Abstract
Development of the nervous system is finely regulated by consecutive expression of cell-specific transcription factors. Here we show that Helios, a member of the Ikaros transcription factor family, is expressed in ectodermal and neuroectodermal-derived tissues. During embryonic development, Helios is expressed by several brain structures including the lateral ganglionic eminence (LGE, the striatal anlage); the cingulated, insular and retrosplenial cortex; the hippocampus; and the accessory olfactory bulb. Moreover, Helios is also expressed by Purkinje neurons during postnatal cerebellar development. Within the LGE, Helios expression follows a dynamic spatio-temporal pattern starting at embryonic stages (E14.5), peaking at E18.5, and completely disappearing during postnatal development. Helios is expressed by a small population of nestin-positive neural progenitor cells located in the subventricular zone as well as by a larger population of immature neurons distributed throughout the mantle zone. In the later, Helios is preferentially expressed in the matrix compartment, where it colocalizes with Bcl11b and Foxp1, well-known markers of striatal projection neurons. In addition, we observed that Helios expression is not detected in Dlx1/2 and Gsx2 null mutants, while its expression is maintained in Ascl1 mutants. These findings allow us to introduce a new transcription factor in the cascade of events that take part of striatal development postulating the existence of at least 4 different neural progenitors in the LGE. An Ascl1-independent but Gsx2- & Dlx1/2-dependent precursor will express Helios defining a new lineage for a subset of matrix striatal neurons.
Introduction
During embryonic development, neural lineages are derived from neural progenitor cells (NPCs) localized in specific brain areas, which consecutively generate both neurons and glial cells [1,2,3]. Within the developing telencephalon, neural stem cells from the wall of the lateral ventricle, the ventricular zone (VZ) [4], undergo successive divisions to expand the pool of NPCs, thereby increasing the volume of the subventricular zone (SVZ). A number of signaling pathways controlled by different factors, such as Notch, Wnt, or FGFs, have been implicated in the self-renewal, proliferation and/or maintenance of the undifferentiated state of NPCs [5]. At certain developmental stages depending on the telencephalic area, NPCs divide for the last time and become specific immature neurons that migrate to postmitotic regions [4,6,7]. Multiple transcription factors finely regulate this neurogenic process [8,9].
The striatum is one of the main telencephalic constituents of the basal ganglia; its embryonic anlage is the lateral ganglionic eminence (LGE) [10,11]. The progenitor zone of the LGE is the germinal zone (GZ) a source of striatal projection neurons and olfactory bulb interneurons [10,12,13]. Within the LGE, separate waves of neurogenesis are coupled to the production of different compartments known as the patches, or striosomes, and the matrix [14]. Neurogenesis between E12 and E13 produces clusters of substance P (SP)-expressing neurons that form the patches or striosomal compartment. The second wave of neurogenesis taking place at E14–E16 in mice will give rise to the matrix compartment, which contains both enkephalin (ENK)- and SP-positive neurons [15,16].
Several transcription factors such as Gsx1 & 2 (formerly named Gsh1 & 2), Ascl1 (formerly named Mash1) and Dlx-family members have distinct but overlapping patterns of expression in the VZ, SVZ, and mantle zone (MZ), and regulate distinct steps in LGE patterning and/or differentiation [17,18,19,20]. In addition to these transcription factors, Ebf-1, Bcl11b (formerly named Ctip2) and Ikaros are exclusively expressed in the striatal MZ where they regulate differentiation of striatal projection neurons [21,22,23,24].
Ikaros-family members are a Krüppel-type zing-finger transcription factors that play essential roles during lymphocyte development [25,26]. Ikaros is the founder member of this small family of DNA-binding proteins which consists of Ikaros, Helios, Aiolos, Eos, and Pegasus [27,28]. Besides the well characterized role of this family of transcription factors during hematopoietic development, Ikaros has also been recently involved in LGE development [24,29]. However, little is known about the expression of Helios in the central nervous system (CNS). In this work on the developing mouse, we present the first description of Helios expression during brain development. We focused on its transient expression in the developing striatum, providing evidence that Helios is a marker of a subpopulation of immature matrix striatal neurons. Further, we defined Helios' expression in several transcription factor mutants, thereby placing it within the known transcriptional hierarchy that drives striatal development.
Material and Methods
Animal Subjects
Mice were maintained in standard conditions with food and water ad libitum. All animal procedures were approved by local committees [Universitat de Barcelona, CEEA (133/10) and Generalitat de Catalunya (DAAM 5712)], in accordance with the European Communities Council Directive (86/609/EU). B6CBA wild-type (wt) mice (from Charles River Laboratories, Les Oncins, France), Dlx-5/6-Cre-IRES-EGFP transgenic mice [12], and Ikaros [30], Gsx2 [31], Ascl1 [32], Dlx-1/2 [33], Helios [34], and Ebf-1 [35] knockout mice (–/–) were used. E18.5 Dlx5/6-cre-IRES-EGFP/gtROSA double transgenic brains were obtained by crossing heterozygous B6;129-Gtrosa26tm1Sho(gtROSA) mice [36] with hemizygous Dlx5/6-cre-IRES-EGFP mice.
All strains were maintained by backcrossing to C57BL/6 mice. Genotypes were determined by polymerase chain reaction (PCR) as described elsewhere [12,30,31,32,33,34,35]. The day of pregnancy, determined by the first detection of a vaginal sperm plug in daily inspection, was considered embryonic day (E)0.5.
Histology
All immunostainings were performed with the following antibodies: anti-Helios (1:1,000; polyclonal; a generous gift of Dr. Stephen T. Smale (University of California Los Angeles (UCLA) School of Medicine [37]), anti-Helios (1:50; polyclonal, Santa Cruz), anti-Ikaros (1:2,000; monoclonal; [24]), anti-nestin (Rat 401; 1:50; polyclonal; Developmental Studies Hybridoma Bank; The University of Iowa), anti-GFAP (1:200; monoclonal; Sigma Chemical Co.), anti-β-III-tubulin (Tuj1; 1:200; monoclonal; Sigma Chemical Co.), anti-MAP2 (1:200; monoclonal; Sternberger Monoclonals), anti-NeuN (1:100; monoclonal; Millipore), anti-dopamine- and cAMP-regulated phosphoprotein of 32 kDa (DARPP-32; 1:500; polyclonal; Chemicon International, Inc.), anti-Ctip2 (1:200; polyclonal; Abcam), anti-calbindin (1:1000; monoclonal; Sigma Chemical Co.) and anti-Foxp-1 (1:200; polyclonal; Abcam).
For histological preparations, brains were removed at specific developmental stages and frozen in dry-ice cooled isopentane. Serial coronal cryostat sections (14 μm) were cut on a cryostat and collected on silane-coated slides and frozen at −20°C. Fluorescent immunolabeling was performed according to the protocol described in [38]. Briefly, tissue sections were blocked for 1 h in PBS containing 0.3% Triton X-100 and 1% bovine serum albumin (BSA) to avoid unspecific binding. Thereafter, they were incubated overnight at 4°C in PBS containing 0.3% Triton X-100 and 1% BSA with the corresponding primary antibodies. After three PBS washes, sections were incubated for 2 h at room temperature with the following secondary antibodies: Cy3-conjugated donkey anti-rabbit IgG (1:500) or Cy2-conjugated donkey anti-mouse or Cy3 donkey anti goat (1:200; all from Jackson Immunoresearch Laboratories Inc.). At the end, tissue sections were counterstained with 4,6'-diamidino-2-phenylindole (DAPI; Sigma Chemical Co.) and mounted in Mowiol (Calbiochem). Fluorescent photomicrographs were taken on a Leica TCS SL laser scanning confocal spectral microscope (Leica Microsystems Heidelberg GmbH). All pictures acquisitions were done as tiff files and adjustments of brightness and contrast were done with Adobe Photoshop 6.0 program (Adobe Systems Incorporated). No signal was detected in control preparations from which the primary antibody was omitted.
Nonisotopic in situ hybridization was performed using a riboprobe for Helios gene (Gene accession No. NM_011770). To obtain the riboprobe, an EcoRI digested fragment of the MSCV plasmid encoding Helios (kindly provided by Dr. Christopher A. Klug, University of Alabama at Birmingham [39]) was cloned into Bluescript SK II vector (Stratagene). To obtain the antisense probe, the plasmid was linearized with SacI, and transcribed with T7 RNA polymerase using the Digoxigenin labeling kit (Roche). Digoxigenin labeled sense probe, linearized with KpnI and transcribed with T3 RNA polymerase, was used as a control. In brief, brains were removed at E18.5 and frozen in dry-ice cooled isopentane. Serial coronal cryostate sections (14 μm) were cut on a cryostat, collected on silane-coated slides, and frozen at −20°C. Frozen tissue sections were air dried, fixed in 4% paraformaldehyde in PBS for 20 min at 4°C, and processed for in situ hybridization as described elsewhere [40].
Production of viral particles and cell transduction
To overexpress Helios, we used the pLV-Helios-IRES-EGFP plasmid or the pLV-IRES-eGFP plasmid, which encode for human Helios and the green fluorescent protein (eGFP) or for eGFP alone, respectively. The pLV-IRES eGFP plasmid was generated as described elsewhere [41]. Briefly, MCS-IRES-eGFP was cloned from the PRV-IRES-eGFP (Genetrix SL, Tres Cantos) using the BamHI and the SalI restriction sites into the pRRLsinPPT plasmid (pRRL) constructed by the Miami Project to Cure Paralysis Viral Vector Core Lab based on the lentiviral transducing plasmid developed by Naldini et al. [42]. To construct the pLV-Helios-IRES-eGFP, the human Helios gene from the SPORT6-Helios plasmid (Invitrogen S.A.) was cloned into pLV-IRES-eGFP between BamHI and XhoI sites.
To overexpress Gsx2, the human Gsx2 gene from the pcDNA-hGsx2 plasmid kindly provided by Dr. Peter Marynen (Université de Leuven, Belgium) was PCR-cloned into the retroviral vector pRV-IRES-eGFP using the MCS BamHI and XhoI sites as described elsewhere [41].
For retroviral or lentiviral production, 293T cells were plated at a density of approximately 6×104 cells per cm2. The following day, cells were transfected by either a 3-plasmid system (pRV-Gsx2-IRES-eGFP or pRV-IRES-eGFP plasmid, the plasmid that expresses gag and pol genes, and the plasmid that expresses vesicular stomatitis virus G) or a 4-plasmid system (pLV-IRES-eGFP or pLV-Helios-IRES-eGFP plasmid, the plasmid that expresses HIV-1 Rev, the plasmid that expresses gag and pol genes, and the plasmid that expresses vesicular stomatitis virus G) using the calcium phosphate/DNA coprecipitation method. Cells were transfected for 7 h and afterwards the medium was replaced with fresh one. The supernatant from vector-producing 293T cells was collected every 22 h during 3 days. Then, the supernatant was passed through a 0.45-μm-pore-size filter to remove producer cells and subjected to 2 centrifugations at 4°C and 25000 g for 120 min to concentrate the virus. The virus-containing pellet was dissolved in BSA 1%. To determine the viral titer, a total of 2×105 3T3 or 293T cells were inoculated with serial dilutions of concentrated retrovirus or lentivirus. Forty-eight hours after infection, eGFP titers (IU/mL) were determined by using a fluorescence-activated cell scanner (FACS).
Neurosphere assay and overexpression
LGE from E14.5 wt mice were dissected out and mechanically disaggregated as previously described [24]. Briefly, LGE-derived neurosphere cultures were obtained by seeding 50,000 cells/cm2 in basal medium [Dubelcco's modified Eagle's medium (DMEM; Sigma Chemical Co.):F12 (Invitrogen S.A.) (1:1), 0.3% glucose (Sigma Chemical Co.), 0.3 mg/mL glutamine (Invitrogen S.A.), 5 mM HEPES (Invitrogen S.A.), 100 IU/mL penicillin, 100 mg/mL streptomycin (Invitrogen S.A.), 4 μg/mL heparin (Sigma Chemical Co.), 4 mg/mL BSA (Sigma Chemical Co.), 1×N2 supplement (Invitrogen S.A.)], supplemented with 20 ng/mL fibroblast growth factor (Sigma Chemical Co.) and 10 ng/mL epidermal growth factor (Invitrogen S.A.). Every 5 days, neurospheres were collected, dissociated by pipetting approximately 40 times with a P100 micropipette, and plated down in fresh media at a density of 10,000 cells/cm2.
To overexpress mouse Dlx-2, we used the pCAGGS-Dlx-2 vector as described previously Studhmer and colleagues [43]. To this end, neurospheres were disaggregated mechanically and 5×106 cells were transfected by nucleofection following the manufacturer's protocol (A33 Nucleofector Program; Amaxa Biosystems). Viable cells were counted by trypan blue exclusion and plated down at a cell density of 50,000 cells/mm2.
To overexpress Helios or Gsx2, neurospheres were transduced with the pLV-Helios-IRES-eGFP or the pRV-Gsx2-IRES-eGFP lentivirus, or the control pLV-IRES-eGFP as described elsewhere [41].
Cell pellets were collected 3 or 5 days after transfection or transduction and frozen at −80°C for mRNA analysis.
Q-PCR assay
Gene expression was evaluated by quantitative PCR (Q-PCR) assays as previously described by Martin-Ibanez and coworkers [44]. The following TaqMan® Gene expression assays (Applied Biosystems) were used: 18s, Hs99999901_s1; Helios, Mm00496108_m1; vimentin, Mm00449208_m1; nestin, Mm00450205_m1; β-tubulin III, Mm00727586_s1; GFAP, Mm00546086_m1; olig1, Mm00497537_s1; olig2, Mm01210556_m1; Dlx2, Mm00438427_m1; and Gsx2, Mm446650_m1. To provide negative controls and exclude contamination by genomic DNA, the reverse transcriptase was omitted in the cDNA synthesis step, and the samples were subjected to the PCR reaction with each TaqMan® gene expression assay.
Analysis and quantification was performed with the Comparative Quantitation Analysis program of the MxPro™ Q-PCR analysis software version 3.0 (Stratagene), using the 18S gene expression as internal loading control. All Q-PCR assays were performed in duplicate and repeated for at least three independent experiments. The results were expressed as relative levels with respect to the expression of the same gene in E14.5 for developmental studies or the control vector overexpressing cells considered as 100%.
Statistical analysis
All results are expressed as the mean of independent experiments±standard error of the mean. Results were analyzed using the Student's t-test or one-way analysis of variance, followed by the Bonferroni post-hoc test.
Results
Helios is expressed in specific brain and sensitive areas during mouse development
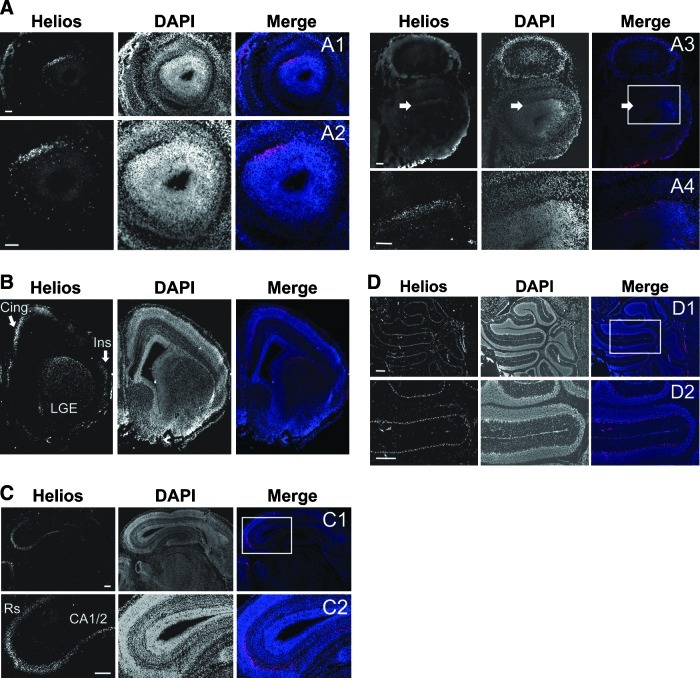
To study the expression and distribution of Helios during mouse brain development, we performed immunohistochemistry analysis at different developmental stages. At E16.5, Helios was expressed in the granule cell layer of the accessory olfactory bulb (Fig. 1A), the LGE, the cingulated (Fig. 1B), insular (Fig. 1B), and retrosplenial (Fig. 1C) cortices, and hippocampal CA1/2 (Fig. 1C). At early postnatal ages, Helios expression was also observed in the Purkinje cell layer of the cerebellum (Fig. 1D), while its expression decreased in the above-mentioned brain areas. In the adult brain, Helios expression was not detected (data not shown).
FIG. 1.
Helios is expressed in specific brain areas during central nervous system development. Helios immunohistochemistry was performed in mouse brain coronal sections at E16.5 and P15. Representative fluorescent photomicrographs corresponding to anterior and posterior sections at E16.5 show Helios expression in: (A) the granular layer of accessory olfactory bulb from the anterior (A1, A2; note that A2 is a high magnification of A1) to the posterior axis (A3, A4; A4 represents the inset in A3); (B) the lateral ganglionic eminence (LGE), the cingulate (Cing) and insular (Ins) cortex; and (C) the retrosplenial cortex (Rs) and CA1/2 layer (CA1/2) of the hippocampus (C1, C2; C2 represents the inset in C1). (D) Helios is expressed by Purkinje neurons in the cerebellum at P15 (D1; D2 represents the inset in D1). Scale bars: (A) 1 mm; and (C, D) 2 mm.
Helios is also expressed outside of the CNS during development (Supplementary Fig. S1; Supplementary materials are available online at http://www.liebertpub.com/). At E16.5, Helios expression was detected in the nasal and oral cavities as well as in the optic vesicles. Within the nasal cavity, this zinc finger transcription factor was detected in the epithelial tissue (Supplementary Fig. S1A). Similarly, in the oral cavity, Helios was also detected in the epithelium of the palate (Supplementary Fig. S1B). Both epithelia showed Helios expression located in the intermediate layers but not in basal cells, which constitute the stem cells of the epithelium. In addition, Helios was also detected in the taste buds of the tongue (Supplementary Fig. S1B). Within the optic vesicles, this transcription factor was detected in the neuro-epithelial layers such as the pigmented epithelium and the neuronal layer of the retina (Supplementary Fig. S1C).
Helios shows a dynamic expression pattern during LGE development
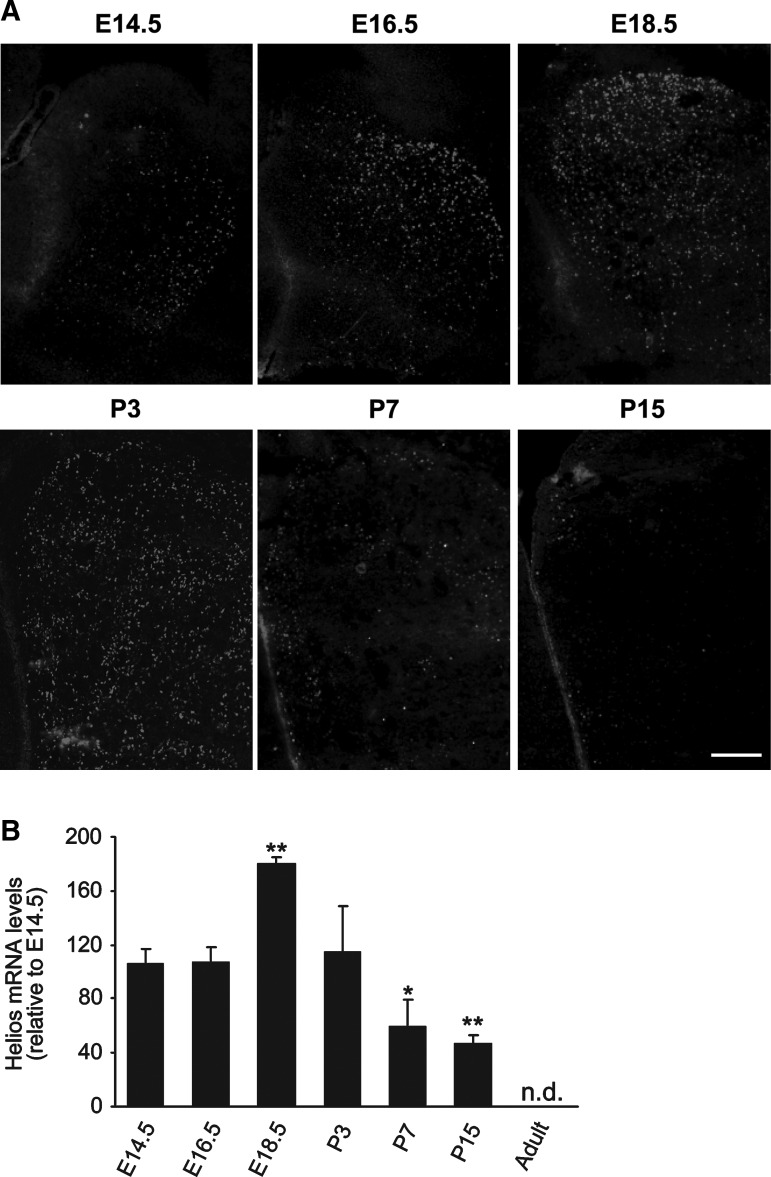
Here we focused our study on Helios expression in the LGE and its derivative the striatum, because of the role of other Ikaros-family members in striatal development [29,24]. Immunohistochemistry studies performed at different LGE developmental stages showed that Helios expression was initially detected at E14.5; its levels increased at later stages, peaking between E18.5 and P3 (Fig. 2A). Subsequently, the levels of expression decreased; after P15 expression was not detectable (Fig. 2A). These results were confirmed by Q-PCR (Fig. 2B). We did not detect Helios expression in the medial ganglionic eminence, the source of striatal interneurons (Supplementary Fig. S2).
FIG. 2.
Helios presents a dynamic spatio-temporal pattern of expression during LGE and striatal development. (A) Helios immunohistochemistry was performed in LGE coronal sections at different stages of embryonic (E14.5, E16.5, and E18.5) and postnatal (P3, P7, and P15) development. Helios is initially expressed at the ventro-lateral area of E14.5 mantle zone (MZ LGE and progressively spreads to the dorsomedial area during development. The expression of Helios is reduced at later postnatal stages and only a few positive cells can be observed at P15. (B) Quantification of Helios mRNA levels by Q-PCR during LGE development showing a peak of expression at E18.5. Results are expressed as the mean of 3 to 5 LGE samples and error bars represents the standard error of the mean (SEM). Statistical analysis was calculated by Student's t-test; *P<0.05; **P<0.005 relative to E14.5 expression. Scale bars: (A) 500 μm. (n.d., not detected).
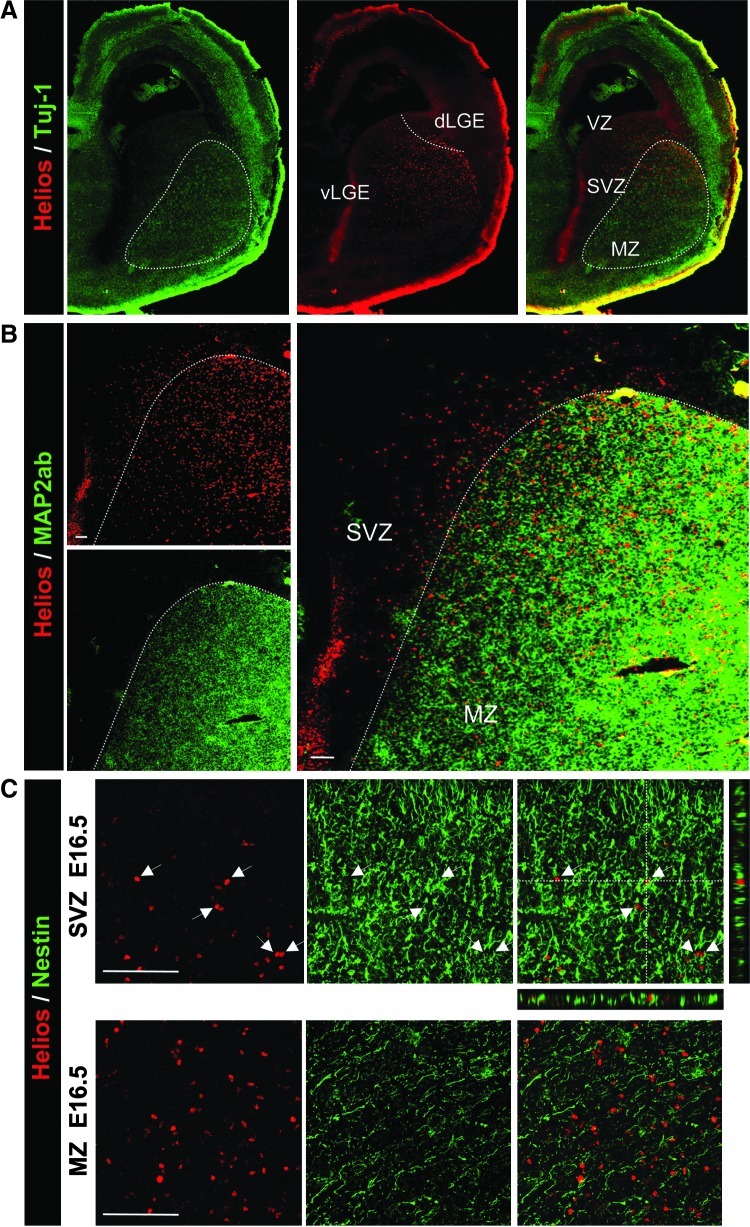
The spatial distribution of Helios expression in the LGE and striatum showed 2 interesting findings. First, the pattern of Helios expression in the striatum changed over time, starting at E14.5 in the ventrolateral area and moving to the dorsomedial region at later stages (Fig. 2A). Secondly, we observed that this transcription factor was detected in 2 different cell populations. During embryonic development, Helios was expressed by a small population of cells in the SVZ (Fig. 3A and B). Within this germinal area, Helios colocalized with nestin-positive NPCs (Fig 3C). In addition, Helios was expressed by a larger population of cells located in the MZ at both embryonic and postnatal stages. However, in this area Helios-expressing cells were nestin-negative but MAP2-positive differentiating neurons.
FIG. 3.
Helios is expressed by nestin-positive cells in the SVZ and MAP2-positive cells in the MZ of the LGE. Double immunohistochemistry was performed in mouse brain coronal sections at E16.5 for Helios and β-III-tubulin (Tuj-1) (A), MAP2 (B), and nestin (C). (A, B). Note the expression of Helios in 2 different cell populations: a small one located in the Tuj-1- and MAP2-negative SVZ of the LGE that is spread through the ventral and dorsal part (A–B), and a larger one distributed along the MZ where it colocalizes with the neuronal marker MAP2 (B). (C) Representative high-magnification photomicrograph showing double immunohistochemistry for nestin and Helios both at the SVZ and MZ. Colocalization of both markers is observed in the SVZ of the LGE. Z-analysis shows the colocalization of Helios positive nuclei (arrows) surrounded by nestin-positive intermediate filaments. VZ, ventricular zone; SVZ, subventricular zone; MZ, mantle zone (delimited by dotted area). Scale bars: (B) 150 μm; (C) 100 μm.
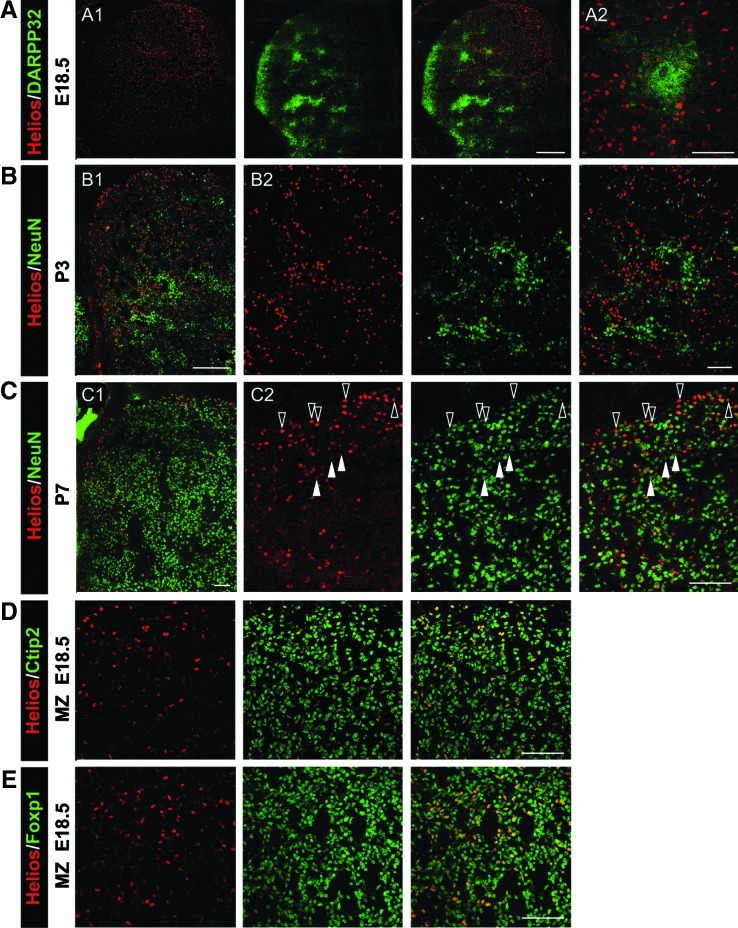
The striatum is compartmentalized into separate matrix and striosomal (or patch) domains, which can be defined by specific markers [45]. The striosomal compartment is positive for DARPP-32 at late embryonic and early postnatal stages. During this period, Helios expressing cells appeared to be DARPP-32-negative (Fig. 4A). This finding is in agreement with the lack of colocalization observed at P3 between Helios and NeuN, showing the later a patchy distribution as well (Fig. 4B). These results suggested that Helios is expressed by matrix cells. This assumption was confirmed by the colocalization of Helios with Bcl11b (Fig. 4D) and Foxp1 (Fig. 4E), 2 transcription factors mainly expressed by matrix cells [22,46]. Interestingly, we did observe double staining for Helios and NeuN at P7, a developmental stage at which the mature neuronal marker NeuN is spread all over the LGE (Fig. 4C). However, the cells that express high levels of Helios still lack NeuN expression, while low Helios-expressing cells are double positive for both markers (Fig. 4C). This dynamic expression of NeuN indicated a progressive maturation of striatal neurons that is in agreement with previous papers demonstrating the early maturation of neurons located in the patches [14].
FIG. 4.
Helios is expressed by neuronal precursors located in the striatal matrix compartment. Double immunohistochemistry was performed in mouse brain coronal sections at several embryonic and postnatal stages for Helios and striatal neuronal markers such as: DARPP-32 (A), NeuN (B, C), Bcl11b (Ctip2) (D), and Foxp1 (E). Representative photomicrographs show that Helios is not expressed in the striosomal compartment, since it has a complementary distribution with DARPP-32, a striosomal marker at this stage (A1, A2; note that A2 is a high magnification of A1). At P3, Helios does not colocalize with NeuN, a marker of mature neurons that at this stage presents a striosomal distribution (B1, B2; note that B2 is a high magnification of B1). However, both markers are coincident in some cells at P7, developmental stage at which NeuN is staining most of the striatal neurons (C1, C2; note that C2 is a high magnification of C1). High magnification picture shows that the cells that express high levels of Helios are negative or express low levels of NeuN (open arrows), while low Helios-expressing cells are positive for both markers (closed arrows) (C1). In addition, Helios is expressed by Bcl11b (Ctip2) (D) and Foxp1-positive neurons (E), two markers of striatal projection neurons. Scale bars: (A1, B1) 500 μm; (A2, B2, C, D, E) 100 μm.
Helios is expressed by a specific type of neuronal precursor in the LGE
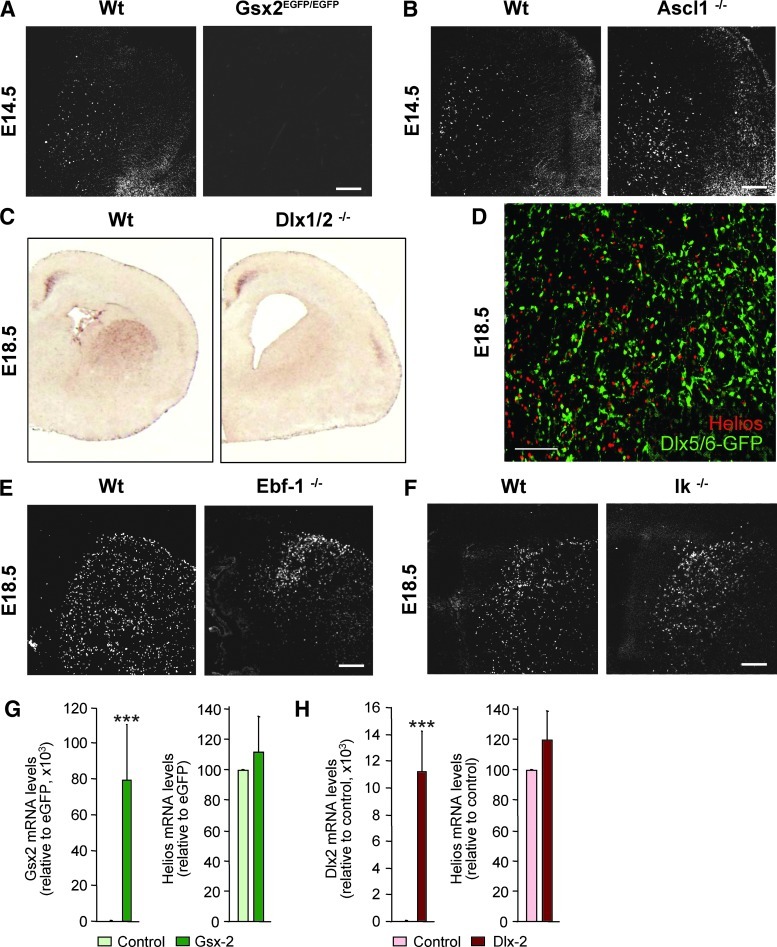
The germinal zone of the LGE is the main source of striatal projection neurons. In order to study the relationship between Helios and other transcription factors involved in the development of these neurons we analyzed the expression of this transcription factor in knockout mice for Gsx2, Ascl1, Dlx-1/2, Ebf1, and Ikaros (Fig. 5). Our results showed that Helios-positive cells completely disappeared in Gsx2 knockout mice (Fig. 5A). However, Helios was normally expressed in Ascl1–/– mice (Fig. 5B). On the other hand, Dlx-1/2–/– mice lacked detectable Helios expression in the LGE, although its expression was not affected in the cerebral cortex (Fig. 5C). These findings indicated that Helios is expressed downstream of Gsx2 and Dlx1/2 specifically in the LGE. However, we demonstrated that Helios is not directly regulated by Gsx2 or Dlx2 since overexpression of these transcription factors in neurosphere cultures did not modify Helios expression (Fig. 5G and H).
FIG. 5.
Helios is expressed by a specific neuronal precursor downstream of Gsx2 and Dlx1/2 during LGE development. Immunohistochemistry and in situ hybridization analysis of Helios expression was performed in several knockout animals for transcription factors involved in striatal projection neurons development. Gsx2 knock-out mice (Gsx2EGFP/EGFP) show a complete lack of Helios expression (A). In contrast, normal Helios protein expression was detected in Ascl1–/– mice (B). In situ hybridization analysis for Helios mRNA in wt and Dlx-1/2–/– mice show that the expression of this transcription factor is lost in the LGE but conserved in cortical areas at E18.5 (C). Double immunohistochemistry for Helios and eGFP performed in recombined Dlx-5/6-Cre-IRES-GFP/gtROSA (Dlx5/6-GFP) mice shows that Helios does not colocalize with eGFP positive cells at E18.5 (D). In addition, Helios immunohistochemistry performed in wt and Ebf-1–/– mice at E18.5, shows that its expression is detected in Ebf-1–/– mice although its distribution changes due to the LGE alterations described in these mice (E). Similarly, Helios protein expression is also preserved in Ikaros–/– mice (IK–/–) at E18.5 (F). Over-expression of Gsx2 (G) or Dlx2 (H) in neurosphere cultures does not affect Helios expression levels 3 days after transduction or transfection, suggesting that this transcription factor is not directly regulated by Gsx2 or Dlx2. Results are expressed as the mean of 4 to 5 neurosphere cultures and error bars represent the s.e.m. Statistical analysis was calculated by Student's t-test; ***p<0.001 relative to neurospheres transduced or transfected with the respective control vectors. Scale bars: (A, B) 500 μm; (D) 150 μm; (E) and (F) 500 μm.
Helios expression levels appeared unchanged in Ebf-1–/– (Fig. 5E) and Ikaros–/– mice (Fig. 5F), although there was a change in the distribution of Helios-expressing cells in both mutants (Fig. 5E and F), probably reflecting striatal phenotypes that have been previously described [24,47].
Interestingly, the analysis of Helios expression in recombinant Dlx-5/6-Cre-IRES-GFP/gtROSA transgenic mice showed a lack of colocalization with GFP-positive cells (Fig. 5D). Thus, Helios-positive cells appear to be in distinct immature striatal neurons, or alternatively, that it could be expressed upstream of Dlx-5/6.
Dimerization could be a prerequisite for DNA binding and function of all Ikaros-family members. Like Aiolos and Ikaros, Helios can dimerize with itself as well as with other family members [48]. To further analyze the relation between Ikaros-family members within the LGE, double immunohistochemistry for Helios and Ikaros was performed at E18.5. A lack of coincidence between these 2 transcription factors is shown in Fig. 6A. The same result was obtained at E14.5 and E16.5 (data not shown). In addition, the expression of Ikaros was not affected in Helios knockout mice (Fig. 6B), indicating a parallel or independent expression of these two transcription factors in different neuronal precursors.
FIG. 6.
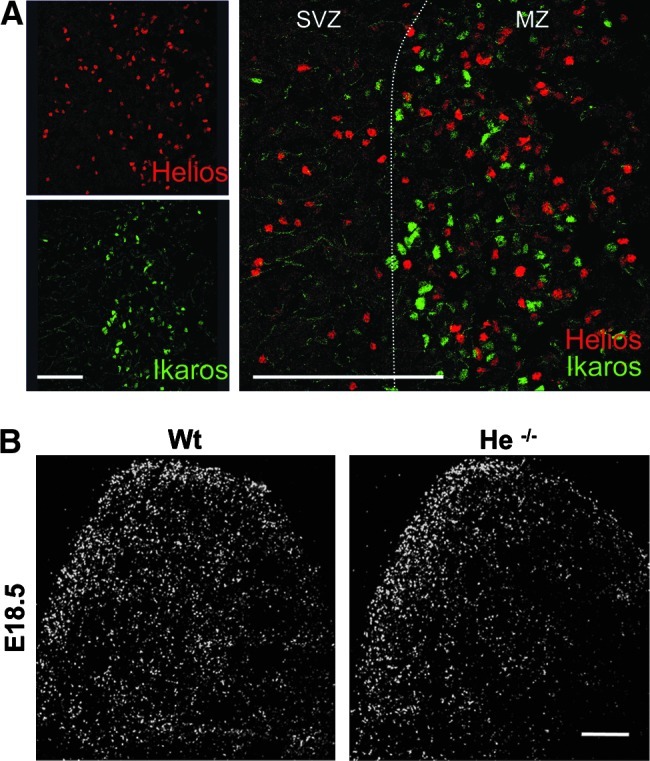
Helios and Ikaros are not expressed in the same LGE cells at E18.5. Double immunohistochemistry for Helios and Ikaros performed in the LGE at E18.5 shows a lack of coincidence between these two transcription factors (A). Note that Helios is expressed in the SVZ and MZ whereas Ikaros is only expressed in the MZ (A). Immunohistochemistry for Ikaros in wt and Helios–/– mice (He–/–) demonstrate no changes in Ikaros expression in the absence of Helios (B). Scale bars: (A) 400 μm; (B) 500 μm.
Helios induces the expression of the neuronal marker β-III-tubulin in neurosphere cultures
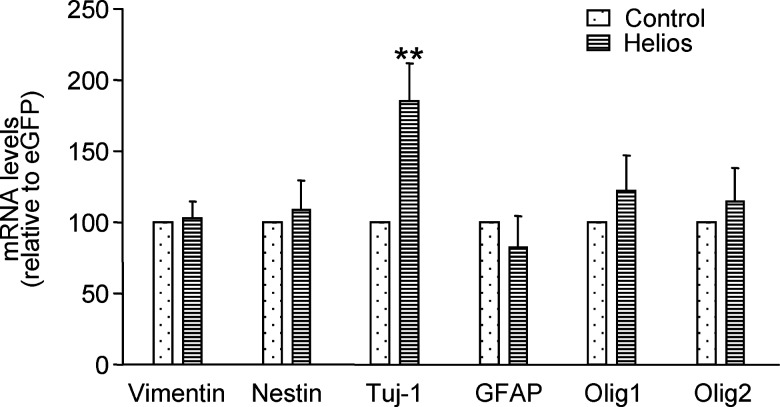
All previous results suggested that Helios might participate in striatal development as a neurogenic factor. To test this hypothesis, we overexpressed Helios in neurosphere cultures, and 5 days later we tested the expression of several neural markers including vimentin, nestin, β-III-tubulin, GFAP, Olig1, and 2. Our results showed that Helios overexpression increased the levels of the neuronal marker β-III-tubulin without affecting the expression of precursors or glial markers (Fig. 7).
FIG. 7.
Helios over-expression increases the levels of the neuronal marker β-III-tubulin. Over-expression of Helios in neurosphere cultures for 5 days induces an increase in the levels of the neuronal marker β-III-tubulin (Tuj-1). However, the expression of markers for neural precursors, vimentin and nestin, or glial cells, GFAP and Olig 1 and 2, are not modified after Helios over-expression in the neurosphere culture. Results are expressed as the mean of 5 neurosphere cultures and error bars represent the s.e.m. Statistical analysis was calculated by Student's t-test; **p<0.005 relative to neurospheres transduced with the control vector.
Present findings point to the hypothesis that Helios is a transcription factor expressed by a specific neuronal precursor in the LGE during striatal development defining a new lineage that might contribute to the generation of a matrix striatal neuron subpopulation (Fig. 8).
FIG. 8.
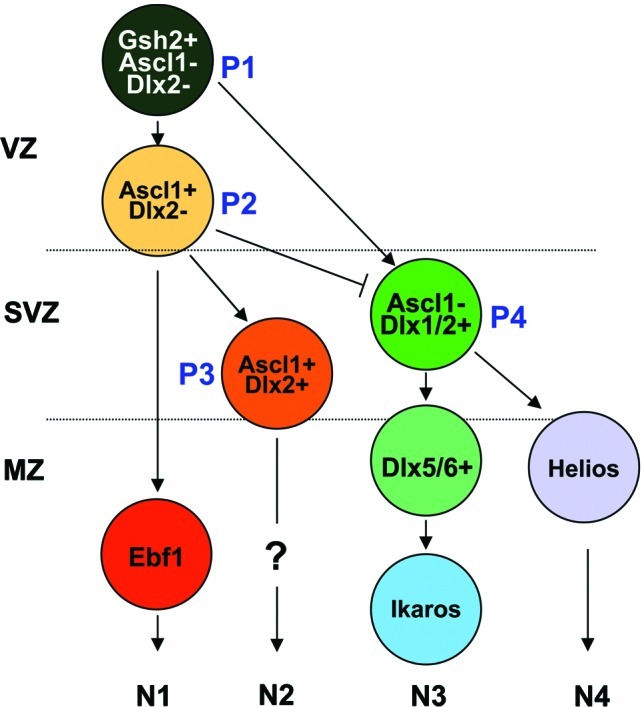
Proposed model for the definition of different lineages contributing to the development of striatal projection neurons. P, NPC; N, immature neuron; VZ, ventricular zone; SVZ subventricular zone; MZ, mantle zone. Color image available online at www.liebertpub.com/scd
Discussion
We demonstrate for the first time that Helios is expressed in specific ectodermal and neuroectodermal-derived tissues. One of these areas is the LGE, the striatum anlage, where we observed that this transcription factor is expressed by both NPCs as well as by a subset of striatal immature neurons.
The relationship between Helios and other transcription factors involved in striatal development allowed us to define a new lineage that could determine the specific development of a subpopulation of matrix striatal projection neurons.
Helios is expressed by a specific precursor of striatal projection neurons during LGE development
Helios was originally described as a cell-type specific hematopoietic factor that is involved in the development of lymphocytes [37]. However, here we show that this transcription factor is expressed in several regions of the CNS and peripheral nervous system (PNS).
In the present work we focused on the characterization of Helios expression in the LGE, the progenitor region for the striatum. It is initially expressed in the ventro-lateral area of the LGE MZ at E14.5. This initial detection spatially coincides with the early pattern of expression of the mature markers of striatal projection neurons, such as proenkephalin [49]. In addition, as embryonic striatal development occurs, Helios-expressing cells appear dorsomedially following the same dynamics that has been previously described for proenkephalin [49]. These results suggest that Helios might be involved in the determination of a neuronal precursor toward a striatal projection neuron that would be committed to an enkephalinergic phenotype. Keeping with this view, we observe that all Helios-expressing cells are positive for Bcl11b, an important transcription factor for striatal projection neurons development that is detected in NPCs at E13.5 [22].
Interestingly, Helios expression also colocalizes with the Forkhead transcription factor Foxp1. Several Foxp-family genes have been detected during telencephalon development [46,50,51]. Although all Foxp examined genes are expressed in the developing LGE, Foxp1 shows a preferential expression for a subset of striatal projection neurons located in the matrix compartment [46] while Foxp2 and Foxp4 are more coincident with markers of the striosomal compartment. Therefore, these findings indicate that Helios and Foxp1 are both expressed by a common neuronal precursor that will be differentiated into a matrix striatal projection neuron. Keeping with this view, Helios does not colocalize with DARPP32, a well known striosomal marker [52,53]. The colocalization of Foxp1 and Helios is not restricted to the LGE; they are also coexpressed in the cortical plate and the CA1/2 subfield of the hippocampus (present results and references 46 and 51]). Therefore, Helios and Foxp1 may share functions in regulating the development of the striatum, cortex, and hippocampus.
Helios is expressed in the dLGE and the vLGE
Similarly to Foxp1, Helios is also detected in a subset of cells localized in the SVZ where it coincides with the NPC marker nestin. In this area, Helios is expressed in both the dorsal (dLGE) and the ventral LGE (vLGE).
The dLGE and vLGE are sources for both striatal projection neurons and olfactory bulb interneurons [12,54,55,56,57, 58]. While Helios was expressed in differentiating striatal neurons, we did not detect Helios-expressing cells migrating to the olfactory bulb, or in interneurons of the main olfactory bulb, during development or in adulthood. On the other hand, we did detect Helios expression in cells of the accessory olfactory bulb, which is responsible for the detection of pheromones [59,60,61]. It has recently been reported that Bcl11b is also expressed in the accessory olfactory bulb [62], where this transcription factor plays an important role during its development [63]. Interestingly, the expression of Helios is also coincident with the expression of Bcl11b in the respiratory epithelium of the nasal cavity (present results and reference 62).
Helios defines a new lineage of neuronal progenitors during LGE development
The expression of Helios in the LGE suggests that it participates as a neurogenic factor for the development of a subset of striatal projection neurons. In fact, we demonstrate that Helios overexpression induces an increase in the levels of neuronal markers without altering the expression of glial markers. Distinct transcription factors acting in cascades underlie the sequential steps of the determination and differentiation of the striatal projection neurons. Specification of the striatum depends on the function of the Gsx1&2 homeobox genes, which are expressed in the VZ and SVZ of the LGE [64,65,54,66,55]. Mice lacking Gsx2 have defects in dLGE specification [64]. Adding to the previously known defects, here we showed that Helios expression is not detectable in Gsx2 knockout mice.
There is clear evidence that Gsx2 drives the expression of Dlx genes and Ascl1 in the LGE [64]. The Dlx homeobox gene family is essential for the development of striatal projection neurons [67,17]. Dlx1 & 2, whose expression are first detected in the VZ and the SVZ, are the earliest Dlx genes expressed during LGE development [17]. Thereafter, SVZ NPCs expressing Dlx1 & 2 start expressing Dlx5 & 6 [17,68]; thereafter, immature striatal neurons maintain Dlx expression [69]. Our results demonstrate that Helios is expressed downstream of Dlx1 & 2 function but upstream or independent of Dlx5 & 6 in the LGE.
Striatal development is not fully blocked in the Dlx1&2−/− mutants [67], demonstrating that parallel and/or redundant pathways continue to promote the generation and migration of some striatal projection neurons. The analysis of gene expression profiles in the double Dlx1/2–/– mice identified Dlx-dependent and Dlx-independent pathways. The Dlx-independent pathway depends in part on the function of the Ascl1 transcription factor [40]. Our present results show that Helios is correctly expressed in Ascl1–/– mice, indicating that this Ikaros-family member is expressed by Dlx-dependent but Ascl1-independent progenitors.
Ascl1 and Dlx2 expression define different lineages of subcortical progenitors [20]. Combinatorial expression of these transcription factors, in conjunction with the expression of proliferation and differentiation markers, provided evidences for at least 3 lineages of NPCs in the germinal area (P1, Ascl1–/Dlx2–; P2, Ascl1+/Dlx2–; and P3, Ascl+/Dlx2+) [20]. In the model proposed in Fig. 8, we introduced a new NPC which will be independent of Ascl1 (P4, Ascl1–/Dlx2+). Our present and previous data also suggest that this new lineage will give rise to 2 different cell progenies that will define subsets of striatal projecting neurons (N3–4). One of these progenies will express Dlx5/6 and thereafter become Ikaros+neuronal precursors (N3) [24], whereas the other neuronal precursors will become Helios+(N4). This is in agreement with our present results demonstrating that Helios and Ikaros are not coexpressed in striatal neurons.
Here we also show that Helios is independent of Ebf1, another LGE specific transcription factor that regulates the development of SP neurons. Our previous data demonstrated that Ebf1 is independent of Ikaros [24] and its expression is preserved in the Dlx1/2–/– mice [24,40]. All these data indicate that Ebf1 might be expressed by the P2 progenitor (Ascl1+/Dlx2–) to mediate the differentiation of SP striatal neurons by a cell-autonomous function exerted in NPCs as described elsewhere [20]. In fact, Ebf1 has been closely related to the development of these striatal projection neurons [23,21]. In contrast, Ascl1 might be indirectly involved in the development of striatal neurons by a non cell-autonomous mechanism that regulates the rate at which adjacent NPCs mature [20]. We propose that these adjacent NPCs could be the P4 that do not express Ascl1.
Many efforts are directed to the identification of specific transcription factors activated temporally and spatially to regulate cell fate determination during LGE development [4,5,70]. Here we describe a new player in striatal development named Helios. This zinc finger transcription factor defines a new lineage of neural precursors that will become a subset of matrix striatal projection neurons. Our results also provide evidences that suggest that Helios might be involved in the development of other specific brain areas.
Supplementary Material
Acknowledgments
We would like to thank Cristina Herranz and Ana López for technical assistance and Dr. Maria Calvo from the confocal microscopy unit of the Serveis Científico-Tècnics (Universitat de Barcelona) for their support and advice on confocal techniques. We are also grateful to Dr. Rudolf Grosschedl for Ebf-1–/– mice. We also thank Dr. Stephen T. Smale (Howard Hughes Medical Institute, Molecular Biology Institute, and Department of Microbiology and Immunology, University of California, Los Angeles School of Medicine) for anti-Helios antibody and Dr. Christopher A. Klug (Department of Microbiology, Division of Developmental, and Clinical Immunology, University of Alabama at Birmingham) for the MSCV plasmid encoding Helios. This study was supported by grants from the Ministerio de Ciencia e Innovación (BFU2007-61230/BFI to C.V.-A; SAF2008-04360, to J. A.; and SAF2009-07774 and PLE2009-0089, to J.M.C.), Spain; Instituto de Salud Carlos III, Ministerio de Ciencia e Innovación [CIBERNED, to C.V.-A. and to J.A.; and RETICS (Red de Terapia Celular, RD06/0010/0006 to J.M.C. and RD06/0010/0023 to S.M.], Spain; and Generalitat de Catalunya (2009SGR-00326 to J.A.), Spain. The Cell Therapy Program is supported by the Centre of Regenerative Medicine in Barcelona (CMRB; Promt-0901 to J.M.C.); Generalitat de Catalunya, Spain. E.C. was a fellow of the Ministerio de Investigación y Ciencia, Spain.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Davis AA. Temple S. A self-renewing multipotential stem cell in embryonic rat cerebral cortex. Nature. 1994;372:263–266. doi: 10.1038/372263a0. [DOI] [PubMed] [Google Scholar]
- 2.Temple S. The development of neural stem cells. Nature. 2001;414:112–117. doi: 10.1038/35102174. [DOI] [PubMed] [Google Scholar]
- 3.Williams BP. Precursor cell types in the germinal zone of the cerebral cortex. Bioessays. 1995;17:391–393. doi: 10.1002/bies.950170506. [DOI] [PubMed] [Google Scholar]
- 4.Merkle FT. Alvarez-Buylla A. Neural stem cells in mammalian development. Curr Opin Cell Biol. 2006;18:704–709. doi: 10.1016/j.ceb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 5.Guillemot F. Cellular and molecular control of neurogenesis in the mammalian telencephalon. Curr Opin Cell Biol. 2005;17:639–647. doi: 10.1016/j.ceb.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Gotz M. Barde YA. Radial glial cells defined and major intermediates between embryonic stem cells and CNS neurons. Neuron. 2005;46:369–372. doi: 10.1016/j.neuron.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 7.Merot Y. Retaux S. Heng JI. Molecular mechanisms of projection neuron production and maturation in the developing cerebral cortex. Semin Cell Dev Biol. 2009;20:726–734. doi: 10.1016/j.semcdb.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 8.Edlund T. Jessell TM. Progression from extrinsic to intrinsic signaling in cell fate specification: a view from the nervous system. Cell. 1999;96:211–224. doi: 10.1016/s0092-8674(00)80561-9. [DOI] [PubMed] [Google Scholar]
- 9.Guillemot F. Cell fate specification in the mammalian telencephalon. Prog Neurobiol. 2007;83:37–52. doi: 10.1016/j.pneurobio.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 10.Campbell K. Dorsal-ventral patterning in the mammalian telencephalon. Curr Opin Neurobiol. 2003;13:50–56. doi: 10.1016/s0959-4388(03)00009-6. [DOI] [PubMed] [Google Scholar]
- 11.Puelles L. Kuwana E. Puelles E. Bulfone A. Shimamura K. Keleher J. Smiga S. Rubenstein JL. Pallial and subpallial derivatives in the embryonic chick and mouse telencephalon, traced by the expression of the genes Dlx-2, Emx-1, Nkx-2.1, Pax-6, and Tbr-1. J Comp Neurol. 2000;424:409–438. doi: 10.1002/1096-9861(20000828)424:3<409::aid-cne3>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 12.Stenman JM. Wang B. Campbell K. Tlx controls proliferation and patterning of lateral telencephalic progenitor domains. J Neurosci. 2003;23:10568–10576. doi: 10.1523/JNEUROSCI.23-33-10568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marin C. Bove J. Bonastre M. Tolosa E. Effect of acute and chronic administration of U50,488, a kappa opioid receptor agonist, in 6-OHDA-lesioned rats chronically treated with levodopa. Exp Neurol. 2003;183:66–73. doi: 10.1016/s0014-4886(03)00107-9. [DOI] [PubMed] [Google Scholar]
- 14.van der Kooy D. Fishell G. Neuronal birthdate underlies the development of striatal compartments. Brain Res. 1987;401:155–161. doi: 10.1016/0006-8993(87)91176-0. [DOI] [PubMed] [Google Scholar]
- 15.Mason HA. Rakowiecki SM. Raftopoulou M. Nery S. Huang Y. Gridley T. Fishell G. Notch signaling coordinates the patterning of striatal compartments. Development. 2005;132:4247–4258. doi: 10.1242/dev.02008. [DOI] [PubMed] [Google Scholar]
- 16.Song DD. Harlan RE. The development of enkephalin and substance P neurons in the basal ganglia: insights into neostriatal compartments and the extended amygdala. Brain Res Dev Brain Res. 1994;83:247–261. doi: 10.1016/0165-3806(94)00145-6. [DOI] [PubMed] [Google Scholar]
- 17.Eisenstat DD. Liu JK. Mione M. Zhong W. Yu G. Anderson SA. Ghattas I. Puelles L. Rubenstein JL. DLX-1, DLX-2, and DLX-5 expression define distinct stages of basal forebrain differentiation. J Comp Neurol. 1999;414:217–237. doi: 10.1002/(sici)1096-9861(19991115)414:2<217::aid-cne6>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 18.Rallu M. Corbin JG. Fishell G. Parsing the prosencephalon. Nat Rev Neurosci. 2002;3:943–951. doi: 10.1038/nrn989. [DOI] [PubMed] [Google Scholar]
- 19.Waclaw RR. Wang B. Pei Z. Ehrman LA. Campbell K. Distinct temporal requirements for the homeobox gene Gsx2 in specifying striatal and olfactory bulb neuronal fates. Neuron. 2009;63:451–465. doi: 10.1016/j.neuron.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yun K. Fischman S. Johnson J. Hrabe de AM. Weinmaster G. Rubenstein JL. Modulation of the notch signaling by Mash1 and Dlx1/2 regulates sequential specification and differentiation of progenitor cell types in the subcortical telencephalon. Development. 2002;129:5029–5040. doi: 10.1242/dev.129.21.5029. [DOI] [PubMed] [Google Scholar]
- 21.Lobo MK. Karsten SL. Gray M. Geschwind DH. Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9:443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- 22.Arlotta P. Molyneaux BJ. Jabaudon D. Yoshida Y. Macklis JD. Ctip2 controls the differentiation of medium spiny neurons and the establishment of the cellular architecture of the striatum. J Neurosci. 2008;28:622–632. doi: 10.1523/JNEUROSCI.2986-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Dominguez M. Poquet C. Garel S. Charnay P. Ebf gene function is required for coupling neuronal differentiation and cell cycle exit. Development. 2003;130:6013–6025. doi: 10.1242/dev.00840. [DOI] [PubMed] [Google Scholar]
- 24.Martin-Ibanez R. Crespo E. Urban N. Sergent-Tanguy S. Herranz C. Jaumot M. Valiente M. Long JE. Pineda JR, et al. Ikaros-1 couples cell cycle arrest of late striatal precursors with neurogenesis of enkephalinergic neurons. J Comp Neurol. 2010;518:329–351. doi: 10.1002/cne.22215. [DOI] [PubMed] [Google Scholar]
- 25.Cobb BS. Smale ST. Ikaros-family proteins: in search of molecular functions during lymphocyte development. Curr Top Microbiol Immunol. 2005;290:29–47. doi: 10.1007/3-540-26363-2_3. [DOI] [PubMed] [Google Scholar]
- 26.Georgopoulos K. Haematopoietic cell-fate decisions, chromatin regulation and ikaros. Nat Rev Immunol. 2002;2:162–174. doi: 10.1038/nri747. [DOI] [PubMed] [Google Scholar]
- 27.John LB. Yoong S. Ward AC. Evolution of the Ikaros gene family: implications for the origins of adaptive immunity. J Immunol. 2009;182:4792–4799. doi: 10.4049/jimmunol.0802372. [DOI] [PubMed] [Google Scholar]
- 28.Rebollo A. Schmitt C. Ikaros, Aiolos and Helios: transcription regulators and lymphoid malignancies. Immunol Cell Biol. 2003;81:171–175. doi: 10.1046/j.1440-1711.2003.01159.x. [DOI] [PubMed] [Google Scholar]
- 29.Agoston DV. Szemes M. Dobi A. Palkovits M. Georgopoulos K. Gyorgy A. Ring MA. Ikaros is expressed in developing striatal neurons and involved in enkephalinergic differentiation. J Neurochem. 2007;102:1805–1816. doi: 10.1111/j.1471-4159.2007.04653.x. [DOI] [PubMed] [Google Scholar]
- 30.Wang JH. Nichogiannopoulou A. Wu L. Sun L. Sharpe AH. Bigby M. Georgopoulos K. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5:537–549. doi: 10.1016/s1074-7613(00)80269-1. [DOI] [PubMed] [Google Scholar]
- 31.Wang B. Waclaw RR. Allen ZJ. Guillemot F. Campbell K. Ascl1 is a required downstream effector of Gsx gene function in the embryonic mouse telencephalon. Neural Dev. 2009;4:5. doi: 10.1186/1749-8104-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guillemot F. Joyner AL. Dynamic expression of the murine Achaete-Scute homologue Mash-1 in the developing nervous system. Mech Dev. 1993;42:171–185. doi: 10.1016/0925-4773(93)90006-j. [DOI] [PubMed] [Google Scholar]
- 33.Qiu M. Bulfone A. Ghattas I. Meneses JJ. Christensen L. Sharpe PT. Presley R. Pedersen RA. Rubenstein JL. Role of the Dlx homeobox genes in proximodistal patterning of the branchial arches: mutations of Dlx-1, Dlx-2, and Dlx-1 and -2 alter morphogenesis of proximal skeletal and soft tissue structures derived from the first and second arches. Dev Biol. 1997;185:165–184. doi: 10.1006/dbio.1997.8556. [DOI] [PubMed] [Google Scholar]
- 34.Cai Q. Dierich A. Oulad-Abdelghani M. Chan S. Kastner P. Helios deficiency has minimal impact on T cell development and function. J Immunol. 2009;183:2303–2311. doi: 10.4049/jimmunol.0901407. [DOI] [PubMed] [Google Scholar]
- 35.Lin H. Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- 36.Mao X. Fujiwara Y. Chapdelaine A. Yang H. Orkin SH. Activation of EGFP expression by Cre-mediated excision in a new ROSA26 reporter mouse strain. Blood. 2001;97:324–326. doi: 10.1182/blood.v97.1.324. [DOI] [PubMed] [Google Scholar]
- 37.Hahm K. Cobb BS. McCarty AS. Brown KE. Klug CA. Lee R. Akashi K. Weissman IL. Fisher AG. Smale ST. Helios, a T cell-restricted Ikaros family member that quantitatively associates with Ikaros at centromeric heterochromatin. Genes Dev. 1998;12:782–796. doi: 10.1101/gad.12.6.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bosch M. Pineda JR. Sunol C. Petriz J. Cattaneo E. Alberch J. Canals JM. Induction of GABAergic phenotype in a neural stem cell line for transplantation in an excitotoxic model of Huntington's disease. Exp Neurol. 2004;190:42–58. doi: 10.1016/j.expneurol.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Z. Swindle CS. Bates JT. Ko R. Cotta CV. Klug CA. Expression of a non-DNA-binding isoform of Helios induces T-cell lymphoma in mice. Blood. 2007;109:2190–2197. doi: 10.1182/blood-2005-01-031930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Long JE. Swan C. Liang WS. Cobos I. Potter GB. Rubenstein JL. Dlx1&2 and Mash1 transcription factors control striatal patterning and differentiation through parallel and overlapping pathways. J Comp Neurol. 2009;512:556–572. doi: 10.1002/cne.21854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Urban N. Martin-Ibanez R. Herranz C. Esgleas M. Crespo E. Pardo M. Crespo-Enriquez I. Mendez-Gomez HR. Waclaw R, et al. Nolz1 promotes striatal neurogenesis through the regulation of retinoic acid signaling. Neural Dev. 2010;5:21. doi: 10.1186/1749-8104-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naldini L. Blomer U. Gage FH. Trono D. Verma IM. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci U S A. 1996;93:11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stühmer T. Anderson SA. Ekker M. Rubenstein JL. Ectopic expression of the Dlx genes induces glutamic acid decarboxylase and Dlx expression. Development. 2002;129:245–252. doi: 10.1242/dev.129.1.245. [DOI] [PubMed] [Google Scholar]
- 44.Martin-Ibanez R. Urban N. Sergent-Tanguy S. Pineda JR. Garrido-Clua N. Alberch J. Canals JM. Interplay of leukemia inhibitory factor and retinoic acid on neural differentiation of mouse embryonic stem cells. J Neurosci Res. 2007;85:2686–2701. doi: 10.1002/jnr.21228. [DOI] [PubMed] [Google Scholar]
- 45.Gerfen CR. The neostriatal mosaic: multiple levels of compartmental organization. Trends Neurosci. 1992;15:133–139. doi: 10.1016/0166-2236(92)90355-c. [DOI] [PubMed] [Google Scholar]
- 46.Tamura S. Morikawa Y. Iwanishi H. Hisaoka T. Senba E. Foxp1 gene expression in projection neurons of the mouse striatum. Neuroscience. 2004;124:261–267. doi: 10.1016/j.neuroscience.2003.11.036. [DOI] [PubMed] [Google Scholar]
- 47.Garel S. Marin F. Grosschedl R. Charnay P. Ebf1 controls early cell differentiation in the embryonic striatum. Development. 1999;126:5285–5294. doi: 10.1242/dev.126.23.5285. [DOI] [PubMed] [Google Scholar]
- 48.Kelley CM. Ikeda T. Koipally J. Avitahl N. Wu L. Georgopoulos K. Morgan BA. Helios, a novel dimerization partner of Ikaros expressed in the earliest hematopoietic progenitors. Curr Biol. 1998;8:508–515. doi: 10.1016/s0960-9822(98)70202-7. [DOI] [PubMed] [Google Scholar]
- 49.Song DD. Harlan RE. Ontogeny of the proenkephalin system in the rat corpus striatum: its relationship to dopaminergic innervation and transient compartmental expression. Neuroscience. 1993;52:883–909. doi: 10.1016/0306-4522(93)90536-o. [DOI] [PubMed] [Google Scholar]
- 50.Takahashi K. Liu FC. Hirokawa K. Takahashi H. Expression of Foxp2, a gene involved in speech and language, in the developing and adult striatum. J Neurosci Res. 2003;73:61–72. doi: 10.1002/jnr.10638. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi K. Liu FC. Oishi T. Mori T. Higo N. Hayashi M. Hirokawa K. Takahashi H. Expression of FOXP2 in the developing monkey forebrain: comparison with the expression of the genes FOXP1, PBX3, and MEIS2. J Comp Neurol. 2008;509:180–189. doi: 10.1002/cne.21740. [DOI] [PubMed] [Google Scholar]
- 52.Ouimet CC. Miller PE. Hemmings HC., Jr. Walaas SI. Greengard P. DARPP-32, a dopamine- and adenosine 3':5'-monophosphate-regulated phosphoprotein enriched in dopamine-innervated brain regions. III. Immunocytochemical localization. J Neurosci. 1984;4:111–124. doi: 10.1523/JNEUROSCI.04-01-00111.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Agnati LF. Fuxe K. Zoli M. Ferraguti F. Benfenati F. Ouimet CC. Walaas SI. Hemmings HC., Jr. Goldstein M. Greengard P. Morphometrical evidence for a complex organization of tyrosine hydroxylase-, enkephalin- and DARPP-32-like immunoreactive patches and their codistribution at three rostrocaudal levels in the rat neostriatum. Neuroscience. 1988;27:785–797. doi: 10.1016/0306-4522(88)90183-2. [DOI] [PubMed] [Google Scholar]
- 54.Toresson H. Campbell K. A role for Gsh1 in the developing striatum and olfactory bulb of Gsh2 mutant mice. Development. 2001;128:4769–4780. doi: 10.1242/dev.128.23.4769. [DOI] [PubMed] [Google Scholar]
- 55.Yun K. Garel S. Fischman S. Rubenstein JL. Patterning of the lateral ganglionic eminence by the Gsh1 and Gsh2 homeobox genes regulates striatal and olfactory bulb histogenesis and the growth of axons through the basal ganglia. J Comp Neurol. 2003;461:151–165. doi: 10.1002/cne.10685. [DOI] [PubMed] [Google Scholar]
- 56.Flames N. Pla R. Gelman DM. Rubenstein JL. Puelles L. Marin O. Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J Neurosci. 2007;27:9682–9695. doi: 10.1523/JNEUROSCI.2750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Long JE. Garel S. varez-Dolado M. Yoshikawa K. Osumi N. varez-Buylla A. Rubenstein JL. Dlx-dependent and -independent regulation of olfactory bulb interneuron differentiation. J Neurosci. 2007;27:3230–3243. doi: 10.1523/JNEUROSCI.5265-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Waclaw RR. Allen ZJ. Bell SM. Erdelyi F. Szabo G. Potter SS. Campbell K. The zinc finger transcription factor Sp8 regulates the generation and diversity of olfactory bulb interneurons. Neuron. 2006;49:503–516. doi: 10.1016/j.neuron.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 59.Halpern M. Martinez-Marcos A. Structure and function of the vomeronasal system: an update. Prog Neurobiol. 2003;70:245–318. doi: 10.1016/s0301-0082(03)00103-5. [DOI] [PubMed] [Google Scholar]
- 60.Brennan PA. Zufall F. Pheromonal communication in vertebrates. Nature. 2006;444:308–315. doi: 10.1038/nature05404. [DOI] [PubMed] [Google Scholar]
- 61.Dulac C. Neuroscience. Charting olfactory maps. Science. 2006;314:606–607. doi: 10.1126/science.1135139. [DOI] [PubMed] [Google Scholar]
- 62.Leid M. Ishmael JE. Avram D. Shepherd D. Fraulob V. Dolle P. CTIP1 and CTIP2 are differentially expressed during mouse embryogenesis. Gene Expr Patterns. 2004;4:733–739. doi: 10.1016/j.modgep.2004.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Enomoto T. Ohmoto M. Iwata T. Uno A. Saitou M. Yamaguchi T. Kominami R. Matsumoto I. Hirota J. Bcl11b/Ctip2 controls the differentiation of vomeronasal sensory neurons in mice. J Neurosci. 2011;31:10159–10173. doi: 10.1523/JNEUROSCI.1245-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Corbin JG. Gaiano N. Machold RP. Langston A. Fishell G. The Gsh2 homeodomain gene controls multiple aspects of telencephalic development. Development. 2000;127:5007–5020. doi: 10.1242/dev.127.23.5007. [DOI] [PubMed] [Google Scholar]
- 65.Toresson H. Potter SS. Campbell K. Genetic control of dorsal-ventral identity in the telencephalon: opposing roles for Pax6 and Gsh2. Development. 2000;127:4361–4371. doi: 10.1242/dev.127.20.4361. [DOI] [PubMed] [Google Scholar]
- 66.Yun K. Potter S. Rubenstein JL. Gsh2 and Pax6 play complementary roles in dorsoventral patterning of the mammalian telencephalon. Development. 2001;128:193–205. doi: 10.1242/dev.128.2.193. [DOI] [PubMed] [Google Scholar]
- 67.Anderson SA. Eisenstat DD. Shi L. Rubenstein JL. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- 68.Nery S. Corbin JG. Fishell G. Dlx2 progenitor migration in wild type and Nkx2.1 mutant telencephalon. Cereb Cortex. 2003;13:895–903. doi: 10.1093/cercor/13.9.895. [DOI] [PubMed] [Google Scholar]
- 69.Wang B. Lufkin T. Rubenstein JL. Dlx6 regulates molecular properties of the striatum and central nucleus of the amygdala. J Comp Neurol. 2011;519:2320–2334. doi: 10.1002/cne.22618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brazel CY. Romanko MJ. Rothstein RP. Levison SW. Roles of the mammalian subventricular zone in brain development. Prog Neurobiol. 2003;69:49–69. doi: 10.1016/s0301-0082(03)00002-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.