Abstract
The complex of lysine-specific demethylase-1 (LSD1/KDM1A) with its corepressor protein CoREST is an exceptionally relevant target for epigenetic drugs. Here, we provide insight into the local and global changes of LSD1/CoREST conformational dynamics that occur upon H3 binding on the basis of a total cumulative time of one microsecond molecular dynamics simulation. The LSD1/CoREST complex functions as an allosteric nanoscale-binding clamp, which is regulated by substrate binding. In the unbound state, LSD1/CoREST reversibly visits clamp states that are more open or significantly more closed compared with the available X-ray crystal structures. The Lys triad of residues Lys355, Lys357, and Lys359 gates the entrance of the H3 pocket. H3 binding shifts the pocket breathing dynamics toward open, higher-volume states while reducing the overall flexibility of the LSD1/CoREST nanoscale clamp. We show that the H3 pocket is an allosteric site for the regulation of the rotation of the amino oxidase domain with respect to the Tower domain. The allosteric mechanism relies on the specific reduction of nanoscale domain rotation upon local H3-tail binding. Instead, clamp opening/closing motions that do not involve domain rotation only reduce in amplitude yet are dominant in the bound state. Overall, our data suggest that the H3 binding pocket is a central target site to (i) switch off LSD1 amino oxidase activity, thus H3-tail demethylation; (ii) block the competitive binding of transcription factors; and (iii) prevent chromatin anchoring to LSD1/CoREST. This study underscores the importance of receptor flexibility for future epigenetic drug discovery.
Keywords: chromatin remodeling, epigenetics, protein dynamics, computer simulation, pharmacology
Epigenetic drugs have promising, potential advantages compared with other cancer treatments (e.g., radiotherapy or DNA-modifying drugs), because they could act on replication and gene expression by modulating DNA access without altering the sequence and structure of DNA itself. Therefore, epigenetic pharmacology holds great promise to reduce the side effects that typically limit chemotherapy and antineoplastic efficacy. For example, this general, transferable concept is at the basis of the histone deacetylase inhibitors recently introduced for treatment of certain types of lymphomas (1, 2). Lysine-specific demethylase-1 (LSD1 or KDM1A) is overexpressed in many solid tumors such as breast, colon, neuroblastoma, bladder, small cell lung, blood, and prostate cancers (3–11) and plays an important role in leukemia (12), attracting steadily increasing attention as a major target for epigenetic drug discovery (13, 14).
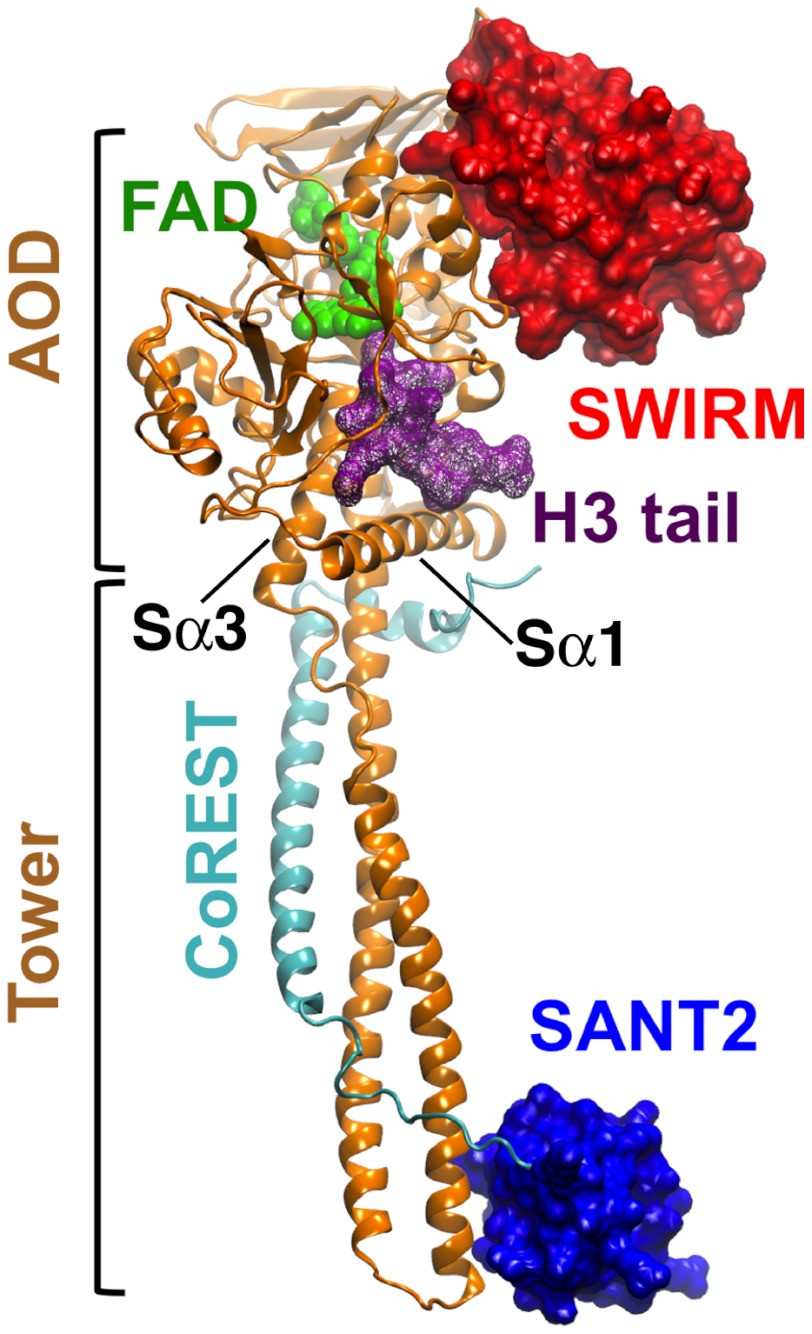
LSD1 associated to its corepressor protein CoREST catalyzes the oxidative, specific demethylation of the H3 histone N-terminal (H3) mono- and di-methylated Lys4 residue, using flavin adenosine dinucleotide (FAD) as a cofactor (15–21). LSD1/CoREST structural biology architecture is summarized in Fig. 1 on the basis of various studies (21–26). Recent X-ray crystallography experiments showed that—in addition to host the H3 N-terminal peptide—LSD1/CoREST can bind, as well, to the N-terminal peptide of the transcription factor SNAIL1 through a molecular mimicry mechanism (22). This observation has outstanding relevance, because SNAIL1 is a master regulator of the epithelial-mesenchymal transition underlying morphogenetic events, such as the establishment of tumor invasiveness (27). Yet, advances in epigenetics face a major critical barrier: the overwhelming complexity of the dynamic interactions that govern LSD1/CoREST substrate recognition. The changes of LSD1/CoREST conformational dynamics upon substrate binding are currently unknown.
Fig. 1.
Structural biology of the LSD1/CoREST complex. The LSD1 (gold cartoons) and CoREST (cyan cartoons) chains are highlighted, together with the FAD cofactor (green spheres) and the H3-histone tail (purple wireframe) based on Protein Data Bank entry 2V1D. LSD1 SWIRM domain (red surface) and CoREST SANT2 domain (blue surface) form the ends of the nanoscale clamp, connected by the amino oxidase domain (AOD) and the Tower domain. The Sα1 and Sα3 helices define the surface at the entrance of the H3-pocket. See also refs. (21–25).
Recently, our group discovered—using computer simulation as a powerful predictive tool—that LSD1/CoREST complex dynamics works as a nanoscale clamp reversibly opening/closing on several hundred nanosecond time scales (28). This nanoscale clamp motion makes LSD1/CoREST particularly suited for binding chromatin and protein partners with varying structural and dynamic complexity. Combined with the H3 histone/SNAIL1 molecular mimicry mechanism supports our working hypothesis that LSD1/CoREST could act as a multi-docking site to competitively anchor an array of structurally and functionally diverse transcription factors that share high N terminus sequence similarity (22, 29, 30). Here, we investigate the changes of LSD1/CoREST dynamics occurring upon binding the H3 N-terminal peptide addressing three key questions. First, does local H3-histone binding result into global, large-amplitude changes of LSD1/CoREST nanoscale clamp dynamics? Second, how does H3 noncovalent binding modulate locally the breathing dynamics of the H3 pocket? Third, are local and global conformational changes correlated?
In the unbound state, LSD1/CoREST reversibly visits clamp states that are significantly more open or closed compared with the available X-ray crystal structures (28). H3-tail binding opens the Lys triad gating the binding pocket, shifts pocket dynamics toward larger volume configurations, and consequently reduces the amplitude of the nanoscale motion in the LSD1/CoREST clamp. We show that the H3 pocket is an allosteric site for the regulation of the rotation of the amino oxidase domain with respect to the Tower domain. Instead, clamp opening/closing motions that do not involve domain rotation reduce in amplitude, yet are dominant in the bound state. Overall, our study underscores the importance of receptor flexibility for future epigenetic drug discovery.
Results
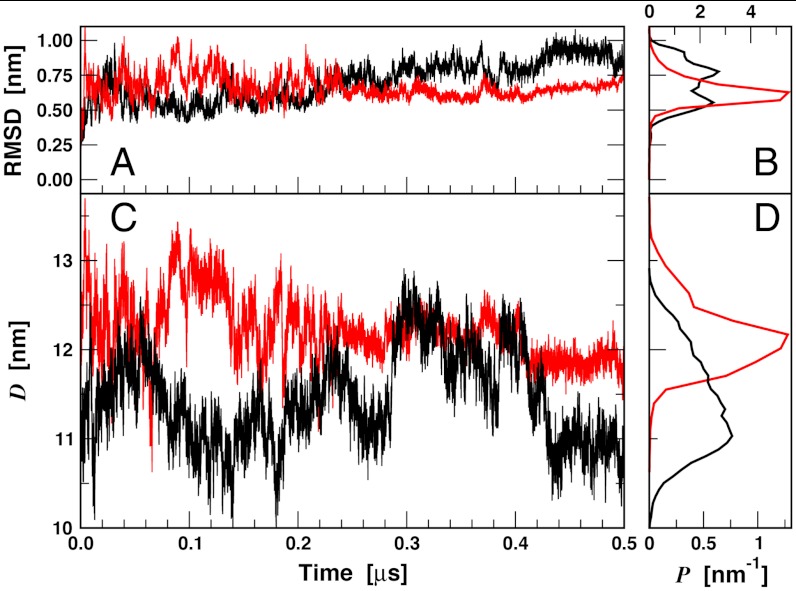
We investigated LSD1/CoREST dynamics in solution based on two independent molecular dynamics (MD) simulation trajectories, each 0.5 μs long. A first MD simulation of LSD1/CoREST was initialized from the X-ray structure by Yang et al. (24). A second MD simulation of LSD1/CoREST bound to the H3-histone 16 N-terminal residues was initialized from the X-ray structure by Forneris et al. (23). Both simulations are consistent with the general stability of the LSD1/CoREST complex in solution, as inferred by the stable fluctuation of the rmsd from the initial X-ray reference structures (Fig. 2A). The amplitude of these fluctuations is smaller for the simulation of LSD1/CoREST bound to the H3 peptide (Fig. 2B; standard deviation of the rmsd values are 0.15 and 0.09 nm from unbound and H3-bound simulations, respectively). The time series of the SWIRM–SANT2 interdomain distance display pronounced fluctuations with amplitudes over 2 nm in both simulations (Fig. 2C), and the corresponding standard deviations are 0.52 and 0.35 nm from unbound and bound simulations, respectively. H3-histone binding shifts pocket dynamics toward open-pocket configurations (Fig. 3).
Fig. 2.
Molecular dynamics of the LSD1/CoREST nanoscale clamp in solution along the 0.5-μs unbound (black lines) and H3-histone-bound (red lines) trajectories. (A) Time series of LSD1/CoREST backbone Cα atom-positional rmsd from the starting reference X-ray structures and (B) corresponding normalized probability distributions. (C) Time series of the SWIRM–SAINT2 interdomain distance, D, and (D) corresponding normalized probability distributions.
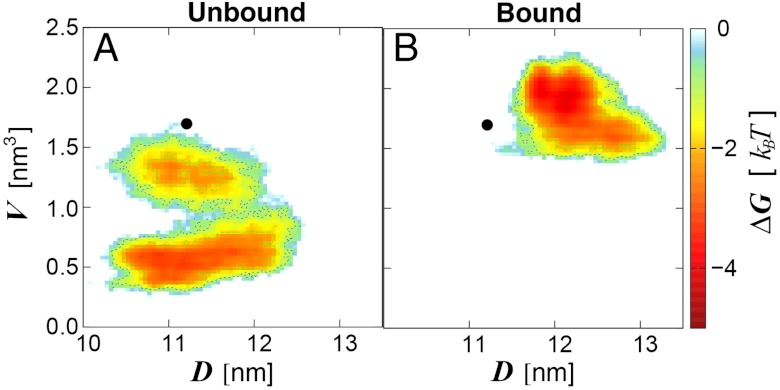
Fig. 3.
Shift of LSD1/CoREST conformational sampling in solution upon H3-histone-tail binding. Data from the 0.5-μs (A) unbound and (B) H3-bound ensembles is summarized in terms of the SWIRM–SAINT2 interdomain distance, D, and the H3-pocket volume, V. Color-coding ranges from light blue (less favorable free energy) to red (most favorable free energy). Sampling starts from the X-ray reference structure (black circle). D and V values from the X-ray reference structure are representative of all X-ray models deposited in the Protein Data Bank to date.
On one hand, these results show that the LSD1/CoREST nanoscale clamp reversible opening/closing dynamics in solution (28) is maintained upon binding to the H3-histone tail. On the other hand, they clearly indicate that the nanoscale clamp opening/closing dynamics is more pronounced in the ubound state, in line with a reduction of overall LSD1/CoREST flexibility upon binding. The reversible nature of this large-amplitude motion is strong evidence that reasonable sampling was achieved along our long trajectories. More computational studies are ongoing in our group to further enhance sampling statistics in the LSD1/CoREST system. These data are consistent with a major opening/closing motion of the LSD1/CoREST clamp in solution compared with the crystal structures currently deposited (average interdomain distance of 11.2 nm was calculated using the X-ray models in the Protein Data Bank to date; maximum value of 11.3 for 2XOL) likely owing to crystal packing effects. Volume variability in the X-ray models deposited is also minimal (1.5–1.7 nm3). We note that most X-ray structures have been solved in the presence of a substrate molecule. Therefore, simulation data is well in line with the general observation that significant reduction of pocket volume is noticed only in the unbound ensemble (Fig. 3A). Yet, the average pocket volume in our simulations is on top of that from the X-ray reference structure (Fig. 3B).
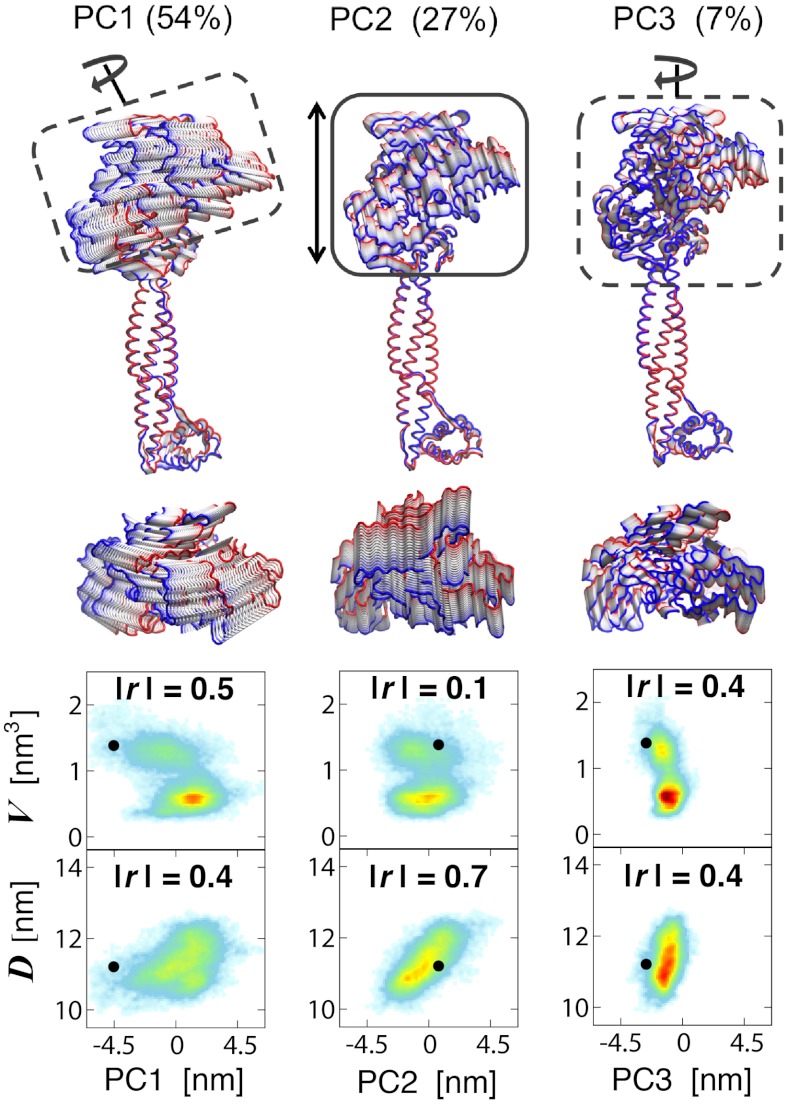
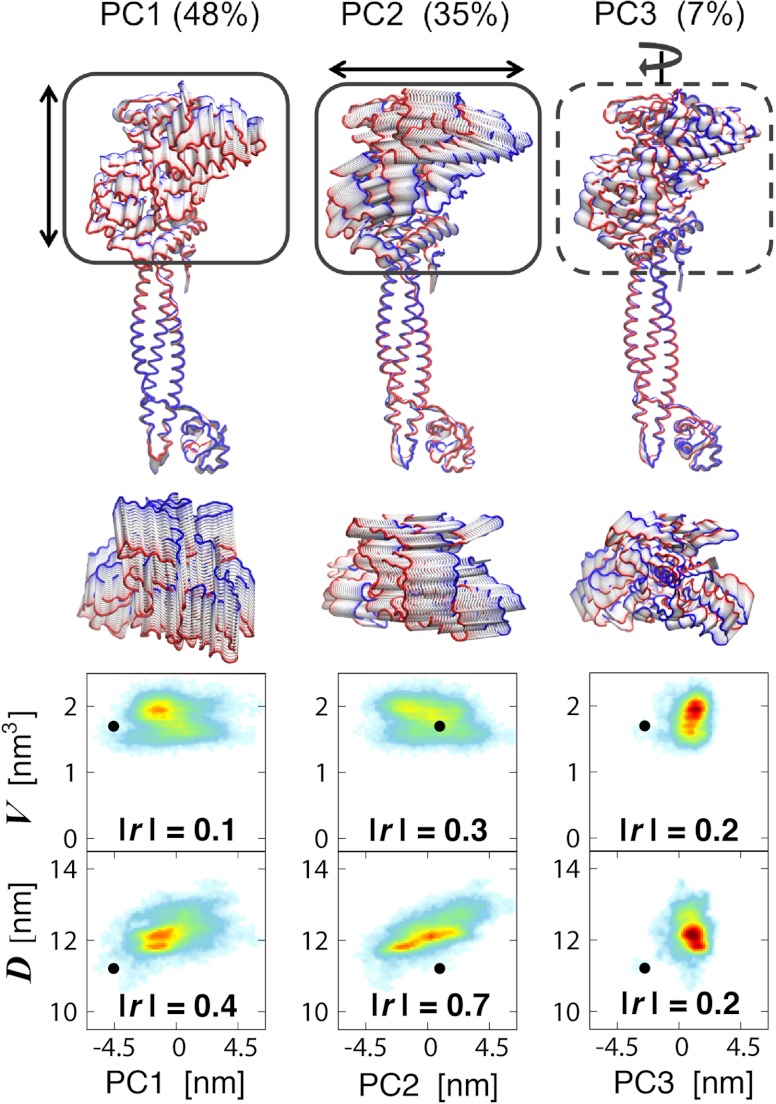
We examined the changes upon H3-tail binding of the LSD1/CoREST nanoscale clamp using principal component analysis (PCA) of protein fluctuations (31, 32). We observe a variety of large-amplitude motions along the two 0.5-μs-long MD trajectories, all involving the rigid-body movment of LSD1 AOD domain relatively to the Tower domain (Fig. 1). The major conformational motion captured through PCA analysis are summarized in Figs. 4 and 5 for the unbound and H3-bound LSD1/CoREST configurational ensembles, respectively. The comparison of these dominant motions between unbound and bound ensembles allows characterizing the large-amplitude comformational changes upon substrate binding. In the unbound state, the three most dominant principal components (PC1, PC2, and PC3) represent about 88% of the overall protein backbone fluctuations (Fig. S1). PC1 (54%) is responsible for the rotational clamp motion of the AOD domain with respect to the Tower domain (Movie S1). PC2 (27%) also describes the opening/closing of the clamp yet with no torsional component (Movie S2). PC3 (7%) captures the rigid-body rotation of the AOD domain around an axis defined along the Tower domain (Movie S3). In the unbound state, over 60% of the principal motions involve remarkable rotation of the AOD domain relatively to the Tower domain. In the H3-bound state, the three most dominant principal components total about 90% of the overall fluctuations (Fig. S1). PC1 (48%) is responsible for the opening/closing of the clamp Movie S4, similarly to PC2 in the unbound state (Movie S2). PC2 (35%) captures a twist of the AOD domain with respect to the Tower domain (Movie S5). PC3 (7%) is a rigid-body rotation of the AOD domain around an axis defined by the Tower domain Movie S6, similarly to PC3 in the unbound state (Movie S3). In the H3-bound state only 7% of the principal motions involve remarkable rotation of the AOD domain relatively to the Tower domain. Are these nanoscale motions and their change upon binding regulated by the local recruitment of the H3-histone tail?
Fig. 4.
Principal motions of the unbound LSD1/CoREST complex from 0.5-μs molecular dynamics simulation. Arrows and boxes highlight the most evident motion of the AOD domain with respect to the Tower domain (dashed lines: rotation; continuous line: clamp opening/closing; see also Fig. 1). Lateral and top view representations are displayed maintaining identical relative orientation of the Tower domain. The backbone chain is colored in red and blue for highlighting the extreme states of the motions. See Movies S1–S3. Bottom: correlation of each principal LSD1/CoREST clamp motion with the SWIRM–SAINT2 interdomain distance, D, and the H3-pocket volume, V. The absolute values of Pearson’s r values are reported as a measure of linear correlation. Color-coding ranges from light blue (low probability) to red (high probability).
Fig. 5.
Principal motions of the LSD1/CoREST complex bound to the H3-histone tail from 0.5-μs molecular dynamics simulation. See Movies S4–S6. See Fig. 4 legend for a description of representations, color-coding, and correlation analysis.
To address this crucial question we analyzed as well the relationship between opening/closing of the LSD1/CoREST nanoscale clamp—as captured by the SWIRM–SANT2 interdomain distance—and the H3-pocket breathing dynamics—well-summarized using the volume of the H3 pocket. We investigated the relevance of each principal motion for determining opening/closing of the LSD1/CoREST nanoscale clamp or changing the H3-pocket volume. Figs. 4 and 5 summarize these results in terms of the Pearson product-moment correlation coefficient, or Pearson’s r-value, as a measure of linear dependence. Highest absolute values of r within a certain ensemble evidence the specific principal component that is most correlated with a given observable, such as the SWIRM–SAINT2 interdomain distance, D, or the H3-pocket volume, V. Our data indicate that in the unbound state the PC1 and PC3 rotational motions of LSD1/CoREST are those most clearly correlated with changes in H3-pocket volume (Fig. 4; absolute r values of 0.5 and 0.4 for PC1 and PC3 vs. 0.1 for PC2). Instead, the PC2 clamp opening/closing motion does not involve AOD domain rotation around the Tower domain, and it is the principal component most evidently correlated with a change of interdomain distance (Fig. 4; absolute r values of 0.7 for PC2 vs 0.4 and 0.2 for both PC1 and PC3). H3-tail binding shifts H3-pocket breathing dynamics toward open-pocket, larger-volume configurations and prevents reversible sampling of open/closed pocket configurations (Fig. 3), previously reported for the unbound state (28). The principal components of LSD1/CoREST dynamics that represent AOD rotation around the Tower domain are those more highly correlated with changes of H3-pocket volume. Thus, upon H3 binding their overall relevance drops from over 60% (Fig. 4, PC1 and PC3; Movies S1 and S3) to 7% (Fig. 5, PC3; Movie S6). Instead, components of LSD1/CoREST dynamics that regulate the opening/closing of the LSD1/CoREST nanoscale clamp are not affected to a similar extent by histone binding. This is confirmed by the observation that PC1 and PC2 in the H3-bound state display high correlation with changes of interdomain distance (Fig. 5; absolute r values of 0.4 and 0.7 vs 0.2 for PC3).
Overall, our data clearly indicates that the major consequence of H3-tail binding is a remarkable population shift toward open, larger pocket volume configurations (Fig. 3). However, this local conformational change affects to different extent the principal components of LSD1/CoREST conformational sampling. The loss of rotation of the AOD domain around the axis defined along the LSD1/CoREST Tower domain is the most evident consequence (Figs. 4 and 5). This result is in line with the discovery that the principal components involving AOD rigid-body rotation are more strictly correlated with changes of H3 pocket volume compared with those in which rotation is absent. Instead, substrate binding does not preclude the opening/closing reversible oscillation of the LSD1/CoREST nanoscale clamp (Figs. 2 and 3), because this motion is influenced by changes of H3 pocket volume to a smaller extent. Yet, it triggers a reduction of overall flexibility in the LSD1/CoREST nanoscale clamp and shifts the nanoscale clamp toward more open clamp states. Based on this data we are able to suggest that the local binding of the H3-histone tail governs the global conformational sampling of the LSD1/CoREST nanoscale clamp, including nanoscale motion and remote regions of the nanoscale complex. These findings based on a total MD sampling of one microsecond are a strong evidence of the allosteric nature of the H3-pocket.
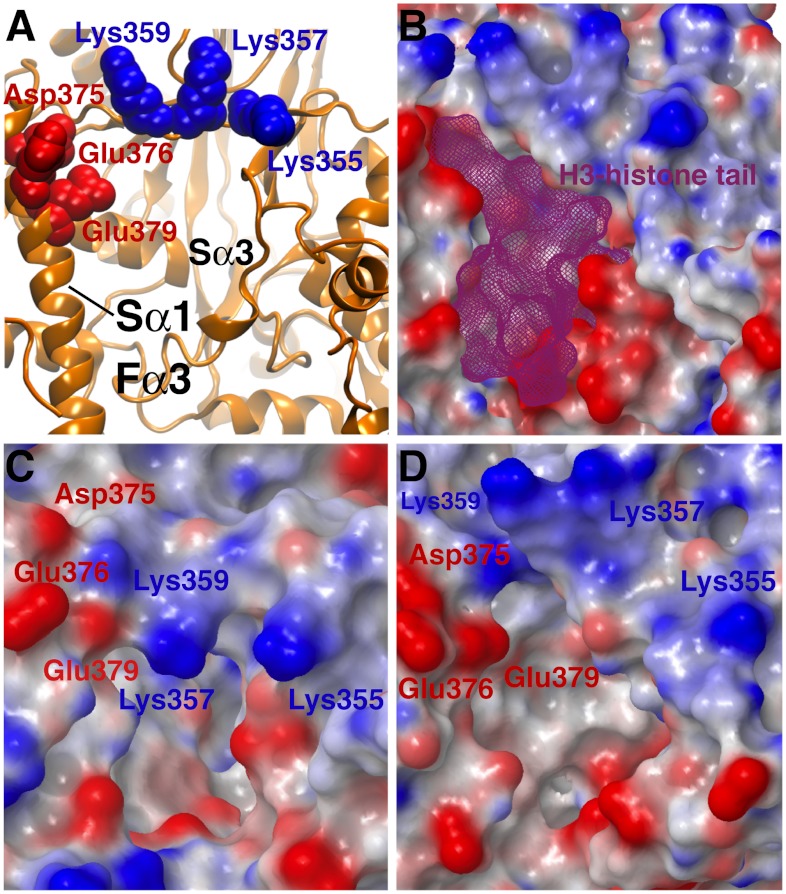
LSD1 has a unique structural feature that sets it apart from other flavin-dependent oxidases: the Sα1 helix, tilted with respect to the Sα3 helix, that defines the surface at the entrance of the H3-pocket (Figs. 1 and 6A). Instead, the majority of the residues forming the catalytic cavity are highly conserved among LSD1 orthologs in different as well as in LSD2, a close homolog of LSD1 (19). The presence of negatively charged residues on the Sα1 helix might favor the recruitment of the positively charged histone H3 N-terminal tail, prior to intercalation of the substrate deep into the 2.3-nm H3-pocket tunnel. However, the molecular mechanism of H3-tail recognition remains unclear. Inspection of the LSD1/CoREST X-ray structure bound to the H3-histone tail shows three regions of prominent electrostatic potential that surround the access to the H3-pocket. The first two regions are dominated by a negative charge, corresponding with the Sα1 helix and the Fα3 loop at the entrance of the H3 pocket (Fig. 6 A and B). Therefore, it is expected to facilitate the recruitment of N-terminal peptides carrying an opposite, positive charge. In line with this hypothesis, mutations reducing the Fα3 loop positive charge, such as Asp555Ala and Asp556Ala, abrogate LSD1 demethylation activity toward the H3-tail substrate dimethylated on Lys4 (24). The third region is dominated by a positive charge due to the presence of the Lys triad formed by Lys355, Lys357, and Lys359 (Fig. 6B). The role of this region has been neglected thus far, as all X-ray structures deposited to date display the Lys triad as fully open, toward the protein surface (21–25). An important point that remains to be addressed in the context of LSD1/CoREST substrate recognition is how substrates with a net positive charge could be favorably attracted toward a binding pocket gated by Lys residues that carry a same, repelling positive charge. This point is extremely relevant not only in the context of H3 histone molecular recognition. LSD1 could recruit a variety of transcription factors using molecular mimicry mechanisms similar to that proposed for SNAIL1 using the H3-pocket as an anchoring site for their N-terminal tails (22, 30). Our MD simulations supply new insight on the gating role of the Lys triad in substrate recognition.
Fig. 6.
Dynamics of the Lys triad gating the access to the LSD1 H3-histone binding pocket. The flexible Lys triad formed by “finger residues” Lys359, Lys357, and Lys355 gates the pocket entrance, which is defined by the Sα1 and Sα3 helices and the Fα3 loop. The protein surface is colored depending on the protein electrostatic potential (blue: positive; red: negative). (A) Reference H3-bound X-ray crystal structure (PDB ID code 2VID); the H3-histone N-terminal tail is not represented for graphical purposes. (B) Surface potential of the H3-bound X-ray crystal structure; the H3-histone tail is shown as a purple wireframe. (C) Example of closed-gate configuration from the unbound LSD1/CoREST simulation; closing of the gate is promoted by electrostatic attraction with Asp375, Glu376, and Glu379. (D) Example of open-gate configuration from the H3-bound simulation. See also Fig. 1 as reference.
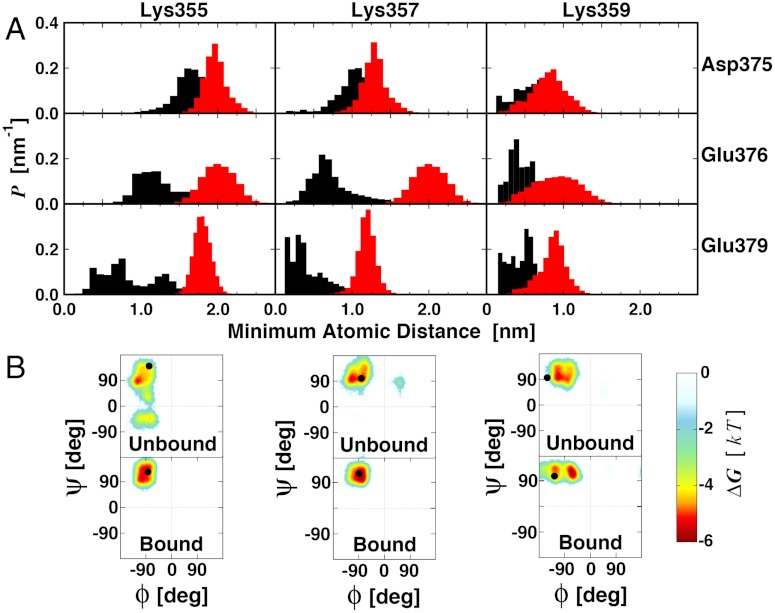
The Lys triad is highly dynamic along the 0.5–μs-long unbound LSD1/CoREST simulation and transiently gates the access to the H3 pocket in the unbound state (28). The three Lys residues act as fingers that reversibly expose their positive charges to the solvent or retract them to cover the entrance of the H3 pocket, helped by electrostatic attraction with residues Asp375, Glu376, and Glu379. Therefore, the Lys gate in the unbound state reversibly visits open- and closed-gated configurations (28). Upon H3-tail binding, the Lys triad gate shifts toward open state configurations, similar to the X-ray reference model. Fig. 6 displays examples of these alternative states of the Lys triad gate and compares the corresponding electrostatic potential maps at the atomic level. LSD1 propensity to recruit the H3 histone tail and other substrates is likely to be at variance with the different electrostatic potential of the protein surface at the entrance of the H3 pocket. The transient nature of the Lys triad gating suggests that a substrate could sneak into the H3 pocket tunnel only when fully open-gate configuration are sampled. In line with this picture, the Lys triad gate shifts toward open state configurations upon H3-tail binding, as inferred by pronounced shift of the distributions of the minimum atomic distances between the Lys triad and the negatively charged residues (Fig. 7A). In the case of Lys355, gate opening involves as well a pronounced rotation of the peptide backbone, and a basin corresponding to negative ψ values becomes inaccessible once the H3-tail is bound (Fig. 7B). However, we do not observe similar changes upon binding for Lys357 and Lys359 peptide backbone sampling. These local conformational changes essentially switch the gate in its open conformation once the substrate occupies the H3 pocket.
Fig. 7.
Gating of the H3-pocket entrance by the Lys triad and changes upon H3-histone binding. (A) Normalized probability distributions of the minimum distance between Lys359, Lys357, and Lys355 with residues Asp375, Glu376, and Glu379 located on the opposite side of the gate are displayed from unbound (black lines) and bound (red lines) 0.5-μs simulations. Minimum distances were calculated using all atoms in each residue pair. (B) Ramachandran’s plots for Lys triad polypeptide backbone sampling. See Fig. 6 for graphical representations of the residues considered.
Discussion
The complex of lysine-specific demethylase-1 (LSD1 or KDM1A) with its corepressor protein CoREST is an exceptionally relevant target for epigenetic drugs because LSD1 is overexpressed in many solid tumors such as breast, colon, neuroblastoma, bladder, small cell lung, blood, and prostate cancers and plays a major role in leukemia (3–9, 12–14). Yet, advances in epigenetics are hampered by our poor understanding of the complex dynamic interactions that govern LSD1/CoREST substrate recognition and other epigenetic targets (11). In this research report, we presented the changes of LSD1/CoREST conformational dynamics upon H3-histone (H3) binding using a total cumulative time of one microsecond molecular dynamics simulation. To our knowledge this study is unique in addressing the changes of large-amplitude motions and dynamics upon substrate binding to an epigenetic target.
The LSD1/CoREST complex functions as an allosteric nanoscale-binding clamp, which is regulated by substrate binding. In the unbound state, LSD1/CoREST reversibly visits clamp states that are more open or significantly more closed compared with the available X-ray crystal structures and the Lys triad of residues Lys355, Lys357, and Lys359 gates the entrance ot the H3 pocket. Substrate binding shifts the pocket breathing dynamics toward open, higher-volume states, while reducing the overall flexibility of the LSD1/CoREST nanoscale clamp and locks the Lys triad into an open configuration, which is more closely similar to that observed in all X-ray structural models currently available (21–25).
We showed that the H3 pocket is an allosteric site for the regulation of the rigid-body rotation of the amino oxidase domain with respect to the Tower domain. The allosteric mechanism relies on the specific reduction of nanoscale domain rotation upon local H3-tail binding. Instead, in the bound state clamp opening/closing motions involving no domain rotation were still dominant with reduced aplitude compared with the unbound state. Overall, our data underscores the importance of receptor flexibility for future epigenetic drug discovery and lead to the hypothesis of an opening/closing gating mechanism that uses electrostatic forces as a major selectivity filter to recruit and anchor a variety of substrate peptides with high sequence similarity to the H3-histone tail (22, 29, 30). Further research is ongoing to confirm whether this mechanism is transferable to other LSD1 substrates.
Our study dramatically expands the relevance of the H3 binding pocket for LSD1 biological function. We suggest that the H3 binding pocket is a central target site not limited to its role to switch off LSD1 amino oxidase activity, thus H3-tail demethylation (15–21), or to block the competitive binding of transcription factors, as supported by recent studies that highlighted the role of molecular mimicry mechanisms for binding substrates with high N-terminal sequence similarity (22, 29, 30). Based on the evidence that the local binding of the H3-tail determines a significant loss of rotation of the AOD domain with respect to the Tower domain, our results suggest a second fascinating hypothesis: that inhibitor binding could govern as well chromatin anchoring to LSD1/CoREST (28). The remarkable rotation movement reported herein for LSD1 around the Tower domain could be a crucial component of the ability of LSD1/CoREST to dock nucleosome units and perform chromatin remodeling. Ongoing research will address as well this second hypothesis.
We hope this computational study will stimulate the design of new experiments for probing LSD1/CoREST nanoscale dynamics and the newly discovered allosteric and gating mechanisms. Immediate choices to probe the large-amplitude motions described herein will include advanced experiments to probe biomolecular dynamics in solution, such as combinations of CRINEPT and TROSY nuclear magnetic resonance that enable the study in solution of systems of the size of LSD1/CoREST. To our knowledge no mutation study of the Lys triad Lys355, Lys357, and Lys359 or other possibly relevant residues (e.g., Asp375, Glu376, and Glu379) has been reported to date. The relevance of these residues could be probed by integrating kinetic rates measurements and structural determination on engineered LSD1 mutants. Complemented by molecular simulations, these experiments could help to shed light as well on atomic-level, dynamic mechanism that play crucial roles for drug discovery and design.
Methods
A first MD simulation of unbound LSD1/CoREST was initialized from the X-ray structure by Yang et al. (24) (PDB ID code 2IW5; 0.26 nm resolution). A second MD simulation of LSD1/CoREST bound to the H3-histone N-terminal tail (16 residues) was initialized from the X-ray structure by Forneris et al. (23) (PDB ID code 2V1D; 0.31 nm resolution). Standard preparation, minimization, heat-up, and equilibration procedures were followed, as described in SI Text. Trajectories of 0.5-μs periods were generated and analyzed in double precision using the GROMACS 4.5.4 software (33). Force field parameters and charges were set from the 53A6 GROMOS parameter set (34) to reproduce the experimental condition of apparent neutral pH. The compatible SPC water model (35) and ion parameters (36) were employed. We refer the reader to SI Text for additional computational details. Whenever else specified, structural fitting was performed by (i) superimposing the centers of mass (to remove overall translation) and (ii) performing an atom-positional least-square fitting procedure (to remove overall rotation) using all Cα atoms of the LSD1/CoREST domain considered (37). We investigated the dominant protein motion connecting these clamp states by principal component analysis (PCA) of protein fluctuations (31, 32) with the bio3d software (38) and displayed using VMD (39) (SI Text, Movies S1–S6). PCA analysis was performed after superimposition of the MD trajectory snapshots on the Cα atoms of LSD1/CoREST Tower domain. Calculations of the H3-tail pocket volume were performed with the POVME program (40) by defining the pocket using residues in ranges Ala354–Tyr363, Val370–Tyr391, and Leu529–His564. In this case, only the Cα atoms of these residues were used for structural fitting, using the X-ray structure by Yang et al. (24) (PDB ID code 2IW5) to allow for direct comparison between the unbound and H3-bound configurational ensembles. The qualitative volume trends reported are independent of alternative definitions of the H3 pocket and the reference structure chosen for fitting.
Supplementary Material
ACKNOWLEDGMENTS.
The authors thank Andrea Mattevi for useful discussions and a critical reading of the manuscript and the Center for High Performance Computing for technical support and computing allocations. This work was supported by the University of Utah and Department of Medicinal Chemistry startup funds to R.B. In part, this project used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation Grant OCI-1053575.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207892109/-/DCSupplemental.
References
- 1.Botrugno OA, Santoro F, Minucci S. Histone deacetylase inhibitors as a new weapon in the arsenal of differentiation therapies of cancer. Cancer Lett. 2009;280:134–144. doi: 10.1016/j.canlet.2009.02.027. [DOI] [PubMed] [Google Scholar]
- 2.Janzen WP, Wigle TJ, Jin J, Frye SV. Epigenetics: Tools and technologies. Drug Discov Today Technol. 2010;7:e59–e65. doi: 10.1016/j.ddtec.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayami S, et al. Overexpression of LSD1 contributes to human carcinogenesis through chromatin regulation in various cancers. Int J Cancer. 2011;128:574–586. doi: 10.1002/ijc.25349. [DOI] [PubMed] [Google Scholar]
- 4.Kahl P, et al. Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence. Cancer Res. 2006;66:11341–11347. doi: 10.1158/0008-5472.CAN-06-1570. [DOI] [PubMed] [Google Scholar]
- 5.Lim S, et al. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis. 2010;31:512–520. doi: 10.1093/carcin/bgp324. [DOI] [PubMed] [Google Scholar]
- 6.Schulte JH, et al. Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: Implications for therapy. Cancer Res. 2009;69:2065–2071. doi: 10.1158/0008-5472.CAN-08-1735. [DOI] [PubMed] [Google Scholar]
- 7.Cho H-S, et al. Demethylation of RB regulator MYPT1 by histone demethylase LSD1 promotes cell cycle progression in cancer cells. Cancer Res. 2011;71:655–660. doi: 10.1158/0008-5472.CAN-10-2446. [DOI] [PubMed] [Google Scholar]
- 8.Singh MM, et al. Inhibition of LSD1 sensitizes glioblastoma cells to histone deacetylase inhibitors. Neurooncology. 2011;13:894–903. doi: 10.1093/neuonc/nor049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu X, et al. LSD1-mediated epigenetic modification is required for TAL1 function and hematopoiesis. Proc Natl Acad Sci USA. 2009;106:10141–10146. doi: 10.1073/pnas.0900437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009;138:660–672. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- 11.Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 12.Schenk T, et al. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat Med. 2012;18:605–611. doi: 10.1038/nm.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Culhane JC, Cole PA. LSD1 and the chemistry of histone demethylation. Curr Opin Chem Biol. 2007;11:561–568. doi: 10.1016/j.cbpa.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rotili D, Mai A. Targeting histone demethylases: A new avenue for the fight against cancer. Genes Cancer. 2011;2:663–679. doi: 10.1177/1947601911417976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forneris F, Binda C, Battaglioli E, Mattevi A. LSD1: Oxidative chemistry for multifaceted functions in chromatin regulation. Trends Biochem Sci. 2008;33:181–189. doi: 10.1016/j.tibs.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Forneris F, Binda C, Vanoni M, Battaglioli E. Human histone demethylase LSD1 reads the histone code. J Biol Chem. 2005;280:41360–41365. doi: 10.1074/jbc.M509549200. [DOI] [PubMed] [Google Scholar]
- 17.Mosammaparast N, Shi Y. Reversal of histone methylation: Biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem. 2010;79:155–179. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- 18.Shi Y. Histone lysine demethylases: Emerging roles in development, physiology and disease. Nat Rev Genet. 2007;8:829–833. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- 19.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 20.Forneris F, Binda C, Vanoni M, Mattevi A. Histone demethylation catalysed by LSD1 is a flavin-dependent oxidative process. FEBS Lett. 2005;579:2203–2207. doi: 10.1016/j.febslet.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 21.Chen Y, et al. Crystal structure of human histone lysine-specific demethylase 1 (LSD1) Proc Natl Acad Sci USA. 2006;103:13956–13961. doi: 10.1073/pnas.0606381103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baron R, Binda C, Tortorici M, McCammon JA, Mattevi A. Molecular mimicry and ligand recognition in binding and catalysis by the histone demethylase LSD1-CoREST complex. Structure. 2011;19:212–220. doi: 10.1016/j.str.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forneris F, Binda C, Adamo A, Battaglioli E, Mattevi A. Structural basis of LSD1-CoREST selectivity in histone H3 recognition. J Biol Chem. 2007;282:20070–20074. doi: 10.1074/jbc.C700100200. [DOI] [PubMed] [Google Scholar]
- 24.Yang M, et al. Structural basis for CoREST-dependent demethylation of nucleosomes by the human LSD1 histone demethylase. Mol Cell. 2006;23:377–387. doi: 10.1016/j.molcel.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 25.Stavropoulos P, Blobel G, Hoelz A. Crystal structure and mechanism of human lysine-specific demethylase-1. Nat Struct Mol Biol. 2006;13:626–632. doi: 10.1038/nsmb1113. [DOI] [PubMed] [Google Scholar]
- 26.Yang M, et al. Structural basis of histone demethylation by LSD1 revealed by suicide inactivation. Nat Struct Mol Biol. 2007;14:535–539. doi: 10.1038/nsmb1255. [DOI] [PubMed] [Google Scholar]
- 27.Lin Y, et al. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 2010;29:1803–1816. doi: 10.1038/emboj.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baron R, Vellore NA. LSD1/CoREST reversible opening-closing dynamics: Discovery of a nanoscale clamp for chromatin and protein binding. Biochemistry. 2012;51:3151–3153. doi: 10.1021/bi300068r. [DOI] [PubMed] [Google Scholar]
- 29.Barrallo-Gimeno A, Nieto MA. Evolutionary history of the Snail/Scratch superfamily. Trends Genet. 2009;25:248–252. doi: 10.1016/j.tig.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Laurent B, et al. A short Gfi-1B isoform controls erythroid differentiation by recruiting the LSD1-CoREST complex through the dimethylation of its SNAG domain. J Cell Sci. 2012;125:993–1002. doi: 10.1242/jcs.095877. [DOI] [PubMed] [Google Scholar]
- 31.Garcia AE. Large-amplitude nonlinear motions in proteins. Phys Rev Lett. 1992;68:2696–2699. doi: 10.1103/PhysRevLett.68.2696. [DOI] [PubMed] [Google Scholar]
- 32.Amadei A, Linssen AB, Berendsen HJC. Essential dynamics of proteins. Proteins. 1993;17:412–425. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- 33.Hess B, Kutzner C, van Der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 34.Oostenbrink C, Villa A, Mark AE, van Gunsteren WF. A biomolecular force field based on the free enthalpy of hydration and solvation: The GROMOS force-field parameter sets 53A5 and 53A6. J Comput Chem. 2004;25:1656–1676. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- 35.Berendsen HJC. In: Interaction Models for Water in Relation to Protein Hydration. Pullman BE, editor. Dordrecht: 1981. [Google Scholar]
- 36.Åqvist J. Ion water interaction potentials derived from free-energy perturbation simulations. J Phys Chem. 1990;94:8021–8024. [Google Scholar]
- 37.McLachlan AD. Gene duplications in the structural evolution of chymotrypsin. J Mol Biol. 1979;128:49–79. doi: 10.1016/0022-2836(79)90308-5. [DOI] [PubMed] [Google Scholar]
- 38.Grant BJ, Rodrigues APC, ElSawy KM, McCammon JA, Caves LSD. Bio3d: An R package for the comparative analysis of protein structures. Bioinformatics. 2006;22:2695–2696. doi: 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
- 39.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 40.Durrant JD, de Oliveira CAF, McCammon JA. POVME: An algorithm for measuring binding-pocket volumes. J Mol Graph Model. 2011;29:773–776. doi: 10.1016/j.jmgm.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.