Abstract
Analyses of the human genome have proven extremely successful in identifying changes that contribute to human disease. Genetically engineered mice provide a powerful tool to analyze these changes, although they are slow and costly and do not always recapitulate human biology. Recent advances in genomic technologies, rodent-modeling approaches, and the production of patient-derived reprogrammed cell lines now provide a plethora of complementary systems to study disease states and test new therapies. Continued evolution and integration of these model systems will be the key to realizing the benefits of the genomic revolution and refining our understanding and treatment of human diseases.
Introduction
There is no broader topic in mammalian genetic research than human disease. Literally thousands of human diseases, disorders, and syndromes have been described and, through advances in genome technologies, are being characterized in much greater molecular detail. The fundamental challenge for medical research is to develop and exploit the most relevant and predictive model systems to understand the physiological impact of this genetic variation, with the ultimate goal of improving patient care and treatment. Already in the past two decades, the range of approaches used to study gene function in normal and diseased states has increased dramatically. However, as our depth of knowledge has grown so too have our expectations—both in the accuracy of experimental models to recapitulate human conditions as well as in the speed and scope at which they can be applied to ask biological questions.
These expectations are now being met through new approaches that integrate existing models systems with powerful new genetic technologies, thereby changing the scale and depth at which we can understand the genetics of disease. Although many effective model systems exist, we focus our discussion predominantly on the laboratory mouse and new technologies and approaches in mouse genetics that will have a major impact on disease research in the next 10 years. In addition, we highlight efforts to translate advances in modeling disease in mice to other mammalian systems and the development of the next generation of rodent and human genetic models. Combined, these efforts will ensure that biologists keep pace with the flood of information arising from genomic data and lead to quantum leaps in our understanding of disease and the development of new therapeutic strategies.
The Powerhouse Mouse
For more than 100 years, the laboratory mouse (Mus musculus) has served as the backbone of mammalian genetic research.
Mice are small, inexpensive to house, breed quickly, are amenable to genetic manipulation, and, importantly, share almost 99% of their genes with humans. To disease research, the mouse serves as both an investigative tool—defining the role of individual genes in disease manifestation and progression—and a discovery tool—allowing the identification of previously uncharacterized genes and molecular interactions that underlie disease states.
Step-wise advances in mouse embryonic stem cell (ESC) culture and homologous recombination-mediated targeting over the past 20 years have led to the creation of an extensive toolkit with which to manipulate the mouse genome (Figure 1). Through the adaptation of regulated promoters and DNA recombinase systems (Flp/FRT and Cre/LoxP), it is now possible to constitutively or inducibly mutate or delete (knockout) individual genes or large genomic regions in a temporal and cell-type-dependent manner (reviewed in Frese and Tuveson, 2007). Deletion of many thousands of genes in mice by this approach has been critical for the functional annotation of genes in normal and disease states and established model systems for preclinical treatment studies.
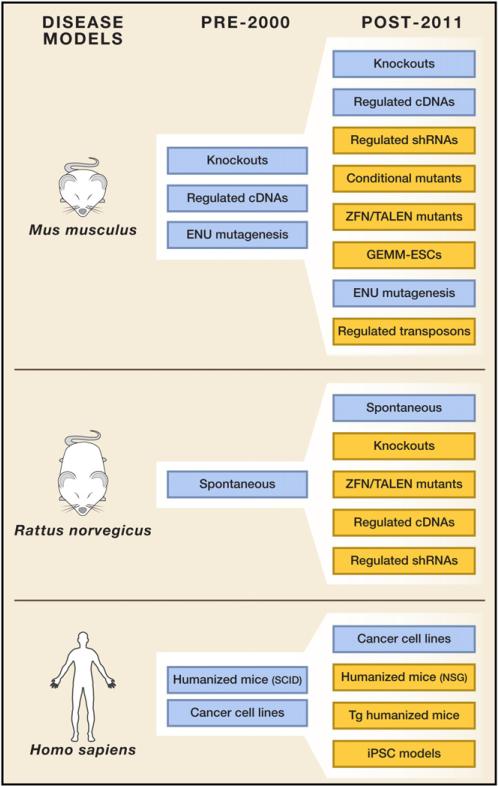
Figure 1. New Tools in Mammalian Disease Models.
Platforms for the genetic manipulation of mammalian model systems (primarily mouse, rat, and human) have developed at an incredible rate in the past 10–15 years. Recent sophisticated adaptations in each of these systems (indicated in yellow) now provide an extremely flexible array of tools for investigating and understanding the underlying causes of human disease.
However powerful, the process to generate a knockout or conditional knockout/mutant mouse is slow and expensive. It has been estimated that the development of a single targeted knockout mouse can cost $30,000–$100,000 and take up to 1 year to complete—many months longer if the resulting allele needs to be recombined in vivo or backcrossed to a specific strain (Figure 2). It is not only the time and cost that is restrictive. Unlike “lower” genetic model systems such as the worm and fly, the labor associated with production of genetically engineered mice (GEM) usually limits the development of new models to only a handful of genes/lab/year, if any at all. In stark contrast, the accessibility of genome-wide analysis tools (micro-array, ChIPseq, CGH), large-scale genome-wide association studies (GWAS), and mutagenesis and complex library screens (discussed later) have produced hundreds if not thousands of potential disease-associated genes that need to be thoroughly vetted in animal models. Thus, despite the impact genetically engineered mice have had on our understanding of human disease, traditional methods lack the speed and scale to handle the onslaught of information arising from genomic studies. Nonetheless, considerable steps are being taken to address the need for faster and more scalable approaches to make mouse models a force in the post-genomics future.
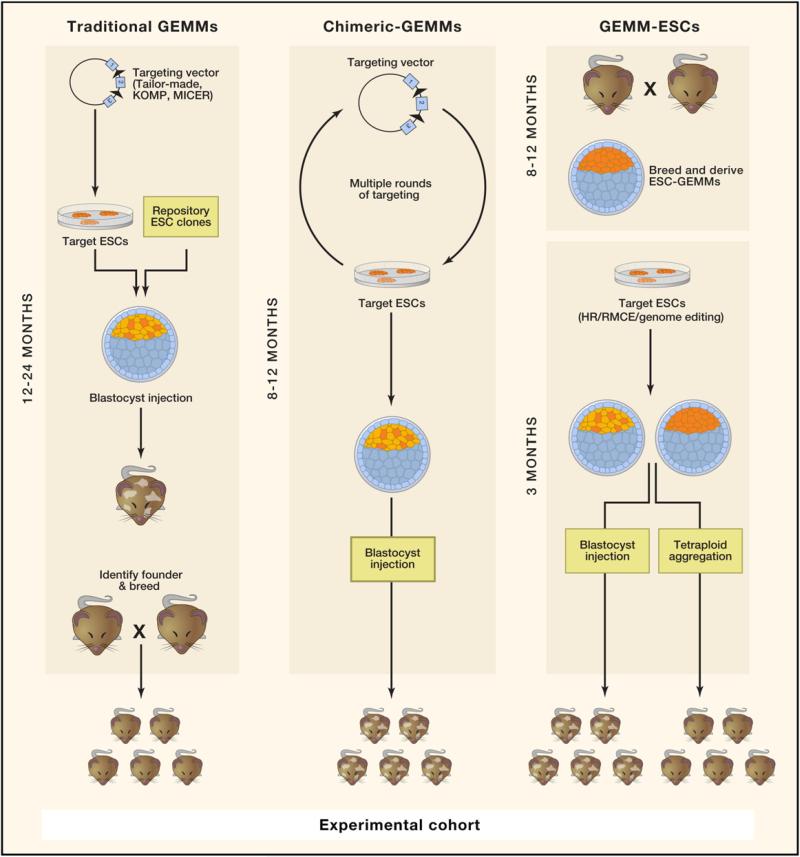
Figure 2. Traditional and “Speedy” Mouse Model Development.
The development of traditional knockout and conditional knockout mice requires the generation of unique targeting vectors, low-efficiency homologous recombination, identification of founders that transmit the targeted allele through the germline, and subsequent breeding and/or backcrossing. ESC-based GEMMs make use of multiallelic ESCs developed through multiple rounds of ESC targeting (chimeric-GEMMs) or rederivation of ESCs carrying established disease alleles (speedy or GEMM-ESCs). The initial establishment phase of GEMM-ESC models demands breeding but, once created, requires little manipulation in vitro. Experimental cohorts can be produced directly from the multiallele ESC and do not require any breeding steps. Experimental mice can be generated by blastocyst injection, by producing chimeric mice (depicted by brown and white coat color), or through tetraploid embryo complementation, generating wholly ESC-derived animals (brown coat).
Stop-and-Shop Mouse Models
A major bottleneck in the development of targeted genetic models is the painstaking, one-by-one production of specific gene-targeting vectors. For the production of knockout mice (constitutive and conditional), this is largely a thing of the past. Both commercial entities and large publicly funded consortia have made significant progress in establishing extensive repos itories of gene-targeting vectors as well as pretargeted ESCs and GEM. The MICER repository (http://www.sanger.ac.uk/resources/mouse/micer) offers thousands of ready-made targeting vectors for generating both constitutive and conditional alleles (Adams et al., 2004). In addition, because MICER provides vectors for targeting thousands of regions throughout the genome, it offers a shortcut for manipulation of large chromosomal regions through chromosome engineering. International, multi-institute initiatives such as the International Knockout Mouse Consortium (IKMC) (http://www.knockoutmouse.org), incorporating the Knockout Mouse Project (KOMP: http://www.nih.gov/science/models/mouse/knockout/) and the European Conditional Mouse Mutagenesis program (EUCOMM), as well as the Gene Trap consortium (http://www.genetrap.org), provide vectors as well as pre-engineered ESC lines. As of February 2012, the IKCM had amassed close to 18,000 deletion, conditional deletion, and gene trap alleles in ESCs, and it is expected that targeting of the majority of the mouse genes and microRNAs (miRNAs) will be completed within a few years (Skarnes et al., 2011). A major part of this effort has been spear-headed by members of the Wellcome Trust Sanger Institute (William Skarnes and Alan Bradley), who use an elegant “knockout first” targeting strategy that, from a single targeting event, allows the creation of transcriptional reporter (gene trap), conditional, and constitutive deletions in ESCs and/or mice (Skarnes et al., 2011). Together, the targeting vector and ESC repositories provide a much-needed resource for the medical research community and will significantly decrease the time associated with the future development of knockout strains. In addition, large-scale and systematic phenotyping efforts (e.g., International Mouse Phenotyping Consortium: http://www.mousephenotype.org/) will provide investigators with much needed baseline information as they begin to explore the biology of poorly characterized genes.
New methods have also been developed to accelerate the process of transgene delivery in mammalian cells. Recombinase-mediated cassette exchange (RMCE), pioneered by Juergen Bode and colleagues (Seibler et al., 1998), is a particularly efficient means to target transgenes to a defined genomic region and avoid the consequences of positional effects on transgene expression. This strategy has been used to derive robust ESC lines carrying RMCE “acceptor sites” at the Rosa26 and Col1A1 loci (Hitz et al., 2007; Hochedlinger et al., 2005; Seibler et al., 2007), and efforts are underway to identify additional “safe-harbor” loci in the mouse that are immune to epigenetic silencing. RMCE-ready ESC lines enable the simple and reliable production of transgenic mice on a new scale. Using ColA1-RMCE (KH2) ESCs developed by the Jaenisch laboratory and our recently described shRNA-targeting platform (Premsrirut et al., 2011), we are now developing an NCI-funded, public repository of ~1,500 ESC lines carrying doxycycline-inducible miRNAs that can be used to study miRNA biology in ESCs or mice (Y. Park, G. Hannon, and S.W.L., unpublished data).
Exploiting RNA Interference
RNA interference (RNAi) is an extremely powerful method of gene regulation that is conserved throughout evolution. Initially most effective for studying gene function in model organisms such as C. elegans and Drosophila, it is now established that RNAi can also be achieved in mice through the transgenic expression of short hairpin RNAs (shRNAs), offering an effective alternative to the traditional knockout approach. shRNAs, expressed as stem-loop RNAs or embedded within an endogenous miRNA fold, induce targeted gene silencing by stably reducing mRNA transcript levels and/or inhibiting translation. Although they do not induce complete gene loss, the key feature of shRNAs that makes them so powerful for in vivo studies is that they function in trans; hence, whereas traditional knockouts require deletion of both gene copies, a single shRNA allele can induce silencing of a gene (or genes) expressed from two alleles. The reduced need for allelic intercrossing enables faster generation of models systems for analysis.
Because shRNAs do not disrupt the endogenous locus of the target gene, their silencing effects are reversible, so the requirement for gene function can be investigated during defined intervals of normal development or disease pathogenesis. Such reversible approaches facilitate evaluation of the consequences of inhibiting a putative drug target on disease progression, where transient target suppression in established disease may accurately mimic the impact of a drug treatment. By contrast, achieving similar capabilities with standard genomic engineering is tedious and requires dedicated allelic variants (Ventura et al., 2007).
Our lab and others have taken advantage of the efficiency of RMCE targeting in combination with shRNA technology to produce a fast and scalable platform that allows production of mice with tissue-specific, inducible, and reversible gene silencing (Dow et al., 2012). As the production and targeting of shRNA cassettes are standardized and efficient, it is reasonable to envisage the creation of a genome-wide repository of ESC lines carrying shRNAs targeting the mouse genome. Although such a collection does not yet exist, the recent development of a high-throughput functional assay for identification of potent shRNAs (Fellmann et al., 2011) will inevitably lead to the production of genome-wide prevalidated shRNA libraries that can be quickly adapted for generation of transgenic mice.
Genome Editing in a Flash
Many disease-associated genetic changes involve point mutations, and large sequencing efforts are identifying ever more single-nucleotide variations of potential disease relevance. Such changes may produce loss of function (akin to knockouts) but may also induce gain of function or altered gene function, and many may be found outside protein-coding regions (see, for example, Bond et al., 2004). With knockout production moving forward at a rapid pace, how will the production of disease-relevant point mutations be systematically addressed? Unfortunately there is currently no coordinated effort to produce, en masse, conditional “endogenous” mutant alleles like LSLTrp53R270H or LSL-KrasG12D (Jackson et al., 2001; Olive et al., 2004). However, new technologies developed and tested for gene disruption and homologous recombination in rat and human cells could prove an invaluable tool for the rapid generation of targeted point mutations in mice. Zinc finger nucleases (ZFNs) and TAL effector nucleases (TALENs) are, in effect, genomic scissors—recombinant, sequence-specific DNA nucleases that can be adapted to recognize specific nucleotide sequences and induce DNA breaks. These double-strand breaks, through homologous repair from exogenous template DNA, can be directed to incorporate anything from a single nucleotide change to a large transgenic insertion (for review, see Bogdanove and Voytas, 2011; Carroll, 2011).
Additionally, repair through nonhomologous end-joining (NHEJ) can be exploited to create random single-base changes, small insertions, or deletions. ZFNs enabled the generation of the first targeted knockout rats at surprisingly high efficiency (Geurts et al., 2009), and the technology of both ZFNs and TALENs promises faster and more precise genomic manipulation or “genome editing” of virtually any organism, including plants, worms, and other mammals (Hockemeyer et al., 2011; Miller et al., 2011; Townsend et al., 2009; Wood et al., 2011). Still, “off-target effects” remain a significant concern for nuclease-mediated therapies, as double-strand breaks induced outside the target region can cause mutations that are almost impossible to detect without whole-genome analysis. Thorough sequencing efforts will be required to define the true off-target “hit rate” of each nuclease pair.
For those wanting to develop a mouse model targeting their favorite gene, the options are faster and more flexible than even 5 years ago. Through both commercial companies and publicly funded repositories, tools for genome manipulation have never been easier to access. As the catalog of mouse knockouts grows and validated genome-wide shRNA and ZFN/TALEN collections become a reality, the question is no longer whether we can produce the mouse models but whether we can leverage the power of each approach to develop models that accurately recapitulate the simple or complex genetics of human disease.
Managing the Multiallelic Nature of Human Disease
Generating multiallelic transgenic mouse models has facilitated understanding of complex diseases. For example, it is clear that the malignant endpoint arises as a combination of genetic mutations. The order and nature of these changes impact disease evolution, influencing which genes are required for disease maintenance and hence encode suitable therapeutic targets. More and more, genetic and genomic studies are highlighting the multifactorial nature of human disease, and it is likely that as we unravel the complexities of this information, such models will become increasingly important, not only as tools to understand disease mechanisms but also as accurate models for preclinical evaluation of potential therapies. Ignoring for a moment the formidable task to produce and characterize mutants, knockouts, and knockdowns of all known genes (currently underway), the process of intercrossing these individual alleles is even more tedious, time-consuming, and thus expensive. This constant barrier severely limits the scale at which complex genetic experiments can be performed and thus the number of genetic combinations that can be examined in vivo.
“Speedy” ESC Models of Disease
To expedite the process of multiallelic mouse model production, we and others have begun to develop so-called “chimeric-GEMM” or “GEMM-ESC” models (Figure 2) (Huijbers et al., 2011; Premsrirut et al., 2011; Zhou et al., 2010). This approach is based on the derivation of ESCs carrying disease-associated alleles that in combination generate a functional mouse model of disease. Like wild-type ESC lines, GEMM-ESCs can be manipulated in vitro, through standard (homologous recombination) targeting, using ZFNs/TALENs or through the introduction of shRNAs/cDNAs by RMCE. In this way, many different genes and genetic contexts can be interrogated in parallel. Generation of chimeric or genetically identical mice (by blastocyst injection or tetraploid complementation, respectively) facilitates the production of large cohorts of experimental animals for analysis and/or preclinical treatment studies. Moreover, animal production does not rely on breeding success and thus can be tailored to the needs of the user and synchronized for streamlined experimental protocols.
We recently showed that rederived ESCs carrying four alleles (LSL-KRasG12D, CCSP-rtTA, Rosa26-LSL-luciferase, and the Col1A1-RMCE cassette) can be used to generate a mouse model of lung adenocarcinoma in only a fraction of the time required to develop and breed the genetic combinations through traditional crossing (Premsrirut et al., 2011). Zhou et al. also showed that similar results can be achieved through consecutive rounds of ESC targeting and selection (Zhou et al., 2010). The ultimate goal of such efforts is the creation of a diverse bank of ESC models that could be accessed as needed to investigate specific biological questions or triage a set of candidate genes identified by patient sequencing or other screening approaches.
To date, ESC-based proof-of-principle studies have been focused on the development of complex genetic combinations for modeling cancer in the mouse, but similar methods can be adapted to create GEMM-ESC models for any genetic disease. For instance, Nichols et al. recently reported the generation of germline-competent ESCs from the nonobese diabetic (NOD) mouse strain—providing a platform for the rapid functional analysis of candidate diabetes modifiers (Nichols et al., 2009). Similarly, GEMM-ESCs could be developed for complex transgenic strains such as the humanized sickle-cell disease mouse (discussed later) or for models of muscular dystrophy (Sacco et al., 2010) and cancer (Maser et al., 2007) that rely on multiple generations of breeding on a telomerase-compromised background.
The GEMM-ESC approach promises a significant reduction in the maintenance of large, expensive breeding colonies but more importantly a dramatic increase in the throughput of functional annotation of disease genes. Taking advantage of the vast array of targeting vectors in public repositories and shRNA and ZFN/ TALEN technology, the possibility to create flexible, tailored, and complex models of disease is incredibly exciting.
Mosaic Mouse Models
An alternative approach for recapitulating complex genetic situations that is particularly relevant for cancer research is the use of “mosaic” or “non-germline” mouse models. We and others have used orthotopic transplantation-based mosaic models in combination with viral delivery of cDNAs and shRNAs to generate mouse models of a variety of cancers, including leukemia, lymphoma, breast and pancreatic cancer, and hepato-cellular carcinoma, among others (for review, see Heyer et al., 2010). Similarly, some laboratories have successfully adapted the avian virus RCAS-Tva system to deliver genes and shRNAs that promote cancer in the adult mouse (Hambardzumyan et al., 2009; Seidler et al., 2008). Recently, Beronja and Livshits et al. described an approach for the efficient delivery of lentivirus (carrying cDNAs and/or shRNAs) to the epidermis of developing embryos, enabling widespread (almost complete) genetic modification of the skin lineages without the need for cell transplantation or germline transgenesis (Beronja et al., 2010).
All of these non-germline approaches share a common theme—the flexibility to quickly alter the genetics of the target cell and vary that of the host (microenvironment) to rapidly interrogate the function of not just single genes but networks or whole pathways. Together the methodologies provide an incredible tool with which to dissect the genetic components of disease. Improved orthotopic transplantation and/or delivery of genetic material (e.g., Cre recombinase, shRNAs, reporters) to different organ systems in vivo will open the door for the use of such mosaic models in a wide range of disease research fields.
Together the evolving repositories and large-scale phenotyping efforts, ESC-GEMMs, and mosaic models provide an array of tools to tackle what is an emerging challenge in biology: making sense of the flood of information streaming out of GWAS, whole-genome sequencing, and functional genomic screens to define those variations that contribute most to human disease. Functional studies are crucial for realizing the fruits of these large-scale but descriptive efforts, yet traditional methods have been too slow to interrogate more than a handful of variants. However, with recent application of more high-throughput methods, it has been possible to filter genomic data from human disease in mouse models, finding genes that act as drivers of disease (Sawey et al., 2011; Zender et al., 2008). When combined with genome editing and shRNA technologies, these platforms will provide a rapid means to mimic the many mutational and gene-dosage alterations identified in human disease. Ultimately, there will be no “one-size-fits-all” disease model, but increasing accessibility to a range of refined experimental systems will play a central role in “functionalizing” the human genome.
Forward Genetics to Tackle Complex Genomes
Identifying the genes that cause or contribute to disease is a significant task, particularly when the disease is caused by multiple genetic factors. Analysis of genomic information from patients is one approach, although defining meaningful gene-disease associations requires incredibly large datasets. Another, unbiased approach that has been the mainstay of lower genetic model systems is disease gene identification through forward genetic screens.
N-ethyl-N-nitrosourea
Implementation of forward genetics in mice, historically through N-ethyl-N-nitrosourea (ENU) mutagenesis (for review, see Kile and Hilton, 2005), has been effective not only in generating monoallelic mouse models of specific human diseases but also in identifying modifiers and genetic interactions through “sensitized” screens in models of diabetes, developmental neuropathies, and disorders such as Waardenburg's syndrome (Matera et al., 2007; Stottmann et al., 2011; Tchekneva et al., 2007). However, until recently, mapping disease-causative mutations following ENU treatment was notoriously time consuming and complex. Now, as the “$1000 genome” is set to become a reality, “next-generation” sequencing is revolutionizing the value and feasibility of large-scale forward genetic screens—and ENU mutagenesis programs all over the world are taking full advantage. For example, in recent years, laboratories at The Scripps Research Institute (B. Beutler) and the Australian National University (C. Goodnow) have generated and cataloged hundreds of novel mutant strains (http://mutagenetix.scripps.edu). An additional by-product of this effort has been the identification of more than 1,800 “incidental mutations,” found through whole-genome sequencing, that offer a tremendous potential resource for the research community. In concept, sensitized ENU screens offer an ideal setting to interrogate the mechanisms underlying complex genetic traits such as learning and behavioral disorders (e.g., autism and schizophrenia), cardiovascular disease, diabetes, and other autoimmune conditions. With future decreased cost and increased throughput and sensitivity of genome sequencing, effectively executing these types of comprehensive screens may no longer be the exclusive domain of dedicated mouse-breeding consortia.
Transposons and shRNAs
For the discovery of genes involved in cancer, viral and transposon mutagenesis and pooled cDNA and shRNA screens have a proven history (Heyer et al., 2010; Kool and Berns, 2009). In recent years, the laboratories of Copeland/Jenkins, Largaespada, and others have developed elegant transgenic mouse strategies to activate highly mutagenic Sleeping Beauty (SB) transposons in a temporal and tissue-specific manner (reviewed in Copeland and Jenkins, 2010). Recently, SB technology has been effectively combined with tissue-specific GEM models of intestinal cancer and melanoma to identify genetic events that contribute to malignant progression (Karreth et al., 2011; March et al., 2011; Starr et al., 2009). Moreover, application of next-generation sequencing to deconvolute these SB screens enables the identification of common insertion sites and is sufficiently sensitive to reveal frequent co-occurrence of insertion sites that may suggest cooperative interaction between specific gene networks (March et al., 2011). It will be interesting to see how the generation of more and more similar datasets will inform our understanding of regulatory networks in disease.
Although the majority of SB integrations result in gain-of-function activity in nearby genes (overexpression), the system, in theory, provides an unbiased approach for identification of any cancer-relevant genetic event. In contrast, cDNA/shRNA screens can be configured to be broad (genome-wide), narrow (directed at a particular pathway or process), or, as has recently proven fruitful, guided by human genomic data (Sawey et al., 2011; Scott et al., 2011; Zender et al., 2008). Our lab and others have used a variety of non-germline mosaic models and cancer-focused shRNA libraries as a screening platform in both lymphoma and hepatocellular carcinoma models to identify numerous novel tumor suppressor genes (for review, see Heyer et al., 2010). Similarly, screening libraries of cDNAs recurrently amplified in human tumors has yielded insights into the biology of malignant progression (Sawey et al., 2011; Scott et al., 2011). The development of large cDNA and shRNA repositories (e.g., Open Biosystems) in the past 5 years has significantly increased the feasibility of these screening approaches for most labs, and continued improvements in organ-directed viral and transposase gene/shRNA delivery to adult mice will further fuel the use of mosaic models as a gene discovery tool in disease research. Together these approaches will identify new cancer drivers and also better model the genetic heterogeneity of cancer that occurs in human patients.
Finding the Weakest Link—Drug Target Discovery and Validation
Understanding the factors that are essential for disease progression and maintenance is key to developing effective treatment strategies. As more information concerning the underlying genetic basis of disease is produced, as well as modifiers of disease penetrance and progression, efforts to consider more personalized therapies are becoming attractive. Although only clinical trials with a drug can truly validate its efficacy in disease treatment, model systems can be highly useful in drug target discovery owing to the ability to precisely control the genetic background and/or in vivo microenvironment. Indeed, traditional knockout mice have been used to assess the consequences of target inhibition in vivo but suffer from the disadvantage that gene deletions examine disease initiation and not maintenance, whereas conditional gene deletion in established disease is often inefficient and cannot mimic a transient treatment regime, as it is not reversible. Moreover, such approaches are sufficiently slow and expensive to allow for interrogation of only one or, at most, a few targets.
Most current drugs act as inhibitors of protein function. As such, inducible RNAi provides an effective tool to interrogate the potential consequences of protein silencing in established disease and, better, can be applied on a high-throughput scale (Zuber et al., 2011a). Indeed, in vivo screens to identify genes that modulate cell survival or drug response have been reported, some leading to unanticipated therapeutic strategies, for example, the utilities of Brd4 inhibitors to treat aggressive forms of acute myeloid leukemia (AML) (Zuber et al., 2011b). Similar in vivo screening strategies have been used to identify components of the actin machinery and metabolome as key regulators of disease maintenance in lymphoma and breast cancer, respectively (Meacham et al., 2009; Possemato et al., 2011). Even for screens performed in vitro, in vivo validation with inducible shRNA systems, such as GEMM-ESC models, is a fast and powerful check that an identified gene/pathway is indeed a valid target. Currently most studies require an in vitro step and transplantation into conditioned recipients; however, improved viral and transposon-based delivery may ultimately enable truly in vivo screens, most likely to capture the precise conditions in which a therapy should be efficacious. Integrating these approaches provides a powerful means for vetting potential therapeutic targets before investing in expensive drug development projects.
Mouse models may be an effective system for drug target discovery, but whether they represent an ideal platform for evaluating potential drug intervention strategies has been a more controversial topic. Historically, drug regimens established in mouse models of various diseases have shown mixed performance in clinical settings (Driver et al., 2011; Olive et al., 2009; Richmond and Su, 2008; Talmadge et al., 2007). Although this signaled to many that mouse models were not appropriate predictive systems, it may reflect technical limitations of those systems rather than the usefulness of the mouse for drug intervention trials. In the past few years, the development of more sophisticated transgenic models, better humanized mice (discussed below), and powerful small animal imaging platforms has shown that the mouse can be an effective gateway for translational research. Importantly, some models can accurately mimic the response and resistance observed in the clinic (Zhou et al., 2010), although models that predict drug-induced relapse have been limited thus far. Additionally, inducible and reversible shRNA mouse models now offer a setting to genetically test the effect of reactivating suppressed gene networks or inhibiting potential drug targets. Notwithstanding the important differences between mice and humans in terms of drug specificity and metabolism, with the increased arsenal of genetic tools, the mouse represents arguably the most flexible system to filter drug candidates before entering enormously expensive clinical trials.
As we move forward with functional annotation of the disease genome, describing the complex genetic interactions that contribute to biological dysfunction will be a challenging process. An important tool will be the integration of technologies to pinpoint disease genes, such as using genomic data to inform the genesis of targeted mouse models and gene sets for forward genetic screens. In addition, continual advances in sequencing technology are driving forward both genome resequencing and mutagenesis screens at an unprecedented scale. Intersecting and cross-referencing the results from these approaches will surely identify high-confidence “hits” that can then be rapidly moved into relevant hypothesis-driven preclinical models.
Moving beyond the Mouse: When Size Does Matter
Although the rat is often overshadowed by its smaller cousin, many researchers turn to it in settings where physiological or anatomical restrictions limit the utility of the mouse. In fact, although genetic tools and ESC manipulations are significantly more sophisticated in the mouse, rats as a model system have many advantages over mice: they have more similar cardiovascular and respiratory physiology (heart and breath rate is significantly slower than the mouse), are considered smarter and thus better for learning and addiction research, and are the preferred preclinical rodent model for drug toxicity evaluation. In addition, their larger size (around ten times that of mice) enables more frequent blood sampling, easier measurement of biometric parameters such as blood pressure, and more straightforward imaging of brain activity for neuronal research.
Paralleling the early days of mouse genetics, disease models in the rat have relied heavily on spontaneous or chemically derived (ENU) mutants. Two spontaneously derived models, the type 1 diabetes-prone BB rat and the spontaneous hypertensive rat, were identified over 30 years ago and remain two of the most heavily used models systems for understanding human metabolic and cardiovascular disease (Mordes et al., 2004; Pinto et al., 1998). Even in cancer research, the traditional domain of the mouse, rats have also proven useful. As one example, Dove and colleagues recently identified an ENU-induced mutant rat strain (pirc) that carries a missense mutation in the adenomatous polyposis coli (APC) gene (Amos-Landgraf et al., 2007) similar to those described in mouse models and patients with familial adenomatous polyposis (FAP). However, in contrast to the mouse, which develops polyps almost exclusively in the intestine, the pirc rat kindred shows a greater burden of polyps in the colon, which is more similar to human FAP. This important distinction also provide a means to monitor tumor growth and response to therapies via colonoscopy, something more challenging in the smaller mouse organ and impossible for tumors that arise in the intestine.
Despite their utility for disease research, rat models have been limited by a lack of tools to manipulate the genome. However, recent advances in rat ESC culture and nuclease-driven genetic manipulation of rat genomes have now opened the door for more advanced genetic models. In 2008, two groups reported the derivation of the first germline-competent rat ESCs from two different inbred strains (Buehr et al., 2008; Li et al., 2008), and subsequently, Ying and colleagues described the generation of a p53 null rat with the same homologous recombination approach that produced the original p53 null mouse 20 years earlier (Tong et al., 2010). The long sought after establishment of rat ESCs through advances in culture conditions also suggests that other, potentially relevant mammalian models could be adapted for the same types of systematic genetic engineering that are currently being accomplished in the mouse.
The second major advance for rat came in 2009 when Guerts et al. used ZFNs to create targeted knockout rats without the need for ESC culture (Geurts et al., 2009). Since that time, many more knockout/mutant strains have been produced, and efforts are currently underway to develop 100 new rat models of hypertension and kidney disease (Dolgin, 2010). In addition, Tesson et al. have reported the use of TALENs to generate knockout rats, adding another tool to the rat geneticist armory (Tesson et al., 2011). Some commercial entities are now also offering custom production of ZFN-derived mutant rats and doxycycline-regulated shRNA transgenic rats—albeit at a significant cost. Given the preference for the larger rodents as a system to evaluate drug efficacy, it is likely that many of these genetically engineered rats will replace mice as front-line preclinical models. However, their (significant) housing costs make it unlikely that rats will supersede the mouse as a workhorse genetic tool in academic research.
A Man or a Mouse?
Species-specific problems with rodent-based research, be they genetic or physiological, will only be overcome by conducting experiments on human cells. Xenograft-based human tumor experiments in immunocompromised host mice have for many years served as a surrogate for tumorigenicity and preclinical modeling. However, as more evidence mounts for the importance of immune cell interaction and the tumor microenvironment, these models are quickly losing favor. Moreover, there are hundreds of diseases besides cancer—such as metabolic and autoimmune disorders and bacterial and viral infectious diseases—that would benefit from direct examination of human tissues.
Humanized Mice
The term “humanized mice” describes both the expression of a human gene or genes in a transgenic context and/or the engraftment of immunocompromised mice with human immune cells. A number of model systems have been developed, based on the expression of disease-specific human alleles. The best example of this is the replacement of mouse globin genes with their human counterparts (including both wild-type and sickle-cell disease [SCD] mutant β-globins) to recreate the disease progression of human SCD. This humanized mouse, now available through the Jax repository, has been essential for evaluating new autologous gene therapy approaches and small-molecule-based treatments for SCD (Chang et al., 2010; Hanna et al., 2007).
The creation of humanized models through engraftment of CD34+ human hematopoetic stem cells (hHSCs) or peripheral blood mononucleocytes (PBMCs) has recently expanded, following the development of severely immunocompromised strains that act as optimal recipients for human stem cells, in particular the NOD-severe combined immunodeficiency (SCID)-Il2rg–/– (NSG) mice (McDermott et al., 2010). Such mice have proven invaluable for understanding the normal function of the human immune system as well as the (human) response to viral and microbial pathogens, such as hepatitis C virus and HIV, that do not infect mouse tissues (Shultz et al., 2007). In some cases, humanized models have also shown efficacy for the development and testing of vaccines (Yu et al., 2008).
Mice humanized by the ectopic expression of human cytokines show improved grafting of hHSCs and the development of more sustained natural killer (NK) and myeloid cell responses (Billerbeck et al., 2011; Chen et al., 2009) presumably by providing more effective crosstalk between mouse and human compartments. In addition, scientists at the Jackson Laboratories have reported that NSG mice expressing human HLA molecules not only provide a good setting for hHSCs engraftment but also allow a stronger antigen-dependent T cell response than current models (Shultz et al., 2010). The progressive advances of “humanizing” technology suggest that with the right combination of factors (in both the host and donor tissue), NSG-based humanized models will provide the best surrogate for human immune studies. However, the reproducible generation of humanized chimeras is technically challenging and, for most labs, not yet cost effective for the production of large cohorts for high-throughput studies.
Reprogramming Human Cell Models
Five years ago, Yamanaka and colleagues reported their seminal discovery that mouse (2006) and human (2007) fibroblasts could be “reprogrammed” (by viral delivery of four transcription factors: Oct4, Sox2, Klf4, and c-Myc) to a state of induced pluripotency, capable of generating all somatic cell types (Takahashi et al., 2007; Takahashi and Yamanaka, 2006). These observations led to the first platform to produce and study nonembryonic and nontransformed human cells. Driven by the hope of autologous gene therapy and regenerative medicine, induced pluripo-tent stem cell (iPSC) technology has evolved extensively (for review, see Stadtfeld and Hochedlinger, 2010). It is now possible to generate human iPSCs with only modified mRNA transcripts or purified proteins (Kim et al., 2009; Warren et al., 2010; Zhou et al., 2009), eliminating the need for disruption of the host genome by viral or plasmid transgenesis and providing “clean” reprogrammed cells for functional analysis.
The implications of iPSC technology for disease research and treatment are profound. The generation of patient-specific iPSCs provides a previously unimagined avenue for the dissection of simple and complex genetic diseases, with almost limitless access to sample. Already, a number of groups have reported the development of patient-specific iPSCs for diverse monogenic disorders such as Long-QT syndrome, SCD, Lesch-Nyan syndrome, and retinitis pigmentosis and complex traits such as Parkinson's disease and schizophrenia (Brennand et al., 2011; Jin et al., 2011; Moretti et al., 2010; Soldner et al., 2009; Zou et al., 2011). Importantly, in many of these cases the iPSC clones can be reproducibly differentiated into the cell types in which the disease manifests.
iPSCs are also being used as a therapeutic tool. For example, Rhee et al. recently showed that human iPSC-derived neurons rescue motor deficits when transplanted into a rat model of Parkinson's disease (Rhee et al., 2011), implying that in vitro differentiation can produce fully functional cell populations. Further, disease-specific differentiated cell types can show predicted phenotypic traits and response to disease-relevant drugs in vitro (Brennand et al., 2011), suggesting that in some cases an in vivo animal system may not be a requisite for disease research. All the early iPSC data look genuinely promising, although most of the successful examples reported thus far have used protocols to differentiate iPSCs into a restricted number of cell types, commonly neurons and cardiac myocytes. The development of standardized protocols for the reproducible derivation of diverse cell types in vitro will be key to harnessing the power of iPSC-based disease models.
The creation of large repositories of disease and “normal” iPSCs will eventually open the door for large-scale functional genetics with shRNA libraries and high-throughput drug sensitivity/toxicity screens, much as is occurring now in cell lines and engineered mice. For cancer studies, production of new tumor cell lines from biopsies could be paralleled by the development of patient-specific fibroblast iPSC lines that would serve as a matched “normal” tissue for functional studies and large-scale drug and shRNA-negative selection screens. Such screens not only will enable dissection of the genetic requirements of disease but could identify potential therapeutic avenues for individualized disease management. Some have also suggested that iPSCs could one day provide a useful surrogate for testing patient-specific response and drug toxicity prior to clinical treatment, although the protocols for iPSC derivation and differentiation are currently too slow for this to become a reality.
iPSCs and Genome Editing in the Clinic
One of the most exciting possibilities of patient-specific iPSC technology is the potential for autologous cell therapy with “gene-corrected” patient cells. The ability to selectively alter the genome of iPSCs would also strengthen the value of iPSCs as a models system, allowing the controlled perturbation of gene function or “correction” of disease-specific mutations in human cells. In this regard, the continued development and application of sequence-specific nucleases such as ZFNs and TALENs provides one path toward achieving this goal.
In the past 2 years, numerous groups have demonstrated the potential for targeted nucleases in genome editing and translational research for human disease, including the correction of mutations causing SCD and the autosomal recessive metabolic disorder a1-antitrypsin deficiency (A1ATD) in iPSCs (Sebastiano et al., 2011; Yusa et al., 2011; Zou et al., 2011). Perhaps the most exciting of these results are data from a humanized mouse model carrying HIV, which show that disruption of the HIV core-ceptor CCR5 with ZFNs could generate resistance to viral infection (Holt et al., 2010; Perez et al., 2008). Early positive signs from phase I clinical trials suggest that this form of genome editing may become a true success story of translational research, although for reasons mentioned above, concerns linger for the potential mutagenicity of such an approach. For additional discussion on iPSCs in the study of human diseases, please see Perspective by Cherry and Daley on page 1110 of this issue.
Getting a Move On: Looking to the Future
Over the past two decades, the development of new model systems to recapitulate and study human disease has progressed at an astonishing rate. A variety of approaches and methodologies have been adapted to manipulate the genomes of mouse, rat, and human cells and thus enable systematic analysis of the genetic requirements of disease initiation and maintenance. The present challenge for the biomedical research community is two-fold: (1) integrate the large array of sophisticated genetic tools to produce the best and most predictive model system or systems to define treatment strategies, and (2) develop these models at a rate so as to accommodate testing and/or validation of the many candidate disease genes being identified by micro-array/ChIPseq/CGH, shRNA and mutagenesis screens, and GWAS. Already, the development of platforms to quickly produce ESC or iPSC-based models shows how interrogation and understanding of disease progression need not be a slow process. With this increased efficiency, the mechanisms underlying disease pathogenesis can now be investigated in a variety of different settings, providing the best chance of finding effective therapeutic avenues in the shortest time possible.
ACKNOWLEDGMENTS
The authors thank Amaia Lujambio for critical input and apologize to those authors whose work was not cited due to space constraints. L.E.D. was supported by an Overseas Biomedical Training fellowship from National Health & Medical Research Council of Australia, and S.W.L. is an investigator of the Howard Hughes Medical Institute. This work was supported by a consortium grant from the National Cancer Institute. S.W.L. is one of the founders of Mirimus Inc., a company that has licensed technology relating to the development of shRNA transgenic mice.
REFERENCES
- Adams DJ, Biggs PJ, Cox T, Davies R, van der Weyden L, Jonkers J, Smith J, Plumb B, Taylor R, Nishijima I, et al. Mutagenic insertion and chromosome engineering resource (MICER). Nat. Genet. 2004;36:867–871. doi: 10.1038/ng1388. [DOI] [PubMed] [Google Scholar]
- Amos-Landgraf JM, Kwong LN, Kendziorski CM, Reichelderfer M, Torrealba J, Weichert J, Haag JD, Chen K-S, Waller JL, Gould MN, Dove WF. A target-selected Apc-mutant rat kindred enhances the modeling of familial human colon cancer. Proc. Natl. Acad. Sci. USA. 2007;104:4036–4041. doi: 10.1073/pnas.0611690104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beronja S, Livshits G, Williams S, Fuchs E. Rapid functional dissection of genetic networks via tissue-specific transduction and RNAi in mouse embryos. Nat. Med. 2010;16:821–827. doi: 10.1038/nm.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billerbeck E, Barry WT, Mu K, Dorner M, Rice CM, Ploss A. Development of human CD4+FoxP3+ regulatory T cells in human stem cell factor-, granulocyte-macrophage colony-stimulating factor-, and interleukin-3-expressing NOD-SCID IL2Rγ(null) humanized mice. Blood. 2011;117:3076–3086. doi: 10.1182/blood-2010-08-301507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, Bargonetti J, Bartel F, Taubert H, Wuerl P, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, Li Y, Mu Y, Chen G, Yu D, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehr M, Meek S, Blair K, Yang J, Ure J, Silva J, McLay R, Hall J, Ying QL, Smith A. Capture of authentic embryonic stem cells from rat blastocysts. Cell. 2008;135:1287–1298. doi: 10.1016/j.cell.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188:773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Patton JT, Sarkar A, Ernst B, Magnani JL, Frenette PS. GMI-1070, a novel pan-selectin antagonist, reverses acute vascular occlusions in sickle cell mice. Blood. 2010;116:1779–1786. doi: 10.1182/blood-2009-12-260513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Khoury M, Chen J. Expression of human cytokines dramatically improves reconstitution of specific human-blood lineage cells in humanized mice. Proc. Natl. Acad. Sci. USA. 2009;106:21783–21788. doi: 10.1073/pnas.0912274106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland NG, Jenkins NA. Harnessing transposons for cancer gene discovery. Nat. Rev. Cancer. 2010;10:696–706. doi: 10.1038/nrc2916. [DOI] [PubMed] [Google Scholar]
- Dolgin E. The knockout rat pack. Nat. Med. 2010;16:254–257. doi: 10.1038/nm0310-254. [DOI] [PubMed] [Google Scholar]
- Dow LE, Premsrirut PK, Zuber J, Fellmann C, McJunkin K, Miething C, Park Y, Dickins RA, Hannon GJ, Lowe SW. A pipeline for the generation of shRNA transgenic mice. Nat. Protoc. 2012;7:374–393. doi: 10.1038/nprot.2011.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driver JP, Serreze DV, Chen Y-G. Mouse models for the study of autoimmune type 1 diabetes: a NOD to similarities and differences to human disease. Semin. Immunopathol. 2011;33:67–87. doi: 10.1007/s00281-010-0204-1. [DOI] [PubMed] [Google Scholar]
- Fellmann C, Zuber J, McJunkin K, Chang K, Malone CD, Dickins RA, Xu Q, Hengartner MO, Elledge SJ, Hannon GJ, Lowe SW. Functional identification of optimized RNAi triggers using a massively parallel sensor assay. Mol. Cell. 2011;41:733–746. doi: 10.1016/j.molcel.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frese KK, Tuveson DA. Maximizing mouse cancer models. Nat. Rev. Cancer. 2007;7:645–658. doi: 10.1038/nrc2192. [DOI] [PubMed] [Google Scholar]
- Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X, et al. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325:433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambardzumyan D, Amankulor NM, Helmy KY, Becher OJ, Holland EC. Modeling Adult Gliomas Using RCAS/t-va Technology. Transl Oncol. 2009;2:89–95. doi: 10.1593/tlo.09100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Wernig M, Markoulaki S, Sun C-W, Meissner A, Cassady JP, Beard C, Brambrink T, Wu L-C, Townes TM, Jaenisch R. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- Heyer J, Kwong LN, Lowe SW, Chin L. Non-germline genetically engineered mouse models for translational cancer research. Nat. Rev. Cancer. 2010;10:470–480. doi: 10.1038/nrc2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitz C, Wurst W, Kühn R. Conditional brain-specific knockdown of MAPK using Cre/loxP regulated RNA interference. Nucleic Acids Res. 2007;35:e90. doi: 10.1093/nar/gkm475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, Crooks GM, Kohn DB, Gregory PD, Holmes MC, Cannon PM. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat. Biotechnol. 2010;28:839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijbers IJ, Krimpenfort P, Berns A, Jonkers J. Rapid validation of cancer genes in chimeras derived from established genetically engineered mouse models. Bioessays. 2011;33:701–710. doi: 10.1002/bies.201100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z-B, Okamoto S, Osakada F, Homma K, Assawachananont J, Hirami Y, Iwata T, Takahashi M. Modeling retinal degeneration using patient-specific induced pluripotent stem cells. PLoS ONE. 2011;6:e17084. doi: 10.1371/journal.pone.0017084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, DeNicola G, Webster KA, Weiss D, Perez-Mancera PA, et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147:382–395. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kile BT, Hilton DJ. The art and design of genetic screens: mouse. Nat. Rev. Genet. 2005;6:557–567. doi: 10.1038/nrg1636. [DOI] [PubMed] [Google Scholar]
- Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, Kim KS. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kool J, Berns A. High-throughput insertional mutagenesis screens in mice to identify oncogenic networks. Nat. Rev. Cancer. 2009;9:389–399. doi: 10.1038/nrc2647. [DOI] [PubMed] [Google Scholar]
- Li P, Tong C, Mehrian-Shai R, Jia L, Wu N, Yan Y, Maxson RE, Schulze EN, Song H, Hsieh CL, et al. Germline competent embryonic stem cells derived from rat blastocysts. Cell. 2008;135:1299–1310. doi: 10.1016/j.cell.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- March HN, Rust AG, Wright NA, ten Hoeve J, de Ridder J, Eldridge M, van der Weyden L, Berns A, Gadiot J, Uren A, et al. Insertional mutagenesis identifies multiple networks of cooperating genes driving intestinal tumorigenesis. Nat. Genet. 2011;43:1202–1209. doi: 10.1038/ng.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser RS, Choudhury B, Campbell PJ, Feng B, Wong K-K, Protopopov A, O'Neil J, Gutierrez A, Ivanova E, Perna I, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera I, Cockroft JL, Moran JL, Beier DR, Goldowitz D, Pavan WJ. A mouse model of Waardenburg syndrome type IV resulting from an ENU-induced mutation in endothelin 3. Pigment Cell Res. 2007;20:210–215. doi: 10.1111/j.1600-0749.2007.00371.x. [DOI] [PubMed] [Google Scholar]
- McDermott SP, Eppert K, Lechman ER, Doedens M, Dick JE. Comparison of human cord blood engraftment between immunocompromised mouse strains. Blood. 2010;116:193–200. doi: 10.1182/blood-2010-02-271841. [DOI] [PubMed] [Google Scholar]
- Meacham CE, Ho EE, Dubrovsky E, Gertler FB, Hemann MT. In vivo RNAi screening identifies regulators of actin dynamics as key determinants of lymphoma progression. Nat. Genet. 2009;41:1133–1137. doi: 10.1038/ng.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- Mordes JP, Bortell R, Blankenhorn EP, Rossini AA, Greiner DL. Rat models of type 1 diabetes: genetics, environment, and autoimmunity. ILAR journal / National Research Council. Institute of Laboratory Animal Resources. 2004;45:278–291. doi: 10.1093/ilar.45.3.278. [DOI] [PubMed] [Google Scholar]
- Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flügel L, Dorn T, Goedel A, Höhnke C, Hofmann F, et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N. Engl. J. Med. 2010;363:1397–1409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- Nichols J, Jones K, Phillips JM, Newland SA, Roode M, Mansfield W, Smith A, Cooke A. Validated germline-competent embryonic stem cell lines from nonobese diabetic mice. Nat. Med. 2009;15:814–818. doi: 10.1038/nm.1996. [DOI] [PubMed] [Google Scholar]
- Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, Crowley D, Jacks T. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, Madhu B, Goldgraben MA, Caldwell ME, Allard D, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto YM, Paul M, Ganten D. Lessons from rat models of hypertension: from Goldblatt to genetic engineering. Cardiovasc. Res. 1998;39:77–88. doi: 10.1016/s0008-6363(98)00077-7. [DOI] [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo H-K, Jang HG, Jha AK, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premsrirut PK, Dow LE, Kim SY, Camiolo M, Malone CD, Miething C, Scuoppo C, Zuber J, Dickins RA, Kogan SC, et al. A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell. 2011;145:145–158. doi: 10.1016/j.cell.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee Y-H, Ko J-Y, Chang M-Y, Yi S-H, Kim D, Kim C-H, Shim J-W, Jo A-Y, Kim B-W, Lee H, et al. Protein-based human iPS cells efficiently generate functional dopamine neurons and can treat a rat model of Parkinson disease. J. Clin. Invest. 2011;121:2326–2335. doi: 10.1172/JCI45794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond A, Su Y. Mouse xenograft models vs GEM models for human cancer therapeutics. Dis Model Mech. 2008;1:78–82. doi: 10.1242/dmm.000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco A, Mourkioti F, Tran R, Choi J, Llewellyn M, Kraft P, Shkreli M, Delp S, Pomerantz JH, Artandi SE, Blau HM. Short telomeres and stem cell exhaustion model Duchenne muscular dystrophy in mdx/mTR mice. Cell. 2010;143:1059–1071. doi: 10.1016/j.cell.2010.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawey ET, Chanrion M, Cai C, Wu G, Zhang J, Zender L, Zhao A, Busuttil RW, Yee H, Stein L, et al. Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by Oncogenomic screening. Cancer Cell. 2011;19:347–358. doi: 10.1016/j.ccr.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KL, Nogueira C, Heffernan TP, van Doorn R, Dhakal S, Hanna JA, Min C, Jaskelioff M, Xiao Y, Wu CJ, et al. Proinvasion metastasis drivers in early-stage melanoma are oncogenes. Cancer Cell. 2011;20:92–103. doi: 10.1016/j.ccr.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiano V, Maeder ML, Angstman JF, Haddad B, Khayter C, Yeo DT, Goodwin MJ, Hawkins JS, Ramirez CL, Batista LFZ, et al. In situ genetic correction of the sickle cell anemia mutation in human induced pluripotent stem cells using engineered zinc finger nucleases. Stem Cells. 2011;29:1717–1726. doi: 10.1002/stem.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibler J, Schübeler D, Fiering S, Groudine M, Bode J. DNA cassette exchange in ES cells mediated by Flp recombinase: an efficient strategy for repeated modification of tagged loci by marker-free constructs. Biochemistry. 1998;37:6229–6234. doi: 10.1021/bi980288t. [DOI] [PubMed] [Google Scholar]
- Seibler J, Kleinridders A, Küter-Luks B, Niehaves S, Brüning JC, Schwenk F. Reversible gene knockdown in mice using a tight, inducible shRNA expression system. Nucleic Acids Res. 2007;35:e54. doi: 10.1093/nar/gkm122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidler B, Schmidt A, Mayr U, Nakhai H, Schmid RM, Schneider G, Saur D. A Cre-loxP-based mouse model for conditional somatic gene expression and knockdown in vivo by using avian retroviral vectors. Proc. Natl. Acad. Sci. USA. 2008;105:10137–10142. doi: 10.1073/pnas.0800487105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz LD, Ishikawa F, Greiner DL. Humanized mice in translational biomedical research. Nat. Rev. Immunol. 2007;7:118–130. doi: 10.1038/nri2017. [DOI] [PubMed] [Google Scholar]
- Shultz LD, Saito Y, Najima Y, Tanaka S, Ochi T, Tomizawa M, Doi T, Sone A, Suzuki N, Fujiwara H, et al. Generation of functional human T-cell subsets with HLA-restricted immune responses in HLA class I expressing NOD/SCID/IL2r gamma(null) humanized mice. Proc. Natl. Acad. Sci. USA. 2010;107:13022–13027. doi: 10.1073/pnas.1000475107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, Mitalipova M, et al. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 2010;24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr TK, Allaei R, Silverstein KAT, Staggs RA, Sarver AL, Bergemann TL, Gupta M, O'Sullivan MG, Matise I, Dupuy AJ, et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323:1747–1750. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stottmann RW, Moran JL, Turbe-Doan A, Driver E, Kelley M, Beier DR. Focusing forward genetics: a tripartite ENU screen for neurodevelopmental mutations in the mouse. Genetics. 2011;188:615–624. doi: 10.1534/genetics.111.126862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Talmadge JE, Singh RK, Fidler IJ, Raz A. Murine models to evaluate novel and conventional therapeutic strategies for cancer. Am. J. Pathol. 2007;170:793–804. doi: 10.2353/ajpath.2007.060929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchekneva EE, Rinchik EM, Polosukhina D, Davis LS, Kadkina V, Mohamed Y, Dunn SR, Sharma K, Qi Z, Fogo AB, Breyer MD. A sensitized screen of N-ethyl-N-nitrosourea-mutagenized mice identifies dominant mutants predisposed to diabetic nephropathy. J. Am. Soc. Nephrol. 2007;18:103–112. doi: 10.1681/ASN.2006020164. [DOI] [PubMed] [Google Scholar]
- Tesson L, Usal C, Ménoret S, Leung E, Niles BJ, Rémy S, Santiago Y, Vincent AI, Meng X, Zhang L, et al. Knockout rats generated by embryo microinjection of TALENs. Nat. Biotechnol. 2011;29:695–696. doi: 10.1038/nbt.1940. [DOI] [PubMed] [Google Scholar]
- Tong C, Li P, Wu NL, Yan Y, Ying QL. Production of p53 gene knockout rats by homologous recombination in embryonic stem cells. Nature. 2010;467:211–213. doi: 10.1038/nature09368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, Voytas DF. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature. 2009;459:442–445. doi: 10.1038/nature07845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- Warren L, Manos PD, Ahfeldt T, Loh Y-H, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AJ, Lo T-W, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CI, Gallegos M, Marches F, Zurawski G, Ramilo O, García-Sastre A, Banchereau J, Palucka AK. Broad influenza-specific CD8+ T-cell responses in humanized mice vaccinated with influenza virus vaccines. Blood. 2008;112:3671–3678. doi: 10.1182/blood-2008-05-157016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusa K, Rashid ST, Strick-Marchand H, Varela I, Liu P-Q, Paschon DE, Miranda E, Ordóñez A, Hannan NRF, Rouhani FJ, et al. Targeted gene correction of a1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 2011;478:391–394. doi: 10.1038/nature10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zender L, Xue W, Zuber J, Semighini CP, Krasnitz A, Ma B, Zender P, Kubicka S, Luk JM, Schirmacher P, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135:852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T, Trauger S, Bien G, Yao S, Zhu Y, et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Rideout WM, 3rd, Zi T, Bressel A, Reddypalli S, Rancourt R, Woo J-K, Horner JW, Chin L, Chiu MI, et al. Chimeric mouse tumor models reveal differences in pathway activation between ERBB family- and KRAS-dependent lung adenocarcinomas. Nat. Biotechnol. 2010;28:71–78. doi: 10.1038/nbt.1595. [DOI] [PubMed] [Google Scholar]
- Zou J, Mali P, Huang X, Dowey SN, Cheng L. Site-specific gene correction of a point mutation in human iPS cells derived from an adult patient with sickle cell disease. Blood. 2011;118:4599–4608. doi: 10.1182/blood-2011-02-335554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, McJunkin K, Fellmann C, Dow LE, Taylor MJ, Hannon GJ, Lowe SW. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat. Biotechnol. 2011a;29:79–83. doi: 10.1038/nbt.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011b;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]