Abstract
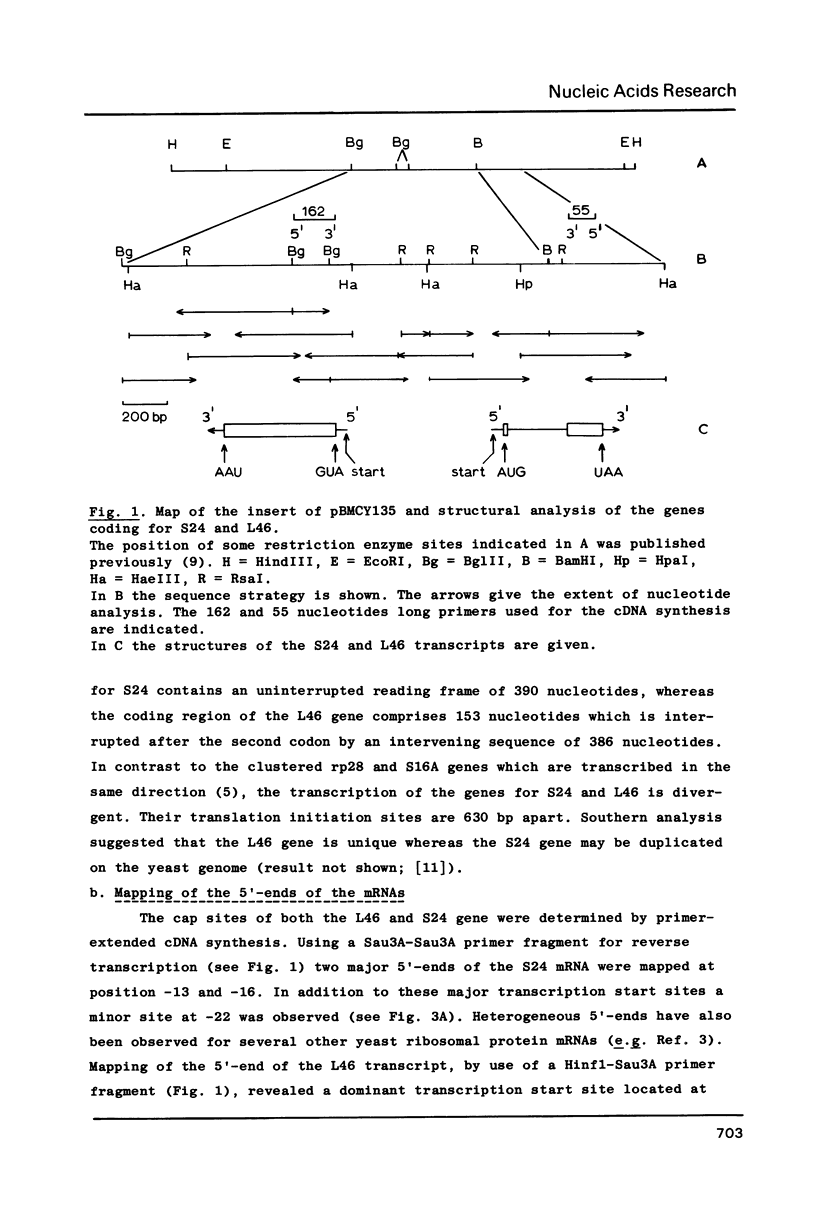
Unlike most yeast ribosomal protein genes studied so far the genes coding for S24 and L46 are adjacent on the genome. Sequence analysis showed that the two genes are transcribed divergently, their initiation codons being 630 bp apart. Taking the respective ATG translation start sites as reference points, the 5'- end of L46 mRNA was mapped at position -26, while the S24 mRNA showed two major 5'-ends mapping at positions -13 and -16 respectively. Unlike most other yeast ribosomal protein genes, the gene for S24 does not contain an intron. Its coding region encompasses 390 nucleotides encoding a protein of 14762 D. The gene for L46 on the other hand is split by an intron of 386 nucleotides starting after its second codon. This gene encodes a small, very basic protein having a molecular weight of 6334 D. Yeast ribosomal proteins S24 and L46 show striking homologies with ribosomal proteins from other organisms. In particular, yeast L46 is clearly the evolutionary counterpart of rat liver L39. A search of the intergenic region for sequence elements previously identified as common to most yeast ribosomal protein genes, revealed the presence of a single conserved box (RPG-box) roughly equidistant from the transcription initiation sites of both genes. We suggest that this box acts as a regulatory signal in either orientation and thus influences the expression of both genes simultaneously.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bollen G. H., Cohen L. H., Mager W. H., Klaassen A. W., Planta R. J. Isolation of cloned ribosomal protein genes from the yeast Saccharomyces carlsbergensis. Gene. 1981 Sep;14(4):279–287. doi: 10.1016/0378-1119(81)90160-8. [DOI] [PubMed] [Google Scholar]
- Bollen G. H., Mager W. H., Planta R. J. High resolution mini-two-dimensional gel electrophoresis of yeast ribosomal proteins. A standard nomenclature for yeast ribosomal proteins. Mol Biol Rep. 1981 Nov 30;8(1):37–44. doi: 10.1007/BF00798383. [DOI] [PubMed] [Google Scholar]
- Bollen G. H., Molenaar C. M., Cohen L. H., van Raamsdonk-Duin M. M., Mager W. H., Planta R. J. Ribosomal protein genes of yeast contain intervening sequences. Gene. 1982 Apr;18(1):29–37. doi: 10.1016/0378-1119(82)90053-1. [DOI] [PubMed] [Google Scholar]
- Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981 Apr;24(1):251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- Langford C. J., Gallwitz D. Evidence for an intron-contained sequence required for the splicing of yeast RNA polymerase II transcripts. Cell. 1983 Jun;33(2):519–527. doi: 10.1016/0092-8674(83)90433-6. [DOI] [PubMed] [Google Scholar]
- Langford C. J., Klinz F. J., Donath C., Gallwitz D. Point mutations identify the conserved, intron-contained TACTAAC box as an essential splicing signal sequence in yeast. Cell. 1984 Mar;36(3):645–653. doi: 10.1016/0092-8674(84)90344-1. [DOI] [PubMed] [Google Scholar]
- Leer R. J., van Raamsdonk-Duin M. M., Hagendoorn M. J., Mager W. H., Planta R. J. Structural comparison of yeast ribosomal protein genes. Nucleic Acids Res. 1984 Sep 11;12(17):6685–6700. doi: 10.1093/nar/12.17.6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leer R. J., van Raamsdonk-Duin M. M., Mager W. H., Planta R. J. The primary structure of the gene encoding yeast ribosomal protein L16. FEBS Lett. 1984 Oct 1;175(2):371–376. doi: 10.1016/0014-5793(84)80771-1. [DOI] [PubMed] [Google Scholar]
- Leer R. J., van Raamsdonk-Duin M. M., Molenaar C. M., Cohen L. H., Mager W. H., Planta R. J. The structure of the gene coding for the phosphorylated ribosomal protein S10 in yeast. Nucleic Acids Res. 1982 Oct 11;10(19):5869–5878. doi: 10.1093/nar/10.19.5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leer R. J., van Raamsdonk-Duin M. M., Schoppink P. J., Cornelissen M. T., Cohen L. H., Mager W. H., Planta R. J. Yeast ribosomal protein S33 is encoded by an unsplit gene. Nucleic Acids Res. 1983 Nov 25;11(22):7759–7768. doi: 10.1093/nar/11.22.7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A., McNally J., Wool I. G. The primary structure of rat liver ribosomal protein L39. J Biol Chem. 1984 Jan 10;259(1):487–490. [PubMed] [Google Scholar]
- Miller A. M. The yeast MATa1 gene contains two introns. EMBO J. 1984 May;3(5):1061–1065. doi: 10.1002/j.1460-2075.1984.tb01927.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar C. M., Woudt L. P., Jansen A. E., Mager W. H., Planta R. J., Donovan D. M., Pearson N. J. Structure and organization of two linked ribosomal protein genes in yeast. Nucleic Acids Res. 1984 Oct 11;12(19):7345–7358. doi: 10.1093/nar/12.19.7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaka E., Higo K., Itoh T. Yeast ribosomal proteins: VII. Cytoplasmic ribosomal proteins from Schizosaccharomyces pombe. Mol Gen Genet. 1983;191(3):519–524. doi: 10.1007/BF00425772. [DOI] [PubMed] [Google Scholar]
- Otaka E., Higo K., Osawa S. Isolation of seventeen proteins and amino-terminal amino acid sequences of eight proteins from cytoplasmic ribosomes of yeast. Biochemistry. 1982 Sep 14;21(19):4545–4550. doi: 10.1021/bi00262a005. [DOI] [PubMed] [Google Scholar]
- Padgett R. A., Konarska M. M., Grabowski P. J., Hardy S. F., Sharp P. A. Lariat RNA's as intermediates and products in the splicing of messenger RNA precursors. Science. 1984 Aug 31;225(4665):898–903. doi: 10.1126/science.6206566. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Krainer A. R., Maniatis T., Green M. R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984 Aug;38(1):317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]



