Abstract
Immunoglobulin (Ig) class switch DNA recombination (CSR) is the crucial mechanism diversifying the biological effector functions of antibodies. Generation of double-strand DNA breaks (DSBs), particularly staggered DSBs, in switch (S) regions of the upstream and downstream CH genes involved in the specific recombination process is an absolute requirement for CSR. Staggered DSBs would be generated through deamination of dCs on opposite DNA strands by activation-induced cytidine deaminase (AID), subsequent dU deglycosylation by uracil DNA glycosylase (Ung) and abasic site nicking by apurinic/apyrimidic endonuclease. However, consistent with the findings that significant amounts of DSBs can be detected in the IgH locus in the absence of AID or Ung, we have shown in human and mouse B cells that AID generates staggered DSBs not only by cleaving intact double-strand DNA, but also by processing blunt DSB ends generated in an AID-independent fashion. How these AID-independent DSBs are generated is still unclear. It is possible that S region DNA may undergo AID-independent cleavage by structure-specific nucleases, such as endonuclease G (EndoG). EndoG is an abundant nuclease in eukaryotic cells. It cleaves single- and double-strand DNA, primarily at dG/dC residues, the preferential sites of DSBs in S region DNA. We show here that EndoG can localize to the nucleus of B cells undergoing CSR and binds to S region DNA, as shown by specific chromatin immunoprecipitation assays. Using knockout EndoG−/− mice and EndoG−/− B cells, we found that EndoG deficiency resulted in defective CSR in vivo and in vitro, as demonstrated by reduced cell surface IgG1, IgG2a, IgG3 and IgA, reduced secreted IgG1, reduced circle Iγ1-Cμ, Iγ3-Cμ, Iε-Cμ, Iα-Cμ transcripts, post-recombination Iμ-Cγ1, Iμ-Cγ3, Iμ-Cε and Iμ-Cα transcripts. In addition to reduced CSR, EndoG−/− mice showed a significantly altered spectrum of mutations in IgH JH-iEμ DNA. Impaired CSR in EndoG−/− B cells did not stem from altered B cell proliferation or apoptosis. Rather, it was associated with significantly reduced frequency of DSBs. Thus, our findings determine a role for EndoG in the generation of S region DSBs and CSR.
Keywords: Activation-induced cytidine deaminase (AID), Antibody, B cell, Endonuclease G (EndoG), Class switch DNA recombination (CSR), Double-strand DNA break (DSB), Immunoglobulin (Ig), Switch region, Knockout mice
1. Introduction
Immunoglobulin (Ig) class switch DNA recombination (CSR) and somatic hypermutation (SHM) are central to the maturation of the antibody response. CSR and SHM diversify antibodies in different ways. CSR substitutes an Ig heavy chain (IgH) constant (CH) region, for instance, Cμ, with a downstream CH region, Cγ, Cα or Cε, thereby endowing an antibody with different biological effector functions without changing the structure/specificity of the antigen-binding site. In CSR, double-strand DNA breaks (DSBs) are introduced in the switch (S) regions of an upstream and a downstream CH gene, followed by intrachromosomal and inter-S-S region recombination, entailing the excision of intervening genomic DNA, and variable introduction of point-mutations around recombined S-S region junctions. SHM inserts single nucleotide changes (point-mutations) in V(D)J DNA, thereby providing the substrate for selection by antigen of high-affinity mutants.
DSBs are obligatory intermediates in CSR (Stavnezer et al., 2008; Zan and Casali, 2008) and likely contribute to SHM as well (Bross et al., 2000; Papavasiliou and Schatz, 2000; Bross and Jacobs, 2003; Wu et al., 2003; Zan et al., 2003; Xu et al., 2005). Both CSR and SHM require activation-induced cytidine deaminase (AID), which is expressed by activated B cells, mainly in germinal centers of peripheral lymphoid organs (Di Noia and Neuberger, 2007; Xu et al., 2007a; Stavnezer et al., 2008; Delker et al., 2009). Like SHM, CSR occurs, in general, in germinal centers in response to T cell-dependent stimuli, including, engagement of CD40 on B cells by CD154 and cytokines expressed by activated CD4+ T cells (Stavnezer et al., 2008; Pone et al., 2010). CSR, however, can also be induced in a T cell-independent fashion, e.g., by ligands of Toll-like receptors (TLRs) (Stavnezer et al., 2008; Pone et al., 2010). CSR requires germline transcription of the intervening (IH), S and CH regions of the upstream (donor) and downstream (acceptor) CH loci (Wuerffel et al., 2007; Stavnezer et al., 2008), leading to chromosomal opening and increased accessibility to the S regions that will be targets of recombination. CSR targets DSBs to the 5′-RGYW-3′/5′-WRCY-3′ motif, mainly in its 5′-AGCT-3′ iteration (Xu et al., 2010). Accordingly, AID is specifically targeted to 5′-AGCT-3′ rich S regions by 14-3-3 adaptor proteins (Xu et al., 2010). It deaminates dC residues to dUs in DNA (Maizels, 2005; Casali et al., 2006; Di Noia and Neuberger, 2007; Xu et al., 2007b; Peled et al., 2008; Stavnezer et al., 2008), more efficiently when phosphorylated by PKA (Cheng et al., 2009; Vuong et al., 2009) at Serine 38 and in the presence of replication protein A (RPA) (Basu et al., 2008; Rada, 2009). dUs are deglycosylated by uracil DNA glycosylase (Ung), yielding abasic sites. These are recognized and nicked by an apurinic/apyrimidic endonuclease (APE). Nicking of abasic sites in close proximity on opposite DNA strands give rise to DSBs to initiate CSR (Maizels, 2005; Di Noia and Neuberger, 2007; Stavnezer et al., 2008).
It has been assumed that the absolute requirement of AID in CSR reflects the indispensability of this enzyme in generating DSBs. AID-dependent DSBs have been widely detected in S region DNA by ligation-mediated (LM)-PCR in human and mouse B cells. However, we and other investigators have shown that S region DSBs can also be generated in an AID-independent fashion in human and mouse B cells (Rush et al., 2004; Zan and Casali, 2008). AID-independent generation of DSBs has been further inferred from the occurrence of intra-S region DNA recombination events in AID-deficient B cell hybridomas (Dudley et al., 2002), and the frequent translocation of c-myc into S regions in AID-deficient mice (Unniraman et al., 2004). These findings could also be explained, at least partially, by residual AID expression from the fusion partner that was used for constructing the hybridomas (Dudley et al., 2002) or the possibility that the DNA amplicons used to reveal translocations (Unniraman et al., 2004) could be amplified also from DNA of cells other than B cells (Ramiro et al., 2005). Nevertheless, a post-cleavage role for AID in CSR has been further suggested by the use of the AID mutant H48A or L49A, the use of Ugi, a specific peptide inhibitor of Ung, and that of an Ung mutant devoid of uracil DNA deglycosylation activity (Begum et al., 2004; Begum et al., 2007; Shivarov et al., 2008; Begum et al., 2009). As we and others have shown, AID-independent DSBs occur also in V(D)J DNA, where they also target preferentially the mutational 5′-RGYW-3′ hotspot (Bross et al., 2000; Papavasiliou and Schatz, 2000; Bross et al., 2002; Papavasiliou and Schatz, 2002a; Bross and Jacobs, 2003; Wu et al., 2003; Zan et al., 2003; Casali and Zan, 2004). These DSBs are blunt and 5′-phosphorylated, and are processed by AID and Ung, yielding staggered DSB ends, which are characteristic of B cells undergoing CSR and/or SHM (Zan et al., 2003; Casali and Zan, 2004; Zan and Casali, 2008). That AID can be recruited to DSBs is suggested by the AID retention rate in the nuclei of B cells treated with DNA break inducers, such as bleomycin, hydrogen peroxide or γ-rays (Brar et al., 2004).
How AID-independent DSBs are generated is still unclear. It is possible that these DSBs result from inherent proneness to breakage of S regions, perhaps related to their repetitive nature, as increased susceptibility to breakage is a property of DNA containing repetitive sequences capable of adopting non-canonical DNA conformations (Kataoka et al., 1983; Baar and Shulman, 1995; Chen et al., 2001; Tashiro et al., 2001; Bacolla et al., 2006). Indeed, CSR is associated with chromatin modifications, including hyperacetylation of histones in S regions and high levels of germline transcriptions through the S regions (Nambu et al., 2003; Wuerffel et al., 2007; Chowdhury et al., 2008; Stavnezer et al., 2008), that can result in DNA strand opening and increase fragility of S regions. This is particularly true for transcription through the highly repetitive S regions that leads to or enhances the formation of secondary DNA structures such as R-loops, G-loops or stem-loops. These structures arise as result of the high dG/dC content of S regions (R- and G-loops) as well as the high occurrence of palindromic 5′-AGCT-3′ (stems of stem-loops) (Baar et al., 1996; Mussmann et al., 1997; Chen et al., 2001; Tashiro et al., 2001; Yu and Lieber, 2003; Maizels, 2005; Xu et al., 2010). R-loops, G-loops and stem-loops contain highly distorted, long and complex, mostly single-strand DNA, which is not only inherently fragile but may undergo cleavage by structure-specific nucleases, such as EndoG, XPF-ERCC1, XPG, GQN1 and exonuclease 1 (Tian and Alt, 2000; Sun et al., 2001; Yu et al., 2003; Duquette et al., 2004; Irvine et al., 2005; Vallur and Maizels, 2008).
EndoG is an abundant nuclease in eukaryotic cells (Irvine et al., 2005). It is known as one of the endonucleases participating in caspase non-associated apoptosis, but its precise function in mammalian cells is still unclear (Irvine et al., 2005; David et al., 2006). EndoG is located primarily in the mitochondrial intermembrane space and is likely processed into a nuclear form and exported from mitochondria to the nucleus (Ohsato et al., 2002). In fact, EndoG was originally identified as a nuclear protein in cells that were not undergoing apoptosis (Ruiz-Carrillo and Renaud, 1987; Gerschenson et al., 1995; Ishihara and Shimamoto, 2006; Yu et al., 2006). It is a homodimer that cleaves both single- and double-strand DNA, with an effective R-loop cutting activity (Widlak et al., 2001; Ohsato et al., 2002; Irvine et al., 2005; Huang et al., 2006b; Wu et al., 2009). EndoG preferentially cleaves DNA at dG/dC residues (Widlak et al., 2001), which are also the preferential sites of DSBs in S regions (Rush et al., 2004; Schrader et al., 2005; Stavnezer and Schrader, 2006). It generates DSBs whose free ends are 5′-phosphorylated, like the DSB ends in S and V(D)J region DNA (Zan et al., 2003; Casali and Zan, 2004; Zan and Casali, 2008). The features of EndoG prompted us to hypothesize that this endonuclease underlies the generation of AID-independent DSBs in CSR. We tested our hypothesis in a variety of in vivo and in vitro experiments using EndoG−/− mice and EndoG−/− B cells. EndoG-deficient B cells did not show altered proliferation or apoptosis, but showed significantly reduced DSBs in S regions and impaired CSR, in association with altered spectrum of point-mutations in the IgH locus. Thus, our findings define an important role for EndoG in the generation of S region DSBs and CSR.
2. Materials and methods
2.1. Mice
The EndoG−/− mice were generated by Dr. Michael R. Lieber (University of Southern California, Los Angeles, CA) by deleting exon 2 of the EndoG gene, which causes exon 3 to be out of frame, when spliced to exon 1 (Irvine et al., 2005). No truncated protein was produced from the disrupted allele and no nuclease activity was detected in EndoG−/− mice (Irvine et al., 2005). These mice were viable and show no age-related or generational abnormalities, nor did they show altered mitochondrial DNA copy number, structure or mutation rate. Their cells showed no obvious changes in nuclear DNA degradation in apoptosis assays (Irvine et al., 2005). All animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of University of California, Irvine, CA.
2.2. B and T cells
The monoclonal human 4B6 B cell line was derived from our CSR- and SHM-inducible human monoclonal IgM+IgD+ CL-01 B cell line (Zan et al., 1998) by sequential subcloning. 4B6 B cells express significant level of AID and undergo spontaneous CSR to IgG and IgA (Zan et al., 2003; Zan and Casali, 2008; Xu et al., 2010). Single cell suspensions were prepared from spleen or Peyer’s patches of EndoG+/+ and EndoG−/− mice and stained with PE-labeled anti-mouse B220 mAb (clone RA3-6B2, BD Biosciences, San Jose, CA), allophycocyanin (APC)-labeled anti-mouse CD3 mAb (145-2C11, eBiosciences, Inc., San Diego, CA), fluorescein isothiocyanate (FITC)-labeled mAb to mouse CD4 (GK1.5, eBiosciences), 7-amino-actinomycin D (7-AAD, BD Biosciences), or FITC-labeled peanut agglutinin (PNA, E-Y Laboratories, San Mateo, CA). The number of B cells (B220+) and T cells (CD3+), the proportions CD4+ T cells, the level of B- and T-cell death and the proportions of germinal center (B220+PNAhi) B cells were determined by flow cytometry analysis. Single B220+ cell suspensions were prepared from the spleen or Peyer’s patches using a B lymphocyte enrichment set (BD Biosciences). For the preparation of germinal center (B220+PNAhi) B cells, Peyer’s patch B cells were stained with phycoerythrin (PE)-labeled anti-mouse CD45R (B220) rat mAb (RA3-6B2, eBiosciences) and FITC-labeled PNA. Labeled lymphocytes were then sorted using a MoFlow™ cell sorter (Dako-Cytomation, Fort Collins, CO), yielding 95% pure B220+PNAhi cells.
2.3. B cell cycle, proliferation and apoptosis
The cell cycle was analyzed by propidium iodide (PI) staining (Zan et al., 2003; Park et al., 2009). Cell proliferation was analyzed using the CellTrace™ carboxyfluorescein succinimidyl ester (CFSE) proliferation kit (Molecular Probes, Eugene, OR) (Park et al., 2009; Mai et al., 2010). Cells were washed in serum-free Hank’s balanced-salt solution (HBSS, Invitrogen Corp., Carlsbad, CA) and resuspended at a density of 1 × 107 cells/ml. After adding an equal volume of 10 μM CFSE, the cell suspension was incubated at 37°C for 12 min and then washed 3 times with RPMI medium containing 10% FBS. Cells were then diluted and cultured with LPS (10 μg/ml) from Escherichia coli (serotype 055:B5; Sigma-Aldrich, Inc., St. Louis, MO) in the presence or absence of recombinant mouse interleukin 4 (rmIL-4, 5 ng/ml; R&D Systems, Inc., Minneapolis, MN). They were harvested 2 or 4 days later and analyzed using a FACSCalibur™ flow cytometer.
To analyze B cell proliferation and apoptosis in vivo, mice were injected intraperitoneally with NP16-CGG. After 10 days, they were injected intraperitoneally twice within 16 hours with bromodeoxyuridine (BrdU) (1 mg) and sacrificed 4 hours after the last injection. Cells from the spleen or Peyer’s patches were stained with PE-anti-B220 mAb (RA3-6B2) or this mAb together with FITC-PNA. Cells were then fixed and incubated with the permeablization buffer for 15 min in the dark at 4°C, and then stained with allophycocyanin (APC)-conjugated anti-BrdU mAb using the BrdU Flow Kit (BD Biosciences) (Zan et al., 2005; Park et al., 2009). Cells were analyzed using a FACSCalibur™ flow cytometer. To measure apoptosis, cells from NP16-CGG immunized mice were stained with 7-AAD in combination with PE-anti-B220 mAb or APC-anti-CD3 mAb and FITC-PNA and incubated for 30 min at 4°C in the dark. Cells were then washed and analyzed using a FACSCalibur™ flow cytometer. Terminal deoxynucleotidyl transferase (TdT) mediated dUTP nick-end-labeling (TUNEL) staining with TdT in situ Apoptosis Detection Kit (R&D Systems, Inc.) was also used to quantify apoptosis by counting TUNEL signals (red dots) using the SPOT Software (Diagnostic Instruments, Inc.).
2.4. Analysis of CSR
To analyze in vivo CSR, serum IgM, IgG1, IgG3 and IgA concentrations were measured by specific ELISAs (Park et al., 2009), and germinal center (PNAhiB220+) B cells expressing IgG1 or IgA were identified using a FACSCalibur™ flow cytometer (BD Biosciences) (Zan et al., 2009). To analyze in vitro CSR, B cells were isolated from red blood cells (RBC)-depleted mice splenocytes using a B cell enrichment kit (StemCell Technologies Inc., Canada) and were cultured at 5 × 105 cell/ml in FCS-RPMI medium (RPMI-1640 medium (Invitrogen) supplemented with 10% (vol/vol) heat-inactivated FBS (Hyclone), 2 mM L-glutamine and 1x antibiotic-antimycotic mixture (penicillin (100 units/ml), streptomycin (100 mg/ml) and amphotericin B fungizone (0.25 mg/ml); Invitrogen)) with 0.05 mM β-mercaptoethanol in 48-well plates and stimulated with LPS (10 μg/ml) with: nil for CSR to IgG3; rmIL-4 (5 ng/ml; R&D Systems, Inc.) for CSR to IgG1; IFN-γ (50 ng/ml; PeproTech, Inc., Rocky Hill, NJ) for CSR to IgG2a; or TGF-β (1 ng/ml; R&D Systems, Inc.), rmIL-4 (5 ng/ml), and rmIL-5 (5 ng/ml; R&D Systems, Inc.) and anti-IgD mAb-dextran (3 ng/ml; provided by Dr. Snapper, C.M., Uniformed Services University of the Health Sciences, Bethesda, MD) for CSR to IgA. Cells were collected on day 3 for analysis of germline IH-CH, circle IH-Cμ and post-recombination Iμ-CH transcripts, and on day 4 for analysis of surface Igs after staining with FITC-labeled rat mAb to mouse IgG1 (A85-1), mouse IgG2a (R19-15), mouse IgG3 (R40-82) or mouse IgA (C10-3), or PE-labeled rat mAb to mouse CD45R (B220; RA3-6B2; all from BD Biosciences). IgG1 concentration in culture supernatants of LPS and rmIL-4-stimulated B cells were measured by specific ELISA (Park et al., 2009).
2.5. Quantitative real-time RT-PCR (qRT-PCR) and semiquantitative RT-PCR
RNA was extracted using the RNeasy Mini Kit (Qiagen Inc., Valencia, CA) according to the manufacturer’s protocol. Residual DNA was removed by treatment with DNase I (Invitrogen Corp., Carlsbad, CA). First strand cDNAs were synthesized from equal amounts of total RNA (2 μg) using the SuperScript™ Preamplification System and oligo (dT) primer (Invitrogen Corp.). The expression of Aicda, germline IH-CH, circle IH-Cμ and post-recombination Iμ-CH transcripts was quantified by real-time qRT-PCR using appropriate primers (Park et al., 2009). Real-time qRT-PCR analysis was performed using an DNA Engine Opticon 2 Real-Time PCR Detection System (Bio-Rad Laboratories, Inc., Hercules, CA) to measure SYBR-green (DyNAmo HS SYBR Green, New England Biolabs, Inc., Ipswich, MA) incorporation with the following protocol: 50 °C for 2 min, 95 °C for 10 min, 40 cycles of 95 °C for 10 sec, 60 °C for 20 sec, 72 °C for 30 sec, 80 °C for 1 sec, and data acquisition at 80 °C, and 72 °C for 10 min. Melting curve analysis was performed from 72°–95 °C and samples were incubated for another 5 min at 72 °C. The ΔΔCt method was used for data analysis. In some cases, VHDJH-Cμ and GAPDH/Gapdh transcripts were analyzed by specific semiquantitative RT-PCR, involving serial twofold diluted template cDNA so there was a nearly linear relationship between the amount of cDNA used and the intensity of the PCR product (Zan and Casali, 2008; Park et al., 2009).
2.6. Cell fractionation and immunoblotting
B cells (3 × 107) were washed with PBS after centrifugation through a Ficoll® gradient to clear debris and dead cells, and resuspended in hypotonic buffer (20 mM Tris pH 7.5, 10 mM KCl, 10 mM MgCl2, 1x protease inhibitors mix (Sigma-Aldrich)) on ice for 10 min. After adding Nonidet P-40 (1% final concentration), the mixture was vortexed and then placed on ice for 1 min. Following centrifugation at 1000 g for 5 min, the supernatant was clarified by centrifugation (5 min at 20,000 g) and collected as cytoplasmic fraction; the pellet was resuspended in S1 solution (0.25 M sucrose, 5.0 mM MgCl2) and layered on a sucrose cushion S2 (0.35 M sucrose, 5 mM MgCl2), before being centrifuged at 3,000 g for 5 min. The pellet was resuspended in S1 solution, re-centrifuged, resolublized and then collected as nuclear fraction.
For immunoblotting analysis, equal amount (25 μg) of proteins were separated through 10 % SDS-PAGE. Proteins were transfered onto polyvinylidene difluoride membranes (Bio-Rad Laboratories) overnight (30 V) at 4 °C. The blocked membranes were incubated overnight at 4 °C with rabbit Ab to Cox4 isoform 1 (NB110-39115, Novus Biologicals), mouse mAb to PCNA (PC-10, BD Biosciences) or mouse mAb to histone H1 (AE-4, Santa Cruz Biotechnology). The membranes were then incubated with horseradish peroxidase (HRP)-conjugated secondary Abs. After washing with PBS-Tween 20 (0.05%), bound HRP-conjugated Abs or mAbs were detected using Western Lightning® Plus–Enhanced Chemiluminescence reagents (PerkinElmer Life and Analytical Sciences).
2.7. Chromatin immunoprecipitation (ChIP) assays
B cells (5 × 107) were treated with 1% formaldehyde for 10 min at 25°C to cross-link chromatin. After washing with cold PBS containing protease inhibitors (Roche, Basel, Switzerland), chromatin was separated using nuclear lysis buffer (10 mM Tris-HCl, 1 mM EDTA, 0.5 M NaCl, 1% Triton-X-100, 0.5% Sodiumdeoxycholate, 0.5% Sarcosyl, pH 8.0) and resuspended in IP-1 buffer (20 mM Tris-HCl, 200 mM NaCl, 2 mM EDTA, 0.1% Sodiumdeoxycholate, 0.1% SDS, protease inhibitors). The chromatin was sonicated to yield approximately 200–1000 bp DNA fragments. DNA fragments were precleared with agarose beads bearing protein G (Santa Cruz Biotechnology, Santa Cruz, CA) and then incubated with rabbit IgG to EndoG (FL-297, Santa Cruz Biotechnology) (Zhao et al., 2010) or mouse IgG1 mAb to AID (clone ZA001, Invitrogen) (Xu et al., 2010). After overnight incubation at 4°C, immune complexes were isolated using beads bearing protein G, eluted with Elution buffer (50 mM Tris-HCl, 0.5% SDS, 200 mM NaCl, 100 μg/ml Proteinase K, pH 8.0), and then incubated at 65°C overnight to reverse the formaldehyde cross-links. DNA was recovered by phenol/chloroform extraction followed by ethanol precipitation, and then resuspended in TE buffer (10 mM Tris-HCl, pH 8.0, 1.0 mM EDTA). Recovered DNA was specified by PCR using the following oligonucleotide primers: human Sμ, forward 5′-ATGGAAGCCAGCCTGGCTGT-3′ and reverse 5′-CAGTTAGTGCAGCCAAGCCCTAGCTCAG-3′; human Sγ1, forward 5′-ACCCTATGCAGTGTCTGGCCCCTC-3′ and reverse 5′-AGTCAGCACAGTCCAGTGTCTCTAG-3′; human Cμ, forward 5′-CACGTGGTGTGCAAAGTCCAGCACC-3′ and reverse 5′-ACGCCAGACCCCACCTGCTT-3′; human VHDJH, forward 5′-CAGGGAAGGGGCTGGAGTGG-3′ and reverse 5′-AGGAAACCCCACAGGCAGTAGCA-3′; mouse Sμ, forward 5′-CTGAGGTGATTACTCTGAGGT-3′ and reverse 5′-CTCCAGAGTATCTCATTTCAG-3′; mouse Sγ1, forward 5′-GAGCCAAGGTAGGTGGAATGTG-3′ and reverse 5′-C TGTGACTGCTTAGAATCCCCAAC-3′; mouse Cμ, forward 5′-GGCTTCTACTTTACCCACAGCATC-3′ and reverse 5′-CATACACAGAGCAACTGGACACCC-3′.
2.8. DNA breaks
The biotin-labeling DNA break assays were performed as reported (Begum et al., 2009; Doi et al., 2009). Live B cells were separated through a Ficoll® gradient, followed by fixation, permeabilization and in situ DNA end-labeling with biotin-16-dUTP (bio-dUTP) using TdT to detect overall DNA cleavages, including single-strand breaks (SSBs) and DSBs, or ligated with the biotinylated double-strand anchor linker BW (bio-linker) with or without pre-treatment of T4 DNA polymerase and dNTP to detect blunt or total (blunt plus staggered) DSBs (Zan et al., 2003; Zan and Casali, 2008; Begum et al., 2009; Doi et al., 2009). Genomic DNA was prepared using QIAamp DNA Mini Kit (Qiagen, Inc.) and digested with Pvu II and Pst I. Biotin-labeled DNA fragments were captured by streptavidin magnetic beads (Promega Corp., Madison, WI). For quantification of broken DNA, biotin-labeled DNA was serially twofold diluted and used as template for Sμ, Sγ1 and Cμ DNA amplification. PCR amplifications were performed at 94°C for 30 sec, 55°C for 45 sec, 72°C for 30 sec for 32 cycles using GoTaq® Hot Start Polymerase (Promega Corp.) and mouse Sμ, Sγ1 or Cμ specific forward and reverse primers described above.
2.9. S-S DNA junction sequences
Genomic DNA was prepared from B cells cultured for 4 days with LPS and rmIL-4. Sμ-Sγ1 junction DNA were amplified using two sequential rounds of specific PCR (Wu et al., 2006) using Phusion™ high-fidelity DNA polymerase (New England Biolabs, Inc.) and the following nested oligonucleotide primers: forward 5′-AACTCTCCAGCCACAGTAATGACC-3′ and reverse 5′-CTGTAACCTACCCAGGAGACC-3′ for the first round; forward 5′-ACGCTCGAGAAGGCCAGCCTCATAAAGCT-3′ and reverse 5′-GTCGAATTCCCCCATCCTGTCACCTATA-3′ for the second round. The first and second rounds of PCR were performed at 94 °C for 45 sec, 55 °C for 45 sec, 72 °C for 4 min, 30 cycles. PCR DNA products were purified using Qiaquick PCR purification kit (Qiagen, Inc.) and cloned into pCR-Blunt II-TOPO® vector (Invitrogen, Corp.) for sequencing. Sequence alignment was done by comparing the sequences of PCR products with Sμ and Sγ1 genomic sequences using NCBI blast http://www.ncbi.nih.gov/BLAST).
2.10. Somatic mutations in intronic VJ558DJH4-iEμ DNA
Peyer’s patches germinal center (PNAhiB220+) B cells were obtained from one pair of 10 week-old and one pair of 28 week-old non-intentionally immunized littermate EndoG−/− and EndoG+/+ mice and were used for analysis of somatic mutations in the intronic DNA downstream of rearranged VJ558DJH4 genes (Zan et al., 2005). The intronic heavy chain region downstream of rearranged VJ558DJH was amplified from genomic DNA by nested PCR using Phusion™ high-fidelity DNA polymerase and primer pairs: forward 5′-AGCCTGACATCTGAGGAC-3′ and reverse 5′-TCTGATCGGCCATCTTGACTC-3′ for the first round PCR and forward: 5′-CATCTGAGGACTCTGCNGTCTAT-3′ and reverse 5′-CCTCACTCCCATTCCTCGGTTAAA -3′ for the second round PCR. Reaction conditions for both the first and second round PCR were 30 cycles of 98°C for 15 sec, 58°C for 45 sec and 72°C for 1 min. PCR products were cloned into the pCR-Blunt II-TOPO® vector (Invitrogen, Corp.) and sequenced. Only sequences from rearrangements involving JH4-iEμ were analyzed. Sequences were analyzed with MacVector software (MacVector, Inc., Cary, NC).
2.11. Statistical analyses
The differences in frequency and spectrum of mutations in EndoG−/− and EndoG+/+ mice were analyzed using the χ2 test. The differences in Ig titers were analyzed using paired t-tests.
3. Results
3.1. EndoG localizes to the nucleus and is specifically recruited to S regions
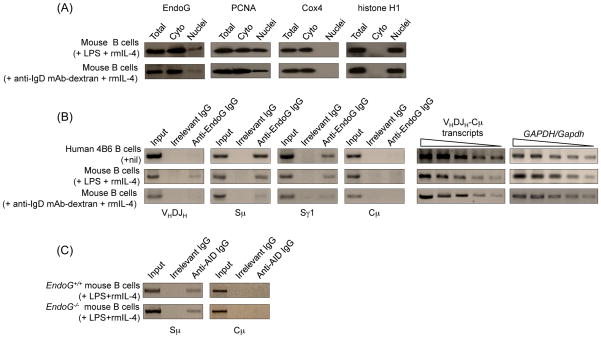
EndoG localizes primarily in the mitochondrial inter-membrane space and can translocate to the nucleus in caspase-independent apoptosis (David et al., 2006). In addition, EndoG can translocate to the nucleus in the absence of significant apoptosis and plays a role in DNA recombination and cellular proliferation (Huang et al., 2006a). To test the hypothesis that EndoG can localize to the nucleus in B cells undergoing CSR, we stimulated mouse B cells with LPS and rmIL-4, which induce CSR to IgG1, or with anti-IgD mAb-dextran and rmIL-4, which induce B cell proliferation but not CSR, and, after a 2-day culture, analyzed EndoG expression and localization by specific immunoblotting. EndoG localized to both the cytoplasm and the nucleus of B cells treated with anti-IgD mAb-dextran and rmIL-4, but its density at both locations increased significantly after stimulation with LPS and rmIL-4 (Fig. 1(A)). The purity of the cytoplasm and nuclear fractions used for immunoblotting was confirmed by the detection of PCNA, which is known to localize to both cytoplasma and nucleus, in both cytoplasm and nucleus fractions, and the absence of cytoplasmic protein Cox4 or nuclear protein histone H1 in the nucleus and cytoplasm fractions, respectively, of B cells. In addition, ChIP assays showed that EndoG was recruited to Sμ and Sγ1 but not Cμ DNA in both spontaneously switching human 4B6 B cells, and mouse B cells stimulated with LPS and rmIL-4 (Fig. 1(B)). In LPS and rmIL-4-stimulated mouse B cells, a small amount of EndoG was also recruited to VHDJH region DNA. The lack of recruitment of EndoG to Cμ DNA did not result from lack of transcription of this region, as indicated by the high VHDJH-Cμ transcript levels. Further, the recruitment of EndoG to S and VHDJH region DNA was significantly less in B cells treated with anti-IgD mAb-dextran and rmIL-4 as compared to B cells stimulated with LPS and rmIL-4. Finally, the absence of EndoG did not affect recruitment of AID to S region DNA, as indicated by lack of alteration in AID recruitment to Sμ in EndoG−/− B cells stimulated with LPS and rmIL-4 (Fig. 1(C)).
Fig. 1.
EndoG is located in both the cytoplasm and the nucleus, and is specifically recruited to S regions in human and mouse B cells that undergo CSR. (A) EndoG, PCNA, Cox4 and histone H1 proteins in whole cell extracts, cytoplasm and nuclei of wild-type C57BL/6 mouse spleen B cells stimulated with LPS and rmIL-4, or anti-IgD mAb-dextran and rmIL-4 for 2 days, as detected by immunoblotting. (B) EndoG is recruited to S region but not CH region DNA. Left and middle panels: cross-linked chromatin was precipitated from human spontaneously switching 4B6 or C57BL/6 mouse spleen B cells stimulated with LPS and rmIL-4, or anti-IgD mAb-dextran and rmIL-4 for 2 days using a rabbit IgG Ab specific for EndoG or preimmune control rabbit IgG. The precipitated DNA was specified by PCR using human or mouse VHDJH, Sμ, Sγ1 or Cμ primers; right panels: VHDJH-Cμ and GAPDH/Gapdh transcripts in 4B6, LPS and rmIL-4- or anti-IgD mAb-dextran and rmIL-4-stimulated mouse spleen B cells were amplified by RT-PCR using two-fold diluted template cDNA. (C) EndoG-deficiency does not alter the recruitment of AID to S regions. Cross-linked chromatin was precipitated from EndoG+/+ and EndoG−/− B cells stimulated with LPS and rmIL-4 for 2 days using a IgG1 mAb specific for AID or a control mouse IgG1. The precipitated DNA was specified by PCR using mouse Sμ or Cμ primers.
3.2. EndoG−/− mice show normal B and T cells, and germinal center formation
To directly address the role of EndoG in CSR, we turned to EndoG−/− mice and EndoG−/− B cells. As EndoG possibly modulates cell division, by virtue of its role in apoptosis, we first examined EndoG−/− mice for any defect in B and T cell development or proliferation and germinal center formation.
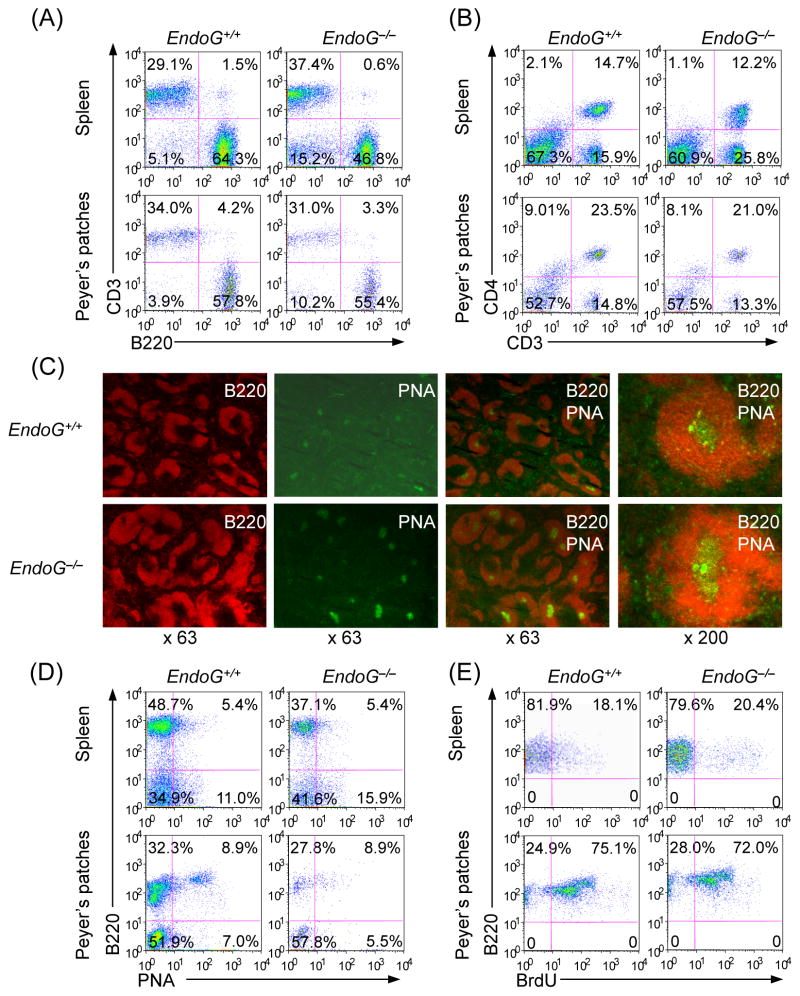
In EndoG−/− mice, the size of the spleen and the number and size of Peyer’s patches were similar to those of EndoG+/+ mice (not shown). Moreover, the number of B and T cells, the proportion of CD4+ T cells in the spleen and Peyer’s patches were also comparable to those of EndoG+/+ mice (Fig. 2(A) and 2(B)). To examine whether EndoG deficiency has effect on germinal center formation, we immunized 8–10 weeks old littermate EndoG−/− and EndoG+/+ mice with NP16-CGG. Mice were sacrificed and spleen sections were stained with FITC-PNA and PE-anti-B220 mAb 14 days later (Park et al., 2009). The number and architecture of germinal centers in the spleen of EndoG−/− mice were comparable to those of EndoG+/+ mice (Fig. 2(C)). In addition, in EndoG−/− mice, the proportion of germinal center (B220+PNAhi) B cells in both spleen and Peyer’s patches, the proportion of proliferating B cells, as shown by in vivo incorporation of the thymidine analog BrdU, were not significantly different from those of EndoG+/+ mice (Fig. 2(D) and 2(E)).
Fig. 2.
EndoG deficiency does not affect B cell, T cell or CD4+ T cell numbers, germinal center formation, germinal center B cells or in vivo B cell proliferation. (A and B) Flow cytometric profiles of spleen and Peyer’s patch cells stained with PE-anti-B220 and FITC-anti-CD3 (left panels) or FITC-anti-CD3 and PerCP-anti-CD4 (right panels). Data are representative of three experiments. (C) Staining of germinal center. Spleen sections prepared from EndoG+/+ and EndoG−/− mice 10 days after immunization with NP16-CGG. Germinal center B cells were identified by staining with PE-anti-B220 mAb and FITC-PNA. The original magnifications are indicated at the bottom of each set of panels. Data are representative of three experiments. (D) Flow cytometric profiles of cells from Peyer’s patches of NP16-CGG immunized EndoG+/+ and EndoG−/− mice stained with PE-labeled anti-B220 mAb and FITC-PNA. Data are from one representative of three pairs of EndoG+/+ and EndoG−/− mice. The proportions of spleens and Peyer’s patches B220+PNA− or B220+PNA+ cells in EndoG−/− mice were not significantly different from that in EndoG+/+ mice (p > 0.2, paired t-test). (E) In vivo B cell proliferation. Three 10 week-old EndoG+/+ and EndoG−/− mice were immunized with NP16-CGG. After 10 days, the mice were injected with BrdU (1 mg) twice within 16 hours and sacrificed 4 hours after the last injection. Cells from spleen and Peyer’s patches were stained with PE-anti-mouse B220 mAb or this mAb together with FITC-PNA. Incorporated BrdU was detected with APC-anti-BrdU mAb and analyzed by flow cytometry. Data are from one representative of three pairs of EndoG+/+ and EndoG−/− mice. The proportions of proliferating (B220+BrdU+) B cells in spleens and Peyer’s patches of EndoG−/− mice were not significantly different from that of EndoG+/+ mice (p > 0.2, paired t-test).
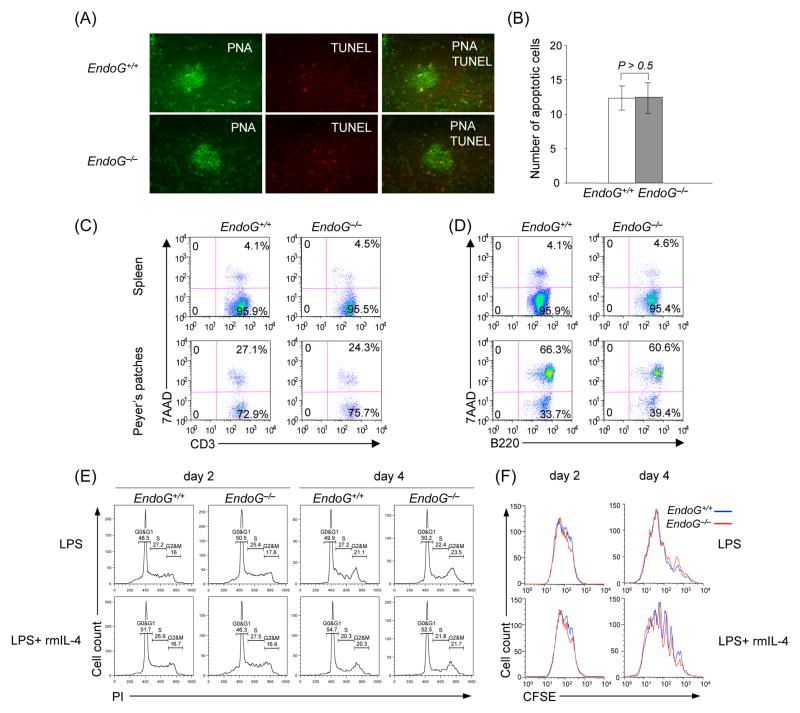
As EndoG can be an apoptosis-related protein, we examined whether EndoG deficiency affects B cell apoptosis/viability, cell cycle or proliferation. The proportion of apoptotic (TUNEL+) cells in the spleen germinal center (PNA+) cells of EndoG−/− mice was comparable to that of EndoG+/+ mice, as shown by the immunofluorescent staining (Fig. 3(A) and 3(B)). Peyer’s patches and spleen cells were further stained with APC-anti-CD3 mAb, PE-anti-B220 mAb and 7-AAD, and live/dead T and B cells were analyzed by flow cytometry. The proportion of apoptosis/necrosis (7-AAD+) T cells (CD3+) and B cells (B220+) in EndoG−/− mice were comparable to those in EndoG+/+ mice (Fig. 3(C) and 3(D)). After stimulation with LPS and IL-4, EndoG−/− B cells behaved similarly to EndoG+/+ cells in terms of cell cycle, as analyzed by staining with propidium iodide (PI), and cell division rate, as measured by incorporation of the vital dye CFSE (Fig. 3(E) and 3(F)). Thus, EndoG−/− mice displayed normal B and T cells numbers and viability, germinal center formation and B cells proliferation.
Fig. 3.
EndoG deficiency does not affect B and T cell apoptosis/viability, B cell cycle or proliferation. (A) Spleen sections harvested from EndoG+/+ and EndoG−/− mice 14 days after immunization with NP16-CGG were stained with FITC-PNA and TUNEL and analyzed by fluorescence microscopy. The number of red dots reflected TUNEL staining of fragmented DNA, which is characteristic of apoptosis (original magnification: 200 X). This was not significantly different in EndoG+/+ and EndoG−/− mice germinal centers as shown by quantitative comparison of TUNEL+ cells (histograms) (B). (C and D) Flow cytometric profiles of cells from spleen and Peyer’s patches stained with 7-AAD and APC-anti-CD3 mAb (left panels), and 7-AAD and PE-anti-B220 mAb (right panels). (E) Cell cycle analysis of EndoG+/+ and EndoG−/− B cells. Spleen EndoG+/+ and EndoG−/− B cells were stimulated with LPS and rmIL-4 and harvested after 2 and 4 days for PI staining and flow cytometry analysis to measure DNA content and enumerate cells in G0/G1, S and G2/M phases. (F) Cell division in EndoG+/+ and EndoG−/− B cells. Spleen EndoG+/+ and EndoG−/− B cells were labeled with CFSE, cultured with LPS and rmIL-4, and harvested 2 and 4 days later for flow cytometry analysis; cell division is indicated by progressive left shift of fluorescence histograms. Data are from one representative of three independent experiments.
3.3. EndoG deficiency impairs CSR in vivo and in vitro
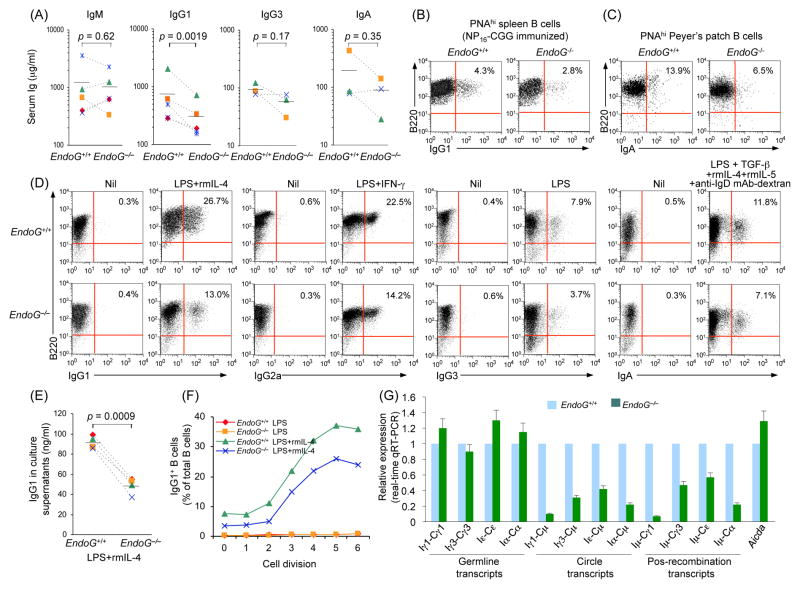
We next examined the in vivo CSR in EndoG−/− mice. In unimmunized EndoG−/− mice, serum IgM concentration were normal (p = 0.62). However, the average serum IgG1 concentration was reduced by almost 60% as compared to their EndoG+/+ counterparts (p = 0.0019, Fig. 4(A)). In addition, serum IgG3 and IgA concentrations were also reduced, albeit less significantly (p = 0.17 and 0.35, respectively), in these mice. Because of the long half-life of plasma cells and antigen-mediated selection, Igs can accumulate in the circulation in significant amounts even when CSR is substantially compromised (Rada et al., 2002). Hence, the lack of reduction of total serum IgG titers in a previous report (Irvine et al., 2005) did not necessarily reflect a lack of CSR impairment. We immunized 10 week-old EndoG−/− and EndoG+/+ littermate mice with NP16-CGG. 14 days later, we harvested the spleen cells and stained them with FITC-conjugated PNA, PE-labeled anti-B220 mAb and APC-labeled anti-IgG1 mAb. We found that the proportion of sIgG1+ B cells among germinal center (PNAhiB220+) B cells in EndoG−/− mice was reduced by more than 34% as compared to their EndoG+/+ littermates (2.8% vs. 4.3%, Fig. 4(B)). Likewise, in the Peyer’s patches of unimmunized 12 week-old EndoG−/− mice, the number of germinal center (PNAhiB220+) B cells switched to IgA was reduced by more than 53% (6.5% vs. 13.9%, Fig. 4(C)).
Fig. 4.
EndoG deficiency impairs CSR in vivo and in vitro. (A) Titers of circulating IgM, IgG1, IgG3 and IgA in unimmunized EndoG+/+ and EndoG−/− littermates. P values, paired t-test. Data are from five (IgM and IgG1) or three (IgG3 and IgA) pairs of EndoG+/+ and EndoG−/− mice. (B) Spleen cells from EndoG+/+ and EndoG−/− mice 14 days after immunization with NP16-CGG were stained with FITC-conjugated PNA, PE-labeled anti-B220 mAb and APC-labeled anti-IgG1 mAb; the sIgG1+ B cells among germinal center (PNAhiB220+) B cells were analyzed by flow cytometry. (C) Cells from Peyer’s patches of 12 week-old unimmunized EndoG+/+ and EndoG−/− mice were stained with Alexa Fluor® 647-labeled PNA, PE-labeled anti-B220 mAb and FITC-labeled anti-IgA mAb; sIgA+ B cells among germinal center (PNAhiB220+) B cells were measured by flow cytometry. Data are representative of three independent experiments. (D) Spleen EndoG+/+ and EndoG−/− B cells were stimulated with LPS in the presence of nil (for CSR to IgG3), rmIL-4 (for CSR to IgG1), IFN-γ (for CSR to IgG2a), or TGF-β1/rmIL-4/rmIL-5/anti-IgD mAb-dextran (for CSR to IgA). After a 4-day culture, the cells were stained with PE-anti-mouse B220 mAb and FITC-anti-mouse IgG1, IgG2a, IgG3, or IgA mAb for cell surface analysis (the number inside each panel indicates the percentage of B220+ cells that are positive for the indicated Ig isotypes). (E) IgG1 titers in supernatants of cultures of EndoG+/+ and EndoG−/− B cells stimulated for 4 days with LPS and rmIL-4. P values, paired t-test. Data are from B cells of four pairs of EndoG+/+ and EndoG−/− mice. (F) EndoG+/+ and EndoG−/− B cells were labeled with cell division-tracking fluorochrome CFSE and stimulated with LPS or LPS and rmIL-4 for 4 days. CFSE intensity and surface IgG1 expression were analyzed by flow cytometry. Proportion of surface IgG1+ B cells at each cell division is indicated. (G) EndoG deficiency does not alter Aicda expression and germline IH-CH transcripts but decreases CSR as indicated by the lower expression of circle IH-Cμ and post-recombination Iμ-CH transcripts. Real-time qRT-PCR analysis of Aicda expression, germline IH-CH, circle IH-Cμ and post-recombination Iμ-CH transcripts in EndoG+/+ and EndoG−/− splenic B cells cultured for 3 days with LPS alone (Iγ3-Cγ3, Iγ3-Cμ and Iμ-Cγ3), or with LPS and rmIL-4 (Aicda, Iγ1-Cγ1, Iγ1-Cμ, Iμ-Cγ1, Iε-Cε, Iε-Cμ and Iμ-Cε) or LPS and TGF-β/rmIL-4/rmIL-5/anti-IgD mAb-dextran (Iα-Cα, Iα-Cμ and Iμ-Cα); expression is normalized to Cd79b expression and is presented relative to the expression in EndoG+/+ B cells, set as 1.
To further determine the impact of the EndoG deficiency on CSR, we stimulated spleen B cells with LPS (to induce switching to IgG3), LPS and rmIL-4 (to induce switching to IgG1), LPS and IFN-γ (to induce switching to IgG2a) or LPS in the presence of TGF-β1, rmIL-5, rmIL-4 and anti-IgD mAb-dextran (to induce switching to IgA). After 4 days of culture, the proportion of live EndoG−/− (7-AAD−B220+) B cells positive for surface IgG1, IgG2a, IgG3 or IgA was reduced by 51%, 37%, 53% or 40%, respectively, as compared to that of the corresponding EndoG+/+ B cells (Fig. 4(D)). Consistent with this, the secretion of IgG1 by EndoG−/− B cells stimulated with LPS and rmIL-4 was 44% lower than their EndoG+/+ counterparts (p = 0.0009, Fig. 4(E)).
Impaired CSR in EndoG−/− B cells was not due to altered proliferation, as after 2 or 4 days of culture with LPS or LPS plus rmIL-4, EndoG−/− B cells completed the same number of divisions as their EndoG+/+ counterparts did (Fig. 3(F)). However, the percentage of IgG1 positive B cells was twofold greater among EndoG+/+ B cells than EndoG−/− B cells that had undergone the same number of divisions (Fig. 4(F)). Further, the reduced CSR in EndoG−/− B cells was not due to impairment of Aicda or germline transcription of the intervening heavy-chain (IH) region and constant heavy-chain (CH) region (IH-CH), which is necessary for CSR. Real-time qRT-PCR showed that the abundance of Aicda and germline Iγ1-Cγ1, Iγ3-Cγ3, Iε-Cε and Iα-Cα transcripts in EndoG−/− B cells stimulated for 3 days with LPS alone or LPS plus cytokines was comparable to that in their EndoG+/+ B cell counterparts (Fig. 4(G)), whereas circle Iγ1-Cμ, Iγ3-Cμ, Iε-Cμ and Iα-Cμ transcripts, and post-recombination Iμ-Cγ1, Iμ-Cγ3, Iμ-Cε and Iμ-Cα transcripts, which are generated only after a CSR event, were significantly reduced in EndoG−/− B cells. Thus, EndoG deficiency impairs CSR without affecting Aicda expression or germline IH-CH transcription.
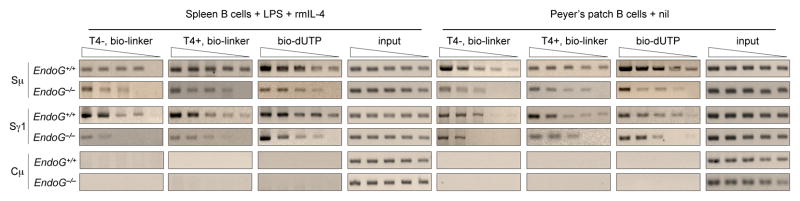
3.4. EndoG deficiency reduces DSBs in S region DNA
To further understand whether EndoG contributes to CSR by generating DNA breaks, we analyzed blunt and total (blunt and staggered) DSBs as well as overall DNA cleavages (SSBs and DSBs) in S regions in LPS and rmIL-4-stimulated EndoG−/− and EndoG+/+ B cells, as well as EndoG−/− and EndoG+/+ Peyer’s patch B cells. For the detection of blunt and total DSBs, DNA ends were in situ ligated with the bio-linker with or without pre-treatment with T4 DNA polymerase and dNTP (Zan et al., 2003; Zan and Casali, 2008; Begum et al., 2009; Doi et al., 2009). To evaluate the overall degree of DNA cleavage, DNA ends were in situ labeled by bio-dUTP using TdT (Begum et al., 2009; Doi et al., 2009). After in situ bio-linker ligation or bio-dUTP labeling, DNA extraction and digestion with restriction endonuclease, the biotin-labeled DNA fragments were isolated with streptavidin magnetic beads, serially twofold diluted and used as templates in specific PCR to amplify Sμ, Sγ1, and, potentially, Cμ regions. Blunt and total DSBs as well as the overall DNA cleavages were readily detected in Sμ and Sγ1 regions EndoG+/+ B cells (Fig. (5)). The S region DNA breaks were significantly reduced in EndoG−/− B cells. Consistent with our previous findings by LM-PCR (Zan et al., 2003; Zan and Casali, 2008), no breaks were detected in Cμ region DNA, further supporting the specificity of these findings. Thus, EndoG plays an important role in the generation of S region DNA breaks in B cells undergoing CSR.
Fig. 5.
DSBs are reduced in EndoG−/− B cells. EndoG+/+ and EndoG−/− spleen B cells stimulated with LPS and rmIL-4 for 2 days or unstimulated Peyer’s patches B cells were analyzed for DNA break ends. DNA was labeled in situ with biotin-dUTP using TdT (to detect overall DNA cleavages, including both SSBs and DSBs), or biotin-labeled BW linker (biotin-linker) with or without pre-treatment of T4 DNA polymerase and dNTP (to detect total, including 5′-phosphorylated blunt and staggered DSBs, or 5′-phosphorylated blunt DSBs, respectively). Sμ, Sγ1 and Cμ DNA were amplified from serially twofold diluted biotin-labeled DNA and the input DNA by PCR. Data are representative of three independent experiments.
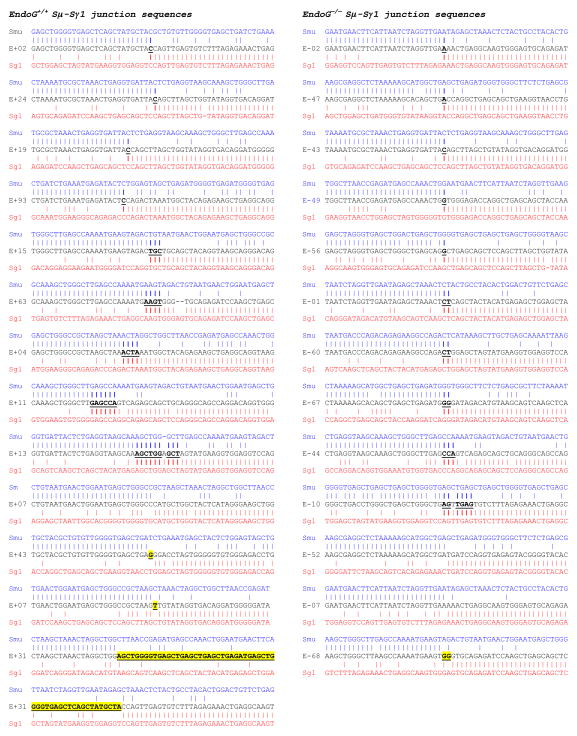
3.5. S-S region DNA junctions in EndoG−/− B cells
To further address the role of EndoG in CSR, we sequenced the recombined Sμ-Sγ1 DNA junctions of LPS and rmIL-4-stimulated EndoG−/− and EndoG+/+ B cells and compared them with the respective Sμ and Sγ1 genomic templates to determine the degree of overlap (microhomology) of the upstream Sμ and downstream Sγ1 regions, and the frequency of inserted nucleotides between the upstream Sμ and downstream Sγ1 DNA ends (insertions), as microhomologies and insertions are associated with CSR (Wu et al., 2006). Analysis of 13 unique Sμ-Sγ1 sequences in B cells from 2 EndoG−/− and 2 EndoG+/+ littermates showed that 3 out of 13 Sμ-Sγ1 DNA junctions from EndoG+/+ B cells and 1 out of 13 Sμ-Sγ1 DNA junctions from EndoG−/− B cells contain insertions; 9 out of 13 Sμ-Sγ1 DNA junctions from EndoG+/+ B cells and 10 out of 13 Sμ-Sγ1 DNA junctions from EndoG−/− B cells contain microhomologies (Fig. (6)). Although the number of insertions and the average length of microhomologies in Sμ-Sγ1 DNA junctions in EndoG+/+ B cells were slightly higher than that of EndoG−/− B cells (1 or 7.7% vs. 3 or 23.1%, and 3.3 nucleotides vs. 2 nucleotides, respectively), the difference in S-S region DNA junctions between EndoG+/+ and EndoG−/− B cells was not significant.
Fig. 6.
Sμ-Sγ1 DNA junctions in B cells. EndoG+/+ and EndoG−/− spleen B cells were stimulated by LPS and rmIL-4 for 4 days. Sμ-Sγ1 junction DNAs from stimulated cells were amplified, cloned, and sequenced. Each sequence is compared with the corresponding germline Sμ (MUSIGCD07) or Sγ1 sequence (MUSIGHANB). Microhomologies are in bold and underlined, insertions are in bold, underlined and highlighted in yellow. Sequences were derived from stimulated B cells of two pairs of EndoG+/+ and EndoG−/− mice.
3.6. EndoG−/− mice display altered spectrum of somatic point-mutations
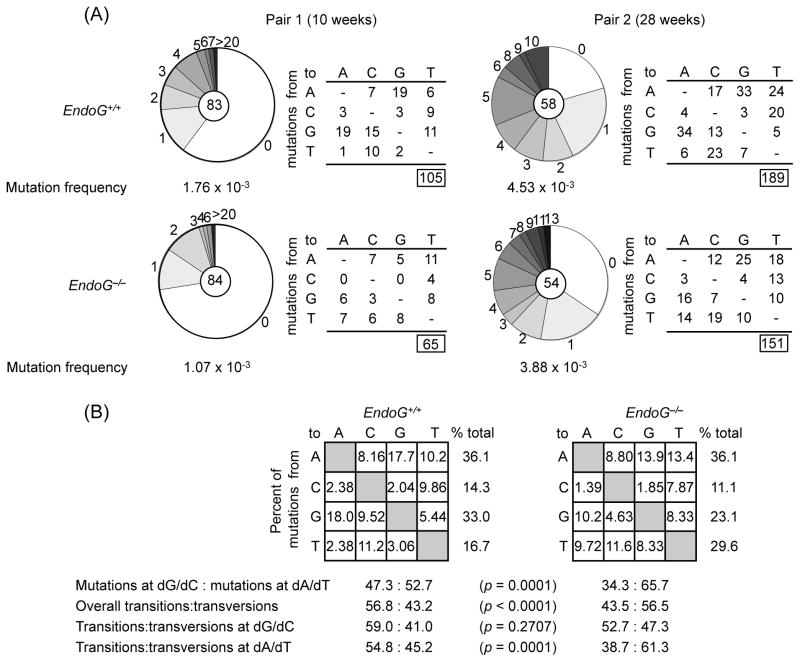
To determine whether the reduced CSR and DNA cleavages in the absence of EndoG were associated with altered somatic point-mutations in the IgH locus, we analyzed the JH4-iEμ sequence downstream of rearranged VJ558DJH4 DNA in germinal center (PNAhiB220+) B cells from Peyer’s patches of one pair of 10 week-old and one pair of 28 week-old non-intentionally immunized littermate EndoG−/− and EndoG+/+ mice. In these mice, the proportions of Peyer’s patch PNAhiB220+ B cells were comparable (Fig. 2(B)). The intronic JH4-iEμ DNA downstream of VJ558DJH4 rearrangement, which is targeted by SHM but is not subjected to positive or negative selection pressure, was amplified using a nested PCR involving VJ558 FR3 forward primers and reverse primers specific for sequences downstream of JH4. We chose to analyze only the sequences downstream of VJ558DJH4 rearrangements, as VHDJH rearrangements involving different JH genes place the JH-iEμ intron at different distances from the VH promoter and may result in different mutation rates.
Analysis of 83 JH4-iEμ intronic DNA sequences (720 bp) from a 10 week-old EndoG−/− mouse and 84 JH4-iEμ intronic DNA sequences from its EndoG+/+ littermate showed a decrease in mutations in the EndoG−/− mouse by 39.2% (1.76 × 10−3 vs. 1.07 × 10−3 change/base, p = 0.0001) (Fig. 7(A)). The decrease of mutation frequency became less significant in older mice, in which the overall mutation frequency is in general greatly increased. Analysis of 58 and 54 JH4-iEμ intronic DNA sequences from a pair of 28 week-old EndoG−/− and EndoG+/+ littermate mice showed a decrease in mutations in EndoG−/− mouse by over 14.3% (4.53 × 10−3 vs. 3.88 × 10−3 change/base, p = 0.0589). Absence of EndoG also resulted in an overall 27.5% reduction in the proportion of mutations at dG/dC (p = 0.0001, Fig. 7(B) ) and a reduction in transitions to transversions ratio (p < 0.0001), mainly due to decreased dA/dT transitions (p = 0.0001). Thus, EndoG deficiency only slightly reduces the frequency of somatic point-mutations, but significantly alters the mutational spectrum.
Fig. 7.
Altered spectrum of somatic point-mutations in the IgH chain intronic JH4-iEμ DNA of Peyer’s patch germinal center (PNAhiB220+) B cells from EndoG−/− mice. (A) Pie charts depict the proportion of sequences carrying various numbers (along margins) of mutations over the 720-bp JH4-iEμ DNA sequences downstream of rearranged VJ558DJH4 DNA in PNAhiB220+ B cells from Peyer’s patches of one pair of 10 week-old and one pair of 28 week-old non-intentionally immunized EndoG−/− and EndoG+/+ littermates. The numbers of the sequences from each mouse analyzed are at the center of the pies. Numbers and nature of independent mutational events scored. Frequencies of point-mutations from each mouse are indicated. (B) Compilations, with the numbers indicating percentages of all mutations scored in the pool of the target sequences. Below the compilations, the ratio of mutations at dG/dC to those at dA/dT is indicated, as is the ratio of transition:transversion substitutions at dG/dC and dA/dT.
4. Discussion
Generation of DSBs, particularly staggered DSBs in S regions is critical for CSR. We and others have shown in the human and mouse IgH locus that AID generates staggered DSBs not only by cleaving intact double-strand DNA, but also by processing blunt DSB ends generated independently of AID (Bross et al., 2002; Papavasiliou and Schatz, 2002b; Bross and Jacobs, 2003; Wu et al., 2003; Zan et al., 2003; Casali and Zan, 2004; Zan and Casali, 2008). How these AID-independent DSBs are generated and what is their role in CSR has been a longstanding question.
We presented here evidence indicating that EndoG introduces a significant proportion of DSBs in S region DNA and contributes to the overall CSR process. We showed that EndoG, which cleaves DNA primarily at dG/dC - the preferential sites of DSBs in S region DNA, can translocate to the nucleus, where it binds to S region DNA in B cells undergoing CSR. EndoG deficiency resulted in decreased CSR. This was associated with significantly reduced DNA cleavages, including DSBs, in S regions. The effects of EndoG deficiency were not limited to the S region DNA, but extended to other areas of the IgH locus, as shown by the altered mutation spectrum in JH4-iEμ intronic DNA, including significant decrease in mutations at dG/dC. Thus, our findings outline an important role of EndoG in CSR and, possibly, SHM. They, however, cannot rule out the possibility that factor(s) other than AID and/or EndoG may also be involved in the generation of DSBs. A more precise role of EndoG in the generation of DSBs would be addressed by using double knockout EndoG−/−Aicda−/− mice.
It has been assumed that the absolute requirement for AID in CSR reflects the indispensability of this enzyme in generating DSBs by deaminating S region DNA. This is further supported by the preferential localization of DSBs at dC:dG base-pairs in the 5′-RGYW-3′/5′-WRCY-3′ motif, the target of AID (Schrader et al., 2005). However, a post-cleavage role for AID in CSR has been indicated by the occurrence of unignorable amounts of “background” DSBs in S regions DNA of AID- or Ung-deficient B cells (Rush et al., 2004; Zan and Casali, 2008). In addition, intra-S region DNA recombination events have been suggested to occur in AID-deficient B cell hybridomas (Dudley et al., 2002) and in vivo c-myc translocations into S regions were putatively detected in AID-deficient mice (Unniraman et al., 2004; Unniraman and Schatz, 2006). Finally, DSBs have been shown to occur in V(D)J DNA of AID-deficient mouse B cells (Bross et al., 2002; Papavasiliou and Schatz, 2002b; Bross and Jacobs, 2003) and in human B cells, in which the AID activity was inhibited by a dominant negative AID mutant (Zan et al., 2003).
EndoG was first identified in the budding yeast S. cerevisiae as Nuc1p; Nuc1p displays endo/exonuclease activities and regulates life and death in yeast (Buttner et al., 2007). In yeast, Nuc1p has been suggested to play roles in caspase-independent apoptosis, mitochondrial DNA replication, DNA recombination and cell proliferation (Burhans and Weinberger, 2007). In mammals, EndoG has been thought to exert similar functions. Knockout of EndoG, however, did not result in obvious abnormalities in mice, thereby questioning the putative vital function(s) of this endonuclease in mammals (Irvine et al., 2005; David et al., 2006). Accordingly, EndoG deficiency did not alter B cell viability, cycle or proliferation in our hands. The apparently normal phenotype of EndoG−/− mice can be explained by the finding that in addition to EndoG, mammalian cells contain other factors, such as members of the paralogous ExoG-protein subfamily, which display complementary and partially overlapping enzymatic activities with EndoG (Cymerman et al., 2008). Such factors could have simply stood in for at least some of the lost EndoG functions in EndoG−/− mice.
As we show here, in the absence of EndoG, overall DNA cleavages, including DSBs, in Sμ and Sγ1 regions were significantly reduced. Reduced DSBs were associated with reduced CSR, indicating that EndoG is involved in the generation of at least some DSBs. EndoG can cleave both single- and double- strand DNA at comparable rates (Cymerman et al., 2008) to generate SSBs and DSBs. Some DSBs may stem from SSBs generated by EndoG and AID on the opposite strands. Besides EndoG, higher eukaryote cells contain its paralog ExoG, which possesses 5′-3′ exonuclease activity; EndoG and ExoG may function together to cleave double-strand DNA (Cymerman et al., 2008). In addition to AID-dependent DSBs, many DSBs in the IgH locus are generated in an AID-independent fashion (Bross et al., 2002; Papavasiliou and Schatz, 2002b; Bross and Jacobs, 2003; Zan et al., 2003; Zan and Casali, 2008). These DSBs are blunt and 5′-phosphorylated, and are processed by AID to yield staggered end DBS in B cells undergoing CSR/SHM (Zan et al., 2003; Casali and Zan, 2004; Zan and Casali, 2008). DSBs arising independently of AID, whether by intervention of EndoG or other factors, would constitute only a first step in the unfolding of the overall CSR process. Such DSBs would be processed by AID (Zan and Casali, 2008) to ensure that the ends are staggered for CSR. Ligation of DSB staggered ends would then generate the microhomologies, nucleotide insertions and point-mutations that characteristically occur in and around intra-S region or inter S-S region recombinations (Chen et al., 2001; Honjo et al., 2002; Wu et al., 2006; Zan and Casali, 2008).
CSR is associated with transcription that may result in R-loop formation to give rise to the (functional) single-strand DNA needed for AID deamination activity. R-loop formation initiates with a R-loop initiation zone (RIZ), which is, in general, a dG-cluster. A SSB at a dG-cluster can function as a strong RIZ (Roy et al., 2010). In addition to DSBs, SSBs can also be generated independently of AID in S regions (Arudchandran et al., 2004; Arudchandran et al., 2008). These SSBs may be created by EndoG and would lead to R-loop formation. EndoG possesses an effective R-loop cutting activity, which can lead to the generation of DNA cleavages (Côté and Ruiz-Carrillo, 1993; Widlak et al., 2001; Ohsato et al., 2002; Irvine et al., 2005). These R-loops can be targeted by EndoG or AID to yield more DNA cleavages, including SSBs and, therefore, more R-loop formation. This may function as a nick and R-loop amplification intermediate. The specific targeting of EndoG to S region DNA would be further established by chromatin modification, such as phosphorylation of histones, in S regions. It has been shown that phosphorylation of histone H2B is necessary for yeast EndoG to attack DNA (Buttner et al., 2007). Indeed, phosphorylation of histone H2B within Ig S and V regions correlates tightly with CSR and SHM (Odegard et al., 2005). The precise mechanisms of this targeting process and the cleavage co-factors involved in the generation of AID-independent DSBs need further analysis.
Acknowledgments
We thank Dr. Michael R. Lieber for EndoG+/− mice; Moon Kang and Elliot Yu for excellent technical assistance. This work was supported by U.S. NIH grants AI 045011, AI 079705 and AI 060573 to P.C.
Abbreviations
- AID
activation-induced cytidine deaminase
- antibody
- B cell
- CSR
class switch DNA recombination
- DSBs
double-strand DNA breaks
- EndoG
Endonuclease G
- Ig
immunoglobulin
- SHM
somatic hypermutation
- S
switch region
- Ung
uracil DNA glycosylase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arudchandran A, Bernstein RM, Max EE. Single-stranded DNA breaks adjacent to cytosines occur during Ig gene class switch recombination. J Immunol. 2004;173:3223–3229. doi: 10.4049/jimmunol.173.5.3223. [DOI] [PubMed] [Google Scholar]
- Arudchandran A, Bernstein RM, Max EE. Single-strand DNA breaks in Ig class switch recombination that depend on UNG but not AID. Int Immunol. 2008;20:1381–1393. doi: 10.1093/intimm/dxn097. [DOI] [PubMed] [Google Scholar]
- Baar J, Pennell NM, Shulman MJ. Analysis of a hot spot for DNA insertion suggests a mechanism for Ig switch recombination. J Immunol. 1996;157:3430–3435. [PubMed] [Google Scholar]
- Baar J, Shulman MJ. The Ig heavy chain switch region is a hotspot for insertion of transfected DNA. J Immunol. 1995;155:1911–1920. [PubMed] [Google Scholar]
- Bacolla A, Wojciechowska M, Kosmider B, Larson JE, Wells RD. The involvement of non-B DNA structures in gross chromosomal rearrangements. DNA Repair. 2006;5:1161–1170. doi: 10.1016/j.dnarep.2006.05.032. [DOI] [PubMed] [Google Scholar]
- Basu U, Wang Y, Alt FW. Evolution of phosphorylation-dependent regulation of activation-induced cytidine deaminase. Mol Cell. 2008;32:285–291. doi: 10.1016/j.molcel.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begum NA, Izumi N, Nishikori M, Nagaoka H, Shinkura R, Honjo T. Requirement of non-canonical activity of uracil DNA glycosylase for class switch recombination. J Biol Chem. 2007;282:731–742. doi: 10.1074/jbc.M607439200. [DOI] [PubMed] [Google Scholar]
- Begum NA, Kinoshita K, Kakazu N, Muramatsu M, Nagaoka H, Shinkura R, Biniszkiewicz D, Boyer LA, Jaenisch R, Honjo T. Uracil DNA glycosylase activity is dispensable for immunoglobulin class switch. Science. 2004;305:1160–1163. doi: 10.1126/science.1098444. [DOI] [PubMed] [Google Scholar]
- Begum NA, Stanlie A, Doi T, Sasaki Y, Jin HW, Kim YS, Nagaoka H, Honjo T. Further evidence for involvement of a noncanonical function of uracil DNA glycosylase in class switch recombination. Proc Natl Acad Sci USA. 2009;106:2752–2757. doi: 10.1073/pnas.0813252106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar S, Watson M, Diaz M. Activation-induced cytosine deaminase, AID, is actively exported out of the nucleus but retained by the induction of DNA breaks. J Biol Chem. 2004;279:26395–26401. doi: 10.1074/jbc.M403503200. [DOI] [PubMed] [Google Scholar]
- Bross L, Fukita Y, McBlane F, Demolliere C, Rajewsky K, Jacobs H. DNA double-strand breaks in immunoglobulin genes undergoing somatic hypermutation. Immunity. 2000;13:589–597. doi: 10.1016/s1074-7613(00)00059-5. [DOI] [PubMed] [Google Scholar]
- Bross L, Jacobs H. DNA double strand breaks occur independent of AID in hypermutating Ig genes. Clin Dev Immunol. 2003;10:83–89. doi: 10.1080/10446670310001626571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bross L, Muramatsu K, Kinoshita K, Honjo H, Jacobs H. DNA double-strand breaks: prior to but not sufficient in targeting hypermutation. J Exp Med. 2002;195:1187–1192. doi: 10.1084/jem.20011749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burhans WC, Weinberger M. Yeast endonuclease G: complex matters of death, and of life. Mol Cell. 2007;25:323–325. doi: 10.1016/j.molcel.2007.01.030. [DOI] [PubMed] [Google Scholar]
- Buttner S, Eisenberg T, Carmona-Gutierrez D, Ruli D, Knauer H, Ruckenstuhl C, Sigrist C, Wissing S, Kollroser M, Frohlich KU, Sigrist S, Madeo F. Endonuclease G regulates budding yeast life and death. Mol Cell. 2007;25:233–246. doi: 10.1016/j.molcel.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Casali P, Pal Z, Xu Z, Zan H. DNA repair in antibody somatic hypermutation. Trends Immunol. 2006;27:313–321. doi: 10.1016/j.it.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casali P, Zan H. Class switching and c-myc translocation: how does DNA break? Nat Immunol. 2004;5:6–8. doi: 10.1038/ni1104-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Kinoshita K, Honjo T. Variable deletion and duplication at recombination junction ends: implication for staggered double-strand cleavage in class-switch recombination. Proc Natl Acad Sci USA. 2001;98:13860–13865. doi: 10.1073/pnas.241524898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HL, Vuong BQ, Basu U, Franklin A, Schwer B, Astarita J, Phan RT, Datta A, Manis J, Alt FW, Chaudhuri J. Integrity of the AID serine-38 phosphorylation site is critical for class switch recombination and somatic hypermutation in mice. Proc Natl Acad Sci U S A. 2009;106:2717–2722. doi: 10.1073/pnas.0812304106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury M, Forouhi O, Dayal S, McCloskey N, Gould HJ, Felsenfeld G, Fear DJ. Analysis of intergenic transcription and histone modification across the human immunoglobulin heavy-chain locus. Proc Natl Acad Sci U S A. 2008;105:15872–15877. doi: 10.1073/pnas.0808462105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté J, Ruiz-Carrillo A. Primers for mitochondrial DNA replication generated by endonuclease G. Science. 1993;261:765–769. doi: 10.1126/science.7688144. [DOI] [PubMed] [Google Scholar]
- Cymerman IA, Chung I, Beckmann BM, Bujnicki JM, Meiss G. EXOG, a novel paralog of Endonuclease G in higher eukaryotes. Nucleic Acids Res. 2008;36:1369–1379. doi: 10.1093/nar/gkm1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David KK, Sasaki M, Yu SW, Dawson TM, Dawson VL. EndoG is dispensable in embryogenesis and apoptosis. Cell Death Differ. 2006;13:1147–1155. doi: 10.1038/sj.cdd.4401787. [DOI] [PubMed] [Google Scholar]
- Delker RK, Fugmann SD, Papavasiliou FN. A coming-of-age story: activation-induced cytidine deaminase turns 10. Nat Immunol. 2009;10:1147–1153. doi: 10.1038/ni.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- Doi T, Kato L, Ito S, Shinkura R, Wei M, Nagaoka H, Wang J, Honjo T. The C-terminal region of activation-induced cytidine deaminase is responsible for a recombination function other than DNA cleavage in class switch recombination. Proc Natl Acad Sci USA. 2009;106:2758–2763. doi: 10.1073/pnas.0813253106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley DD, Manis JP, Zarrin AA, Kaylor L, Tian M, Alt FW. Internal IgH class switch region deletions are position-independent and enhanced by AID expression. Proc Natl Acad Sci USA. 2002;99:9984–9989. doi: 10.1073/pnas.152333499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerschenson M, Houmiel KL, Low RL. Endonuclease G from mammalian nuclei is identical to the major endonuclease of mitochondria. Nucleic Acids Res. 1995;23:88–97. doi: 10.1093/nar/23.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo T, Kinoshita K, Muramatsu M. Molecular mechanism of class switch recombination: linkage with somatic hypermutation. Annu Rev Immunol. 2002;20:165–196. doi: 10.1146/annurev.immunol.20.090501.112049. [DOI] [PubMed] [Google Scholar]
- Huang F-T, Yu K, Hsieh C-L, Lieber MR. Downstream boundary of chromosomal R-loop at murine switch regions: Implication for the mechanism of class switch recombination. Proc Natl Acad Sci USA. 2006a;103:5030–5035. doi: 10.1073/pnas.0506548103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KJ, Ku CC, Lehman IR. Endonuclease G: a role for the enzyme in recombination and cellular proliferation. Proc Natl Acad Sci U S A. 2006b;103:8995–9000. doi: 10.1073/pnas.0603445103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine RA, Adachi N, Shibata DK, Cassell GD, Yu K, Karanjawala ZE, Hsieh CL, Lieber MR. Generation and characterization of endonuclease G null mice. Mol Cell Biol. 2005;25:294–302. doi: 10.1128/MCB.25.1.294-302.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara Y, Shimamoto N. Involvement of endonuclease G in nucleosomal DNA fragmentation under sustained endogenous oxidative stress. J Biol Chem. 2006;281:6726–6733. doi: 10.1074/jbc.M510382200. [DOI] [PubMed] [Google Scholar]
- Kataoka T, Takeda S, Honjo T. Escherichia coli extract-catalyzed recombination in switch regions of mouse immunoglobulin genes. Proc Natl Acad Sci USA. 1983;80:2666–2670. doi: 10.1073/pnas.80.9.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai T, Zan H, Zhang J, Hawkins JS, Xu Z, Casali P. Estrogen receptors bind to and activate the promoter of the HoxC4 gene to potentiate HoxC4-mediated AID induction. Immunoglobulin class-switch DNA recombination and somatic hypermutation. J Biol Chem. 2010 doi: 10.1074/jbc.M110.169086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizels N. Immunoglobulin gene diversification. Annu Rev Genet. 2005;39:23–46. doi: 10.1146/annurev.genet.39.073003.110544. [DOI] [PubMed] [Google Scholar]
- Mussmann R, Courtet M, Schwager J, Du Pasquier L. Microsites for immunoglobulin switch recombination breakpoints from Xenopus to mammals. Eur J Immunol. 1997;27:2610–2619. doi: 10.1002/eji.1830271021. [DOI] [PubMed] [Google Scholar]
- Nambu Y, Sugai M, Gonda H, Lee CG, Katakai T, Agata Y, Yokota Y, Shimizu A. Transcription-coupled events associating with immunoglobulin switch region chromatin. Science. 2003;302:2137–2140. doi: 10.1126/science.1092481. [DOI] [PubMed] [Google Scholar]
- Odegard VH, Kim ST, Anderson SM, Shlomchik MJ, Schatz DG. Histone modifications associated with somatic hypermutation. Immunity. 2005;23:101–110. doi: 10.1016/j.immuni.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Ohsato T, Ishihara N, Muta T, Umeda S, Ikeda S, Mihara K, Hamasaki N, Kang D. Mammalian mitochondrial endonuclease G. Digestion of R-loops and localization in intermembrane space. Eur J Biochem. 2002;269:5765–5770. doi: 10.1046/j.1432-1033.2002.03238.x. [DOI] [PubMed] [Google Scholar]
- Papavasiliou FN, Schatz DG. Cell-cycle-regulated DNA double-stranded breaks in somatic hypermutation of immunoglobulin genes. Nature. 2000;408:216–221. doi: 10.1038/35041599. [DOI] [PubMed] [Google Scholar]
- Papavasiliou FN, Schatz DG. Somatic hypermutation of immunoglobulin genes; merging mechanisms for genetic diversity. Cell. 2002a;109:s35–s44. doi: 10.1016/s0092-8674(02)00706-7. [DOI] [PubMed] [Google Scholar]
- Papavasiliou FN, Schatz DG. The activation-induced deaminase functions in a postcleavage step of the somatic hypermutation process. J Exp Med. 2002b;195:1193–1198. doi: 10.1084/jem.20011858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-R, Zan H, Zhang J, Al-Qahtani A, Pone EJ, Xu Z, Mai T, Casali P. HoxC4 binds to the promoter of the cytidine deaminase AID gene to induce AID expression, class-switch DNA recombination and somatic hypermutation. Nat Immunol. 2009;10:540–550. doi: 10.1038/ni.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peled JU, Kuang FL, Iglesias-Ussel MD, Roa S, Kalis SL, Goodman MF, Scharff MD. The biochemistry of somatic hypermutation. Annu Rev Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- Pone EJ, Zan H, Zhang J, Al-Qahtani A, Xu Z, Casali P. Toll-like receptors and B-cell receptors synergize to induce immunoglobulin class-switch DNA recombination: relevance to microbial antibody responses. Crit Rev Immunol. 2010;30:1–29. doi: 10.1615/critrevimmunol.v30.i1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada C. AID and RPA: PKA makes the connection local. Nat Immunol. 2009;10:367–369. doi: 10.1038/ni0409-367. [DOI] [PubMed] [Google Scholar]
- Rada C, Williams GT, Nilsen H, Barnes DE, Lindahl T, Neuberger MS. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr Biol. 2002;12:1748–1755. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- Ramiro AR, Jankovic M, Nussenzweig MC. Amplifying Igh translocations. Nat Immunol. 2005;6:117. doi: 10.1038/ni0205-117. [DOI] [PubMed] [Google Scholar]
- Roy D, Zhang Z, Lu Z, Hsieh CL, Lieber MR. Competition between the RNA transcript and the nontemplate DNA strand during R-loop formation in vitro: a nick can serve as a strong R-loop initiation site. Mol Cell Biol. 2010;30:146–159. doi: 10.1128/MCB.00897-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Carrillo A, Renaud J. Endonuclease G: a (dG)n X (dC)n-specific DNase from higher eukaryotes. EMBO J. 1987;6:401–407. doi: 10.1002/j.1460-2075.1987.tb04769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush JS, Fugmann SD, Schatz DG. Staggered AID-dependent DNA double strand breaks are the predominant DNA lesions targeted to Smu in Ig class switch recombination. Int Immunol. 2004;16:549–557. doi: 10.1093/intimm/dxh057. [DOI] [PubMed] [Google Scholar]
- Schrader CE, Linehan EK, Mochegova SN, Woodland RT, Stavnezer J. Inducible DNA breaks in Ig S regions are dependent on AID and UNG. J Exp Med. 2005;202:561–568. doi: 10.1084/jem.20050872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivarov V, Shinkura R, Honjo T. Dissociation of in vitro DNA deamination activity and physiological functions of AID mutants. Proc Natl Acad Sci USA 105. 2008;105:15866–15871. doi: 10.1073/pnas.0806641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J, Schrader CE. Mismatch repair converts AID-instigated nicks to double-strand breaks for antibody class-switch recombination. Trends Genet. 2006;22:23–28. doi: 10.1016/j.tig.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Sun H, Yabuki A, Maizels N. A human nuclease specific for G4 DNA. Proc Natl Acad Sci USA. 2001;98:12444–12449. doi: 10.1073/pnas.231479198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro J, Kinoshita K, Honjo T. Palindromic but not G-rich sequences are targets of class switch recombination. Int Immunol. 2001;13:495–505. doi: 10.1093/intimm/13.4.495. [DOI] [PubMed] [Google Scholar]
- Tian M, Alt FW. Transcription-induced cleavage of immunoglobulin switch regions by nucleotide excision repair nucleaseses. J Biol Chem. 2000;275:24163–24172. doi: 10.1074/jbc.M003343200. [DOI] [PubMed] [Google Scholar]
- Unniraman S, Schatz DG. AID and Igh switch region-Myc chromosomal translocations. DNA Repair. 2006;5:1259–1264. doi: 10.1016/j.dnarep.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Unniraman S, Zhou S, Schatz DG. Identification of an AID-independent pathway for IgH switch region-c-myc chromosomal translocations. Nat Immunol. 2004;5:1117–1123. doi: 10.1038/ni1127. [DOI] [PubMed] [Google Scholar]
- Vallur AC, Maizels N. Activities of human exonuclease 1 that promote cleavage of transcribed immunoglobulin switch regions. Proc Natl Acad Sci USA. 2008;105:16508–16512. doi: 10.1073/pnas.0805327105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong BQ, Lee M, Kabir S, Irimia C, Macchiarulo S, McKnight GS, Chaudhuri J. Specific recruitment of protein kinase A to the immunoglobulin locus regulates class-switch recombination. Nat Immunol. 2009;10:420–426. doi: 10.1038/ni.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widlak P, Li LY, Wang X, Garrard WT. Action of recombinant human apoptotic endonuclease G on naked DNA and chromatin substrates. J Biol Chem. 2001;276:48404–48409. doi: 10.1074/jbc.M108461200. [DOI] [PubMed] [Google Scholar]
- Wu SL, Li CC, Chen JC, Chen YJ, Lin CT, Ho TY, Hsiang CY. Mutagenesis identifies the critical amino acid residues of human endonuclease G involved in catalysis, magnesium coordination, and substrate specificity. J Biomed Sc. 2009;16:6. doi: 10.1186/1423-0127-16-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Feng J, Komori A, Kim EC, Zan H, Casali P. Immunoglobulin somatic hypermutation: double-strand DNA breaks, AID and error-prone DNA repair. J Clin Immunol. 2003;23:235–246. doi: 10.1023/a:1024571714867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Tsai CY, Patam MB, Zan H, Chen JP, Lipkin SM, Casali P. A role for the MutL mismatch repair Mlh3 protein in immunoglobulin class switch DNA recombination and somatic hypermutation. J Immunol. 2006;176:5426–5437. doi: 10.4049/jimmunol.176.9.5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuerffel R, Wang L, Grigera F, Manis J, Selsing E, Perlot T, Alt FW, Cogne M, Pinaud E, Kenter AL. S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity. 2007;27:711–722. doi: 10.1016/j.immuni.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Fulop Z, Wu G, Pone EJ, Zhang J, Mai T, Thomas LM, Al-Qahtani A, White CA, Park SR, Steinacker P, Li Z, Yates JI, Herron B, Otto M, Zan H, Fu H, Casali P. 14-3-3 adaptor proteins are recruited to 5′-AGCT-3′-rich switch regions, interact with activation-induced cytidine deaminase AID and mediate immunoglobulin class switch DNA recombination. Nat Struct Mol Biol. 2010;17:1124–1135. doi: 10.1038/nsmb.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Fulop Z, Zhong Y, Evinger AJ, Zan H, Casali P. DNA lesions and repair in immunoglobulin class switch recombination and somatic hypermutation. Ann NY Acad Sci. 2005;1050:146–162. doi: 10.1196/annals.1313.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Pone EJ, Al-Qahtani A, Park SR, Zan H, Casali P. Regulation of aicda expression and AID activity: relevance to somatic hypermutation and class switch DNA recombination. Crit Rev Immunol. 2007a;27:367–397. doi: 10.1615/critrevimmunol.v27.i4.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Zan H, Pal Z, Casali P. DNA replication to aid somatic hypermutation. Adv Exp Med Biol. 2007b;596:111–127. doi: 10.1007/0-387-46530-8_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Sugawara T, Nishi T, Liu J, Chan PH. Overexpression of SOD1 in transgenic rats attenuates nuclear translocation of endonuclease G and apoptosis after spinal cord injury. J Neurotrauma. 2006;23:595–603. doi: 10.1089/neu.2006.23.595. [DOI] [PubMed] [Google Scholar]
- Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loop at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- Yu K, Lieber MR. Nucleic acid structures and enzymes in the immunoglobulin class switch recombination mechanism. DNA Repair. 2003;2:1163–1174. doi: 10.1016/j.dnarep.2003.08.010. [DOI] [PubMed] [Google Scholar]
- Zan H, Casali P. AID- and Ung-dependent generation of staggered double-strand DNA breaks in immunoglobulin class switch DNA recombination: A post-cleavage role for AID. Mol Immunol. 2008;46:45–61. doi: 10.1016/j.molimm.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan H, Cerutti A, Schaffer A, Dramitinos P, Casali P. CD40 engagement triggers switching to IgA1 and IgA2 in human B cells through induction of endogenous TGF-β. Evidence for TGF-β but not IL-10-dependent direct Sμ Sα and sequential Sμ Sγ, Sγ Sα DNA recombination. J Immunol. 1998;161:5217–5225. [PMC free article] [PubMed] [Google Scholar]
- Zan H, Shima N, Xu Z, Evinger AJ, Zhong Y, Schimenti JC, Casali P. The translesion DNA polymerase θ plays a major role in immunoglobulin gene somatic hypermutation. EMBO J. 2005;24:3757–3769. doi: 10.1038/sj.emboj.7600833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan H, Wu X, Komori A, Holloman WK, Casali P. AID-dependent generation of resected double-strand DNA breaks and recruitment of Rad52/Rad51 in somatic hypermutation. Immunity. 2003;18:727–738. doi: 10.1016/s1074-7613(03)00151-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan H, Zhang J, Ardeshna S, Xu Z, Park S-R, Casali P. Lupus-prone MRL/faslpr/lpr mice display increased AID expression and extensive DNA lesions, comprising deletions and insertions, in the immunoglobulin locus: Concurrent upregulation of somatic hypermutation and class switch DNA recombination. Autoimmunity. 2009;42:89–103. doi: 10.1080/08916930802629554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao QL, Fujiwara Y, Kondo T. Synergistic induction of apoptosis and caspase-independent autophagic cell death by a combination of nitroxide Tempo and heat shock in human leukemia U937 cells. Apoptosis. 2010 doi: 10.1007/s10495-010-0522-8. [DOI] [PubMed] [Google Scholar]