Abstract
Phlebotomine sand flies are vectors of Leishmania that are acquired by the female sand fly during blood feeding on an infected mammal. Leishmania parasites develop exclusively in the gut lumen during their residence in the insect before transmission to a suitable host during the next blood feed. Female phlebotomine sand flies are blood feeding insects but their life style of visiting plants as well as animals, and the propensity for larvae to feed on detritus including animal faeces means that the insect host and parasite are exposed to a range of microorganisms. Thus, the sand fly microbiota may interact with the developing Leishmania population in the gut. The aim of the study was to investigate and identify the bacterial diversity associated with wild adult female Lutzomyia sand flies from different geographical locations in the New World. The bacterial phylotypes recovered from 16S rRNA gene clone libraries obtained from wild caught adult female Lutzomyia sand flies were estimated from direct band sequencing after denaturing gradient gel electrophoresis of bacterial 16 rRNA gene fragments. These results confirm that the Lutzomyia sand flies contain a limited array of bacterial phylotypes across several divisions. Several potential plant-related bacterial sequences were detected including Erwinia sp. and putative Ralstonia sp. from two sand fly species sampled from 3 geographically separated regions in Brazil. Identification of putative human pathogens also demonstrated the potential for sand flies to act as vectors of bacterial pathogens of medical importance in addition to their role in Leishmania transmission.
Introduction
Phlebotomine sand flies are responsible for the spread of the medically important Leishmania parasites that populate the female sand fly gut. In Brazil Lutzomyia longipalpis is the main vector for Leishmania infantum, the causative agent of severe visceral leishmaniasis [1]. Visceral Leishmaniasis (VL) in Corumbá, central Brazil [2] is transmitted by Lu. cruzi. The macro-ecology of the infection cycle for Leishmania infantum is complex; in this case Lu. longipalpis is the vector, domestic dogs are the main reservoir hosts in urban environments with humans as the accidental hosts [3]. Very little is known about the microbial ecology of Lutzomyia spp. but they are exposed to an unusually diverse range of microbial communities. Although commonly termed ‘blood feeding insects’, adult sand flies are also plant-feeders; only the female requires occasional blood as a protein source for egg production. Both males and females visit plants to acquire carbohydrates where they will also acquire plant phyllosphere microbiota [4] which may be ingested directly into the gut after piercing leaves and stems [5]. The juvenile larval stages feed on animal faeces and plant material that are undergoing microbial biodegradation. Many of the microorganisms ingested by the larvae will be killed during the pupal stage (when the larval gut is reabsorbed) but some may survive pupation to re-colonise the adult gut [6].
Bacteria in other medically important insect vectors may confer a degree of colonisation resistance upon the host insect, reducing parasite populations within the insect’s gut and interfering with disease transmission [7]–[11]. While Plasmodium sp. cross through the gut barrier the Leishmania parasite is confined to the gut lumen of the female sand fly. The acquired gut bacteria are therefore potentially more important in sand flies because some bacterial species might compete with Leishmania.
In communities where there is an epidemic of leishmaniasis the prevalence of Leishmania in sand flies seldom exceeds 2% of the female sand fly population [12]–[14]. There are many macro-ecological factors governing the prevalence of sand flies with transmissible infections including the number of mammals with infective leishmaniasis. The presence of a robust resident microbiota within the female sand flies may also impede development of a transmissible parasite population. This would therefore need to be considered when predicting the effect of ecological factors on Leishmania transmission. The microbial ecology of phlebotomine sand flies has been the subject of only a few studies [6], [9], [15]–[18] and these have mainly focused on culturable bacteria. One study of cultivatable bacteria associated with the gut of Lutzomyia longipalpis [17] included a pre-enrichment step and the identification of isolates by 16S rDNA sequence analysis. The range of species included members of Enterobacteriaceae, such as Citrobacter, Enterobacter, Serratia, Pantoea, Morganella and other genera such as Acinetobacter, Burkholderia, Flavimonas, Pseudomonas and Stenotrophomonas. A bacterial cultivation approach was used to identify bacteria from Phlebotomus argentipes in India with potential for paratransgenetic strategy [18]. A comprehensive metagenomic approach done by pyrosequencing analysis from field-caught male and female L. longipalpis identified several bacteria from environmental sources, as well as fungi and protists [19]. Characterization based on a non-cultivatable approach is a pre-requisite for understanding the functional role of the microbiota of phlebotomine sand flies in survival and Leishmania transmission. The aim of the study was to examine the bacterial biota and potential endosymbionts associated with female Lu. longipalpis and Lu. cruzi captured in 4 distantly located regions of Brazil and Colombia.
Results
Analysis of the 16S Gene Clone Library from Field-collected Lu. longipalpis and Lu. cruzi
Characterization of bacteria associated with Lutzomyia by sequencing a portion of the bacterial 16S rRNA gene from clone libraries led to the identification of 38 bacterial sequences through BLAST searches, with 19 distinct bacterial phylotypes (Table 1). The relative abundance of different phylotypes in the clone libraries was estimated by band pattern comparison of PCR-amplified bacterial 16S rRNA gene fragments derived from single clones run on DGGE, giving a measure of relative abundance for each bacterial species within the gut of the female Lutzomyia sand fly (Table 2). Between 26 to 28 clones were analysed from bacterial 16S rRNA clone libraries obtained from 4 pools of field-collected females of Lu. longipalpis and Lu. cruzi. These wild-caught sand flies were captured from four distantly located regions in Brazil and Colombia. On the basis of sequence similarity to existing Genbank database sequences, the majority of the bacterial sequences obtained from Lu longipalpis collected in Lapinha belonged to the Betaproteobacteria class. Ralstonia spp. (plant associated species) were the most abundant bacterial phylotype, present in 65.5% of the randomly picked clones (Table 2). The second most frequent bacteria also belong to Betaproteobacteria (Leptothrix sp. −17.2%), followed by the Alphaproteobacteria Bradyrhizobium japonicum (10.3%), with the Gram positive Firmicutes Clostridium disporicum and the Alphaproteobacteria Caulobacter sp. being the least abundant in the Lapinha’s field samples (3.4%).
Table 1. Bacterial phylotypes associated with Lutzomyia sand flies sampled from 4 field sites.
| Clone number | Length (bp) | Bacterial Division | Database match (accession no.) | Score Similarity (%) |
| Lu. longipalpis , Lapinha | ||||
| 1–1 | 750 | Betaproteobacteria | Ralstonia sp. (AY216797) | 1455 (99) |
| 1–3 | 781 | Betaproteobacteria | Leptothrix sp. (AF385528) | 1501 (99) |
| 1–4 | 557 | Firmicutes | Clostridium disporicum (Y18176) | 985 (98) |
| 1–6 | 851 | Betaproteobacteria | Ralstonia pickettii (AY268180.1) | 1631 (99) |
| 1–9 | 914 | Betaproteobacteria | Ralstonia sp. (AY216797) | 1741 (99) |
| 1–11 | 731 | Alphaproteobacteria | Caulobacter sp. (AY807064.1) | 1419 (99) |
| 1–13 | 620 | Alphaproteobacteria | Bradyrhizobium japonicum (AB195988) | 1229 (100) |
| 1–15 | 913 | Betaproteobacteria | Ralstonia sp. (AY216797) | 1717 (99) |
| 1–20 | 918 | Betaproteobacteria | Ralstonia sp. (AY191853) | 1764 (99) |
| 1–23 | 861 | Alphaproteobacteria | Bradyrhizobium japonicum (AF208517) | 1657 (99) |
| 1–26 | 913 | Betaproteobacteria | Ralstonia sp. (AY216797) | 1746 (99) |
| Lu. longipalpis , Alagoas | ||||
| 2–7 | 898 | Actinobacteridae | Propionibacterium acnes (AY642051) | 1764 (99) |
| 2–8 | 918 | Gammaproteobacteria | Erwinia billingiae (AM055711) | 1663 (98) |
| 2–11 | 913 | Betaproteobacteria | Ralstonia sp. (AY216797) | 1762 (100) |
| 2–14 | 914 | Gammaproteobacteria | Enterobacter sp. (NJ-1 AM396909) | 1616 (98) |
| 2–17 | 918 | Gammaproteobacteria | Pantoea sp. (NJ-87 AM419023) | 1618 (98) |
| 2–18 | 892 | Actinobacteridae | Nocardioides albus (AF004997) | 1703 (99) |
| 2–19 | 914 | Betaproteobacteria | Ralstonia sp. (AY216797) | 1748 (99) |
| 2–23 | 850 | Gammaproteobacteria | Serratia sp. (AY827577) | 1655 (99) |
| 2–24 | 909 | Gammaproteobacteria | Klebsiella pneumoniae (AY043391) | 1618 (98) |
| 2–25 | 913 | Gammaproteobacteria | Acinetobacter sp. (DQ227342) | 1784 (99) |
| 2–27 | 916 | Bacteroidetes | Sphingobacterium daejeonense (AB249372) | 1536 (96) |
| 2–30 | 916 | Gammaproteobacteria | Klebsiella pneumoniae (AY043391) | 1659 (97) |
| Lu. cruzi , Corumbá | ||||
| 3–2 | 858 | Alphaproteobacteria | Caulobacter sp. (AY807064) | 1427 (99) |
| 3–4 | 844 | Firmicutes | Clostridium glycolicum (AY007244) | 1481 (97) |
| 3–5 | 925 | Firmicutes; Bacillales | Staphylococcus xylosus (D83374) | 1731 (99) |
| 3–15 | 917 | Betaproteobacteria | Ralstonia sp. (AY216797) | 1756 (99) |
| 3–17 | 945 | Firmicutes | Lactobacillus zymae (AJ632157) | 1699 (98) |
| 3–19 | 894 | Firmicutes | Clostridium disporicum (Y18176) | 1667 (99) |
| 3–22 | 913 | Betaproteobacteria | Ralstonia sp. (AY216797) | 1754 (99) |
| 3–24 | 906 | Bacteroidetes Flavobacteria | Chryseobacterium meningosepticum (AY468445) | 1360 (94) |
| 3–27 | 402 | Alphaproteobacteria | Bradyrhizobium japonicum (AB231926) | 733 (98) |
| 3–30 | 907 | Bacteroidetes Flavobacteria | Chryseobacterium meningosepticum (AY468445) | 1340 (93) |
| Lu. longipalpis , El Callejón | ||||
| 4–1 | 906 | Alphaproteobacteria | Saccharibacter floricola (AB110421) | 1562 (97) |
| 4–7 | 919 | Gammaproteobacteria | Stenotrophomonas maltophilia (AJ293470) | 1570 (99) |
| 4–13 | 907 | Actinobacteria | Propionibacterium acnes (AB041617) | 1756 (99) |
| 4–18 | 907 | Alphaproteobacteria | Saccharibacter floricola (AB110421) | 1566 (98) |
| 4–22 | 898 | Bacteroidetes | Dyadobacter ginsengisoli (AB245369) | 1532 (97) |
Insects collected in field locations were suspended in 70% ethanol. DNA extracts from pooled samples of insects were subjected to PCR using primers specific for a 900 bp region of the 16S rRNA gene. The PCR products were cloned and colonies randomly selected. Purified plasmids from the clones were subjected to specific PCR, the products separated with DGGE and representative clones were selected on the basis of the band pattern of their DGGE profiles.
Table 2. Abundance of bacterial phylotypes recovered from 16S rRNA gene clone libraries constructed from adult Lutzomyia sand flies.
| Bacterial Division | Database Match | Abundance (%) |
| Lu. longipalpis , Lapinha n = 29 | ||
| Betaproteobacteria | Ralstonia sp. AY216797 | 65.5 |
| Betaproteobacteria | Leptothrix sp. | 17.2 |
| Alphaproteobacteria | Bradyrhizobium japonicum | 10.3 |
| Firmicutes | Clostridium disporicum | 3.4 |
| Alphaproteobacteria | Caulobacter sp. | 3.4 |
| Lu. longipalpis , Alagoas n = 28 | ||
| Betaproteobacteria | Ralstonia sp. AY216797 | 50.0 |
| Gammaproteobacteria | Erwinia billingiae AM055711 | 17.8 |
| Gammaproteobacteria | Klebsiella pneumoniae AY043391 | 14.2 |
| Actinobacteridae | Nocardioides albus AF004997 | 3.6 |
| Gammaproteobacteria | Serratia sp. AY827577 | 3.6 |
| Gammaproteobacteria | Acinetobacter sp. DQ227342 | 3.6 |
| Bacteroidetes | Sphingobacterium daejeonense | 3.6 |
| Actinobacteridae | Propionibacterium acnes AY642051 | 3.6 |
| Lu. cruzi , Corumbá n = 26 | ||
| Betaproteobacteria | Ralstonia sp. AY216797 | 40.7 |
| Firmicutes; Bacillales | Staphylococcus xylosus D83374 | 25.9 |
| Bacteroidetes | Chryseobacterium meningosepticum | 11.1 |
| Alphaproteobacteria | Caulobacter sp. AY807064 | 7.4 |
| Firmicutes | Clostridium sp. | 7.4 |
| Firmicutes | Lactobacillus zymae AJ632157 | 3.7 |
| Alphaproteobacteria | Bradyrhizobium japonicum AB231926 | 3.7 |
| Lu. longipalpis , El Callejón n = 28 | ||
| Alphaproteobacteria | Saccharibacter floricola AB110421 | 89.2 |
| Gammaproteobacteria | Stenotrophomonas maltophilia AJ293470 | 4.0 |
| Actinobacteridae | Propionibacterium acnes AB041617 | 4.0 |
| Bacteroidetes Sphingobacteria | Dyadobacter ginsengisoli AB245369 | 4.0 |
Between 26 and 28 clones were randomly selected each of the 4 pools of samples. The PCR products from GC clamped primers [44] were run on DGGE and clones were selected for sequencing on the basis of the gel profile. Gel profiles were grouped according to identical band distribution.
The 16S rRNA gene sequencing data derived from Lu. longipalpis collected in Alagoas state (Brazil) showed that the majority of bacterial species identified belonged to Gammaproteobacteria (Table 1). However, BLAST searches confirmed that clones from this library were also dominated by sequences related to the genus Ralstonia (50%). The most abundant Gammaproteobacteria were Erwinia billingiae (another well known plant pathogen) (17.8%) and Klebsiella pneumoniae (14.2%). Notocardioides albus, Serratia sp., Acinetobacter sp., Sphingobacterium daejeonense and Propionibacterium acnes were identified in 3.6% of the bacterial clones.
The 16S rRNA sequence analysis of Lu. cruzi from Corumbá (Mato Grosso – Brazil) identified 8 bacterial genera across 5 different divisions. Again, Ralstonia spp. was the most abundant phylotype (40.7%), with the Gram-positive Staphyloccoccus xylosus and Cryseobacterium meningosepticum being identified in 25.9 and 11.1% of the clones, respectively. The Caulobacter sp. (Alphaproteobacteria) and the Gram-positive Clostridium sp. (Firmicutes) were found in 7.4% of the individual clones analysed and the Gram-positive bacteria Lactobacillus zymae and the Bradyrhizobium japonicum (Alphaproteobacteria) were detected in 3.7% of the bacterial clones screened.
The vast majority of the bacterial phylotypes identified from Lu. longipalpis collected from cattle in Callejón (Colombia) belonged to the Alphaproteobacteria class: Saccharibacter floricola was present in 89.2% of the clones. Stenotrophomonas floricola (Gammaproteobacteria), Propionibacterium acnes (Actinobacteria) and Dyadobacter ginsengisoli (Bacterioidetes) were present in 4% of the bacterial samples analysed.
Neighbour-joining Phylogram of Bacterial 16S Sequences Obtained from Lu. longipalpis Collected from the Field
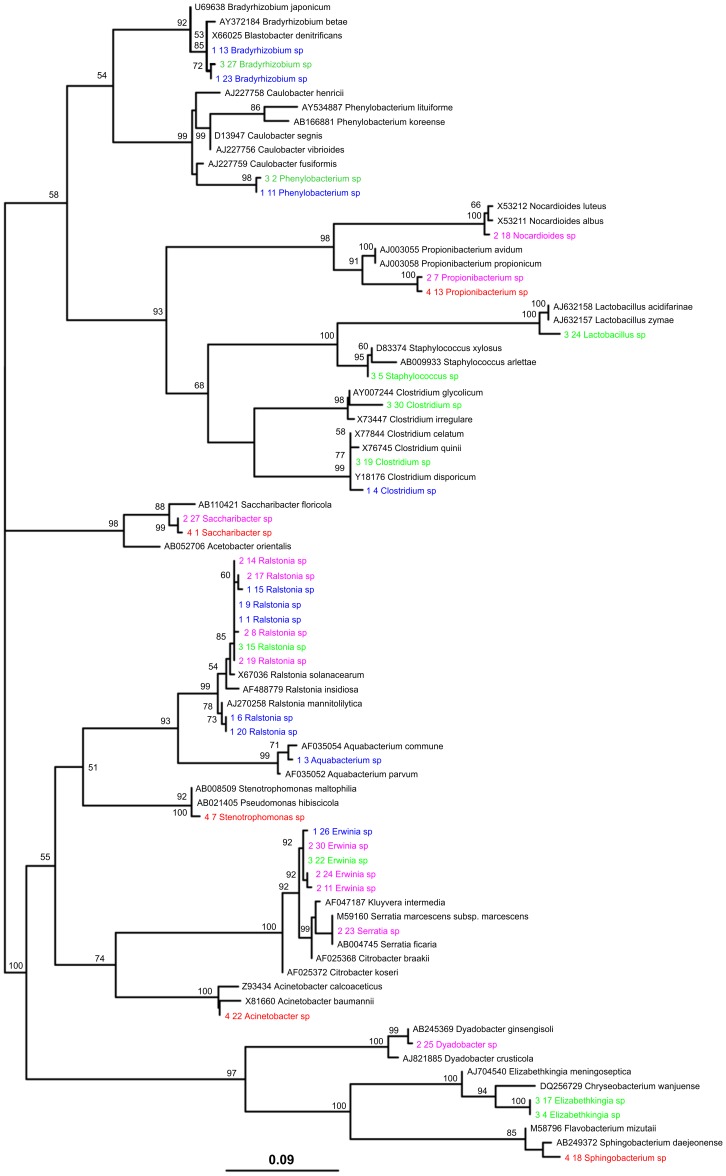
The 16S gene sequences generated from the clone libraries from Lu. longipalpis collected from the field were used to construct a neighbour-joining tree. The topology of the ML and NJ trees (Fig. 1) were identical and showed good correspondence with the RDP classifier results. The phylogenetic analysis showed robust clustering of bacterial species isolated from the different sand fly species and different geographic locations. This is particularly true for the Ralstonia and Erwinia isolates that showed both robust clades and a high degree of similarity (70% bootstrap support) despite the fact that they were from different sand fly species and from different geographical locations.
Figure 1. Neighbour-joining phylogram of bacterial 16S sequences obtained from field collected Lutzomyia species.
Numbering refers to clones described in Table 1. Bacterial species from Lapinha (Minas Gerais-Brazil) are presented in purple; species identified from sand flies collected in Alagoas (Brazil) are presented in green; species identified in Corumbá (Brazil) are presented in pink and red represents bacterial species identified from Lu. longipalpis collected in Callejón (Colombia). Bacterial sequences extracted from Genbank for comparison are presented in black.
DGGE Profiles of Field Caught Lu. longipalpis and Lu. cruzi
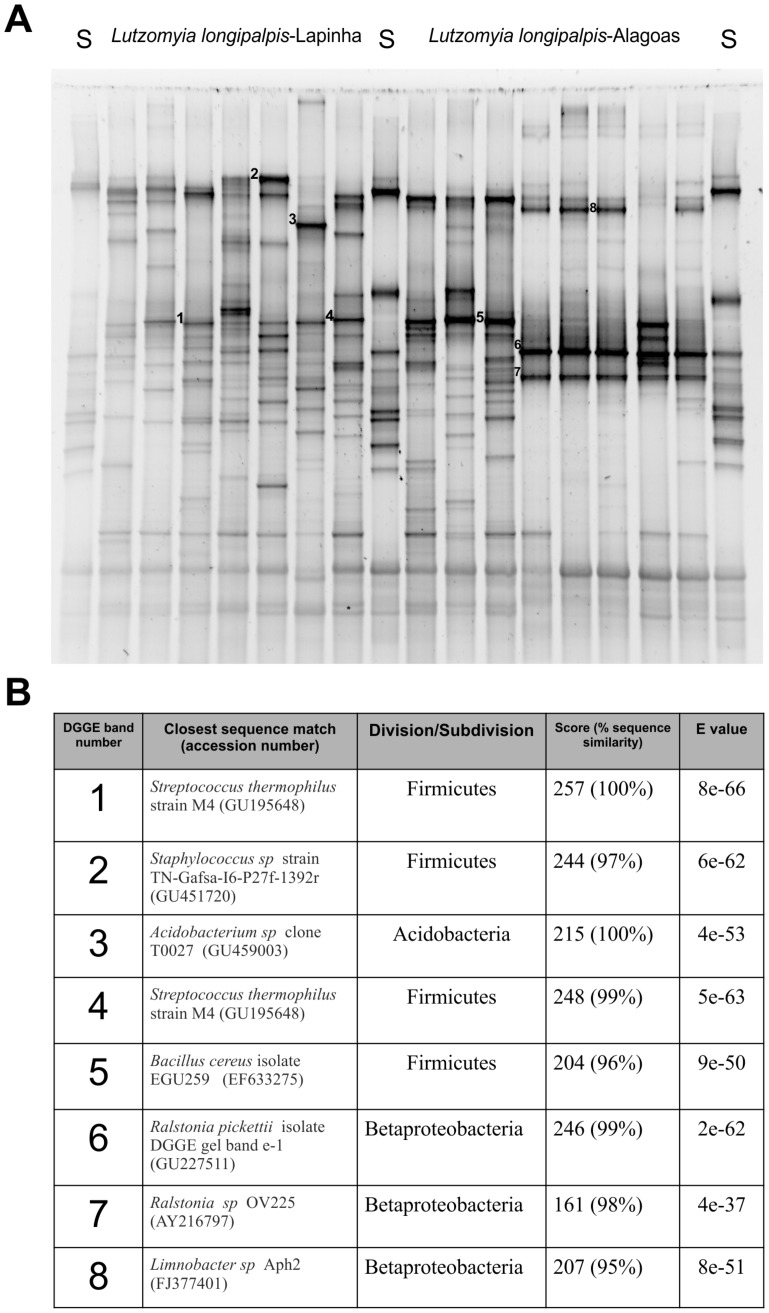
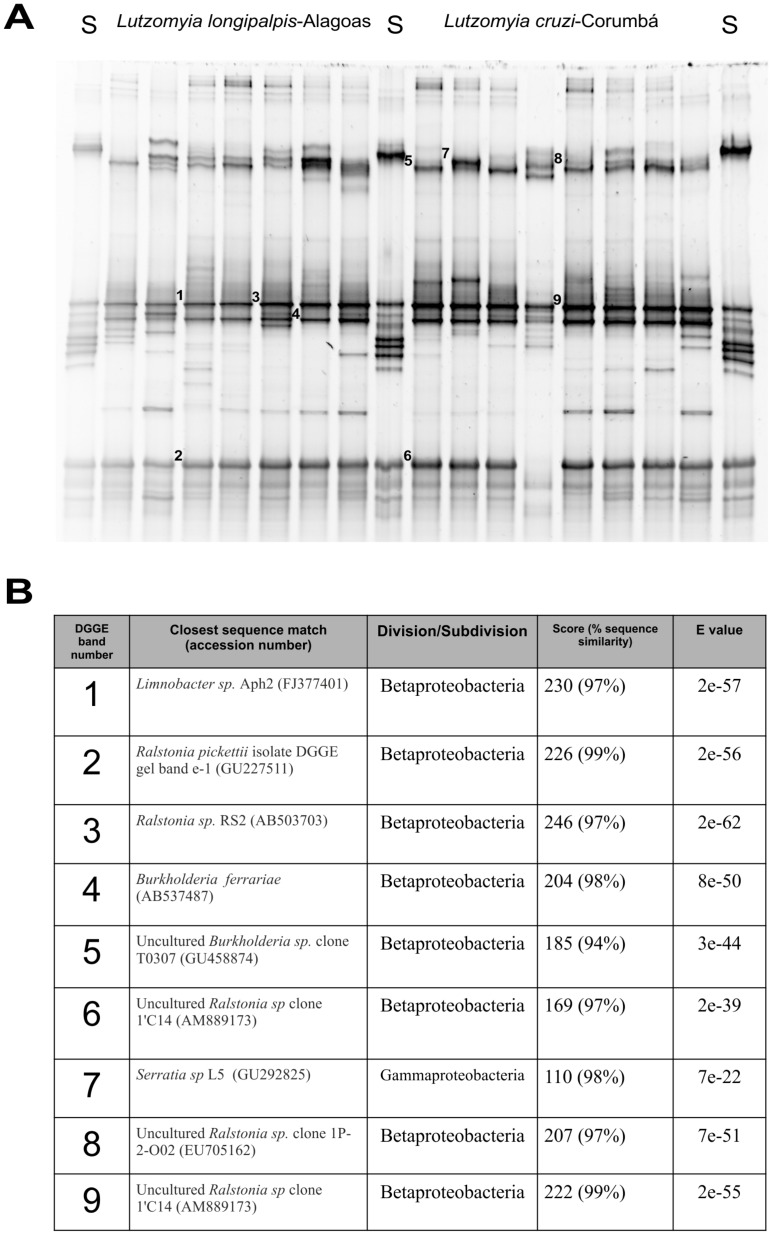
Bacterial DGGE profiles were generated using PCR-amplified bacterial 16S rRNA gene fragments from individual female Lu. longipalpis collected in Lapinha cave and Alagoas and Lu. cruzi collected in Corumbá, Mato Grosso do Sul (Brazil). DGGE band profiles indicated the presence of several putative bacterial phylotypes in Lu. longipalpis from different geographic populations and Lu. cruzi from Corumbá (Fig. 2 and 3). Random sequencing of DGGE bands identified bacterial species that were not previously detected in our clone libraries for Lu. longipalpis from Lapinha cave and Alagoas. DGGE band sequencing also confirmed the presence of Ralstonia in individual Lu. longipalpis from Alagoas and Lu. cruzi from Corumbá (Brazil). It is possible that some bands at identical positions may not be homologous and the diversity of phylotypes may be higher than the profile suggests.
Figure 2. Denaturing gradient gel electrophoresis (DGGE) profiles of PCR-amplified bacterial gene fragments derived from field-collected individual female Lu. longipalpis.
Lanes L1 to L10 corresponds to DGGE profiles of Lu. longipalpis collected from Lapinha cave-Minas Gerais; A1 to A5 correspond to DGGE profiles of Lu. longipalpis collected from Alagoas state/Brazil. Lanes labelled S correspond to standard DGGE markers prepared from a selection of bacterial 16S rRNA gene products to enable gel to gel comparison.
Figure 3. Denaturing gradient gel electrophoresis (DGGE) profiles of PCR-amplified bacterial gene fragments derived from field-collected individual female Lu. longipalpis.
Lanes A6 to A12 corresponds to DGGE profiles of Lu. longipalpis collected from Alagoas state/Brazil; C1 to C8 correspond to DGGE profiles of Lu. cruzi collected from Corumbá-Mato Grosso-Brazil. Lanes labelled S corresponds to DGGE markers prepared from a selection of bacterial 16S rRNA gene products to enable gel-to-gel comparison.
Discussion
The present study used DGGE and 16S rRNA sequence analysis to identify a relatively low range of bacterial phylotypes across several divisions within female adult phlebotomine sand flies. Other insects such as cockroaches, termites and crickets have a much more complex microbiota reflecting their greater contribution to digestion [20]. Studies using classical microbiological techniques demonstrated that bacteria can be frequently isolated from the gut of female Phlebotomus [9] and Lutzomyia [15]–[17]. The approach of cultivating the bacteria prior to identification, however, may have introduced an unknown bias into the sampling as the uncultivated bacteria will not have been considered. Dillon et al. [9] isolated bacteria from the midgut of field-collected P. papatasi caught in Egypt and found that in the sand flies with blood in their midguts, 59% contained bacteria and some of them contained potential human pathogens. Hillesland et al. [18] identified 28 distinct gut bacteria from Phlebotomus argentipes, with Staphyloccoccus and members of the family Enterobacteriaceae being isolated in the greatest abundance. In Lu. longipalpis, a previous study using 3 Brazilian populations identified Serratia marcescens, Pseudomonas aeruginosa and Pantoea agglomerans as the most frequent bacteria associated with all field populations [17]. The present study was an attempt to investigate the microbiota associated with sand flies of the genus Lutzomyia using a molecular approach to identification. A metagenomic analysis of all taxa associated with adult Lutzomyia longipalpis from Brazil identified only ten different bacterial types including sequences for Ralstonia pickettii, Anoxybacillus flavithermus, Geobacillus kaustophilus, Streptomyces coelicolor, Propionibacterium acnes, Acinetobacter baumannii and Veillonella sp. [19].
The phylotypes in this study were similar to those found in an apparently disparate range of insect species representing different orders of insects. This includes Orthoptera (locusts [21]), Lepidoptera (cabbage white butterfly [22]), Diptera (mosquitoes [23] and tephritid fruit flies [24]). We suggest that they represent insects with low diversity, facultative microbiota that are drawn from the reservoir of phylotypes in the local environment of the insect. It should be noted that the identifications are tentative and some clones may represent novel phylotypes. The relative abundances of different phylotypes in the clone libraries give a measure of relative abundance in the female Lutzomyia sand fly. However, larger sample numbers may provide a more accurate comparison of relative abundance.
A notable observation was the apparent abundance of putative Ralstonia spp. from 2 sand fly species sampled from 3 geographically separated regions. The association of Ralstonia sp. with Lu. longipalpis was also found in a previous metagenomic analysis of all taxa associated with this sand fly species from one of the same locations, Lapinha, in Brazil [19]. The most common species is Ralstonia solanacearum, the causal agent of bacterial wilt in solanaceous crops. R. solanacearum is a soil-borne bacterium originating from the tropics, subtropics, and warm temperate regions [25]. Another potential plant pathogen genus Erwinia was also noted. In addition to their plant visiting habits [26]–[28], the female sand flies deposit eggs in terrestrial habitats and the larvae feed on either decaying vegetation or animal faeces. Therefore, there is ample opportunity for these insects to come into contact with plant pathogens. The discovery of Ralstonia sp. in our sand fly samples raises the question whether Phlebotomine sand flies may act as vectors of plant pathogens (see also [19]). Future experiments will establish whether sand flies can harbour or mechanically transmit pathogens from a diseased plant to a healthy individual.
The identification of the bacterial sequences from female sand flies with similarity to Bradyrhizobium sp. (a bacteria found intracellularly in root nodules) is notable, although a number of nitrogen fixing bacteria have been associated with insects and blood feeding invertebrates such as leeches where a new lineage of α-proteobacterial endosymbionts, related to Rhizobiaceae, have been identified [29]. Is there a role for nitrogen fixing bacteria in sand flies that are unable to find a bloodmeal? There are large populations of diazotrophic enterobacteria that express dinitrogen reductase within the gut of fruit flies (Ceratitis capitata) and nitrogen fixation may significantly contribute to the fruit fly nitrogen intake [24], [30]. When a blood source is scarce, the presence of nitrogen fixing bacteria may also be beneficial for the female sand fly. Chryseobacterium meningosepticum is an emerging pathogen, an agent of neonatal meningitis and is involved to a lesser extent in cases of pneumonia and bacterial sepsis in neonates and adults [31], [32]. The preliminary identification of this bacterial species in Lu. cruzi from Corumbá (Brazil) highlights the potential for identifying sand flies as vectors of bacterial pathogens of medical importance. Other potential human and plant pathogens identified in our field samples are bacterial clones with sequence similarity to genus Burkholderia (soil-borne, Gram-negative, motile, obligately aerobic, rod-shaped bacteria). Burkholderia has been previously identified from the digestive tract microbiota in female Lutzomyia longipalpis [17]. Burkholderia mallei is the causative agent of the predominately equine disease glanders and is very closely related to the much more diverse species Burkholderia pseudomallei, an opportunistic human pathogen and the primary cause of melioidosis [33].
The nested PCR screening of the sand fly microbiota did not result in identification of endosymbionts Wolbachia or Cardinium despite successful positive results from control insect material known to contain these species (results not shown). The absence of Wolbachia endosymbionts in most Lutzomyia species [34] including our own colony raises the question as to whether there are other, as yet, undescribed endosymbionts in these insects. One study [6] has shown that Ochrobactrum intermedium survives pupation in Lu. longipalpis to appear in the adult gut. This species is also present throughout our Lu. longipalpis 25 year old colony originally from Jacobina (Bahia-Brazil). Some of the above mentioned phylotypes could represent endosymbionts. The negative result of Wolbachia specific PCR probably means that Wolbachia does not naturally infect Lutzomyia and this is the first time that the sand fly genus has been tested for Cardinium. The lack of these reproductive symbionts in the system may be valuable for the design and deployment of symbiont control strategies. It is also interesting as the lack of these bacteria may suggest either a barrier to their invasion of sand flies or a mismatch in symbiont/host biology.
The molecular microbial ecology approach has resulted in an upsurge in understanding about the relationships of insects with microorganisms; the importance of gut microbes is gaining increased recognition [20], [35]–[38]. Of particular relevance is the hypothesis that resident bacteria of insects may prevent the development of insect pathogens in the gut either directly [7] or indirectly by regulating the host immune response [11]. Adler and Theodor [39] were the first to suggest that the presence of commensal microorganisms might impair Leishmania development in the sand fly midgut. Schlein et al. [40] observed yeast-like fungi in lab reared and field caught Phlebotomus papatasi and P. tobbi and suggested that gut contamination might interfere with Leishmania transmission. Only approximately 1.5% of the adult female population contain Leishmania parasites in the gut [14] and not all these insects may contain parasites at the mammalian infective (metacyclic) stage. The reasons for the low number of Leishmania infected sand flies are related to a number of factors; the main factors being the availability of infected mammalian reservoirs and the short life of the adult sand fly. The presence of competing microbes in the sand flies’ gut could contribute and perhaps reduce even further the chances of a sand fly becoming infected with Leishmania.
Materials and Methods
Insect Collection Sites
Wild caught Lu. longipalpis collected in Brazil were obtained from 3 different localities separated from each other by over 2,000 km: sand flies captured in Estrela de Alagoas (a town located in a rural area of Alagoas State-Brazil) were collected using CDC light traps near the livestock with the presence of adjacent fruit trees; Sand flies collected in Lapinha cave (situated in the municipality of Lagoa Santa-Minas Gerais, Brazil) were collected using a CDC light trap placed near a caged chicken; Lutzomyia cruzi were collected in Mato Grosso do Sul State- Corumbá (Brazil) from a rural area next to a pigpen in the backyard of a small house. Lu. longipalpis from Colombia came from the small rural community of El Callejón (municipality of Ricaute) and were collected by aspiration from cattle.
Ethics Statement
No specific permits were required for the insect collections. Specific permission was obtained from the private owners to collect the sand flies from all sites apart from Lapinha where RPB has a long standing agreement to collect sand flies. The field studies did not involve endangered or protected species.
Extraction and Purification of DNA
Insects were surface sterilized by dipping in 70% ethanol for 30 seconds and then washed twice in sterile saline (NaCl 0.15 M) followed by storage in 90% ethanol. Whole insects were suspended in homogenisation buffer with lysing matrix. Homogenisation was done using a bead beater (Fastprep instrument, MP Biomedicals) for 40 sec. DNA extraction was carried out using the FastDNA Spin Kit for soil (MP Biomedicals) according to the manufacturer’s instructions with the exception that material was centrifuged for 8 minutes at 14,000 g following cell lysis before transferring the supernatant to a clean tube with protein precipitation solution [41]. DNA was stored in a 50 µl aliquot of DNase/pyrogen free water at –70°C. In addition, DNA template was also prepared from pure bacterial cultures as described previously [42]. Essentially, 1 ml of log phase culture was centrifuged for 4 min at 14,000x g. Supernatant was discarded and the cell pellet was re-suspended in 100 µl of 5% (w/v) Chelex 100 (Sigma-Aldrich). The suspension was then heated at 100°C for 5 min prior to placing in ice for 5 min followed by a further heating and cooling step. The crude DNA lysate was then centrifuged as above and used directly for PCR amplification.
Lutzomyia Bacterial Clone Library Production and Sequencing
Partial sequences of the 16S rRNA gene were amplified from DNA extracts from pooled samples of insects (n = 20) using primers 27F and 1492R [43]. Four clone libraries (Lapinha, Alagoas, Corumba, El Callejón) were derived from the amplified products using a cloning kit (TOPO TA, Invitrogen, CA, USA). Purified plasmids from randomly selected clones (n = 26–29) were subjected to DGGE (denaturing gradient gel electrophoresis) specific PCR and the products were run on DGGE (data not shown), representative clones were selected for sequencing on the basis of their DGGE profiles and the abundance of the putative phylotypes estimated on the basis of identical band profiles in comparison to the sequenced clones. Bacterial sequences obtained during this study were deposited at EMBL as HE611532-69.
DGGE Analysis of PCR Amplified Products from Individual Females
For the DGGE approach amplification of the 16S rRNA genes of bacteria were performed using PCR-DGGE primers 357FGC-518R [44] Amplifications were carried out with 4 pmol ul−1 primers, 1 µl of DNA template, 1 x reaction buffer (Promega), 1.5 mM MgCl2, 1.5U Taq DNA polymerase (Promega), 0.25 mM each dNTP in a 50 µl PCR reaction mixture with molecular grade water. Positive (pure culture bacterial DNA) and negative controls (water) were routinely included. PCR conditions were 95°C for 5 mins, 10 cycles of 94°C/30 s; 55°C/30 s; 72°C/60 s, 25cycles of 92°C/30 s; 52°C/30 s; 72°C/60 s, followed by 10 min at 72°C. DGGE was conducted according to previously described methods [21], [45]. DGGE marker was prepared from a selection of bacterial 16S rRNA gene products to enable gel to gel comparison. PCR products prepared from single females were separated (ca 200 ng of each product) using a combined polyacrylamide and denaturant gradient between 6% acrylamide/30% denaturant and 12% acrylamide/60% denaturant. A 100% denaturing condition is equivalent to 7 M urea and 40% (v/v) formamide. Gels were poured with the aid of a 50 ml gradient mixer (Fisher Scientific,) and electrophoresis run at 200 V for 5 h at 60°C. Polyacrylamide gels were stained with SYBRGold nucleic acid gel stain (Molecular Probes) for 30 min and viewed under UV.
Bands were excised with sterile razor blades immediately after staining and visualisation of the gels. Gel bands were stored at –70°C, washed with 100 µl distilled water and DNA extracted with 10–20 µl of water depending on band intensity. DNA was re-amplified using the PCR-DGGE primers and products checked by agarose gel electrophoresis. The PCR products were purified using the QIAGEN PCR purification kit (Qiagen Ltd). The products were directly sequenced with the 518R primer. Partial bacterial 16S rRNA gene sequences (approximately 160 bp) were subjected to a NCBI nucleotide blast search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to identify sequences of the highest similarity.
Endosymbiont Analysis
Bacterial 16S rDNA was amplified by PCR. The PCR reagents comprised 2 mM MgCl2, 0.5 mM each dNTP, 0.5 µM each primer, PCR buffer and 1 U Taq polymerase (Promega, Southampton, UK), and the reaction conditions were 24 cycles of 94°C for 1 min, primer pair annealing temperature for 1 min, 72°C for 1 min, but with the extension time increased to 2 min and 8 min for the first and last cycle, respectively. Universal bacterial 16S rDNA primers were used together with primers specific to Wolbachia 16S rRNA gene; primers 99F and 994R [46] and Cardinium with related Bacteroidetes primers, (forward) Ch-F 5′-TACTGTAAGAATAAGCACCGGC-3′ and (reverse) Ch-R 5′-GTGGATCACTT AACGCTTTCG-3′ [47].
Nested PCR reactions used specific primers and 1 µl cleaned PCR product (QIAquick PCR purification kit, Qiagen, Crawley, UK) from a PCR amplification with the universal bacterial primers and total fly DNA template. All PCR reactions included template-free samples as negative control. PCR products were separated by electrophoresis in a 1.6% agarose gel and visualized under UV following staining with ethidium bromide. Sequences were checked for chimeric artefacts using the CHECK_CHIMERA program of Ribosomal Database Project II (RDP II) [48] and compared with similar rDNA sequences in the DNA databases using the BLAST search program of the National Centre for Biotechnology Information (NCBI) and RDP classifier [49].
For the phylogenetic analysis the Muscle alignment program [50] was used to generate multiple sequence alignment (which was adjusted manually). The neighbour-joining (DnaDist-Neighbour) [51] and maximum likelihood (PhyML) [52] analysis, gaps and ambiguous bases were excluded from the analysis using Gblock [53]. The two methods of tree drawing neighbour-joining (NJ) used the Kimura two-parameter model with correction for multiple substitutions and maximum likelihood (ML) using the HKY85 model that included an estimate of 0.49 for the proportion of sites assumed to be invariable and a transition/transversion ratio of 1.7 (estimated from the data). Bootstrap analyses with 100 replications were performed to estimate the reliability of the resulting gene phylogenies.
Acknowledgments
We thank Jan Lewis and Davina Moor for technical assistance.
Funding Statement
The work was supported by the Wellcome Trust http://www.wellcome.ac.uk (ref 073405) and the Leverhulme Trust http://www.leverhulme.ac.uk (ref F/00 808/C). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lainson R, Rangel EF (2005) Lutzomyia longipalpis and the eco-epidemiology of American visceral leishmaniasis, with particular reference to Brazil: a review. Mem Inst Oswaldo Cruz 100: 811–827. [DOI] [PubMed] [Google Scholar]
- 2. de Pita-Pereira D, Cardoso MAB, Alves CR, Brazil RP, Britto C (2008) Detection of natural infection in Lutzomyia cruzi and Lutzomyia forattinii (Diptera : Psychodidae : Phlebotominae) by Leishmania infantum chagasi in an endemic area of visceral leishmaniasis in Brazil using a PCR multiplex assay. Acta Trop 107: 66–69. [DOI] [PubMed] [Google Scholar]
- 3. Quinnell RJ, Courtnay O (2009) Transmission, reservoir hosts and control of zoonotic visceral leishmaniasis. Parasitology 136: 1915–1934. [DOI] [PubMed] [Google Scholar]
- 4. Lindow SE, Brandl MT (2003) Microbiology of the phyllosphere. Appl Environ Microbiol 69: 1875–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schlein Y, Jacobson RL (1999) Sugar meals and longevity of the sandfly Phlebotomus papatasi in an arid focus of Leishmania major in the Jordan Valley. Med Vet Entomol 13: 65–71. [DOI] [PubMed] [Google Scholar]
- 6. Volf P, Kiewegova A, Nemec A (2002) Bacterial colonisation in the gut of Phlebotomus duboscqi (Diptera : Psychodidae): transtadial passage and the role of female diet. Folia Parasit 49: 73–77. [DOI] [PubMed] [Google Scholar]
- 7. Cirimotich CM, Ramirez JL, Dimopoulos G (2011) Native microbiota shape insect vector competence for human pathogens. Cell Host Microbe 10: 307–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pumpuni CB, Beier MS, Nataro JP, Guers LD, Davis JR (1993) Plasmodium falciparum - inhibition of sporogonic development in Anopheles stephensi by Gram negative bacteria. Exp Parasitol 77: 195–199. [DOI] [PubMed] [Google Scholar]
- 9. Dillon RJ, El Kordy E, Shehata M, Lane RP (1996) The prevalence of a microbiota in the digestive tract of Phlebotomus papatasi . Ann of Trop Med and Parasit 90: 669–673. [DOI] [PubMed] [Google Scholar]
- 10. Gonzalez-Ceron L, Santillan F, Rodriguez MH, Mendez D, Hernandez-Avila JE (2003) Bacteria in midguts of field-collected Anopheles albimanus block Plasmodium vivax sporogonic development. J Med Entomol 40: 371–374. [DOI] [PubMed] [Google Scholar]
- 11. Dong Y, Manfredini F, Dimopoulos G (2009) Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog 5: e1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Silva J, Werneck GL, Cruz M, Costa CHN, de Mendoca IL (2007) Natural infection of Lutzomyia longipalpis by Leishmania sp in Teresina, Piaui State, Brazil. Cad Saude Publica 23: 1715–1720. [DOI] [PubMed] [Google Scholar]
- 13. Acardi SA, Liotta DJ, Santini MS, Romagosa CM, Salomón OD (2010) Detection of Leishmania infantum in naturally infected Lutzomyia longipalpis (Diptera: Psychodidae: Phlebotominae) and Canis familiaris in Misiones, Argentina: the first report of a PCR-RFLP and sequencing-based confirmation assay. Mem Inst Oswaldo Cruz 105: 796–799. [DOI] [PubMed] [Google Scholar]
- 14. Felipe IM, Aquino DM, Kuppinger O, Santos MD, Rangel ME, et al. (2011) Leishmania infection in humans, dogs and sandflies in a visceral leishmaniasis endemic area in Maranhão, Brazil. Mem Inst Oswaldo Cruz 106: 207–211. [DOI] [PubMed] [Google Scholar]
- 15. de Oliveira SMP, de Moraes BA, Gonçalves CA, Giordano-Dias CM, d’Almeida JM, et al. (2000) Prevalence of the microbiota in the digestive tract of wild-caught females of Lutzomyia longipalpis . Rev Soc Bras Med Tro 33: 319–322. [DOI] [PubMed] [Google Scholar]
- 16. Perira de Oliveira SM, de Morais BA, Gonçalves CA, Giordano-Dias CM, Vilela ML, et al. (2001) Digestive tract microbiota in female Lutzomyia longipalpis (Lutz & Neiva, 1912) (Diptera: Psychodidae) from colonies feeding on blood meal and sucrose plus blood meal]. Cad Saude Publica 17: 229–232. [PubMed] [Google Scholar]
- 17. Gouveia C, Asensi MD, Zahner V, Rangel EF, de Oliveira SMP (2008) Study on the bacterial midgut microbiota associated to different Brazilian populations of Lutzomyia longipalpis (Lutz & Neiva) (Diptera: Psychodidae). Neotrop Entomol 37: 597–601. [DOI] [PubMed] [Google Scholar]
- 18. Hillesland H, Read A, Subhadra B, Hurwitz I, McKelvey R, et al. (2008) Identification of aerobic gut bacteria from the Kala Azar vector, Phlebotomus argentipes: A platform for potential paratransgenic manipulation of sand flies. Am J Trop Med Hyg 79: 881–886. [PubMed] [Google Scholar]
- 19. McCarthy CB, Diambra LA, Rivera Pomar RV (2011) Metagenomic analysis of taxa associated with Lutzomyia longipalpis, vector of visceral leishmaniasis, using an unbiased high-throughput approach. PLoS Negl Trop Dis 5: e1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dillon RJ, Dillon VM (2004) The gut bacteria of insects: Nonpathogenic interactions. Ann Rev Entomol 49: 71–92. [DOI] [PubMed] [Google Scholar]
- 21. Dillon RJ, Webster G, Weightman AJ, Dillon VM, Blanford S, et al. (2008) Composition of Acridid gut bacterial communities as revealed by 16S rRNA gene analysis. J Invertebr Pathol 97: 265–272. [DOI] [PubMed] [Google Scholar]
- 22. Robinson C, Schloss P, Ramos Y, Raffa K, Handelsman J (2010) Robustness of the bacterial community in the cabbage white butterfly larval midgut. Microb Ecol 59: 199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ramirez JL, Souza-Neto J, Cosme RT, Rovira J, Ortiz A, et al. (2012) Reciprocal tripartite interactions between the Aedes aegypti midgut microbiota, innate immune system and Dengue virus influences vector competence. PLoS Negl Trop Dis 6: e1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Behar A, Yuval B, Jurkevitch E (2005) Enterobacteria-mediated nitrogen fixation in natural populations of the fruit fly Ceratitis capitata . Mol Ecol 14: 2637–2643. [DOI] [PubMed] [Google Scholar]
- 25. Schonfeld J, Heuer H, van Elsas JD, Smalla K (2003) Specific and sensitive detection of Ralstonia solanacearum in soil on the basis of PCR amplification of fliC fragments. Appl Environ Microbiol 69: 7248–7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Alexander B, Usma MC (1994) Potential sources of sugar for the phlebotomine sandfly Lutzomyia-youngi (diptera, psychodidae) in a colombian coffee plantation. Ann Trop Med Parasitol 88: 543–549. [DOI] [PubMed] [Google Scholar]
- 27. Cameron MM, Davies CR, Monje J, Villaseca P, Ogusuku E, et al. (1994) Comparative activity of phlebotomine sandflies (Diptera, Psychodidae) in different crops in the Peruvian Andes. B Entomol Res 84: 461–467. [Google Scholar]
- 28. Schlein Y, Jacobson RL (1994) Mortality of Leishmania major in Phlebotomus papatasi caused by plant-feeding of the sand flies. Am J Trop Med Hyg 50: 20–27. [PubMed] [Google Scholar]
- 29. Siddall ME, Perkins SL, Desser SS (2004) Leech mycetome endosymbionts are a new lineage of alphaproteobacteria related to the Rhizobiacea. Mol Phylogenet Evol 30: 178–186. [DOI] [PubMed] [Google Scholar]
- 30. Murphy KM, Teakle DS, Macrae IC (1994) Kinetics of colonization of adult Queensland fruit flies (Bactrocera tryoni) by dinitrogen-fixing alimentary tract bacteria. Appl Environ Microbiol 60: 2508–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bloch KC, Nadarajah R, Jacobs R (1997) Chryseobacterium meningosepticum: An emerging pathogen among immunocompromised adults - Report of 6 cases and literature review. Medicine 76: 30–41. [DOI] [PubMed] [Google Scholar]
- 32. Tekerekoglu MS, Durmaz R, Ayan M, Cizmeci Z, Akinci A (2003) Analysis of an outbreak due to Chryseobacterium meningosepticum in a neonatal intensive care unit. Microbiologica 26: 57–63. [PubMed] [Google Scholar]
- 33. Losada L, Ronning CM, DeShazer D, Woods D, Fedorova N, et al. (2010) Continuing evolution of Burkholderia mallei through genome reduction and large-scale rearrangements. Genome Biol Evol 2: 102–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ono M, Braig HR, Munstermann LE, Ferro C, O’Neill SL (2001) Wolbachia infections of phlebotomine sand flies (Diptera : Psychodidae). J Med Entomol 38: 237–241. [DOI] [PubMed] [Google Scholar]
- 35. Dillon RJ, Vennard CT, Charnley AK (2000) Pheromones - Exploitation of gut bacteria in the locust. Nature 403: 851–851. [DOI] [PubMed] [Google Scholar]
- 36. Dillon RJ, Vennard CT, Buckling A, Charnley AK (2005) Diversity of locust gut bacteria protects against pathogen invasion. Ecol Lett 8: 1291–1298. [Google Scholar]
- 37. Brummel T, Ching A, Seroude L, Simon AF, Benzer S (2004) Drosophila lifespan enhancement by exogenous bacteria. Proc Natl Acad Sci U S A 101: 12974–12979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kashima T, Nakamura T, Tojo S (2006) Uric acid recycling in the shield bug, Parastrachia japonensis (Hemiptera: Parastrachiidae), during diapause. J Insect Physiol 52: 816–825. [DOI] [PubMed] [Google Scholar]
- 39. Adler S, Theodor O (1957) Transmission of disease agents by Phlebotomine sand flies. Ann Rev Entomol 2: 203–226. [Google Scholar]
- 40. Schlein Y, Warburg A, Yuval B (1986) On the system by which sandflies maintain a sterile gut. Insect Sci Appl 7: 231–234. [Google Scholar]
- 41. Webster G, Newberry CJ, Fry JC, Weightman AJ (2003) Assessment of bacterial community structure in the deep sub-seafloor biosphere by 16S rDNA-based techniques: A cautionary tale. J Microbiol Meth 55: 155–164. [DOI] [PubMed] [Google Scholar]
- 42. McCaig AE, Glover LA, Prosser JI (2001) numerical analysis of grassland bacterial community structure under different land management regimens by using 16S Ribosomal DNA sequence data and denaturing gradient gel electrophoresis banding patterns. Appl Environ Microbiol 67: 4554–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Webster G, John Parkes R, Cragg BA, Newberry CJ, Weightman AJ, et al. (2006) Prokaryotic community composition and biogeochemical processes in deep subseafloor sediments from the Peru Margin. FEMS Microbiology Ecology 58: 65–85. [DOI] [PubMed] [Google Scholar]
- 44. Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59: 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Webster G, Embley TM, Prosser JI (2002) Grassland management regimens reduce small-scale heterogeneity and species diversity of proteobacterial ammonia oxidizer populations. Appl Environ Microbiol 68: 20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. O’Neill SL, Giordano R, Colbert AM, Karr TL, Robertson HM (1992) 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc Natl Acad Sci U S A 89: 2699–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zchori-Fein E, Perlman SJ (2004) Distribution of the bacterial symbiont Cardinium in arthropods. Mol Ecol 13: 2009–2016. [DOI] [PubMed] [Google Scholar]
- 48. Cole J CB, Farris RJ, Wang Q, Kulam SA, McGarrell DM, et al. (2005) The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res 33: D294–D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang Q GGM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Felsenstein (2005) PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author. Department of Genome Sciences, University of Washington, Seatle. [Google Scholar]
- 52. Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biol 52: 696–704. [DOI] [PubMed] [Google Scholar]
- 53. Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol and Evol 17: 540–552. [DOI] [PubMed] [Google Scholar]