Abstract
Lipidomics, a major part of metabolomics, constitutes the detailed analysis and global characterization, both spatial and temporal, of the structure and function of lipids (the lipidome) within a living system. As with proteomics, mass spectrometry has earned a central analytical role in lipidomics, and this role will continue to grow with technological developments. Currently, there exist two mass spectrometry-based lipidomics approaches, one based on a division of lipids into categories and classes prior to analysis, the “comprehensive lipidomics analysis by separation simplification” (CLASS), and the other in which all lipid species are analyzed together without prior separation, shotgun. In exploring the lipidome of various living systems, novel lipids are being discovered, and mass spectrometry is helping characterize their chemical structure. Deuterium exchange mass spectrometry (DXMS) is being used to investigate the association of lipids and membranes with proteins and enzymes, and imaging mass spectrometry (IMS) is being applied to the in situ analysis of lipids in tissues.
Keywords: CLASS, DXMS, imaging mass spectrometry, lipidomics, novel lipids, shotgun
INTRODUCTION
The exponential growth in the number of fully sequenced genomes available in the public domain has provided biologists with the challenge of connecting genes to gene function, that is, genotype to phenotype, and to determine the actual role of genes. The drive for understanding the function of newly discovered genes has led biologists toward the systematic analysis of expression levels of the components that constitute a biological system, i.e., the mRNA, the proteins, and the metabolites, and the global cataloging of these components has given rise to the various “OMEs” (the genome and the proteome). Additionally, understanding networks and how these different components interact with one another - be it within a specific OME or between them - is the basis of a systems biology approach (1, 2).
Metabolomics is the systematic study of the complete set of the nonproteinaceous, low-molecular-weight, endogenously synthesized intermediates (the metabolome) contained in the cell (3, 4) and, it can be argued, represents the end product of gene expression. The metabolome most closely correlates to an organism's actual phenotype and hence has disease implications. Of all the molecules contained in the metabolome, the fats (or lipids) constitute the largest subset, including tens of thousands of distinct lipid molecular species existing in the cells and tissues. Lipidomics, a dominant part of metabolomics, is the detailed analysis and global characterization, both spatial and temporal, of the structure and function of lipids (the lipidome) within a living system. Comparing the lipidome of healthy versus diseased states can provide information helpful in correlating the role of lipids in various diseases, such as cancer, atherosclerosis, and chronic inflammation. Additionally, a large variety of lipid species comprise cellular membranes, and investigating their interactions with membrane-associating proteins/enzymes can provide insight into such areas as drug/inhibitor interactions.
The very first mass spectrometer was built by Nobel Laureate Sir J.J. Thomson (5) in the early part of the 20th century and was used to analyze marsh gas. In his analysis, Thomson observed the mass-to-charge ratios (m/z) of 16 and 26, which he identified as the positive ions of methane and acetylene, respectively. Since its very first inception, mass spectrometry has played a central analytical role in the various sciences, and its contribution during the past few decades to the fields of proteomics and metabolomics cannot be overstated.
A number of excellent reviews featuring mass spectrometry as well as its applications in the various “OMIC” disciplines already exist (6-10), and the reader is encouraged to explore these resources. Griffiths & Wang (11) recently wrote a particularly informative review that covers mass spectrometry applications in proteomics, metabolomics, and lipidomics.
Our present review presents a critical discussion of the current status of applications of mass spectrometry to the field of lipidomics, and we do not wish to simply repeat what is already available. Thus, four main areas are covered. First, we compare and contrast two current mass spectrometry-based lipidomics approaches, one where the lipidome is divided into lipid categories and classes and each analyzed separately and the other where all lipid species are analyzed essentially together. Second, we review the mass spectrometry-based structure determination of novel lipids. Third, the current state of deuterium exchange mass spectrometry (DXMS) used to study the location and orientation of proteins associated with lipid membranes is covered. Lastly, we review advances in the imaging of lipids in tissues, including matrix-assisted laser desorption ionization (MALDI) mass spectrometry.
MASS SPECTROMETRY--BASED LIPIDOMICS
Lipid Definition and Classification
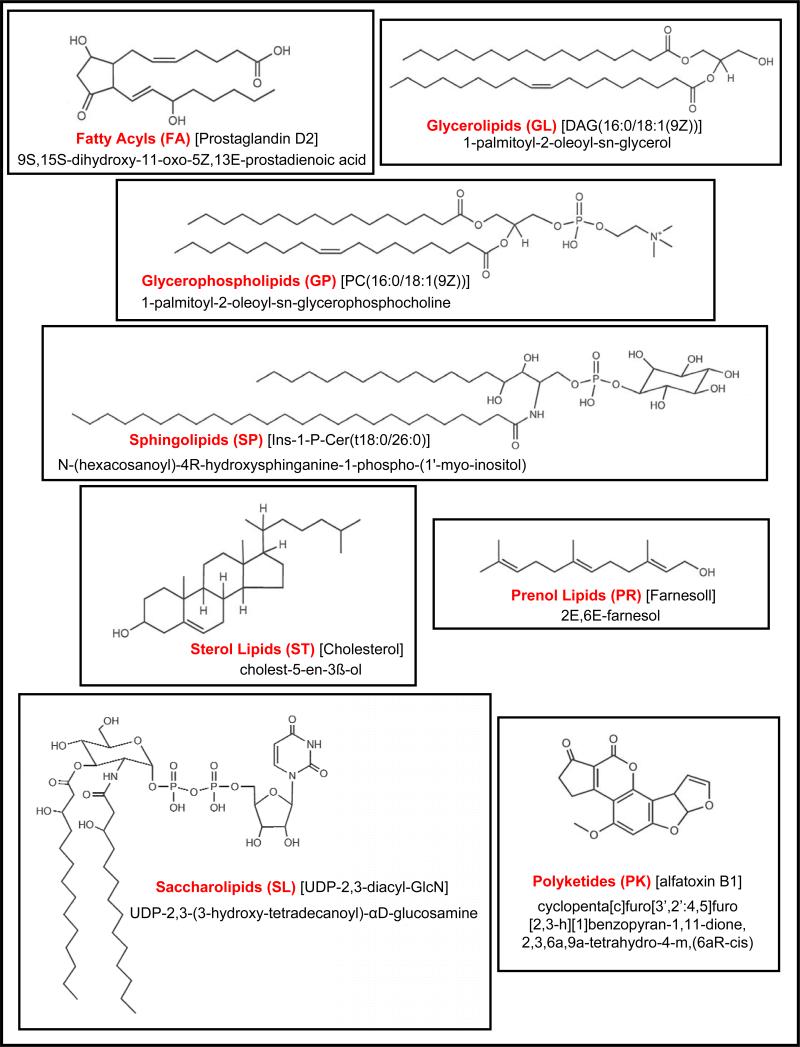
Examination of a typical biochemistry dictionary (12), university biochemistry textbook (13), or volume on lipid analysis (14) for a description of lipids, yields a broadly defined group of organic compounds, including the fats, oils, waxes, sterols, and triglycerides, which are insoluble in water but soluble in nonpolar organic solvents, are oily to the touch, and together with carbohydrates and proteins constitute the principal structural material of living cells. In reality, there are many examples of lipids that do not adhere to this lose definition; this has recently led some in the scientific community to define this most diverse set of molecules on the basis of their biosynthetic origin. The LIPID MAPS Consortium (15) has defined lipids as hydrophobic or amphipathic small molecules that originate by carbanion-based condensation of thioesters (fatty acids, polyketides, etc.) and/or by carbocation-based condensation of isoprene units (prenols, sterols, etc.). Adhering to this definition and in conjunction with the International Committee for the Classification and Nomenclature of Lipids, the LIPID MAPS Consortium has thus defined eight categories of lipids based on their chemically functional backbone - fatty acyls, glycerolipids, glycerophospholipids, sphingolipids, sterol lipids, prenol lipids, saccharolipids, and polyketides - along with numerous classes and subclasses to allow one to describe all lipid molecular species (15, 16). Also, see the LIPID MAPS - Nature Lipidomics Gateway at http://www.lipidmaps.org. Figure 1 illustrates examples of molecular species for each of these eight lipid categories.
Figure 1.
Examples of each of the eight categories of lipids as defined by the Lipid Metabolites and Pathways Strategy (LIPID MAPS) Consortium.
Sample Preparation for Mass Spectrometry-Based Lipidomics Studies
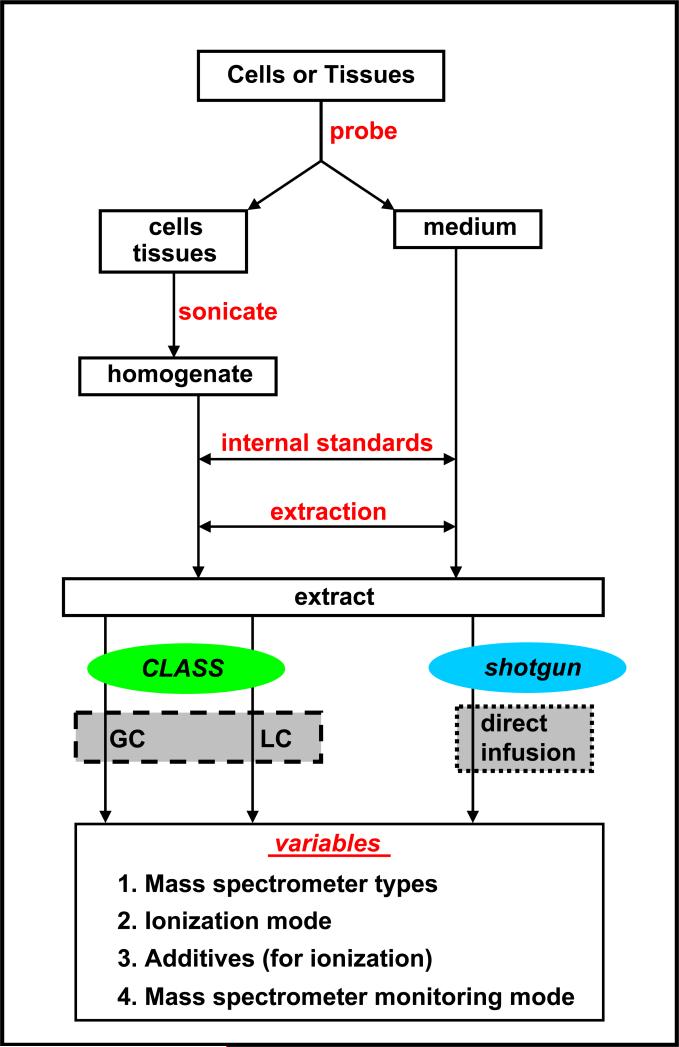
Two fundamentally different approaches exist for the mass spectrometry-based identification and quantification of lipids within cells and tissues. The first approach, a more traditional “comprehensive lipidomics analysis by separation simplification” (CLASS) strategy and the platform used by the LIPID MAPS Consortium, is based on separation of different lipid categories using extraction and chromatographic separation prior to mass analysis and then optimizing the mass spectrometer to perform in a lipid class-specific fashion. The second approach, sometimes termed shotgun lipidomics, omits chromatographic separation and essentially analyzes all the lipid classes together, directly infusing them into the mass spectrometer, while employing different ion source polarities (to form positive or negative ions) and infusing ionization solution additives, which serve to provide a kind of lipid class-specific favored analysis. More details regarding each approach are provided below. For either approach, a general cell or tissue preparation is followed as outlined in Figure 2.
Figure 2.
Flow chart depicting the lipid sample preparation and analysis protocol. After extraction, the comprehensive lipidomics analysis by separation simplification (CLASS) or shotgun lipidomics approach is carried out. GC, gas chromatography; LC, liquid chromatography.
Cells or tissues are cultured and can be subjected to a probe (activation or perturbation), as shown in Figure 2, and their lipidome is compared to a control unperturbed sample. For example, a macrophage cell line can be subjected to a bacterial endotoxin in a time/dose-response study (17-19), or yeast cells can have lipid synthesis genes altered or subjected to different growth temperatures (20). Similarly, human blood plasma (21) or tissue biopsies, for example, from ear tissue from normal versus stressed states, can be analyzed (22). The lipids stored within the cell wall and internal organelles or tissue matrix need to be released employing a disruption mechanism, such as sonication, to produce a uniform homogenate. Some lipids, such as eicosanoids (23), are actually not stored in the cell but secreted into the growth medium, so the medium also needs to be saved and analyzed for a complete lipidomics analysis (17, 19). Prior to extraction, it is necessary to add a mixture of internal standards to enable the absolute quantitation of lipids in the sample. These internal standards are typically deuterium-labeled lipid analogs of the molecular species one wishes to quantitate. When such deuterium-labeled analogs are not available, similar lipid class odd-carbon chain molecular species can be used. In either case, the goal is to use an internal standard that the organism under study cannot possibly synthesize. Recently, advances in the number and commercial availability of such lipid standards (24) have been a critical cornerstone to the growth and successes of mass spectrometry-based lipidomics. Table 1 summarizes the lipid class-specific extraction protocols optimized and used by the LIPID MAPS Consortium (25-35). A recent impressive shotgun lipidomics study, which measured the yeast lipidome, actually employed a two-step extraction procedure that separated relatively apolar and polar lipids (20). In all these studies the solvent containing extracts were then evaporated, and the remaining lipids were resuspended in a liquid medium optimal for direct infusion into the mass spectrometer (shotgun approach) or in a medium compatible with chromatographic separation prior to on line mass spectrometric analysis (CLASS approach). The types of mass spectrometers being used for lipid analysis along with their various modes of operations are mostly common to both techniques and are described next.
TABLE 1.
Extraction and chromatographic separation protocols for lipid sub-classes
| Lipid class | Lipid sub-class a | Extraction b | Chromatographic b separation | Lipid species quantitated | ||
|---|---|---|---|---|---|---|
| study A c | study B d | study C e | ||||
| fatty acyls (FA) | fatty acids eicosanoids |
methanolic HCl/isooctane C18 SPE cartridge |
GC HPLC/RP C18 column |
–
– |
31 76 |
44 9 |
| glycerolipids (GL) | DAG TAG |
ethyl acetate/isooctane | HPLC/NP silica column |
–
– |
55 18 |
107 105 |
| glycerophospholipids (GP) | PA PC PE PS PI PG ether-linked PC ether-linked PE N-acyl PS cardiolipins |

|
HPLC/NP silica column | 22 26 19 24 19 22 5 10 – 16 |
15 36 32 20 19 16 7 13 2 – |
14 26 25 26 21 21 9 9 – 15 |
| sphingolipids (SP) | ceramides dihydroceramides hexosylceramides dihydrohexosylceramides sphingomyelin dihydrosphingomyelin sphingoid bases |
|
HPLC/NP NH2 column | 8 8 8 8 8 8 – |
41 – 56 – 101 – 6 |
21 12 11 9 11 9 – |
| sterol lipids (ST) | free form fatty acyl esters |
methanol/chloroform ethyl acetate/isooctane |
HPLC/RP C18 column HPLC/NP silica column |
13 – |
14 22 |
11 23 |
| prenol lipids (PR) | ubiquinones dolichols | methanol/chloroform | HPLC/RP C8 column | 2 3 |
2 6 |
2 3 |
| TOTAL NUMBER OF SPECIES QUANTITATED: | 229 | 588 | 543 | |||
Mass Spectrometers Optimal for Lipid Analysis
A mass spectrometric analysis of a molecule requires that the molecule must be charged - either as a positive or a negative molecular ion. As such, the ion source is a fundamental component of all mass spectrometers. The development of electrospray ionization (ESI) in the late 1980s (36), for which John Fenn was awarded part of the 2002 Nobel Prize in Chemistry, allows many molecules, including proteins, carbohydrates, and lipids, to be ionized in a liquid medium without the need for prior derivitization. ESI indeed revolutionized the field of mass spectrometry, allowing it to be used for the analysis of molecules that could not be analyzed in the past. ESI is used to form either positive or negative ions, and some lipid classes can form either, both having their advantages for determining molecular detail. It should be noted that ESI is a “soft” ionization technique, and unlike electron impact ionization used for many years prior to the emergence of ESI, the molecule is left whole and unfragmented in the ionization process. ESI sources most commonly form positive molecular ions through the addition of a proton to a molecule [M+H]+ or negative molecular ions through the removal of a proton from a molecule [M-H]–. Some large molecules, such as proteins, can be multiply protonated (e.g., [M+H40]40+) using ESI. In such a case, a 40-kDa protein would have an m/z of 1,000, well within the mass scan range of most mass spectrometers. To increase ionization efficiency for certain molecules, in some cases, a small amount of additive, such as ammonium acetate, is added to the ESI solution to allow the formation of ammonium adduct molecular cations [M+NH4]+ or acetate adduct molecular anions [M+CH3CO2]–. Because of its versatility, ESI is widely used for all the lipid classes shown in Table 1 except for the free fatty acids; details regarding fatty acid analysis are presented in a section below.
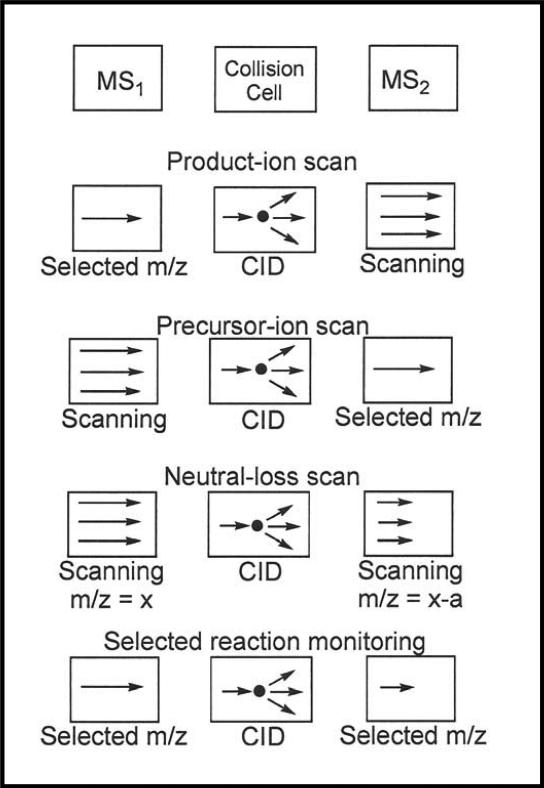
The triple quadrupole mass spectrometer, a very common workhorse in the mass spectrometry field, is schematically diagramed in the top row of Figure 3. The mass spectrometer can be used in single-scan (MS) mode to provide information about precursor molecular masses within a specified mass range, but very often operating the mass spectrometer in a tandem-scan (MS/MS) mode can provide much more useful and additional information. It should be noted that the triple quadrupole instrument is, but not all mass spectrometers are, capable of operating in the MS/MS mode. In a typical MS/MS experiment, a specified precursor ion is selected according to its mass-to-charge ratio in the first filter of the instrument and is fragmented into its product ions in a collision cell. Then, the resulting product ions are scanned according to their mass-to-charge ratio in a second filter, and a mass-to-charge ratio spectrum is recorded. This particular MS/MS scan mode is commonly termed product-ion scan and is depicted in the second row of Figure 3. Structural isomers of some of the lipids, which provide the same precursor molecular mass in MS mode and are impossible to resolve, many times yield different fragment product ions, allowing them to be differentiated using the MS/MS mode.
Figure 3.
Various modes of tandem mass spectrometric analyses available on the triple quadrupole instrument. Reprinted with permission from Reference 11.
The triple quadrupole instrument is capable of additional specialized MS/MS scan modes, see Figure 3, and some of these are particularly useful in the analysis of specific lipid classes (25-35). In the precursor-ion scan, the second filter is set to transmit only a specified ion fragment, e.g., the [PH2O4C2H4N(CH3)3]+ phosphocholine headgroup at m/z = 184, which is characteristic of glycerophosphocholine lipids, while the first filter scans and produces a mass-to-charge ratio spectrum for all precursor molecular species producing this specific fragment. Another example of the precursor-ion scan is when fatty acid cholesteryl esters are analyzed as ammonium adduct cations. The second filter is set to transmit only the [cholesterol - H2O]+-dehydrated cholesterol cation at m/z = 369, the only fragment produced and a characteristic of fatty acid cholesteryl esters, while the first filter produces a mass-to-charge ratio spectrum of all precursor cholesteryl esters producing this fragment (37). In a neutral-loss scan, the first and second filters are scanned in unison with a specified offset mass difference set between them. As an example, a neutral-loss scan of 141 or 185 is used to monitor glycerophosphoethanolamine or glycerophosphoserine lipids, respectively, accounting for the neutral loss of phosphoethanolamine or phosphoserine headgroups.
An ultrasensitive MS/MS mode on the triple quadrupole instrument is called selected reaction monitoring (SRM). In SRM, the first filter is “locked’ on a specified precursor ion, and the second filter is locked on its specified product ion. As described below, the SRM mode is particularly useful when coupled with chromatographic separations.
Although the triple quadrupole mass spectrometer offers these specialized scan modes, it is limited in its mass accuracy and resolution to ≈100 ppm and M/ΔM ≈100, respectively. Other instruments used in lipidomics, such as the time-of-flight (TOF) mass spectrometer and the Fourier transform orbitrap mass spectrometer, can provide mass accuracies and resolutions on the order of 5-20 ppm and 105, which in some cases is useful when determining the elemental composition of lipid molecular species, such as C42H81O9P (760.5618 Da) versus C43H85O8P (760.5982 Da). Reference 6 provides a helpful comparison of mass spectrometer specifications (cost, size, mass accuracy, mass resolution, etc.).
Mass Spectrometric Analysis: The Shotgun Approach
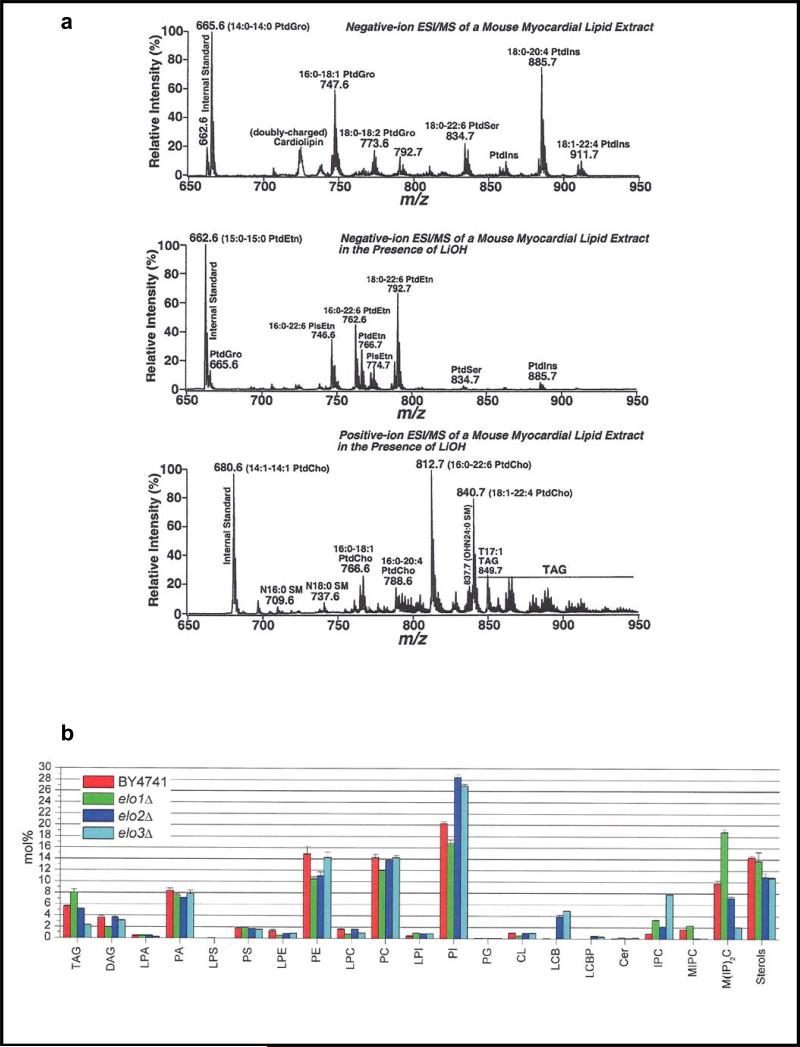
The shotgun lipidomics approach (20, 38) - the name adapted from earlier shotgun genomics and shotgun proteomics approaches - has produced impressive results to profile lipids, including glycerophospholipids, glycerolipids, sphingolipids, and sterols, in biological extracts by cleverly selecting the optimal ESI polarity, ESI solution additives, and tandem mass spectrometer monitoring modes, as well as by developing and utilizing sophisticated analysis software to decipher the complicated data output (39). Although the shotgun approach is biased toward the ionization of molecular species that are most abundant or those having the highest ionization efficiencies which can cause ion suppression resulting in low-level species not being detected, Han & Gross (38, 40) sought to minimize this drawback by introducing the technique of “intrasource separation” in which a crude biological extract is initially analyzed by ESI in the negative ionization mode, which favors ionization of anionic glycerophospholipids that are negatively charged at neutral pH (glycerophosphoinositols, glycerophosphoglycerols, glycerophosphoserines, glycerophosphates, cardiolipins). Next, a base is added, such as LiOH in methanol, which favors ionization of weakly anionic glycerophospholipids (glycerophosphoethanolamines). Lastly, the extract is analyzed in the positive ESI mode, which favors polar lipids (glycerophosphocholines, sphingolipids). Intrasource separation greatly expands the dynamic range of lipidome observations, and using this technique, the recorded mass spectra of a crude extract from a mouse myocardium (40) is shown in Figure 4a, where many classes of glycerolipids, glycerophospholipids, and sphingolipids were observed.
Figure 4.
Examples of shotgun lipidomics. (a) Demonstration of intrasource separation, which is used to minimize ion suppression and improve detection of low-level species. Reprinted with permission from Reference 40. (b) Comparison of the lipid composition of yeast cells for wild-type (BY4741) cells versus those with a fatty acid elongase gene mutation (elo1Δ, elo2Δ, elo3Δ. Reprinted with permission from Reference 17.
In another example of shotgun lipidomics, although yeast cells are only capable of synthesizing a limited number of fatty acids and other more complex lipids compared to mammalian cells, Ejsing et al. (20) carried out an impressive yeast lipidome study that quantified 250 molecular lipid species covering 21 types of lipid and is shown in Figure 4b.
Comprehensive Lipidomics Analysis by Separation Simplification
The mammalian lipidome is estimated to contain some hundreds of thousands of molecular species (2, 11), and compared to the yeast lipidome, it is far more complex. To explore such an enormous lipidome, the LIPID MAPS Consortium chose to develop a comprehensive lipidomics analysis by separation simplification known as the CLASS approach, which simplifies what is put into the mass spectrometer by first chromatographically separating the components on line and coupling the chromatographic effluent directly into the mass spectrometer. In short, this consitutes a “divide-and-conquer” strategy. An advantage of CLASS is the minimization of ion suppression which improves the ability to detect very low-level lipid species, should they be present. Table 1 summarizes the LIPID MAPS CLASS approach and lists the lipid class-specific chromatographic separation protocols they have employed. Details for each of these protocols are found in References 25-35.
All of the lipid classes, except the fatty acids, in Table 1 were separated prior to mass analysis, employing high-performance liquid chromatography (HPLC). Historically, gas chromatography (GC) has been the method of choice for separating mixtures of fatty acids, and the combination of GC-MS methods allows the resolution of molecular details, such as the number and position of double bonds within the fatty acid molecule. In measuring the mouse macrophage lipidome, Quehenberger et al. (25) reported a stable isotope dilution GC-MS method capable of quantitating 30 saturated and unsaturated fatty acids contained in the mouse macrophage in a single analysis. The methodology employs a rapid extraction procedure of free fatty acids from the cell medium, followed by a relatively quick and simple derivatization, permitting a limit of detection in the femtomole range. GC-MS instruments do not employ an ESI source, so derivatization of the free fatty acid is required to make it volatile in the chemical ion source used with this instrument.
For the other lipid classes listed in Table 1, HPLC effluent is coupled directly into the mass spectrometer and analyzed employing the various MS/MS scan modes and the high mass accuracy/resolution capabilities described above. Impressive lipidomic analyses of the subcellular organelles of the mouse macrophage (18), the mouse macrophage (19) and human blood plasma (21) have been carried out. Table 1 lists (in the last three columns) the number of lipid molecular species accurately quantitated in each of these three studies.
In comparing the two lipidomics approaches, it is important to note that structural information can be inferred using the elution order from the HPLC column, which can aid in the characterization of lipids that cannot be inferred by mass spectrometric analysis alone. One such case is the occurrence of ether-linked triglycerides in mixtures of triacylglycerols (41). Another such case involves the SRM scan mode described above. A method has been created in which the mass spectrometer can cycle through numerous SRM pairs repeatedly; it is referred to as multiple reaction monitoring (MRM). Coupling a liquid chromatographic separation to the mass spectrometer and employing the MRM mode, the mass spectrometer is used as a highly selective and highly sensitive HPLC detector. If a specific molecule is eluting from the chromatographic column at any time during the analysis, its specified MRM pair can be detected and recorded. Sophisticated methods have been developed that can monitor hundreds of lipid mediators, such as eicosanoids, through their defined MRM pairs in a single analysis lasting only 20 min (42). Figure 5 is an example where such a method was employed for measuring temporal genomics and lipidomics changes in eicosanoid biosynthesis in mouse macrophages in response to endotoxin stimulation (43); this allowed investigators to determine the flux of metabolites (44). As a tool in constructing such MRM methods for other lipid classes, a highly valuable resource for many hundreds of MS/MS spectra for lipid standards is freely available at http://www.lipidmaps.org/data/standards/index.html.
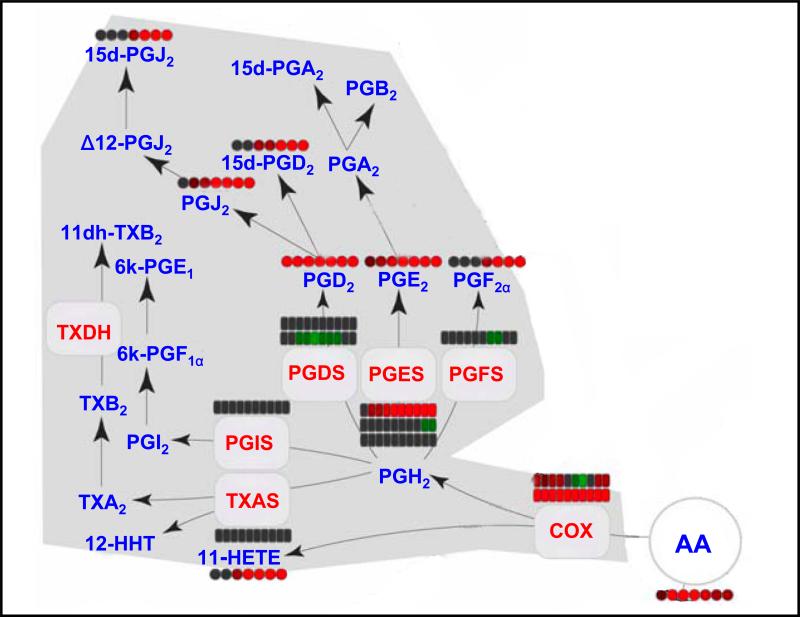
Figure 5.
Example of a comprehensive lipidomics analysis by separation simplification (CLASS) approach. A heat map shows the temporal changes of various lipid metabolites (prostaglandins), as well as gene expression, in endotoxin-stimulated mouse macrophage cells. Enzymes are shown in red font and the prostaglandin lipid metabolites are shown in blue font. The arrows depict the synthesis pathways, including various metabolite intermediates and corresponding enzymes. Changes are represented as a function of time (left to right), where rectangles indicate mRNA levels and circles indicate lipid metabolite levels. Greater intensity of red indicates increasing levels; greater intensity of green indicates decreasing levels; and gray represents no change in levels relative to unstimulated cells. When enzyme activity can result from multiple genes, each is represented as a separate line. Redrawn and reprinted with permission from Reference 43.
MASS SPECTROMETRY--BASED STRUCTURE DETERMINATION OF NOVEL LIPIDS
Targeted versus Untargeted Lipidomics
Since its inception nearly 100 years ago, mass spectrometry has played a central role in the discovery of new molecules. Technological developments that followed its initial inception, such as tandem mass spectrometry, high mass accuracy and resolution capabilities, various ionization methods, and the on line coupling of HPLC, have served well to increase its utility in the molecule discovery area. The emerging field of lipidomics has provided fertile ground to discover and characterize novel lipid species, and mass spectrometry has and will continue to play a vital role in this area.
Lipidomics-based assays can be described as being “targeted,” where lipid species to be monitored are known before commencing the analysis. An example of such a targeted assay is the MRM method described above. Although MRM-based assays can offer superb sensitivity and are ideal for accurate quantitation, basically one only finds what one is looking for. Lipidomics-based assays that are described as “untargeted” are more of an exploratory, qualitative survey of the lipid landscape. Operating the mass spectrometer in the full-scan mode to search for new mass-to-charge ratio peaks is an option for an untargeted assay, where if a novel lipid species is present, one hopes to detect it without previous knowledge, that is, assuming the species ionizes efficiently and the sensitivity of the mass spectrometer is sufficient. However, once a potentially novel lipid is observed, further investigation can be conducted (e.g., via the MS/MS mode) to confirm its novelty.
Examples of a few of the novel lipids observed recently in untargeted assays are presented below. Additionally, a protocol for an untargeted assay that can “flag” low-level, unexpected novel lipids is described.
Examples of Novel, Unexpected Lipids
The discovery and interest in lipids began well before the early 20th century invention of the mass spectrometer. Impressively, in 1823, the French chemist Michel Eugène Chevreul published “Recherches chimiques sur les corps gras d'origine animale,” which culminated his work investigating the structure and properties of lipids. Chevreul was the first lipid specialist to discover the concept of fatty acids, including several chemical species such as oleic, butyric, caproic, and steric fatty acids. He also discovered cholesterol and glycerol. Amazingly, even today, new lipid compounds are being discovered, and mass spectrometry is playing a central role in elucidating their chemical structure.
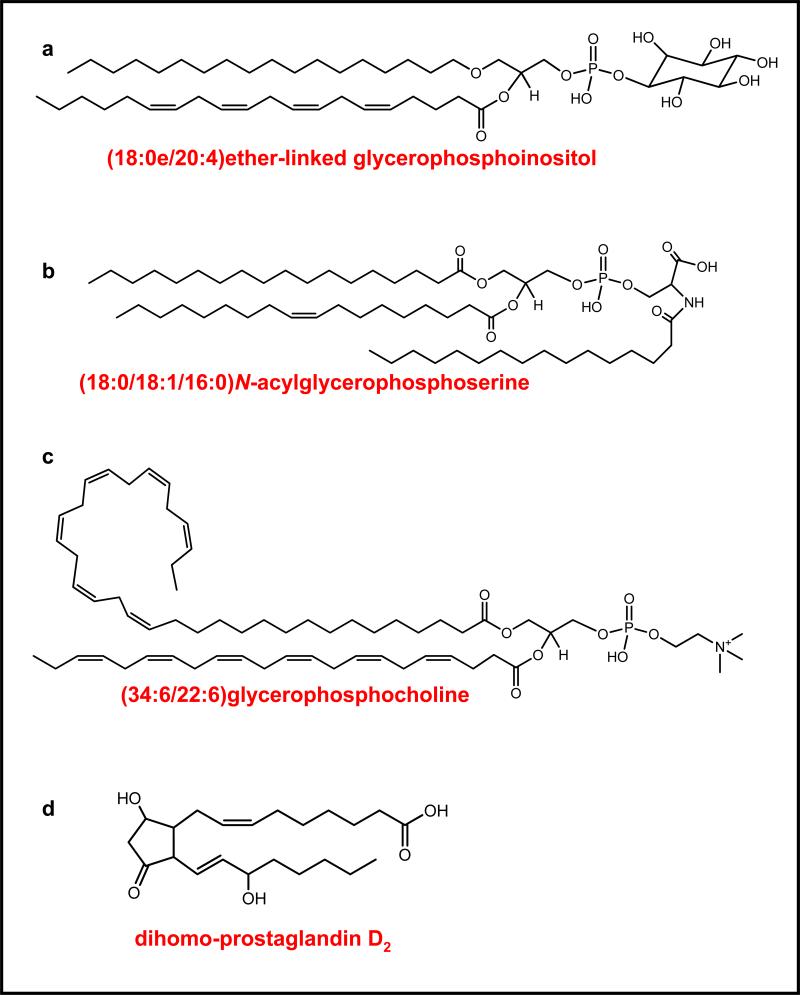
Figure 6 shows several examples of some of the novel, unexpected lipid species that have been discovered in living systems during the past few years. Employing HPLC and tandem mass spectrometry, the chemical structures were obtained for a number of ether-linked glycerophospholipids (45); an example of these molecules is shown in Figure 6a. While recently profiling the lipidome of the mouse brain using mass spectrometry, Guan et al. (46) discovered a novel family of N-acylphosphoserine derivatives; an example of these molecules is shown in Figure 6b.
Figure 6.
Examples of a few of the novel species observed recently in untargeted lipidomics mass spectrometry-based assays. References 45, 46, 48-51, 52.
Although it was observed nearly 30 years ago that the retina was highly enriched with very long-chain polyunsaturated fatty acids (47), more recent work employing both GC-MS and HPLC-ESI-MS/MS (48-51) methods has led to a more full characterization of these as glycerophosphocholine species containing the very long-chain polyunsaturated fatty acids at the sn-1 position and docosahexaenoic acid at the sn-2 position (see the example shown in Figure 6c).
Lastly, the in vivo formation of 22-carbon dihomoprostaglandins from a 20-carbon arachidonic acid (AA, 20:4, n-6) precursor was unexpectedly observed in endotoxin-stimulated mouse macrophage cells (52), and an example of one of these is shown in Figure 6d. A sensitive, untargeted lipidomics approach that was critical in this discovery is described below.
Untargeted–Flagged Lipidomics
As mentioned above, the ultrasensitive MRM mode has the capability of monitoring numerous molecular species in a single analysis; however, assumptions have to be made in advance regarding the species that might be present to provide MRM pairs for the detection scheme. If the goal of untargeted lipidomics is to conduct a global survey of lipids in a system under study, then the concern must be raised as to whether biologically significant lipid species are being overlooked. Are there possibly unexpected species and hence no available MRM pairs that would be required for their detection?
To overcome such a dilemma, a mass spectrometry-based stable isotope labeling strategy coined DIMPLES/MS (52) for diverse isotope metabolic profiling of labeled exogenous substrates using mass spectrometry was developed and is illustrated in Figure 7. Briefly, in these experiments, mouse macrophage cells were incubated in a medium supplemented with deuterium-labeled arachidonic acid (AA-d8) and then stimulated with endotoxin. Separately, this was also carried out with cells that were not supplemented with AA-d8. The medium from each was extracted and individually analyzed with the mass spectrometer operated in full-scan mode. Two sets of eicosanoid generation resulted, one set from endogenous AA and the other from the supplemented exogenous AA-d8. This results in a characteristic and obvious doublet pattern or flag, resolvable with mass spectrometry, allowing for a sensitive and comprehensive eicosanoid search without any previous knowledge or assumptions as to what species may be present.
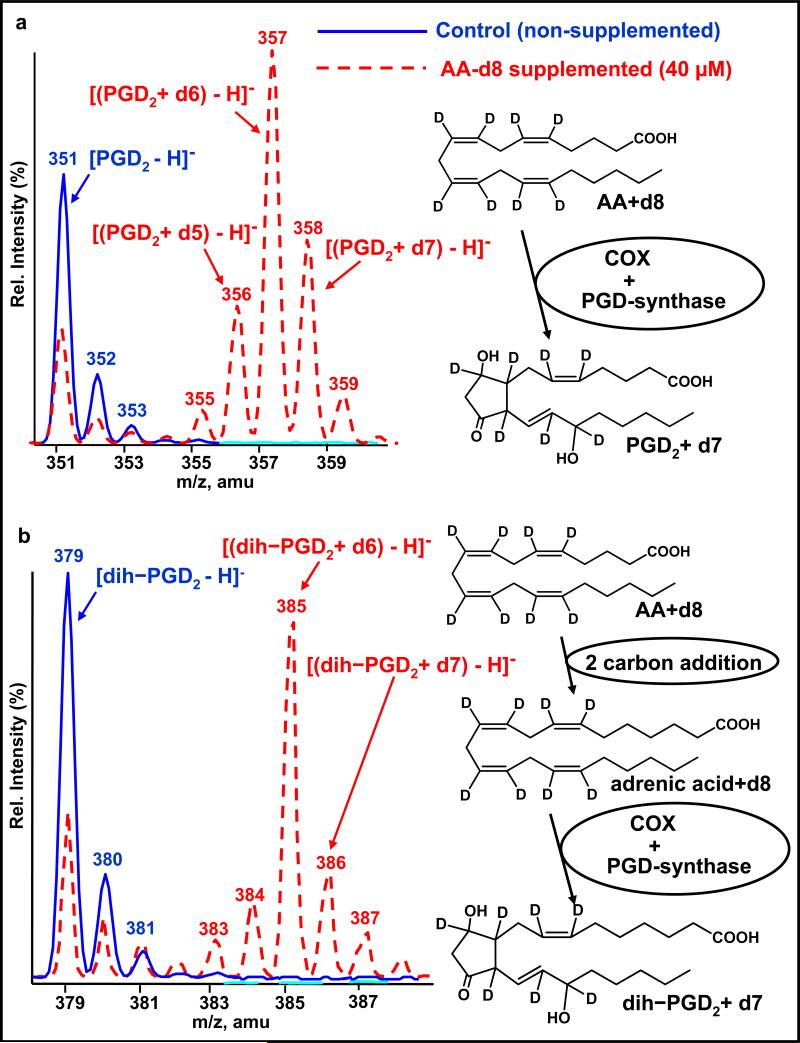
Figure 7.
Example of the diverse isotope metabolic profiling of labeled exogenous substrates using mass spectrometry (DIMPLES/MS) approach. (a) The characteristic doublet pattern, the observation of prostaglandin D2 (PGD2), and deuterium-labeled PGD2 produced by macrophage cells supplemented with deuterium-labeled arachidonic acid (AA-d8). (b) The unexpected observation of 22-carbon dihomo-prostaglandin D2 (dih-PGD2), resulting from the 2-carbon elongation of arachidonic acid (AA). Redrawn and reprinted with permission from Reference 52. amu, atomic mass unit; COX, cyclooxygenase; m/z, mass-to-charge ratio.
As shown in the nonsupplemented sample (Figure 7a), prostaglandin D2 (PGD2) was observed at m/z = 351 (the solid blue trace) as expected for endotoxin-stimulated macrophages. For the AA-d8-supplemented sample (the dotted red trace, data collected separately and overlaid), a mass offset pattern, not observed in the nonsupplemented sample, is clearly observed, resulting from the action of eicosanoid producing enzymes, cyclooxygenase (COX) and PGD-synthase, on the AA-d8 substrate. Note that, even in the supplemented sample, some nondeuterated PGD2 is observed, indicating some endogenous AA within the cell remains to serve as a substrate.
In the above example, the production of PGD2 was expected and shown for the purpose of the DIMPLES/MS demonstration. Using the DIMPLE/MS strategy, and as shown in Figure 7b, an unexpected and particularly interesting observation was the production of 22-carbon dihomoprostaglandins, products of adrenic acid (22:4, n-6), resulting from the 2-carbon elongation of AA by the stimulated mouse macrophage (52). Although it had been previously observed that adrenic acid could serve as a COX substrate when added to cells, resulting in the production of dihomoprostaglandins (53, 54), the DIMPLES/MS strategy revealed its formation de novo from an arachidonate precursor. Even though this DIMPLES/MS demonstration was for AA and prostaglandin production, similar labeling strategies should prove equally valuable for other substrates and lipid species.
PROTEIN/ENZYME ASSOCIATION WITH LIPIDS AND MEMBRANES
Peptide Amide Hydrogen/Deuterium Exchange Mass Spectrometry
DXMS is a useful technique for studying protein dynamics and folding, ligand binding, and protein-protein interactions; it strongly complements and adds to the X-ray crystallography and nuclear magnetic resonance methods for the exploration of protein structure (55-58). Recently, DXMS has proven to be a particularly valuable technique for exploring the location and orientation of proteins and enzymes associated with individual lipid molecule membrane phospholipids (59-61).
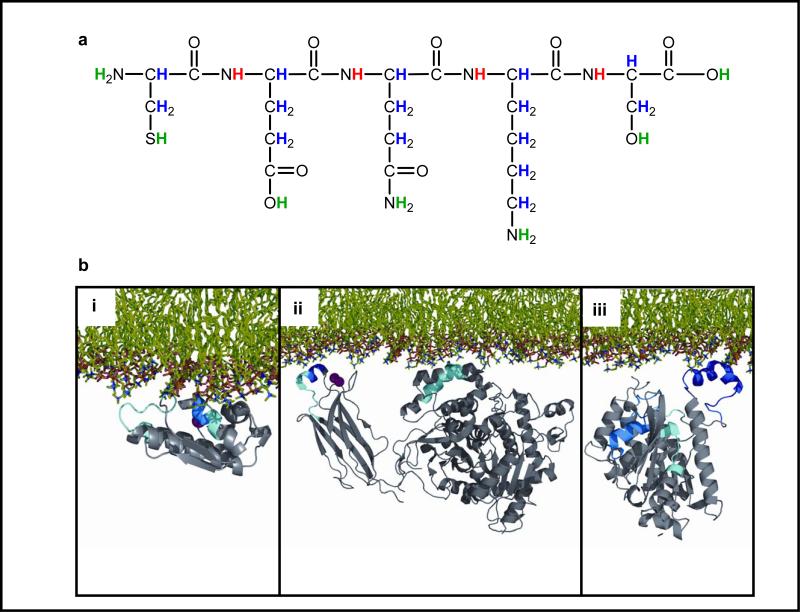
Utilizing DXMS as a tool for studying protein structure and interactions is based on the principle of hydrogen exchange with a solvent. As illustrated in Figure 8a, hydrogen atoms contained on a protein molecule can be divided into three classes based on their rate of exchange with an aqueous solvent: those that hardly ever exchange, those that exchange extremely quickly, and those whose exchange rate depends on their local environment. As such, amide hydrogen atom exchange rates in a protein vary depending upon their environment/location and can be used to investigate the structural dynamics of the protein.
Figure 8.
Deuterium exchange mass spectrometry (DXMS) used to investigate protein/enzyme associations with lipids. (a) Hydrogen atoms contained on a protein molecule can be divided into three classes based on their rate of exchange with the aqueous solvent. Those attached directly to carbon atoms (blue) hardly ever exchange; those attached to amino acid side chain atoms, the N-terminal amine, and the C-terminal carboxylic acid (green) exchange extremely rapidly; and amide hydrogen atoms (red) have variable exchange rates from seconds to months depending on the protein conformation and solvent accessibility. (b) Depictions of the membrane interactions for representatives of the three main kinds of phospholipase A2 (PLA2), (i) the small secreted sPLA2 (reprinted with permission from Reference 59), (ii) the cytosolic cPLA (reprinted with permission from Reference 60), and (iii) the calcium-independent iPLA2 (reprinted with permission from Reference 61). Each has a different and distinct interaction with the phospholipid membrane.
Basic DXMS methodology is as follows: The protein of interest is incubated in deuterated water for different periods of time, during which the protein can be subjected to various additional substances, probes, or perturbations to explore how this may influence its water accessibility and conformation and, hence, local amide hydrogen exchange rates. Amide hydrogen atoms that are in the hydrophobic regions of the protein need to be incubated longer to exchange, or may not exchange at all, whereas those on the exposed surface exchange quickly at the start of the incubation. The deuterium atoms can then be “locked in place” or quenched and prevented from further exchange by lowering the solution pH to ≈2.5 and temperature to ≈2°C. An HPLC-mass spectrometry analysis is then conducted that is similar to a typical “bottom-up” proteomics analysis (62, 63) wherein a protease is used to digest the protein into its corresponding peptides, each generally on the order of 5 to 15 amino acids in length. These peptides may also be fragmented into smaller pieces in the mass spectrometer (using the MS/MS scan mode), which helps in their identification and in the sequencing of the protein. In DXMS experiments, the instrumentation is often customized to include a component such as an online chilled pepsin protease digestion HPLC column, which minimizes deuterium back exchange and helps automate the analysis (59-61, 64).
The data collected from various deuterium incubation times are then used to construct maps that indicate which regions of the protein contain amide hydrogens that exchange quickly, slowly, or not at all, as well as the degree of surface exposure. This technique has now been used successfully to examine the interaction between individual lipid molecules and phospholipids in membrane vesicles to determine which peptides interact. One limitation to this approach is that collision-induced dissociation (the usual fragmentation mechanism used with most mass spectrometers, including triple quadrupole instruments) causes “scrambling,” whereby, upon fragmentation, the deuterium atom changes position within the peptide (65). Electron transfer dissociation, a more recently developed fragmentation mechanism, appears to produce minimal scrambling under the correct experimental conditions (66), suggesting it will indeed be possible to achieve single-residue resolution of hydrogen/deuterium exchange reactions (67). So in the future, even more precise information on lipid conformations when bound to proteins should be possible.
Association of Proteins/Enzymes with Lipids and Membranes
The use of DXMS to study the interaction of proteins with lipids and membranes has been sparse (68, 69), but this technique has recently been applied to several members of the phospholipase A2 (PLA2) enzyme superfamily (59-61) interacting with lipid molecular species in their active sites as well as with phospholipid vesicle membranes. One of the major phospholipase A2s, the group IVA cytosolic PLA2 (cPLA2) (60), is very important in cell signaling and in the release of AA, the precursor of numerous inflammatory mediators (e.g. prostaglandins). DXMS studies with two potent and specific reversible lipid inhibitors suggested specific docking modalities, each in a different specific location at or near the catalytic site (70). When these inhibitors were docked and then subjected to molecular dynamics calculations, their precise binding conformations were further defined giving a precise picture of the protein's interaction with these two lipid inhibitors and their bound conformations. One of these is a substrate analog, so it can provide an excellent picture of the conformation of a phospholipid molecular species bound in the active site of this enzyme (70).
When this same cPLA2 is presented with a phospholipid vesicle membrane, changes in several peptide deuterium exchange rates were observed, suggesting the peptidic locations of interaction with the vesicle. Both the C2 domain and the α/β hydrolase fold domain showed such changes, leading to the conclusion that the large cap region, which includes a lid normally covering the active site, undergoes a broad interaction with the membrane phospholipids (60). Each has a different and distinct interaction with the phospholipid membrane. Although the 85-kDa cPLA2 interacts with both its C2 domain and its large cap region, in contrast, the smaller 13-kDa secreted sPLA2 inserts a limited number of amino acid side chains into the phospholipid surface (59). In further contrast, the 85-kDa iPLA2, which has both a large ankyrin repeat domain (not shown in the figure) and an α/β hydrolase fold domain, contains an exposed helix, which inserts into the membrane with little or no effect on the active site (61). With the further refinements discussed above, DXMS has the potential for providing even more precise pictures of the conformation of lipids when interacting with proteins in active sites and in membranes.
CELL/TISSUE IMAGING OF LIPIDS BY MASS SPECTROMETRY
Localization of Lipids in Tissues and Cells
The mass spectrometry-based lipidomics analyses described above have greatly advanced our knowledge regarding the diversity and number of unique molecular lipid species contained in biological systems. Knowledge of the subcellular localization of individual lipids can provide additional detailed insight into lipid functions, such as those involved in disease and cell death. Employing differential centrifugation for subcellular organelle isolation, Andreyev et al. (18) recently provided the first mapping of the subcellular mouse macrophage lipidome in basal and activated states, focusing on the major lipid species and the major organelles. A total of 229 individual/isobaric species were identified, and these are shown according to lipid class in Table 1. Although highly impressive, it is possible that this isolation technique might chemically alter the lipids under study or their location. Additionally, if one is studying tissue samples, the lipid extraction procedure clearly destroys information regarding the location of the lipid within the tissue. Certainly, carrying out a mass spectrometric lipid analysis of tissue samples in situ and, ultimately, individual cells might avoid such pitfalls and provide us with an unperturbed picture of the cellular and subcellular lipidome.
Matrix-Assisted Laser Desorption Ionization-Imaging Mass Spectrometry
A relatively new technique, pioneered by Caprioli and colleagues (71) within the past decade, is imaging mass spectrometry (IMS), which provides spatially resolved ion intensity maps corresponding to the mass-to-charge ratio of intact molecular species at specific locations in a prepared tissue sample. A most common mode of IMS employs an ultraviolet (UV) laser to form ions from molecules contained in a tissue sample that is coated with a UV-absorbing matrix, and as such, this is known as matrix assisted laser desorption ionization (MALDI). MALDI was pioneered in the 1980s by Karas et al. (72) and Tanaka et al. (73); Koichi Tanaka shared the 2002 Nobel Prize in Chemistry for this contribution (as noted above, John Fenn was awarded part of this same Nobel Prize for his contribution to ESI). Briefly, MALDI-IMS involves preparing a thin (≈10-μm thick) slice of tissue sample, which is mounted onto a small plate (MALDI target, approximately 3 cm × 3 cm) and then coated with a film of a UV-absorbing compound (matrix), such as dihydroxybenzoic acid. Firing a small UV laser spot onto the matrix/tissue sample generates an energetic plasma, resulting in the formation of both positive and negative biomolecular ions from the tissue, which are ejected from the target surface and coupled directly into the entrance of a mass spectrometer. As with ESI, MALDI is a soft ionization technique, leaving the molecule whole and unfragmented in the ionization process. A mass-to-charge ratio spectrum is recorded for each X/Y grid coordinate on the target the MALDI laser is focused upon.
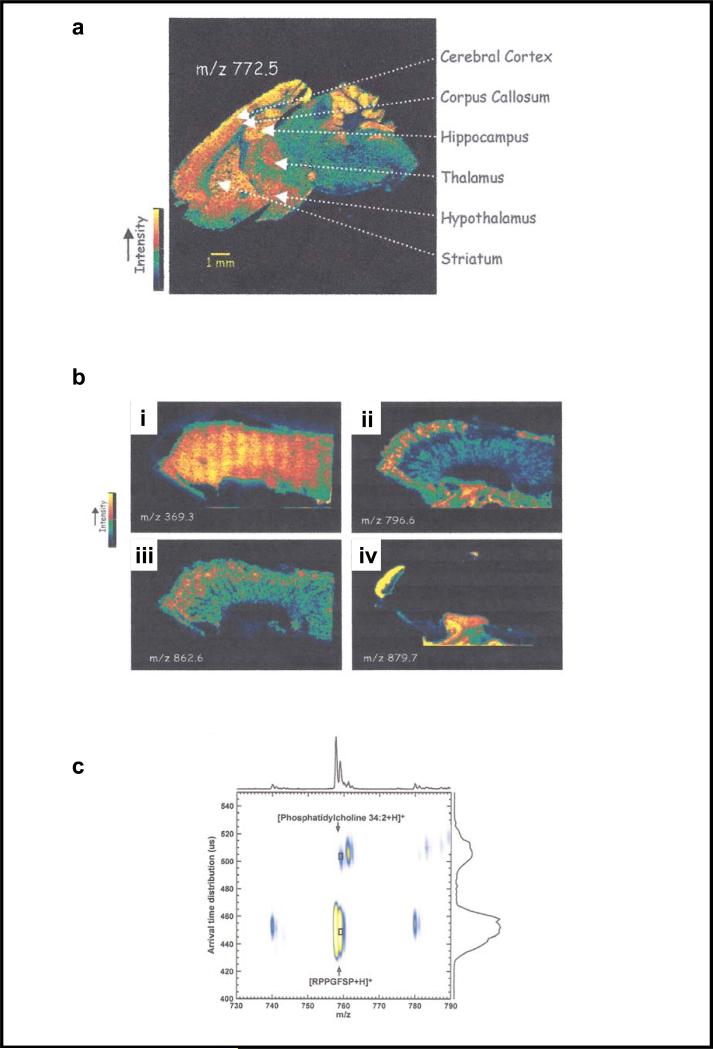
The prepared target is placed atop a mechanical stage, which is programmed to be moved incrementally such that an array of mass-to-charge ratio spectra are recorded for the entire tissue sample. Software is then used to generate mass-to-charge ratio images as a function of the X/Y grid location for the entire tissue sample. While data is recorded for all mass-to-charge ratio values within the specified mass scan range, software permits visualization of designated (extracted) values, enabling lipid species-specific images and comparisons. Current laser technology permits MALDI-IMS to have a lateral resolution on the order of 25-100 μm. Owing to its fast duty cycle, which allows it to be compatible with a high-repetition-rate MALDI laser, and its high mass accuracy capability, the TOF mass spectrometry is the instrument most frequently used for imaging. Most of the early MALDI-IMS work involved peptide and protein imaging in tissue samples (71, 74-77); however, recently, impressive results from imaging lipids have been reported (78-82). Figure 9a is an extracted ion image for m/z = 772.5 from a mouse brain slice that was obtained using a TOF mass spectrometer scan range of m/z 500 to 900. The 16:0/16:0-PC + K+ (potassium cation adduct) species has a corresponding mass of 772.5 Da. Using their appropriate corresponding accurate masses, similar extracted ion images are possible for other gylcerophospholipid species collected in this data set. Although such an image suggests that regional differences in concentrations for specific molecular species are present, additional work is still needed to determine if ion abundance truly reflects local concentrations (81). Observed signal intensities may be skewed owing to local intracellular concentrations of sodium and potassium, local environments in tissue slices, as well as other factors.
Figure 9.
Examples of matrix-assisted laser desorption ionization imaging mass spectrometry used to investigate lipids in tissue samples. (a) Extracted ion image for m/z = 772.5 from a mouse brain slice. The mass of 772.5 Da corresponds to the 16:0/16:0-PC + K+ (potassium cation adduct) species. (b) Images from a section of mouse kidney. Using MS/MS scan mode and high mass accuracy to confirm identity, cholesterol (i), 16:0/18:2-PC (ii), 40:6-PC (iii), and 16:0/20:4/18:1-TAG (iv) were some of the lipid species observed. Panels a and b reprinted with permission from Reference 78. (c) Example of an ion mobility-mass spectrometry (IM-MS) two-dimensional plot for a nominally isobaric peptide (RPPGGFSP) and lipid (32:4-PC). The mass-to-charge ratio (m/z) is plotted on the x-axis, and ion mobility drift time is plotted on the y-axis. The mass-to-charge ratio signals overlap and cannot be resolved; however, they clearly have different ion mobility drift times. Reprinted with permission from Reference 88.
The specialized MS/MS scan modes, which are very useful for monitoring various lipid classes (described above), using the triple quadrupole mass spectrometer can also be utilized with MALDI-IMS. The tandem quadrupole-TOF instrument is a particularly useful instrument for such work, as the first two sectors of this instrument are the same as the triple quadrupole and capable of the many scan modes shown in Figure 3. However, the third sector is a TOF unit with high mass accuracy capabilities.
Future Directions in Imaging Mass Spectrometry
Although IMS is still in its relative infancy, it has already provided a highly valuable tool to the lipidomics investigational toolchest, and we can expect its contributions and capabilities will only continue to grow in the future. One area that is aggressively being pushed forward is the ability to conduct single-cell, intracellular imaging. This is not possible with the current lateral resolution capabilities available with MALDI-IMS (25-100 μm); however, the use of focused ion beam clusters as a means of initiating ionization is showing much promise. Using highly focused ion beam clusters, such as Buckminsterfullerenes (C60 clusters), lateral resolutions on the nanometer scale have been reported (83, 84) putting this technique well within the range required to allow single-cell imaging. As lateral resolution is improved, however, the amount of ablated imaging target material decreases; thus, improvements in mass spectrometer sensitivity need to keep step. Additionally, as lateral resolution improves so does the size of the collected data set. This will require concomitant improvements in data processing and storage.
Another area of development in IMS involves coupling an ion mobility spectrometer between the ionization source (MALDI or ion beam cluster) and the mass spectrometer. Ion mobility spectrometry separates analyte ions on the basis of their ion-neutral collision cross section (85). This is accomplished by having ions, upon slight acceleration, drift through a tube maintained at ≈3-5 torr of helium gas before being directed into a coupled mass spectrometer. Ions having the same mass-to-charge ratio yet different three-dimensional shapes, and therefore different collision cross sections, have different drift times throughthe helium gas and can be resolved. For example, different molecular classes exhibit significant differences in their collision cross section at a given mass-to-charge ratio, e.g., oligonucleotides/carbohydrates < peptides < lipids (86, 87). In the imaging of complex biological samples, IM-MS can provide qualitative identification of the analyte molecular class, suppression of chemical noise, and the potential for high mass measurement accuracy by using internal calibrating standards that do not interfere with the analyte of interest on the basis of structure. This form of imaging is also showing promise in the area of lipidomics (87, 88).
SUMMARY AND OUTLOOK.
The extraordinary achievements in genomics during the past few decades have given birth to the other notable -OMIC disciplines: proteomics, the systematic mapping of protein and enzyme gene products; and metabolomics, the systematic study of the unique chemical fingerprints that cellular processes leave behind, specifically the study of their small-molecule metabolite profiles. Lipidomics, a central part of metabolomics, is the detailed analysis and global characterization, both spatial and temporal, of the structure and function of lipids (the lipidome) within a living system. A comprehensive understanding of an organism's awesomely complex lipidome (the mammalian lipidome is estimated to contain some hundreds of thousands of distinct molecular species) offers both an analytical challenge and a tremendous opportunity for correlating the role of lipids in various diseases, such as cancer, atherosclerosis, and chronic inflammation.
Mass spectrometry is playing a central analytical role in lipidomics by providing sensitive and quantitative assays of hundreds of lipid species in a single analysis; these assays are leading to the molecular characterization of novel lipid species, allowing for unperturbed spatial lipidome images in tissue samples, and showing promise for similar images on the subcellular level. Concomitant advances in mass spectrometry hardware and data analysis will continue to grow with advances in the molecular biological sciences to further our understanding of lipidomics and the central role of lipids in living systems and disease.
SUMMARY POINTS.
The LIPID MAPS Consortium, in conjunction with the International Committee for Classification and Nomenclature of LIPIDS, has defined eight categories of lipids on the basis of their biosynthetic origin. Currently there exist two fundamentally different approaches for mass spectrometry-based lipidomics analyses. One approach (CLASS) is based upon the separation of different lipid categories using optimal extraction and chromatographic techniques prior to mass analysis. The second approach (shotgun lipidomics) omits chromatographic separation and essentially analyzes all the lipid classes together, directly infusing them into the mass spectrometer, while selectively utilizing different mass spectrometer parameters for optimal lipid class-specific optimization. Both approaches have merit and have produced impressive results.
The emerging field of lipidomics has provided fertile ground to discover and characterize novel lipid species, and mass spectrometry has and will continue to play a vital role in this area. A few of the novel lipids observed recently in untargeted assays include ether-linked glycerophospholipids, N-acylphosphoserine derivatives, glycerophosphocholine species containing very long-chain polyunsaturated fatty acids, and the 22-carbon dihomoprostaglandins.
Deuterium exchange mass spectrometry (DXMS) is a powerful tool for investigating the location and orientation of proteins and enzymes associated with lipid substrates and membranes. DXMS has been particularly useful for categorizing the bound lipid conformations and membrane interactions for members of the phospholipase A2 (PLA2) enzyme superfamily.
Imaging mass spectrometry (IMS) is proving useful for the in situ lipid analysis of tissue samples, providing an unperturbed spatial picture of the lipidome. Ongoing technology developments to improve spatial resolution may allow for future imaging on the subcellular level.
FUTURE ISSUES.
How will technological advances in mass spectrometry hardware and data analysis carry over to lipidomics? Will one lipidomics approach - CLASS versus shotgun - prove to be the superior analytical platform for global lipidomics research? Will IMS allow for a true, undisturbed picture of the subcellular lipidome? What technology advances will be necessary to achieve this goal?
As the number of explored lipidomes for various living systems increases, how many and of what variety of novel lipid species will be discovered? Will genetic engineering for living systems lead to dramatically exotic novel lipids?
How will our understanding of protein/enzyme interactions with lipid membranes improve as advances occur in DXMS technology, such as the realization of single-residue peptide resolution?
How will our growing understanding of lipidomics translate into our understanding of disease, drug discovery, and biomarkers for therapeutic treatment outcomes? What new -OMICs disciplines will emerge in the future?
ACKNOWLEDGMENTS
We wish to thank all of our LIPID MAPS colleagues for their contributions, generous interactions, and all that they have taught us about mass spectrometry. We especially recognize the contributions of Professors Alex Brown, Christopher Glass, Alfred Merrill, Robert Murphy, Christian Raetz, David Russell, Shankur Subramaniam, Stephen White, Nicholas Winograd and Joseph Witztum; and Doctors Eoin Fahy and Walter Shaw. Funding for this effort was provided by the National Institute of General Medical Sciences Large-Scale Collaborative “Glue” Grant U54 GM069338 as well as R01 GM020501 and R01 GM064611.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–64. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 2.Dennis E. Lipidomics joins the omics evolution. Proc. Natl. Acad. Sci. USA. 2009;106:2089–90. doi: 10.1073/pnas.0812636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daviss B. Growing pains for metabolomics. Scientist. 2005;19(8):25–28. [Google Scholar]
- 4.Gomase VS, Changbhale SS, Patil SA, Kale KV. Metabolomics. Curr. Drug Metab. 2008;9:89–98. doi: 10.2174/138920008783331149. [DOI] [PubMed] [Google Scholar]
- 5.Thomson JJ. Rays of positive electricity. Phil. Mag. 1910;20(118):752–767. [Google Scholar]
- 6.McLuckey SA, Wells JM. Mass analysis at the advent of the 21st century. Chem. Rev. 2001;101:571–606. doi: 10.1021/cr990087a. [DOI] [PubMed] [Google Scholar]
- 7.Yates JR., III Mass spectrometry and the age of the proteome. J. Mass Spectrom. 1998;33:1–19. doi: 10.1002/(SICI)1096-9888(199801)33:1<1::AID-JMS624>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 8.Aebersold R, Goodlett DR. Mass spectrometry in proteomics. Chem. Rev. 2001;101:269–95. doi: 10.1021/cr990076h. [DOI] [PubMed] [Google Scholar]
- 9.Murphy RC, Fiedler J, Hevko J. Analysis of nonvolatile lipids by mass spectrometry. Chem. Rev. 2001;101:479–526. doi: 10.1021/cr9900883. [DOI] [PubMed] [Google Scholar]
- 10.Dettmer K, Aronov PA, Hammock BD. Mass spectrometry--based metabolomics. Mass Spectrom. Rev. 2007;26:51–78. doi: 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffiths WJ, Wang Y. Mass spectrometry: from proteomics to metabolomics and lipidomics. Chem. Soc. Rev. 2009;38:1882–96. doi: 10.1039/b618553n. [DOI] [PubMed] [Google Scholar]
- 12.Smith A. Oxford Dictionary of Biochemistry and Molecular Biology. 2nd ed. Oxford Univ. Press; Oxford, UK: 2000. [Google Scholar]
- 13.Voet D, Voet JG, Pratt CW. Fundamentals of Biochemistry. Life at the Molecular Level. 3rd ed. Wiley; New York: 2008. [Google Scholar]
- 14.Christie WW. Lipid Analysis. 3rd ed. Oily Press; Oxford, UK: 2003. [Google Scholar]
- 15.Fahy E, Subramanium S, Brown HA, Glass CK, Merrill AH, Jr, et al. A comprehensive classification system for lipids. J. Lipid Res. 2005;46:839–61. doi: 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 16.Fahy E, Subramanium S, Murphy RC, Nishijima M, Raetz CR, et al. Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 2009;50:S9–14. doi: 10.1194/jlr.R800095-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raetz CRH, Garrett TA, Reynolds CM, Shaw WA, Moore JD, et al. Kdo2-Lipid A of Escherichia coli, a defined endotoxin that activates macrophages via TLR-4. J. Lipid Res. 2006;47:1097–111. doi: 10.1194/jlr.M600027-JLR200. [DOI] [PubMed] [Google Scholar]
- 18.Andreyev AY, Fahy E, Guan Z, Kelly S, Li X, et al. Subcellular organelle lipidomics in TLR-4-activated macrophages. J. Lipid Res. 2010;51:2785–97. doi: 10.1194/jlr.M008748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dennis EA, Deems RA, Harkewicz R, Quehenberger O, Brown HA, et al. A mouse macrophage lipidome. J. Biol. Chem. 2010;285:39976–85. doi: 10.1074/jbc.M110.182915. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, et al. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. USA. 2009;106:2136–41. doi: 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quehenberger O, Armando AM, Brown HA, Milne SB, Myers DS, et al. Lipidomics reveals a remarkable diversity of lipids in human plasma. J. Lipid Res. 2010;51:3299–305. doi: 10.1194/jlr.M009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Serhan CN, Takano T, Chiang N, Gronert K, Clish CB. Formation of endogenous “anti-inflammatory” lipid mediators by transcellular biosynthesis. Am. J. Respir. Crit. Care Med. 2000;161:S95–101. doi: 10.1164/ajrccm.161.supplement_1.ltta-19. [DOI] [PubMed] [Google Scholar]
- 23.Funk CD. Prostaglandins and leukotrienes: advances in eicosanoid biology. Science. 2001;294:1871–75. doi: 10.1126/science.294.5548.1871. [DOI] [PubMed] [Google Scholar]
- 24.Moore JD, Caufield WV, Shaw WA. Quantitation and standardization of lipid internal standards for mass spectrometry. Methods Enzymol. 2007;432:351–67. doi: 10.1016/S0076-6879(07)32014-4. [DOI] [PubMed] [Google Scholar]
- 25.Quehenberger O, Armando A, Dumlao D, Stephens DL, Dennis EA. Lipidomics analysis of essential fatty acids in macrophages. Prostaglandins Leukot. Essent. Fat. Acids. 2008;79:123–29. doi: 10.1016/j.plefa.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deems R, Buczynski MW, Bowers-Gentry R, Harkewicz R, Dennis EA. Detection and quantitation of eicosanoids via high performance liquid chromatography-electrospray ionization-mass spectrometry. Methods Enzymol. 2007;432:59–82. doi: 10.1016/S0076-6879(07)32003-X. [DOI] [PubMed] [Google Scholar]
- 27.Krank J, Murphy RC, Barkley RM, Duchoslav E, McAnoy A. Qualitative analysis and quantitative assessment of changes in neutral glycerol lipid molecular species within cells. Methods Enzymol. 2007;432:1–20. doi: 10.1016/S0076-6879(07)32001-6. [DOI] [PubMed] [Google Scholar]
- 28.Hutchins PM, Barkley RM, Murphy RC. Separation of cellular nonploar neutral lipids by normal-phase chromatography and analysis by electrospray ionization mass spectrometry. J. Lipid Res. 2008;49:S9–14. doi: 10.1194/jlr.M700521-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivanova PT, Milne SB, Byrne MO, Xiang Y, Brown HA. Glycerophospholipid identification and quantitation by electrospray mass spectrometry. Methods Enzymol. 2007;432:21–57. doi: 10.1016/S0076-6879(07)32002-8. [DOI] [PubMed] [Google Scholar]
- 30.Milne S, Ivanova P, Forrester J, Brown HA. Lipidomics: an analysis of cellular lipids by ESI-MS. Methods. 2006;39:92–103. doi: 10.1016/j.ymeth.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 31.Sullards MC, Allegood JC, Kelly S, Wang E, Haynes CA, et al. Structure-specific, quantitative methods for analysis of sphingolipids by liquid chromatography-tandem mass spectrometry: “inside-out” sphingolipidomics. Methods Enzymol. 2007;432:83–115. doi: 10.1016/S0076-6879(07)32004-1. [DOI] [PubMed] [Google Scholar]
- 32.Shaner RL, Allegood JC, Park E, Wang E, Kelly S, et al. Quantitative analysis of sphingolipids for lipidomics using triple quadrupole and quadrupole linear ion trap mass spectrometers. J. Lipid Res. 2009;50:1692–707. doi: 10.1194/jlr.D800051-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guan Z, Li S, Smith DC, Shaw WA, Raetz CR. Identification of N-acylphosphatidylserine molecules in eukaryotic cells. Biochemistry. 2007;46:14500–13. doi: 10.1021/bi701907g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garrett TA, Guan Z, Raetz CR. Analysis of ubiquinones, dolichols, and dolichol diphosphate-oligosaccharides by liquid chromatography-electrospray ionization-mass spectrometry. Methods Enzymol. 2007;432:117–43. doi: 10.1016/S0076-6879(07)32005-3. [DOI] [PubMed] [Google Scholar]
- 35.Garrett TA, Raetz CR, Richardson T, Kordestani R, Son JD, et al. Identification of phosphatidylserylglutamate: a novel minor lipid in Escherichia coli. J. Lipid Res. 2009;50:1589–99. doi: 10.1194/jlr.M800549-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 37.Harkewicz R, Hartvigsen K, Almazan F, Dennis EA, Witztum JL, et al. Cholesteryl ester hydroperoxides are biologically active components of minimally oxidized LDL. J. Biol. Chem. 2008;283:10241–51. doi: 10.1074/jbc.M709006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han X, Gross RW. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom. Rev. 2005;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- 39.Ejsing CS, Duchoslav E, Sampaio J, Simons K, Bonner R, et al. Automated identification and quantification of glycerophospholipid molecular species by multiple precursor ion scanning. Anal. Chem. 2006;78:6202–14. doi: 10.1021/ac060545x. [DOI] [PubMed] [Google Scholar]
- 40.Han X, Gross RW. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J. Lipid Res. 2003;44:1071–79. doi: 10.1194/jlr.R300004-JLR200. [DOI] [PubMed] [Google Scholar]
- 41.Hutchins PM, Barkley RM, Murphy RC. Separation of cellular nonpolar neutral lipids by normal-phase chromatography and analysis by electrospray ionization mass spectrometry. J. Lipid Res. 2008;49:804–13. doi: 10.1194/jlr.M700521-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blaho VA, Buczynski MW, Brown CR, Dennis EA. Lipidomic analysis of dynamic eicosanoid reponses during the induction and resolution of Lyme arthritis. J. Biol. Chem. 2009;284:21599–612. doi: 10.1074/jbc.M109.003822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buczynski MW, Dumlao DS, Dennis EA. An integrated omics analysis of eicosanoid biology. J. Lipid Res. 2009;50:1015–38. doi: 10.1194/jlr.R900004-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gupta S, Maurya MR, Stephens DL, Dennis EA, Subramaniam S. An integrated model of eicosanoid metabolism and signaling based on lipidomics flux analysis. Biophys. J. 2009;96:4542–51. doi: 10.1016/j.bpj.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivanova PT, Milne SB, Brown HA. Identification of atypical ether-linked glycerophospholipid sopecies in macrophages by mass spectrometry. J. Lipid Res. 2010;51:1581–90. doi: 10.1194/jlr.D003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guan Z, Li S, Smith DC, Shaw WA, Raetz CRH. Identification of N-acylphosphosphatidylserine molecules in eukaryotic cells. Biochemistry. 2007;46:14500–13. doi: 10.1021/bi701907g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aveldano MI, Bazin NG. Molecular species of phosphatidylcholine, -ethanolamine, - serine, and -inositol in microsomal and photoreceptor membranes of bovine retina. J. Lipid Res. 1983;24:620–27. [PubMed] [Google Scholar]
- 48.Agbaga M-P, Brush RS, Mandal MNA, Hentry K, Elloitt MH, et al. Role of Stargardt-3 macular dystrophy protein (ELOVL4) in the biosynthesis of very long chain fatty acids. Proc. Natl. Acad. Sci. USA. 2008;105:12843–48. doi: 10.1073/pnas.0802607105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agbaga M-P, Mandal MNA, Anderson RE. Retinal very long-chain PUFAs: new insights from studies on ELOVL4 protein. J. Lipid Res. 2010;51:1624–42. doi: 10.1194/jlr.R005025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bedell M, Harkewicz R, Wang X, Zhang K. Focus on molecules: ELOVL4. Exp. Eye Res. 2010;90:476–77. doi: 10.1016/j.exer.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 51.McMahon A, Jackson SN, Woods AS, Kedzierski W. A Stargardt disease-3 mutation in the mouse Elovl4 gene causes retinal deficiency of C32--C36 acyl phosphatidylcholines. FEBS Lett. 2007;581:5459–63. doi: 10.1016/j.febslet.2007.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harkewicz R, Fahy E, Andreyev A, Dennis EA. Arachidonate-derived dihomoprostaglandin production observed in endotoxin-stimulated macrophage-like cells. J. Biol. Chem. 2007;282:2899–910. doi: 10.1074/jbc.M610067200. [DOI] [PubMed] [Google Scholar]
- 53.Sprecher H, VanRollins M, Sun F, Wyche A, Needleman P. Dihomoprostaglandins and -thromboxane. A prostaglandin family from adrenic acid that may be preferentially synthesized in the kidney. J. Biol. Chem. 1982;257:3912–18. [PubMed] [Google Scholar]
- 54.Campbell WB, Falck JR, Okita JR, Johnson AR, Callahan KS. Synthesis of dihomoprostaglandins from adrenic acid (7,10,13,16-docosatetraenoic acid) by human endothelial cells. Biochim. Biophys. Acta. 1985;837:67–76. doi: 10.1016/0005-2760(85)90086-4. [DOI] [PubMed] [Google Scholar]
- 55.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom. Rev. 2006;25:158–70. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 56.Konermann L, Tong X, Pan Y. Protein structure and dynamics studied by mass spectrometry: H/D exchange, hydroxyl radical labeling, and related approaches. J. Mass Spectrom. 2008;43:1021–36. doi: 10.1002/jms.1435. [DOI] [PubMed] [Google Scholar]
- 57.Engen JR. Analysis of protein conformation and dynamics by hydrogen/deuterium exchange MS. Anal. Chem. 2009;81:7870–75. doi: 10.1021/ac901154s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaltashov IA, Bobst CE, Abzalimov RR. H/D exchange and mass spectrometry in the studies of protein conformation and dynamics: Is there a need for a top-down approach? Anal. Chem. 2009;81:7892–99. doi: 10.1021/ac901366n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burke JE, Karbarz MJ, Deems RA, Li S, Woods VL, Jr, et al. Interaction of group IA phospholipase A2 with metal ions and phospholipid vesicles probed with deuterium exchange mass spectrometry. Biochemistry. 2008;47:6451–59. doi: 10.1021/bi8000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burke JE, Hsu YH, Deems RA, Li S, Woods VL, Jr, et al. A phospholipid substrate molecule residing in the membrane surface mediates opening of the lid region in group IVA cytosolic phospholipase A2. J. Biol Chem. 2008;283:31227–36. doi: 10.1074/jbc.M804492200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hsu YH, Burke JE, Li S, Woods VL, Jr, Dennis EA. Localizing the membrane binding region of group VIA Ca2+-independent phospholipase A2 using peptide amide hydrogen/deuterium exchange mass spectrometry. J. Biol. Chem. 2009;284:23652–61. doi: 10.1074/jbc.M109.021857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aebersold R, Mann M. Mass spectrometry--based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 63.Chait BT. Mass spectrometry: bottom-up or top-down. Science. 2006;314:65–66. doi: 10.1126/science.1133987. [DOI] [PubMed] [Google Scholar]
- 64.Englander JJ, Del Mar C, Li W, Kim JS, Stranz DD, et al. Protein structure change studied by hydrogen-deuterium exchange, functional labeling, and mass spectrometry. Proc. Natl. Acad. Sci. USA. 2003;100:7057–62. doi: 10.1073/pnas.1232301100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jørgensen TJ, Gårdsvoll H, Ploug M, Roepstorff P. Intramolecular migration of amide hydrogens in protonated peptides upon collisional activation. J. Am. Chem. Soc. 2005;127:2785–93. doi: 10.1021/ja043789c. [DOI] [PubMed] [Google Scholar]
- 66.Zehl M, Rand KD, Jensen ON, Jørgensen TJ. Electron transfer dissociation facilitates the measurement of deuterium incorporation into selectively labeled peptides with single residue resolution. Am. Chem. Soc. 2008;130:17453–59. doi: 10.1021/ja805573h. [DOI] [PubMed] [Google Scholar]
- 67.Rand KD, Zehl M, Jensen ON, Jørgensen TJ. Protein hydrogen exchange measured at single-residue resolution by electron transfer dissociation mass spectrometry. Anal. Chem. 2009;81:5577–84. doi: 10.1021/ac9008447. [DOI] [PubMed] [Google Scholar]
- 68.Demmers JA, Haverkamp J, Heck AJ, Koeppe RE, 2nd, Killian JA. Electrospray ionization mass spectrometry as a tool to analyze hydrogen/deuterium exchange kinetics of transmembrane peptides in lipid bilayers. Proc. Natl. Acad. Sci. USA. 2000;97:3189–94. doi: 10.1073/pnas.050444797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Man P, Montagner C, Vernier G, Dublet B, Chenal A, et al. Defining the interacting regions between apomyoglobin and lipid membrane by hydrogen/deuterium exchange coupled to mass spectrometry. J. Mol. Biol. 2007;368:464–72. doi: 10.1016/j.jmb.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 70.Burke JE, Babakhani A, Gorfe AA, Kokotos G, Li S, et al. Location of inhibitors bound to group IVA phospholipase A2 determined by molecular dynamics and deuterium exchange mass spectrometry. J. Am. Chem. Soc. 2009;131:8083–91. doi: 10.1021/ja900098y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Charuand P, Schwartz SA, Caprioli RM. Imaging mass spectrometry: a new tool to investigate the spatial organization of peptides and proteins in mammalian tissue sections. Curr. Opin. Chem. Biol. 2002;6:676–81. doi: 10.1016/s1367-5931(02)00370-8. [DOI] [PubMed] [Google Scholar]
- 72.Karas M, Bachmann D, Hillenkamp F. Influence of the wavelength in high-irradiance ultraviolet laser desorption mass spectrometry of organic molecules. Anal. Chem. 1985;57:2935–39. [Google Scholar]
- 73.Tanaka K, Waki H, Ido Y, Akita S, Yoshida Y, et al. Protein and polymer analyses up to m/z 100,000 by laser ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 1988;2:151–53. [Google Scholar]
- 74.Charuand P, Schwartz SA, Caprioli RM. Assessing protein patterns in disease using imaging mass spectrometry. J. Proteome Res. 2004;3:245–52. doi: 10.1021/pr0341282. [DOI] [PubMed] [Google Scholar]
- 75.Chaurand P, Norris JL, Cornett DS, Mobley JA, Caprioli RM. New developments in profiling and imaging of proteins from tissue sections by MALDI mass spectrometry. J. Proteome Res. 2006;5:2889–900. doi: 10.1021/pr060346u. [DOI] [PubMed] [Google Scholar]
- 76.Reyzer ML, Caprioli RM. MALDI-MS-based imaging of small molecules and proteins in tissues. Curr. Opin. Chem. Biol. 2007;11:29–35. doi: 10.1016/j.cbpa.2006.11.035. [DOI] [PubMed] [Google Scholar]
- 77.Schwartz SA, Weil RJ, Thompson RC, Shyr Y, Moore JH, et al. Proteomic-based prognosis of brain tumor patients using direct-tissue matrix-assisted laser desorption ionization mass spectrometry. Cancer Res. 2005;65:7674–81. doi: 10.1158/0008-5472.CAN-04-3016. [DOI] [PubMed] [Google Scholar]
- 78.Hankin JA, Barkley RM, Murphy RC. Sublimation as a method of matrix application for mass spectrometric imaging. J. Am. Soc. Mass Spectrom. 2007;18:1646–52. doi: 10.1016/j.jasms.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Puolitaival SM, Burnum KE, Cornett DS, Caprioli RM. Solvent-free matrix dry-coating for MALDI imaging of phosholipids. J. Am. Soc. Mass Spectrom. 2008;19:882–86. doi: 10.1016/j.jasms.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen Y, Allegood J, Liu Y, Wang E, Cachón-Gonzalez B. Imaging MALDI mass spectrometry using an oscillating capillary nebulizer matrix coating system and its application to analysis of lipids in brain from a mouse model of Tay-Sachs/Sandhoff disease. Anal. Chem. 2008;80:2780–88. doi: 10.1021/ac702350g. [DOI] [PubMed] [Google Scholar]
- 81.Murphy RC, Hankin JA, Barkley RM. Imaging of lipid species by MALDI mass spectrometry. J. Lipid Res. 2009;50:S317–22. doi: 10.1194/jlr.R800051-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reyzer ML, Chaurand P, Angel PM, Caprioli RM. Direct molecular analysis of whole-body animal tissue sections by MALDI imaging mass spectrometry. Methods Mol. Biol. 2010;656:285–301. doi: 10.1007/978-1-60761-746-4_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Winograd N. The magic of cluster SIMS. Anal. Chem. 2005;77:A143–49. 2005. [Google Scholar]
- 84.Carado A, Passarelli MK, Kozole J, Wingate JE, Winograd N, et al. C60 secondary ion mass spectrometry with a hydrid-quadrupole orthogonal time-of-flight mass spectrometer. Anal. Chem. 2008;80:7921–29. doi: 10.1021/ac801712s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Clemmer DE, Jarrold MF. Ion mobility measurements and their applications to clusters and biomolecules. J. Mass Spectrom. 1997;32:577–92. [Google Scholar]
- 86.Koomen JM, Ruotolo BT, Gillig KJ, McLean JA, Russell DH, et al. Oligonucleotide analysis with MALDI-ion-mobility-TOFMS. Anal. Bioanal. Chem. 2002;373:612–17. doi: 10.1007/s00216-002-1363-2. [DOI] [PubMed] [Google Scholar]
- 87.Woods AS, Ugarov M, Egan T, Koomen J, Gillig KJ, et al. Lipid/peptide/nucleotide separation with MALDI-ion mobility-TOF MS. Anal. Chem. 2004;76:2187–95. doi: 10.1021/ac035376k. [DOI] [PubMed] [Google Scholar]
- 88.McLean JA, Ridenour WB, Caprioli RM. Profiling and imaging of tissues by imaging ion mobility-mass spectrometry. J. Mass Spectrom. 2007;42:1099–105. doi: 10.1002/jms.1254. [DOI] [PubMed] [Google Scholar]