Abstract
CD1d molecules are major histocompatibility complex (MHC) class I-like molecules that present lipid antigens to Natural Killer T (NKT) cells. Although we have previously shown that several different cell signaling molecules can play a role in the control of antigen presentation by CD1d, a defined mechanism by which a cell signaling pathway regulates CD1d function has been unclear. In the current study, we have found that the Rho kinases, ROCK1 and ROCK2, negatively regulate both human and mouse CD1d-mediated antigen presentation. Inhibition of ROCK pharmacologically, through specific ROCK1 and ROCK2 shRNA, or by using dendritic cells generated from ROCK1-deficient mice all resulted in enhanced CD1d-mediated antigen presentation compared to controls. ROCK regulates the actin cytoskeleton by phosphorylating LIM kinase which, in turn, phosphorylates cofilin, prohibiting actin fiber depolymerization. Treatment of antigen presenting cells with the actin filament depolymerizing agent, cytochalasin D, as well as knockdown of LIM kinase by shRNA, resulted in enhanced antigen presentation to NKT cells by CD1d, consistent with our ROCK inhibition data. Therefore, our overall results reveal a model whereby CD1d-mediated antigen presentation is negatively regulated by ROCK via its effects on the actin cytoskeleton.
Keywords: Antigen Presentation, Signal Transduction, T cells
Introduction
CD1d is a major histocopatibility complex (MHC) class-I like molecule. However, unlike MHC class-I that presents peptide antigens, CD1d presents lipid antigens (1). CD1d presents these lipids to a novel T lymphocyte subset called natural killer T (NKT) cells (2). These NKT cells can be subdivided into two groups based on their T cell receptor (TCR) usage. Type I NKT cells express a TCR with a specific α chain rearrangement and β chains of limited diversity whereas type II NKT cells contain TCRs with diverse α and β chain rearrangements (3). Upon stimulation of NKT cells by lipid-loaded CD1d, the NKT cells rapidly produce both Th1 and Th2 cytokines (1). CD1d-mediated antigen presentation has been shown to be important for both antiviral and antitumor immunity (2, 4, 5). Thus, lipid antigen presentation by CD1d molecules to NKT cells is an important arm of the host’s innate immune response (1, 2, 6, 7).
Due to the importance of CD1d in the host’s immune response, it is critical to understand the cellular signaling pathways that can modulate CD1d-mediated antigen presentation. We have previously reported that individual cell signaling molecules such as ERK1/2 (5, 8), p38 (5), and protein kinase C δ (PKCδ) (9), are involved in the regulation of antigen presentation by CD1d. However, the precise mechanism by which a signaling pathway controls antigen presentation by CD1d is unknown.
Actin is an important structural component of cells that is involved in many cellular functions such as proliferation and motility (10). One pathway that regulates actin dynamics is the Rho kinase/LIM kinase/cofilin pathway. The serine/threonine kinase Rho kinase (ROCK) can phosphorylate and activate the serine/threonine kinase LIM kinase (LIMK) (10, 11). LIMK can phosphorylate and thereby inactivate the actin depolymerizing protein cofilin (10, 11). Thus, the ROCK signaling pathway affects actin dynamics in the cell.
Here we show that ROCK is a negative regulator of CD1d-mediated antigen presentation through its effects on the cytoskeleton. Specific knockdown of ROCK or its downstream effector LIMK by short hairpin RNA, results in enhanced CD1d-mediated antigen presentation. Disruption of the actin cytoskeleton results in a similar effect on antigen presentation. Thus, these results reveal that the ROCK/LIMK/actin pathway plays a critical role in the control of innate immunity vis-à-vis CD1d-mediated antigen presentation to NKT cells, by dynamic regulation of the actin cytoskeleton in antigen presenting cells.
Materials and Methods
Cell lines, reagents and mice
Murine LMTK-CD1d1, LMTK-VC, L-CD1d-DR4, human HEK293-hCD1d, HEK293-VC, murine 17.9 T cell hybridoma (kind gift of J. Blum, Indiana Univ.), and the murine NKT cell hybridomas DN32.D3, N37-1A12 and N38-3C3 (kind gifts from K. Hayakawa, Fox Chase Cancer Center) have all been previously described (4, 12–14). Anti-ROCK1 and anti-ROCK2 antibodies were purchased from BD Biosciences (San Jose, CA). Anti-GAPDH antibody was purchased from Cell Signaling Technology (Danvers, MA). Anti-hCD1d antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and eBioscience (San Diego, CA). The peroxidase conjugated anti-mouse (Sigma-Aldrich, St. Louis, MO) and anti-rabbit (Bio-Rad; Hercules, CA) IgG secondary antibodies were used for Western blotting. For flow cytometry, anti-mouse CD1d (1H6), anti-mouse MHC class 1 (TIB126; ATCC, Manassas, VA), and isotype control for 1H6 [anti-vaccinia protein (TW2.3)] were used. CD1d tetramers and CD1d tetramers loaded with α-galactosylceramide (α-GalCer; PBS-57) were from the NIH Tetramer Core Facility (Atlanta, GA). PE-conjugated anti-mouse (Dako, Carpenteria, CA) and anti-rat antibodies (Jackson ImmunoResearch, West Grove, PA) were used as secondary antibodies. Purified and biotinylated anti-mouse IL-2 (BD Biosciences), purified and biotinylated antihuman IL-4 and GM-CSF (BioLegend, San Diego, CA), and avidin-peroxidase (Sigma-Aldrich), were used to measure cytokine production by ELISA. Recombinant murine IL-2, human IL-4 and GM-CSF (Peprotech, Rocky Hill, NJ) were used as standards. ROCK1 knockout mice have been previously described (15). Wild type littermates were used as controls. All animal studies were performed in accordance with protocols approved by the Indiana University School of Medicine’s IACUC.
shRNA constructs and cell line generation
ROCK1, ROCK2, and negative control (NC) vectors were generated using the pLKO.1 vector (kindly provided by Dr. David Riese, Auburn University). Oligonucleotides for the target sequences were purchased from Integrated DNA Technologies, Inc. (Coralville, IA), annealed, phosphorylated and ligated into the pLKO.1 vector. shRNA (21-mer) constructs targeting LIMK2 and mouse LIMK1 in the pLKO.1 vector were purchased from Sigma-Aldrich. The shRNA sequences were: NC (negative control: 5’-TCAGTCACGTTAATGGTCGTT-3’); ROCK1 (5’-GCTCGAATTACATCTTTACAA-3’); ROCK2 (5’-GCCTTGCATATTGGTCTGGAT-3’); LIMK1 (5’-GATGGTGATGAAGGAACTTAT-3’); LIMK2 (5’-GATGCACATCAGTCCCAACAA-3’). LMTK-CD1d1 and HEK293-hCD1d cells were transfected with these plasmids using polyethylenimine as previously described (14). Transfected cells were selected in puromycin (Sigma-Aldrich; 2 µg/ml for HEK293-CD1d cells and 10–25 µg/ml for LMTK-CD1d1 cells). Drug-resistant cells were pooled and used as stable cell lines.
NKT co-culture assays
Mouse LMTK-CD1d1 cell co-culture assays have been previously described (12). All drug treatments of cells were performed at 37°C at the indicated concentrations. Treatments were for the following lengths of time: Y-27632 (Ascent Scientific, Princeton, NJ) for 2 h with LMTK-CD1d1 cells or 16 h with L-CD1d-DR4 cells; cytochalasin D (Thermo Fisher Scientific, Waltham, MA) for 2 h with LMTK-CD1d1 cells or 24 h with L-CD1d-DR4 cells. L-CD1d-DR4 cells were also incubated with Human Serum Albumin (HSA; Sigma-Aldrich) for the entire drug treatment unless otherwise indicated. Co-cultures of HEK293-hCD1d cells with human NKT cells have been previously described (14). Generation of mouse BMDCs for use as APCs was as described (16). APCs were washed and fixed in 0.05% paraformaldehyde prior to co-culturing.
Mass spectroscopic analysis
A tandem affinity purification (TAP) tag protocol was adapted and used to purify proteins bound to the 13 carboxy-terminal amino acids of hCD1d (17). Briefly, HEK293 cells were transfected with a construct expressing the TAP tag sequence with the 13 AA tail of hCD1d or the TAP tag sequence alone. Transfected cells were selected in G418 and GFP+ cells were electronically sorted and adapted to growth in suspension media (Pro293s-CDM; Lonza, Walkersville, MD). Suspension cultures were expanded and approximately 1.5×109 cells were lysed and the lysate treated as described (17). Protein bands were excised and analyzed by the Purdue Proteomics Facility (West Lafayette, IN) via mass spectrometry. Protein identification was performed by database searching with GPS Explorer software (Applied Biosystems, Foster City, CA) utilizing the Mascot algorithm and the NCBI database. Alternatively, human CD1d was immunoprecipitated and the eluates digested using triethylphosphine and iodoethanol for cysteine modification using described methods (18) and analyzed by mass spectrometry. Spectrum Mill software (Agilent Technologies, Santa Clara, CA) using the NCBI database was utilized for protein search identification.
Western blotting
Cells were lysed in 2% CHAPS lysis buffer [10 mM Tris (pH 7.5), 150 mM NaCl, 0.5 mM EDTA, 2% CHAPS, 0.02% sodium azide, and protease inhibitors; Roche, Indianapolis, IN]. Sodium fluoride and sodium orthovanadate (1 mM each) were included for phosphoprotein blotting. Protein concentrations of lysates were determined using Coomassie Protein Assay Reagent (Pierce, Rockfored, IL) and equal amounts (or amounts indicated) of protein lysate were mixed with protein sample buffer, boiled, and resolved on an SDS-PAGE gel (16). Following transfer onto a polyvinylidene difluoride membrane (Millipore, Billerica, MA), blots were probed with the indicated antibodies and developed using chemiluminescence and exposure on film. Images were quantified using ImageJ (1.37v, National Institutes of Health, Bethesda, MD).
Liver mononuclear cells
The harvesting and preparation of mouse liver mononuclear cells, as well as their staining and analysis by flow cytometry has been previously described (19).
Confocal microscopy
Analysis by confocal microscopy consisted of treatment of LMTK-CD1d1 cells with TGF-β (20 ng/ml), the ROCK inhibitor Y-27632 (100 µM), cytochalasin D (2 µM), or mock treatment (2 mg/ml BSA/PBS and DMSO) for 24 h in 35 mm collagen-coated glass bottom dishes (MatTek, Ashland, MA). After treatment, cells were washed twice with PBS and fixed for 20 min on ice with 4% paraformaldehyde. Cells were permeabilized and stained with specific monoclonal antibodies as described (8). To evaluate the actin cytoskeleton, mock- and TGF-β-treated cells were stained for 1 h with 0.14 µM Rhodamine-Phalloidin (Cytoskeleton, Inc., Denver, CO), washed and placed in PBS for analysis at room temperature with a Carl Zeiss 510-UV confocal microscope with the 63X water objective and 78 µm aperature. LSM Image Browser acquisition software was used. Untreated NC and shRNA cells were stained for CD1d using neat 1H6 antibody hybridoma (12) supernatant/Alexa488 anti-mouse IgG antibody (Invitrogen, Carlsbad, CA) and for LAMP-1 using neat 1D4B antibody hybridoma (Developmental Studies Hybridoma Bank, Iowa City, IA) supernatant/Texas Red anti-rat IgG antibody (Jackson ImmunoResearch). Colocalization of CD1d and LAMP-1 from images collected from an Olympus 2 FV1000 confocal microscope with the 60X water objective, 103 µm aperature, and Olympus Fluoview FV1000 acquisition software, was measured using Metamorph (Molecular Devices, Sunnyvale, CA). All images were collected as a series of slices into Z-stacks. Slice sizes for images collected from the Carl Zeiss confocal microscope were 0.5 µm, whereas those from the Olympus 2 were 0.2 µm.
CD1d internalization assay
HEK293-hCD1d cells were incubated with the 42.1 mAb (5 µg/ml) at 4°C for 30 min. After washing in media, cells were incubated at 37°C for 0, 5, 10 and 30 min. All cells were then incubated with a PE-anti-mouse antibody (1:100) at 4°C. The cells were then fixed for analysis by flow cytometry.
Exogenous lipid treatment
LMTK-CD1d1 cells containing NC or ROCK1 shRNA were treated with α-GalCer (Enzo Life Sciences, Farmingdale, NY) or galactose(α1→2)galactosylceramide (α-GalGalCer) (kindly provided by Dr. Paul Savage, Brigham Young University) for three hours at 37°C. Cells were washed, fixed and used in co-culture experiments with NKT cells as described above.
In vitro kinase assays
ROCK1 and ROCK2 kinases were purchased from Cell Signaling Technology. Peptides corresponding to the carboxy-terminal tail of CD1d1, hCD1b, hCD1c and hCD1d were kindly provided by J. Hastie (Univ. of Dundee, UK). Control peptides (CREB and PLK) were purchased from Cell Signaling Technology. A 40 µl kinase assay reaction was performed [30 µl of 6 µM peptide, 4 µl of 10X kinase buffer (Cell Signaling), 1 µl of kinase (100 µg/ml stock) and 5 µl of 0.16 µCi/µl [32P]-ATP (PerkinElmer, Waltham, MA) diluted in 250 µM ATP (Sigma-Aldrich)] for 15 min at 30°C. The entire 40 µl of the reaction was spotted onto a circle of P81 paper (Whatman, Piscataway, NJ) and allowed to air dry. P81 papers were then washed 3 times for 5 min each with 75 mM phosphoric acid. P81 papers were then washed once for 2 min with acetone then allowed to air dry. Each P81 paper was transferred to a scintillation vial with 5 ml scintillation fluid and radioactivity was measured using a Beckman LS 6000IC.
hCD1d mutants
HEK293-hCD1d cells in which the two putative sites of ROCK phosphorylation (T329/S330) were mutated to alanines (T329A/S330A) were generated using previously described methods (14). These cells were used in human NKT cell co-culture assays as previously reported (14).
C3 treatment
LMTK-CD1d1 cells were treated +/− the cell permeable C3 toxin (2 µg/ml) (Cytoskeleton, Inc.) for six hours at 37°C. The cells were used in a NKT co-culture assay as previously described (12).
Statistics and data analysis
Graphs were generated and statistics calculated using GraphPad Prism 5 (GraphPad Software, LaJolla, CA). The mean of triplicates of a representative assay is shown with error bars representing the S.E.M. A one-way ANOVA with Bonferroni’s post-test or Student’s t-test was used as appropriate. A P value less than 0.05 was considered significant.
Results
ROCK1 and ROCK2 negatively regulate CD1d-mediated antigen presentation
The mechanisms by which the functional expression of CD1d-mediated antigen presentation is regulated by cell signaling pathways have not yet been clearly elucidated. As a starting point to identify possible cell signaling pathways that might be important in the regulation of antigen presentation by CD1d, we performed a tandem affinity (TAP-tag) purification (17) of the human CD1d tail in HEK293 cells (Supplemental Fig. S1A, S1B). Using this methodology, we identified the Rho GTPase effector and serine/threonine kinase, Rho kinase 2 (ROCK2) (Supplemental Table I). Additionally, when we immunoprecipitated CD1d and analyzed the bound proteins by mass spectrometry, we identified ROCK1 (Supplemental Table I). The Rho GTPase family of molecules consists of important cell signaling pathways critical for actin cytoskeleton dynamics, cell trafficking, as well as the intracellular transport of molecules playing key roles throughout the endocytic pathway (10, 20). As we and others have shown that CD1d traffics through compartments of the endocytic pathway (2, 12, 21–25), this observation was clearly of potential importance. It was thus important to then determine whether ROCK proteins affect CD1d-mediated Ag presentation.
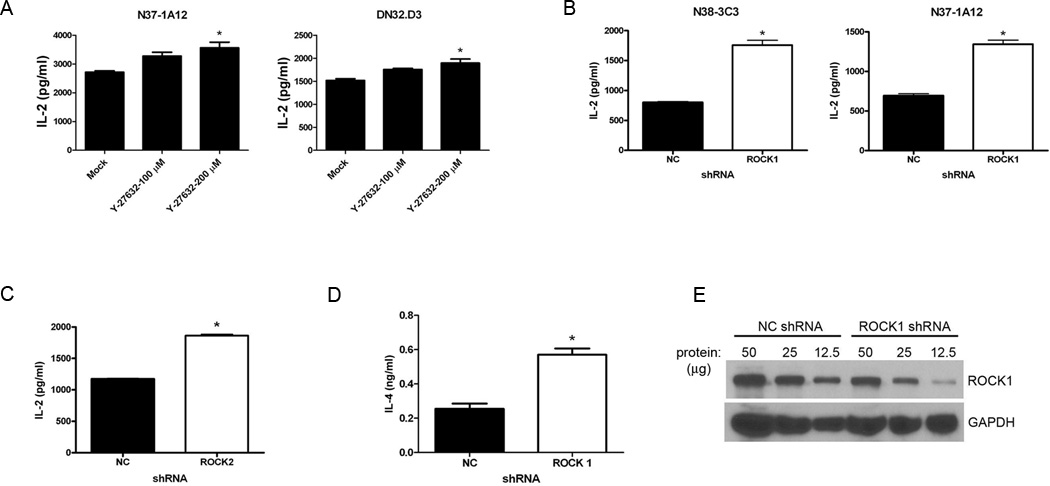
Two different ROCK proteins are encoded by homologous, yet distinct genes, ROCK1 and ROCK2 (10). Although we found that formed a complex with CD1d in the TAP-tag experiments, this did not necessarily mean they. To address whether ROCK1 and ROCK2 play a role in CD1d-mediated Ag presentation, we pretreated murine LMTK-CD1d1 cells (4) with the ROCK-specific inhibitor, Y-27632, and then cocultured these cells with a panel of mouse NKT cell hybridomas. IL-2 production by the NKT cells was used as a readout for antigen presentation by CD1d. Interestingly, in LMTK-CD1d1 cells treated with the ROCK inhibitor, antigen presentation was increased over those cells treated with vehicle alone (Fig. 1A). This result suggested that ROCK serves as a negative regulator of antigen presentation by CD1d.
FIGURE 1.
ROCK negatively regulates CD1d-mediated Ag presentation. A. Murine LMTK-CD1d1 cells were treated with the ROCK inhibitor Y-27632 or vehicle only and co-cultured with the indicated NKT cell hybridomas. Antigen presentation was measured by an IL-2 ELISA; *, P < 0.05 compared to mock. This experiment was repeated four times. B, LMTK-CD1d1 cells were transfected with a ROCK1-specific shRNA and CD1d-mediated Ag presentation to NKT cells was measured as above. NC: control shRNA; *, P = 0.0003. This result is representative of five independent experiments. C, LMTK-CD1d1 cells were transfected with a ROCK2-specific shRNA and CD1d-mediated Ag presentation to the representative N38-3C3 NKT cell hybridoma was measured as above; *, P < 0.001. This result is representative of five independent experiments. D, HEK293-hCD1d cells were transfected with a ROCK1-specific shRNA and CD1d-mediated Ag presentation to human NKT cells was measured by an IL-4 ELISA; *, P = 0.0027. This result is representative of four independent experiments. E, Western blot of lysates showing decreased ROCK1 expression in HEK293-hCD1d cells expressing the ROCK1 shRNA. Three different amounts of protein (indicated in µg per well) were loaded for each cell line. This result is representative of four independent experiments.
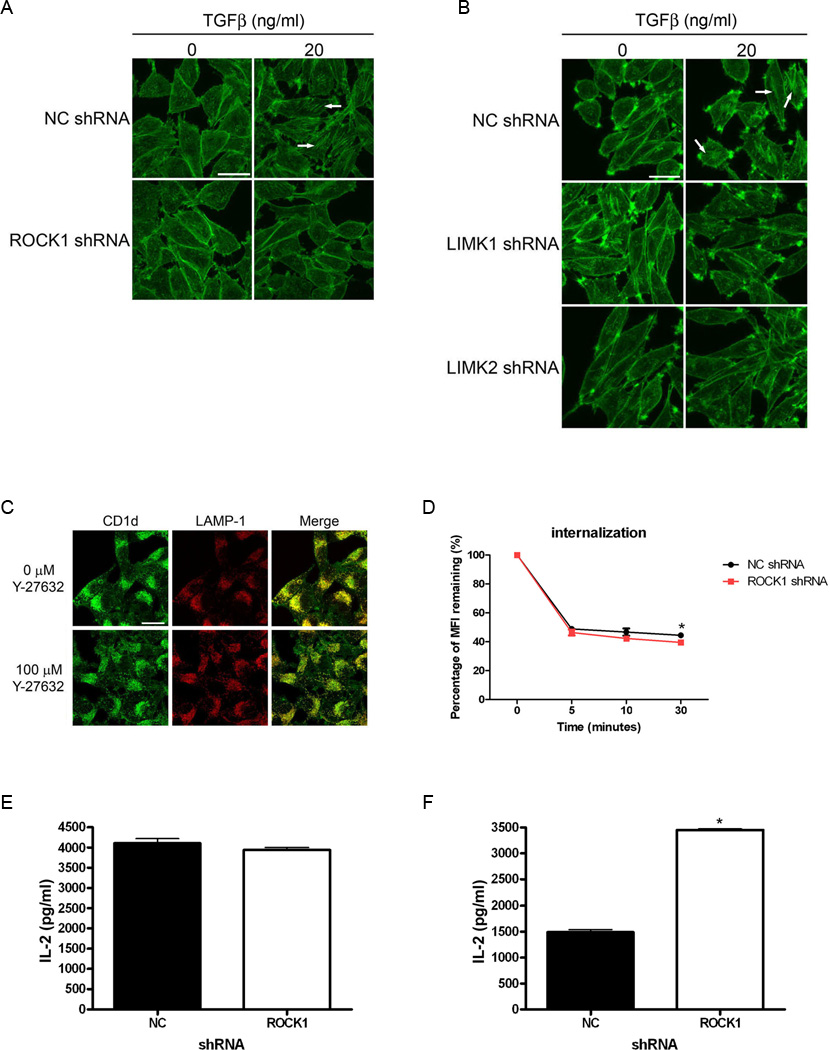
Because pharmacological inhibitors can have off-target effects, we employed two genetic approaches to confirm the role of ROCK in CD1d-mediated antigen presentation. In the first, ROCK-specific short hairpin RNA (shRNA) was used to knockdown ROCK1 or ROCK2 expression in both murine LMTK-CD1d1 and human HEK293-hCD1d cells. Control cells consisted of the appropriate CD1d+ cell lines transfected with a control shRNA (NC shRNA) sequence. These cells were then cocultured with their respective NKT cells. In both murine (Fig. 1B, 1C) and human (Fig. 1D) systems, reducing ROCK1 or ROCK2 protein expression resulted in an increase in antigen presentation by CD1d to NKT cells. Thus, as was observed in the mouse, inhibition of ROCK also enhanced human CD1d-mediated antigen presentation. A significant decrease in ROCK protein expression occurred in those cells transfected with the ROCK-specific shRNA (Fig. 1E and data not shown). Quantification of the bands and normalizing to GAPDH shows that there was a greater than 50% decrease in ROCK1 expression in the cells containing ROCK1-specific shRNA.
ROCK1 knock-out mice have increased CD1d-mediated antigen presentation and liver NKT cells
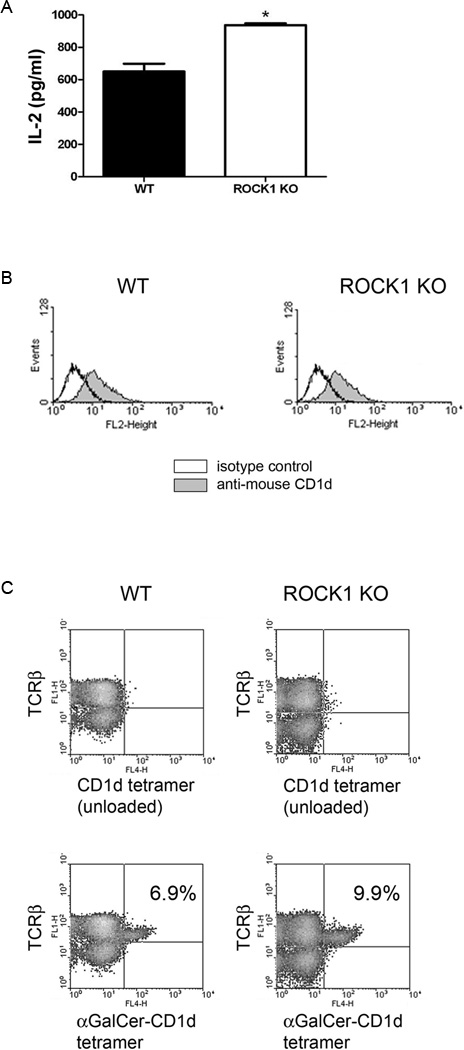
As an alternative approach, we generated bone marrow-derived dendritic cells (BMDCs) from ROCK1-deficient (15) and wildtype (WT) mice, and co-cultured them with murine NKT cells as above. Dendritic cells are very efficient antigen presenting cells to NKT cells (3, 26, 27). As expected, BMDCs from WT mice were able to stimulate NKT cells (Fig. 2A). However, DCs from ROCK1-deficient mice (ROCK1 KO) were actually better able at stimulating NKT cells than those from wildtype mice (Fig. 2A). As detected by flow cytometry, there were comparable levels of CD1d on the cell surface of the BMDCs of WT and ROCK1 KO mice (Fig. 2B). This suggests that the mechanism by which inhibition of ROCK results in increased CD1d-mediated antigen presentation is not merely due to an increased number of cell surface CD1d molecules. Of additional note, ROCK1-deficient mice had more liver NKT cells than WT mice (Fig. 2C), serving as an important in vivo confirmation of our in vitro antigen presentation assays. ROCK1 KO mice also had more NKT cells in the spleen, although the increase compared to WT mice was not as great as that seen in the liver, yet the number of mononuclear cells were within the normal range (data not shown). Therefore, these data strongly suggest that ROCK is a negative regulator of CD1d-mediated antigen presentation.
FIGURE 2.
Enhanced CD1d functional expression and NKT cell numbers in ROCK1 KO mice. A, Bone marrow-derived dendritic cells (BMDCs) from control (WT) and ROCK1 KO mice were co-cultured with the representative N37-1A12 NKT cell hybridoma; *, P = 0.0043. B, BMDCs from WT and ROCK1 KO mice were stained for surface CD1d and analyzed by flow cytometry. C, Liver mononuclear cells from WT and ROCK1 KO mice were harvested and NKT cells were identified by flow cytometry [TCRβ+/α-GalCer (PBS-57)-loaded CD1d tetramer+]. The percentage of NKT cells is shown in the upper right quadrant of the lower two-color plots. The experiments shown are representative of two performed. Two mice per group were analyzed in each experiment.
Disrupting actin polymerization results in increased CD1d-mediated antigen presentation
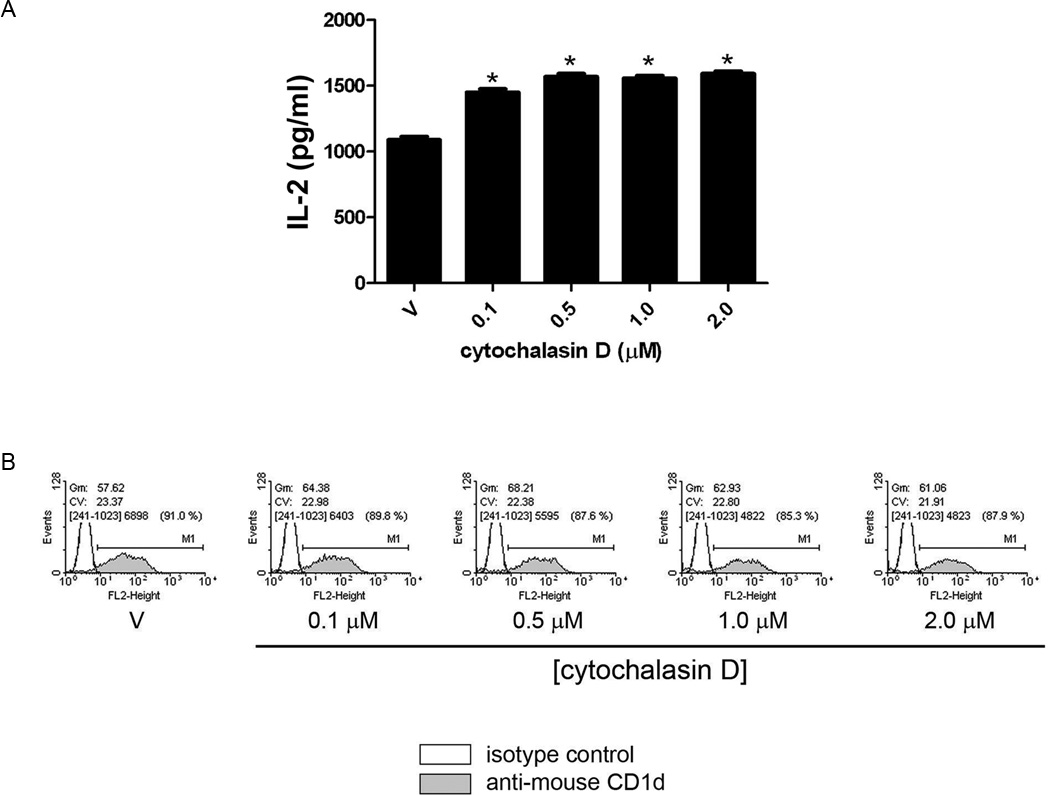
ROCK plays an important role in the dynamic regulation of the actin cytoskeleton (10, 20). In particular, ROCK is important in actin polymerizing into its filamentous form, F-actin. In order to determine if ROCK is able to control antigen presentation by CD1d due to effects on F-actin, we first disrupted the actin cytoskeleton in LMTK-CD1d1 cells using the actin depolymerizing agent, cytochalasin D. If indeed the ROCK-mediated effects on CD1d were through the regulation of actin, it would be expected that cytochalasin D treatment of CD1d+ cells would enhance antigen presentation to NKT cells. As predicted, cytochalasin D-treated LMTK-CD1d1 cells were better able to stimulate NKT cells than vehicle-treated cells (Fig. 3A). No change in CD1d expression on the cell surface of LMTK-CD1d1 cells treated with cytochalasin D was observed (Fig. 3B).
FIGURE 3.
Modulation of the actin cytoskeleton alters CD1d-mediated Ag presentation. A, Disruption of actin by cytochalasin D results in increased CD1d-mediated Ag presentation to the representative DN32.D3 NKT cell hybridoma; *, P < 0.05 compared to vehicle control (DMSO, V). This result is representative of two independent experiments. B, Treatment of LMTK-CD1d1 cells with cytochalasin D does not affect surface levels of CD1d as measured by flow cytometry. The experiment shown is representative of two performed.
LIMK1 and LIMK2 also negatively regulate CD1d-mediated antigen presentation
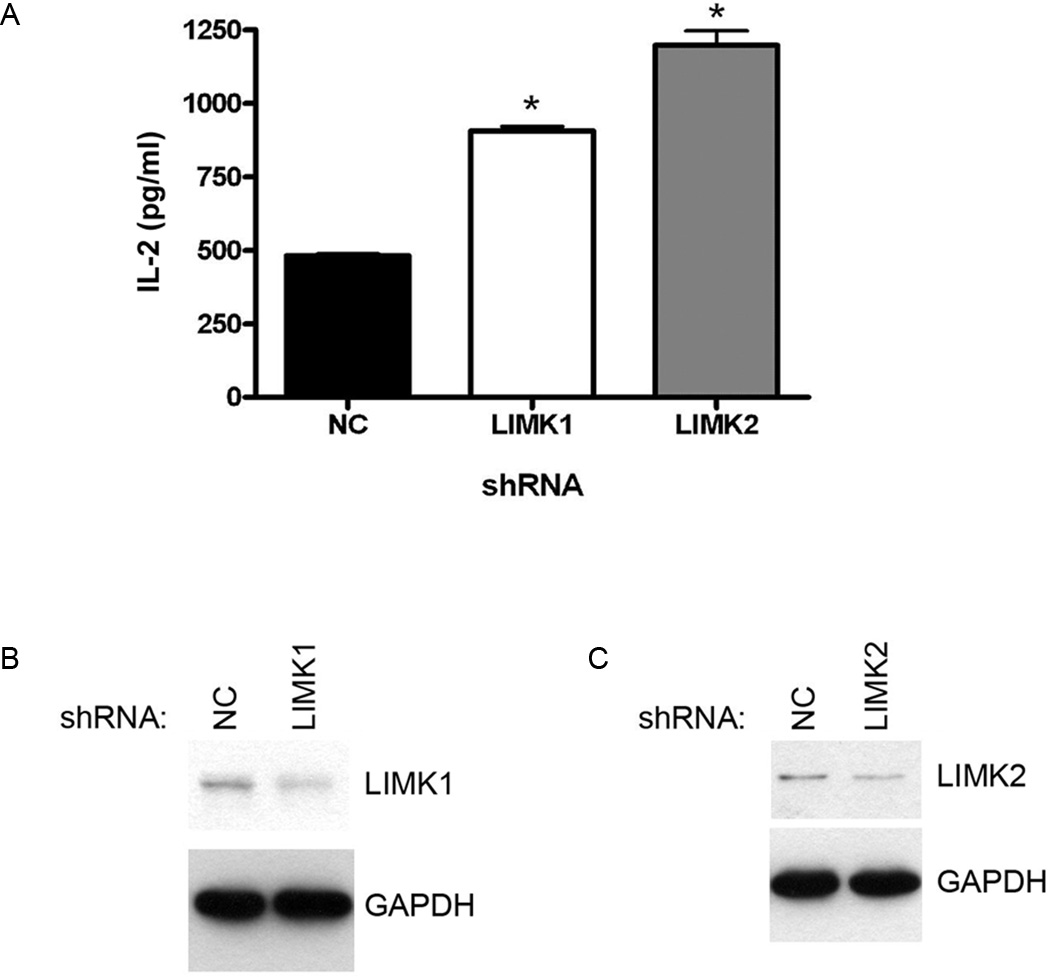
With the observations that ROCK and F-actin serve as negative regulators of CD1d-mediated antigen presentation, it was important to determine if these two results were related. It is very well known that ROCK controls actin polymerization through a series of steps (10). First, ROCK phosphorylates the LIM kinases (LIMK) 1 and LIMK2. These LIMKs are serine/threonine kinases that regulate actin cytoskeleton rearrangement (11). In order to determine if this pathway is important in the ability of ROCK to reduce CD1d-mediated antigen presentation, we used shRNA to knockdown LIMK1 and LIMK2 in LMTK-CD1d1 cells. These CD1d+ cells with LIMK1, LIMK2 or control (NC) shRNA were cocultured with murine NKT cells. It was found that knockdown of either LIMK1 or LIMK2 resulted in an increase in antigen presentation by CD1d to NKT cells (Fig. 4A). These results suggest that, like ROCK, LIMK is a negative regulator of CD1d-mediated antigen presentation. Expression of LIMK1 (Fig. 4B) and LIMK2 (Fig. 4C) was decreased by the shRNAs targeting these genes. Quantification and normalization to GAPDH shows that each were reduced by 50% as compared to the control shRNA.
FIGURE 4.
LIMKs negatively regulate CD1d-mediated Ag presentation. A, shRNA against either LIMK1 or LIMK2 results in increased CD1d-mediated Ag presentation to the representative N38-3C3 NKT cell hybridoma; *, P < 0.001 as compared to control shRNA (NC). This experiment is representative of five performed. B, LMTK-CD1d1 NC shRNA and LMTK-CD1d1 LIMK1 shRNA cells were lysed, equal amounts of protein were resolved by SDS-PAGE and LIMK1 expression was detected via Western blotting. The membrane was stripped and reprobed for GAPDH to confirm equal loading. C, LMTK-CD1d1 NC shRNA and LMTK-CD1d1 LIMK2 shRNA cells were lysed, equal amounts of protein were resolved by SDS-PAGE and LIMK2 expression was detected via Western blotting. The membrane was stripped and reprobed for GAPDH to confirm equal loading.
Actin polymerization is impaired in ROCK and LIMK shRNA cells
To further connect ROCK and LIMK signaling to actin polymerization, we examined actin stress fiber formation in our LMTK-CD1d1 cells containing the shRNAs described above. Actin stress fibers were induced by treating the cells with TGFβ. Rhodamine-phalloidin was used to stain the stress fibers. Confocal microscopic analyses confirmed alterations in the actin cytoskeleton in the CD1d+ cells in which ROCK or LIMK was knocked down by shRNA (Fig. 5A, 5B; Supplemental figures S2 and S3 contain larger versions of these images to better visualize the stress fibers). Those images show TGFβ-induced stress fiber formation (examples are indicated with white arrows) in control cells (NC shRNA). However, under the same conditions, the cells expressing the ROCK- or LIMK-specific shRNA did not readily form stress fibers. Therefore, these results lend further support to indicate that ROCK is a negative regulator of CD1d-mediated antigen presentation, through its effects on the actin cytoskeleton via the ROCK/LIMK pathway.
FIGURE 5.
Inhibition of ROCK or LIMK affects actin polymerization, CD1d internalization, and CD1d antigen processing, but not intracellular CD1d localization. A–B, LMTK-CD1d1 cells with control (NC), ROCK1, LIMK1, or LIMK2 shRNA were treated +/– TGFβ (20 ng/ml for 24 h) and stained with Rhodamine-phalloidin to visualized stress fibers by confocal microscopy. Cells with ROCK1, LIMK1, or LIMK2 shRNA do not form as many TGFβ-induced stress fibers as compared to control shRNA (NC). Examples of stress fibers are indicated by the white arrows. This result is representative of two independent experiments. C, LMTK-CD1d1 cells were treated +/− with the ROCK inhibitor Y-27632. CD1d/LAMP-1 co-localization was determined by confocal microscopy and showed no difference in the drug-treated cells. Scale bars in (A–C) represent 20 µm. D, HEK293-hCD1d cells expressing ROCK1 shRNA show increased internalization as compared to control shRNA (NC); *, P < 0.05. This result is representative of three independent experiments. E, LMTK-CD1d1 NC shRNA and ROCK1 shRNA cells were pulsed with α-GalCer for 3 h, washed and fixed. Antigen presentation was assayed by co-culture with the N38-3C3 NKT cell hybridoma and measurement of IL-2 secretion by ELISA. This result is representative of two independent experiments. F, LMTK-CD1d1 NC shRNA and ROCK1 shRNA cells were pulsed with α-GalGalCer for 3 h, washed and fixed. Antigen presentation was assayed by co-culture with the N38-3C3 NKT cell hybridoma and measurement of IL-2 secretion by ELISA. *, P < 0.0001. This result is representative of two independent experiments.
Inhibition of ROCK does not alter CD1d localization but does have a modest effect on its internalization
As mentioned above, CD1d internalization and endocytic trafficking are critical to its ability to present antigen. Thus, we examined whether ROCK was exerting its effect on CD1d through altering these processes. Experiments aimed at understanding other consequences of inhibiting ROCK showed that there were no obvious effects on the intracellular localization of CD1d. For example, the amount of CD1d co-localized with LAMP-1 was not affected when ROCK was inhibited with Y-27632 (Fig. 5C). The same results were observed when CD1d and LAMP-1 co-localization was measured in control and ROCK1 shRNA cells (data not shown). We also examined the rate at which CD1d internalizes from the cell surface. We did find that human CD1d internalization was slightly faster in HEK293 cells with reduced ROCK expression (Fig. 5D), consistent with the enhanced antigen presentation observed. However, the rate of recycling of CD1d in these cells was not affected (data not shown). Nonetheless, it seems likely that the ROCK pathway affects the loading (or perhaps selection) of the CD1d ligand(s) presented to NKT cells (1). This would result in a qualitative change which was apparent in our NKT cell assays, but not necessarily by confocal microscopy analyses.
ROCK affects the processing of exogenous lipid antigens
Because there was not a large difference in CD1d recycling, internalization or LAMP-1 co-localization observed, we examined the ability of ROCK inhibition to affect exogenous lipid antigen processing and ultimate presentation by CD1d. LMTK-CD1d1 cells transfected with control or ROCK1 shRNA were treated for three hours with either α-galactosylceramide (α-GalCer) or galactose(α1→2)galactosylceramide (α-GalGalCer). These two molecules have been shown to be presented to NKT cells as CD1d ligands, but α-GalGalCer differs from α-GalCer in that the disaccharide requires processing to be able to stimulate NKT cells (23, 28, 29). As shown in Figure 5E, both control and ROCK1 shRNA cells treated with α-GalCer stimulated similar levels of IL-2 secretion from NKT cells. However, ROCK1 shRNA cells induced much more IL-2 secretion from NKT cells than control shRNA-expressing cells when α-GalGalCer was used as the exogenous ligand (Fig. 5F). These results strongly suggest that the processing of CD1d-presented ligands is negatively regulated by ROCK1.
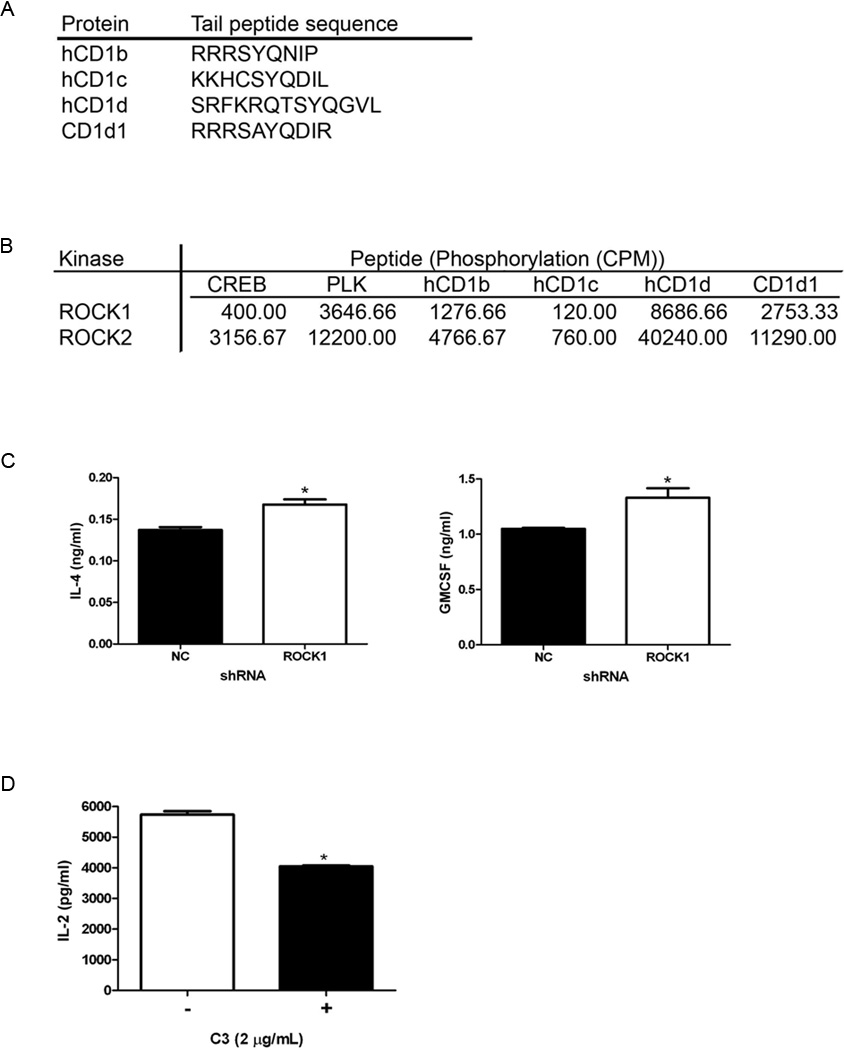
ROCK can phosphorylate CD1d, but it is not necessary for affecting antigen presentation
Because ROCK was observed by preliminary MS analysis to bind the CD1d cytoplasmic tail which contains serine and threonine residues, we wanted to examine if CD1d could be a substrate for ROCK. Interestingly, the cytoplasmic tails of human and mouse CD1d (as well as human CD1b) contain the consensus motif for phosphorylation by ROCK [RXXS/T or RXS/T; ref. (30)]. In fact, we found that both ROCK1 and ROCK2 can phosphorylate synthetic peptides corresponding to the cytoplasmic tails of murine and human CD1d (and human CD1b), but not human CD1c, which does not contain the ROCK phosphorylation motif (Fig. 6A, 6B). Thus, it was possible ROCK-mediated phosphorylation of serine and/or threonine residues in the cytoplasmic tail contributed to its control of antigen presentation. We have previously reported that phosphorylation of the human CD1d T322 residue impairs its functional expression (14); however, this threonine is not the one present in the ROCK phosphorylation motif. Knocking down ROCK in HEK293-hCD1d cells expressing CD1d with an altered ROCK phosphorylation motif, still resulted in enhanced recognition by human NKT cells (Fig. 6C). Therefore, these data therefore suggest that ROCK does not regulate antigen presentation by phosphorylating the CD1d cytoplasmic tail.
FIGURE 6.
Potential roles for ROCK-mediated CD1d phosphorylation and RhoGTPases in CD1d-mediated Ag presentation. A, Amino acid sequences of the intracellular, carboxy-terminal tails of human CD1b, CD1c, CD1d and mouse CD1d1. B, ROCK1 and ROCK2 kinase were used in in vitro kinase assays with the indicated CD1 peptides. CREB and PLK peptides served as positive controls. Incorporation of radioactive phosphate into the peptides was measured by scintillation counting and presented in cpm. This result is representative of two independent experiments. C, HEK293-hCD1d cells in which T329 and S330 (part of the ROCK phosphorylation motif) have been mutated to alanines were transfected with ROCK1-specific or control (NC) shRNA, and used in a co-culture assay with human NKT cells. IL-4 and GM-CSF production by human NKT cells was measured by ELISA; *, P = 0.0125 for IL-4 and P = 0.0306 for GM-CSF. This result is representative of three independent experiments. D, LMTK-CD1d1 cells were treated +/− the Rho-specific inhibitor C3 toxin and co-cultured with the representative N37-1A12 NKT cell hybridoma. NKT cell production of IL-2 was measured by ELISA; *, P = 0.0001. This result is representative of two independent experiments.
ROCK effects on CD1d are Rho independent
We showed above that, downstream of ROCK, LIMK and actin form part of an important signaling pathway that regulates CD1d-mediated antigen presentation (Fig. 3 & 4). The GTPase Rho is a major activator of ROCK (31). Therefore, we wanted to determine whether ROCK is regulated by Rho GTPases in the modulation of CD1d-mediated antigen presentation. We used the Rho GTPase-specific inhibitor C3 toxin (32) to measure the effect on CD1d-mediated antigen presentation. Thus, LMTK-CD1d cells were treated with vehicle or the C3 toxin and used as targets for mouse NKT cells. Interestingly, the effects of ROCK on CD1d-mediated antigen presentation appear to be Rho GTPase-independent, as treatment of CD1d+ cells with the Rho-specific C3 toxin actually resulted in a decrease in CD1d-dependent NKT cell activation (Fig. 6D).
Inhibition of ROCK and actin polymerization differentially affects CD1d- and MHC class II-mediated antigen presentation
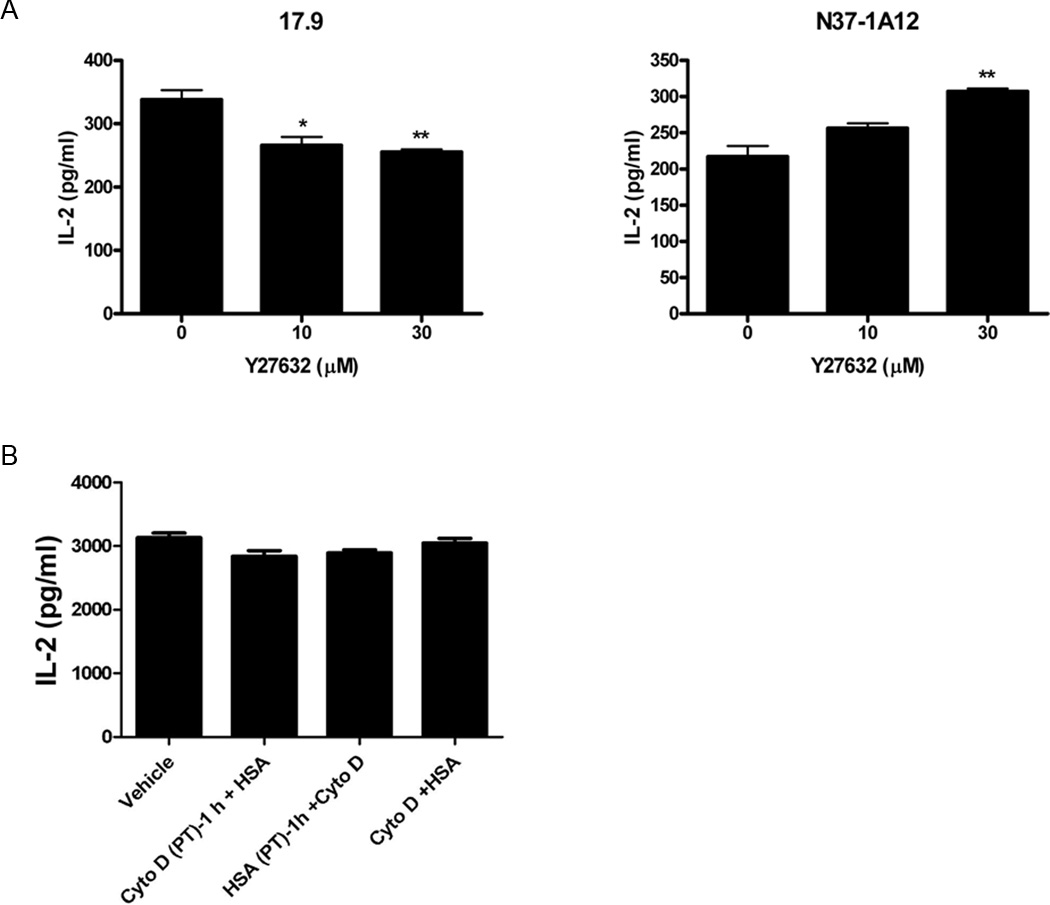
CD1d traffics through the same intracellular compartments as MHC class II molecules (1, 2) (e.g., endosomes, lysosomes). Moreover, similar cell signaling cascades have been shown to be involved in the control of antigen presentation by both CD1d and MHC class II (5, 8, 9, 16, 33). Thus, to determine if ROCK also regulates antigen presentation by MHC class II molecules, L-CD1d-DR4 cells [mouse L-CD1d cells transfected with the human HLA-DR4 cDNA; ref. (4)] were pulsed with the human serum albumin (HSA) protein in the presence of various concentrations of the ROCK-specific inhibitor Y-27632 (or vehicle) as above. In contrast to that observed with CD1d, inhibiting ROCK actually decreased MHC class II-mediated antigen presentation (Fig. 7A). These data indicate that, although ROCK negatively regulates antigen presentation by CD1d, it is a positive regulator of MHC class II-mediated antigen presentation. This is the first system whereby these two antigen presenting molecules can be functionally distinguished in terms of cell signaling pathway requirements. Disrupting actin filaments with cytochalasin D did not significantly alter antigen presentation by MHC class II molecules under the conditions used here, again in contrast to CD1d (Fig. 7B). Therefore, this suggests that the ROCK/actin pathway that negatively regulates CD1d-mediated antigen presentation (see Fig. 8 model) does not affect all antigen presentation pathways in the same way.
FIGURE 7.
ROCK and actin polymerization inhibition differentially affects Ag presentation by CD1d and MHC class II. A, L-CD1-DR4 cells were pulsed with vehicle or HSA in the presence or absence of the ROCK-specific inhibitor Y-27632. The cells were then fixed and co-cultured with the HLA-DR4-specific 17.9 T cell hybridoma (left panel) for analysis of Ag presentation by MHC class II, or with the NKT cell hybridoma N37-1A12 for measuring CD1d-mediated Ag presentation (right panel); *, P < 0.05; **, P < 0.01 as compared to mock-treated. This result is representative of two independent experiments. B, HSA was added to L-CD1d-DR4 cells before, after, or simultaneously with cytochalasin D. The cells were then fixed and co-cultured with the 17.9 T cell hybridoma. IL-2 production was measured by ELISA. This result is representative of two independent experiments.
FIGURE 8.
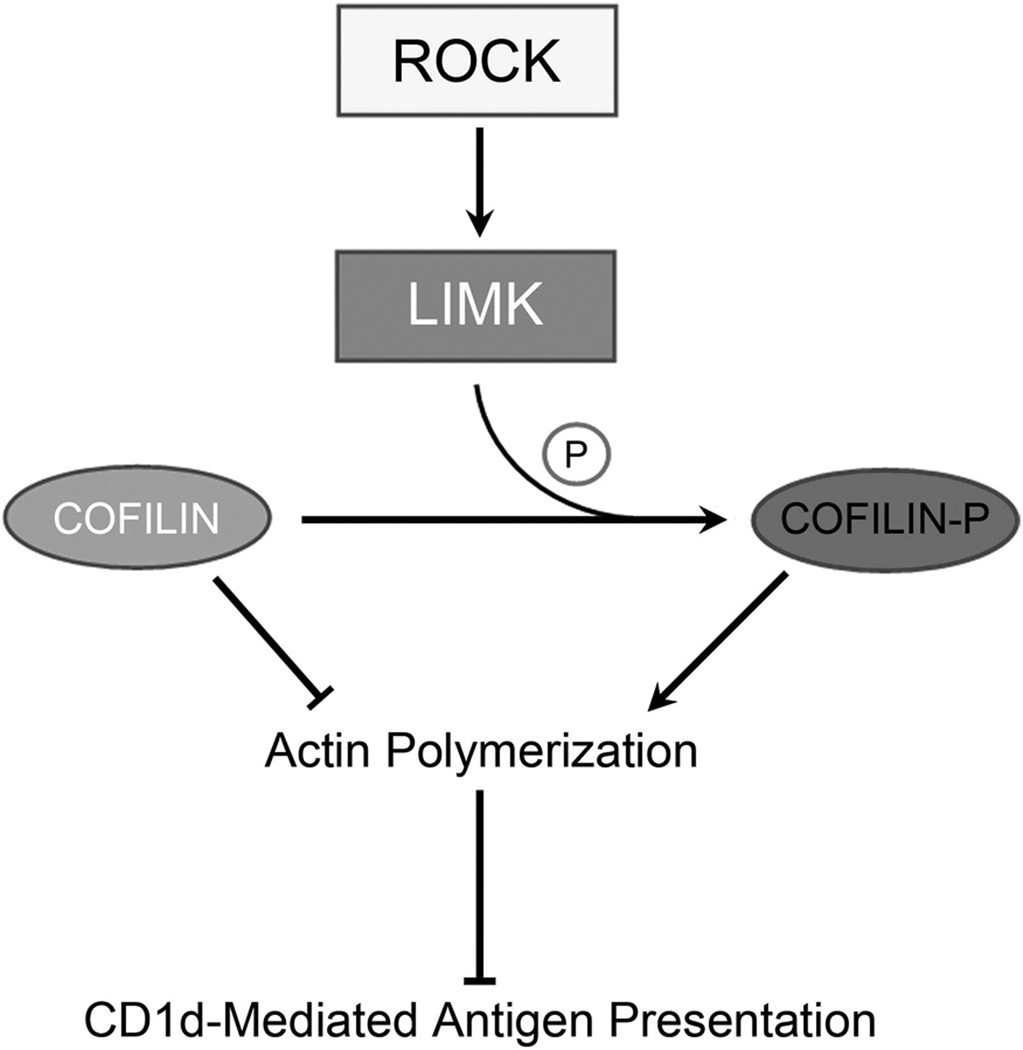
Proposed model of ROCK- and actin cytoskeleton-dependent regulation of Ag presentation by CD1d. The activation of ROCK (in a RhoGTPase-independent manner) phosphorylates LIMK which, in turn, inactivates cofilin. This results in actin polymerization and a concomitant decrease in CD1d-mediated Ag presentation to NKT cells.
Discussion
Here we show that the serine/threonine kinase ROCK is a negative regulator of CD1d-mediated antigen presentation using both pharmacological and genetic approaches. Additionally, we identified the signaling pathway through which ROCK exerts its effects on CD1d. By activating LIMK and thus affecting actin polymerization, ROCK is able to negatively regulate Ag presentation by CD1d. Several bacteria and viruses affect this pathway during infection [reviewed in (34)]. Although it is not the likely reason for these pathogens to target this pathway, CD1d-mediated antigen presentation is consequently affected in the process. Additionally, the ROCK/actin pathway has been shown to be important in several types of cancer including hepatocellular carcinoma (35), lung cancer (36) and glioblastoma (37). It is likely that CD1d-mediated antigen presentation is altered in these tumors. Drugs that inhibit this pathway may have the added benefit of increasing CD1d-mediated antigen presentation and thus potentially enhance immune recognition of the tumor.
We used mass spectrometry screening methods to identify potentially relevant pathways that affect CD1d. Although these screens identified ROCK proteins, this does not necessarily demonstrate a confirmed interaction with CD1d. Subsequent co-immunoprecipitation and Western blotting experiments were not able to clearly demonstrate a complex between CD1d and ROCK. Nonetheless, the mass spectrometry analyses led us to determine the functional role of ROCK proteins in the regulation of Ag presentation by CD1d. Using pharmacological inhibitors, ROCK1-deficient mice, and shRNA methods, we were able to definitively demonstrate that the ROCK pathway is a negative regulator of CD1d-mediated Ag presentation.
We showed that ROCK, LIMK and actin all play a role in the regulation of CD1d-mediated antigen presentation. As seen in the model presented in Figure 8, there is one additional component to this pathway, cofilin. To further link the effects of the inhibition of ROCK and LIMK to actin, we used shRNA to knock down expression of the LIMK target, cofilin. Upon phosphorylation by LIMK, cofilin is inactivated, permitting the polymerization of F-actin (11). Although we were able to decrease cofilin expression in cells by shRNA, this resulted in cells with an abnormal morphology (data not shown). This is likely due to the critical role of cofilin in normal actin dynamics. Thus, we could not directly test the role of cofilin in CD1d-mediated antigen presentation. However, it is well-established that cofilin links LIMK to actin dynamics (11), suggesting that it too is involved in the proposed pathway for the regulation of CD1d-mediated antigen presentation by ROCK.
It is interesting that we did not see changes in CD1d localization or recycling and only minor effects on internalization. Due to the importance of the actin cytoskeleton in trafficking, one would predict that disrupting the ROCK/actin pathway would affect CD1d localization and/or trafficking. It is possible that the effect on CD1d localization is altered in another endocytic compartment other than those that are LAMP-1+. Another possibility is that CD1d localization is not altered; rather, the ROCK/actin pathway is affecting other molecules that are critical for CD1d antigen loading or processing in endosomes. Several lipid-modulating enzymes have been shown to be important for CD1d lipid antigen processing and presentation (38–43). It would be interesting to attempt to determine their intracellular localization and activity under conditions that we have used here to study CD1d. The slight difference in internalization observed when inhibiting ROCK suggests that there is some effect on CD1d directly. It is possible that small changes are below the limit of detection of the assays or do not affect steady-state levels. However, they can have profound effects, as measured by the very sensitive biological assays of antigen presentation. Additional work is required to further dissect where this pathway is causing these changes in CD1d-mediated antigen presentation.
It has been shown that membrane subdomains such as lipid rafts can affect CD1d-mediated antigen presentation (44–46). Although we did not notice any differences in the surface pattern of CD1d in confocal micrographs, it does not rule out the possibility that there are changes that were not detectable in the assays used in this study. However, detailed fractionation of membrane domains or use of additional reagents to study lipid raft colocalization may reveal whether this plays a role in the ROCK/actin effects on the functional expression of CD1d. Additionally, the localization of CD1d (46) as well as the lipid presented (47–50) by it can induce a Th1 or Th2 cytokine bias in the NKT cells. The ROCK/actin pathway may be able to affect CD1d function via altering the lipid presented, resulting in a bias towards the production of Th1 or Th2 cytokines. This could have very important implications for the role of ROCK/actin effects on CD1d-mediated antigen presentation in the context of disease and infection. Additionally, it is possible that the ROCK/actin pathway may play a role in NKT cells as well. Future studies could examine the functionality of NKT cells from ROCK1 KO mice, that would allow us to determine if the major effects of ROCK are only on the antigen presenting cell side, or also on NKT cells.
We showed that mouse and human CD1d, as well as human CD1b, are capable of being phosphorylated by both ROCK1 and ROCK2 kinases. As these sequences contain the ROCK phosphorylation motif, it is not surprising that they are ROCK substrates. By using mutants of the CD1d tail that cannot be phosphorylated, we found that inhibition of ROCK still enhances CD1d-mediated antigen presentation. This suggests that ROCK does not need to phosphorylate CD1d to exert its effects; rather, the mechanism whereby antigen presentation by CD1d is controlled by ROCK is through regulating actin cytoskeleton dynamics.
It was somewhat surprising that the effects of ROCK on CD1d-mediated antigen presentation are Rho-independent. Rho is a canonical activator of ROCK (10). However, there are several other activators and inhibitors of ROCK [reviewed in (51)]. One of these, or some novel regulator, may be upstream of ROCK in this pathway. Nonetheless, there is precedence for Rho-independent, ROCK-mediated effects on cells (52, 53). Most of the known regulators of ROCK other than Rho are isoform-specific (i.e., affecting either ROCK1 or ROCK2). Because we found that both ROCK1 and ROCK2 negatively control CD1d-mediated antigen presentation, it may not be one of these. Rho also can regulate proteins other than ROCK (31). Thus, it is possible that while ROCK negatively regulates CD1d, another Rho effector is a positive regulator. For example, when Rho is inhibited, that may be affecting multiple opposing pathways. The result one would observe would be the net effect of inhibiting these multiple pathways simultaneously. Further studies are required to identify why a reduction in CD1d-mediated antigen presentation is observed when Rho is inhibited.
Recently, it has been reported that another small GTPase, Arl8b, affects CD1d-medited antigen presentation (54). This was via effects on lysosomal trafficking, which is different than what we see with the ROCK/actin pathway. However, this work further shows the importance of these small GTPase signaling pathways in CD1d-mediated antigen presentation.
It is interesting that ROCK and actin do not affect CD1d- and MHC class II-mediated antigen presentation in the same way. We have previously shown that signaling molecules that affect CD1d also affect MHC class II (8, 9, 16). Here, we identify a signaling pathway that distinguishes between the two. Additional work is required to elucidate how and why these two antigen presenting molecules are regulated differently by the ROCK pathway. This result is less surprising in light of the results of the α-GalCer and α-GalGalCer experiments, as these data suggest it may be the antigen processing molecules that are being affected by the ROCK/actin pathway; CD1d and MHC class II are distinct in the molecules they require for antigen processing and presentation (38, 41, 55–57).
In summary, we have shown here that ROCK can have profound effects on antigen presentation via CD1d, by modulating the ROCK-LIMK-actin signaling pathway, leading to changes in the actin cytoskeleton (Fig. 8). A number of pathogens and disease states can alter the actin cytoskeleton; such changes can have a substantial impact on CD1d-mediated antigen presentation in terms of immune evasion as evident in our prior studies with viral infections (5). Thus, this cell signaling pathway is of fundamental importance to a host in controlling a critical component of the innate immune response.
Supplementary Material
Acknowledgements
We thank P. Cohen for the HEK293 cells, C. Mill and D. Riese for the pLKO.1 vector, J. Blum for the 17.9 cells, K. Hayakawa for the N37-1A12 and N38-3C3 cells, P. Savage for his kind gift of galactose(α1→2)galactosylceramide and D. Inerowicz for help with proteomics analyses. We also gratefully acknowledge the NIH Tetramer Facility for α-GalCer (PBS-57)-loaded (and empty) CD1d tetramers.
Abbreviations used
- BMDC
bone marrow-derived dendritic cells
- hCD1d
human CD1d
- HEK293
human embryonic kidney cell line
- HSA
human serum albumin
- KO
knock-out
- LAMP
lysosomal-associated membrane protein
- LIMK
LIM domain kinase
- NC
negative control
- NKT
natural killer T cell
- PKC
protein kinase C
- ROCK
Rho-associated, coiled-coil containing protein kinase
- WT
wild type
Footnotes
This work was supported by NIH grants R01 AI46455 and P01 AI056097 to R.R.B. R.M.G. was the recipient of a NIH T32 postdoctoral fellowship (T32 HL07910).
References
- 1.Brutkiewicz RR. CD1d ligands: the good, the bad, and the ugly. J Immunol. 2006;177:769–775. doi: 10.4049/jimmunol.177.2.769. [DOI] [PubMed] [Google Scholar]
- 2.Brutkiewicz RR, Lin Y, Cho S, Hwang YK, Sriram V, Roberts TJ. CD1d-mediated antigen presentation to natural killer T (NKT) cells. Crit Rev Immunol. 2003;23:403–419. doi: 10.1615/critrevimmunol.v23.i56.30. [DOI] [PubMed] [Google Scholar]
- 3.Kronenberg M. Toward an understanding of NKT cell biology: progress and paradoxes. Annu Rev Immunol. 2005;23:877–900. doi: 10.1146/annurev.immunol.23.021704.115742. [DOI] [PubMed] [Google Scholar]
- 4.Sriram VS, Cho S, Li P, O'Donnell PW, Dunn C, Hayakawa K, Blum JS, Brutkiewicz RR. Inhibition of glycolipid shedding rescues recognition of a CD1+ T cell lymphoma by natural killer T (NKT) cells. Proc Natl Acad Sci U S A. 2002;99:8197–8202. doi: 10.1073/pnas.122636199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Renukaradhya GJ, Roberts Webb TJ, Khan MA, Lin YL, Du W, Gervay- Hague J, Brutkiewicz RR. Virus-induced inhibition of CD1d1-mediated antigen presentation: reciprocal regulation by p38 and ERK. J Immunol. 2005;175:4301–4308. doi: 10.4049/jimmunol.175.7.4301. [DOI] [PubMed] [Google Scholar]
- 6.Brigl M, Brenner MB. How invariant natural killer T cells respond to infection by recognizing microbial or endogenous lipid antigens. Semin Immunol. 2010;22:79–86. doi: 10.1016/j.smim.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Salio M, Silk JD, Cerundolo V. Recent advances in processing and presentation of CD1 bound lipid antigens. Curr Opin Immunol. 2010;22:81–88. doi: 10.1016/j.coi.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 8.Khan MA, Gallo RM, Brutkiewicz RR. Anthrax lethal toxin impairs CD1d-mediated antigen presentation by targeting the ERK1/2 MAPK pathway. Infect Immun. 2010;78:1859–1863. doi: 10.1128/IAI.01307-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brutkiewicz RR, Willard CA, Gillett-Heacock KK, Pawlak MR, Bailey JC, Khan MA, Nagala M, Du W, Gervay-Hague J, Renukaradhya GJ. Protein kinase C δ is a critical regulator of CD1d-mediated antigen presentation. Eur J Immunol. 2007;37:2390–2395. doi: 10.1002/eji.200737124. [DOI] [PubMed] [Google Scholar]
- 10.Riento K, Ridley AJ. ROCKS: multifunctional kinases in cell behaviour. Nat Rev Mol Cell Biol. 2003;4:446–456. doi: 10.1038/nrm1128. [DOI] [PubMed] [Google Scholar]
- 11.Bernard O. Lim kinases, regulators of actin dynamics. Int J Biochem Cell Biol. 2007;39:1071–1076. doi: 10.1016/j.biocel.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 12.Roberts TJ, Sriram V, Spence PM, Gui M, Hayakawa K, Bacik I, Bennink JR, Yewdell JW, Brutkiewicz RR. Recycling CD1d1 molecules present endogenous antigens processed in an endocytic compartment to NKT cells. J Immunol. 2002;168:5409–5414. doi: 10.4049/jimmunol.168.11.5409. [DOI] [PubMed] [Google Scholar]
- 13.Sriram V, Willard CA, Liu J, Brutkiewicz RR. Importance of N-linked glycosylation in the functional expression of murine CD1d1. Immunology. 2008;123:272–281. doi: 10.1111/j.1365-2567.2007.02696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Shaji D, Cho S, Du W, Gervay-Hague J, Brutkiewicz RR. A threonine-based targeting signal in the human CD1d cytoplasmic tail controls its functional expression. J Immunol. 2010;184:4973–4981. doi: 10.4049/jimmunol.0901448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang YM, Bo J, Taffet GE, Chang J, Shi J, Reddy AK, Michael LH, Schneider MD, Entman ML, Schwartz RJ, Schwartz L. Targeted deletion of ROCK1 protects the heart against pressure overload by inhibiting reactive fibrosis. FASEB J. 2006;20:916–925. doi: 10.1096/fj.05-5129com. [DOI] [PubMed] [Google Scholar]
- 16.Khan MA, Gallo RM, Renukaradhya GJ, Du W, Gervay-Hague J, Brutkiewicz RR. Statins impair CD1d-mediated antigen presentation through the inhibition of prenylation. J Immunol. 2009;182:4744–4750. doi: 10.4049/jimmunol.0804311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 18.Hale JE, Butler JP, Gelfanova V, You JS, Knierman MD. A simplified procedure for the reduction and alkylation of cysteine residues in proteins prior to proteolytic digestion and mass spectral analysis. Anal Biochem. 2004;333:174–181. doi: 10.1016/j.ab.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 19.Larkin J, Renukaradhya GJ, Sriram V, Du W, Gervay-Hague J, Brutkiewicz RR. CD44 differentially activates mouse NK T cells and conventional T cells. J Immunol. 2006;177:268–279. doi: 10.4049/jimmunol.177.1.268. [DOI] [PubMed] [Google Scholar]
- 20.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 21.Rodionov DG, Nordeng TW, Pedersen K, Balk SP, Bakke O. A critical tyrosine residue in the cytoplasmic tail is important for CD1d internalization but not for its basolateral sorting in MDCK cells. J Immunol. 1999;162:1488–1495. [PubMed] [Google Scholar]
- 22.Jayawardena-Wolf J, Benlagha K, Chiu YH, Mehr R, Bendelac A. CD1d endosomal trafficking is independently regulated by an intrinsic CD1d-encoded tyrosine motif and by the invariant chain. Immunity. 2001;15:897–908. doi: 10.1016/s1074-7613(01)00240-0. [DOI] [PubMed] [Google Scholar]
- 23.Prigozy TI, Naidenko O, Qasba P, Elewaut D, Brossay L, Khurana A, Natori T, Koezuka Y, Kulkarni A, Kronenberg M. Glycolipid antigen presentation by CD1d molecules. Science. 2001;291:664–667. doi: 10.1126/science.291.5504.664. [DOI] [PubMed] [Google Scholar]
- 24.Brigl M, Brenner MB. CD1: antigen presentation and T cell function. Annu Rev Immunol. 2004;22:817–890. doi: 10.1146/annurev.immunol.22.012703.104608. [DOI] [PubMed] [Google Scholar]
- 25.Chen X, Wang X, Keaton JM, Reddington F, Illarionov PA, Besra GS, Gumperz JE. Distinct endosomal trafficking requirements for presentation of autoantigens and exogenous lipids by human CD1d molecules. J Immunol. 2007;178:6181–6190. doi: 10.4049/jimmunol.178.10.6181. [DOI] [PubMed] [Google Scholar]
- 26.Barral DC, Brenner MB. CD1 antigen presentation: how it works. Nat Rev Immunol. 2007;7:929–941. doi: 10.1038/nri2191. [DOI] [PubMed] [Google Scholar]
- 27.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Deng S, Bai L, Freigang S, Mattner J, Teyton L, Bendelac A, Savage PB. Synthesis of diglycosylceramides and evaluation of their iNKT cell stimulatory properties. Bioorg Med Chem Lett. 2008;18:3052–3055. doi: 10.1016/j.bmcl.2007.12.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Veerapen N, Brigl M, Garg S, Cerundolo V, Cox LR, Brenner MB, Besra GS. Synthesis and biological activitiy of α-galactosyl ceramide KRN7000 and galactosyl (α1-->2) galactosyl ceramide. Bioorg Med Chem Lett. 2009;19:4288–4291. doi: 10.1016/j.bmcl.2009.05.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang JH, Jiang Y, Toita R, Oishi J, Kawamura K, Han A, Mori T, Niidome T, Ishida M, Tatematsu K, Tanizawa K, Katayama Y. Phosphorylation of Rho-associated kinase (Rho-kinase/ROCK/ROK) substrates by protein kinases A and C. Biochimie. 2007;89:39–47. doi: 10.1016/j.biochi.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 31.Hall A. Rho GTPases and the control of cell behaviour. Biochem Soc Trans. 2005;33:891–895. doi: 10.1042/BST20050891. [DOI] [PubMed] [Google Scholar]
- 32.Vogelsgesang M, Pautsch A, Aktories K. C3 exoenzymes, novel insights into structure and action of Rho-ADP-ribosylating toxins. Naunyn Schmiedebergs Arch Pharmacol. 2007;374:347–360. doi: 10.1007/s00210-006-0113-y. [DOI] [PubMed] [Google Scholar]
- 33.Majewski M, Bose TO, Sille FC, Pollington AM, Fiebiger E, Boes M. Protein kinase C δ stimulates antigen presentation by Class II MHC in murine dendritic cells. Int Immunol. 2007;19:719–732. doi: 10.1093/intimm/dxm034. [DOI] [PubMed] [Google Scholar]
- 34.Munter S, Way M, Frischknecht F. Signaling during pathogen infection. Sci STKE. 2006 doi: 10.1126/stke.3352006re5. re5. [DOI] [PubMed] [Google Scholar]
- 35.Chen Y, Wang D, Guo Z, Zhao J, Wu B, Deng H, Zhou T, Xiang H, Gao F, Yu X, Liao J, Ward T, Xia P, Emenari C, Ding X, Thompson W, Ma K, Zhu J, Aikhionbare F, Dou K, Cheng SY, Yao X. Rho kinase phosphorylation promotes Ezrin-mediated metastasis in hepatocellular carcinoma. Cancer Res. 2011;71:1721–1729. doi: 10.1158/0008-5472.CAN-09-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang X, Zhang Y, Wang S, Shi W. Effect of fasudil on growth, adhesion, invasion, and migration of 95D lung carcinoma cells in vitro. Can J Physiol Pharmacol. 2010;88:874–879. doi: 10.1139/y10-047. [DOI] [PubMed] [Google Scholar]
- 37.Deng L, Li G, Li R, Liu Q, He Q, Zhang J. Rho-kinase inhibitor, fasudil, suppresses glioblastoma cell line progression in vitro and in vivo. Cancer Biol Ther. 2010;9:875–884. doi: 10.4161/cbt.9.11.11634. [DOI] [PubMed] [Google Scholar]
- 38.Zhou D, Cantu C, 3rd, Sagiv Y, Schrantz N, Kulkarni AB, Qi X, Mahuran DJ, Morales CR, Grabowski GA, Benlagha K, Savage P, Bendelac A, Teyton L. Editing of CD1d-bound lipid antigens by endosomal lipid transfer proteins. Science. 2004;303:523–527. doi: 10.1126/science.1092009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van den Elzen P, Garg S, Leon L, Brigl M, Leadbetter EA, Gumperz JE, Dascher CC, Cheng TY, Sacks FM, Illarionov PA, Besra GS, Kent SC, Moody DB, Brenner MB. Apolipoprotein-mediated pathways of lipid antigen presentation. Nature. 2005;437:906–910. doi: 10.1038/nature04001. [DOI] [PubMed] [Google Scholar]
- 40.Sagiv Y, Hudspeth K, Mattner J, Schrantz N, Stern RK, Zhou D, Savage PB, Teyton L, Bendelac A. Cutting edge: impaired glycosphingolipid trafficking and NKT cell development in mice lacking Niemann-Pick type C1 protein. J Immunol. 2006;177:26–30. doi: 10.4049/jimmunol.177.1.26. [DOI] [PubMed] [Google Scholar]
- 41.Yuan W, Qi X, Tsang P, Kang SJ, Illaniorov PA, Besra GS, Gumperz J, Cresswell P. Saposin B is the dominant saposin that facilitates lipid binding to human CD1d molecules. Proc Natl Acad Sci U S A. 2007;104:5551–5556. doi: 10.1073/pnas.0700617104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schrantz N, Sagiv Y, Liu Y, Savage PB, Bendelac A, Teyton L. The Niemann-Pick type C2 protein loads isoglobotrihexosylceramide onto CD1d molecules and contributes to the thymic selection of NKT cells. J Exp Med. 2007;204:841–852. doi: 10.1084/jem.20061562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Darmoise A, Teneberg S, Bouzonville L, Brady RO, Beck M, Kaufmann SH, Winau F. Lysosomal alpha-galactosidase controls the generation of self lipid antigens for natural killer T cells. Immunity. 2010;33:216–228. doi: 10.1016/j.immuni.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lang GA, Maltsev SD, Besra GS, Lang ML. Presentation of α-galactosylceramide by murine CD1d to natural killer T cells is facilitated by plasma membrane glycolipid rafts. Immunology. 2004;112:386–396. doi: 10.1111/j.1365-2567.2004.01896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park YK, Lee JW, Ko YG, Hong S, Park SH. Lipid rafts are required for efficient signal transduction by CD1d. Biochem Biophys Res Commun. 2005;327:1143–1154. doi: 10.1016/j.bbrc.2004.12.121. [DOI] [PubMed] [Google Scholar]
- 46.Im JS, Arora P, Bricard G, Molano A, Venkataswamy MM, Baine I, Jerud ES, Goldberg MF, Baena A, Yu KO, Ndonye RM, Howell AR, Yuan W, Cresswell P, Chang YT, Illarionov PA, Besra GS, Porcelli SA. Kinetics and cellular site of glycolipid loading control the outcome of natural killer T cell activation. Immunity. 2009;30:888–898. doi: 10.1016/j.immuni.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miyamoto KS, Miyake S, Yamamura T. A synthetic glycolipid prevents autoimmune encephalomyelitis by inducing TH2 bias of natural killer T cells. Nature. 2001;413:531–534. doi: 10.1038/35097097. [DOI] [PubMed] [Google Scholar]
- 48.Schmieg J, Yang G, Frank RW, Tsuji M. Superior protection against malaria and melanoma metastases by a C-glycoside analogue of the natural killer T cell ligand α-galactosylceramide. J Exp Med. 2003;198:1631–1641. doi: 10.1084/jem.20031192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu KO, Im JS, Molano A, Dutronc Y, Illarionov PA, Forestier C, Fujiwara N, Arias I, Miyake S, Yamamura T, Chang YT, Besra GS, Porcelli SA. Modulation of CD1d-restricted NKT cell responses by using N-acyl variants of - α galactosylceramides. Proc Natl Acad Sci U S A. 2005;102:3383–3388. doi: 10.1073/pnas.0407488102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Velmourougane G, Raju R, Bricard G, Im JS, Besra GS, Porcelli SA, Howell AR. Synthesis and evaluation of an acyl-chain unsaturated analog of the Th2 biasing, immunostimulatory glycolipid, OCH. Bioorg Med Chem Lett. 2009;19:3386–3388. doi: 10.1016/j.bmcl.2009.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi J, Wei L. Rho kinase in the regulation of cell death and survival. Arch Immunol Ther Exp (Warsz) 2007;55:61–75. doi: 10.1007/s00005-007-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coleman ML, Sahai EA, Yeo M, Bosch M, Dewar A, Olson MF. Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nat Cell Biol. 2001;3:339–345. doi: 10.1038/35070009. [DOI] [PubMed] [Google Scholar]
- 53.Sebbagh MC, Renvoize C, Hamelin J, Riche N, Bertoglio J, Breard J. Caspase-3-mediated cleavage of ROCK I induces MLC phosphorylation and apoptotic membrane blebbing. Nat Cell Biol. 2001;3:346–352. doi: 10.1038/35070019. [DOI] [PubMed] [Google Scholar]
- 54.Garg S, Sharma M, Ung C, Tuli A, Barral DC, Hava DL, Veerapen N, Besra GS, Hacohen N, Brenner MB. Lysosomal trafficking, antigen presentation, microbial killing are controlled by the Arf-like GTPase Arl8b. Immunity. 2011;35:182–193. doi: 10.1016/j.immuni.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watts C. Capture and processing of exogenous antigens for presentation on MHC molecules. Annu Rev Immunol. 1997;15:821–850. doi: 10.1146/annurev.immunol.15.1.821. [DOI] [PubMed] [Google Scholar]
- 56.Kang SJ, Cresswell P. Saposins facilitate CD1d-restricted presentation of an exogenous lipid antigen to T cells. Nat Immunol. 2004;5:175–181. doi: 10.1038/ni1034. [DOI] [PubMed] [Google Scholar]
- 57.Cohen NR, Garg S, Brenner MB. Antigen presentation by CD1 lipids, T cells, NKT cells in microbial immunity. Adv Immunol. 2009;102:1–94. doi: 10.1016/S0065-2776(09)01201-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.