Abstract
ClpB is a ring-forming, ATP-dependent protein disaggregase that cooperates with the cognate Hsp70 system to recover functional protein from aggregates. How ClpB harnesses the energy of ATP binding and hydrolysis to facilitate the mechanical unfolding of previously aggregated, stress-damaged proteins remains unclear. Here, we present crystal structures of the ClpB D2 domain in the nucleotide-bound and -free states, and the fitted cryoEM structure of the D2 hexamer ring, which provide a structural understanding of the ATP power stroke that drives protein translocation through the ClpB hexamer. We demonstrate that the conformation of the substrate-translocating pore loop is coupled to the nucleotide state of the cis subunit, which is transmitted to the neighboring subunit via a conserved but structurally distinct intersubunit-signaling pathway common to diverse AAA+ machines. Furthermore, we found that an engineered, disulfide cross-linked ClpB hexamer is fully functional biochemically, suggesting that ClpB deoligomerization is not required for protein disaggregation.
Keywords: ATPase, chaperone, Hsp100, protein unfoldase
ClpB is an ATP-dependent protein-remodeling machine that has the remarkable ability to rescue stress-damaged proteins from a previously aggregated state. As the major protein disaggregase in cells, bacterial ClpB and its yeast (Hsp104) and plant (Hsp101) homologs are essential for thermotolerance development (1–3), and for cell survival from acute stress conditions (4).
At the molecular level, ClpB is a multidomain protein composed of two tandem Walker-type ATP-binding domains (AAA+ domains), termed D1 and D2, which drive ClpB’s chaperone activity. The D1 domain features the ClpB-specific M-domain, which forms a long coiled-coil (5) and is essential for protein disaggregation (6, 7). Like other type II AAA+ ATPases, ClpB forms a double-ring structure, with six copies of the D1 and D2 domains each making up a homohexamer ring (5, 8). Although ClpB shares similar quaternary structure with ClpA (9), ClpC (10), and the single-ring ClpX (11) and HslU (12, 13) AAA+ ATPases, which function as the protein unfoldase components of energy-dependent proteases, ClpB does not associate with a chambered peptidase to degrade proteins. Instead, ClpB cooperates with the cognate Hsp70 system (DnaKJ/GrpE) in a species-specific manner (14, 15) to recover functional protein from aggregates (16–18).
The prevailing model suggests that DnaKJ/GrpE targets the ClpB motor activity to aggregates (19, 20), which is consistent with an upstream role of the DnaK system in protein disaggregation (21–23). Once targeted, ClpB disaggregates protein aggregates by extracting unfolded polypeptides (24) and threading them through the ClpB hexamer ring (21, 25). In support of a direct ClpB–DnaKJ/GrpE interaction, it was reported that ClpB interacts with DnaK via the ClpB M-domain (15, 26). Notably, replacing the M-domain of bacterial ClpB with that of its yeast homolog Hsp104 switched the species specificity of the bichaperone system so that ClpB now cooperated with the eukaryotic Hsp70/40 system and vice versa (7, 27). The role of the M-domain in mediating DnaKJ/GrpE interaction is consistent with the M-domain being on the outside of the ClpB hexamer (5, 8), but incompatible with the previously proposed structure of yeast Hsp104 (28, 29), which placed the M-domains on the interior or intercalated between subunits.
Although we recently showed that ClpB and Hsp104 share a similar 3D structure (30), functional differences exist (19, 31–33). Moreover, it remains unclear how ClpB exerts the ATP power stroke to thread substrates through the ClpB hexamer ring, and how ClpB unfolds substrates that are typically much larger than the ClpB hexamer itself. The latter might involve ClpB deoligomerization of a substrate-bound chaperone complex (15, 25, 34), or perhaps other mechanical activities that remodel aggregates prior to substrate translocation (5, 35).
Here, we combine structural and biochemical studies to provide mechanistic insights into the functional role of ClpB in protein disaggregation. Notably, we demonstrate using a disulfide cross-linked ClpB variant that deoligomerization of the ClpB hexamer is not required for protein disaggregation. Furthermore, we found that the active conformation of the D2 substrate-translocating pore loop is controlled in cis by the nucleotide state of the same D2 domain, and demonstrate the existence of a conserved but structurally distinct intersubunit-signaling pathway found in diverse AAA+ ATPases. Our findings suggest that AAA+ machines may utilize a common mechanism to harness the energy of ATP binding and hydrolysis to fuel their diverse biological activities.
Results
Crystal Structure of the ClpB D2 Domain.
To visualize the structural basis for the ATP-driven power stroke, we crystallized the isolated D2 domain (residues 542–854) of Thermus thermophilus ClpB (Fig. S1), featuring an E668A mutation in the Walker B motif. This Walker B mutant can bind nucleotide and, when combined with the analogous E271A mutation in the Walker B motif of the D1 domain, renders ClpB ATP-hydrolysis deficient (8, 36). Three different crystal forms were obtained in the presence or absence of nucleotide, representing a total of seven independent structures of the D2 domain (Fig. 1A and Fig. S2A and Table S1). Notably, crystals obtained in the presence of nucleotide diffracted consistently to higher resolution, but were merohedrally twinned (form I). To determine the crystal structure of the nucleotide-bound conformer, we prepared a selenomethionine (SeMet) derivative of a D2E668A variant that featured three additional point mutations (I683M, L706M, and L770M). Fortuitously, in addition to twinned crystals, this D2 variant also produced a new, untwinned crystal form (form II) in the same drop, which was used for structure determination by molecular replacement. Although the best crystal of the nucleotide-bound D2 domain diffracted to 2.35-Å resolution (form I), the apo structure could not be resolved beyond 4 Å despite extensive efforts to improve the diffraction quality of those crystals (Table S1).
Fig. 1.
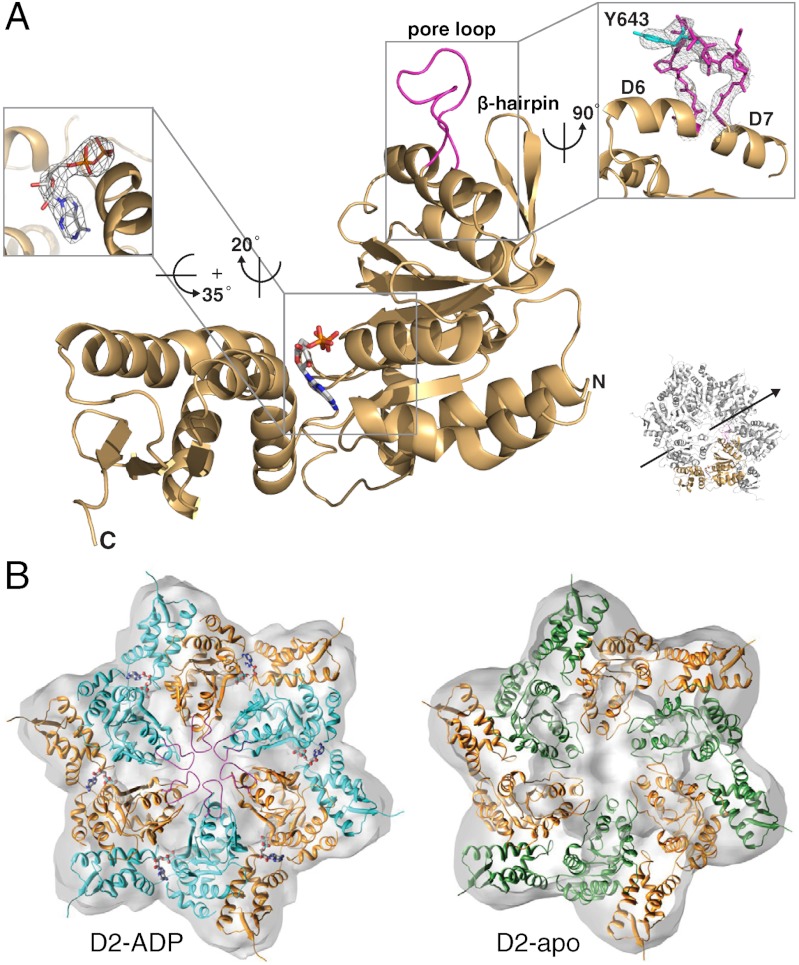
Crystal structure the intact ClpB D2 domain. (A) Ribbon diagram of the D2 domain (gold) with the pore loop colored in magenta (PDB entry code: 4FCV_A). ADP is shown as stick model. The orientation of the D2 domain in the hexamer and relative to the sixfold axis (arrow) is indicated. (Inset, Left) Enlarged view of the nucleotide-binding pocket. The simulated annealed omit map was calculated with ADP omitted and is contoured at 1.5σ. (Inset, Right) Enlarged view of the pore loop, with the substrate-interacting Tyr643 highlighted in cyan. The 2Fo - Fc map is contoured at 1.0σ. (B) Top-down view of the fitted cryoEM structure of the D2-ADP (Left) and apo D2 hexamer (Right). The cryoEM maps are shown as semitransparent surfaces. Neighboring subunits are colored differently for clarity.
As anticipated, the crystal structures of the isolated D2 domain resemble that of the D2 domain in the full-length protein (5) and form a clamp around the nucleotide (Fig. 1A). Surprisingly, despite lacking ATPase activity, all six copies of the nucleotide-bound conformer unequivocally showed bound ADP in simulated annealed omit maps (Fig. 1A), resulting from either spontaneous ATP hydrolysis or presence of ADP in commercial ATP preparations. Indeed, it was previously reported that the ClpB D2 domain binds ADP two orders of magnitude more tightly than ATP (37). The higher affinity for ADP could explain the bound ADP molecule in our crystal structure.
Structural comparison of the six independent representations of the ADP-bound conformer gave a rmsd of 0.54 ± 0.16 Å when superimposed pairwise through their Cα atoms (excluding residues 637–650 and 719–725), indicating that the overall structure and relative orientation of the large α/β and small α-helical subdomains are essentially the same. However, unlike previously determined ClpB structures, the substrate-translocating pore loop (residues 637–650) is ordered in four copies and adopts essentially the same conformation (Fig. 1A and Fig. S2E). The other two copies feature a pore loop that is partially ordered and adopts an alternate conformation because of crystal packing interactions (Fig. S2F). However, the pore loop was completely disordered in the apo structure (Fig. S2 A and B; also, compare Fig. S2 C with D).
Fitted CryoEM Structure of the D2 Hexamer Ring.
To examine the structure of the D2 subunit interface in the ring assembly, we fitted the X-ray structures of the D2-ADP and apo D2 domain into the mass densities that were previously attributed to the D2 ring in the cryoEM reconstructions of full-length ClpB (8). The resulting atomic structure fits are fully compatible with the cryoEM maps (Fig. 1B and Figs. S3 and S4). Our new fits differ from our previous ClpB–AMPPNP model (8) by a 17° rotation along an axis that is almost perpendicular to the sixfold axis of the ClpB hexamer and a 2.5-Å translation from its center of mass for the D2-ADP hexamer, and an 8° rotation and a 2.8-Å translation for the apo D2 hexamer. Notably, in the D2-ADP hexamer structure, the pore loops line the axial channel (Fig. 1B and Fig. S3), which is consistent with their proposed role in substrate translocation (21). The α/β domain of one D2 subunit and the small α-helical domain of its neighbor contribute primarily to the subunit interface, which is similar to the subunit arrangement in the recently determined crystal structure of the ClpC hexamer complex (10) (Fig. S5).
To confirm our hexamer model, we used structure-based mutagenesis to introduce pairs of cysteines at the D2 domain interface in full-length ClpB (Fig. 2A). The rationale behind this approach was to engineer cysteine pairs, which could potentially form a disulfide bond between neighboring subunits to generate a cross-linked ClpB hexamer. It is entirely fortuitous that Thermus thermophilus ClpB lacks cysteines in its amino acid sequence, allowing us to introduce unique cysteine pairs at different locations. Three disulfide-forming ClpB variants were engineered, namely: ClpBR576C/A821C, ClpBL581C/R776C, and ClpBN746C/R810C, as well as a single-cysteine mutant (ClpBR576C) and one that featured a mismatched cysteine pair (ClpBL581C/R726C) (Fig. 2A).
Fig. 2.
Functional analysis of a disulfide cross-linked ClpB hexamer. (A) Position of engineered disulfide pairs, which were introduced into full-length ClpB. Only the D2 ring is shown. Mutation sites are depicted as spheres, and cysteine pairs are colored in different hues: ClpBR576C/A821C in pink and magenta, ClpBL581C/R776C in light and dark green, and ClpBN746C/R810C in light and dark brown. Neighboring subunits are shown in different gray shades for clarity. Bound nucleotide is depicted as stick model. (B) ClpBR576C/A821C, ClpBL581C/R776C, and ClpBN746C/R810C form high molecular weight, cross-linked oligomers in the presence of ATP after 10 min of cross-linking reaction. In contrast, ClpB, ClpBR576C, and ClpBL581C/R726C that feature a mismatched cysteine pair do not. An asterisk marks the product when performing the cross-linking reaction on ice. (C) Time course of catalyzed cross-linking reaction of ClpBR576C/A821C in the presence of ATP. M, marker; C, disulfide cross-linked ClpBR576C/A821C hexamer after 20 min; G, glutaraldehyde crosslinked ClpBE271A/E668A hexamer. (D) Size-exclusion chromatograms of wild-type (wt) ClpB and crosslinked (xl) ClpBR576C/A821C. (E–G) ATPase (E) and coupled chaperone activities (F, G) of cross-linked ClpBR576C/A821C relative to wild-type (wt) ClpB, non-crosslinked ClpBR576C/A821C, and ClpBR576C. Error bars represent standard deviations of three independent experiments. (H) Coupled chaperone activities of cross-linked ClpBR576C/A821C and cysteine-free ClpB variants featuring the L460A or L396A mutation, or both. Error bars represent standard deviations of three independent experiments.
Upon oxidation, all three disulfide-forming ClpB variants formed discrete cross-linked oligomers ranging from dimers to hexamers (Fig. 2 B and C). As anticipated, our single- and mismatched-cysteine mutants remained monomeric or only formed dimers (Fig. 2B). Notably, ClpBR576C/A821C formed stable oligomers with the cross-linked ClpB hexamer as the final product (Fig. 2 C and D and Fig. S6). Cross-linked oligomers were also observed in the absence of nucleotide (Fig. S7A), although ATP facilitated the formation of cross-linked hexamers, which is consistent with its known role in stabilizing the ClpB oligomer (38, 39). We found that our crosslinked ClpBR576C/A821C hexamer, like wild-type ClpB, has ATPase activity, which is further stimulated by κ-casein (Fig. 2E), and, most remarkably, cooperated with DnaKJ/GrpE to rescue heat-aggregated protein substrates (Fig. 2 F and G). The ability to recover functional protein was not caused by dissociation of the cross-linked hexamer, because ClpBR576C/A821C remained cross-linked even after the refolding assay was complete (Fig. S7B).
To ensure that the cross-linking procedure did not alter the structure or function of ClpB, we generated ClpB variants in the ClpBR576C/A821C mutant background by changing Leu396, Leu460, or both, to alanine. We previously showed that mutations of Leu396 and Leu460 reduced ClpB's chaperone activity, presumably by destabilizing the position of the M-domain (5). Consistently, mutating Leu396 and Leu460 to alanine impaired the protein disaggregation activity of cross-linked ClpBR576C/A821C to a similar extent, which was completely abolished when both leucine mutations were present (Fig. 2H).
Taken together, our findings suggest that the cross-linked ClpBR576C/A821C hexamer is structurally and functionally similar to the wild-type ClpB hexamer, and, perhaps more importantly, that ClpB deoligomerization is not required for protein disaggregation.
Arg747 Is a Critical Trans-Acting Element Required for ATP Hydrolysis.
The structure of the ClpB D2 hexamer provides the structural framework to examine how ATP binding and hydrolysis are coupled to conformational changes that drive protein disaggregation. It is widely accepted that the ATPase activities in the ClpB ring complex are regulated by conserved structural elements, which include the cis-acting Walker A and B and sensor 1 and 2 motifs, and the trans-acting arginine-finger (Arg-finger) residue (6, 39–41).
It has been suggested that Arg747 is the Arg-finger residue in T. thermophilus ClpB D2 (41, 42). In our fitted D2 hexamer structure, Arg747 is found at the domain interface and contacts the bound nucleotide in the neighboring subunit (Fig. 3A). To confirm whether Arg747 is the trans-acting Arg-finger residue, we mutated Arg747 to alanine in a full-length ClpB mutant background in which the catalytic glutamate (Glu271) of the first Walker B motif had been additionally mutated to prevent ATP turnover by the D1 domain. We found that the R747A mutation nearly abolished the remaining ATPase activity of ClpBE271A (Fig. 3B), whereas mutating a close-by arginine (Arg741) to alanine did not, confirming that Arg747 is the Arg-finger residue in D2 (41), which is required for the ATPase activity in the D2 ring.
Fig. 3.
Structural determinants coordinating ATP binding and hydrolysis in the D2 ring. (A) Arg747 reaches out from one subunit (gold) into the ATP-binding site of its neighbor (cyan). ADP is shown as stick model. (B) The R747A mutation abolishes the residual ATPase activity of ClpBE271A, whereas the R741A mutation did not, indicating that Arg747 is the Arg-finger. Error bars represent standard deviations of three independent experiments. (C) Stereo diagram of the interaction between the pore loop (magenta) and the β-hairpin (stick model) in the D2-ADP structure. Asp685 is highlighted in green and the β-hairpin loop deletion in blue. The orientation of the pore loop relative to the hexamer and sixfold axis (arrow) is indicated. (D–F) Basal and casein-stimulated ATPase (D) and coupled chaperone activities (E, F) of ClpB, ClpBY643A, and ClpB variants in which structural elements of the ISS motif (ClpBD685A) or β-hairpin (ClpBH693A, ClpBΔ691–695, ClpBΔ690–696/GGG) were mutated.
Structural Basis for the Allosteric Mechanism of ATP Binding and Hydrolysis in ClpB.
Structural comparison of the nucleotide-bound and -free structures showed that the D2 pore loop is ordered when ADP is bound (Fig. 1A and Fig. S2 D and E), but disordered in the absence of nucleotide (Fig. S2 A and C), suggesting that the pore loop conformation is coupled to the nucleotide state of the cis subunit.
In our structure, the D2 pore loop is stabilized by a β-hairpin (residues 688–699), which buttresses the D2 pore loop (Fig. 3C). To examine the functional role of this hairpin, we either deleted the β-hairpin loop (ClpBΔ691–695) or replaced the loop with three glycines (ClpBΔ690–696/GGG). We found that deleting or replacing the loop impaired ClpB’s ATPase and chaperone function to a similar extent as the Y643A mutation (Fig. 3 D–F), which nearly abolished substrate translocation through the ClpB hexamer (21). Our structure also shows that His693 in the β-hairpin loop forms stacking interaction with Tyr646 of the pore loop (Fig. 3C). Notably, mutating His693 to alanine significantly reduced the ATPase activity to 43% of wild-type level (Fig. 3D), concomitant with a small but measurable impact on protein disaggregation (Fig. 3 E–F).
Closer inspection further revealed that the β-hairpin extends from a network of residues, which are part of a signaling cascade responsible for sensing the nucleotide state of the neighboring subunit in the m-AAA protease (43), a distantly related AAA+ machine. An important element of this intersubunit-signaling (ISS) motif is an aspartic acid residue, which, in the m-AAA protease, prevents a subunit from hydrolyzing ATP if the neighboring subunit is also in the ATP-bound state (43). In ClpB, this residue is Asp685 (Fig. 3C). Notably, mutating Asp685 to alanine not only impaired ClpB’s ability to turn over ATP (Fig. 3D), but also significantly reduced its ability to recover heat-aggregated protein substrates (Fig. 3 E and F), indicating that the ISS motif is conserved and has similar function in diverse AAA+ machines.
Discussion
Bacterial ClpB and its eukaryotic homologs are ATP-dependent protein disaggregases, which have the remarkable ability to rescue stress-damaged proteins from a previously aggregated state. Here, we have presented the complete structure of the ClpB D2 domain, featuring an intact D2 pore loop, which couples the ATP power stroke to substrate translocation in cis. Moreover, we demonstrate that deoligomerization of the ClpB hexamer is not required for protein disaggregation, because our engineered, disulfide cross-linked ClpB hexamer is fully functional in recovering active protein from aggregates.
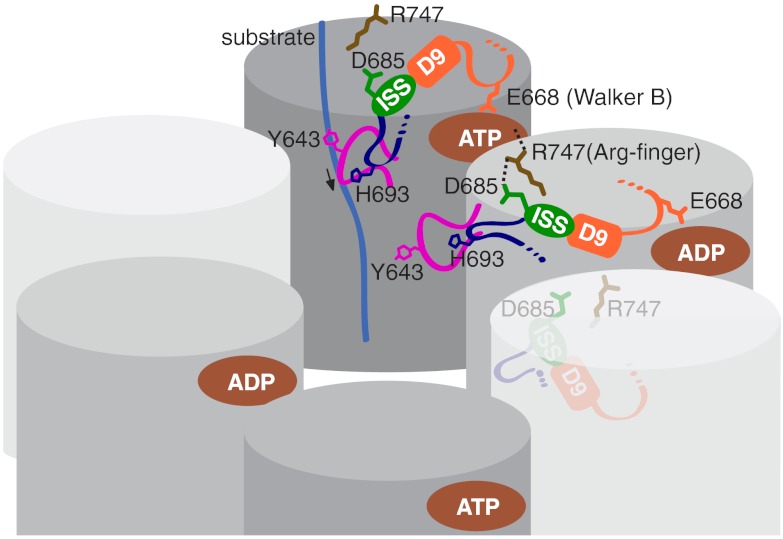
Although AAA pore loops can differ in length and sequence (Fig. S8), they feature a conserved Φ–Xxx–Gly tripeptide motif (21, 44) and share a common function in substrate translocation. In our structure, the conformation of the D2 pore loop is controlled by the cis subunit that senses the nucleotide state of the neighboring subunit through the Arg-finger, Arg747, and transmits this signal to the ISS motif via Asp685 (Fig. 4). From there, the signal traverses to the Walker B motif through helix D9, and to the D2 pore loop via the β-hairpin loop that includes His693. We propose that this signaling network is crucial to sense the nucleotide state in the adjacent subunit and to reset the nucleotide cycle in the ClpB ring following ATP hydrolysis.
Fig. 4.
Model of the intersubunit-signaling cascade in the ClpB D2 ring. Proposed model of the ISS pathway, which regulates the ATP power stroke that drives substrate translocation through the D2 ring. The D2 pore loop is colored magenta and the ISS network orange/green/blue.
The existence of an ISS network that regulates ATP hydrolysis in diverse AAA+ ring complexes is also consistent with a sequential ATP-hydrolysis mechanism proposed for ClpB (45, 46) and Hsp104 (47), with four out of six subunits in the ClpB homohexamer occupied by nucleotides at any one time (46). This model is similar to the staircase mechanism proposed for the T7 gene 4 ring helicase (48), and is consistent with the nucleotide occupancy observed in the crystal structure of an engineered, covalently linked ClpX hexamer (11). In our model (Fig. 4), the unfolded polypeptide is bound to ClpB in the ATP-bound state, which displays the highest substrate-binding affinity (8, 36). ATP hydrolysis triggers substrate translocation along the hexamer axis. Once hydrolyzed, release of ADP results in substrate dissociation from the cis subunit, concomitant with binding of the unfolded polypeptide by the ATP-bound neighboring subunit. The cycle repeats itself until the substrate is fully translocated through the hexamer ring.
Finally, the conservation of the ISS motif in diverse AAA+ ATPases also suggests that similar structural elements may drive the ATP power stroke in other AAA+ machines, and support a common, ATP-fueled mechanism underlying the distinct cellular activities of diverse AAA+ ATPases.
Materials and Methods
Site-Directed Mutagenesis.
T. thermophilus ClpBE271A, ClpBE271A/R747A, ClpBE271A/R741A, ClpBR741A, ClpBR576C, ClpBR576C/A821C, ClpBL581C/R776C, ClpBL581C/R726C, ClpBN746C/R810C, ClpBL396A, ClpBL460A, ClpBL396A/L460A, ClpBL396A/R576/A821C, ClpBL460A/R576/A821C, ClpBL396A/L460A/R576C/A821C, ClpBD685A, ClpBY643A, ClpBH693A, ClpBΔ691–695, ClpBΔ690–696/GGG, and D2E668A/I683M/L706M/L770M were generated by site-directed mutagenesis from ClpB, ClpBE271A, or D2E668A (residues 542–854). Constructs were cloned into pET24a, which harbors a noncleavable C-terminal His6-tag to facilitate protein purification. In cases where multiple mutations were introduced, site-directed mutagenesis was carried out stepwise. All constructs were verified by DNA sequencing.
Protein Expression and Purification.
Plasmids harboring full-length ClpB, ClpB variants, or D2E668A were transformed into Escherichia coli BL21 CodonPlus (DE3)-RIL cells, and were cultured in 2xYT media supplemented with 50 μg/mL kanamycin and 34 μg/mL chloramphenicol to select for plasmids. Cells were grown at 37 °C to midlog phase, induced with 0.5 mM isopropyl β-d-thiogalactopyranoside (IPTG), and harvested after 12 h of continuous growth at 24 °C. Proteins were purified by column chromatography using Ni-NTA sepharose followed by Superdex-200 for full-length ClpB and Q-sepharose for D2E668A. SeMet-substituted D2E668A/I683M/L706M/L770M was prepared by transforming E. coli B834 (DE3) cells. Cells were grown at 37 °C for 16 h in minimal media containing 40 μg/ml SeMet, induced with 0.5 mM IPTG, and allowed to grow for another 8 h at 37 °C. Proteins were purified by Ni-NTA sepharose followed by Q-sepharose. Protein concentrations were determined as described (49), using a molar extinction coefficient of 51,780 M-1·cm-1 for full-length ClpB and 14,650 M-1·cm-1 for D2 variants. T. thermophilus DnaKJ/DafA and GrpE were purified essentially as described (16).
Crystallization.
The nucleotide-bound D2 complex was crystallized by the hanging-drop vapor-diffusion technique by mixing 4 μl of D2E668A or SeMet-labeled D2E668A/I683M/L706M/L770M protein (10 mg/ml in 40 mM Tris-HCl pH 7.5, 100 mM KCl, 10% glycerol, 5 mM ATP, 5 mM MgCl2) with 4 μl of reservoir solution consisting of 100 mM sodium citrate pH 5.5 and 10% isopropanol (form I), or 200 mM NH4OAc and 20% PEG 3350 (form II). Crystals of the apo form were obtained similarly by mixing 4 μl of D2E668A protein (10 mg/ml in 40 mM Tris-HCl pH 7.5, 100 mM KCl, 10% glycerol) with 4 μl reservoir solution [200 mM (NH4)2SO4, 100 mM Tris-HCl pH 8.0, 25% PEG 3350, 20 mM 1,6-hexanediol].
Data Collection, Processing, and Refinement.
For data collection, crystal form I and form II were harvested in mother liquor supplemented with 50% (vol/vol) PEG 200, and 30% (vol/vol) glycerol, respectively. Apo crystals were flash-frozen in mother liquor containing an additional 10% (vol/vol) glycerol. X-ray diffraction data were collected at the NSLS X25 (form I, D2E668A/I683M/L706M/L770M), and APS–SBC ID-19 beamlines (form I, D2E668A, form II, and Apo). Data were processed and scaled using HKL2000 (50). Crystal structures of the nucleotide-bound and -free D2 domains were determined by molecular replacement using Phaser in CCP4 (51). Structure refinement and map calculations were done using CCP4 (51) and CNS (52), and were interspersed with manual rebuilding of the model using Coot (53). Finally, structures were refined by TLS and gradient minimization, using multigroup TLS models generated by the TLSMD web server (54). Figures were generated with PyMOL (55) and UCSF Chimera (56).
CryoEM Fit.
The crystal structure of D2-ADP was superimposed onto the D2 domain of the previous ClpB–AMPPNP hexamer fit (8). This arrangement resulted in steric clashes between neighboring D2 pore loops. Therefore, the fit of the D2 monomer was adjusted manually, keeping the D2 hexamer ring within the boundary of the cryoEM density (EMD-1244). The apo D2 domain was fitted similarly into the apo ClpB cryoEM density (EMD-1241).
Disulfide Cross-Linking.
Engineered cysteine-containing ClpB mutants (1.5 mg/ml) in 40 mM Tris-HCl pH 7.5, 100 mM KCl, 10% glycerol were incubated at 55 °C in the presence of 5 mM ATP, 5 mM MgCl2, 10 μM CuCl2, and 10 μM 1,10-phenanthroline for 20 min unless indicated otherwise. Reactions were quenched by adding 20 mM (final) EDTA. Formation of disulfide bonds was followed by nonreducing 3–8% Tris-acetate gradient PAGE.
Analytical Size-Exclusion Chromatography.
ClpB and ClpBR576C/A821C were analyzed on a Superdex 200 10/300 size-exclusion column preequilibrated with 40 mM MOPS-NaOH pH 7.5 and 100 mM KCl.
ATPase Assay.
ATPase activities were measured with or without 0.1 mg/mL κ-casein at 50 °C in 40 mM MOPS–NaOH pH 7.5, 150 mM KCl, 10 mM MgCl2 using 0.1–0.5 μM ClpB (monomer), 10 mM ATP, and a coupled ATP-regenerating system consisting of 0.2 μM pyruvate kinase, 0.5 μM lactate dehydrogenase, 0.5 mM phosphoenolpyruvate, and 0.2 mM NADH. ATP-hydrolysis rates were calculated from the oxidation of NADH, which was monitored by absorbance at 340 nm.
Protein Disaggregation Assays.
Recombinant Bacillus stearothermophilus α-glucosidase (0.2 μM) was heat-denatured in refolding buffer A (25 mM HEPES–NaOH pH 7.5, 150 mM KCl, 10 mM MgCl2, 5 mM ATP) at 73 °C for 12 min in the presence of 0.2 μM DnaKJ–DafA complex (trimer) and 0.1 μM GrpE (monomer). The aggregate was immediately diluted 20-fold into refolding buffer A containing 0.3 μM ClpB (monomer), 0.2 μM DnaKJ-DafA complex (trimer), and 0.1 μM GrpE (monomer), and incubated at 55 °C for 40 min. To measure the recovered α-glucosidase activity, p-nitrophenyl glucopyranoside was added to a final concentration of 2 mM. The reaction mixture was incubated at 55 °C for 20 min and stopped with 0.4 M (final) sodium carbonate. The amount of p-nitrophenol released was measured by absorbance at 400 nm.
Purified enhanced GFP (EGFP, 3 μM) was heat-denatured in refolding buffer A at 80 °C for 10 min in the presence of 0.2 μM DnaKJ–DafA complex (trimer) and 0.1 μM GrpE (monomer). The aggregate was immediately diluted 12-fold into buffer A containing 0.3 μM ClpB (monomer), 0.2 μM DnaKJ-DafA complex (trimer), and 0.1 μM GrpE (monomer). The amount of refolded EGFP was monitored continuously at 25 °C for 40 min using a LS55 fluorescence spectrometer at an excitation wavelength of 488 nm and emission wavelength of 510 nm.
Recombinant firefly luciferase (1 μM) was heat-denatured in refolding buffer B [25 mM MOPS–NaOH pH 7.5, 150 mM KOAc, and 10 mM Mg(OAc)2] at 42 °C for 30 min in the presence of 5 mM ATP and 1 μM DnaKJ/GrpE (monomer). The aggregate was immediately diluted 10-fold into refolding buffer B containing 1 μM ClpB (monomer), 1 μM DnaKJ/GrpE (monomer), and an ATP-regenerating system consisting of 5 mM ATP, 20 mM phosphoenolpyruvate, and 0.2 μM pyruvate kinase. Reactions were incubated at 25 °C for 2 h, and the amount of recovered enzymatic activity was measured at 560 nm after addition of 0.1 μM luciferin.
Supplementary Material
ACKNOWLEDGMENTS.
We thank J. Lee for purified EGFP, J. Choi for the ClpBE271A and D2E668A constructs, and A. Reger and the staff of NSLS X25 and APS-SBC 19ID for help with data collection. This work was supported by grants from National Institutes of Health (R01AI076239, P41RR012408, P41GM103473) and the Welch Foundation (Q-1530). Use of the NSLS and APS was supported by the Department of Energy, Office of Biological and Environmental Research, under Contract No. DE-AC02-98CH10886 and DE-AC02-06CH11357, respectively. A.B.B. was a Welch Predoctoral Fellow.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207040109/-/DCSupplemental.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4FCT, 4FCV, 4FCW, and 4FD2).
References
- 1.Sanchez Y, Lindquist S. Hsp104 required for induced thermotolerance. Science. 1990;248:1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- 2.Eriksson MJ, Clarke AK. The heat shock protein ClpB mediates the development of thermotolerance in the cyanobacterium Synechococcus sp. strain PCC. J Bacteriol. 1996;178:4839–4846. doi: 10.1128/jb.178.16.4839-4846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Queitsch C, Hong SW, Vierling E, Lindquist S. Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell. 2000;12:479–492. doi: 10.1105/tpc.12.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanchez Y, Taulien J, Borkovich KA, Lindquist S. Hsp104 is required for tolerance to many forms of stress. EMBO J. 1992;11:2357–2364. doi: 10.1002/j.1460-2075.1992.tb05295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee S, et al. The structure of ClpB: A molecular chaperone that rescues proteins from an aggregated state. Cell. 2003;115:229–240. doi: 10.1016/s0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- 6.Mogk A, et al. Roles of individual domains and conserved motifs of the AAA+ chaperone ClpB in oligomerization, ATP hydrolysis, and chaperone activity. J Biol Chem. 2003;278:17615–17624. doi: 10.1074/jbc.M209686200. [DOI] [PubMed] [Google Scholar]
- 7.Sielaff B, Tsai FTF. The M-domain controls Hsp104 protein remodeling activity in an Hsp70/Hsp40-dependent manner. J Mol Biol. 2010;402:30–37. doi: 10.1016/j.jmb.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee S, Choi JM, Tsai FTF. Visualizing the ATPase cycle in a protein disaggregating machine: Structural basis for substrate binding by ClpB. Mol Cell. 2007;25:261–271. doi: 10.1016/j.molcel.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Effantin G, Ishikawa T, De Donatis GM, Maurizi MR, Steven AC. Local and global mobility in the ClpA AAA+ chaperone detected by cryo-electron microscopy: Functional connotations. Structure. 2010;18:553–562. doi: 10.1016/j.str.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang F, et al. Structure and mechanism of the hexameric MecA-ClpC molecular machine. Nature. 2011;471:331–335. doi: 10.1038/nature09780. [DOI] [PubMed] [Google Scholar]
- 11.Glynn SE, Martin A, Nager AR, Baker TA, Sauer RT. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell. 2009;139:744–756. doi: 10.1016/j.cell.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song HK, et al. Mutational studies on HslU and its docking mode with HslV. Proc Natl Acad Sci USA. 2000;97:14103–14108. doi: 10.1073/pnas.250491797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sousa MC, et al. Crystal and solution structure of an HslUV protease-chaperone complex. Cell. 2000;103:633–643. doi: 10.1016/s0092-8674(00)00166-5. [DOI] [PubMed] [Google Scholar]
- 14.Krzewska J, Langer T, Liberek K. Mitochondrial Hsp78, a member of the Clp/Hsp100 family in Saccharomyces cerevisiae, cooperates with Hsp70 in protein refolding. FEBS Lett. 2001;489:92–96. doi: 10.1016/s0014-5793(00)02423-6. [DOI] [PubMed] [Google Scholar]
- 15.Schlee S, Beinker P, Akhrymuk A, Reinstein J. A chaperone network for the resolubilization of protein aggregates: Direct interaction of ClpB and DnaK. J Mol Biol. 2004;336:275–285. doi: 10.1016/j.jmb.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 16.Motohashi K, Watanabe Y, Yohda M, Yoshida M. Heat-inactivated proteins are rescued by the DnaK.J-GrpE set and ClpB chaperones. Proc Natl Acad Sci USA. 1999;96:7184–7189. doi: 10.1073/pnas.96.13.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goloubinoff P, Mogk A, Zvi AP, Tomoyasu T, Bukau B. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc Natl Acad Sci USA. 1999;96:13732–13737. doi: 10.1073/pnas.96.24.13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zolkiewski M. ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. J Biol Chem. 1999;274:28083–28086. doi: 10.1074/jbc.274.40.28083. [DOI] [PubMed] [Google Scholar]
- 19.Tipton KA, Verges KJ, Weissman JS. In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol Cell. 2008;32:584–591. doi: 10.1016/j.molcel.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Acebrón SP, Martín I, del Castillo U, Moro F, Muga A. DnaK-mediated association of ClpB to protein aggregates. A bichaperone network at the aggregate surface. FEBS Lett. 2009;583:2991–2996. doi: 10.1016/j.febslet.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 21.Weibezahn J, et al. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–665. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 22.Zietkiewicz S, Krzewska J, Liberek K. Successive and synergistic action of the Hsp70 and Hsp100 chaperones in protein disaggregation. J Biol Chem. 2004;279:44376–44383. doi: 10.1074/jbc.M402405200. [DOI] [PubMed] [Google Scholar]
- 23.Zietkiewicz S, Lewandowska A, Stocki P, Liberek K. Hsp70 chaperone machine remodels protein aggregates at the initial step of Hsp70-Hsp100-dependent disaggregation. J Biol Chem. 2006;281:7022–7029. doi: 10.1074/jbc.M507893200. [DOI] [PubMed] [Google Scholar]
- 24.Schlieker C, Tews I, Bukau B, Mogk A. Solubilization of aggregated proteins by ClpB/DnaK relies on the continuous extraction of unfolded polypeptides. FEBS Lett. 2004;578:351–356. doi: 10.1016/j.febslet.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 25.Haslberger T, et al. Protein disaggregation by the AAA+ chaperone ClpB involves partial threading of looped polypeptide segments. Nat Struct Mol Biol. 2008;15:641–650. doi: 10.1038/nsmb.1425. [DOI] [PubMed] [Google Scholar]
- 26.Kedzierska S, Chesnokova LS, Witt SN, Zolkiewski M. Interactions within the ClpB/DnaK bi-chaperone system from Escherichia coli. Arch Biochem Biophys. 2005;444:61–65. doi: 10.1016/j.abb.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 27.Miot M, et al. Species-specific collaboration of heat shock proteins (Hsp) 70 and 100 in thermotolerance and protein disaggregation. Proc Natl Acad Sci USA. 2011;108:6915–6920. doi: 10.1073/pnas.1102828108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wendler P, et al. Atypical AAA+ subunit packing creates an expanded cavity for disaggregation by the protein-remodeling factor Hsp104. Cell. 2007;131:1366–1377. doi: 10.1016/j.cell.2007.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wendler P, Saibil H. Cryo electron microscopy structures of Hsp100 proteins: Crowbars in or out? Biochem Cell Biol. 2010;88:89–96. doi: 10.1139/o09-164. [DOI] [PubMed] [Google Scholar]
- 30.Lee S, Sielaff B, Lee J, Tsai FTF. CryoEM structure of Hsp104 and its mechanistic implication for protein disaggregation. Proc Natl Acad Sci USA. 2010;107:8135–8140. doi: 10.1073/pnas.1003572107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shorter J, Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 2004;304:1793–1797. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- 32.Abbas-Terki T, Donze O, Briand PA, Picard D. Hsp104 interacts with Hsp90 cochaperones in respiring yeast. Mol Cell Biol. 2001;21:7569–7575. doi: 10.1128/MCB.21.22.7569-7575.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moosavi B, Wongwigkarn J, Tuite MF. Hsp70/Hsp90 co-chaperones are required for efficient Hsp104-mediated elimination of the yeast [PSI(+)] prion but not for prion propagation. Yeast. 2010;27:167–179. doi: 10.1002/yea.1742. [DOI] [PubMed] [Google Scholar]
- 34.Werbeck ND, Schlee S, Reinstein J. Coupling and dynamics of subunits in the hexameric AAA+ chaperone ClpB. J Mol Biol. 2008;378:178–190. doi: 10.1016/j.jmb.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 35.Glover JR, Tkach JM. Crowbars and ratchets: Hsp100 chaperones as tools in reversing protein aggregation. Biochem Cell Biol. 2001;79:557–568. [PubMed] [Google Scholar]
- 36.Weibezahn J, Schlieker C, Bukau B, Mogk A. Characterization of a trap mutant of the AAA+ chaperone ClpB. J Biol Chem. 2003;278:32608–32617. doi: 10.1074/jbc.M303653200. [DOI] [PubMed] [Google Scholar]
- 37.Werbeck ND, Kellner JN, Barends TR, Reinstein J. Nucleotide binding and allosteric modulation of the second AAA+ domain of ClpB probed by transient kinetic studies. Biochemistry. 2009;48:7240–7250. doi: 10.1021/bi900880c. [DOI] [PubMed] [Google Scholar]
- 38.Zolkiewski M, Kessel M, Ginsburg A, Maurizi MR. Nucleotide-dependent oligomerization of ClpB from Escherichia coli. Protein Sci. 1999;8:1899–1903. doi: 10.1110/ps.8.9.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watanabe Y-H, Motohashi K, Yoshida M. Roles of the two ATP binding sites of ClpB from Thermus thermophilus. J Biol Chem. 2002;277:5804–5809. doi: 10.1074/jbc.M109349200. [DOI] [PubMed] [Google Scholar]
- 40.Barnett ME, Zolkiewski M. Site-directed mutagenesis of conserved charged amino acid residues in ClpB from Escherichia coli. Biochemistry. 2002;41:11277–11283. doi: 10.1021/bi026161s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamasaki T, Nakazaki Y, Yoshida M, Watanabe YH. Roles of conserved arginines in ATP-binding domains of AAA+ chaperone ClpB from Thermus thermophilus. FEBS Lett. 2011;278:2395–2403. doi: 10.1111/j.1742-4658.2011.08167.x. [DOI] [PubMed] [Google Scholar]
- 42.Ogura T, Whiteheart SW, Wilkinson AJ. Conserved arginine residues implicated in ATP hydrolysis, nucleotide-sensing, and inter-subunit interactions in AAA and AAA+ ATPases. J Struct Biol. 2004;146:106–112. doi: 10.1016/j.jsb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 43.Augustin S, et al. Intersubunit signaling coordinates ATP-hydrolysis and controls force generation by m-AAA protease ring complexes. Mol Cell. 2009;35:574–585. doi: 10.1016/j.molcel.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J, et al. Crystal structure of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism. Structure. 2001;9:177–184. doi: 10.1016/s0969-2126(01)00570-6. [DOI] [PubMed] [Google Scholar]
- 45.Hoskins JR, Doyle SM, Wickner S. Coupling ATP utilization to protein remodeling by ClpB, a hexameric AAA+ protein. Proc Natl Acad Sci USA. 2009;106:22233–22238. doi: 10.1073/pnas.0911937106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fernández-Higuero JÁ, et al. Allosteric communication between the nucleotide binding domains of caseinolytic peptidase B. J Biol Chem. 2011;286:25547–25555. doi: 10.1074/jbc.M111.231365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Franzmann TM, Czekalla A, Walter SG. Regulatory circuits of the AAA+ disaggregase Hsp104. J Biol Chem. 2011;286:17992–18001. doi: 10.1074/jbc.M110.216176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–599. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 49.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 50.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 51.CCP4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 52.Brünger AT, et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 53.Emsley P, Kevin C. Coot: Model-building tools for molecular graphics. Acta Cryst D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 54.Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. J Appl Cryst. 2006;39:109–111. [Google Scholar]
- 55.DeLano W. San Carlos, CA: DeLano Scientific; 2002. The PyMOL Molecular Graphics System. [Google Scholar]
- 56.Pettersen EF, et al. UCSF Chimera: A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.