Abstract
Like many diseases, diabetic nephropathy is defined in a histopathological context and studied using reductionist approaches that attempt to ameliorate structural changes. Novel technologies in mass spectrometry-based proteomics have the ability to provide a deeper understanding of the disease beyond classical histopathology, redefine the characteristics of the disease state, and identify novel approaches to reduce renal failure. The goal is to translate these new definitions into improved patient outcomes through diagnostic, prognostic and therapeutic tools. Here, we review progress made in studying the proteomics of diabetic nephropathy and provide an introduction to the informatics tools used in the analysis of systems biology data, while pointing out statistical issues for consideration. Novel bioinformatics methods may increase biomarker identification, and other tools, including selective reaction monitoring, may hasten clinical validation.
Keywords: bioinformatics, diabetes, diabetic nephropathy, mass spectrometry, proteomics, sytems biology
Introduction
Our knowledge of the pathogenesis and progression of chronic diabetic complications historically has been based on functional and structural assessments of the disease. Renal functional assessments have included glomerular filtration rate and albuminuria, with the latter being used as a biomarker of disease progression for several decades. Many perceive the disease from a glomerulocentric view and characterize the disease based on histopathological findings such as glomerular enlargement with mesangial expansion, glomerular capillary basement membrane thickening, podocyte structural changes, and Kimmelstiel-Wilson nodules. Consequently, research into the pathophysiology, as well as the development of therapeutics to treat diabetic complications, have focused on ameliorating this histopathology. An important goal of this review is to provide a current understanding of how proteomics has contributed to a systems biology view of diabetic nephropathy based on disease pathophysiology, not solely on the structural characteristics that are produced by the disease process.
The Diabetes Control and Complications Trial [1] provided strong documentation of a causal link between tight control of hyperglycemia with insulin and the rate of development of diabetic nephropathy in type 1 diabetic patients. Despite this clinical evidence, the mechanism of how diabetes leads to the functional and structural changes that currently define diabetic nephropathy still remains controversial [2]. Over the last four decades, reductionistic strategies focused on individual proteins or metabolic pathways have been used to understand the pathophysiology of diabetic complications, and numerous biochemical and metabolic pathways have emerged as predominant mechanisms for glucose-induced tissue injury [3-16]. While it is recognized that there are numerous points of crosstalk between these different mechanisms [17-19], strategies that try to integrate the numerous biomolecules into a systems understanding of diabetic complications have not yet been fully developed.
Identification of the entire human and rodent genomes has led to the development of web-based databases containing the amino acid sequence of all expressed proteins derived from the genes identified in these genome projects (Mascot, if the MS/MS spectra of peptides are used for protein identification; ProteinProspector if the mass of the charge ratios of peptides are used for protein identification). The field of proteomics — a description of all expressed proteins within particular subcellular compartments, cells, tissues, organs, or model organisms — has grown tremendously due to a convergence of these databases with technical advances in protein fractionation, high throughput mass spectrometry, protein identification, and web-based data resources. These advances in mass spectrometry and web data resources now enable the expression level of thousands of proteins to be quantified simultaneously, much like the genomic approaches that allow thousands of genes to be quantified in one experiment. To date, descriptions of these proteomes have resulted in large datasets of proteins identified in normal tissues, and large sets of dysregulated proteins identified in disease states. However, it is difficult to compare proteomic studies due to a variety of reasons, including: differences in cells and tissues evaluated, differences in methods used for protein extraction and separation, differences in the mass spectrometers that are used, lack of standardization of what constitutes a significantly dysregulated protein, and uncertain application of bioinformatics.
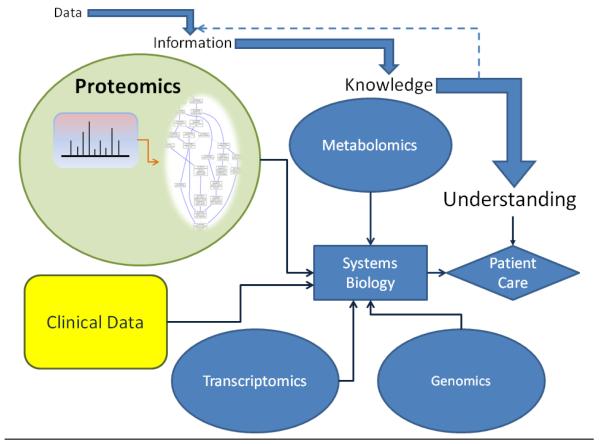
Proteomics contributes to systems biology, a process of knowledge discovery from large sets of biological data created by omics approaches, regardless of the source. These omics datasets that are parts of systems biology include genomics (genes), transcriptomics (message), proteomics (proteins), and metabolomics (metabolites) that can be integrated into higher level understandings of a disease process (Figure 1). In a systems biology-based approach of proteomics (to which we will restrict our attention for this review), individual protein elements (e.g., angiotensin-II or the angiotensin receptor) are subsets of higher order cassettes of information processing (e.g., angiotensin receptor signaling). The function of the individual elements must be integrated to describe the overall function of the cassette (e.g. regulation of systemic arterial blood pressure by renin and angiotensin). More importantly, individual cassettes function within the context of all activities to which cells and tissues are responding, and higher order relationships must be integrated based on the interactions of multiple cassettes (e.g., regulation of renal sodium excretion, regulation of inflammation). Consequently, the overall process of developing a proteomics-focused, systems biology-based understanding of diabetic nephropathy requires information about the expression level of thousands of individual proteins, their function, and how their function is altered by the diabetes-driven metabolic activities within cells and tissues. This is diametrically opposed to reductionistic strategies focused on an individual protein or pathway affected by diabetes. A goal of systems biology is to integrate all of these interconnected, dysregulated proteins and metabolites to build a model that provides novel pathophysiological perspectives based on the functional activities of all these intrinsic proteins. This is an immense challenge, but ultimately, will redefine diabetic nephropathy based on the pathophysiology of the disease, not solely on the structural characteristics that are produced by the disease process.
Figure 1.
Data, information, and knowledge in systems biology. Clinical data is in a systems view of human disease that is incorporated with the additional data and information from other “omics” experiments. Mass spectrometry data from proteomics experiments is given meaning (often with the application of previous knowledge) to form information that is accumulated in systems biology to discover knowledge that can provide understanding for clinical care decisions. The information takes the form of Gene Ontology analysis in this illustration but may include other bioinformatic tools.
As technology advances in the quality and quantity of protein identification, it is imperative that parallel advances be made in the standardization and bioinformatic analysis of data. An important goal in this review, in addition to describing new biological knowledge being developed through proteomics, is to provide an update on novel strategies being developed in this area. Proteomics, as a part of systems biology, requires methods for the analysis of a multitude of proteins at once. Since a major application of proteomics is biomarker identification, systems biology will play an important role in identifying appropriate protein targets. While most of the diabetes-related proteomic effort has focused on biomarker discovery and identification of new therapeutic targets, here our focus will be on reviewing the status of literature in the use of systems approaches to proteomics. We will review some concepts in mass spectrometry-based proteomics to provide a background to explore the status of efforts to understand the proteome of diabetic kidney disease as well as cover issues relating to analysis of such data.
Proteomic technologies used for studying diabetic nephropathy
Numerous reviews over the last 10 years have detailed the technical aspects of proteomics, including advantages and disadvantages of the different mass spectrometers available for this work, technical advances in the mass spectrometers, the inherent complexity of the proteome due to cellular and tissue protein expression patterns that change over time and with disease, and the extensive posttranslational modification of proteins that govern function and that are disturbed by disease [20-22]. It is beyond the scope of this review to provide a detailed assessment of this literature. Conceptually, proteomic research starts with protein extraction from cells, tissues and organs, and in some cases, degradation into peptides with concurrent isotope labeling. Then, proteins or peptides are separated using either gel electrophoresis or liquid chromatography (LC) before being measured by mass spectrometry. Because the majority of proteomic studies in diabetes have used gel-based separation, this approach, with its inherent disadvantages, will be briefly described, and then contrasted with non gel-based approaches that have been developed to overcome some of these weaknesses.
Gel-based methods
Two-dimensional polyacrylamide gel electrophoresis (2D-GE) followed by mass spectrometry initially has been the most widely used proteomics technique to explore diabetic nephropathy. This gel-based approach requires that complex mixtures of proteins be separated by isoelectric point (pI) in the first dimension and by molecular weight in the second dimension. Despite its widespread application in diabetic studies using tissue culture [23,24], animal [24-27], and human [28] renal tissues, relatively few dysregulated proteins have been identified. Down regulation of podocyte annexins III and VI were observed out of 39 proteins identified in an attempt to elucidate the podocyte proteome [23]. Using proteins derived from whole kidney extracts of OVE26 mice, only 41 proteins were identified, with expression of monocyte/neutrophil elastase inhibitor increased and elastase IIIB decreased, suggesting that elastin expression is altered in diabetic kidneys [27]. Similarly, in our work, we used 2D-GE to identify 147 nonredundant proteins dysregulated in the renal cortex of db/db mice, and used Gene Ontology classification to map molecular functions dysregulated by diabetes [25].
This inability of 2D-GE to identify large numbers of proteins is due primarily to several shortcomings of this experimental approach, including: limited loading capacity of the 2D gels, inability of hydrophobic membrane proteins to enter the gel used for isoelectric focusing, poor resolution of proteins at the extreme range of pI and molecular weight. The need to individually pick, extract, digest and analyze spots on 2D gels creates a time-consuming process not readily amenable to high throughput. Another major challenge using 2D-GE is associated with issues of quantitation and reproducibility [29-31], given the number of replicate gels necessary to give adequate detection of differentially expressed proteins [25]. An additional caveat is the observation that individually resolved spots may contain multiple distinct proteins of nearly identical molecular weight and pI, thus making protein quantification in that spot problematic. This limitation is increasingly an issue as significant advances are made in the resolving capability of mass spectrometers. Finally, separation by charge in the first dimension of 2D gels can result in significant charge training, in which post-translational modifications of a single protein result in multiple spots with slight variations in molecular weight and pI. While the intensity of individual spots within the charge train can differ significantly between experimental groups, the total protein amount may not differ. Recent advances, including prefractionation and sequential extraction with improved detergents to enrich subpopulations of proteins [32], improved dyes for spot staining, and DIGE (difference gel electrophoresis) that allows multiplexing for higher throughput [33,34], have aided 2D-GE.
Non gel-based, shotgun methods
The aforementioned shortcomings of 2D-GE have resulted in the development of alternative methods for protein separation prior to mass spectrometry. Since mass spectrometry characterizes peptides and proteins based on molecular mass, stable isotope labeling of peptides has been an ideal choice for quantification of proteins by mass spectrometry. A variety of metabolic (isotopically enriched nutrients incorporated into proteins at specific amino acid sites during growth), chemical, or enzymatic methods have been used for stable isotope labeling [35]. A less expensive approach than metabolic labeling is the incorporation of isotope-coded affinity tags (ICAT) on particular components of total peptide digests, such as cysteine-containing proteolytic peptides [36]. An even simpler approach is the incorporation of stable tags on N- and C-terminal sites during enzymatic proteolysis, and 18O incorporation on the C-terminal end of cleaved peptides is one of the most promising approaches [37]. In this method, when cleavage is performed in heavy water (H 182O) with proteases such as trypsin, Glu-C, or Lys-C, two 18O atoms will be incorporated into the C-terminal carboxylic acid of all peptides, thereby increasing the mass of the peptide by 4 Da compared to those peptides cleaved in normal water (H 162O). With this mass difference, a high-resolution mass spectrometer can distinguish the labeled and unlabeled peptides, and because 18O labeling occurs on all the protease-digested peptides, proteome coverage and quantitative accuracy are improved. Additionally, 18O labels in the carboxylate group of peptides are resistant to back exchange, and under routine conditions used for electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI), covalent bonds between oxygen atoms and carbonyl carbon in a C-terminal carboxylate group are stable. Importantly, due to the increased 4 Da mass of the 18O-labeled peptides, all peptide fragments from a control and experimental group can be mixed together and run simultaneously to quantify differentially expressed proteins using LC-MS/MS [37]. Despite the slight difference in mass, the peptides elute from LC columns that separate by charge or hydrophobicity together since their physicochemical properties remain unchanged. These shotgun proteomics approaches have developed into powerful techniques to identify large numbers of proteins in complex biological samples, but also suffer from a poor relative quantification of the identified proteins due to limited sensitivity of the approach, and poor reproducibility of protein identification when measured repeatedly from the same or similar complex protein samples (technical replicates; discussed below).
New methods in quantitative proteomics research
Although the non-gel, shotgun approaches in which proteins are enzymatically degraded to peptides and labeled prior to analysis by mass spectrometry are rapidly becoming the gold standard, methods to more precisely and reliably quantify proteins of interest are being developed. Selected reaction monitoring (SRM) has been used for decades with triple quadrupole mass spectrometers to quantitatively analyze small molecules [38,39], but is increasingly being used to overcome the shortcomings of the shotgun label approaches [40,41]. As with the quantification of small molecules, SRM use in proteomics takes advantage of the triple quadrupole mass spectrometer for the quantitative analysis. In contrast to the shotgun approaches, SRM analyses target specific, predetermined sets of peptides that represent proteins of interest, and depend on specific SRM transitions for each targeted peptide. Prior information is required in SRM. First, the specific proteins of interest must be identified. Second, for each protein of interest, multiple tryptic peptides must be identified. Suitable peptides must uniquely identify the protein and have good mass spectrometry responses (no missed cleavages, good ionization efficiency, reproducibly detected in the mass spectrometer, good mass to charge ratio within the mass range of the instrument). Third, the fragment ions of these tryptic peptides must have excellent signal intensities to discriminate these peptides from all other peptides in the complex mixture. As discussed below, there is assistance provided by web-based databases that can improve the identification of appropriate peptides. SRM has the potential to assist in the biomarker validation process as an alternative to ELISA methods.
Defining the renal proteome in normal and diabetic states
Normal renal and urine proteomes
As detailed above, numerous proteomic studies have used 2D-GE and LC-MS/MS approaches to characterize the renal proteome, including proteomes of renal cortex [42,43], glomerular cells [44], and tubular epithelial cells [45,46]. In addition, proteomic methodologies increasingly have focused on the identification and quantification of proteins found in urine [47-51], primarily to identify potential biomarkers of renal disease [52]. These studies essentially have resulted in large lists of proteins that can be identified in these tissues. The identified proteins are dependent upon protein abundance that is determined by how the tissues are evaluated (whole or fractionated tissues; eg, glomeruli and tubules), how proteins are extracted, what fractionation strategies are used immediately prior to submitting the sample to a mass spectrometer, and what type of mass spectrometer is used. While systems biology of renal diseases has been discussed [53], to date, there have been no attempts to use bioinformatic approaches to combine these datasets into higher order cassettes describing organ function using systems biology strategies.
Diabetic renal proteome
Construction of dysregulated proteome maps from the quantification of diabetes-induced global changes in renal protein expression patterns has rarely been attempted. Investigators have used the db/db mouse model of type 2 diabetes combined with 2-DE to identify large increases in 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA), a key enzyme in ketogenesis [26], and to identify a number of anti-oxidant (peroxiredoxin 1 and 3, glutathione peroxidase 1, SOD-1) and glycation (glyoxalase 1) enzymes that were increased [54]. The OVE26 mouse model of type 1 diabetes combined with 2-DE has been used to identify 30 dysregulated proteins, with the calcium binding protein calbindin-D28k demonstrating the largest increase [55]. In a second study using 2-DE of whole kidney extracts from the same animal model, these authors describe increased expression of monocyte/neutrophil elastase inhibitor and decreased expression of elastase IIIB [27]. Using immunohistochemistry of mouse and human renal tissue, they documented increased elastin expression in tubular epithelium and interstitium but not in glomeruli. Other investigators, using the same OVE26 mouse model, utilized spectral counting techniques to derive a protein abundance factor, and identified a much larger number of dysregulated proteins in purified renal tubules (476 differentially expressed proteins) [56]. This number of proteins was sufficiently large to perform bioinformatics analyses, and dysregulated functional groups included TGF-β signaling, tight junction maintenance, oxidative stress, and glucose metabolism. Grb2-related adaptor protein (GRAP) was identified as a novel regulator of TGF-β signaling in renal tubule cells. An interesting approach combined 2-DE followed by western blotting using antibodies against advanced glycation endproducts to identify glycated proteins. Mass spectrometry of these antibody-tagged spots identified a number of glycated proteins involved in metabolic pathways, oxidative stress, cell signaling, and cell transport in kidneys sampled from streptozotocin-diabetic rats [57]. While these animal studies have documented novel proteins of interest in diabetic nephropathy, the varying results obtained, even using the same animal model and proteomics techniques, emphasize the difficulty in comparing proteomic studies.
In contrast, a much larger number of studies have addressed the urinary proteome of diabetic nephropathy [58-60]. However, most studies have been used for biomarker identification purposes, rather than to define the renal proteome under pathophysiological (i.e. diabetic kidney disease) conditions [61-70]. The discussion will focus on clinical studies of biomarker identification in diabetic nephropathy and, in one example, some movement from identification to validation of protein biomarkers.
Using proteomics for biomarker and therapeutic target discovery in diabetes
For 30 years, microalbuminuria has been used as a biomarker for increased risk for developing diabetic nephropathy based on the initial studies documenting that ~80 % of patients progressed from microalbuminuria to full proteinuria in approximately a decade. Overt proteinuria, defined as >300 mg/day of urine albumin excretion, is a major hallmark of diabetic nephropathy [71-74]. However, more recent studies fail to show that changes in microalbuminuria predict nephropathy progression, and there is a contentious, ongoing debate about the predictive value of microalbuminuria, and the relative importance of using microalbuminuria as a renal endpoint in clinical trials [75-77]. This debate centers around three concerns, including 1) a large number of patients with microalbuminuria revert to normal albumin excretion, 2) only a small percentage of patients with microalbuminuria progress to proteinuria, and 3) progressive renal functional decline is already present in one third of patients that progress into microalbuminuria [78,79]. Nevertheless, urine has increasingly emerged as an important source for proteomic-based, biomarker evaluation. This is based on the ease of collection and pre-analytical handling, as well as the stability of proteins in urine. Urinary proteins are now recognized as composed of the following: filtered and secreted plasma proteins, proteins secreted by renal tubular epithelium, proteolytic degradation products of extracellular matrix, and proteins derived from shedded cells along the urinary tract. Importantly, the ~70 % of urinary proteins derived from the kidney [80,81] provide a useful biomarker source for evaluating the health status of the kidney under normal and diseased conditions. While virtually every gel- and non gel-based mass spectrometry approach has been utilized to assess the urinary proteome of human and experimental animal models of diabetes, most studies have used 2D-GE and capillary electrophoresis-mass spectrometry (CE-MS) techniques. The latter appears to be ideally suited to the low molecular weight range of urinary proteins (<20 kDa) that are well represented in the normal urine proteome [82]. Numerous recent reviews, including practical points on sample collection, protein fractionation, and mass spectrometery are available [50,61,80,83-88], and will not be repeated here.
The possibility that tubulointerstitial inflammation could contribute to the progression of diabetic nephropathy [89-91] and could be measured in urine by assessing pro-inflammatory cytokines, led to a multiplex-based assessment of urine chemokines and cytokines [62]. In this study, five inflammatory markers (IL-6, IL-8, MCP-1, interferon gamma-inducible protein (IP-10, and macrophage inflammatory protein -1δ) were assayed in urine obtained from the First Joslin Study of the Natural History of Microalbuminuria in Type 1 Diabetes, that started in 1991 and was completed in 2005. 43 patients with microalbuminuria and stable renal function were compared to 28 patients with microalbuminuria and early progressive renal function decline, and 74 controls with no microalbuminuria and stable renal function. All 5 markers were higher in decliners vs nondecliners, and those with more than two were 5 times more likely to have early progressive renal decline. While there are caveats in the experimental design that temper a conclusion of causality, the potential importance of assessing panels of biomarkers is strengthened by this study.
Merchant et al., [92] compared patients with microalbuminuria and early renal functional decline to patients with microalbuminuria but stable renal function for 10-12 years. They identified three peptides that were increased, including inositol pentakisphosphate 2 kinase (IPP2K), zona occludens 3 (ZO-3), and cadherin-like protein FAT tumor suppressor 2, and three peptides that were decreased, including α-1(IV) and α-1(V) collagen and tenascin-X. Once again, their results suggest that panels of proteins may provide the best prognostic value.
Alkhalaf et al. [69] showed some movement from biomarker identification to eventual validation using patients with overt nephropathy compared to age and sex-matched controls. The putative biomarkers were previously identified in a similar study [61]. Hopefully, there will be a clinical return on investment into these biomarker studies, but to date, shotgun proteomics has not led to a single biomarker in clinical use.
Bioinformatics and systems biology for proteomics
As first presented in Figure 1, the idea that data and information lead to knowledge which leads to understanding is an important concept when using systems biology (for more information see [93]). Proteomics generate large volumes of data and, when combined with shared information in the form of the identification and quantification of the protein, can lead to the discovery of new knowledge and provide understanding of disease processes. In regards to the process of adding meaning to MS data, it is beyond the scope of this article to include a full review of analysis of MS signals for peptide and protein identification and excellent recent reviews are available [94]. Instead, this review will focus on analysis of protein data once appropriate information, namely the protein identification, has been annotated to MS data. First, we will explore some of the computational issues surrounding proteomics research. Then, an introduction to application of systems biology to proteomics data will be covered, including the following topics: controlled vocabularies, biological pathways, and network analysis.
Computational issues
There is a need to balance biological and technical replicates for proteomics studies. Technical replicates increase the depth of proteome coverage by increasing the probability that a protein with lower protein abundance (as can be measured by the spectral abundance) is found in a particular sample. Biological replicates, on the other hand, are necessary due to attempts to minimize within-class subject variability and inherent noise of proteomic studies.
Urinary proteomic inter- and intra-sample variability in biological and technical replicates, respectively, was addressed by evaluating urinary proteomic changes in a single subject with technical replicates, differently timed samples, and subject to subject (i.e. biologic) variability [95]. The data suggest that the vast majority of variability comes from inter-sample (biological) variability and not intra-sample (technical) variability. That is, the coefficient of variation (CV) for technical replicates was found to be 0.18 and for biological replicates as high as 0.66 between different individuals [95]. This is similar to results found by others [96]. The large variance in biological replicates is a serious obstacle for gaining significant knowledge and identifying biomarkers from proteomic studies. Statistical power is often an issue for proteomic studies given the biological variance [97], and pilot studies, in general, are often underpowered and fail to reach valid statistical conclusions largely due to this biological variance [98].
There are two components to this problem of inherent biological variance that are logistical and statistical. From a logistical perspective, there is limited time and funding for these proteomic studies. One way to overcome this is the sharing of proteomic studies through online databases, including: PRIDE, TRANCHE and PeptideAtlas. PRIDE is especially useful for sharing data as it allows access to unprocessed data for potential meta-analysis of multiple studies [99]. Other databases are resources to aid in peptide sequencing and protein identification; this is important for selective reaction monitoring to provide the prior knowledge for assay development. Indeed, these initiatives have joined together to form the ProteomeXchange as a way to access all three databases under a single query, although it is still in development. Also, Kolker et al. [100] recently developed an easy way to access multiple databases that contain raw data from proteomics experiments and includes methods for meta-analysis of data. Data standards have been created for use by the Proteomics Standards Initiative (PSI), a work group of the Human Proteome Organization (HUPO) [101]. This allows for improved communication in reporting data from proteomic studies similar to the Health Level 7 (HL7) process for electronic medical records.
From a statistical perspective, it is important that results be analyzed in a meaningful way. One method is to incorporate the use of random effects models (using the methods of Dersimonian and Laird) to combine replicate experiments (i.e. biological replicates) [102]. Although currently used by a few researchers for this purpose [103-105], in theory the method is extensible across multiple instrument types and labeling techniques. The mining of data from multiple related proteomic experiments can then be analyzed using random effects models similar to the meta-analysis of clinical trials. That is, studies may have different sources of determinate and indeterminate error that can be accounted for through the use of random effects as has been effectively used in evidence-based medicine to gain knowledge through analysis of heterogeneous trials of similar purpose. The combination of standardized data warehousing/sharing and random effects models should overcome the statistical issues when fully implemented. Once a list of the proteomic changes associated with diabetic kidney disease is established, bioinformatics allows for knowledge discovery based on the data and information. We will discuss with some examples, a few of the key resources available for bioinformatics and summarized in Table 1.
| Tool | Web-based Resource |
Advantages | Limitations | Examples of Use |
|---|---|---|---|---|
| Gene Ontology (GO) |
www.geneontology.org |
|
|
|
| Pathway Analysis |
www.genome.jp/kegg/ |
|
|
|
| Network Analysis |
www.ingenuity.com
www.genego.com www.cytoscape.org |
|
|
Controlled vocabularies
Controlled vocabularies provide a predefined dictionary of terms that can be used for descriptive purpose. One common example of a controlled vocabulary is the Medical Subject Heading (MeSH) used by the National Library of Medicine to describe scientific literature for indexing and searching on Medline. In terms of analysis of systems biology data (in general, as well as proteomics data, specifically), the Gene Ontology (GO) project (www.geneontology.org) was created to provide a controlled vocabulary to describe biological data. Ontologies are, by definition, organized and typically hierarchical in nature and GO is no exception. GO allows for description of biological data utilizing a controlled vocabulary that also contains some relations between terms. In general there are parent and child terms that are structured to have increasing levels of granularity among terms in the more specific child terms.
The mapping of genes and gene products to GO terminology provides a qualitative abstraction of terms to more generalized molecular functions, cellular components and biological functions. In proteomics, it is easy to perform GO analysis using a variety of sources available through the GO website. Identifiers such as the UniProt accession can be directly imported and the GO terminology assigned along with the statistical probability (in certain applications) that this list of proteins would be assigned to that GO term versus a randomly selected list of UniProt accession numbers. For example, Figure 1 contains information in terms of GO classification of the term “angiotensin mediated vasoconstriction” (within the proteomics domain) involved in regulation of systemic arterial blood pressure and was generated using AmiGO software [106]. In fact, the terms of protein cassettes and their functional elements in the Introduction refer to GO terminology (e.g. regulation of systemic arterial blood pressure by renin and angiotensin). The concept that differentially expressed proteins could then be mapped to specific GO terminology allows one to understand the overall molecular functions, cellular processes, and biological functions of the differentially expressed proteome. Yoshida, et al. [44] documented GO biological processes and molecular functions of the human glomerular proteome. The primary molecular function was cytoskeletal and the primary biological process was cell organization. This compares to the evaluation of the total mouse cortical proteome that contains a large number of enzymes that are mitochondrial [107].
Biological Pathways
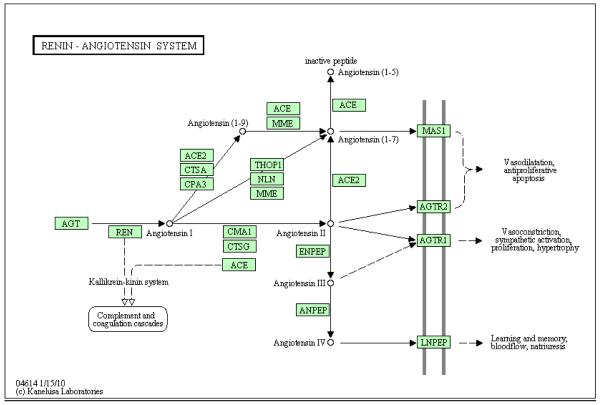
The Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/) is another invaluable asset that contains a knowledge base of proteins in biochemical pathways that are organized by biochemical function. Figure 2 shows the associated KEGG pathway for angiotensin II formation and signaling [108]. Notice that as opposed to GO terminology, KEGG demonstrates relationships among proteins that map to a pathway rather than the interaction and hierarchy of descriptive terms. For example, Cummins, et al. used pathway analysis to identify the importance of Grb2-related adaptor protein (GRAP) in the TGF-β signaling in response to diabetes in OVE26 mice [56]. This knowledge was used to show the involvement of GRAP in vitro tubular cells in response to hyperglycemia [56].
Figure 2.
KEGG pathway of the renin-angiotensin system. The generation of angiotensin II by angiotensin converting enzyme (ACE) and signaling through the AGTR1 and AGTR2 are depicted as well as the metabolite, angiotensin 1-7, that activates mas. It demonstrates metabolic pathways containing intermediaries and associated enzymes.
Network Analysis
The interactions between biomolecules can similarly be established from the data itself, as opposed to mapping to existing pathways, using network analysis. IPA (Ingenuity® Systems, www.ingenuity.com) is a commercial product that, in addition to GO and pathway (often following KEGG for metabolic pathways but also including proprietary signaling pathways) analyses, generates interaction networks de novo from user-submitted data. This is opposed to GO and pathway analysis in which user-submitted data are mapped to pre-existing terms and pathways, respectively. IPA networks are generated using a knowledge base that is extensively curated from the literature (semi-automated). Extensive information is available on the IPA website (www.ingenuity.com) as to the methods of network generation and scoring. GeneGo (Thompson Reuters, www.genego.com) is an additional commercial product for bioinformatic analysis that also includes many of the same features as IPA.
An open-source product, Cytoscape, (www.cytoscape.org), has been developed to serve as an alternative for creation of networks using user data and information with plugin-based knowledge comprised of shared protein interaction networks from other open-source web initiatives, including GO and KEGG (among others) [109]. VisANT also provides network visualization that can integrate both GO and KEGG pathways as an alternative to Cytoscape [110]. In both cases, the size of the knowledge bases has the potential for substantial growth due to the open-source environment and the use of plugins that are separate from the visualization platform.
All network analyses rely on knowledge bases that contain protein-protein interactions (PPI). There are three major sources of the protein interactions: scientific literature (typically using reductionist strategies as IPA contains), high throughput experiments (e.g. yeast two hybrid screens) and computational prediction [111]. Each PPI discovery method is fraught with type I and type II errors, generating false positives and negatives, respectively. For example, a criticism of the IPA knowledge base is the potential for literature bias (i.e. the most studied proteins are most likely to have known interactions). Predictions based upon high throughput experiments suffer from high false positive rates due to spurious interaction that is not an in vivo interaction but a product of the experiment. Computational prediction provides only indirect evidence of interactions. In the realm of proteomics, utilizing mass spectrometry for large scale identification of protein interaction networks is becoming more common [112].
Once established, network representations provide an understanding of both interconnecting molecules and allows the establishment of signaling ‘hubs’ that show potential points of regulation. The flexibility of networks is that it can differentiate different biomolecules and provide bridging between ‘omics technologies. For example, we recently published a proteomics study that provided a comparison of db/db mice to their C57 db/m counterparts [113]. This led to the generation of a network that contained a metabolite, all-trans retinoic acid, as a potential hub. Therefore, we undertook a targeted metabolomics study on the status of renal retinoid concentration in this model of type 2 diabetes. Similarly, using the plasma of type 1 diabetic patients, Overgaard, et al. used IPA network analysis to demonstrate interactions among apolipoproteins [114]. This highlights the process of gaining understanding and insight into a disorder through the knowledge discovery process on a proteomics dataset.
Conclusions
Systems biology provides a platform for the discovery of biological knowledge from large amounts of data and information. Although significant progress has been made towards ensuring the quality of data obtained by mass spectrometry-based proteomics, there still remains a lot of work to provide meaningful results that can be translated to improved patient outcomes. The identification of biomarkers is currently the primary purpose of most proteomics experiments in diabetic nephropathy. However, there is a need to shift to systems biology for a more comprehensive view of the disease and gain understanding of the pathophysiological processes. The tools presented here are a way to start thinking of the disease in a more complete, integrated manner instead of relying on histopathological characteristics to define disease processes. Proteomics has immense potential to improve patient outcomes by translating the large amounts of data and information obtained by MS into knowledge of diagnostic, prognostic and therapeutic biomarkers as well as provide understanding of the pathophysiology of diabetic kidney disease.
Acknowledgments
Work in our laboratory was supported by the Juvenile Diabetes Research Foundation, International (JDRF) grant award #1-2008-202 and grant Award 39-2009-643.
Footnotes
Ethical standards The authors declare that all experiments over which the authors have control in this review article comply with all current laws of the country in which these experiments were performed (USA).
Conflict of interest The authors do not have a financial relationship with the organization that sponsored the research. We have full control of all primary data and agree to allow the journal to review these data if requested.
Reference List
- 1.The Diabetes Control and Complications Trial Research Group The Effect of Intensive Treatment of Diabetes on the Development and Progression of Long-Term Complications in Insulin-Dependent Diabetes Mellitus. 1993;329:977–986. doi: 10.1056/NEJM199309303291401. [DOI] [PubMed] [Google Scholar]
- 2.Welsh GI, Hale LJ, Eremina V, Jeansson M, Maezawa Y, Lennon R, Pons DA, Owen RJ, Satchell SC, Miles MJ, Caunt CJ, McArdle CA, Pavenstadt H, Tavare JM, Herzenberg AM, Kahn CR, Mathieson PW, Quaggin SE, Saleem MA, Coward RJ. Insulin signaling to the glomerular podocyte is critical for normal kidney function. Cell Metab. 2010;12:329–340. doi: 10.1016/j.cmet.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oates PJ, Mylari BL. Aldose reductase inhibitors: therapeutic implications for diabetic complications. Expert Opin Investig Drugs. 1999;8:2095–2119. doi: 10.1517/13543784.8.12.2095. [DOI] [PubMed] [Google Scholar]
- 4.Brownlee M, Cerami A, Vlassara H. Advanced products of nonenzymatic glycosylation and the pathogenesis of diabetic vascular disease. Diabetes Metab Rev. 1988;4:437–451. doi: 10.1002/dmr.5610040503. [DOI] [PubMed] [Google Scholar]
- 5.Bucala R, Vlassara H. Advanced glycosylation end products in diabetic renal and vascular disease. Am J Kidney Dis. 1995;26:875–888. doi: 10.1016/0272-6386(95)90051-9. [DOI] [PubMed] [Google Scholar]
- 6.Singh R, Barden A, Mori T, Beilin L. Advanced glycation end-products: a review. Diabetologia. 2001;44:129–146. doi: 10.1007/s001250051591. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt AM, Hori O, Chen JX, Li JF, Crandall J, Zhang J, Cao R, Yan SD, Brett J, Stern D. Advanced glycation endproducts interacting with their endothelial receptor induce expression of vascular cell adhesion molecule-1 (VCAM-1) in cultured human endothelial cells and in mice. A potential mechanism for the accelerated vasculopathy of diabetes. J Clin Invest. 1995;96:1395–1403. doi: 10.1172/JCI118175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bierhaus A, Schiekofer S, Schwaninger M, Andrassy M, Humpert PM, Chen J, Hong M, Luther T, Henle T, Kloting I, Morcos M, Hofmann M, Tritschler H, Weigle B, Kasper M, Smith M, Perry G, Schmidt AM, Stern DM, Haring HU, Schleicher E, Nawroth PP. Diabetes-associated sustained activation of the transcription factor nuclear factor-kappaB. Diabetes. 2001;50:2792–2808. doi: 10.2337/diabetes.50.12.2792. [DOI] [PubMed] [Google Scholar]
- 9.Craven PA, Studer RK, Negrete H, DeRubertis FR. Protein kinase C in diabetic nephropathy. J Diabetes Complications. 1995;9:241–245. doi: 10.1016/1056-8727(95)80012-4. [DOI] [PubMed] [Google Scholar]
- 10.DeRubertis FR, Craven PA. Activation of protein kinase C in glomerular cells in diabetes. Mechanisms and potential links to the pathogenesis of diabetic glomerulopathy. Diabetes. 1994;43:1–8. doi: 10.2337/diab.43.1.1. [DOI] [PubMed] [Google Scholar]
- 11.Koya D, King GL. Protein kinase C activation and the development of diabetic complications. Diabetes. 1998;47:859–866. doi: 10.2337/diabetes.47.6.859. [DOI] [PubMed] [Google Scholar]
- 12.Baynes JW, Thorpe SR. Role of oxidative stress in diabetic complications: a new perspective on an old paradigm. Diabetes. 1999;48:1–9. doi: 10.2337/diabetes.48.1.1. [DOI] [PubMed] [Google Scholar]
- 13.Szabo C. Roles of poly(ADP-ribose) polymerase activation in the pathogenesis of diabetes mellitus and its complications. Pharmacol Res. 2005;52:60–71. doi: 10.1016/j.phrs.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 14.Harding HP, Ron D. Endoplasmic reticulum stress and the development of diabetes: a review. Diabetes. 2002;51(Suppl 3):S455–S461. doi: 10.2337/diabetes.51.2007.s455. [DOI] [PubMed] [Google Scholar]
- 15.Araki E, Oyadomari S, Mori M. Endoplasmic reticulum stress and diabetes mellitus. Intern Med. 2003;42:7–14. doi: 10.2169/internalmedicine.42.7. [DOI] [PubMed] [Google Scholar]
- 16.Wolf G. Molecular mechanisms of angiotensin II in the kidney: emerging role in the progression of renal disease: beyond haemodynamics. Nephrol Dial Transplant. 1998;13:1131–1142. doi: 10.1093/ndt/13.5.1131. [DOI] [PubMed] [Google Scholar]
- 17.Nishikawa T, Edelstein D, Du XL, Yamagishi S, Matsumura T, Kaneda Y, Yorek MA, Beebe D, Oates PJ, Hammes HP, Giardino I, Brownlee M. Normalizing mitochondrial superoxide production blocks three pathways of hyperglycaemic damage. Nature. 2000;404:787–790. doi: 10.1038/35008121. [DOI] [PubMed] [Google Scholar]
- 18.Hammes HP, Du X, Edelstein D, Taguchi T, Matsumura T, Ju Q, Lin J, Bierhaus A, Nawroth P, Hannak D, Neumaier M, Bergfeld R, Giardino I, Brownlee M. Benfotiamine blocks three major pathways of hyperglycemic damage and prevents experimental diabetic retinopathy. Nat Med. 2003;9:294–299. doi: 10.1038/nm834. [DOI] [PubMed] [Google Scholar]
- 19.Williamson JR, Chang K, Frangos M, Hasan KS, Ido Y, Kawamura T, Nyengaard JR, Van den EM, Kilo C, Tilton RG. Hyperglycemic pseudohypoxia and diabetic complications. Diabetes. 1993;42:801–813. doi: 10.2337/diab.42.6.801. [DOI] [PubMed] [Google Scholar]
- 20.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 21.Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 22.Cox J, Mann M. Quantitative, high-resolution proteomics for data-driven systems biology. Annu Rev Biochem. 2011;80:273–299. doi: 10.1146/annurev-biochem-061308-093216. [DOI] [PubMed] [Google Scholar]
- 23.Schordan S, Schordan E, Endlich N, Lindenmeyer MT, Meyer-Schwesinger C, Meyer TN, Giebel J, Cohen CD, Endlich K, Maurer MH. Alterations of the podocyte proteome in response to high glucose concentrations. Proteomics. 2009;9:4519–4528. doi: 10.1002/pmic.200800214. [DOI] [PubMed] [Google Scholar]
- 24.Li Z, Zhang H, Dong X, Burczynski FJ, Choy P, Yang F, Liu H, Li P, Gong Y. Proteomic profile of primary isolated rat mesangial cells in high-glucose culture condition and decreased expression of PSMA6 in renal cortex of diabetic rats. Biochem Cell Biol. 2010;88:635–648. doi: 10.1139/O09-185. [DOI] [PubMed] [Google Scholar]
- 25.Tilton RG, Haidacher SJ, LeJeune WS, Zhao Y, Kurosky A, Brasier AR, Denner LA. Diabetes-induced changes in the renal cortical proteome assessed with two-dimensional gel electrophoresis and mass spectrometry. 2007;7:1729–1742. doi: 10.1002/pmic.200700017. [DOI] [PubMed] [Google Scholar]
- 26.Zhang D, Yang H, Kong X, Wang K, Mao X, Yan X, Wang Y, Liu S, Zhang X, Li J, Chen L, Wu J, Wei M, Yang J, Guan Y. Proteomics analysis reveals diabetic kidney as a ketogenic organ in type 2 diabetes. Am J Physiol Endocrinol Metab. 2011;300:E287–E295. doi: 10.1152/ajpendo.00308.2010. [DOI] [PubMed] [Google Scholar]
- 27.Thongboonkerd V, Barati MT, McLeish KR, Benarafa C, Remold-O’Donnell E, Zheng S, Rovin BH, Pierce WM, Epstein PN, Klein JB. Alterations in the renal elastin-elastase system in type 1 diabetic nephropathy identified by proteomic analysis. J Am Soc Nephrol. 2004;15:650–662. doi: 10.1097/01.asn.0000115334.65095.9b. [DOI] [PubMed] [Google Scholar]
- 28.Miyamoto M, Yoshida Y, Taguchi I, Nagasaka Y, Tasaki M, Zhang Y, Xu B, Nameta M, Sezaki H, Cuellar LM, Osawa T, Morishita H, Sekiyama S, Yaoita E, Kimura K, Yamamoto T. In-depth proteomic profiling of the normal human kidney glomerulus using two-dimensional protein prefractionation in combination with liquid chromatography-tandem mass spectrometry. J Proteome Res. 2007;6:3680–3690. doi: 10.1021/pr070203n. [DOI] [PubMed] [Google Scholar]
- 29.Almeida JS, Stanislaus R, Krug E, Arthur JM. Normalization and analysis of residual variation in two-dimensional gel electrophoresis for quantitative differential proteomics. Proteomics. 2005;5:1242–1249. doi: 10.1002/pmic.200401003. [DOI] [PubMed] [Google Scholar]
- 30.Merril CR, Creed GJ, Joy J, Olson AD. Identification and use of constitutive proteins for the normalization of high resolution electrophoretograms. Appl Theor Electrophor. 1993;3:329–333. [PubMed] [Google Scholar]
- 31.Nishihara JC, Champion KM. Quantitative evaluation of proteins in one- and two-dimensional polyacrylamide gels using a fluorescent stain. Electrophoresis. 2002;23:2203–2215. doi: 10.1002/1522-2683(200207)23:14<2203::AID-ELPS2203>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 32.Molloy MP, Herbert BR, Walsh BJ, Tyler MI, Traini M, Sanchez JC, Hochstrasser DF, Williams KL, Gooley AA. Extraction of membrane proteins by differential solubilization for separation using two-dimensional gel electrophoresis. Electrophoresis. 1998;19:837–844. doi: 10.1002/elps.1150190539. [DOI] [PubMed] [Google Scholar]
- 33.Marouga R, David S, Hawkins E. The development of the DIGE system: 2D fluorescence difference gel analysis technology. Anal Bioanal Chem. 2005;382:669–678. doi: 10.1007/s00216-005-3126-3. [DOI] [PubMed] [Google Scholar]
- 34.Friedman DB, Lilley KS. Optimizing the difference gel electrophoresis (DIGE) technology. Methods Mol Biol. 2008;428:93–124. doi: 10.1007/978-1-59745-117-8_6. [DOI] [PubMed] [Google Scholar]
- 35.Tao WA, Aebersold R. Advances in quantitative proteomics via stable isotope tagging and mass spectrometry. Curr Opin Biotechnol. 2003;14:110–118. doi: 10.1016/s0958-1669(02)00018-6. [DOI] [PubMed] [Google Scholar]
- 36.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 37.Heller M, Mattou H, Menzel C, Yao X. Trypsin catalyzed 16O-to-18O exchange for comparative proteomics: tandem mass spectrometry comparison using MALDI-TOF, ESI-QTOF, and ESI-ion trap mass spectrometers. J Am Soc Mass Spectrom. 2003;14:704–718. doi: 10.1016/S1044-0305(03)00207-1. [DOI] [PubMed] [Google Scholar]
- 38.Kostiainen R, Kotiaho T, Kuuranne T, Auriola S. Liquid chromatography/atmospheric pressure ionization-mass spectrometry in drug metabolism studies. J Mass Spectrom. 2003;38:357–372. doi: 10.1002/jms.481. [DOI] [PubMed] [Google Scholar]
- 39.Yost RA, Enke CG. Triple quadrupole mass spectrometry for direct mixture analysis and structure elucidation. Anal Chem. 1979;51:1251–1264. doi: 10.1021/ac50048a002. [DOI] [PubMed] [Google Scholar]
- 40.Gallien S, Duriez E, Domon B. Selected reaction monitoring applied to proteomics. J Mass Spectrom. 2011;46:298–312. doi: 10.1002/jms.1895. [DOI] [PubMed] [Google Scholar]
- 41.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cutillas PR, Biber J, Marks J, Jacob R, Stieger B, Cramer R, Waterfield M, Burlingame AL, Unwin RJ. Proteomic analysis of plasma membrane vesicles isolated from the rat renal cortex. Proteomics. 2005;5:101–112. doi: 10.1002/pmic.200400886. [DOI] [PubMed] [Google Scholar]
- 43.Magni F, Sarto C, Valsecchi C, Casellato S, Bogetto SF, Bosari S, Di FA, Perego RA, Corizzato M, Doro G, Galbusera C, Rocco F, Mocarelli P, Galli KM. Expanding the proteome two-dimensional gel electrophoresis reference map of human renal cortex by peptide mass fingerprinting. Proteomics. 2005;5:816–825. doi: 10.1002/pmic.200401077. [DOI] [PubMed] [Google Scholar]
- 44.Yoshida Y, Miyazaki K, Kamiie J, Sato M, Okuizumi S, Kenmochi A, Kamijo K, Nabetani T, Tsugita A, Xu B, Zhang Y, Yaoita E, Osawa T, Yamamoto T. Two-dimensional electrophoretic profiling of normal human kidney glomerulus proteome and construction of an extensible markup language (XML)-based database. Proteomics. 2005;5:1083–1096. doi: 10.1002/pmic.200401075. [DOI] [PubMed] [Google Scholar]
- 45.Barile M, Pisitkun T, Yu MJ, Chou CL, Verbalis MJ, Shen RF, Knepper MA. Large scale protein identification in intracellular aquaporin-2 vesicles from renal inner medullary collecting duct. Mol Cell Proteomics. 2005;4:1095–1106. doi: 10.1074/mcp.M500049-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dihazi H, Asif AR, Agarwal NK, Doncheva Y, Muller GA. Proteomic analysis of cellular response to osmotic stress in thick ascending limb of Henle’s loop (TALH) cells. Mol Cell Proteomics. 2005;4:1445–1458. doi: 10.1074/mcp.M400184-MCP200. [DOI] [PubMed] [Google Scholar]
- 47.Pieper R, Gatlin CL, McGrath AM, Makusky AJ, Mondal M, Seonarain M, Field E, Schatz CR, Estock MA, Ahmed N, Anderson NG, Steiner S. Characterization of the human urinary proteome: a method for high-resolution display of urinary proteins on two-dimensional electrophoresis gels with a yield of nearly 1400 distinct protein spots. Proteomics. 2004;4:1159–1174. doi: 10.1002/pmic.200300661. [DOI] [PubMed] [Google Scholar]
- 48.Oh J, Pyo JH, Jo EH, Hwang SI, Kang SC, Jung JH, Park EK, Kim SY, Choi JY, Lim J. Establishment of a near-standard two-dimensional human urine proteomic map. Proteomics. 2004;4:3485–3497. doi: 10.1002/pmic.200401018. [DOI] [PubMed] [Google Scholar]
- 49.Thongboonkerd V, McLeish KR, Arthur JM, Klein JB. Proteomic analysis of normal human urinary proteins isolated by acetone precipitation or ultracentrifugation. Kidney Int. 2002;62:1461–1469. doi: 10.1111/j.1523-1755.2002.kid565.x. [DOI] [PubMed] [Google Scholar]
- 50.Castagna A, Cecconi D, Sennels L, Rappsilber J, Guerrier L, Fortis F, Boschetti E, Lomas L, Righetti PG. Exploring the hidden human urinary proteome via ligand library beads. J Proteome Res. 2005;4:1917–1930. doi: 10.1021/pr050153r. [DOI] [PubMed] [Google Scholar]
- 51.Adachi J, Kumar C, Zhang Y, Olsen JV, Mann M. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 2006;7:R80. doi: 10.1186/gb-2006-7-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hewitt SM, Dear J, Star RA. Discovery of protein biomarkers for renal diseases. J Am Soc Nephrol. 2004;15:1677–1689. doi: 10.1097/01.asn.0000129114.92265.32. [DOI] [PubMed] [Google Scholar]
- 53.He JC, Chuang PY, Ma’ayan A, Iyengar R. Systems biology of kidney diseases. Kidney Int. 2012;81:22–39. doi: 10.1038/ki.2011.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barati MT, Merchant ML, Kain AB, Jevans AW, McLeish KR, Klein JB. Proteomic analysis defines altered cellular redox pathways and advanced glycation end-product metabolism in glomeruli of db/db diabetic mice. Am J Physiol Renal Physiol. 2007;293:F1157–F1165. doi: 10.1152/ajprenal.00411.2006. [DOI] [PubMed] [Google Scholar]
- 55.Thongboonkerd V, Zheng S, McLeish KR, Epstein PN, Klein JB. Proteomic identification and immunolocalization of increased renal calbindin-D28k expression in OVE26 diabetic mice. Rev Diabet Stud. 2005;2:19–26. doi: 10.1900/RDS.2005.2.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cummins TD, Barati MT, Coventry SC, Salyer SA, Klein JB, Powell DW. Quantitative mass spectrometry of diabetic kidney tubules identifies GRAP as a novel regulator of TGF-beta signaling. Biochim Biophys Acta. 2010;1804:653–661. doi: 10.1016/j.bbapap.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chougale AD, Bhat SP, Bhujbal SV, Zambare MR, Puntambekar S, Somani RS, Boppana R, Giri AP, Kulkarni MJ. Proteomic analysis of glycated proteins from streptozotocin-induced diabetic rat kidney. Mol Biotechnol. 2012;50:28–38. doi: 10.1007/s12033-011-9409-3. [DOI] [PubMed] [Google Scholar]
- 58.Sharma K, Lee S, Han S, Lee S, Francos B, McCue P, Wassell R, Shaw MA, RamachandraRao SP. Two-dimensional fluorescence difference gel electrophoresis analysis of the urine proteome in human diabetic nephropathy. Proteomics. 2005;5:2648–2655. doi: 10.1002/pmic.200401288. [DOI] [PubMed] [Google Scholar]
- 59.Meier M, Kaiser T, Herrmann A, Knueppel S, Hillmann M, Koester P, Danne T, Haller H, Fliser D, Mischak H. Identification of urinary protein pattern in type 1 diabetic adolescents with early diabetic nephropathy by a novel combined proteome analysis. J Diabetes Complications. 2005;19:223–232. doi: 10.1016/j.jdiacomp.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 60.Dihazi H, Muller GA, Lindner S, Meyer M, Asif AR, Oellerich M, Strutz F. Characterization of diabetic nephropathy by urinary proteomic analysis: identification of a processed ubiquitin form as a differentially excreted protein in diabetic nephropathy patients. Clin Chem. 2007;53:1636–1645. doi: 10.1373/clinchem.2007.088260. [DOI] [PubMed] [Google Scholar]
- 61.Rossing K, Mischak H, Rossing P, Schanstra JP, Wiseman A, Maahs DM. The urinary proteome in diabetes and diabetes-associated complications: New ways to assess disease progression and evaluate therapy. Proteomics Clin Appl. 2008;2:997–1007. doi: 10.1002/prca.200780166. [DOI] [PubMed] [Google Scholar]
- 62.Wolkow PP, Niewczas MA, Perkins B, Ficociello LH, Lipinski B, Warram JH, Krolewski AS. Association of urinary inflammatory markers and renal decline in microalbuminuric type 1 diabetics. J Am Soc Nephrol. 2008;19:789–797. doi: 10.1681/ASN.2007050556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Otu HH, Can H, Spentzos D, Nelson RG, Hanson RL, Looker HC, Knowler WC, Monroy M, Libermann TA, Karumanchi SA, Thadhani R. Prediction of diabetic nephropathy using urine proteomic profiling 10 years prior to development of nephropathy. Diabetes Care. 2007;30:638–643. doi: 10.2337/dc06-1656. [DOI] [PubMed] [Google Scholar]
- 64.Rao PV, Lu X, Standley M, Pattee P, Neelima G, Girisesh G, Dakshinamurthy KV, Roberts CT, Jr., Nagalla SR. Proteomic identification of urinary biomarkers of diabetic nephropathy. Diabetes Care. 2007;30:629–637. doi: 10.2337/dc06-2056. [DOI] [PubMed] [Google Scholar]
- 65.Jiang H, Guan G, Zhang R, Liu G, Cheng J, Hou X, Cui Y. Identification of urinary soluble E-cadherin as a novel biomarker for diabetic nephropathy. Diabetes Metab Res Rev. 2009;25:232–241. doi: 10.1002/dmrr.940. [DOI] [PubMed] [Google Scholar]
- 66.Papale M, Di PS, Magistroni R, Lamacchia O, Di Palma AM, De MA, Rocchetti MT, Furci L, Pasquali S, De CS, Cignarelli M, Gesualdo L. Urine proteome analysis may allow noninvasive differential diagnosis of diabetic nephropathy. Diabetes Care. 2010;33:2409–2415. doi: 10.2337/dc10-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ben AR, Molina L, Bolvin C, Kifagi C, Jarraya F, Ayadi H, Molina F, Granier C. Proteomic approaches for discovering biomarkers of diabetic nephropathy. Nephrol Dial Transplant. 2010;25:2866–2875. doi: 10.1093/ndt/gfq258. [DOI] [PubMed] [Google Scholar]
- 68.Zhi W, Purohit S, Carey C, Wang M, She JX. Proteomic technologies for the discovery of type 1 diabetes biomarkers. J Diabetes Sci Technol. 2010;4:993–1002. doi: 10.1177/193229681000400431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alkhalaf A, Zurbig P, Bakker SJ, Bilo HJ, Cerna M, Fischer C, Fuchs S, Janssen B, Medek K, Mischak H, Roob JM, Rossing K, Rossing P, Rychlik I, Sourij H, Tiran B, Winklhofer-Roob BM, Navis GJ. Multicentric validation of proteomic biomarkers in urine specific for diabetic nephropathy. PLoS One. 2010;5:e13421. doi: 10.1371/journal.pone.0013421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maahs DM, Siwy J, Argiles A, Cerna M, Delles C, Dominiczak AF, Gayrard N, Iphofer A, Jansch L, Jerums G, Medek K, Mischak H, Navis GJ, Roob JM, Rossing K, Rossing P, Rychlik I, Schiffer E, Schmieder RE, Wascher TC, Winklhofer-Roob BM, Zimmerli LU, Zurbig P, Snell-Bergeon JK. Urinary collagen fragments are significantly altered in diabetes: a link to pathophysiology. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Parving HH, Oxenboll B, Svendsen PA, Christiansen JS, Andersen AR. Early detection of patients at risk of developing diabetic nephropathy. A longitudinal study of urinary albumin excretion. Acta Endocrinol (Copenh) 1982;100:550–555. doi: 10.1530/acta.0.1000550. [DOI] [PubMed] [Google Scholar]
- 72.Viberti GC, Jarrett RJ, Keen H. Microalbuminuria as prediction of nephropathy in diabetics. Lancet. 1982;2:611. doi: 10.1016/s0140-6736(82)90688-2. [DOI] [PubMed] [Google Scholar]
- 73.Mogensen CE, Christensen CK. Predicting diabetic nephropathy in insulin-dependent patients. N Engl J Med. 1984;311:89–93. doi: 10.1056/NEJM198407123110204. [DOI] [PubMed] [Google Scholar]
- 74.Mogensen CE. Microalbuminuria predicts clinical proteinuria and early mortality in maturity-onset diabetes. N Engl J Med. 1984;310:356–360. doi: 10.1056/NEJM198402093100605. [DOI] [PubMed] [Google Scholar]
- 75.Weir MR, Bakris GL. Editorial perspective. Should microalbuminuria ever be considered as a renal endpoint in any clinical trial? Am J Nephrol. 2010;31:469–470. doi: 10.1159/000292500. [DOI] [PubMed] [Google Scholar]
- 76.Lambers Heerspink HJ, de ZD. Debate: PRO position. Should microalbuminuria ever be considered as a renal endpoint in any clinical trial? Am J Nephrol. 2010;31:458–461. doi: 10.1159/000292501. [DOI] [PubMed] [Google Scholar]
- 77.Glassock RJ. Debate: CON position. Should microalbuminuria ever be considered as a renal endpoint in any clinical trial? Am J Nephrol. 2010;31:462–465. doi: 10.1159/000313553. [DOI] [PubMed] [Google Scholar]
- 78.Perkins BA, Ficociello LH, Silva KH, Finkelstein DM, Warram JH, Krolewski AS. Regression of microalbuminuria in type 1 diabetes. N Engl J Med. 2003;348:2285–2293. doi: 10.1056/NEJMoa021835. [DOI] [PubMed] [Google Scholar]
- 79.Perkins BA, Ficociello LH, Ostrander BE, Silva KH, Weinberg J, Warram JH, Krolewski AS. Microalbuminuria and the risk for early progressive renal function decline in type 1 diabetes. J Am Soc Nephrol. 2007;18:1353–1361. doi: 10.1681/ASN.2006080872. [DOI] [PubMed] [Google Scholar]
- 80.Thongboonkerd V, Songtawee N, Sritippayawan S. Urinary proteome profiling using microfluidic technology on a chip. J Proteome Res. 2007;6:2011–2018. doi: 10.1021/pr060586+. [DOI] [PubMed] [Google Scholar]
- 81.Thongboonkerd V. Recent progress in urinary proteomics. Proteomics Clin Appl. 2007;1:780–791. doi: 10.1002/prca.200700035. [DOI] [PubMed] [Google Scholar]
- 82.Coon JJ, Ueberheide B, Syka JE, Dryhurst DD, Ausio J, Shabanowitz J, Hunt DF. Protein identification using sequential ion/ion reactions and tandem mass spectrometry. Proc Natl Acad Sci U S A. 2005;102:9463–9468. doi: 10.1073/pnas.0503189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zerefos P, Prados J, Kossida S, Kalousis A, Vlahou A. Sample preparation and bioinformatics in MALDI profiling of urinary proteins. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;853:20–30. doi: 10.1016/j.jchromb.2007.02.063. [DOI] [PubMed] [Google Scholar]
- 84.Zerefos PG, Vougas K, Dimitraki P, Kossida S, Petrolekas A, Stravodimos K, Giannopoulos A, Fountoulakis M, Vlahou A. Characterization of the human urine proteome by preparative electrophoresis in combination with 2-DE. Proteomics. 2006;6:4346–4355. doi: 10.1002/pmic.200500671. [DOI] [PubMed] [Google Scholar]
- 85.Kushnir MM, Mrozinski P, Rockwood AL, Crockett DK. A depletion strategy for improved detection of human proteins from urine. J Biomol Tech. 2009;20:101–108. [PMC free article] [PubMed] [Google Scholar]
- 86.Kolch W, Neususs C, Pelzing M, Mischak H. Capillary electrophoresis-mass spectrometry as a powerful tool in clinical diagnosis and biomarker discovery. Mass Spectrom Rev. 2005;24:959–977. doi: 10.1002/mas.20051. [DOI] [PubMed] [Google Scholar]
- 87.Fliser D, Novak J, Thongboonkerd V, Argiles A, Jankowski V, Girolami MA, Jankowski J, Mischak H. Advances in urinary proteome analysis and biomarker discovery. J Am Soc Nephrol. 2007;18:1057–1071. doi: 10.1681/ASN.2006090956. [DOI] [PubMed] [Google Scholar]
- 88.Mischak H, Julian BA, Novak J. High-resolution proteome/peptidome analysis of peptides and low-molecular-weight proteins in urine. Proteomics Clin Appl. 2007;1:792. doi: 10.1002/prca.200700043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gilbert RE, Cooper ME. The tubulointerstitium in progressive diabetic kidney disease: more than an aftermath of glomerular injury? Kidney Int. 1999;56:1627–1637. doi: 10.1046/j.1523-1755.1999.00721.x. [DOI] [PubMed] [Google Scholar]
- 90.Bohle A, Wehrmann M, Bogenschutz O, Batz C, Muller CA, Muller GA. The pathogenesis of chronic renal failure in diabetic nephropathy. Investigation of 488 cases of diabetic glomerulosclerosis. Pathol Res Pract. 1991;187:251–259. doi: 10.1016/s0344-0338(11)80780-6. [DOI] [PubMed] [Google Scholar]
- 91.Wada T, Furuichi K, Sakai N, Iwata Y, Yoshimoto K, Shimizu M, Takeda SI, Takasawa K, Yoshimura M, Kida H, Kobayashi KI, Mukaida N, Naito T, Matsushima K, Yokoyama H. Up-regulation of monocyte chemoattractant protein-1 in tubulointerstitial lesions of human diabetic nephropathy. Kidney Int. 2000;58:1492–1499. doi: 10.1046/j.1523-1755.2000.00311.x. [DOI] [PubMed] [Google Scholar]
- 92.Merchant ML, Perkins BA, Boratyn GM, Ficociello LH, Wilkey DW, Barati MT, Bertram CC, Page GP, Rovin BH, Warram JH, Krolewski AS, Klein JB. Urinary peptidome may predict renal function decline in type 1 diabetes and microalbuminuria. J Am Soc Nephrol. 2009;20:2065–2074. doi: 10.1681/ASN.2008121233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bernstam EV, Smith JW, Johnson TR. What is biomedical informatics? J Biomed Inform. 2010;43:104–110. doi: 10.1016/j.jbi.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nesvizhskii AI. A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. J Proteomics. 2010;73:2092–2123. doi: 10.1016/j.jprot.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nagaraj N, Mann M. Quantitative analysis of the intra- and inter-individual variability of the normal urinary proteome. J Proteome Res. 2011;10:637–645. doi: 10.1021/pr100835s. [DOI] [PubMed] [Google Scholar]
- 96.Hunt SM, Thomas MR, Sebastian LT, Pedersen SK, Harcourt RL, Sloane AJ, Wilkins MR. Optimal replication and the importance of experimental design for gel-based quantitative proteomics. J Proteome Res. 2005;4:809–819. doi: 10.1021/pr049758y. [DOI] [PubMed] [Google Scholar]
- 97.Levin Y. The role of statistical power analysis in quantitative proteomics. Proteomics. 2011;11:2565–2567. doi: 10.1002/pmic.201100033. [DOI] [PubMed] [Google Scholar]
- 98.Moore CG, Carter RE, Nietert PJ, Stewart PW. Recommendations for planning pilot studies in clinical and translational research. Clin Transl Sci. 2011;4:332–337. doi: 10.1111/j.1752-8062.2011.00347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Martens L, Hermjakob H, Jones P, Adamski M, Taylor C, States D, Gevaert K, Vandekerckhove J, Apweiler R. PRIDE: the proteomics identifications database. Proteomics. 2005;5:3537–3545. doi: 10.1002/pmic.200401303. [DOI] [PubMed] [Google Scholar]
- 100.Kolker E, Higdon R, Haynes W, Welch D, Broomall W, Lancet D, Stanberry L, Kolker N. MOPED: Model Organism Protein Expression Database. Nucleic Acids Res. 2012;40:D1093–D1099. doi: 10.1093/nar/gkr1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Orchard S, Hermjakob H. The HUPO proteomics standards initiative--easing communication and minimizing data loss in a changing world. Brief Bioinform. 2008;9:166–173. doi: 10.1093/bib/bbm061. [DOI] [PubMed] [Google Scholar]
- 102.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 103.Jorge I, Navarro P, Martinez-Acedo P, Nunez E, Serrano H, Alfranca A, Redondo JM, Vazquez J. Statistical model to analyze quantitative proteomics data obtained by 18O/16O labeling and linear ion trap mass spectrometry: application to the study of vascular endothelial growth factor-induced angiogenesis in endothelial cells. Mol Cell Proteomics. 2009;8:1130–1149. doi: 10.1074/mcp.M800260-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bonzon-Kulichenko E, Martinez-Martinez S, Trevisan-Herraz M, Navarro P, Redondo JM, Vazquez J. Quantitative in-depth analysis of the dynamic secretome of activated Jurkat T-cells. J Proteomics. 2011;75:561–571. doi: 10.1016/j.jprot.2011.08.022. [DOI] [PubMed] [Google Scholar]
- 105.Fogle RL, Hollenbeak CS, Stanley BA, Vary TC, Kimball SR, Lynch CJ. Functional proteomic analysis reveals sex-dependent differences in structural and energy-producing myocardial proteins in rat model of alcoholic cardiomyopathy. Physiol Genomics. 2011;43:346–356. doi: 10.1152/physiolgenomics.00203.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhao Y, Denner L, Haidacher SJ, LeJeune WS, Tilton RG. Comprehensive analysis of the mouse renal cortex using two-dimensional HPLC - tandem mass spectrometry. Proteome Sci. 2008;6:15. doi: 10.1186/1477-5956-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, vila-Campilo I, Creech M, Gross B, Hanspers K, Isserlin R, Kelley R, Killcoyne S, Lotia S, Maere S, Morris J, Ono K, Pavlovic V, Pico AR, Vailaya A, Wang PL, Adler A, Conklin BR, Hood L, Kuiper M, Sander C, Schmulevich I, Schwikowski B, Warner GJ, Ideker T, Bader GD. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hu Z, Hung JH, Wang Y, Chang YC, Huang CL, Huyck M, DeLisi C. VisANT 3.5: multi-scale network visualization, analysis and inference based on the gene ontology. Nucleic Acids Res. 2009;37:W115–W121. doi: 10.1093/nar/gkp406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vidal M, Cusick ME, Barabasi AL. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pflieger D, Gonnet F, de la Fuente van Bentem, Hirt H, de la FA. Linking the proteins--elucidation of proteome-scale networks using mass spectrometry. Mass Spectrom Rev. 2011;30:268–297. doi: 10.1002/mas.20278. [DOI] [PubMed] [Google Scholar]
- 113.Starkey JM, Zhao Y, Sadygov RG, Haidacher SJ, LeJeune WS, Dey N, Luxon BA, Kane MA, Napoli JL, Denner L, Tilton RG. Altered retinoic acid metabolism in diabetic mouse kidney identified by O isotopic labeling and 2D mass spectrometry. PLoS One. 2010;5:e11095. doi: 10.1371/journal.pone.0011095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Overgaard AJ, Thingholm TE, Larsen MR, Tarnow L, Rossing P, McGuire JN, Pociot F. Quantitative iTRAQ-Based Proteomic Identification of Candidate Biomarkers for Diabetic Nephropathy in Plasma of Type 1 Diabetic Patients. Clin Proteomics. 2010;6:105–114. doi: 10.1007/s12014-010-9053-0. [DOI] [PMC free article] [PubMed] [Google Scholar]