Abstract
Deamidation of asparagine and glutamine is the most common non-enzymatic, post-translational modification. Deamidation can influence the structure, stability, folding, and aggregation of proteins and has been proposed to play a role in amyloid formation. However there are no structural studies of the consequences of deamidation on amyloid fibers, in large part because of the difficulty of studying these materials using conventional methods. Here we examine the effects of deamidation on the kinetics of amyloid formation by amylin, the causative agent of type 2 diabetes. We find that deamidation accelerates amyloid formation and the deamidated material is able to seed amyloid formation by unmodified amylin. Using site-specific isotope labeling and two-dimensional infrared (2D IR) spectroscopy, we show that fibers formed by samples that contain deamidated polypeptide contain reduced amounts of β-sheet. Deamidation leads to disruption of the N-terminal β-sheet between Ala-8 and Ala-13, but β-sheet is still retained near Leu-16. The C-terminal sheet is disrupted near Leu-27. Analysis of potential sites of deamidation together with structural models of amylin fibers reveals that deamidation in the N-terminal β-sheet region may be the cause for the disruption of the fiber structure at both the N- and C-terminal β-sheet. Thus, deamidation is a post-translational modification that creates fibers which have an altered structure, but can still act as a template for amylin aggregation. Deamidation is very difficult to detect with standard methods used to follow amyloid formation, but isotope labeled IR spectroscopy provides a means for monitoring sample degradation and investigating the structural consequences of deamidation.
Keywords: amylin, islet amyloid polypeptide, deamidation, two dimensional IR, amyloid, Type 2 diabetes, post-translational modification
Introduction
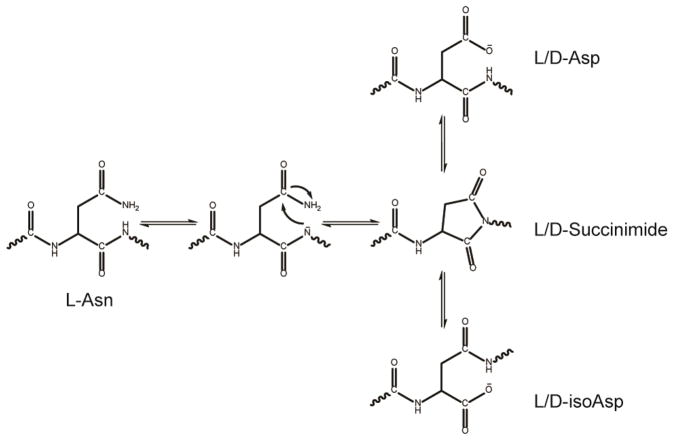
Post-translational modifications are common steps in polypeptide and protein biosynthesis. They include phosphorylation, glycoslyation, lipidation, amidation of the C-terminus and acetylation, as well as disulfide bond formation.1–3 These post-translational modifications occur in vivo in a myriad of proteins and are under careful biological control. Deamidation of Asn and Gln side chains is a common spontaneous, non-enzymatic post translation modification in vivo and in vitro, and is caused by a reaction of asparagine or glutamine side chains with the protein backbone.4–7 Deamidation can influence the structure, stability, folding, and aggregation of proteins and is proposed to play a role in amyloid formation.6,8–11 Deamidation is also a concern with protein and polypeptide based pharmaceuticals.12 Under physiological conditions deamidation proceeds through a 5-membered succinimide intermediate. The ring subsequently hydrolyzes into either aspartic or isoaspartic acid (from asparagine), while for glutamine, either glutamic or isoglutamic acid is formed (Figure 1). Deamidation favors the production of the L-forms of the products, but can racemize to the D-forms via the succinimide intermediate. The ratio of the iso-products to normal products is 3:1.5 Deamidation can cause major perturbations to the structure and stability of a protein because it adds a negative charge. When the iso-product is formed, deamidation extends the length of the backbone by a CH2 group and thus introduces another rotatable bond into the peptide backbone.
Figure 1.
Mechanism for deamidation of L-Asn.
Deamidation has been implicated in amyloid formation by several proteins involved in human disease, including amyloid-β (Aβ) from Alzheimer’s6,8,11,13–15 and the crystallins9,16,17 from cataracts. α-synuclein, which causes Parkinson’s and other neurodegenerative diseases, also deamidates.18,19 Some otherwise non-amyloidogenic peptides have been shown to become amyloidogenic upon deamidation.20 Although not strictly deamidation, all three Asp residues in the Aβ polypeptide of Alzheimer’s disease patients have been found to isomerize to isoAsp, thus adding an additional bond to the peptide backbone and causing structural disorder. It has been reported that 55% of Asp-7 in Aβ isomerizes to isoAsp-7 in senile plaques.11 In addition, two common Aβ mutations, the Iowa mutation (Asn-23) and the Tottori-Japanese mutation (Asn-7) have both been found to deamidate to isoAsp in Alzheimer’s disease patients.6,13 While it is still not explicitly understood the role deamidation has in forming Alzheimer plaques, one hypothesis is that the mutation at Asp-23 to Asn-23 and the subsequent deamidation to isoAsp-23, in the turn region of Aβ, might cause a structural change to Aβ that initiates folding into β-sheets.15 Deamidation may contribute to plaque formation through the isomerization to isoAsp that presumably prevents proper degradation of damaged peptides.15 Regarding the crystallin proteins, King and coworkers showed that deamidation of Gln residues, which appear at the interface of two domains in D-crystallin, lowered the barrier to unfolding, thus increasing the probability of populating partially unfolded states.16 It is suggested that these partially unfolded states could be more likely to aggregate. Using non-cataractous lenses from human patients of varying ages, ten different residues of D-crystallin were determined to deamidate (out of 167) and, in general, the amount of deamidation at each site increased with age of the protein. Thus, deamidation is thought to be an important cause of cataract formation.
Similar in sequence to human amylin is pramlintide, the synthetic drug used as replacement therapy in diabetics. The two peptides differ slightly in structure with mutations at A25P, S28P, and S29P.21 A deamidation study was performed on pramlintide in which it was thermally stressed at 40°C for 45 days.22,23 The results show that five of the six asparagine residues, with the exception of Asn-31 showed some amount of deamidation to either aspartic or isoaspartic acid. The residues with the most observed deamidation were Asn-21 and Asn-35. High levels of deamidation at Asn-21 were attributed to the hydrophilic Ser-Asn-Asn sequence and residues 20 and 22 aid in deamidation of Asn-21. Asn-35 was the second-most deamidated residue. It is thought that Asn-Ser sequences are more prone to deamidation, and in fact have been measured to have deamidation half-lives has short as 16 days under physiological conditions.24 It was speculated that Thr-36, being similar in structure to Ser, might allow Asn-35 to have higher amounts of deamidation than other Asn residues,22,23 and the deamidation half-life for Asn-Thr is 47 days.24 Deamidation rates are also closely correlated with peptide structure, and typically the deamidation rate slows with increasing three-dimensional structure.25 Overall, the C-terminus of pramlintide had higher levels of deamidation than the N-terminus, with Asn-3, located in the sequence between the disulfide bond, having the lowest amount of deamidation products.
In this paper, we focus on the effects of deamidation on the in vitro fiber structure and aggregation kinetics of human amylin (human islet amyloid polypeptide-hIAPP) the major component of the islet amyloid associated with type 2 diabetes. Amylin has been identified in all mammals examined and it is secreted together with insulin. The polypeptide normally acts as a partner to insulin26–30, but it is responsible for islet amyloid in type-2 diabetes.28–37 The factors which trigger amyloid formation by human amylin are not completely understood, but it is one of the most amyloidogenic polypeptides known and many studies have been done to understand its aggregation.38–44 Aggregates of human IAPP are toxic to pancreatic β-cells, strongly suggesting that the process of amyloid formation by amylin contributes to islet cell death in type 2 diabetes. Islet amyloid has also been proposed to play an important role in the failure of transplanted islet cell grafts.45,46 Human IAPP is a 37 residue peptide hormone that has a disulfide bridge between Cys-2 and Cys-7 and an amidated C-terminus (Figure 2a). The polypeptide contains six Asn residues and one Gln. Interactions involving Asn side chains have been proposed to be important in amyloid formation by human amylin and mutation of certain Asn residues to Leu, an approximately isoteric substitution, has been shown to have large effects on the kinetics of assembly.47 While it is not known if amylin is deamidated in the plaques of the β-islet cells of type 2 diabetics, we report here that it readily deamidates under physiological conditions in vitro. Moreover, we discover that there is a major change to the fiber β-sheet structure which is not resolved with TEM, CD, or standard linear or 2D IR spectroscopy, all of which produce images or spectra that are indistinguishable from those of polypeptides that are not deamidated. We are able to identify the change in secondary structure by the combination of isotope labeling and 2D IR spectroscopy. 13C18O labeling allows the backbone amide I vibrational modes of individual residues to be spectroscopically identified. As described in more detail below, the isotope labeled amino acids are very strongly coupled to one another in the parallel β-sheet structure of amyloid fibers, which produces distinctive frequency shifts and cross peaks. These spectral signatures are a sensitive measure of structure change. While it is clear that the addition of a charge and the conversion of a standard amino acid to a β-amino acid could have repercussions for amyloid fiber structure and aggregation kinetics, ascertaining the location and extent of the structure change is difficult. Isotope edited 2D IR spectroscopy enables one to probe, residue-by-residue, the effects of deamidation.
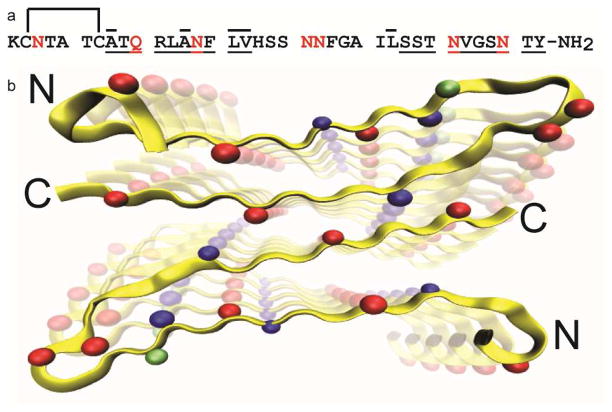
Figure 2.
a. Sequence of hIAPP. Potential sites for deamidation are shown in red and residues that were isotope labeled for this study are shown with an overbar. There is a disulfide bond between Cys-2 and Cys-7, and the C-terminus is amidated. Underlined amino acids have parallel β-sheet structure, according to Tycko.43 b. Structural model for an isomorphic structure of amylin fiber from solid-state NMR. Asparagines and glutamine are shown in red. The four isotope labeled residues incorporated into deamidated amylin are shown in blue. Val-17 is shown in green. The perspective of the figure is looking down the fiber axis.
Overview of how isotope labeling and 2D IR spectroscopy allow for the identification of parallel β-sheets with residue-specificity
Many amyloid polypeptides and proteins, including hIAPP, whose sequence and structure is shown in Figure 2, form parallel β-sheets when folded into amyloid fibers.38,48,49 In particular, hIAPP fibers consist of two columns of hIAPP monomers that contain one turn. Intermolecular hydrogen bonds form between the stacked monomers, essentially creating a structure of four parallel β-sheets that run along the fiber axis. There are no intra-peptide hydrogen bonds between backbone groups. Amyloid fibers have a distinctive absorption band in their infrared spectra at about 1620 cm−1, which is caused by vibrational coupling between the amide groups of the amino acids.50 The strongest couplings are between in-register residues that span from one strand to the next, although the coupling is caused by electrostatics, not hydrogen bonding.51 The couplings cause the vibrational modes to delocalize over several amino acids, so that they all vibrate in unison. Delocalized normal modes occur in β-sheets in globular proteins as well, but they are particularly evident in amyloid fibers, because the parallel β-sheets are very large and structurally well-ordered.
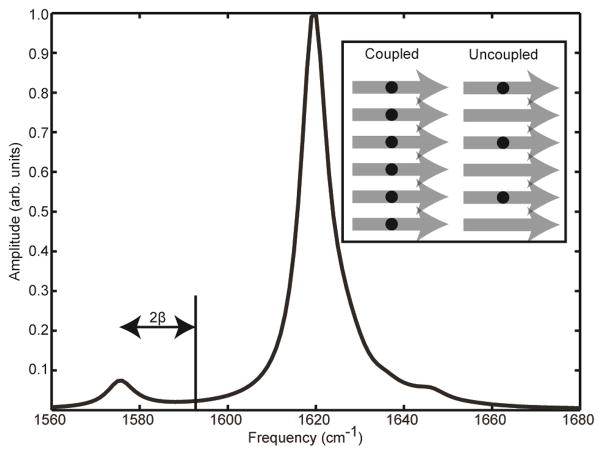
The strong coupling between β-strands provides an effective means of using isotope labeling to test for parallel β-sheet structure. If an amino acid is isotope labeled with a 13C18O backbone carbonyl, the frequency of that particular amide I mode is shifted ~54 cm−1 from the frequency of the unlabeled (random coil) amide I modes due to the change in its reduced mass.50 In hIAPP, each polypeptide contributes one strand to each of the β-sheets. Thus, in the parallel β-sheets of the amyloid fiber, the isotope labeled amino acids align in register (Figure 3).50,52 As mentioned above, the coupling between in-register residues is particularly strong, and so the vibrational modes of the labeled residues become delocalized into a chain that spans many β-strands. The result is a shift in frequency that is less than or equal to twice the coupling constant, β.53,54 Typically, the coupling constant between two in-register residues in adjacent parallel β-strands is -9 to -11 cm−1, and thus the frequency shift due to excitonic coupling can be as large as 20 cm−1 (Figure 3).48 The frequency shift varies from residue to residue, because each amino acid experiences a different amount of structural disorder, which localizes the vibrations. Residues at the ends of the peptide where the β-sheet frays and residues in the turn region are less ordered and the effects of the coupling are small.50 Moreover, the absolute frequency for each chain of residues may be different, even if the coupling strengths are the same, because of differences in electrostatic environments.
Figure 3.
Model slice through the diagonal of a 2D IR spectrum of an isotopically labeled parallel β-sheet. The unlabeled amide I mode absorbs at 1620 cm−1, while the strongly coupled, in-register isotope labeled amino acid absorbs at ~1575 cm−1.
Electrostatic environments influence the frequencies of individual amino acids (local mode frequencies) and thus the diagonal linewidth. Measuring diagonal linewidths and overall lineshapes of isotope labeled amino acids in a peptide can be a useful experiment to determine the local environment of the labeled residue. For example, a narrower diagonal linewidth can be ascribed to a more homogeneous local environment and less electrostatics, whereas a larger diagonal linewidth means the residue lies in a more inhomogeneous environment and is subject to stronger electrostatic forces. This has been shown to be particularly useful when studying membrane peptides where the diagonal linewidths of residues that are located outside the membrane versus inside the membrane can vary by ~15 cm−1.39,55
Thus, to obtain the frequency shift caused by coupling, we also need to know the local mode frequency of the individual amino acid, which we find by measuring fibers formed by a ~1:3 mixture of labeled and unlabeled peptides.48,56 In isotope diluted samples, the coupling is less effective across the linear chains, and so the vibrational modes localize. The frequency of the isotope labeled peak now reports the frequency when no coupling exists at that residue and is the intrinsic frequency of the labeled amino acid, which is a measure of its electrostatic environment. By measuring the frequency of fibers formed with 100% isotope labeled polypeptides and comparing it to the frequency of fibers formed from isotope diluted polypeptides, we obtain the frequency shift caused by coupling from which we can determine if the labeled amino acid residues in a parallel β-sheet. We have used this characteristic of parallel β-sheets to study the residue-specific aggregation kinetics, the stability, and the drug binding to amyloid fibers of hIAPP.56,57
Materials and Methods
Peptide synthesis and preparation of isotope labeling amino acids
hIAPP is a 37 residue polypeptide whose sequence and structure is shown in Figure 2. Peptides were prepared using standard FMOC solid phase peptide synthesis with a CEM Liberty 12-Channel Automated Peptide Synthesizer.58 To produce an amidated C-terminus, PAL-PEG-PS resin was used. Pseudoproline dipeptides were incorporated to facilitate the synthesis as described elsewhere.59 Standard trifluoroacetic acid (TFA) cleavage and deprotection protocols were used, and 1,2-ethanedithiol, anisole, and thioanisole served as scavengers. Crude peptide was dissolved overnight in dimethylsulfoxide (DMSO) at ~5 mg/mL to induce disulfide bond formation between Cys-2 and Cys-7. 13C18O isotope labeled alanine, leucine, and valine were individually incorporated into hIAPP at positions Ala-8, Ala-13, Leu-16, Leu-27, and Val-17. Fmoc-protected amino acids with the backbone carbonyl labeled with 13C18O were prepared as described starting from 13C C1 labeled amino acids (Cambridge Isotope Laboratories) and 18O water.57 The labeling levels were confirmed with LC-MS and greater than 90% labeling efficiency was achieved. Peptides were purified using reversed-phase HPLC with a C18 preparative column (Vydac) and absorbance was monitored at 200 nm and 280 nm. A two buffer system was used: buffer A consisted of H2O and 0.045% HCl (v/v), and buffer B consisted of 80% acetonitrile (ACN), 20% H2O, and 0.045% HCl (v/v). The gradient varied from 0–90% B over 90 minutes. TFA was not used as the counter ion to allow for the complete removal of residual TFA, which is required for IR spectroscopy. The molecular weight of all peptides was confirmed using MALDI-MS.
Detection of deamidation
Following synthesis, the peptides were allowed to age. Deamidation occurred spontaneously over the course of several months. Throughout this paper, all peptides that were allowed to age will be referred to as ‘deamidated’, whereas peptides that had not degraded and undergone deamidation will be referred to as ‘unmodified’. To assess the degree of deamidation for each of the four samples, reverse-phase HPLC was performed with a C18 analytical column (Vydac) and absorbance was monitored at 220 nm and 280 nm. A two buffer gradient was used: buffer A consisted of H2O and 0.1% (TFA) and buffer B consisted of 80% ACN, 20% H2O, and 0.1% TFA. The gradient varied from 0–90% B over 90 minutes. Under these conditions (pH 2), hIAPP with native asparagine and deamidated hIAPP with isoaspartic acid will have identical retention times, while hIAPP that has had asparagine deamidated to aspartic acid will elute later.20 Additionally, HPLC was performed at pH 5. Except for different buffers, HPLC at pH 5 was performed identically to that at pH 2. A two buffer gradient was used: buffer A consisted of 10 mM phosphate in H2O and buffer B consisted of 10 mM phosphate in 80% ACN and 20% H2O. Under these less acidic conditions, deamidated hIAPP with aspartic acid or isoaspartic acid has a shorter retention time than hIAPP with native asparagine.
Sample preparation
Samples for spectroscopy experiments were prepared by dissolving hIAPP in deuterated hexafluoroisopropanol (d-HFIP) to a final concentration of 1 mM. After four hours, hIAPP was completely deuterated. d-HFIP was evaporated under a stream of nitrogen and fiber formation was initiated by adding 20 mM deuterated potassium phosphate buffer at ~pH 7.4 to a final peptide concentration of 1 mM. The sample was then placed between two 2 mm thick CaF2 windows separated by a 56 μm Teflon spacer. All samples were kept in a purge box until and during use. This method of fiber formation is known to create polymorphic fibers that differ near the turn region but have similar β-sheet forming regions.43,56
Two-dimensional infrared spectroscopy
The 2D IR spectroscopy experimental setup consists of a home-built Ti:sapphire oscillator that seeds a Nd:YLF-pumped regenerative amplifier as described in more detail elsewhere.57 Briefly, the 800 nm output is sent to a home-built BBO-based optical parametric amplifier (OPA). The signal and idler beams generated from the OPA are difference frequency mixed in a AgGaS2 crystal, producing broad-bandwidth 6 μm pulses. The resulting mid-infrared light is split into pump and probe pulses, and the pump pulse passes through a Ge acousto-optic modulator (AOM)-based pulse shaper.60,61 Finally, pump and probe beams are focused onto the sample. Steady-state spectra were collected 4 hours after aggregation was initiated at which time the fiber structure has equilibrated according to ThT and 2D IR kinetics.38,62,63 No differences in 2D IR spectra are found at longer times.
Transmission electron microscopy
TEM was performed at the University of Wisconsin Medical School Electron Microscope Facility. Peptides were prepared as for spectroscopy experiments, except all samples were diluted from 1 mM to 0.1 mM with deuterated potassium phosphate buffer. The peptides were allowed to aggregate for 12 hours prior to TEM. The samples were then prepared using a two-step negative staining procedure. Nano-W (methylamine tungstate) (Nanoprobes, Inc.) was used as the negative stain. The samples were then placed on a pioloform coated 300 mesh thinbar Cu grid, and viewed on a Philips CM120 electron microscope at 80 kV. The images were documented with a SIS (Olympus-Soft Imaging Systems) MegaView III digital camera.
Multiple images were collected for each sample. Apparent widths of the fibers were measured using Adobe Illustrator.56 Lines were drawn across the fibers and the lengths of the lines were converted to pixels and then into nanometers using the resolution of the image. Random sampling of the images was achieved. However, due to poor contrast, some fibers may not have been measured. For publication purposes, the contrast of the images was enhanced.
Circular dichroism spectroscopy
CD spectra of unmodified and deamidated 13C18O labeled Ala-13 hIAPP were collected using an Aviv 420 circular dichroism spectrophotometer at room temperature. Spectra were collected at 200–300 nm with 1 nm intervals and a 3 second sampling time. A 0.1 cm path length cuvette was used, and the total sample volume was 300 μL. Peptide samples were diluted from 1 mM d-HFIP stock solutions, dried under a stream of nitrogen, and redissolved into 5 mM deuterated potassium phosphate buffer to a final concentration of 25 μM (deamidated peptide) and 75 μM (unmodified peptide). Following addition of buffer, all peptides were allowed to aggregate for 48 hours. Prior to collecting spectra of peptides, a background spectrum was collected of deuterated potassium phosphate buffer. This spectrum was subtracted from all subsequent peptide spectra.
Aggregation kinetics and seeding experiments
Samples of unmodified and deamidated peptides were prepared for kinetics as described above. However, rather than waiting four hours to allow the peptides to fold following aggregation initiation with buffer, 2D IR spectra were collected immediately after initiation with buffer, leading to a dead time of ~10 minutes. The seeding experiment was performed by mixing deamidated hIAPP fibers with unmodified hIAPP monomers. Unmodified and deamidated peptides were prepared in separate tubes as described above. Deamidated Ala-13 was allowed to aggregate for 6 hours prior to mixing with unmodified Val-17. After 6 hours, the unmodified peptide monomers were rehydrated with buffer and quickly mixed with the deamidated fibers. Ala-13 fibers were formed at 1 mM and Val-17 monomers were added for final concentrations of 0.1 mM Ala-13 deamidated fibers and 1 mM Val-17 unmodified monomers in buffer. 2D IR spectra were collected immediately following mixing, with a deadtime of ~5 minutes.
Methyl esterification of deamidated peptides
The methyl esterification procedure used to detect the presence of deamidation in the peptides was described by Raleigh and Tuong.20,64 Methanolic HCl was prepared by the addition of 5 mL acetyl chloride to 30 mL anhydrous methanol under nitrogen at 0°C. Following stirring for five minutes, 500 μL methanolic HCl was added to 10 nmol of dried peptide. The mixture was briefly vortexed and brought to room temperature. Aliquots of the reaction were removed at various time intervals and quenched with 4 times excess volume of water. After quenching, all aliquots were stored at 0°C. Samples were then dried by speed-vac and dissolved in methanol for MALDI-MS preparation. All aliquots were measured using MALDI-MS and the ratio of the peak intensity of the native peptide to the peak intensity of the methylated peptide allowed for determination of deamidation.
Results
Amylin deamidation shown by HPLC and methyl esterification reaction
Table 1 lists the percent deamidation calculated for each peptide sample. These quantities are taken from Figures S1a-d in the Supporting Information that show HPLC chromatograms of Ala-8, Ala-13, Leu-16, and Leu-27 when purified using acidic buffers. The characteristic hIAPP retention time is approximately 45 minutes on a 0–90%B gradient over 90 minutes, and the shoulder attributed to asparagine deamidated to aspartic acid elutes 1–2 minutes after the main peak. Figure S2a–d in the Supporting Information shows the peptide peaks enlarged. The presence of these two peaks suggests that hIAPP monomers that contained asparagine and isoaspartic acid separated from monomers containing aspartic acid. From the HPLC chromatograms of each peptide, the percent deamidation of each sample is measured. The ratio of the peak heights was used to compare the amount of aspartic acid products to the total amount of asparagine and isoaspartic acid products. Using the known literature ratio of isoaspartic acid:aspartic acid deamidation products, the percent deamidation of each of the four peptides was calculated.5 The rate of deamidation is not uniform for all residues in most proteins.5,24,65,66 The calculated percentage is the average over all the residues and we estimate 5% error in the calculations resulting from integration of the HPLC peaks. Figure S3 in the Supporting Information shows the HPLC chromatogram of Ala-13 purified using buffers at pH 5. Between 40 and 50 minutes, there are two small peaks, the second being the shoulder on the main peak at 50 minutes. These two smaller peaks are two different deamidation products, containing Asp and isoAsp in place of Asn in the peptide. The presence of two deamidated peaks is indicative that some of the Ala-13 monomers could have more deamidated residues than others, resulting in multiple deamidation peaks in the chromatogram.
Table 1.
Frequency of various features in 2D IR spectra and calculated percent deamidation from HPLC.
| Label | Frequency unmodified peptide isotope peak (cm−1) | Frequency unmodified peptide unlabeled peak (cm−1) | Frequency deamidated peptide isotope peak (cm−1) | Frequency deamidated peptide unlabeled peak (cm−1) | Frequency unmodified peptide unlabeled peak without coupling (cm−1)56 | % Deamidated (from HPLC) |
|---|---|---|---|---|---|---|
| A8 | 1590 | 1618 | ~1600 | 1617 | 1600 ±3 | 85 ±5 |
| A13 | 1579 | 1615 | ~1595 | 1620 | 1598 ±1 | 95 ±5 |
| L16 | 1581 | 1617 | 1585 | 1619 | 1588 ±1 | 69 ±5 |
| L27 | 1577 | 1614 | NA | 1620 | 1598±2 | 64 ±5 |
The methyl esterification reaction of amylin containing isotope labeled Ala-13 verifies the presence of a deamidated residue by converting any aspartic acid or isoaspartic acid residues to their methyl ester, which results in an increase in mass of 15 Da for each methyl group added.20,64 Human amylin contains no aspartic acid or glutamic acid residues and the C-terminus is amidated; therefore, the only sites at which this reaction can occur are at Asn or Gln residues that have deamidated. Although MALDI-MS cannot be used as a quantitative method for measuring the percent of peptide that is deamidated, the ratio of the intensity of the native peptide peak to the intensity of the methylated peptide peak should increase over the course of the reaction. In Supporting Information Figure S4 shows MALDI-MS spectra collected from peptide aliquots after 1 and 3 hours of the esterification reaction. In Supporting Information Figure S4a, the ratio of the peak at 3923 m/z (singly methylated hIAPP) to the peak at 3908 m/z (native hIAPP) after 1 hour of methylation is 0.29. In Supporting Information Figure S4b, the ratio of the peak at 3922 m/z to the peak at 3907 m/z after 3 hours is 0.64. In addition, after 3 hours, a peak appears at 3937, suggesting that given enough time, hIAPP becomes doubly methylated. The ratio of the intensities of the doubly methylated peak to the native hIAPP peak is 0.31. MALDI-MS spectra were collected for an aliquot that was given 7.5 hours to react, however, after that amount of time; the peptide was degraded (not shown). In addition, under these conditions and given enough time, the peptide may have started to aggregate, thus preventing some deamidation sites from becoming methylated.
Neither TEM nor CD spectroscopy reveal a change in fibril structure upon deamidation
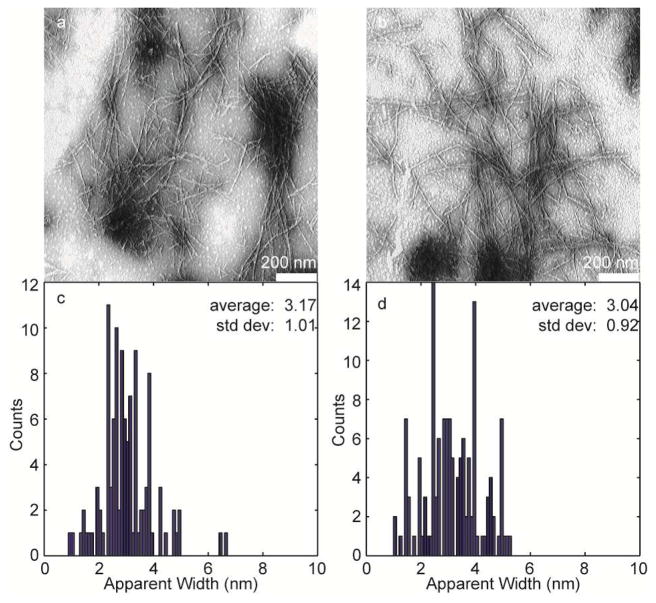
Deamidated hIAPP still aggregates into amyloid fibers as determined by TEM, which is shown in Figure 4a. These images were collected using samples of Ala-13 hIAPP, for which 95% of the peptides are deamidated (Table 1). For comparison, TEM of non-degraded hIAPP is shown in Figure 4b, prepared under identical conditions. To within the resolution of these TEM, we do not observe differences in morphologies between the two sets of fibers. A statistical analysis of the fiber full-width-at-half-maxima, place the widths of the deamidated fibers at 3.17 nm ± 1.01 nm (Figure 4c) and unmodified fibers at 3.04 nm ± 0.92 nm (Figure 4d).
Figure 4.
a. TEM image of deamidated Ala-13 fibers. b. TEM image of unmodified Ala-13 fibers. c. Statistical analysis of the apparent width of deamidated Ala-13 fibers. d. Statistical analysis of the apparent width of unmodified Ala-13 fibers. Scale bars on TEM represent 200 nm.
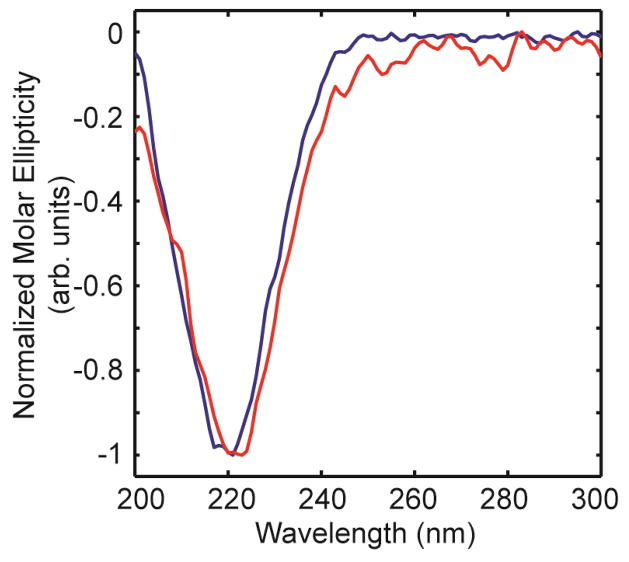
Circular dichroism (CD) spectroscopy is often used to measure the secondary structure content of amyloid proteins. Normalized CD spectra of fibers formed from unmodified and deamidated hIAPP fibers are shown in Figure 5. These fibers are made from the same Ala-13 labeled stock as used for the TEM. The CD spectra of the unmodified and deamidated fibers exhibit a single peak at 220 nm and no shoulder at 208 nm. The experimental spectra are typical of those reported for amylin fibers and indicate β-sheet secondary structure.
Figure 5.
Normalized CD spectra of deamidated Ala-13 (blue) and unmodified Ala-13 (red).
Aggregation kinetics experiments reveal a difference in aggregation times
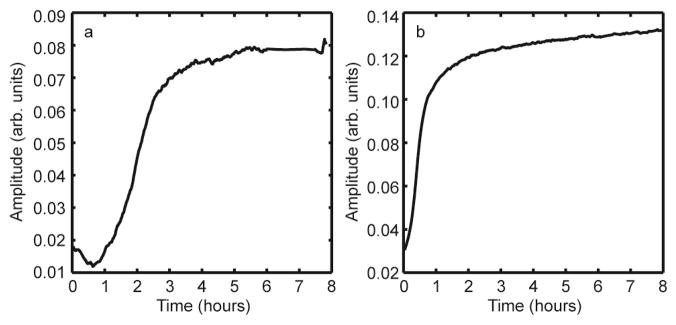
Shown in Figure 6 are kinetic traces for the aggregation of unmodified (Figure 6a) and deamidated (Figure 6b) Ala-13 hIAPP. The kinetic traces plot the increasing intensity of the fundamental of the amide I β-sheet peak that appears at 1615–1620 cm−1 in the 2D IR spectra. This peak intensity matches that of aggregation kinetics measured using the fluorescent dye ThT, so long as the dye binds to all β-sheets in the fiber.56,63 In the unmodified peptides, there is consistently a lag time of 30 minutes to 1 hour before folding commences, for all samples measured from multiple peptide syntheses. In contrast, there is no lag time in the deamidated sample. The half-rise time of the unmodified samples are >1 hour and for deamidated peptides it is 30 minutes.
Figure 6.
a. Aggregation kinetics of human amylin monitored by following the amide I peak intensity in unmodified amylin. b. Aggregation kinetics of human amylin monitored by following the amide I peak in deamidated amylin.
Isotope edited 2D IR reveals that deamidation alters the structure of the N-terminal β-sheet
The spectral features discussed in the introduction are illustrated in the 2D IR spectrum and diagonal slice in Figure 7a and b, which was collected on a sample of Ala-13 labeled hIAPP fibers formed from polypeptides that have not deamidated. The peak at ωpump=ωprobe=1615 cm−1 is due to the unlabeled amide I band of the parallel β-sheet amyloid fiber. The frequency of this peak can be as high as 1618 cm−1 for isotope labels that appear in the middle of the amyloid β-sheets.38,56 The peak also appears higher for structurally disordered fibers in which the excitonic coupling is reduced. The intensity around 1645 cm−1 is mostly a measure of the random coil regions of the fiber, such as from residues 1–7 that are prevented from forming β-structure by the disulfide bond and residues ~18–22 in the turn region.43,67 The isotope label for Ala-13 appears at 1579 cm−1. Spectra of isotope diluted fibers absorb at 1598 cm−1 (not shown).56 Thus, β=−10 cm−1, which is consistent with a well-ordered β-sheet.
Figure 7.
a. Diagonal slice through the 2D IR spectrum of unmodified Ala-13. b. 2D IR spectrum of unmodified Ala-13. c. Diagonal slice through the 2D IR spectrum of deamidated Ala-13. d. 2D IR spectrum of deamidated Ala-13.
Having outlined the spectral signatures for isotope labeled hIAPP, we now compare 2D IR spectra collected for fibers formed from normal and deamidated peptides. Shown in Figure 7, c and d, are a 2D IR spectrum and its corresponding diagonal slice for Ala-13 labeled hIAPP fibers formed from a sample in which 95% of the peptides are deamidated, as measured by HPLC (see Table I). In the 2D spectrum, the strong β-sheet mode from the unlabeled residues appears at 1620 cm−1. The higher frequency for the unlabeled β-sheet peak indicates that the β-sheets in the deamidated fibers are particularly structurally disordered. The exact interpretation of the unlabeled features is difficult because the location of the isotope labels alters the unlabeled vibrational modes.53 As for the Ala-13 isotope labeled peak in deamidated samples, it appears at 1595 cm−1. 1595 cm−1 matches the frequency of the isotopically diluted sample, which indicates that the isotope labeled amide I peak is not shifted due to excitonic coupling.56 Consequently, the strong coupling caused by parallel β-sheets is not present at the Ala-13 position in the deamidated sample. Therefore, even though amyloid fibers are formed as indicated by the 1620 cm−1 mode (and TEM and CD spectroscopy), the lack of the isotope frequency shift indicates that Ala-13 does not contribute to in-register parallel β-sheets in the deamidated sample. Exact values of the isotope and unlabeled β-sheet peaks in both the unmodified and deamidated spectra are given in Table 1. The linewidths and lineshapes of the isotope label were not measured in this set of experiments. The frequency assignments in Table 1 were made by inspection.
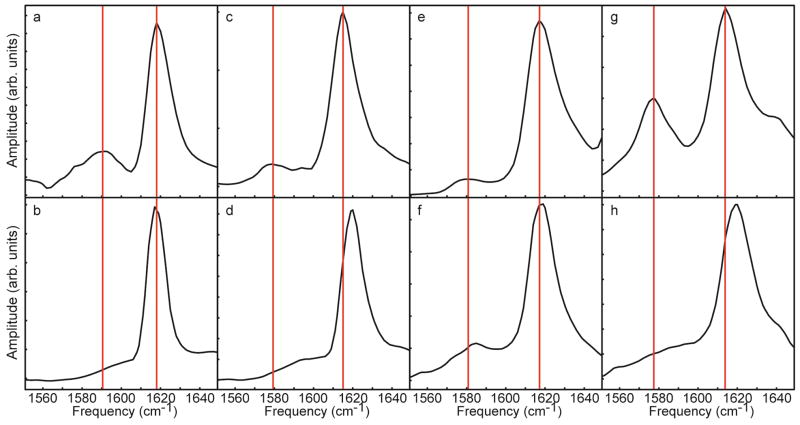
Similar experiments were performed for peptides isotope labeled at Ala-8, Leu-16 and Leu-27. Figure 8 shows slices through the diagonal of 2D IR spectra taken of fibers formed from both unmodified and deamidated samples. The full 2D IR spectra are given in Supporting Information Figures S5 and S6. As discussed for Ala-13 above, all the samples have a strong amide I band near 1620 cm−1 that is the result of the unlabeled β-sheet while the isotope labels appear at 1579–1590 cm−1 when coupled and 1585–1600 cm−1 when not coupled. The range for these values spans ~15 cm−1 due to the variety of locations of the isotope labels in the peptide. Ala-13 and Leu-27 are some of the most ordered amino acids in the entire peptide, while Ala-8, adjacent to the disulfide bond is very disordered. The difference in the structure of hIAPP at these residues explains the large span of values for the isotope label frequency in the unmodified and deamidated spectra.
Figure 8.
Slices through 2D IR spectra. a. Unmodified Ala-8. b. Deamidated Ala-8. c. Unmodified Ala-13. d. Deamidated Ala-13. e. Unmodified Leu-16. f. Deamidated Leu-16. g. Unmodified Leu-27. h. Deamidated Leu-27. Red lines are included as a guide to the eye for the frequencies of the unlabeled amide I and isotope label peaks.
The red lines in Figure 8 mark the frequencies at which the isotope label and unlabeled β-sheet appear in the unmodified samples. For Ala-8 and Leu27, the data are similar to Ala-13. That is, the frequency of the isotope label in the deamidated samples appears at a much higher frequency and the peak is much weaker than in the spectrum of the unmodified peptide. Rather than the isotope label forming a discrete peak, it now becomes a shoulder on the side of the 1620 cm−1 β-sheet mode. The higher frequency and weaker intensity for these residues is a clear indication that the coupling between the isotope labels is much weaker than in the unmodified samples, which is again consistent with the loss of parallel β-sheet structure at these positions. That is not the case for Leu-16, for which there is only a 5 cm−1 frequency shift and the isotope label is still intense and well resolved in the deamidated spectrum. Thus, the coupling between Leu-16 residues is still significant for fibers formed from deamidated samples and the data are consistent with the deamidated samples containing parallel β-sheets at Leu-16. Comparing the unlabeled β-sheet peaks, in each sample there is a large amount of amyloid β-sheet secondary structure, evidenced by the lack of large random coil features at 1640 cm−1. However, as in the case of Ala-13, the unlabeled β-sheet peak in Leu-27 is shifted to higher frequency (see Table 1), indicating that the β-sheets in the Leu-27 fibers are more structurally disordered.
Deamidated fibers seed amyloid formation by unmodified amylin
A seeding experiment was performed to determine if deamidated fibers could nucleate unmodified monomers.10,20 These experiments were carried out using seeds created from deamidated peptides mixed with unmodified peptides isotope labeled at Val-17 in a ratio of 1:10. The Val-17 label was chosen in order to probe the structure of the seeded fibers near the location in which the deamidated fibers are known to possess parallel β-sheets (Figure 2). The seeds were created from deamidated peptides labeled at Ala-13, for which we know from HPLC that 95% of the peptides are deamidated and that the Ala-13 peak is very weak and broad (Figure 7c and d). Thus, the Ala-13 peak intensity is negligible in the interpretation of the 2D IR spectra presented below.
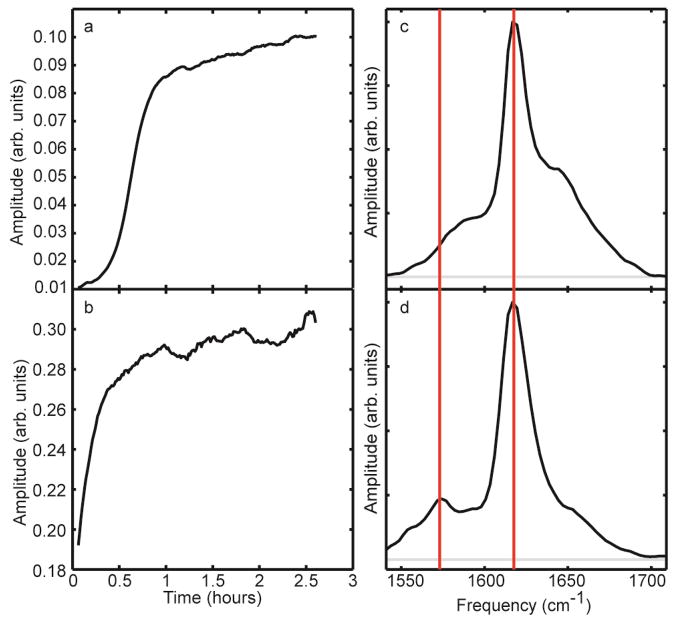
Shown in Figure 9a and b are kinetic traces that measure β-sheet formation of seeded and unseeded aggregation. These traces are made by plotting the intensity of the unlabeled β-sheet at 1620 cm−1 over the course of aggregation. Seeding eliminates the lag time, which indicates that the seeds from deamidated peptides do indeed nucleate unmodified peptides into amyloid fibers. Figure 9c and d shows slices through the 2D IR spectrum immediately after seeding and when the fibers have equilibrated at 2.5 hours. In the time-zero spectra, when less than half of the peptides are incorporated into fibers, the Val-17 isotope label is very broad and absorbs near 1590 cm−1, consistent with most peptides adopting random coil structures in solution. In contrast, a very prominent and sharp isotope labeled peak appears in the equilibrated spectra at 1574 cm−1, indicating that Val-17 residues, once they are nucleated into amyloid fibers, lie in-register by adopting a parallel β-sheet formation. Thus, unmodified peptides can be nucleated to form fibers by deamidated seeds, and the seeded fibers have amyloid β-sheets at Val-17, which is in close proximity to the β-sheet region in deamidated fibers.
Figure 9.
a. Aggregation kinetics of unseeded Val-17 amide I folding monitored by following the amide I peak intensity in unmodified amylin. b. Aggregation kinetics trace of seeded Val-17 amide I folding monitored by following the amide I peak intensity. c. Slice through the diagonal of a 2D IR spectrum of unmodified Val-17 monomers seeded with deamidated Ala-13 fibers, averaged over the first 8 minutes following mixing. d. Slice through the diagonal of a 2D IR spectrum averaged over 8 minutes 2.5 hours after mixing.
Discussion
Ascertaining the effect of deamidation on the structure of amyloid fibers is difficult. In fact, even recognizing that a polypeptide is deamidated is not easy. Deamidated amylin still aggregates into amyloid fibers, and the CD maximum of fibers formed from deamidated polypeptides are indistinguishable from unmodified peptides, as are the fiber widths measured by TEM. Deamidated polypeptides can be identified by HPLC purification and by mass spectrometry, but the differences are slight since the HPLC retention time is only slightly diminished and deamidation only produces a mass change of 1 Da.20 Using 2D IR spectroscopy and isotope labeling, we are able to identify the changes deamidation induces to the amylin fibril structure. Moreover, we are able to probe the effect on fibril structure residue-by-residue as well as measure differences in fiber kinetics. Thus, we obtain insight into the effect of deamidation on the structure and folding kinetics of amylin fibers.
Shown in Figure 2b is a model for the fibrillar structure as determined by Tycko and co-workers using solid-state NMR.43 Possible deamidation sites are shown in red and isotope labeled positions are shown in blue. The isotope labeled amino acids have been chosen to probe the inner and outer β-sheets, as well as near the disordered loop. The fibers in the Tycko study and ours probably differ in the number of stacked columns of peptides, but not in the locations of the β-sheets in each column. The parallel β-sheet structure of the fibers at Ala-8 and Ala-13 are broken, which lie adjacent to or 1 residue away from a potential deamidation site. HPLC traces show that in samples of Ala-8 and Ala-13, nearly all of the peptides have some degree of deamidation. Thus, deamidation may cause a local loss of β-sheet structure due to puckering of the backbone with the addition of the extra CH2 group. However, deamidation may be occurring in the C-terminal β-sheet as well. Leu-27 is positioned 3 or 4 residues away from the nearest possible deamidation site in the sequence (Figure 2). The study of deamidation of pramlintide showed that the residues with the highest degree of deamidation are Asn-21 and Asn-35.22 Although those residues lie further from Leu-27, they may be more likely to deamidate. Deamidation of asparagine causes a negative charge, which may be the cause of this long-range effect.
The difference in lag times between unmodified Ala-13 and deamidated Ala-13 (Figure 6) is consistent with experiments on deamidated Aβ. ThT fluorescence experiments were performed on four different Aβ variants and compared to the fluorescence of native Aβ.13 While one mutation, Aβ1–42(L-isoAsp-23), produced more extensive aggregation over 24 hours, as shown as being almost twice as fluorescent at the end of the experiment, that and two other mutations, Aβ1–42(isoAsp-7) and Aβ1–42(E22Q), aggregated faster than wild-type Aβ. Although the three variants aggregated faster than wild-type Aβ, the E22Q mutation was by far the fastest. This mutant maximized ThT fluorescence after only 10 hours, compared to 20 hours for that of the wild-type. It is thought that the modifications at residues 22 and 23, located directly in the turn region, enhance aggregation due to the conformational changes that they cause. Asn-21 and Asn-22, in the turn region of hIAPP could be deamidated, thus promoting a shorter lag time and faster folding in the deamidated sample.
While deamidation eliminates the N-terminal β-sheet between Ala-8 and Ala-13, as well as the C-terminal sheet near Leu-27, parallel β-sheets are still present in the deamidated fibers near Leu-16. Moreover, deamidated fibers can seed β-sheet formation. When unmodified Val-17 monomers are added to deamidated Ala-13 seeds, Val-17 fibers form faster than if they were unseeded. However, the structure of the deamidated fibers does not necessarily propagate to the unmodified monomers. After equilibration of Val-17 fiber formation, Val-17 is strongly coupled (Figure 9d), suggesting that at Val-17, ordered, parallel β-sheets are formed. Val-17 lies adjacent to Leu-16, the residue that still retains some parallel β-sheet structure upon deamidation. Thus, while the β-sheet regions of deamidated fibers are much reduced, the β-sheet content is still sufficient to nucleate additional monomers. In addition, the unmodified fibers that are formed are more ordered than the deamidated seeds. This finding is interesting in light of the mechanism of amylin fiber formation. Previous studies have shown that amylin fibers nucleate somewhere in the disordered loop between Val-17 and Ala-25, with nucleation at Val-17 occurring faster than Ala-25.38 Thus, even though deamidation decreases overall β-sheet content, deamidated peptides still retain the most critical region for nucleation. It is also interesting to note that the 20–29 fragment of amylin that shares a similar sequence range is induced to aggregate by as little as 5% deamidation impurity.20 Thus, we hypothesize that in vivo post translational deamidation, if it occurs, still leads to amyloid deposits with much the same structure.
Conclusions
We find that deamidation has a major structural influence of fiber structure, by eliminating half of the N-terminal β-sheet and either disrupting the C-terminal β-sheet near Leu-27 or eliminating it entirely. The one region that it does not disrupt is the β-sheet structure is near Leu-16 and Val-17, which is why fibers formed from deamidated samples can still seed fiber formation of unmodified peptides. As we show, deamidation is very difficult to detect by either, CD spectroscopy or TEM. It is also not readily apparent in the FTIR or 2D IR spectra of un-isotope labeled peptides. Deamidation occurs spontaneously, even when peptides are carefully stored. As shown by HPLC, the peptides used in this study all displayed varying amounts of deamidation. However, all samples were synthesized, purified, stored, and aged in the same way. We therefore assume that while some samples may be more deamidated than others, the peptides are all undergoing the same deamidation process and the same residues are deamidating in each sample. Thus, our studies emphasize the need for careful monitoring of sample purity and the utility of isotope edited IR spectroscopy to not only easily identify sample degradation, but to also provide site-specific structural information on the structural consequences.
Supplementary Material
Acknowledgments
Support for this research was provided by the NSF GRFP through grant DGE-0718123 for L.E.B., and the NIH through grant GM078114 for P.M., P.C., and D.P.R. and DK79895 for E.B.D. and M.T.Z. We thank Randall Massey at the University of Wisconsin Medical School Electron Microscope Facility for help with TEM and Lisa Johnson for help with CD spectroscopy. We also thank Dr. Robert Tycko for the coordinates for his structural model of human amylin fibers.
Footnotes
Associated Content
Supporting information for this article includes HPLC chromatograms of deamidated peptides. Also included are MALDI-MS spectra from the methyl esterification reaction. 2D IR spectra and associated diagonal slices are also included for all unmodified and deamidated samples. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Walsh CT, Garneau-Tsodikova S, Gatto GJ. Angew Chem Int Ed. 2005;44:7342–7372. doi: 10.1002/anie.200501023. [DOI] [PubMed] [Google Scholar]
- 2.Mann M, Jensen ON. Nat Biotechnol. 2003;21:255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 3.Witze ES, Old WM, Resing KA, Ahn NG. Nat Methods. 2007;4:798–06. doi: 10.1038/nmeth1100. [DOI] [PubMed] [Google Scholar]
- 4.Bischoff R, Kolbe HVJ. J Chromatogr B: Biomed Sci App. 1994;662:261–78. doi: 10.1016/0378-4347(94)00203-7. [DOI] [PubMed] [Google Scholar]
- 5.Geiger T, Clarke S. J Biol Chem. 1987;262:785–94. [PubMed] [Google Scholar]
- 6.Shimizu T, Matsuoka Y, Shirasawa T. Biol Pharm Bull. 2005;28:1590–1596. doi: 10.1248/bpb.28.1590. [DOI] [PubMed] [Google Scholar]
- 7.Abedini A, Gupta R, Marek P, Meng F, Raleigh DP, Taskent H, Tracz S. Protein Misfolding Diseases. John Wiley & Sons, Inc; 2010. pp. 131–144. [Google Scholar]
- 8.Shimizu T, Watanabe A, Ogawara M, Mori H, Shirasawa T. Arch Biochem Biophys. 2000;381:225–234. doi: 10.1006/abbi.2000.1955. [DOI] [PubMed] [Google Scholar]
- 9.Takata T, Oxford JT, Demeler B, Lampi KJ. Protein Sci. 2008;17:1565–1575. doi: 10.1110/ps.035410.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nilsson MR, Dobson CM. Protein Sci. 2003;12:2637–2641. doi: 10.1110/ps.0360403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roher AE, Lowenson JD, Clarke S, Wolkow C, Wang R, Cotter RJ, Reardon IM, Zürcher-Neely HA, Heinrikson RL, Ball MJ. J Biol Chem. 1993;268:3072–3083. [PubMed] [Google Scholar]
- 12.Xie M, Schowen RL. J Pharm Sci. 1999;88:8–13. doi: 10.1021/js9802493. [DOI] [PubMed] [Google Scholar]
- 13.Shimizu T, Fukuda H, Murayama S, Izumiyama N, Shirasawa T. J Neurosci Res. 2002;70:451–461. doi: 10.1002/jnr.10350. [DOI] [PubMed] [Google Scholar]
- 14.Fonseca MI, Head E, Velazquez P, Cotman CW, Tenner AJ. Exp Neurol. 1999;157:277–288. doi: 10.1006/exnr.1999.7058. [DOI] [PubMed] [Google Scholar]
- 15.Sargaeva NP, Lin C, O’Connor PB. Anal Chem. 2009;81:9778–9786. doi: 10.1021/ac901677t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flaugh SL, Mills IA, King J. J Biol Chem. 2006;281:30782–30793. doi: 10.1074/jbc.M603882200. [DOI] [PubMed] [Google Scholar]
- 17.Hanson SRA, Smith DL, Smith JB. Exp Eye Res. 1998;67:301–312. doi: 10.1006/exer.1998.0530. [DOI] [PubMed] [Google Scholar]
- 18.Robinson NE, Robinson ML, Schulze SES, Lai BT, Gray HB. Protein Sci. 2009;18:1766–1773. doi: 10.1002/pro.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nilsson MR. Amyloid Proteins. Wiley-VCH Verlag GmbH; 2008. pp. 81–109. [Google Scholar]
- 20.Nilsson MR, Driscoll M, Raleigh DP. Protein Sci. 2002;11:342–349. doi: 10.1110/ps.48702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nonoyama A, Laurence JS, Garriques L, Qi H, Le T, Middaugh CR. J Pharm Sci. 2008;97:2552–2567. doi: 10.1002/jps.21197. [DOI] [PubMed] [Google Scholar]
- 22.Hekman C, DeMond W, Dixit T, Mauch S, Nuechterlein M, Stepanenko A, Williams JD, Ye M. Pharm Res. 1998;15:650–658. doi: 10.1023/a:1011934829263. [DOI] [PubMed] [Google Scholar]
- 23.Hekman CM, DeMond WS, Kelley PJ, Mauch SF, Williams JD. J Pharm Biomed Anal. 1999;20:763–772. doi: 10.1016/s0731-7085(99)00075-8. [DOI] [PubMed] [Google Scholar]
- 24.Robinson NE, Robinson AB. Proc Natl Acad Sci U S A. 2001;98:944–949. doi: 10.1073/pnas.98.3.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson NE, Robinson AB. Proc Natl Acad Sci U S A. 2001;98:4367–4372. doi: 10.1073/pnas.071066498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hutton JC. Diabetologia. 1989;32:271–281. doi: 10.1007/BF00265542. [DOI] [PubMed] [Google Scholar]
- 27.Cooper GJS. Endocrine Reviews. 1994;15:163–201. doi: 10.1210/edrv-15-2-163. [DOI] [PubMed] [Google Scholar]
- 28.Cooper GJS, Willis AC, Clark A, Turner RC, Sim RB, Reid KBM. Proc Natl Acad Sci U S A of the United States of America. 1987;84:8628–8632. doi: 10.1073/pnas.84.23.8628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Westermark P, Wernstedt C, Wilander E, Hayden DW, Obrien TD, Johnson KH. Proc Natl Acad Sci U S A. 1987;84:3881–3885. doi: 10.1073/pnas.84.11.3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishi M, Sanke T, Nagamatsu S, Bell GI, Steiner DF. J Biol Chem. 1990;265:4173–4176. [PubMed] [Google Scholar]
- 31.Kahn SE, Andrikopoulos S, Verchere CB. Diabetes. 1999;48:241–253. doi: 10.2337/diabetes.48.2.241. [DOI] [PubMed] [Google Scholar]
- 32.Clark A, Cooper GJS, Morris JF, Lewis CE, Willis AC, Reid KBM, Turner RC. Lancet. 1987;2:231–234. doi: 10.1016/s0140-6736(87)90825-7. [DOI] [PubMed] [Google Scholar]
- 33.Hull RL, Westermark GT, Westermark P, Kahn SE. J Clin Endocrinol Metab. 2004;89:3629–3643. doi: 10.1210/jc.2004-0405. [DOI] [PubMed] [Google Scholar]
- 34.Marzban L, Park K, Verchere CB. Exp Gerontol. 2003;38:347–351. doi: 10.1016/s0531-5565(03)00004-4. [DOI] [PubMed] [Google Scholar]
- 35.Lorenzo A, Razzaboni B, Weir GC, Yankner BA. Nature. 1994;368:756–760. doi: 10.1038/368756a0. [DOI] [PubMed] [Google Scholar]
- 36.Clark A, Wells CA, Buley ID, Cruickshank JK, Vanhegan RI, Matthews DR, Cooper GJS, Holman RR, Turner RC. Diabetes Res Clin Exp. 1988;9:151–159. [PubMed] [Google Scholar]
- 37.Butler AE, Janson J, Bonner-Weir S, Ritzel R, Rizza RA, Butler PC. Diabetes. 2003;52:102–110. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- 38.Shim SH, Gupta R, Ling YL, Strasfeld DB, Raleigh DP, Zanni MT. Proc Natl Acad Sci U S A. 2009;106:6614–6619. doi: 10.1073/pnas.0805957106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L, Middleton CT, Singh S, Reddy AS, Woys AM, Strasfeld DB, Marek P, Raleigh DP, de Pablo JJ, Zanni MT, Skinner JL. J Am Chem Soc. 2011;133:16062–16071. doi: 10.1021/ja204035k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dupuis NF, Wu C, Shea JE, Bowers MT. J Am Chem Soc. 2011;133:7240–7243. doi: 10.1021/ja1081537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dupuis NF, Wu C, Shea JE, Bowers MT. J Am Chem Soc. 2009;131:18283–18292. doi: 10.1021/ja903814q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Apostolidou M, Jayasinghe SA, Langen R. J Biol Chem. 2008;283:17205–17210. doi: 10.1074/jbc.M801383200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luca S, Yau WM, Leapman R, Tycko R. Biochemistry. 2007;46:13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sellin D, Yan LM, Kapurniotu A, Winter R. Biophys Chem. 2010;150:73–79. doi: 10.1016/j.bpc.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 45.Potter KJ, Abedini A, Marek P, Klimek AM, Butterworth S, Driscoll M, Baker R, Nilsson MR, Warnock GL, Oberholzer J, Bertera S, Trucco M, Korbutt GS, Fraser PE, Raleigh DP, Verchere CB. Proc Natl Acad Sci U S A. 2010;107:4305–4310. doi: 10.1073/pnas.0909024107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Westermark GT, Westermark P, Berne C, Korsgren O. N Engl J Med. 2008;359:977–979. doi: 10.1056/NEJMc0802893. [DOI] [PubMed] [Google Scholar]
- 47.Koo BW, Hebda JA, Miranker AD. Protein Eng, Des Sel. 2008;21:147–154. doi: 10.1093/protein/gzm076. [DOI] [PubMed] [Google Scholar]
- 48.Kim YS, Liu L, Axelsen PH, Hochstrasser RM. Proc Natl Acad Sci U S A. 2008;105:7720–7725. doi: 10.1073/pnas.0802993105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moran SD, Woys AM, Buchanan LE, Bixby E, Decatur SM, Zanni MT. Proc Natl Acad Sci U S A. 2012;109:3329–3334. doi: 10.1073/pnas.1117704109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buchanan L, Dunkelberger EB, Zanni MT, editors. Examining Amyloid Structure and Kinetics with 1D and 2D Infrared Spectroscopy and Isotope Labeling. 1. Vol. 1 Springer; Heidelberg: 2011. [Google Scholar]
- 51.Moran A, Mukamel S. Proc Natl Acad Sci U S A. 2004;101:506–510. doi: 10.1073/pnas.2533089100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Decatur SM. Acc Chem Res. 2006;39:169–175. doi: 10.1021/ar050135f. [DOI] [PubMed] [Google Scholar]
- 53.Strasfeld DB, Ling YL, Gupta R, Raleigh DP, Zanni MT. J Phys Chem B. 2009;113:15679–15691. doi: 10.1021/jp9072203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim YS, Liu L, Axelsen PH, Hochstrasser RM. Proc Natl Acad Sci U S A. 2009;106:17751–17756. doi: 10.1073/pnas.0909888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Woys AM, Lin YS, Reddy AS, Xiong W, de Pablo JJ, Skinner JL, Zanni MT. J Am Chem Soc. 2010;132:2832–2838. doi: 10.1021/ja9101776. [DOI] [PubMed] [Google Scholar]
- 56.Middleton CT, Marek P, Cao P, Chiu C-c, Singh S, Woys AM, de Pablo JJ, Raleigh DP, Zanni MT. Nat Chem. 2012;4:355–360. doi: 10.1038/nchem.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Middleton CT, Woys AM, Mukherjee SS, Zanni MT. Methods. 2010;52:12–22. doi: 10.1016/j.ymeth.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marek P, Woys AM, Sutton K, Zanni MT, Raleigh DP. Org Lett. 2010;12:4848–4851. doi: 10.1021/ol101981b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abedini A, Raleigh DP. Org Lett. 2005;7:693–696. doi: 10.1021/ol047480+. [DOI] [PubMed] [Google Scholar]
- 60.Shim SH, Strasfeld DB, Fulmer EC, Zanni MT. Opt Lett. 2006;31:838–840. doi: 10.1364/ol.31.000838. [DOI] [PubMed] [Google Scholar]
- 61.Shim SH, Strasfeld DB, Zanni MT. Opt Express. 2006;14:13120–13130. doi: 10.1364/oe.14.013120. [DOI] [PubMed] [Google Scholar]
- 62.Marek P, Gupta R, Raleigh DP. ChemBioChem. 2008;9:1372–1374. doi: 10.1002/cbic.200800052. [DOI] [PubMed] [Google Scholar]
- 63.Strasfeld DB, Ling YL, Shim SH, Zanni MT. J Am Chem Soc. 2008;130:6698–6699. doi: 10.1021/ja801483n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tuong A, Maftouh M, Ponthus C, Whitechurch O, Roitsch C, Picard C. Biochemistry. 1992;31:8291–8299. doi: 10.1021/bi00150a024. [DOI] [PubMed] [Google Scholar]
- 65.Robinson NE, Robinson AB. Proc Natl Acad Sci U S A. 2001;98:12409–12413. doi: 10.1073/pnas.221463198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tyler-Cross R, Schirch V. J Biol Chem. 1991;266:22549–22556. [PubMed] [Google Scholar]
- 67.Wiltzius JJW, Sievers SA, Sawaya MR, Cascio D, Popov D, Riekel C, Eisenberg D. Protein Sci. 2008;17:1467–1474. doi: 10.1110/ps.036509.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.