Abstract
Thymopoiesis depends on recruitment and expansion of bone marrow-derived progenitors, tight regulation of which is required to maintain T-lineage homeostasis. Lyl1, a transcription factor regulating hematopoietic progenitors, is expressed in thymocyte progenitors until T cell commitment. Here we demonstrate a requirement for Lyl1 in lymphoid specification and the maintenance of early T lineage progenitors (ETPs). Lyl1 deficiency resulted in profound defects in generation of lymphoid primed multipotent progenitors (LMPPs), common lymphoid progenitors (CLPs) and ETPs. Lyl1-deficient ETPs and DN2 thymocyte progenitors showed increased apoptosis, blocked differentiation and impaired expansion. We identified Gfi1 as a critical transcriptional target of Lyl1-mediated T-lymphopoiesis. Thus, Lyl1 is a pivotal component of a transcriptional program that controls lymphoid specification and maintenance of ETPs.
Introduction
The generation of T cells in the thymus crucially depends on the recruitment and expansion of multi-potent bone marrow (BM) derived progenitor cells1. Although the specific identity of the thymus-seeding progenitor is still a matter of debate, studies have identified extensive T lineage potential within a subset of lymphoid-primed multi-potent progenitors (LMPPs) whose hallmark is high expression of Flt32, 3. LMPPs are thought to differentiate into early T lineage progenitors (ETPs), which represent the earliest and most efficient intra-thymic T cell progenitors. ETPs share fundamental characteristics with BM progenitors, such as high expression of c-kit and CD44, absence of mature cell markers and multi-lineage developmental potential4. Upon thymic arrival, ETPs lose the potential to generate B cells whereas myeloid, NK- and dendritic-cell potential is retained within the next thymocyte developmental stage (DN2a)4, 5. ETPs and DN2 thymocytes undergo 1000-fold expansion6, leading to the DN2b stage, at which point developmental potential is T-lineage restricted. At the DN3a stage, proliferation of fully committed T cell progenitors decelerates and the αβ and γδ T lineages diverge.
Although much is known about the transcriptional regulation of lineage fate choices in multi-potent progenitor cells7–11, the mechanisms controlling and maintaining stage-specific progenitors are just beginning to be understood. The distinct transcriptional program observed in thymocyte progenitors at the ETP and DN2 stage suggests sustained regulatory inputs from a core group of transcription factors associated with hematopoietic stem cells (HSCs), including Pu.1, Scl/Tal1, Mef2c, Gata2, Cebpα and Lyl1, which all become sharply down-regulated before the DN3 stage12. These data indicate that the transcriptional circuitry that maintains adult HSC function may also be utilized to sustain intrinsic control over uncommitted thymic progenitors throughout the critical stages of pro-T cell expansion13.
Lyl1 (lymphoblastic leukemia 1) is a basic-HLH transcription factor critically involved in the homeostasis of immature hematopoietic cells14–16. While broadly expressed in the hematopoietic system, expression is highest in progenitors (Lin−Sca1+cKit+, or LSK) and pro B cells, but undetectable in mature T cells17–19. Notably, Lyl1−/− BM cells selectively fail to engraft all lymphoid lineages after transplantation, suggesting a role for Lyl1 in early lymphoid differentiation15, 17.
LYL1 was originally discovered due to its ectopic expression in human t(7;19)(q35;p13)-positive T cell acute lymphoblastic leukemia (T-ALL)20. However, high LYL1 expression is independent of specific genomic alterations and highly correlated with an immature “ETP-like” T-ALL pheno- and genotype and poor prognosis21. Despite this clinical relevance, the function of LYL1 in normal and malignant hematopoiesis is unknown. Murine studies identified a weak oncogenic potential for Lyl1 in T- and B-cell lymphomas, but the mechanism of Lyl1-mediated transformation remains elusive16, 22.
In this study, we demonstrate that Lyl1 is required for lymphoid specification in multi-potent progenitors and for the expansion and survival of ETPs. Collectively our support a model in which Lyl1 regulates a transcriptional program required to control the maintenance of uncommitted T cell progenitors during their expansion in the thymus.
Methods
Mice
Transgenic mice (C57Bl/6-Lyl1-Mg15) were bred and maintained in pathogen free conditions in the animal facility at Baylor College of Medicine (Houston, Texas). For controls we used C57Bl/6 -CD45.1 and CD45.2 isotype mice. All mice were 8–10 weeks of age at the time of analysis. Housing, breeding and experimental use of animals was performed according to the Institutional Animal Care and Use Committee (IACUC) guidelines.
Antibodies
Please see Supplementary Table 3.
Flow cytometry, cell sorting, and population definitions
Single cell suspensions were prepared from spleen, thymus and bone marrow (femoral and tibial bone) by passage through a 70µl cell strainer (Fischer Scientific). Blood was obtained by retro-orbital puncture after isoflourane treatment. Cells were resuspended in HBSS containing 2% FBS. For progenitor analysis cells were stained on ice for 20 minutes with the following fluorochrome antibodies: ETP-DN2-DN3 (thymus): Lineage: (CD3ε, CD8α, TCRβ, TCRγδ, NK1.1, CD11c, Ter119, CD11b, Ly-6G, B220, CD19), c-kit, CD25; HSC-MPP-LMPP-CLP (BM): Lineage: (CD3ε, CD4, CD8α, NK1.1, Ter119, CD11b, Ly-6G, B220), c-kit, sca-1, Flt3, IL-7Rα; CMP-MEP-GMP (BM): Lineage: (CD3ε, CD4, CD8α, NK1.1, Ter119, CD11b, Ly-6G, B220, CD19, IG-M), IL-7Rα, c-kit, sca-1, FcγRII/III and CD34. After the staining, cells were washed and resuspended in HBSS/2%FBS containing propidium iodide for exclusion of dead cells.
For sorting, cells were first stained with biotin conjugated lineage antibodies and then depleted by Magnetic-Activated Cell Separation (Auto-MACS) using streptavidin conjugated MicroBeads (Miltenyi Biotec). Gating strategies were performed as described before44, 45. Analyses were performed using a LSRII (BD) and FlowJo (Tree star) or FACS Diva (BD) software, for cell sorting we used FACSAriaII (BD).
Populations are defined as: HSCs (LSK Flt3neg), MPPs (LSK Flt3low), LMPPs (LSK Flt3high), ETPs (Linneg c-kitpos CD25neg), DN2 (Linnegc-kitposCD25pos) and DN3 (Linnegc-kitnegCD25pos).
Determination of progenitor population size
Absolute viable cell numbers of BM (2 tibias and 2 femurs) and thymuses were determined by Trypan blue exclusion. To determine the absolute number of progenitors in each mice this number was multiplied by the percent of each sub-gate in the viable cell gate (PIneg). To minimize differences in cell numbers caused by animal size, the number of BM progenitors was further normalized to 50×106 total BM cells.
Real-time PCR
Total RNA was extracted using RNAqueous from FACS-sorted populations and treated with DNaseI (Invitrogen). Hereafter, RNA was reverse transcribed using SuperScript III (Invitrogen) with random hexamer primers. CDNA input was standardized and amplifications were performed using either Taqman maser Mix (Applied Biosystems), 18s-rRNA probe (VIC-MGB; Applied Biosystems) and gene specific probes (FAM-MGB; Applied Biosystems) or SYBR Green Mastermix (Applied Biosystems), GAB-DH and gene specific primers for 40 cycles on a AbiPrism 7900HT (Applied Biosystems). Samples were analyzed in triplicate reactions and were normalized to either 18S or GAP-DH expression. Fold-change was determined by the ΔΔCT method. All PCR primers are listed in Supplementary Table 4.
In vitro methylcellulose colony-forming cell assay
For the CFU-E, CFU-GM and CFU-GEMM multi-lineage colony assay, single LMPPs from Lyl1+/+ and Lyl1−/− mice were sorted into 96 well plates containing Methocult M3434 (supplemented with rh erythropoietin (Epo), rm IL-3, rh IL-6 and rm stem cell factor (SCF); StemCell Technologies) and incubated for 12d at 37C° with 5% CO2 and >95% humidity. Colonies were screened at day 9 for BFU-E colonies and analyzed at day 12 of culture using an inverted microscope (Olympus IX70) according to described criteria46. The identity and proportions of cell types represented in individually picked multi-lineage colonies was confirmed by flow cytometry (megakaryocyte (CD41pos), granulocyte (Mac-1pos, Gr1pos, F4/80neg) macrophage (Mac1pos, F4/80pos) and monocytes (Mac1pos, Gr1neg, F4/80neg).
In vitro differentiation assays
OP9-GFP and OP9-DL1 cells were maintained and co-cultured as described47. Sorted LMPPs (250 per well) or transduced sca1pos progenitors (5×105 per well) from Lyl1+/+ and Lyl1−/− were cultured for 12 or 14 days on OP9-GFP and OP9-DL1 cells in the presents of rm Flt3-ligand (20ng/ml) and rm IL-7 (20ng/ml). In addition, LMPPs were also cultured without stromal support in Iscove´s modified Dulbecco´s media supplemented with rm IL-7 (20ng/ml), rm Flt3-ligand (100ng/ml) and rm SCF (50 ng/ml) (Peprotech). After incubation cells were stained for myeloid, B and T cell specific markers and analyzed using a LSRII.
LMPP Transplants
CD45.2 positive Lyl1+/+ and Lyl1−/− LMPPs were sorted (5000 cells per recipient) and transplanted by retroorbital injection into sublethally (1× 5.25 Gy) irradiated recipient mice. At day +14 after transplantation bone marrow, spleens and thymuses were harvested and analyzed for donor output by flow cytometry. Thymuses were depleted of CD45.1 host cells before staining.
Intrathymic Injections
CD45.2 positive Lyl1+/+ and Lyl1−/− LMPPs were sorted (10,000 cells per recipient) and transplanted by intrathymic injection into non-irradiated recipient mice as previously described48. Thymuses were harvested at day +9 after injections and analyzed for T cell linage output by flow cytometry.
Engraftment analysis
T cell output was analyzed in the thymus, so thymocytes were first depleted from host cells by Magnetic-Activated Cell Separation (Auto-MACS) using streptavidin conjugated MicroBeads (Miltenyi Biotec) and CD45.1 biotin conjugated antibodies. After depletion, thymocytes were stained and all remaining cells analyzed by flow cytometry. Spleens and BM were harvested, processed into single cell suspensions and treated with lysis buffer (9× 0.16M NH4CL + 1× 0.17M TRIS pH 7.65) to remove red blood cells prior to staining with myeloid and B cell specific fluorochrome antibodies. Bone marrow- spleen- and thymus-chimerisms were expressed as the percentage of CD45.2 cells in the viable cell gate (PIneg). All other values were listed as percentage of donor cells.
Retroviral transduction of BM cells
Retroviral transduction of BM cells was described previously22. Briefly, sca1-enriched progenitors from 5-FU treated C57Bl/6-CD45.2 Lyl1−/− and Lyl1+/+ mice were transduced with MIG-GFP, MIG-Bcl2 or MIG-Lyl1 (expressing wildtype mouse Bcl2 or Lyl1) and transplanted into lethally irradiated recipients (10.5 Gy in two doses); or cultured in Stempro34 media (Invitrogen). After 36h, cultured GFP expressing cells were sorted and prepared for RT-PCR. From transplant recipient’s peripheral blood, BM and thymuses were analyzed after staining with fluorochrome antibodies by flow cytometry. In addition, complete blood counts were performed using Hemavet HV959FS.
In vivo BrdU Incorporation Analysis
Mice received one intra-peritoneally injection with BrdU (1mg per 6g of mouse weight; Sigma Aldrich) 20h before analysis of BM and thymocyte progenitor populations by flow cytometry. Samples were stained for analysis of BrdU incorporation using the APC-BrdU Flow Kit (BD Pharmingen) according to the manufacturer’s instruction.
In vivo and in vitro apoptosis assays
LSK populations HSCs, MPPs and LMPPs were sorted and cultured for 20h in StemPro media (Stem Cell Technologies) lacking both nutrient supplies and serum. After culture, cells were stained with AnnexinV-APC (Invitrogen) and PI following the manufacturer´s instruction. Thymocytes were harvested and immediately stained with fluorochrome antibodies and AnnexinV-APC for detection of apoptotic cells.
Microarray analysis
LMPPs (2× 104) from Lyl1+/+ and Lyl1−/− mice were sorted by flow cytometry in replicates and RNA was isolated using RNAqueous extraction kit (Ambion). Then, RNA was treated with DNAse I (Invitrogen) and precipitated with phenol:chloroform:isoamyl alcohol (Invitrogen). Next, the RNA was linearly amplified within two rounds of in vitro transcription (T7, MessageAmp, Ambion). During the second round of amplification RNA was labeled with biotin-UTP and –CTP (Enzo Biotech). Labeled RNA was hybridized to Affymetrix MOE430.2 chips following standard protocols. Microarray chips passed quality control test and were further analyzed using DNASTAR Arraystar software. Differentially expressed genes between samples were defined as fold-change >1.6 and adjusted p-value <0.05.
Chromatin Immunoprecipitation
ChIP was performed following standard protocols. Briefly, 1% formaldehyde crosslinked DNA of 1×106 c-kit+ cells from Lyl1+/+ and Lyl1−/− mice was sheared to approximately 200–500 bp fragments using a Bioruptor UCD-200TM-EX (Diagenode). All immunoprecipitations were performed using 2 µg Rabbit IgG (Santa Cruz) or anti-Lyl1 antibody (Goodell lab) aside with Protein A Dynabeads (Invitrogen). Quantification of precipitated genomic DNA relative to input was performed in triplicates after real-time PCR using SYBR Green mastermix (Applied Biosystems). Primer sequences are listed in Supplementary Table 2.
Luciferase Transactivation Assay
The mouse Gfi1 −35kb enhancer element was cloned into pGL3-promotor luciferase vector (Promega). 293T cells were cultured in Dulbecco's modified minimum essential medium (DMEM) supplemented with 10% FBS (fetal bovine serum), penicillin and streptomycin (both 100 µg/ml). Cells were seeded into 24-well plates at 20,000 cells/well and 24h later transiently transfected with luciferase reporter and DNA plasmids using Lipofectamine (Invitrogen) according to the manufacturer's instructions. The total amount of transfected DNA was adjusted to 300 ng/well using the following expression plasmids: pGL3- reporter 100ng, pCDNA-DEST40-Lyl1 100ng, pCDNA-DEST40-Lmo2 30ng, pCDNA-DEST40-Ldb1 30ng and pCDNA-DEST40-E12 30 ng. DNA amounts were maintained constant by adding control plasmid: pCDNA-DEST40-empty. Renilla-TS (10 ng/well) served as an internal control. The cells were lysed after 48h and reporter activity readout was prepared using the dual luciferase assay reagents according to the manufacturer's instructions (Promega). Actual Luciferase activity was measured using a Microlumat LB 96P luminometer (EG&G Berthold's). All assays were at least performed twice in triplicate and the data was normalized to Renilla null luciferase activity and plotted ±s.e.m..
Results
Lyl1 dosage-dependent generation of LMPPs and ETPs
We first determined the level of Lyl1 expression in purified HSCs (LSK Flt3neg), MPPs (LSK Flt3low), LMPPs (LSK Flt3high), ETPs (Linneg c-kitpos CD25neg), DN2 (Linnegc-kitposCD25pos) and DN3 (Linnegc-kitnegCD25pos) thymocytes by quantitative real-time PCR. Lyl1 was highly expressed in BM progenitors MPPs, LMPPs and in ETPs. In contrast, HSCs and DN2 thymocytes expressed 3-fold less Lyl1, while transcripts were undetectable in T-lineage committed DN3 cells (Fig. 1a). We next quantified the BM HSC, MPP and LMPP populations in wild-type, Lyl1+/− and Lyl1−/− mice. The frequency and total number of BM LSK cells is decreased in Lyl1+/− and Lyl1−/− compared to wild-type. However, detailed analyses showed only a mild decrease of HSCs (1.3-fold; p<0.05) and no significant difference in MPPs. In contrast, we found a pronounced decrease in the frequency and absolute number of LMPPs in Lyl1−/− mice (3.9-fold; p<0.0001) (Fig. 1b,c,d), with heterozygous mice showing an intermediate difference, suggesting a Lyl1 dosage-dependent effect.
Figure 1. Lyl1 dosage-dependent generation of LMPPs and ETPs.
(a) Quantitative PCR analysis of Lyl1 expression in sorted HSC, MPP, LMPP, ETP, DN2 and DN3 thymocytes. Data are representative of two experiments with 3 independently sorted populations each analyzed in triplicate (mean and s.d.). (b) Representative flow cytometry analysis of BM from 8-week old wild-type, Lyl1+/− and Lyl1−/− mice. The LSK compartment (upper plots; gated on Linneg cells lineage: CD3, CD4, CD8, NK1.1, TER119, CD11b, Ly-6G, B220) was fractionated on the basis of Flt3 expression (middle plots) for analysis of HSCs, MPPs and LMPPs. Expression of VCAM1 in LMPPs is shown in lower plots. Numbers represent the percent of cells in the indicated gate. (c) Cell numbers of HSCs, MPPs, LMPPs in Lyl1+/− and Lyl1−/− mice relative to wild-type (displayed as 1). Data are representative of 2 experiments with at least 6 mice in each group; bars show the mean ± s.e.m. (d) Absolute cell numbers of LSKs, HSCs, MPPs, LMPPs, ETPs, DN2 and DN3 thymocytes in wild-type, Lyl1+/− and Lyl1−/−; bars show the mean ± s.e.m. (e) Numbers of ETPs, DN2 and DN3 thymocytes in Lyl1+/− and Lyl1−/− mice relative to wild-type (displayed as 1). Data representative of 2 experiments with ≥ 6 mice per group; bars show the mean ± s.e.m. (f) Expression of c-kit and CD25 on Lin− thymocytes (lineage: CD3, CD8, TCRβ, TCRγδ, NK1.1, CD11c, Ter119, CD11b, Ly-6G, B220, CD19). Numbers represent the percent of cells in the indicated gate. (e) Cell numbers of ETPs, DN2 and DN3 thymocytes in Lyl1+/− and Lyl1−/− mice relative to wild-type (displayed as 1). Data representative of 2 experiments with ≥ 6 mice per group; bars show the mean ± s.e.m.; (a–f) * p<0.05, ** p<0.01, *** p<0.001. See methods for population definitions.
Because LMPPs give rise to both CLPs and ETPs, we also quantified these progenitors. We observed significant and Lyl1 dosage-dependent decreases in the percent and total numbers of CLPs (Supplementary Fig. 1), ETPs, DN2 as well as DN3 thymocytes (Fig. 1d,e,f). While BM CLPs were only moderately decreased (Lyl1+/−: 1.5-fold, p<0.05; Lyl1−/−: 1.75-fold; p<0.01), in the thymus, we observed a severe reduction of ETPs (17.5-fold, p<0.001) and almost a complete loss of DN2 thymocytes (97-fold, p<0.001). Additional analysis of erythro-myeloid progenitors (CMP, GMP, MEP) revealed no differences between Lyl1−/− and wild-type mice (Supplementary Fig. 1). These observations demonstrate that Lyl1 is required in a gene dose-dependent manner for the generation of LMPPs, CLPs and ETPs.
Lyl1 restricts MPP and LMPP proliferation and promotes ETP and DN2 survival
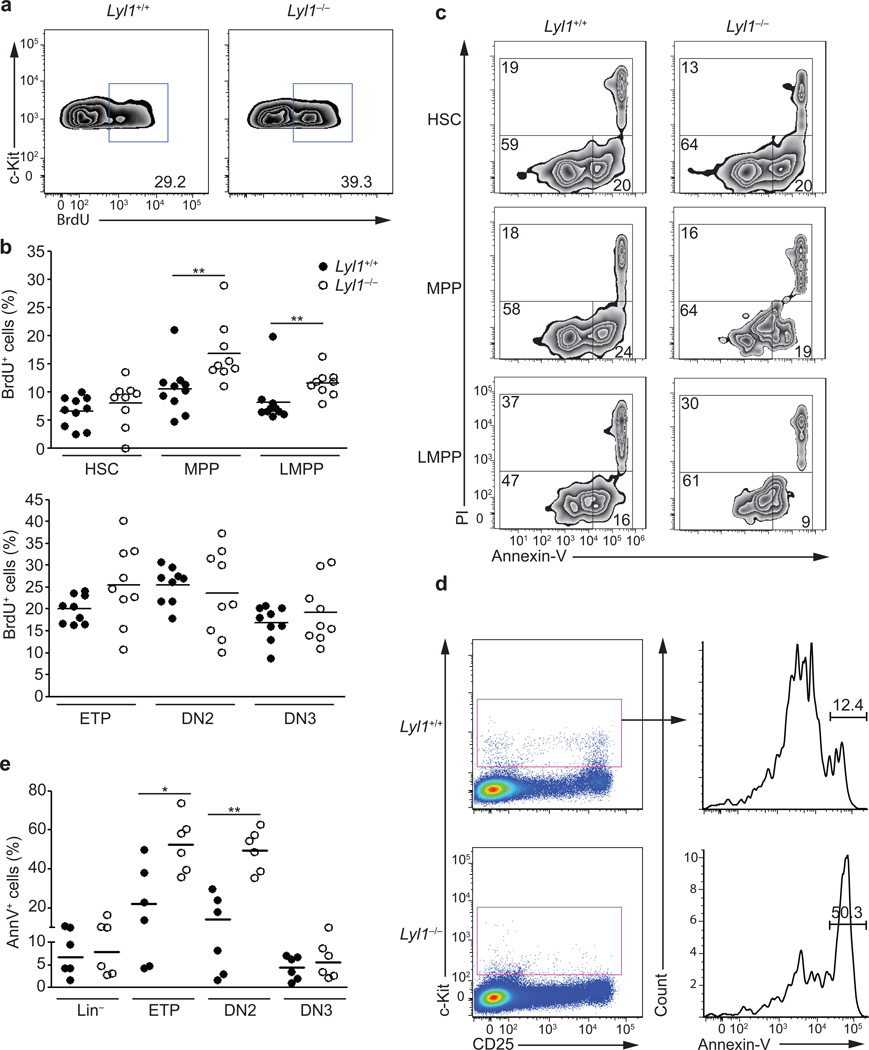
Because LYL1 has oncogenic potential21 and over-expression of Lyl1 in murine BM increased proliferation and restrained apoptosis22, we tested whether Lyl1 was required for the survival and/or expansion of these multi-potent progenitors. BrdU incorporation in Lyl1−/− and wild-type BM and thymic subpopulations showed higher BrdU incorporation in all Lyl1−/− BM subsets examined (HSCs (p=0.06); LMPPs (p=0.02) and MPPs (p<0.001); Fig. 2a,b). In contrast, Lyl1−/− thymocyte subsets (ETPs, DN2 and DN3) revealed a proliferative state comparable to wild-type (Fig. 2b).
Figure 2. Lyl1 restricts MPP and LMPP proliferation and promotes ETP and DN2 survival.
(a) Representative flow cytometry analysis for BrdU incorporation in wild-type and Lyl1−/− BM LSKs are shown after 20h of in vivo exposure to BrdU. The data summarize of 2 independent experiments with 5 mice per group. (b) Fraction of BrdU+ HSCs, MPPs, LMPPs, ETPs, DN2- and DN3-thymocytes after 20h of in vivo exposure to BrdU. Ten wild-type (●) and 9 Lyl1−/− (○) mice were analyzed. (c) Representative flow cytometry analysis for Annexin-V and PI staining on sorted wild-type and Lyl1−/− HSCs, MPPs and LMPPs after 20h culture in serum-free media. Data are representative of 2 independent experiments with ≥ 5 mice/group. Numbers show the percent of cells in the indicated gates: alive (PIneg / Annexin- Vneg); apoptotic (PIneg / Annexin-Vpos); dead (PIpos). (d) Representative flow cytometry analysis for Annexin-V staining of Linneg / c-Kitpos wild-type and Lyl1−/− thymocyte progenitors. (e) Thymocytes from wild-type (●) and Lyl1−/− (○) mice were stained for expression of lineage markers (CD3, CD8, TCRβ, TCRγδ, NK1.1, CD11c, Ter119, CD11b, Ly-6G, B220, CD19), c-kit and CD25. Indicated subsets are displayed for the percentage of Annexin-V+ cells. ETPs, DN2 and DN3 subsets were gated as indicated in Figure 1d. Data represent 2 independent experiments with 3 mice per group. (b and e) Bars indicate the mean; * indicates p<0.01 and ** indicates p<0.001.
Annexin-V binding in in vitro cultures revealed similar frequencies of apoptotic HSCs and MPPs in wild-type and Lyl1-deficient cells, while Lyl1−/− LMPPs showed even lower frequencies of apoptotic cells than wild-type LMPPs (p=0.08; Fig. 2c), suggesting that at the LMPP stage Lyl1 does not control cell death. However, Annexin-V staining was significantly higher in the Lyl1−/− ETP (p=0.01) and DN2 (p<0.001) subpopulations (Fig. 2d,e; Supplementary Fig. 2).
These observations indicate that the reduced number of LMPPs in Lyl1−/− mice is not a consequence of restricted proliferation or increased cell death of the LMPPs or their precursors. Instead, the almost complete loss of ETPs and DN2s in Lyl1−/− mice is at least partially attributable to a role for Lyl1 in survival of these cells.
Impaired T cell development from LMPPs in Lyl1-deficient mice
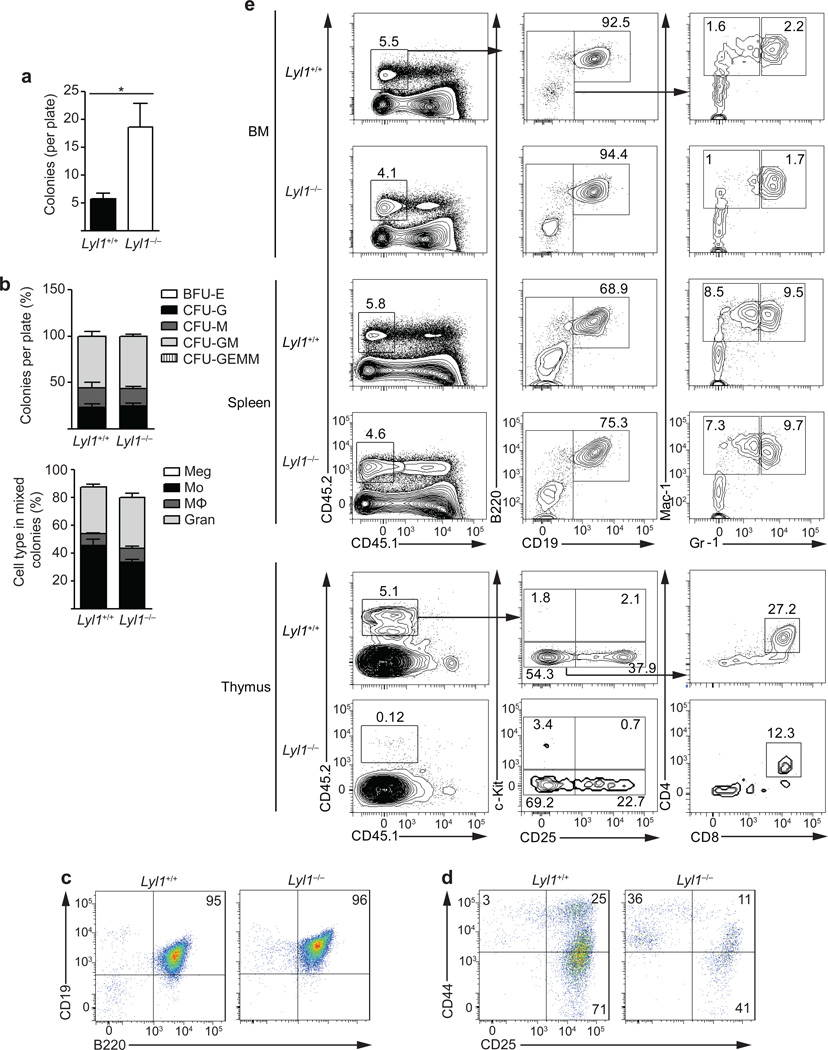
Our observations together with previous data15, 17 indicate that Lyl1 is critical for lymphoid priming of multi-potent progenitor cells. The pronounced depletion of T cell progenitors after Lyl1 loss led us to test its involvement in lineage specification of multi-potent progenitors. The erythro-myeloid differentiation potential of single wild-type and Lyl1−/− LMPPs was compared in methylcellulose assays. Lyl1−/− LMPPs generated larger and significantly more colonies than wild-type LMPPs (Fig. 3a). In these conditions, LMPPs from both genotypes exclusively generated CFU-G, CFU-M or CFU-GM colonies (Fig. 3b). Flow-cytometric analysis verified their MkE lineage restriction via absence of megakaryocytes (Fig. 3b).
Figure 3. Impaired T cell development from LMPPs in Lyl1-deficient mice.
(a) Single wild-type and Lyl1−/− LMPPs were sorted into 96-well plates containing methylcellulose media and colonies were counted and analyzed by microscopy for erythro-myeloid differentiation at day +9 and +12. Data are representative of 3 independent experiments with a total of nine 96-well plates analyzed per group. Bars show the mean ± s.e.m.; * indicates p<0.01. (b) Flow cytometry analysis of cell types represented in wild-type and Lyl1−/− CFU-GM colonies. Data presented are averages of two individual experiments each comprising analysis of 10 individual CFU-GM colonies. (c and d) Lymphoid development potential of 250 sorted wild-type and Lyl1−/− LMPPs after 14 days of culture on OP9-GFP (c) or OP9-DL1 (d) cells in the presence of IL-7 and Flt3-ligand. Numbers indicate the percentage of cells in each quadrant; data are representative of two independent studies performed in triplicate. (e) In vivo lineage potential of 5000 sorted wild-type and Lyl1−/− LMPPs 14 days after intravenous transplantation into sublethally irradiated recipient mice. T-Cell output was examined in the thymus (lower plots), whereas myeloid and B-cell output was measured in both spleens and BM (upper and middle plots). The left column describes the BM, spleen and thymic chimerisms as the percentage of live cells. All other values are listed as percentage of donor cells. Numbers shown in representative plots are a merge of 2 independent experiments with ≥ 6 mice per group.
The B and T cell differentiation potential of wild-type and Lyl1−/− LMPPs was compared on OP9-GFP and OP9-DL1 cells. On OP9-GFP cells, LMPPs from both genotypes generated CD19+B220+ B cells (Fig. 3c). In contrast, Lyl1−/− LMPPs showed limited expansion and delayed differentiation kinetics in comparison to wild-type cells in OP9-DL1 co-cultures. At day +14, wild-type LMPPs had generated DN3 thymocytes, while very few Lyl1−/− LMPPs had reached this stage, exhibiting a partial developmental block at the DN1 to DN2 transition (Fig. 3d).
To examine the in vivo lineage potential of Lyl1−/− LMPPs, 5000 wild-type or Lyl1−/− LMPPs were transplanted intravenously into sublethally irradiated recipients. Fourteen days later, LMPPs from both strains had a similar engraftment in the BM and the spleen of recipients (4–5%) and showed a similar lineage output biased towards B cells (BM >90%; Spleen 68–75%) and low numbers of granulocytes or macrophages. Wild-type LMPPs showed substantial thymic engraftment (5.1%) and robust generation of CD4+CD8+ DP thymocytes, whereas Lyl1−/− LMPPs gained only marginal thymic engraftment (0.12%) and failed to generate T cells (Fig. 3e).
These data suggest that loss of Lyl1 does not affect the myeloid and B lymphoid developmental ability of LMPPs, but severely impairs their potential to develop into T lymphocytes.
Lyl1 is required for ETP to DN2 transition
Because we observed a significantly higher rate of apoptotic ETPs and DN2 thymocytes in Lyl1−/− mice and because Lyl1−/− LMPPs were defective in their ability to differentiate and expand on OP9-DL1 stromal cells, we tested whether Lyl1 is required for thymic progenitor homeostasis. To assess whether the observed phenotypes in Lyl1-deficient progenitors had an impact on later developmental stages in thymopoiesis, we compared the absolute cell numbers of DP and SP thymocytes as well as the frequencies of non-T cell lineages in the thymuses of Lyl1−/− and wild-type mice. On average, the total cell numbers per thymus as well as the DP populations were reduced to 85% of levels found in wild-type mice, whereas the CD4 and CD8 SP populations were unaffected. Comparable to our in OP9-DL1 cultures, we found a partial developmental block in the thymus of Lyl1−/− mice at the DN1 to DN2 transition. Additionally, the frequencies of myeloid (Gr-1+) and NK (NK1.1+) cells in the thymuses of Lyl1−/− mice were slightly higher compared to controls (Supplementary Fig. 3). These indicate a complete restoration of T cell development in Lyl1−/− cells after the DN3 stage, suggesting active compensatory mechanisms in Lyl1−/− thymopoiesis during later T cell development.
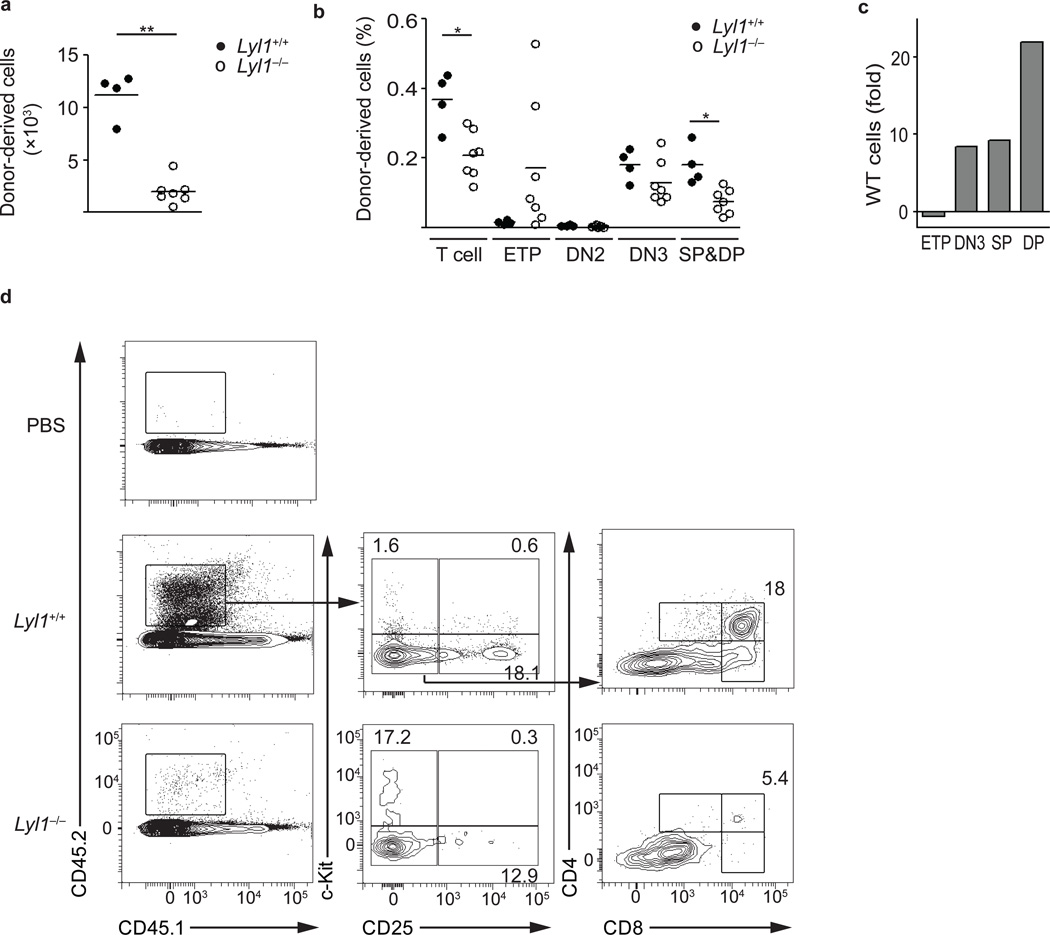
The selective engraftment defect observed in the thymus after intravenous transplantation of Lyl1−/− LMPPs could be due to defective homing of Lyl1-deficient progenitors to the thymus. To directly assess the T-lineage differentiation and expansion potential of Lyl1−/− LMPPs in the absence of homing requirements, we injected sorted LMPPs into the thymus of non irradiated recipients and analyzed their T cell output. Injection of wild-type LMPPs resulted in a 5-fold higher recovery of total donor cells compared to injections of Lyl1−/− LMPPs (p<0.001) (Fig. 4a). Approximately 40% of the donor cells derived from wild-type LMPPs were of T cell lineage (DN3, DP, SP), whereas T-lineage commitment among Lyl1−/− cells was only seen in 20% of the recovered cells (p<0.01). By absolute numbers, wild-type LMPPs had generated more DN3 (8-fold), DP (21-fold) and SP (9-fold) T-lineage committed thymocytes, and fewer wild-type cells remained in the c-kit+ ETP-LMPP-like stage (−0.5-fold) (Fig. 4b,c,d).
Figure 4. In vivo T-lineage potential of Lyl1−/− LMPPs by intrathymic transplantation.
10,000 wild-type and Lyl1−/− LMPPs were sorted and transplanted by intrathymic injection into non-irradiated recipients. Thymuses were harvested at day +9 after transplantation and analyzed for T-lineage output by flow-cytometry. Shown are the combined of two independent experiments. Bars indicate the mean; * indicates p<0.01 and ** indicates p<0.001. (a) Absolute number of donor (CD45.2) cells recovered after intrathymic transplantation. (b) Distribution of donor T-lineage cells listed as a fraction of total thymic donor cells. T-lineage cells comprise the populations DN3, single positive (SP) CD4, SP CD8 and double positive (DP). (c) Fold increase of thymocyte population by absolute cell number after injection of wild-type compared to Lyl1−/− LMPPs. (d) Representative stains and gating strategy for intrathymic analysis. Numbers shown in representative plots are listed as percentage of total thymic donor cells.
To test a requirement of Lyl1 in suppressing alternative lineage fates in ETPs, we also evaluated the myeloid, NK, T and B cell potential of Lin− (CD3ε, CD4, CD8α, NK1.1, Ter119, CD11b, Ly-6G, B220) Lyl1−/− BM cells 3 weeks after intrathymic injection into sublethally irradiated mice. Injection of Lyl1−/− Lin− BM cells resulted in significantly lower T cell engraftment, while slightly inducing intrathymic myeloid development (Supplementary Fig. 4). Although we also confirmed an enhanced myeloid output of Lyl1−/− cells in vitro under conditions promoting both myeloid and lymphoid differentiation (Supplementary Fig. 4), the in vivo effect was too weak to account for the almost complete loss of thymic T cell development observed in transplantation assays. Collectively these data confirm the in vitro OP9-DL1 co-cultures findings, showing delayed differentiation and impaired expansion of Lyl1−/− LMPPs after intrathymic injection and suggest a defect in the ability of Lyl1−/− progenitors to undergo the ETP to DN2 transition. Although a small number of ETP alternatively adopted a myeloid fate, loss of Lyl1 did not support alternative lineage fate choices in the thymus.
Reintroduction of Lyl1 restores the thymic progenitors T lineage fate
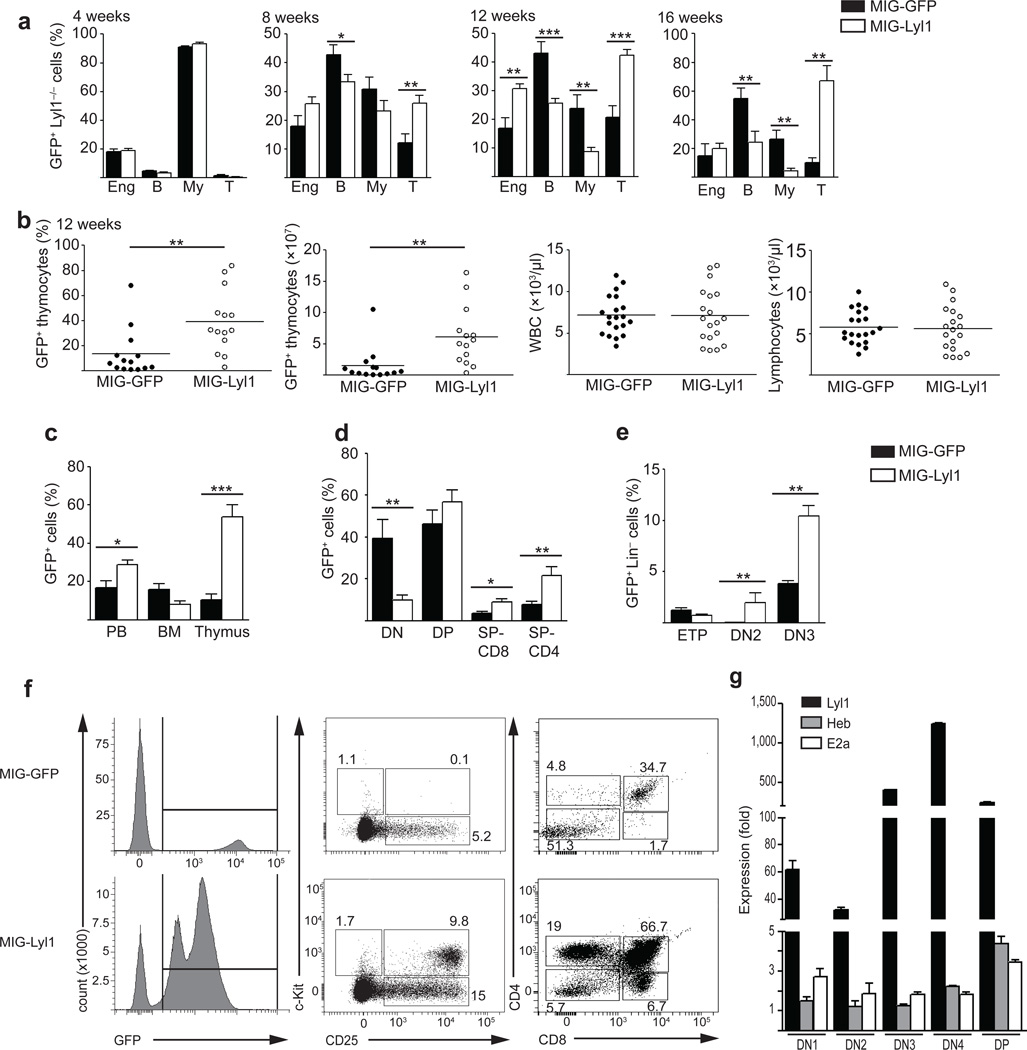
To assess whether the defects in T-lineage differentiation and expansion observed in the Lyl1−/− mice were a direct consequence of the loss of Lyl1 in hematopoietic progenitors, we attempted to rescue this phenotype with retroviral vectors expressing Lyl1. BM progenitors from Lyl1−/− mice (CD45.2) were transduced with retroviral vectors expressing either green fluorescent protein only (MIG-GFP) or Lyl1 and GFP (MIG-Lyl1) and transplanted into lethally irradiated wild-type recipients (CD45.1). After four weeks, the transduction efficiencies and lineage output of MIG-GFP and MIG-Lyl1 retroviruses were comparable (Fig. 5a). After 8 to 12 weeks, there was a significant expansion of MIG-Lyl1 (GFP+) transduced cells that was solely attributable to expansion of T cells (Fig. 5a). At 16 weeks, T cell expansion was saturated and accounted for 70% of the GFP+ white blood cells (Fig 5a). Notably, the peripheral T cells from MIG-Lyl1 transduced transplants appeared normal and mature showing similar percentages of CD4+, CD8+ and TCR_+ subpopulations compared to non-transduced wild-type controls (data not shown), indicating enhanced but normal T cell development.
Figure 5. Reintroduction of Lyl1 restores the thymic progenitors T lineage fate.
(a) Flow cytometry of PB lineage 4-, 8-, 12- and 16-weeks after transplantation of MIG-GFP (control) or MIG-Lyl1 transduced Lyl1−/− progenitors (100,000 Sca1pos cells/recipient). Bars show the mean ± s.e.m. of 1 representative experiment with 20 mice per group. (b) Flow cytometry and absolute cell number of GFPpos thymocytes (61.4 ± 12.5 x106 vs. 15.6 ± 7.3 x106; p=0.003) and complete blood counts of MIG-GFP (●) and MIG-Lyl1 (○) transplants at 12 weeks. Bars indicate the mean. (c, d and e) Distribution of GFPpos cells in the PB, BM and thymus 12 weeks after transplantation of MIG-GFP (black bars) or MIG-Lyl1 (white bars) transduced Lyl1−/− cells. Data are representative of 2 experiments with ≥ 6 mice per group; bars show the mean ± s.e.m.. Populations defined as in 5f. (f) Representative FACS plots of GFPpos thymocytes 12 weeks after transplantation for expression of lineage marker, c-kit, CD25, CD4 and CD8. Left plots are gated on live cells, middle plots on GFP+/Lin− and right plots on GFP+ cells. Numbers in plots indicate percent of GFPpos cells. (g) Quantitative PCR analysis of Lyl1, Heb and E2a mRNA expression from sorted GFPpos thymocyte populations 12 weeks after injection of MIG-Lyl1 transduced BM progenitors. Expression levels are presented relative to endogenous levels determined in 12 week-old wild-type mice. Data are representative of two experiments comprising 4 independently sorted populations for each subset. Samples were analyzed in triplicate and presented as mean ±SD. (a–g) * indicates p<0.05, ** indicates p<0.01, *** indicates p<0.001.
At 12 weeks, thymuses of MIG-Lyl1 transplanted recipients showed significantly greater overall cellularity attributable to a significantly higher proportion of GFP+ thymocytes compared to the MIG-GFP transplanted control group (Fig 5b). Hence, the absolute number of GFP+ thymocytes was also significantly higher in recipients of MIG-Lyl1 transduced cells compared to MIG-GFP recipients. However, the peripheral lymphocyte- and white blood cell counts of these groups were indistinguishable (Fig. 5b). When we compared the contribution of GFP+ cells from MIG-Lyl1 and MIG-GFP transduced BM progenitors to the peripheral blood, BM and the thymus, recipients of MIG-Lyl1 transduced cells had a significantly higher percentage of GFP+ cells in the PB, which was attributable to the enhanced thymic T cell output (Fig. 5c). In contrast, the percent of GFP+ cells in the BM was lower in the MIG-Lyl1 group compared to the MIG-GFP control group, suggesting that Lyl1 overexpression does not enhance expansion of progenitor cells in general, but specifically in the thymus (Fig. 5c). The majority of GFP+ thymocytes in both groups were DP (MIG-GFP 46.2 ± 6.7%; MIG-Lyl1 56.8 ± 5.8%). MIG-GFP transduced Lyl1−/− cells generated significantly lower proportions of CD4 and CD8 SP cells but a higher percentage of DN thymocytes (MIG-GFP 39.5 ± 8.8% vs. MIG-Lyl1 10.1 ± 2.1%; p=0.009) than MIG-Lyl1 transduced cells, confirming that T cell differentiation is impaired in the absence of Lyl1 (Fig. 5d).
While all GFP+ thymocytes in the MIG-GFP recipients were c-kit−, MIG-Lyl1 transduced cells were c-kit+CD25+ in the majority of recipients (Fig. 5e), suggesting that Lyl1-transduced progenitors had undergone the ETP-DN2 transition. In re-transplantation assays, GFP+ thymocytes from MIG-Lyl1 and MIG-GFP failed to engraft after transplantation into irradiated wild-type recipients (data not shown), suggesting that over-expression of Lyl1, unlike Lmo2, cannot induce thymocyte self-renewal.
Quantitative real-time PCR analyses of Lyl1 mRNA in sorted thymocyte subsets (DN1, DN2, DN3, DN4 and DP) from MIG-Lyl1 and MIG-GFP transduced Lyl1−/− thymocytes confirmed high expression of Lyl1 at all stages of thymocyte maturation in MIG-Lyl1 transduced thymocytes, suggesting that superabundant levels of Lyl1 are fully compatible with the T cell developmental program. In contrast, transcripts of potential hetero-dimerization partners of Lyl1, such as E2a and Heb, were only moderately up-regulated (Fig. 5g).
Our demonstrate that reintroduction of Lyl1 in BM progenitors promotes the T cell lineage fate in Lyl1-overexpressing cells due to a complete restoration of the thymic progenitor populations.
Gfi1 is directly regulated by Lyl1
To gain understanding of the underlying mechanisms of Lyl1-mediated lymphoid specification, we performed global gene expression profiling of wild-type and Lyl1−/− LMPPs. We identified 91 genes that were differentially expressed (i.e. > 2-fold difference; p<0.05) with 29 genes up- and 62 genes down-regulated in Lyl1−/− LMPPs (Supplementary Table 1); quantitative real-time PCR on a subset corroborated these data (not shown). To identify direct targets of Lyl1 in LMPPs we looked for differentially expressed genes that contained Lyl1 binding-signals identified previously using ChIP-Seq in murine HPC-7 hematopoietic progenitor cells23. Of the 91 genes with a >2-fold expression difference, 34 contained a Lyl1 binding peak, and of genes with a >1.6-fold differential expression, 81 direct candidates were identified (Supplementary Table 2). Chromatin immunoprecipitation (ChIP) assays on some of these genes (Gfi1, Il15, Selp, Havcr2, Infgr2 and Rap1α) using c-kit-enriched wild-type BM cells confirmed Lyl1 enrichment in vivo (Supplementary Fig. 5).
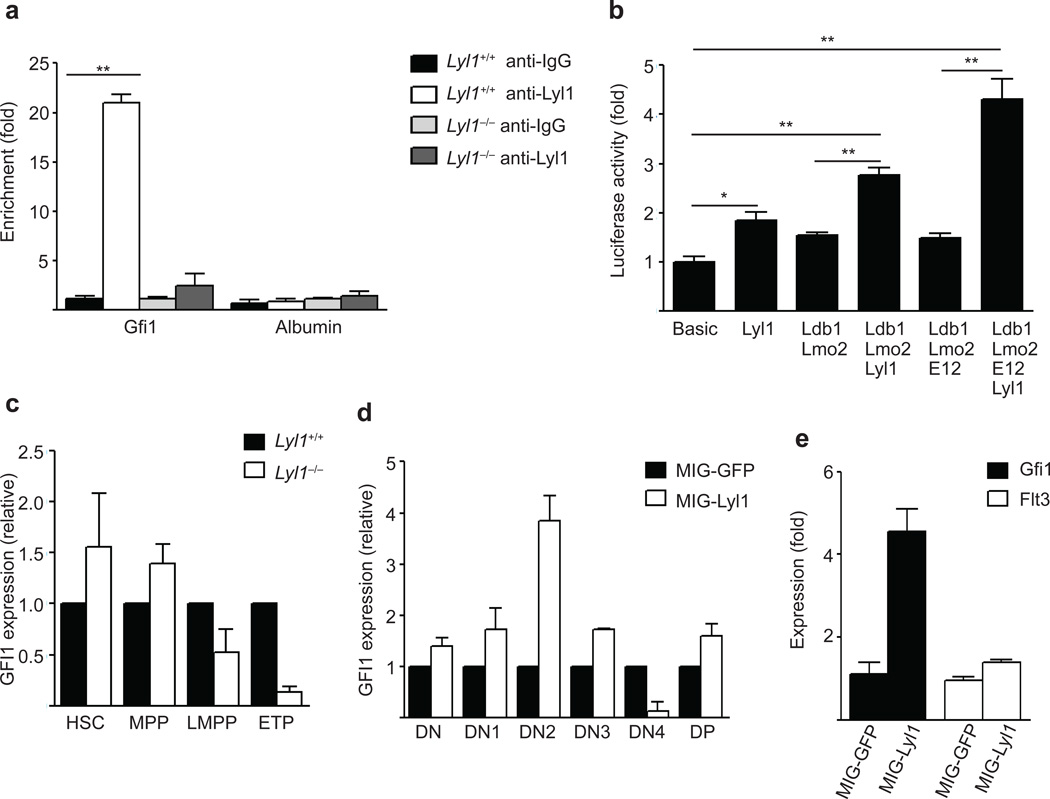
A potential Lyl1 binding peak at a known Gfi1 enhancer located 35kb upstream of Gfi1 (Supplementary Fig. 5), shown to exhibit consistent activity in transgenic embryos24, was of particular interest because Gfi1 is known to have a role in both HSC and early thymocyte homeostasis25, 26. ChIP assays in c-kit+ BM cells revealed a strong enrichment of Lyl1 in wild-type, but not in Lyl1−/− cells. (Fig. 6a). Transactivation assays demonstrated that Lyl1 increased the activity of this enhancer alone or in cooperation with known members of the Scl complex27 including E12, Lmo2, and Ldb1 (Fig. 6b), establishing that Lyl1 can regulate Gfi1 expression.
Figure 6. Gfi1 is directly regulated by Lyl1.
(a) ChIP assay in c-kit positive BM cells from wild-type and Lyl1−/− mice revealed strong binding of Lyl1 at the 35kb Gfi1 enhancer region in-vivo. are representative of 2–4 independent experiments. Quantitative PCR data are presented as means ± SD. (b) Luciferase activity of a pGL3 Gfi1 (−35kb) promoter reporter construct in 293T cells after transfection with control, Lyl1, Lmo2, Ldb1 and E12 expressing plasmids. are representative of 3 independent experiments. Data are presented as means ± SD (c) Quantitative PCR analysis of Gfi1 mRNA expression in sorted wild-type and Lyl1−/− HSCs, MPPs, LMPPs and ETPs. Data are representative of two experiments comprising 4 independently sorted populations for each subset. All samples were analyzed in triplicate, data are presented in mean ± SD. (d) Quantitative PCR analysis of Gfi1 mRNA expression in sorted GFPpos thymocyte populations (DN, DN1, DN2, DN3, DN4 and DP) 12 weeks after injection of MIG-GFP or MIG-Lyl1 transduced Lyl1−/− BM progenitors. Data are representative of two experiments comprising 4 independently sorted populations for each subset. Samples were analyzed in triplicate and presented as mean ±SD. (e) Quantification of Gfi1 and Flt3 mRNA transcripts in wild-type BM progenitors 36 hours after transduction with MIG-GFP (control) or MIG-Lyl1. Data are representative of three experiments and a total of 6 samples per group (mean ± SD). (a–e) * indicates p<0.01, ** indicates p<0.001.
To determine whether Lyl1 promotes T cell lymphopoiesis by regulating Gfi1 expression at the LMPP and ETP stages, we performed quantitative real-time PCR of Gfi1 mRNA in highly purified wild-type and Lyl1−/− subsets of HSCs, MPPs, LMPPs, ETPs, DN1, DN2 and DN3 thymocytes. Gfi1 expression was higher in Lyl1−/− than wild-type HSCs and MPPs, but was decreased by 2- and 7.5- fold respectively in Lyl1−/− LMPPs and ETPs (Fig. 6c). Consistent with these data, Gfi1 expression was also decreased in Lyl1−/− DN1 to DN3 thymocytes by 2- to 3-fold compared to wild-type (data not shown). In addition, Gfi1 expression was augmented in thymocytes derived from MIG-Lyl1 transduced Lyl1−/− cells compared to the MIG-GFP recipients 12 weeks after transplantation (Fig. 6d). Likewise, transduction of wild-type BM progenitors with MIG-Lyl1 induced Gfi1 mRNA expression by >4-fold after 36h in culture, suggesting immediate induction of Gfi1 expression upon Lyl1 dosage boost. In contrast, overexpression of Lyl1 did not induce significant Flt3 expression (Fig. 6e).
Lyl1 controls thymocyte progenitors in part through Gfi1
Next we tested whether Gfi1 controls T cell development downstream of Lyl1 by over-expressing Gfi1 in wild-type and Lyl1−/− cells. However, following BM transplantation of MIG-Gfi1-transduced wild-type or Lyl1−/− cells, abundant levels of Gfi1 were not compatible with lymphoid development, likely due to an immediate induction of both myeloid differentiation (data not shown). Therefore, we intra-thymically injected equal numbers of Lyl1−/− BM progenitor cells immediately after transduction with MIG-GFP, MIG-Gfi1 and MIG-Lyl1 retroviruses. Although the total numbers of GFP+ thymocytes were unaffected, retroviral expression of Gfi1 in Lyl1−/− progenitors permitted lymphoid development and showed a trend towards increased frequencies of T-lineage cells compared to GFP-transduced controls (p=0.1) suggesting that Gfi1 is a critical target of Lyl1 at the ETP stage, enabling thymic T cell lineage development. A more complete rescue was noted after Lyl1 retroviral expression (p<0.001) (Supplementary Fig. 6).
We also used the OP9-DL1 co-culture system to assess the ability of Gfi1 to rescue the Lyl1 deficiency. Transduction of Lyl1−/− cells with retrovirus expressing Gfi1 or Lyl1 promoted lymphocyte expansion and generated DN3 thymocytes after 12 days of culture (Fig. 7a). We also quantified the absolute number of rescued cells following transduction of equal numbers of wild-type and Lyl1−/− cells (Fig. 7 b,c). Although over-expression of Gfi1 increased the generation of T cells derived from Lyl1−/− progenitors significantly (p<0.05), the absolute number of T cells was significantly lower compared to those from Lyl1-transduced Lyl1−/− cells or Gfi1-transduced wild-type cells (Fig. 7b). Interestingly, overexpression of Gfi1 in wild-type progenitors significantly decreased the absolute number of T cells compared to overexpression of GFP or Lyl1, indicating a dose-limiting role for Gfi1 in T lymphoid development (Fig. 7c). Collectively, these data clearly support our finding that Gfi1 acts as a key down-stream target of Lyl1 mediating T cell development.
Figure 7. Lyl1 controls the thymocyte progenitor pool in part through Gfi1.
Rescue of T cell development derived from Lyl1−/− progenitors on OP9-DL1 cells after expression of Gfi1, Bcl2 and Lyl1 (a) 5×105 Sca1pos BM progenitors from Lyl1+/+ and Lyl1−/− mice pretreated with 5-fluorouracil were transduced with MIG-GFP, MIG-Lyl1, MIG-Gfi1 or MIG-Bcl2 retroviruses and cultured on OP9-DL1 stromal cells in the presence of IL-7 and Flt3-L and were analyzed at day 12 for CD44 and CD25 expression. Numbers indicate the percentage of cells in each quadrant; data are representative of three independent studies. (b + c) Shown is the absolute number of rescued CD45.2/GFP+ T cell subsets per well, when equal numbers of transduced Lyl11−/− and Lyl1+/+ progenitors were cultured for day+12 on OP9-DL1 cells in a 24-well plate. Data are representative of three independent experiments comprising at least 3 wells per genotype and viral construct in each experiment. Data are presented as means ± SD.
Because some of our data suggested a role for Lyl1 in protection from cell death (Fig. 2), we also tested whether retroviral over-expression of the pro-survival factor Bcl2 could rescue the Lyl1−/− T-lineage phenotype. In transplantation assays and OP9-DL cultures, Bcl2 promoted development and expansion of T-cells, but flow cytometry analysis of thymocytes in vivo revealed that Bcl2 could not overcome the DN2 progression defect (Supplementary Fig. 7).
Discussion
In this study we show that Lyl1 is a crucial player in the transcriptional network that regulates lymphoid specification of multi-potent BM progenitors and the maintenance of uncommitted T cell progenitors. Our data suggest that Lyl1 gains control over the survival and expansion of thymic progenitors during the critical stages of pro-T cell expansion.
Loss of Lyl1 in the hematopoietic system diminishes the capacity to generate LMPPs, most likely accounting for the profound deficiency in lymphoid engraftment seen after transplantation of Lyl1−/− BM cells15, 17. However, unlike loss of PU.17, 9, 19 or Ikaros11, 17, ablation of Lyl1 still permits generation of LMPPs, although in reduced numbers. Consistent with this, we observed relatively mild changes in gene expression in Lyl1-deficient LMPPs.
These characteristics are reminiscent of LMPPs after loss of Tcfe2a (E2a)8; both Lyl1 and E2A regulate the LMPP population in a dosage-dependent manner and loss of either is associated with reduced apoptosis of LMPPs and increased Bcl2 mRNA8. Since E2a and Lyl1 are basic-HLH TFs, which heterodimerize in BM progenitors28, they may interact during lymphoid priming of MPPs to regulate a set of common target genes. On the other hand, several known E2a target genes10, 29, 30 (e.g. Rag1, Dntt, Notch1 and Notch3) were not differentially expressed in Lyl1−/− LMPPs31, 32, implying that the relationships of Lyl1 and E-proteins change during hematopoietic development31, 32. Accordingly, loss of E2a, in contrast with loss of Lyl1, only in mild HSC defects, suggesting that Lyl1-E2a interactions are not critical in HSCs14, 15, 33. Similarly, during lymphoid differentiation, Ikaros, Pu.1 and Gfi1 are not dependent on E2a for their expression, signifying distinct roles for Lyl1 and E2a in early thymocyte progenitors8, 34. Fully delineating these dynamic relationships would require further analyses.
Our data indicating that Lyl1 is critical to maintain thymopoiesis raises the question of how Lyl1 negatively impacts ETP development. Because ETP numbers are correlated with LMPP numbers35, one simple explanation could be the reduced number of LMPPs, or a requirement of Lyl1 for thymic homing. However, our data demonstrate a key role for Lyl1 after thymic entry in part through activation of Gfi1. The overlap of the phenotypes observed after loss of either Gfi1 or Lyl1– reduced numbers of Flt3high LSKs as well as lymphoid engraftment defects– supports this conclusion25, 26. Moreover, T cell development in both Gfi1- or Lyl1-deficient mice is severely impaired due to increased apoptosis of c-kit+ thymocyte progenitors26, 35. Nevertheless, Lyl1−/− HSCs and MPPs exhibit normal Gfi1 expression and lack the defects seen in Gfi1−/− mice 15, 17, establishing that Lyl1 is not essential for Gfi1 expression before the LMPP stage. Therefore, control of Gfi1 expression in different progenitor populations is likely mediated by multiple transcription factors in addition to Lyl1 in a context-dependent manner.
Interestingly, Scl/Tal1 was also identified by ChIP-Seq to bind the Gfi1 35kb enhancer element24. In BM HSCs, Scl and Lyl1 act redundantly to enable HSC survival14 and may be interchangeable in terms of regulating Gfi1 expression. The down regulation of Scl/Tal1 earlier than Lyl1 during thymocyte development may explain the specific sensitivity of T-progenitors to loss of Lyl1 and the non-redundant function of Lyl1. Other regulators such as Pu.1 and Gata2 that control Gfi1 during myeloid development are also not able to compensate for Lyl1 loss at the LMPP-ETP stage. Collectively, these findings support a model in which Lyl1 becomes increasingly important in lymphoid progenitor development and finally indispensible at the ETP-DN2 stage to maintain T progenitor survival and homeostasis via Gfi1.
Proliferation and survival of ETPs is also highly dependent on IL-7-IL-7R pathway activation of Jak-Stat via Bcl2 expression36, 37. During B-cell development, Gfi1 modulates IL-7 receptor signaling through Socs3, a negative Jak regulator, as well as through direct regulation of IL-7R expression38. Therefore it is possible that similar to B-cell development, Lyl1 controls survival of early thymocytes through Gfi1-dependent regulation of IL-7-IL-7R signaling. This would explain our findings that over-expression of both Gfi1 and Bcl2 partially rescued the Lyl1−/− T cells.
Since its initial description in human T-ALL, LYL1 has been linked to hematologic transformation, but the underlying mechanisms are elusive. Because loss of E2a leads to T cell lymphomas39, Scl/Tal1 and Lyl1 over-expression have been assumed to effect transformation primarily through disrupting E2a homodimers40. However, our offering a distinct mechanism for the involvement of LYL1 in T-ALL, via control of the T-progenitor pool. Given that super-abundant levels of Lyl1 were fully compatible with T cell development also refutes the competitive inhibition model. The role of Lyl1 in expression of Gfi1 and Rap1α suggests a new potential mechanism for Lyl1-mediated malignant transformation. Activation of Rap1α promotes thymocyte proliferation and transformation41, whereas Gfi1 inhibits apoptosis42 and enhances cell-cycle entrance43. These data suggest that LYL1 contributes to transformation via deregulation of critical target genes rather than disruption of E protein function.
Collectively, our data demonstrate that pro-T cell expansion and survival is regulated through intrinsic control of thymic progenitors that employ a transcriptional program already established in hematopoietic stem and progenitor cells. Lyl1 is a critical component of this regulatory network, vital for the maintenance of T lineage homeostasis. Identification of downstream mediators of Lyl1 function illuminates molecular mechanisms underlying early T cell development and suggests previously unrecognized pathways likely to play a role in LYL1-mediated development of leukemia and lymphoma.
Supplementary Material
Acknowledgements
We thank M.K. Brenner for comments on our manuscript. We thank J.C. Zuniga-Pflücker (University of Toronto) for the OP9 cell lines. This work was supported by National Institute of Health (NIH) Grants DK58192, DK092883, CA126752, P30 CA125123, AI007495, the Dan L. Duncan Cancer Center, the UK Medical Research Council and Leukaemia and Lymphoma Research. FZ was supported by a Dr. Mildred Scheel Foundation for Cancer Research fellowship and by a Cancer Prevention and Research Institute of Texas (CPRIT) scholarship (RP101499).
Footnotes
Author contributions
FZ designed and performed most of the experiments, analyzed and interpreted data, and wrote the manuscript. GPS and GLL performed experiments, provided intellectual input and contributed to writing the manuscript. MRI, ML, UG and NKW performed experiments. BG analyzed and interpreted data, provided intellectual input and contributed to writing the manuscript. MAG provided financial support, discussed experimental design, data, and interpretation, and wrote the manuscript.
The authors have no conflicting financial interest to declare.
Reference List
- 1.Donskoy E, Goldschneider I. Thymocytopoiesis is maintained by blood-borne precursors throughout postnatal life. A study in parabiotic mice. J. Immunol. 1992;148:1604–1612. [PubMed] [Google Scholar]
- 2.Krueger A, Willenzon S, Lyszkiewicz M, Kremmer E, Forster R. CC chemokine receptor 7 and 9 double-deficient hematopoietic progenitors are severely impaired in seeding the adult thymus. Blood. 2010;115:1906–1912. doi: 10.1182/blood-2009-07-235721. [DOI] [PubMed] [Google Scholar]
- 3.Schwarz BA, et al. Selective thymus settling regulated by cytokine and chemokine receptors. J. Immunol. 2007;178:2008–2017. doi: 10.4049/jimmunol.178.4.2008. [DOI] [PubMed] [Google Scholar]
- 4.Allman D, et al. Thymopoiesis independent of common lymphoid progenitors. Nat. Immunol. 2003;4:168–174. doi: 10.1038/ni878. [DOI] [PubMed] [Google Scholar]
- 5.Balciunaite G, Ceredig R, Rolink AG. The earliest subpopulation of mouse thymocytes contains potent T significant macrophage, and natural killer cell but no B-lymphocyte potential. Blood. 2005;105:1930–1936. doi: 10.1182/blood-2004-08-3087. [DOI] [PubMed] [Google Scholar]
- 6.Petrie HT, Zuniga-Pflucker JC. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu. Rev. Immunol. 2007;25:649–679. doi: 10.1146/annurev.immunol.23.021704.115715. [DOI] [PubMed] [Google Scholar]
- 7.Dakic A, et al. PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. J. Exp. Med. 2005;201:1487–1502. doi: 10.1084/jem.20050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dias S, Mansson R, Gurbuxani S, Sigvardsson M, Kee BL. E2A proteins promote development of lymphoid-primed multipotent progenitors. Immunity. 2008;29:217–227. doi: 10.1016/j.immuni.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwasaki H, et al. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood. 2005;106:1590–1600. doi: 10.1182/blood-2005-03-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kee BL, Murre C. Induction of early B cell factor (EBF) and multiple B lineage genes by the basic helix-loop-helix transcription factor E12. J. Exp. Med. 1998;188:699–713. doi: 10.1084/jem.188.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshida T, Ng SY, Zuniga-Pflucker JC, Georgopoulos K. Early hematopoietic lineage restrictions directed by Ikaros. Nat. Immunol. 2006;7:382–391. doi: 10.1038/ni1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rothenberg EV, Zhang J, Li L. Multilayered specification of the T-cell lineage fate. Immunol. Rev. 2010;238:150–168. doi: 10.1111/j.1600-065X.2010.00964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat. Rev. Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Souroullas GP, Salmon JM, Sablitzky F, Curtis DJ, Goodell MA. Adult hematopoietic stem and progenitor cells require either Lyl1 or Scl for survival. Cell Stem Cell. 2009;4:180–186. doi: 10.1016/j.stem.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Souroullas GP, Goodell MA. A new allele of Lyl1 confirms its important role in hematopoietic stem cell function. Genesis. 2011 doi: 10.1002/dvg.20743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhong Y, Jiang L, Hiai H, Toyokuni S, Yamada Y. Overexpression of a transcription factor LYL1 induces T- and B-cell lymphoma in mice. Oncogene. 2007;26:6937–6947. doi: 10.1038/sj.onc.1210494. [DOI] [PubMed] [Google Scholar]
- 17.Capron C, et al. The SCL relative LYL-1 is required for fetal and adult hematopoietic stem cell function and B-cell differentiation. Blood. 2006;107:4678–4686. doi: 10.1182/blood-2005-08-3145. [DOI] [PubMed] [Google Scholar]
- 18.Chambers SM, et al. Hematopoietic fingerprints: an expression database of stem cells and their progeny. Cell Stem Cell. 2007;1:578–591. doi: 10.1016/j.stem.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Visvader J, Begley CG, Adams JM. Differential expression of the LYL, SCL and E2A helix-loop-helix genes within the hemopoietic system. Oncogene. 1991;6:187–194. [PubMed] [Google Scholar]
- 20.Mellentin JD, Smith SD, Cleary ML. lyl-1, a novel gene altered by chromosomal translocation in T cell leukemia, codes for a protein with a helix-loop-helix DNA binding motif. Cell. 1989;58:77–83. doi: 10.1016/0092-8674(89)90404-2. [DOI] [PubMed] [Google Scholar]
- 21.Ferrando AA, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1:75–87. doi: 10.1016/s1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- 22.Lukov GL, Rossi L, Souroullas GP, Mao R, Goodell MA. The expansion of T-cells and hematopoietic progenitors as a result of overexpression of the lymphoblastic leukemia gene, Lyl1 can support leukemia formation. Leuk. Res. 2011;35:405–412. doi: 10.1016/j.leukres.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson NK, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 24.Wilson NK, et al. Gfi1 expression is controlled by five distinct regulatory regions spread over 100 kilobases, with Scl/Tal1, Gata2, PU.1, Erg, Meis1, and Runx1 acting as upstream regulators in early hematopoietic cells. Mol. Cell Biol. 2010;30:3853–3863. doi: 10.1128/MCB.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hock H, et al. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature. 2004;431:1002–1007. doi: 10.1038/nature02994. [DOI] [PubMed] [Google Scholar]
- 26.Yucel R, Karsunky H, Klein-Hitpass L, Moroy T. The transcriptional repressor Gfi1 affects development of early, uncommitted c-Kit+ T cell progenitors and CD4/CD8 lineage decision in the thymus. J. Exp. Med. 2003;197:831–844. doi: 10.1084/jem.20021417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lecuyer E, et al. The SCL complex regulates c-kit expression in hematopoietic cells through functional interaction with Sp1. Blood. 2002;100:2430–2440. doi: 10.1182/blood-2002-02-0568. [DOI] [PubMed] [Google Scholar]
- 28.Miyamoto A, Cui X, Naumovski L, Cleary ML. Helix-loop-helix proteins LYL1 and E2a form heterodimeric complexes with distinctive DNA-binding properties in hematolymphoid cells. Mol. Cell Biol. 1996;16:2394–2401. doi: 10.1128/mcb.16.5.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kee BL, Murre C. Transcription factor regulation of B lineage commitment. Curr. Opin. Immunol. 2001;13:180–185. doi: 10.1016/s0952-7915(00)00202-8. [DOI] [PubMed] [Google Scholar]
- 30.Mansson R, et al. Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity. 2007;26:407–419. doi: 10.1016/j.immuni.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 31.Beck K, Peak MM, Ota T, Nemazee D, Murre C. Distinct roles for E12 and E47 in B cell specification and the sequential rearrangement of immunoglobulin light chain loci. J. Exp. Med. 2009;206:2271–2284. doi: 10.1084/jem.20090756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agata Y, et al. Regulation of T cell receptor beta gene rearrangements and allelic exclusion by the helix-loop-helix protein, E47. Immunity. 2007;27:871–884. doi: 10.1016/j.immuni.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 33.Semerad CL, Mercer EM, Inlay MA, Weissman IL, Murre C. E2A proteins maintain the hematopoietic stem cell pool and promote the maturation of myelolymphoid and myeloerythroid progenitors. Proc. Natl. Acad. Sci. U. S. A. 2009;106:1930–1935. doi: 10.1073/pnas.0808866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu W, Kee BL. Growth factor independent 1B (Gfi1b) is an E2A target gene that modulates Gata3 in T-cell lymphomas. Blood. 2007;109:4406–4414. doi: 10.1182/blood-2006-08-043331. [DOI] [PubMed] [Google Scholar]
- 35.Louis I, et al. The signaling protein Wnt4 enhances thymopoiesis and expands multipotent hematopoietic progenitors through beta-catenin-independent signaling. Immunity. 2008;29:57–67. doi: 10.1016/j.immuni.2008.04.023. [DOI] [PubMed] [Google Scholar]
- 36.Ciofani M, Zuniga-Pflucker JC. A survival guide to early T cell development. Immunol. Res. 2006;34:117–132. doi: 10.1385/IR:34:2:117. [DOI] [PubMed] [Google Scholar]
- 37.Jiang Q, et al. Cell biology of IL-7, a key lymphotrophin. Cytokine Growth Factor Rev. 2005;16:513–533. doi: 10.1016/j.cytogfr.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 38.Moroy T, Khandanpour C. Growth factor independence 1 (Gfi1) as a regulator of lymphocyte development and activation. Semin. Immunol. 2011;23:368–378. doi: 10.1016/j.smim.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 39.Bain G, et al. E2A deficiency leads to abnormalities in alphabeta T-cell development and to rapid development of T-cell lymphomas. Mol. Cell Biol. 1997;17:4782–4791. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murre C. Intertwining proteins in thymocyte development and cancer. Nat. Immunol. 2000;1:97–98. doi: 10.1038/77881. [DOI] [PubMed] [Google Scholar]
- 41.Wang SF, et al. Development of Notch-dependent T-cell leukemia by deregulated Rap1 signaling. Blood. 2008;111:2878–2886. doi: 10.1182/blood-2007-07-103119. [DOI] [PubMed] [Google Scholar]
- 42.Grimes HL, Gilks CB, Chan TO, Porter S, Tsichlis PN. The Gfi-1 protooncoprotein represses Bax expression and inhibits T-cell death. Proc. Natl. Acad. Sci. U. S. A. 1996;93:14569–14573. doi: 10.1073/pnas.93.25.14569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karsunky H, Mende I, Schmidt T, Moroy T. High levels of the onco-protein Gfi-1 accelerate T-cell proliferation and inhibit activation induced T-cell death in Jurkat T-cells. Oncogene. 2002;21:1571–1579. doi: 10.1038/sj.onc.1205216. [DOI] [PubMed] [Google Scholar]
- 44.Adolfsson J, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 45.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 46.Nakahata T, Ogawa M. Clonal origin of murine hemopoietic colonies with apparent restriction to granuclocyte-macrophage-megakaryocyte (GMM) differentiation. J. Cell Physiol. 1982;111:239–246. doi: 10.1002/jcp.1041110304. [DOI] [PubMed] [Google Scholar]
- 47.Schmitt TM, Zuniga-Pflucker JC. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17:749–756. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- 48.Inlay MA, et al. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Dev. 2009;23:2376–2381. doi: 10.1101/gad.1836009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.