutralizing antibodies. AIDS Res Hum Retroviruses. 2005;21:58–67. doi: 10.1089/aid.2005.21.58. [DOI] [PubMed] [Google Scholar]
- 25.Beddows S, et al. A comparative immunogenicity study in rabbits of disulfide-stabilized, proteolytically cleaved, soluble trimeric human immunodeficiency virus type 1 gp140, trimeric cleavage-defective gp140 and monomeric gp120. Virology. 2007;360:329–340. doi: 10.1016/j.virol.2006.10.032. [DOI] [PubMed] [Google Scholar]
- 26.Grundner C, et al. Analysis of the neutralizing antibody response elicited in rabbits by repeated inoculation with trimeric HIV-1 envelope glycoproteins. Virology. 2005;331:33–46. doi: 10.1016/j.virol.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 27.Melchers M, et al. Targeting HIV-1 envelope glycoprotein trimers to B cells by using APRIL improves antibody responses. J Virol. 2012;86:2488–2500. doi: 10.1128/JVI.06259-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng Y, et al. Biochemically defined HIV-1 Env variant immunogens display differential binding and neutralizing specificities to the CD4 binding site. J Biol Chem. 2012;287:5673–5686. doi: 10.1074/jbc.M111.317776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lavine CL, et al. High-mannose glycan-dependent epitopes are frequently targeted in broad neutralizing antibody responses during infection of human immunodeficiency virus type 1. J Virol. 2012;86:2153–2164. doi: 10.1128/JVI.06201-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pejchal R, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pantophlet R, Burton DR. GP120: Target for neutralizing HIV-1 antibodies. Annu Rev Immunol. 2006;24:739–769. doi: 10.1146/annurev.immunol.24.021605.090557. [DOI] [PubMed] [Google Scholar]
- 32.Rizzuto CD, et al. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 33.Kwong PD, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen B, et al. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature. 2005;433:834–841. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- 35.Li Y, et al. Mechanism of neutralization by the broadly neutralizing HIV-1 monoclonal antibody VRC01. J Virol. 2011;85:8954–8967. doi: 10.1128/JVI.00754-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gorny MK, et al. Identification of a new quaternary neutralizing epitope on human immunodeficiency virus type 1 virus particles. J Virol. 2005;79:5232–5237. doi: 10.1128/JVI.79.8.5232-5237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davenport TM, et al. Binding interactions between soluble HIV envelope glycoproteins and quaternary-structure-specific monoclonal antibodies PG9 and PG16. J Virol. 2011;85:7095–7107. doi: 10.1128/JVI.00411-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonsignori M, et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol. 2011;85:9998–10009. doi: 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McLellan JS, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frey G, et al. Distinct conformational states of HIV-1 gp41 are recognized by neutralizing and non-neutralizing antibodies. Nat Struct Mol Biol. 2010;17:1486–1491. doi: 10.1038/nsmb.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ioannou XP, Griebel P, Hecker R, Babiuk LA, van Drunen Littel-van den Hurk S. The immunogenicity and protective efficacy of bovine herpesvirus 1 glycoprotein D plus Emulsigen are increased by formulation with CpG oligodeoxynucleotides. J Virol. 2002;76:9002–9010. doi: 10.1128/JVI.76.18.9002-9010.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li M, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, et al. Characterization of antibody responses elicited by human immunodeficiency virus type 1 primary isolate trimeric and monomeric envelope glycoproteins in selected adjuvants. J Virol. 2006;80:1414–1426. doi: 10.1128/JVI.80.3.1414-1426.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dey AK, David KB, Lu M, Moore JP. Biochemical and biophysical comparison of cleaved and uncleaved soluble, trimeric HIV-1 envelope glycoproteins. Virology. 2009;385:275–281. doi: 10.1016/j.virol.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srivastava IK, et al. Comparative evaluation of trimeric envelope glycoproteins derived from subtype C and B HIV-1 R5 isolates. Virology. 2008;372:273–290. doi: 10.1016/j.virol.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 46.Dey AK, David KB, Klasse PJ, Moore JP. Specific amino acids in the N-terminus of the gp41 ectodomain contribute to the stabilization of a soluble, cleaved gp140 envelope glycoprotein from human immunodeficiency virus type 1. Virology. 2007;360:199–208. doi: 10.1016/j.virol.2006.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu X, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim M, et al. The stoichiometry of trimeric SIV glycoprotein interaction with CD4 differs from that of anti-envelope antibody Fab fragments. J Biol Chem. 2001;276:42667–42676. doi: 10.1074/jbc.M104166200. [DOI] [PubMed] [Google Scholar]
- 49.Zhang CW, Chishti Y, Hussey RE, Reinherz EL. Expression, purification, and characterization of recombinant HIV gp140. The gp41 ectodomain of HIV or simian immunodeficiency virus is sufficient to maintain the retroviral envelope glycoprotein as a trimer. J Biol Chem. 2001;276:39577–39585. doi: 10.1074/jbc.M107147200. [DOI] [PubMed] [Google Scholar]
- 50.Zhu P, et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 51.Zhou T, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barouch DH, et al. Vaccine protection against acquisition of neutralization-resistant SIV challenges in rhesus monkeys. Nature. 2012;482:89–93. doi: 10.1038/nature10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edmonds TG, et al. Replication competent molecular clones of HIV-1 expressing Renilla luciferase facilitate the analysis of antibody inhibition in PBMC. Virology. 2010;408:1–13. doi: 10.1016/j.virol.2010.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.