Abstract
Understanding differentiation, a biological process from a multipotent stem or progenitor state to a mature cell is critically important. We developed a theoretical framework to quantify the underlying potential landscape and pathways for cell development and differentiation. We proposed a new mechanism of differentiation and found the differentiated states can emerge from the slow binding/unbinding of regulatory proteins to gene promoters. With slow promoter binding/unbinding, we found multiple meta-stable differentiated states, which can explain the origin of multiple states observed in recent experiments. The kinetic time for the differentiation and reprogramming strongly depends on the time scale of the promoter binding/unbinding processes. We discovered an optimal speed for differentiation for certain promoter binding/unbinding rates. Future experiments might be able to tell if cells differentiate at that optimal speed. We also quantified irreversible kinetic pathways for the differentiation and reprogramming, which captures the non-equilibrium dynamics in multipotent stem or progenitor cells.
During cell differentiation, the cell evolves from undifferentiated phenotypes in a multipotent stem or progenitor state to differentiated phenotypes in a mature cell. In this process, the gene regulatory network, which governs the progressive changes of gene expression patterns of the cell, forces the cell to adopt the cell type-specific phenotypes. Cells can have states with the higher probability of appearance, which leads to different cell phenotypes. Different cell phenotypes correspond to different basins of attractions on the potential landscape1,2,3. Therefore the differentiation and developmental process of the cell can be thought as the evolution of the underlying landscape topography from one basin to another. One grand challenge is to explain how this occurs, what the underlying mechanism is and how to quantify the differentiation and developmental process. Furthermore, the unidirectional developmental process poses another challenge to explain the origin of the arrow of time.
In the cell, intrinsic fluctuations are unavoidable due to the limited number of protein molecules. There have been increasing numbers of studies on how the gene regulatory networks can be stable and functional under such highly fluctuating environments4,5,6. In addition, the gene state fluctuations from the regulatory proteins binding/unbinding to the promoters can be significant for gene expression dynamics. Conventionally, it was often assumed that the binding/unbinding is significantly faster than the synthesis and degradation (adiabatic limit)7,8. This assumption may hold in some prokaryotic cells in certain conditions, in general there is no guarantee it is true. In fact, one expects in eukaryotic cells and some prokaryotic cells, binding/unbinding can be comparable or even slower than the corresponding synthesis and degradation (non-adiabatic limit). This can lead to nontrivial stable states and coherent oscillations appearing as a result of new time scales introduced due to the non-adiabaticity9,10,11,12,13,14,15,16,17,18. Therefore, the challenge for us is to understand how the biological differentiation and reprogramming can be functional under both intrinsic fluctuations and non-adiabatic fluctuations.
Previous studies showed that the change in the self activation regulatory strengths can cause the differentiation of phenotypes2,3,19. In this article, we used a canonical gene regulatory circuit module to study cell fate decision and commitment in multipotent stem or progenitor cells2,3,19. We will study a model of cell developmental circuit (Fig. 1)20, which is composed of a pair of mutually inhibiting but self activating genes. This gene regulatory motif has been found in various tissues where a pluri/multipotent stem cell has to undergo a binary cell fate decision21,22. For example, in the multipotent common myeloic progenitor cell (CMP) facing the binary cell fate decision between the myeloid and the erythroid fate, the fate determining transcription factors (TF), PU.1, and GATA1, which promote the myeloid or the erythroid fates, respectively, form such a gene network circuit. The relative expression levels A (PU.1) and B (GATA1) of these two reciprocal TFs can bias the decision toward either lineage20,22.
Figure 1. Network diagram of canonical gene regulatory circuit of two mutually opposing proteins that positively self-regulate themselves.
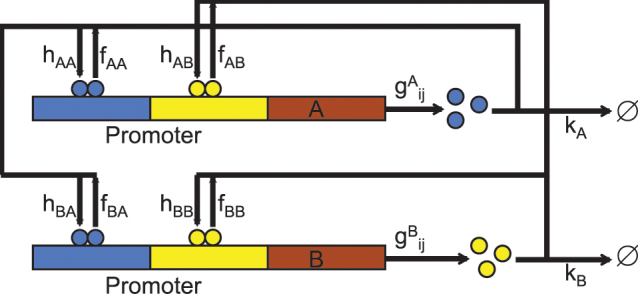
Two types of genes, A and B are translated into proteins A and B respectively. The proteins A(B) can bind to the promoter of the gene A(B) to activate the synthesis rate of A(B), which makes a self-activation feedback loop. The proteins A(B) can bind to the gene B(A) to repress the synthesis rate of B(A), which makes a mutual repression loop. Both protein A and protein B bind to promoters as a dimer with the binding rate 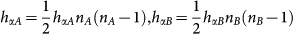 respectively, and the unbinding rate fαA, fαB respectively, with α = (A, B).
respectively, and the unbinding rate fαA, fαB respectively, with α = (A, B).
We found that the change in the time scale of the binding/unbinding of regulatory proteins to the promoters may provide a new important mechanism for the process of cell differentiation. We studied the underlying potential landscapes associated with differentiation and development, and found that the underlying landscapes developed from un-differentiated multipotent state to the differentiated states as the binding/unbinding rate decreased to the slow non-adiabatic binding region. In addition, in the slow non-adiabatic binding region, we predicted the emergence of multiple meta-stable states in the development of multipotent stem cells and explained the origin of this observation in experiments19. We also calculated the mean first passage transition time for the differentiation and reprogramming. We found that the mean first passage transition time strongly depends on the time scale of the promoter binding/unbinding processes. There is an optimal speed for differentiation and development with certain promoter binding/unbinding rates. It will be natural to ask whether the differentiation and development happens at this optimal speed? Future experimental and bioinformatics studies might be able to give the answer. In addition, we quantified the kinetic pathways for the differentiation and reprogramming, and found that they are irreversible. This captures the non-equilibrium prosperities for the biological processes of the underlying gene regulatory networks in multipotent stem or progenitor cells. It may provide the origin of the arrow of time for development.
Results
In our calculations, we only consider the A-B symmetric case:
We define the normalized binding/unbinding rate of the gene states: ωA = fA/k, ωR
= fR/k, and equilibrium constants: 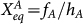 ,
, 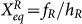 , which indicate the ratio between unbinding and binding speed. There are four gene states for each gene and the synthesis rates from gene A and B are also symmetric:
, which indicate the ratio between unbinding and binding speed. There are four gene states for each gene and the synthesis rates from gene A and B are also symmetric: 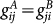 . When gene A is bound by protein A (self activation) while not bound by protein B (mutual repression), the synthesis rate of protein for protein A is the largest:
. When gene A is bound by protein A (self activation) while not bound by protein B (mutual repression), the synthesis rate of protein for protein A is the largest: 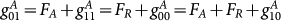 , where FA is the activation strength and FR is the repression strength. Here, we chose equilibrium constants
, where FA is the activation strength and FR is the repression strength. Here, we chose equilibrium constants 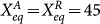 , symmetric binding/unbinding speed ωA = ωR = ω, the repression strength FR = 60 and scale the time to make k = 1.
, symmetric binding/unbinding speed ωA = ωR = ω, the repression strength FR = 60 and scale the time to make k = 1.
The potential Landscapes and two mechanisms for cell fate decision of development and differentiation
Such circuits with above control parameters can generate asymmetric attractors representing the differentiated states with almost mutually excluding expression of protein A (i.e. GATA1) and B (i.e. PU.1). In addition, central symmetric attractor states characterized by approximately equal levels of nA and nB expression can also be generated, which represent the multipotent state that exhibits the characteristic balanced or promiscuous expression of the two opposing, fate-determining concentrations-a hallmark of the indeterminacy of the undecided multipotent stem cell.
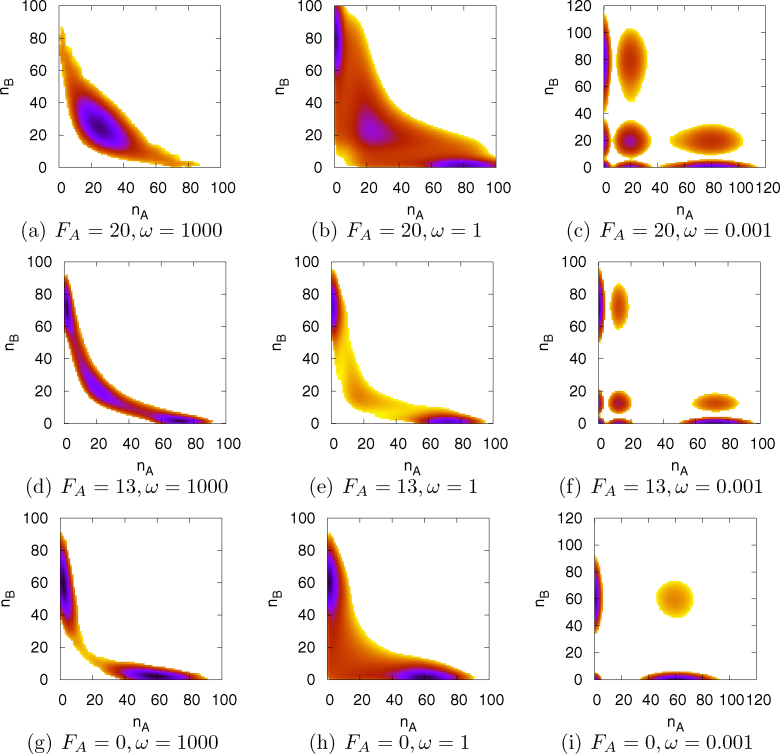
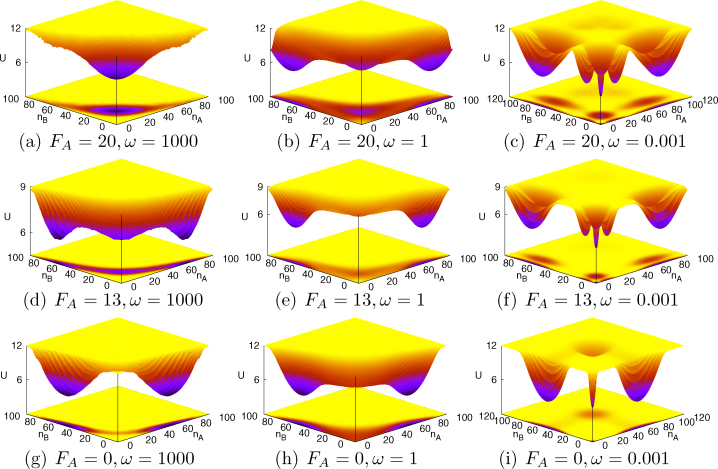
Through Gillespie simulations, probability distributions for the master equations and associated potential landscapes can be calculated23. We plotted the potential landscapes in nA-nB plane for different activation strength FA and binding/unbinding speed ω in Fig. 2(a), 2(b), 2(c), 2(d), 2(e), 2(f), 2(g), 2(h), 2(i) for the contour view, and Fig. 2(a), 2(b), 2(c), 2(d), 2(e), 2(f), 2(g), 2(h), 2(i) for the 3-dimensional view. In these figures, we found two kinds of mechanisms for cell differentiation.
Figure 2. The potential landscape (contour view) in the nA-nB plane for different self activation strength FA and binding/unbinding speed ω.
During the developmental process, the self activation regulation coming from an effective regulation and its change is due to the regulations on these transcription factors mediated by other regulators such as Klf4. When the self activation is strong (large FA), the system is mono-stable with one un-differentiated central basin, as in Fig. 2(a) (or 3(a)). As self activation strength FA decreases, the central basin gets weaker and differentiated basins on both sides start to develop, which results tri-stability as in Fig. 2(d) (or 3(d)). When self activation strength FA → 0, the circuit will reduce to a normal symmetric toggle switch. For the toggle switch, nA and nB can not be both large in adiabatic limit, because they suppress each other. Then the un-differentiated central basin disappeares and two differentiated basins on both sides survives, which gives bi-stability as in Fig. 2(g) (or 3(g)). Therefore, decreasing the self activation regulatory strength FA will lead the cell system to differentiate. Changing of the effective self activation regulatory strengths of transcription factors binding to the genes therefore provides a possible differentiation mechanism which is currently under investigation2,3,20,22.
Figure 3. The potential landscape (3 dimensional view) in the nA-nB plane for different self activation strength FA and binding/unbinding speed ω.
We would like to point out that there is another possible mechanism of the cell differentiation from the slow binding/unbinding of protein regulators to gene promoters. We noticed that for a fixed activation strength FA, cells can develop more stable differentiated states on both sides. As shown in Fig. 2(a) (or 3(a)), 2(b) (or 3(b)), 2(c) (or 3(c)) and Fig. 2(d) (or 3(d)), 2(e) (or 3(e)), 2(f) (or 3(f)), when the binding/unbinding rate ω decreases, the un-differentiated central basin becomes weaker and less stable, while differentiated basins on both sides become stronger and more stable. We also noticed that in the non-adiabatic slow binding limit (small binding/unbinding rate ω), multiple meta-stable basins show up. In addition, in the non-adiabatic slow binding limit, cells have chances to extinct and there are “extinct basins” near (nA = 0, nB = 0), as shown in Fig. 2(c) (or 3(c)), 2(f) (or 3(f)), 2(i) (or 3(i)). These behaviors are directly due to the non-adiabatic effect: slow binding/unbinding of protein regulators to promoters. When the binding/unbinding rate ω is small, the interactions (either repressions or activations) between gene states are weak and different gene states statistically co-exist in cells. Each gene state will give a basin in the concentration and the sum of these basins will give a multiple stable potential landscape. This leads to development and differentiation with slow binding from the original undifferentiated equally populated single basin of attraction with fast binding. Slow binding provides another possible mechanism for differentiation and development.
Kinetic and optimal speed for development and differentiation
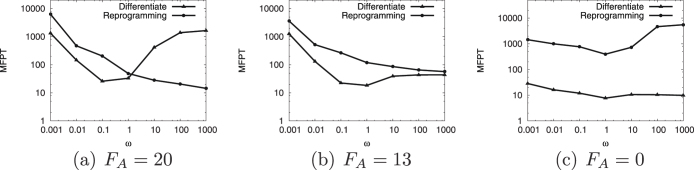
To quantitatively characterize the dynamics of differentiation and the reverse process as reprogramming, we study the speed of differentiation and reprogramming in terms of mean first passage time (MFPT) , as shown in Fig. 4(a), (4b) and (4c). In an attractor landscape, the lifetime of an attractor reflects its stability, which can be measured by MFPT. MFPT is the average transition time induced by intrinsic statistical fluctuations of molecule numbers between attractors on a landscape, since the traversing time represents how easy to switch from one place to another. When the binding/unbinding rate ω is relatively large, the un-differentiated central basin becomes more stable, as in Fig. 2(a) (or 3(a)), 2(d) (or 3(d)), 2(g) (or 3(g)), and cells have more chances to stay in the undifferentiated state. Therefore, the differentiation process will be more difficult and MFPT is longer for faster binding. For the differentiation process, it is noticed that, as the binding/unbinding rate ω increases, the MFPT decreases first, and then increases. In the nonadiabatic limit (small binding/unbinding rate ω), the differentiation limiting steps are the binding/unbinding events. Therefore, increasing binding/unbinding speed ω will accelerate the kinetics from the un-differentiated central basin to the differentiated side basins. So for the differentiation process, resulting from the shift from faster binding to slower binding of regulatory proteins to the genes, we notice that the speed for differentiation is slow when the landscape is dominated by the undifferentiated state for faster binding. In addition, it is also slower for slower binding, which is due to the occasional binding being the rate limiting step for differentiation. There is an optimal speed for differentiation. As binding becomes faster from low speed end (non-adiabatic limit), the speed of differentiation is controlled by the binding speed and therefore increases. As the binding becomes even faster, the differentiation is dominated by the escape from the undifferentiated basin of attraction and therefore is significantly slowed down. This creates an optimal speed for differentiation and development.
Figure 4. The MFPT of the differentiation and reprogramming for different self activation strength FA and binding/unbinding speed ω.
The reverse process of cell differentiation is the reprogramming of differentiated cells back to a multi- or pluripotent state. In Fig. 4(a), 4(b) and 4(c), the MFPT for the reprogramming for different self activation strength FA and binding/unbinding speed ω is plotted. We observed that, for a typical differentiated system, as in Fig. 2(c) (or 3(c)) and Fig. 2(g) (or 3(g)), the reprogramming chance is very low and requires a very long time. For self activation strength FA = 20 (Fig. 4(a)) and FA = 13 (Fig. 4(b)), the MFPT for the reprogramming decreases as the increasing of the binding/unbinding speed ω, because the stability of un-differentiated symmetric central state increases with the binding/unbinding speed ω as we can see in potential landscapes, Fig. 2(a) (or 3(a)), 2(b) (or 3(b)), 2(c) (or 3(c)), 2(d) (or 3(d)), 2(e) (or 3(e)) and 2(f) (or 3(f)). While in Fig. 4(c), since there is no self-activation and no stable symmetric central basin in the landscape, the reprogramming is difficult and the MFPT is very long for different the binding/unbinding speed ω.
Biological dynamic pathways of differentiation and reprogramming
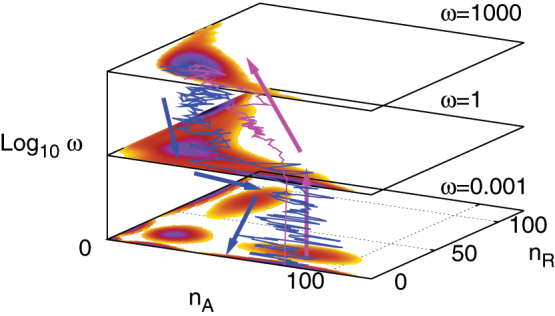
Both the differentiation and reprogramming can be caused by the change of genetic regulations during the developmental process. Here we consider the evolution of the binding/unbinding rate from fast to slow ω(t) = 1000e−κt + 0.001 from 1000 → 0.001 for the differentiation and the evolution of the binding/unbinding rate ω(t) = 1000[1 − e−κt]+0.001 from 0.001 → 1000 for the reprogramming from slow to fast. The transition paths from Gillespie simulation are plotted in Fig. 5, accompanied with the potential landscapes for the binding/unbinding speed ω = 0.001, 1, 1000. It is interesting to observe that the biological dynamic paths are irreversible, i.e. the differentiation path and reprogramming path are totally different. In the differentiation process, the system remains on the multipotent undifferentiated state for a while until binding becomes slower. As the binding becomes slower, the undifferentiated state becomes less stable. Furthermore, the gene state can be switched through binding/unbinding event of regulatory proteins to the promoters and the system will then be evolved from the undifferentiated basin to the differentiated basin of attraction. In the reprogramming process, the system will be gradually attracted into the undifferentiated basin as the increasing of the binding/unbinding rate ω. The paths of differentiation do not follow the gradient steepest descent of the potential landscape, nor do they follow the paths of the reprogramming (the reverse differentiation process). This irreversibility reflects the underlying non-equilibrium nature of the differentiation and developmental network systems3. It can give us the fundamental understanding of the biological origin of the arrow of time in cell development.
Figure 5. Transition paths for differentiation (blue) and reprogramming (purple) with κ = 0.1, with self activation strength FA = 20.
Discussion
We developed a theoretical framework to quantify the potential landscape and biological paths for cell development and differentiation. We found a new mechanism for differentiation. The differentiated state can emerge from the slow promoter binding/unbinding processes. We found under slow promoter binding, there can be many meta-stable differentiated states. This has been observed experimentally19. Our theory gives a possible explanation for the origins of those meta-stable states in the experiments.
We show that the developmental process can be quantitatively described and uncovered by the biological paths on the potential landscape and the dynamics of the developmental process is controlled by a combination of the intrinsic fluctuations of protein concentrations and gene state fluctuations through promoter binding. We also show that the biological paths of the reverse differentiation process or reprogramming are irreversible and different from those ones of the differentiation process.
We explored the kinetic speed for differentiation. We found that the cell differentiation and reprogramming dynamics strongly depends on the binding/unbinding rate of the regulatory proteins to the gene promoters. We found an optimal speed for differentiation and development with certain binding/unbinding rates of regulatory proteins to the gene promoters. We found an optimal speed for differentiation and development with certain binding/unbinding rates of regulatory proteins to the gene promoters. Such optimal speed for differentiation is in the non-adiabatic region with relatively slow promoter binding/unbinding processes. It can be hardly found in previous framework of Waddington landscapes in the adiabatic limit, where the binding/unbinding rate is not tunable3. Therefore, there are two mechanisms of the stem cell differentiation and reprograming. For previous work, the stem cell differentiation and reprograming are through the change of the self-regulation strengths. For the present work, the stem cell differentiation and reprograming are through the change of the binding/unbinding of regulators to the genes relative to the synthesis and degradation of the proteins. An interesting question we may ask is whether differentiation and development happen at their optimal speed? More experimental and bioinformatics studies might be able to pin down the answer. Furthermore, the irreversibility in cell development gives biological examples, which can be easily observed in experiments, for the understanding of the origin of the arrow of time in general non-equilibrium systems.
Methods
As shown in Fig. 1, the gene regulatory circuit that governs the binary cell fate decision module consists of mutual regulation of two opposing fate determining master TFs A and B. The module has been shown to control developmental cell fate decision and commitment in several instances of multipotent stem or progenitor cells that faces a binary fate decision (i.e. GATA1 and PU.1)21,22. A and B are coexpressed in the multipotent undecided cell and commitment to either one of the two alternative lineages is associated with one factor dominating over the others, leading to mutually exclusive expression patterns21,24. Importantly, in many cases the genes A and B also self-activate (positive autoregulate) themselves (Fig. 1). Here, the hybrid promoter α can be bound by the regulatory protein β with the binding rate hαβ and dissociation rate fαβ (both hαβ and fαβ can depend on protein concentration nβ). The synthesis of protein α is controlled by the gene state of promoter α. There are two types of genes, A and B, to be translated into proteins A and B respectively. The proteins A(B) can bind to the promoter of gene A(B) to activate the synthesis rate of A(B), which makes a self-activation feedback loop. Protein A(B) can bind to the gene B(A) to repress the synthesis rate of B(A), which makes a mutual repression loop. Here, both protein A and protein B bind to promoters as a dimer with the binding rate 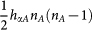 and
and 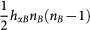 respectively. Therefore, each gene has 4 states with self activator binding or non-binding and with mutual repression from another gene binding or non-binding (assuming we have two different binding sites, one for the self activator and one for the other gene). The whole system has 16 gene states in total. For simplicity, we neglect the roles of mRNAs by assuming translation processes are very fast. The model can be expressed by the following chemical reactions:
respectively. Therefore, each gene has 4 states with self activator binding or non-binding and with mutual repression from another gene binding or non-binding (assuming we have two different binding sites, one for the self activator and one for the other gene). The whole system has 16 gene states in total. For simplicity, we neglect the roles of mRNAs by assuming translation processes are very fast. The model can be expressed by the following chemical reactions:
with α = A(B) for the hybrid promoter of gene A(B). For the gene state index ij of gene Oα, the first index i = 1(0) stands for the activator protein A unbound(bound) to the promoter α; the second index j = 1(0) stands for the repressor protein R unbound(bound) to the promoter α. 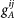 (
(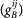 ) is the synthesis rate of the protein A(B) when the gene A(B) is in state ij. The probability distribution of the microstate is indicated as Pijkl(nA, nB) where nA and nB are the concentration of the activator A and the repressor B respectively. The index i(j) represents the gene A occupation state by the protein A(B) and the index k(l) represents the gene B occupation state by the protein A(B). This results sixteen master equations for the probability distribution which are shown explicitly in Supporting Material (SM).
) is the synthesis rate of the protein A(B) when the gene A(B) is in state ij. The probability distribution of the microstate is indicated as Pijkl(nA, nB) where nA and nB are the concentration of the activator A and the repressor B respectively. The index i(j) represents the gene A occupation state by the protein A(B) and the index k(l) represents the gene B occupation state by the protein A(B). This results sixteen master equations for the probability distribution which are shown explicitly in Supporting Material (SM).
The steady state probability distribution satisfies 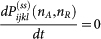 for all i, j, k, l. The total probability distribution is
for all i, j, k, l. The total probability distribution is 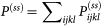 . The generalized potential function U of the non-equilibrium network can be quantified as: U(nA, nB) = −ln P(ss). It maps to the potential landscape, which gives a quantitative measure of the global stability and function of the underlying network25. Above equations are difficult to deal with because each one actually represents an infinite number of equations (n range from 0 to ∞). A direct way to find the steady state Pss is through kinetic simulations following Gillespie algorithm23. There are four steps in the Gillespie algorithm for kinetic Monte Carlo simulations23: 1. initialize the state in the system; 2. generate random numbers to determine the next reaction to occur as well as the time interval. The probability of a given reaction to be chosen is based on the current state; 3. increase the time step by the time interval generated in step 2 and update the state based on the reaction that occurred; 4. iterate step 2 unless the simulation time has been exceeded. Here, we used Gillespie simulation to find the stead state distribution of master equations (see Supporting Material (SM)).
. The generalized potential function U of the non-equilibrium network can be quantified as: U(nA, nB) = −ln P(ss). It maps to the potential landscape, which gives a quantitative measure of the global stability and function of the underlying network25. Above equations are difficult to deal with because each one actually represents an infinite number of equations (n range from 0 to ∞). A direct way to find the steady state Pss is through kinetic simulations following Gillespie algorithm23. There are four steps in the Gillespie algorithm for kinetic Monte Carlo simulations23: 1. initialize the state in the system; 2. generate random numbers to determine the next reaction to occur as well as the time interval. The probability of a given reaction to be chosen is based on the current state; 3. increase the time step by the time interval generated in step 2 and update the state based on the reaction that occurred; 4. iterate step 2 unless the simulation time has been exceeded. Here, we used Gillespie simulation to find the stead state distribution of master equations (see Supporting Material (SM)).
Author Contributions
JW designed the project. HD performed the research. All authors wrote and reviewed the manuscript.
Supplementary Material
Supplementary Informations
Acknowledgments
This work was supported by National Science Foundation.
References
- Waddington C. H. The Strategy of the Genes. London: Allen and Unwin pages 1-262 (1957). [Google Scholar]
- Wang J., Xu L., Wang E. K. & Huang S. The Potential Landscape of Genetic Circuits Imposes the Arrow of Time in Stem Cell Differentiation. Biophys. J. 99, 29–39 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhang K., Xu L. & Wang E. K. Quantifying the Waddington landscape and biological paths for development and differentiation. Proc. Natl. Acad. Sci. 108, 8257–8262 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz M. B. & Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature 403, 335–338 (2000). [DOI] [PubMed] [Google Scholar]
- Kim K., Lepzelter D. & Wang J. Single Molecule Dynamics and Statistical Fluctuations of Gene Regulatory Networks: A Repressilator. J. Chem. Phys. 126, 034702 (2007). [DOI] [PubMed] [Google Scholar]
- Arkin A., Ross J. & McAdams H. H. Stochastic kinetic analysis of developmental pathway bifurcation in phage lambda-infected Escherichia coli cells. Genetics 149, 1633–1648 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackers G. K., Johnson A. D. & Shea M. A. Quantitative model for gene regulation by lambda phage repressor. Proc. Natl. Acad. Sci. 79, 1129–1133 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin D. W. et al. Gene Network Shaping of Inherent Noise Spectra. Nature 439, 608–611 (2006). [DOI] [PubMed] [Google Scholar]
- Hornos J. E. M. et al. Self-Regulating gene: An Exact Solution. Phys. Rev. E 72, 051907 (2005). [DOI] [PubMed] [Google Scholar]
- Walczak A. M., Onuchic J. N. & Wolynes P. G. Absolute rate theories of epigenetic stability. Proc. Natl. Acad. Sci. 102, 18926–18931 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz D., Onuchic J. N. &Wolynes P. G. Understanding stochastic simulations of the smallest genetic networks. J. Chem. Phys. 126, 245102 (2007). [DOI] [PubMed] [Google Scholar]
- Kepler T. B. & Elston T. C. Stochasticity in Transcriptional Regulation: Origins, Consequences, and Mathematical Representations. Biophys. J. 81, 3116–3136 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das J., Kardar M. & Chakraborty A. K. Purely stochastic binary decisions in cell signaling models without underlying deterministic instabilities. Proc. Natl. Acad. Sci. 104, 18598–18963 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H., Han B. & Wang J. Dominant Kinetic Paths of Complex Systems: Gene Networks. J. Phys. Chem. Lett. 1, 1836–1840 (2010). [Google Scholar]
- Feng H., Han B. & Wang J. Adiabatic and Non-Adiabatic Non-Equilibrium Stochastic Dynamics of Single Regulating Genes. J. Phys. Chem. B 115, 1254–1261 (2011). [DOI] [PubMed] [Google Scholar]
- Feng H., Han B. & Wang J. Landscape and Global Stability of Nonadiabatic and Adiabatic Oscillations in a Gene Network. Biophys. J. 102, 1001–1010 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A. & Weinberger L. S. Stochastic gene expression as a molecular switch for viral latency. Current Opinion in Microbiology 12, 460–466 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi P. J., Cai L., Friedman K. & Xie S. A Stochastic Single-Molecule Event Triggers Phenotype Switching of a Bacterial Cell. Science 322, 442–446 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmar T. et al. Regulated Fluctuations in Nanog Expression Mediate Cell Fate Decisions in Embryonic Stem Cells. PLoS Biol. 7, e1000149 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf T. & Enver T. Forcing cells to change lineages. Nature 462, 587–594 (2009). [DOI] [PubMed] [Google Scholar]
- Zhou J. X. & Huang S. Understanding gene circuits at cell-fate branch points for rational cell reprogramming. Trends Genet 27, 55–62 (2010). [DOI] [PubMed] [Google Scholar]
- Huang S., Guo Y. P., May G. & Enver T. Bifurcation dynamics of cell fate decision in bipotent progenitor cells. Dev. Biol. 305, 695–713 (2007). [DOI] [PubMed] [Google Scholar]
- Gillespie D. T. Exact Stochastic Simulation of Coupled Chemical Reactions. J. Phys. Chem. 81, 2340–2361 (1977). [Google Scholar]
- Hu M. et al. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 11, 774–785 (1997). [DOI] [PubMed] [Google Scholar]
- Wang J., Xu L. & Wang E. K. Potential Landscape and Flux Framework of Non-Equilibrium Networks: Robustness, Dissipation and Coherence of Biochemical Oscillations. Proc. Natl. Acad. Sci. 105, 12271–12276 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Informations