Abstract
The gene B lymphocyte kinase (BLK) is associated with rheumatoid arthritis, systemic lupus erythematosus and several other autoimmune disorders. The disease risk haplotype is known to be associated with reduced expression of BLK mRNA transcript in human B cell lines; however, little is known about cis-regulation of BLK message or protein levels in native cell types. Here, we show that in primary human B lymphocytes, cis-regulatory effects of disease-associated single nucleotide polymorphisms in BLK are restricted to naïve and transitional B cells. Cis-regulatory effects are not observed in adult B cells in later stages of differentiation. Allelic expression bias was also identified in primary human T cells from adult peripheral and umbilical cord blood (UCB), thymus and tonsil, although mRNA levels were reduced compared with B cells. Allelic regulation of Blk expression at the protein level was confirmed in UCB B cell subsets by intracellular staining and flow cytometry. Blk protein expression in CD4+ and CD8+ T cells was documented by western blot analysis; however, differences in protein expression levels by BLK genotype were not observed in any T cell subset. Blk allele expression differences at the protein level are thus restricted to early B cells, indicating that the involvement of Blk in the risk for autoimmune disease likely acts during the very early stages of B cell development.
INTRODUCTION
Genetic evidence for cis-regulation of gene expression has been commonly observed in genome-wide studies of cell lines (1) as well as peripheral blood cells in humans (2); these expression differences are likely to underlie many human disease associations, particularly autoimmune disorders. With a few exceptions, genome-wide analysis of both cis- and trans-regulation of gene expression has most often been carried out in reference panels of B lymphoblastoid cell lines, using either microarray technology or, more recently, deep RNA sequencing (3). The B lymphocyte kinase gene, BLK, has consistently shown significant cis-regulation in these studies, and is often highlighted in discussions of this phenomenon (4). It is generally assumed that cis-regulatory effects that are observed in B cell lines are indicative of similar patterns of regulation in primary cells, but this does not appear to be the case for the majority of regulatory variation that has been observed when comparing B cell lines and primary cells from different tissues (5). Thus, for gene regulation that is implicated in disease risk, it is clearly important to establish the specific cell types where such expression differences are present, both at the transcript and protein levels.
In this report, we have carried out a detailed analysis of cis-regulation of BLK expression in the major cell types of the human immune system. BLK is of particular interest because of the widely replicated association of BLK polymorphisms with systemic lupus erythematosus and rheumatoid arthritis, in both Caucasian (6,7) and Asian populations (8,9). Associations with other autoimmune disorders have also been reported [systemic sclerosis (10), Sjögren's syndrome (11) and anti-phospholipid syndrome (12)]. The disease-related BLK haplotype is associated with significantly lower expression of BLK transcript in B lymphoblastoid cell lines (4,7), suggesting that lower production of BLK in human B cells confers risk for autoimmunity. The single nucleotide polymorphism (SNP) rs922483 is in high linkage disequilibrium with both systemic lupus erythematosus and rheumatoid arthritis-associated SNPs and has been shown to regulate expression of BLK (4 and Robert Graham, personal communication). Similarly, SNP-associated regulation of BLK expression has also been found in human CD4+ T cells (2), suggesting that non-B-lineage cell types may contribute to the risk for autoimmunity.
BLK encodes a non-receptor protein tyrosine kinase and is a member of the Src family of tyrosine kinases (13). BLK is named for its high expression in B-lineage cells (13), but it is also expressed in non-B-lineage cell types, such as human thymocytes and pancreatic β-cells (14,15). Blk is activated upon B cell receptor (BCR) stimulation (16–18), and phosphorylates other molecules in the BCR signaling pathway (18,19). A knockout (−/−) mouse for Blk was reported over a decade ago, and exhibited no obvious defect in B cell development or function (20). In contrast, transgenic mice expressing a constitutively active form of Blk have an expanded pre-B cell population (21,22), a phenomenon that is dependent on pre-BCR signaling (22). BCR signaling is also regulated by Lyn and Fyn, two other Src kinase family members expressed in B cells (23). The partially redundant nature of these Src kinases is emphasized by the phenotype of mice deficient for the triad of Blk, Fyn and Lyn. These triple knockout mice develop severe B cell lymphopenia in contrast to single knockout mice (23). Revisiting the immune phenotypes in the Blk single knockout mouse in the C57BL/6 background recently revealed a role for Blk in the development and function of marginal zone (MZ) B cells and γδ TCR+ T cells (24,25). The function of Blk in human B cells or other hematopoietic cells has not been extensively studied, and there is little information concerning the genetic regulation of BLK at the mRNA and protein levels in primary human cells.
RESULTS
Allelic imbalance of BLK is not detected in B cells from adult peripheral blood
In B cell lines, expression of the disease-associated allele rs922483 A is reduced relative to the expression of transcript containing rs922483 G (4,7). To confirm the association of this SNP with other autoimmune disease-associated SNPs, we analyzed the linkage disequilibrium of rs922483 with the disease-associated variants: rs2736340 (6,26), rs13277113 (7), rs2736345 (4,11) and rs2618476 (27) (Supplementary Material, Fig. S1). As neither of the published associated variants is present in mature mRNA, we used the rs922483 SNP, which is present in mRNA transcript, for our studies. To measure allele expression bias, we developed a pyrosequencing assay in which the relative expression of each SNP allele in mRNA is quantified in heterozygous cells. Relative SNP expression levels are normalized to the allelic representation observed when genomic DNA is utilized as the substrate for pyrosequencing (Fig. 1A). This approach avoids the potential confounding that can result from correlating expression with the genotype between different cell lines or individuals in which other factors may influence gene expression. We first tested five heterozygous B lymphoblastoid cell lines and confirmed previous observations with four out of five of the B cell lines displaying allelic imbalance (P = 0.0284), as shown in Figure 1A.
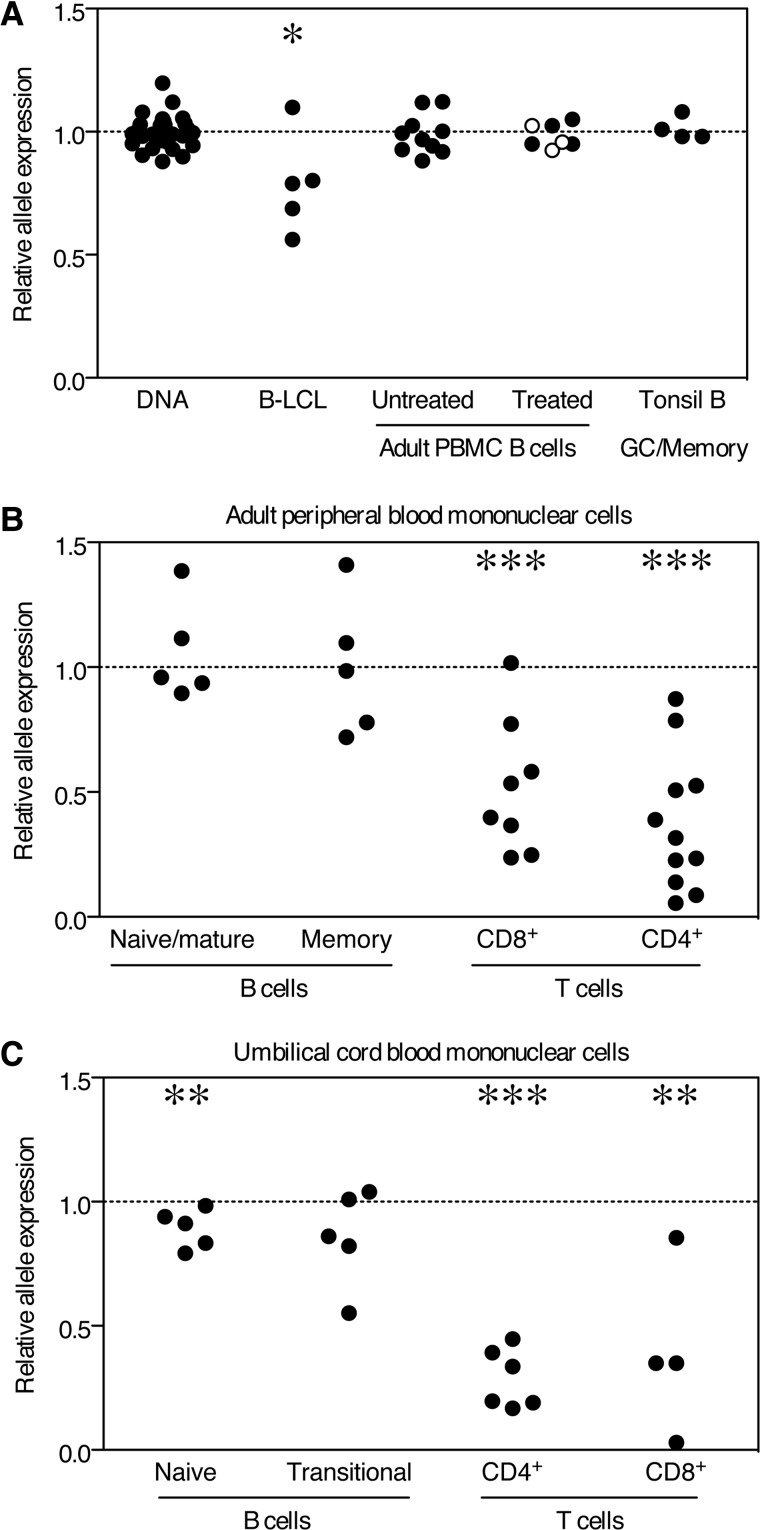
Figure 1.
Allelic imbalance of rs922438 is restricted to T cells and UCB B cells. Relative expression of SNP alleles was assayed and relative expression of SNP alleles normalized to DNA are shown for (A) DNA, B lymphoblastoid B cell lines (B-LCL), adult peripheral blood B cells untreated, anti-CD40 treated (open circles) and IL4/anti-CD40/anti-IgM treated (closed circles) adult peripheral blood B cells and tonsil germinal center (GC)/memory B cells (CD19+CD3−CD38+IgD−) (40,41). (B) Single lymphocytes were gated on CD3 and CD19 in order to examine allelic expression of BLK in sorted cell subsets. CD19+ cells were sorted on the basis of CD27-positive or -negative staining; naïve/mature (CD19+CD3−CD27−) and memory (CD19+, CD3−, CD27+) B cells. CD3+ T cells were divided into CD4+, CD8+ and CD4−CD8−γδ TCR+ cells. (C) Sorted lymphocyte populations in UCB were examined for allelic expression levels of BLK. Single CD14− lymphocytes were gated on CD3 and CD19. CD19+ CD27− cells were sorted on the basis of CD10 and CD38 expression; naïve (CD19+CD3−CD14−CD27−CD10−CD38int) and transitional (CD19+CD3−CD14−CD27−CD10+CD38hi) B cells. CD3+ T cells were divided into CD4+ and CD8+. A value of one (dotted line) represents equal allele expression and statistically significant differences as determined by the Mann–Whitney non-parametric U-test to compare DNA values to other groups are denoted as *P <0.05, **P <0.01 and ***P <0.001.
Also shown in Figure 1A, we tested total B cells isolated from adult peripheral blood and, in contrast to B cell lines, we did not observe any evidence of allelic imbalance. In order to investigate whether activation would reveal a difference in allelic expression of BLK, peripheral blood B cells were stimulated with anti-CD40 or anti-IgM, IL4 and anti-CD40. However, allelic imbalance was not observed in either resting or in vitro-stimulated B cells. Moreover, tonsillar B cells (germinal center/memory B cells) also failed to exhibit allelic imbalance for BLK mRNA expression (Fig. 1A).
Evidence for allelic imbalance of BLK in umbilical cord blood B cells and in T cells
Since our initial results did not provide evidence of differential allelic expression in total circulating human B cells, we expanded our experiments to investigate the relative expression of rs922483 SNP alleles in B cell and T cell subsets in both adult peripheral and umbilical cord blood (UCB). Similar to what was seen in unseparated adult peripheral blood B cells, no allelic imbalance was observed in CD27− (naïve/mature) or CD27+ (memory) B cell subsets (Fig. 1B). Strikingly, however, both CD4+ and CD8+ T cells showed prominent evidence of allelic imbalance in cells isolated from peripheral blood of normal heterozygote carriers (P < 0.001).
Given that Blk is implicated in early B cell development in the mouse, we reasoned that allelic imbalance may only be detectable in immature B cells. Therefore, we analyzed human UCB, which is a rich source of naïve B cells and transitional B cells [B cells transitioning from immature B cells to mature naive stage B cells (28)]. Relative allele expression was measured in UCB naïve and transitional B cells (Fig. 1C). The values were slightly lower in both of these B cell subsets for the risk-associated allele of rs922483, although these changes were not statistically significant in transitional B cells. As observed in adult peripheral blood, both CD4+ and CD8+ T cells showed obvious allelic imbalance in cord blood (P < 0.01, Fig. 1C), as did thymocytes and tonsil T follicular helper (Tfh) cells (P < 0.001, Supplementary Material, Fig. S1). Monocytes and dendritic cells showed no evidence of allelic imbalance (Supplementary Material, Fig. S1).
Quantitative analysis of BLK mRNA in subjects homozygous for the rs922483 allele
The unexpectedly prominent evidence of allelic imbalance in T cells prompted us to compare the quantitative expression of BLK in human primary B and T cell subsets, and to examine whether mRNA expression in these cells differed based on the rs922483 genotype. Real-time quantitative PCR (qPCR) was used to measure BLK transcript expression and values were normalized to GAPDH. The highest expression of BLK was detected in naïve B cells from adult peripheral blood (Fig. 2A). Similar to what was observed in the allelic imbalance assays, there was no difference in the expression of BLK between rs922483 G/G (non-risk) and A/A (risk) groups in either naïve/mature or memory B cells from adult peripheral blood. Given that there was a trend towards allele expression bias in UCB B cells, we also measured BLK expression in umbilical cord transitional (CD10+CD38hi) and naïve (CD10−CD38int) B cells. Expression of BLK was significantly (P < 0.05) lower in cells from A/A subjects in both B cell subsets.
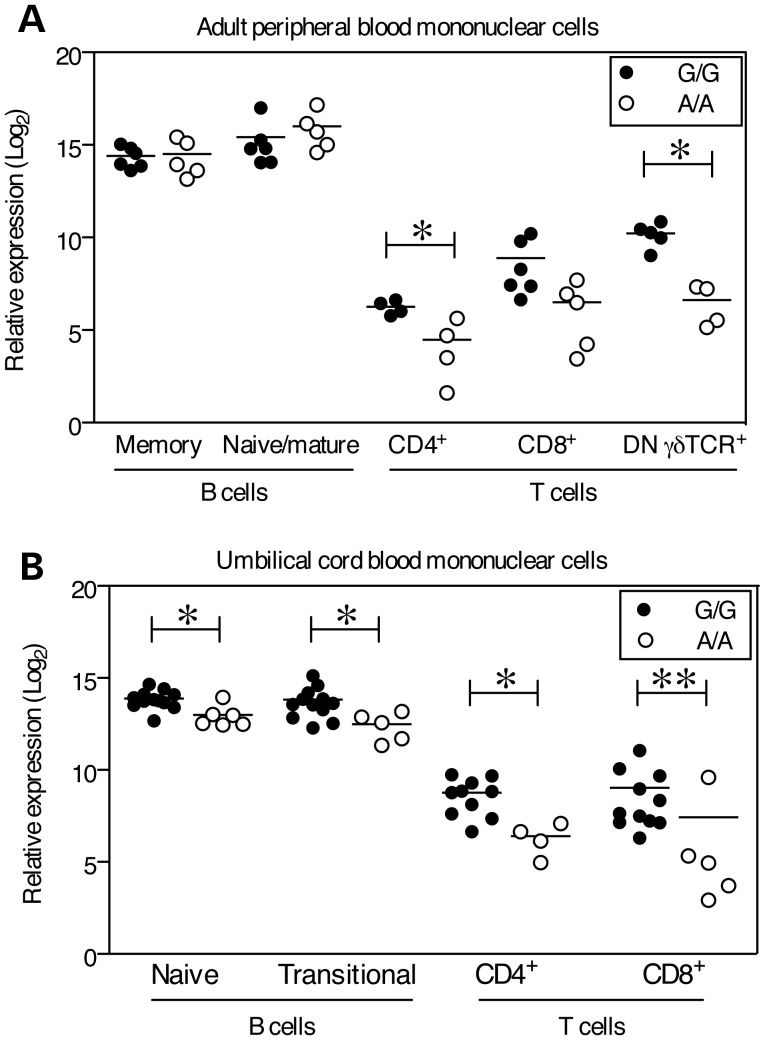
Figure 2.
Lower expression of BLK mRNA in T cells and UCB B cells in rs922483 homozygous risk cells. BLK transcript was measured by qPCR in primary human B and T cell subsets and normalized to GAPDH. rs922438 homozygous A (risk allele) subjects are shown as open circles and homozygous G subjects are shown as filled circles. (A) Adult peripheral blood cell subsets: memory (CD19+CD3−CD27+) and mature/naïve (CD19+CD3−CD27−) B cells, CD4+, CD8+ and CD4−CD8− γδ TCR+ T (CD3+CD19−) cells, (B) UCB cell subsets: naïve (CD19+CD3−CD27−CD10−CD38int) and transitional (CD19+CD3−CD27−CD10+CD38hi) B cells, CD4+ and CD8+ T cells. Statistically significant differences as determined by the Mann–Whitney non-parametric U-test to compare indicated groups are denoted as *P <0.05 and **P <0.01 and the mean is represented by a horizontal line.
The expression of BLK in T cell subsets was substantially lower (5–10 logs, base 2) than in B cells from adult peripheral blood. The highest expression of BLK in T cell subsets was observed in subjects homozygous for the non-risk (G) allele (Fig. 2A). The expression of BLK in γδ TCR+ T cells from A/A subjects was significantly lower than in G/G subjects (P = 0.03). BLK expression was also lower in CD4+ (P = 0.029) and CD8+ T cells (NS) from A/A subjects compared with G/G subjects (Fig. 2A). Similar to the observations in adult peripheral blood T cell subsets, BLK expression in UCB was significantly lower in CD4+ and CD8+ T cells from A/A subjects in comparison to G/G subjects (P = 0.031 and =0.004, respectively, Fig. 2).
Blk protein expression in B and T cells
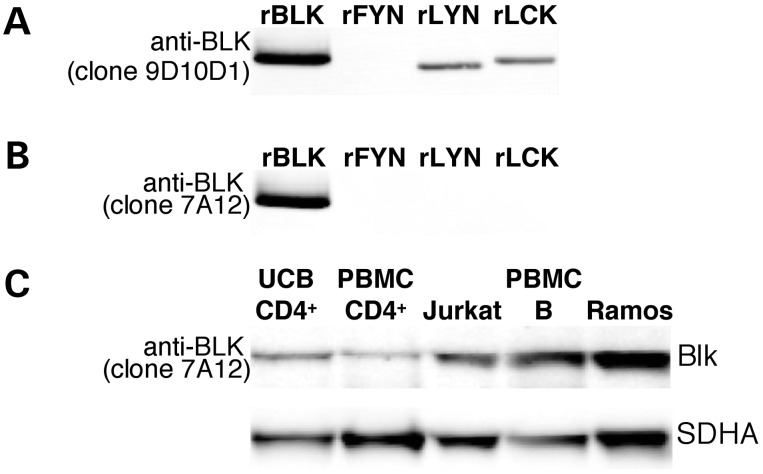
In order to establish whether these patterns of BLK expression are reflected in the levels of protein in the various cell types, we endeavored to analyze Blk protein expression using western blot analysis. Our initial studies with commercially available monoclonal reagents for Blk suggested that there might be substantial Blk protein present in many types of lymphocytes. However, given the close sequence similarity between Blk and other Src kinases (29), we carried out a detailed analysis of antibody reactivity using recombinant protein substrates by western blot. Several commonly utilized commercial antibodies including clones 9D10D1 (Fig. 3A) and 9D10B7H6 (data not shown) exhibit cross reactivity with one or more Src kinases, and thus are not specific for Blk. On the other hand, we identified some antibody clones (7A12, 1E6 and 3E5-3A8) with convincing evidence of specificity for Blk as no binding to recombinant Fyn, Lyn and Lck was observed by the western blot analysis (Fig. 3B). Using the 7A12 monoclonal antibody, we have shown that Blk is easily detected in lysates from the Ramos B cell line, as well as from total adult peripheral blood B cells. Blk can also be detected in the Jurkat T cell line, as well as in CD4+ T cells, albeit at lower levels (Fig. 3C).
Figure 3.
Western blot analysis of Blk antibody specificity. Recombinant Blk (lane 1) and Src kinases Fyn, Lyn and Lck (lanes 2, 3 and 4, respectively) were run on an SDS–PAGE gel. Western blot analysis was performed using anti-Blk monoclonal antibody clones 9D10D1 (A) and 7A12 (B). (A) Anti-Blk clone 9D10D1 clearly detects rBlk, rLyn and rLck and is thus not specific for Blk. (B) The anti-Blk antibody clone 7A12 recognizes rBlk and does not label rFyn, rLyn or rLck. (C) Whole cell lysates were analyzed by western blot: lanes; 1, UCB CD4+ T cells; 2, adult peripheral blood CD4+ T cells; 3, Jurkat cells; 4, adult peripheral blood total B cells separated using MACS; 5, Ramos cells. Blk was detected using anti-Blk clone 7A12. The lower panel shows SDHA as a control for the amount of protein loaded.
Blk protein expression is reduced in umbilical cord B cells in subjects with the BLK risk allele
The anti-Blk antibody 7A12 was used in intracellular staining by flow cytometry in order to measure relative levels of Blk protein in cell subsets without the need for physical cell separation. This assay was validated by showing that the level of Blk protein detected by flow in Jurkat cells transduced with a lentiviral expression vector encoding Blk was significantly greater than the level in Jurkat cells transduced with the control plasmid or mock-infected cells (Supplementary Material, Fig. S2a–d). Another validation was performed to demonstrate that relative protein levels detected by western blot correlated with relative protein levels measured by flow; Blk protein levels measured by western blot (normalized to GAPDH) were highly correlated with relative Blk protein levels measured by flow (fluorescent geometric means normalized to an isotype control) (Supplementary Material, Fig. S2e–f).
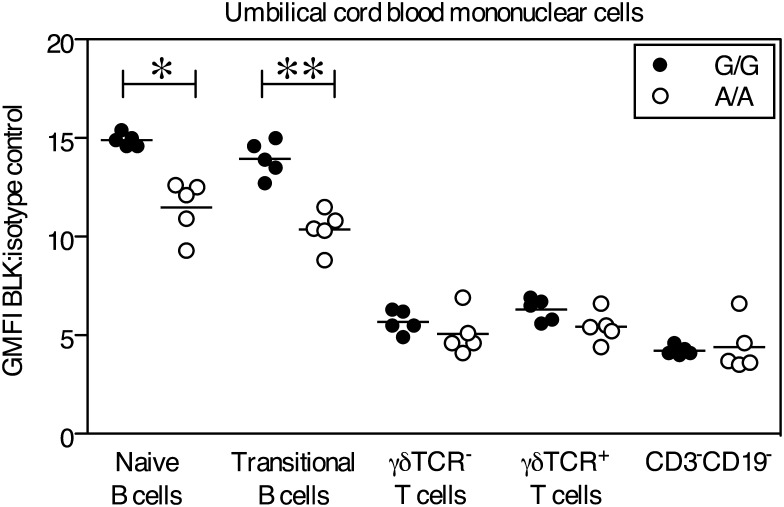
The intracellular flow assay was used to measure Blk protein levels in adult peripheral blood and UCB. Blk protein level values are expressed as fluorescent geometric means normalized to the geometric mean of isotype controls. Relative Blk protein levels were significantly lower in UCB naïve (CD10−CD38int) and transitional (CD10+CD28hi) B cell populations in homozygous risk subjects (A/A) than in subjects homozygous for the rs922483 non-risk allele (Fig. 4). In contrast, no differences in Blk protein levels between subjects based on genotype were observed in any T cell subset by flow cytometry or western blot analysis (Fig. 4 and data not shown), despite the clear evidence for allelic imbalance at the transcript level. Neither B cells nor T cells from adult peripheral blood or thymus showed significant differences in protein expression by the genotype (Supplementary Material, Fig. S3a and b).
Figure 4.
Blk protein levels are significantly reduced in umbilical cord naïve and transitional B cells from subjects homozygous for the risk allele. Blk was measured by intracellular staining and flow cytometry. The GMFI of cells labeled with anti-Blk was normalized to the GMFI of isotype labeled cells. Shown is the relative expression of Blk in cells gated using the following surface marker phenotypes: UCB naïve B cells (CD3−CD19+IgD+CD27−CD10−CD38int), UCB transitional B cells (CD3−CD19+IgD+CD27−CD10+CD38hi), UCB T cells (CD3+CD19−γδTCR− and CD3+CD19−γδTCR+) and UCB CD3−CD19− cells. Samples homozygous for rs922483 A (risk allele) are shown as open circles and homozygous G samples are shown as filled circles. Statistically significant differences as determined by the Mann–Whitney non-parametric U-test to compare indicated groups are denoted as *P <0.05 and **P <0.01 and the mean is represented by a horizontal line. The data are representative of three independent experiments.
DISCUSSION
These data allow us to report for the first time that the autoimmune disease risk variant rs922483 in the BLK gene is strongly associated with regulation of both BLK mRNA and protein expression in primary human transitional and naïve B cells from UCB. Strikingly, this genetic effect on expression was not observed in adult B cell subsets, even after B cell activation in vitro, or in freshly isolated tonsillar B cells engaged in an ongoing immune response. These data suggest that the mechanisms by which low BLK expression predisposes to autoimmunity are likely to be mediated early in B cell development.
A difference in BLK regulation in B cells isolated from adult peripheral blood compared with UCB at birth is not entirely unexpected. ‘Naïve’ and ‘transitional’ cell subsets are captured using a combination of surface markers: CD19, IgD, CD38 and CD10. However, these phenotyping tools may be too simple to discriminate between cell populations within these two subsets. Umbilical cord B cells are considered a more primitive and immature population (for example, ∼50% of UCB B cells express CD5, a marker of B cell immaturity) (30). The investigation of IgD+ CD27− B cells in umbilical cord and adult peripheral blood has revealed that naïve cells can be further divided into true naïve (CD38lo), intermediate (CD38int) and transitional (CD38hi) B cell subsets (28). In cord blood, intermediate and transitional cells represent ∼70% of CD27− IgD+ B cells, whereas these two populations represent only 14% of adult peripheral blood CD27− B cells. These subpopulations have been demonstrated to differ in maturity and function with respect to calcium mobilization following BCR cross-linking and cell death (28). These differences in B cell subpopulations are likely to explain the differences we observe in BLK regulation when comparing adult and cord blood naïve B cell populations. Although early studies clearly indicate that Blk is directly involved in BCR activating events (16–18), the published data suggest that Blk may act as either a positive or negative regulator of BCR activation, depending on the stage of B cell development (22,25,31). Early investigation of Blk knockout animals did not reveal any striking phenotypes, although in the absence of Lyn and Fyn, the lack of Blk resulted in severe attenuation of B cell differentiation from the pro B to pre B cell stage (23). In addition, expression of a constitutively active transgene of Blk can enhance this developmental progression by mimicking the effects of BCR signaling (22). If Blk positively affects BCR signaling events later in development, reduction in the levels of Blk conferred by the disease-associated haplotype may alter B cell selection leading to a more autoimmune prone receptor repertoire, similar to what has been proposed for PTPN22, another autoimmune associated gene (32).
Despite the early negative studies in mice, more subtle phenotypes have recently been reported in studies of Blk knockout animals, consistent with a potential negative regulatory effect of Blk. Thus, Blk knockout as well as haplodeficient (+/−) animals exhibit evidence of autoimmunity with higher levels of anti-nuclear antibodies than wild-type mice; interestingly, this phenotype was in fact more common in Blk+/− animals (25). The presence of autoimmunity was ascribed to the increased number of MZ and B1 B cells in these animals. In addition, Blk−/− MZ B cells appeared relatively hyper-responsive to BCR signaling with enhanced ERK activation after BCR cross-linking. This raises the possibility that the BLK risk haplotype in humans is acting via MZ or B1 cells, an issue that is difficult to address, given the challenges of identifying these cells in human peripheral blood.
Finally, BLK is expressed in T cells, albeit at considerably lower levels than in B cells. Expression of BLK in human thymus and certain human T cell leukemia has been reported previously (15,33), and our data confirm that Blk is present at both the message and protein level in the major subsets of human T cells. A recent study in the mouse has suggested a role for Blk expression in γδ TCR+ T cells (24). Blk was reportedly expressed in mouse γδ TCR+ T cells and Blk deficiency led to the absence of IL-17 producing γδ TCR+ T cells in Blk−/− γδ TCR transgenic mice (24).
These murine studies of Blk function in T cells raise the possibility that the BLK risk allele is acting via an effect on T cells. However, while we have observed evidence of allelic regulation of mRNA levels in human T cells, we do not observe significant genotype-dependent differences in protein expression in the T cell compartment, nor in the thymus. Furthermore, the contribution of Blk to the physiological function of T cells is uncertain given that Blk protein levels are markedly lower in T cells than in B cells. It is unknown why Blk protein levels in T cells do not reflect transcript levels; however, it is also possible that very subtle protein differences are beyond the detection limit of the assay. Thus, it is hard to make a strong case for a T cell mechanism at this time, although it remains possible that there are specific human T cell subsets that are particularly affected by SNP-regulated alterations in Blk protein levels.
At the molecular level, it is important to note that Blk protein has been shown to physically associate with a number of proteins important in B cell signaling: mIgM and mIgD (34), phospholipase Cγ2, p21ras-GTPase activating protein and microtubule-associated protein kinase (35). Furthermore, the BLK gene exhibits a gene–gene interaction with several genes [MMEL1 (36), TNFSF4 and TNFAIP3 (37)], some of which are important in immune signaling. Interestingly, BLK was recently shown to exhibit a gene–gene interaction with the B cell scaffolding protein BANK1 (38), and co-expression of Blk and BANK1 leads to retention of Blk in the cytoplasm. BANK1 also interacts with the Src family kinase Lyn, is tyrosine phosphorylated by Syk and is involved in BCR-induced calcium mobilization (39). These data suggest that BANK1 may function to sequester Blk and Lyn from the BCR to restrict BCR signaling. Moving forward, further functional studies are required to address how the interaction of Blk and BANK1 may affect umbilical cord B cell signaling and B cell function in double homozygous risk subjects.
Our data provide a cautionary tale regarding the use of monoclonal antibody reagents for the detection of protein expression when dealing with protein targets that are members of closely related gene families. It is striking that a large number of commercially available reagents that are allegedly specific for Blk in fact cross-react with other Src kinases, including Lck, which is predominantly expressed in T cells. Nevertheless, the identification and confirmation of a Blk-specific antibody reagent that is amenable to flow analysis should greatly simplify the future exploration of Blk expression in rare subsets of lymphocytes.
Overall, our data emphasize the importance of examining regulatory variation of putative risk genes in the full range of cell types that may be relevant to disease pathogenesis. While screening for genetic effects on gene expression in transformed B cell lines is useful, these studies do not necessarily reflect the regulatory features of these risk alleles in native cell types, even within the same lineage. The relatively limited sample sizes we have examined make it possible that modest genetic effects on Blk expression may exist in more mature stages of B cell development. However, our data emphasize that future studies investigating dosage effects of Blk on B cell function will be most fruitfully focused on early B cell subsets from UCB (and possibly bone marrow) samples of defined genotype. These studies should include a detailed analysis of BCR signaling and an analysis of the BCR repertoire.
MATERIALS AND METHODS
Human blood and tissue samples
Fresh, de-identified human blood was collected from healthy control subjects from the Tissue Donation Program (TDP) at The Feinstein Institute for Medical Research (FIMR) through an Institutional Review Board (IRB)-approved protocol ‘Genes and Phenotype (GAP) Registry (IRB# 09-081), a national resource for genotype–phenotype studies.’ Fresh, anonymous human UCB was obtained through the TDP of the FIMR using an IRB exempt protocol. Tonsil cells were obtained from discarded anonymous tissue obtained during tonsillectomies through the IRB protocol ‘Collection of solid tissues from tonsil, spleen and lymph nodes' (IRB# 05-034). Fresh, anonymous, thymus tissue was obtained through the TDP of the FIMR through an IRB approved protocol ‘Functional analysis of polymorphisms in BLK and other autoimmune disease risk alleles in human thymocytes’ (IRB# 10-041).
Mononuclear cells were purified from buffy coats of adult peripheral blood and UCB using a Ficoll (Cellgro) density gradient. The mononuclear cell layer was extracted and washed and cells were stored in FCS containing 10% DMSO. Single cell suspensions were made from thymus and tonsil tissues by mincing the tissue with a scalpel and forcing the tissue through a metal sieve. The cell suspensions were then washed and mononuclear cells were separated using a Ficoll density gradient.
RNA from the CEPH B lymphoblastoid cell lines was a kind gift from Robert Graham at Genentech.
Genotyping and pyrosequencing
Samples were genotyped for the BLK SNP rs922483 by pyrosequencing using the following primers: biotinylated forward primer: 5′ GGACTGGGGAGGCTCTGAT 3′, reverse primer: 5′ CCTAGCGACAACGAGCAACTT 3′ and sequencing primer: 5′ CCGGCAGCAGACAGAGGT 3′.
Cell culture
Total CD19+ B cells, CD14+ monocytes and CD4+ T cells were isolated from peripheral blood mononuclear cells (PBMC) using MACS Magnetic Separation (Miltenyi Biotec). Macrophages were obtained by culturing CD14+ monocytes with 100 ng/ml mCSF (R&D) for 7 days. Immature dendritic cells were generated by culturing CD14+ monocytes in 800 U/ml GMCSF and 1000 U/ml IL-4 (R&D) for 6 days. These cells were further cultured for 2 days in 10 ng/ml TNF, 10 ng/ml IL-1β, 1000 U/ml IL-6 (R&D) and 1 μg/ml PGE (Sigma) to develop into mature dendritic cells. CD19+ B cells were stimulated with 200 ng/ml anti-CD40 (Coulter mAb89), or with anti-CD40, 10 μg/ml anti-IgM (Southern Biotech) and 5 ng/ml IL-4 (R&D). Monocytes were activated with IFN-γ (1000 U/ml) (R&D) and LPS (200 ng/ml) (Sigma) for 48 h.
Ramos and Jurkat cell lines were cultured in RPMI supplemented with 10% FCS. Recombinant lentivirus particles were generated by co-transfecting 293Ta cells with a lentiviral expression plasmid and a Lenti-Pac HIV Expression Packaging Kit (GeneCopoeia). Lentivirus was harvested 48 h post-transfection. Jurkat cells were transduced with lentivirus particles expressing Blk-EGFP or EGFP alone and cells were cultured in the presence or absence of Puromycin (InvivoGen) at 600 ng/ml.
Flow cytometry
Antibodies used in surface labeling for flow cytometry were various combinations of the following: CD3-PE CF594 (BD, clone UCHT1), CD3-QDOT605 (Invitrogen, clone UCHT1), CD4-Alexa Fluor 700 (Biolegend, clone RPA-T4), CD4-Pacific Blue (BD, clone RPA-T4), CD4-FITC (BD, clone SK3), CD8-APC (BD, clone RPA-T8), CD8-APC (BD, clone CD28.2), CD8-PE (BD, clone SK1), CD10-PE Cy7 (Biolegend, clone HI10A), CD14-PerCP (BD, clone MφP9), CD19-APC Cy7 (Biolegend, clone HIB19), CD25-PE (BD, clone M-A251), CD27-APC (BD, clone L128), CD27-FITC (BD, clone M-T271), CD38-PE (BD, clone HB7), CD14-PE/CD45- FITC (BD, clones MφP9 and 2D1), CD45RO-FITC (Invitrogen, clone UCHL1), γδ TCR-PE (BD, clone 11F2), γδ TCR-Tri-Color (Invitrogen, clone 5A6.E9), IgD-FITC (Southern Biotech), ICOS-Biotin (eBioscience) and Streptavidin-APC (Molecular Probes, Invitrogen).
For intracellular Blk staining, frozen PBMC were thawed at 37°C and transferred immediately to pre-warmed RPMI supplemented with 10% FCS. The cells were washed, filtered, counted and fixed in 32% PFA for 5 min at 37°C. Post-fixation, the cells were permeabilized in a Perm Wash I Buffer (BD® Phosflow) containing 1% FCS. Once pelleted, 1 × 106 total mononuclear cells were labeled with either unconjugated anti-Blk (Novus, 7A12) or mouse IgG1 (BD, clone X40) and incubated at 4°C for 30 min. Cells were washed in Perm Wash buffer and incubated in either V450-labeled Rat anti-mouse IgG1 (clone A85-1) or PE-labeled Goat anti-mouse IgG (Southern Biotech) for 30 min at 4°C. Cells were washed again and surface labeled with various combinations of surface markers. Cells were examined using an LSRII (BD Biosciences).
Cell populations were isolated from purified PBMC by fluorescent activated cell sorting using a FACSAria IIU (BD Biosciences). Post-sort purity was performed to determine that cell populations were >95% pure.
qPCR and allelic imbalance assays
RNA was isolated from cell suspensions using RNeasy (Qiagen). Up to 200 ng of total RNA was used in a cDNA reaction using Multiscribe (Applied Biosystems). Allelic imbalance assays were performed using a pyrosequencing assay developed for the rs922483 SNP. cDNA or DNA of heterozygous samples were used as a template for pyrosequencing and the reaction peaks generated when nucleotides were incorporated into the sequence were measured. To determine the relative expression of SNP alleles, the ratio between the peaks corresponding to the alleles of the SNP from pyrosequencing of cDNA were calculated. Ratios were normalized to the allele ratios generated by pyrosequencing DNA samples.
For qPCR analysis, BLK mRNA transcripts were measured in cDNA using TaqMan assays (Applied Biosystems) in a Lightcycler 480 II (Roche). BLK expression was measured using hs00176441_m1 which amplifies a region spanning the exon1-2 border of BLK. Results are expressed as ΔΔCp values normalized to GAPDH (HuGAPDH, Applied Biosystems).
Western blot
Protein lysate was extracted from cell pellets using RIPA reagent containing Proteinase Inhibitor (Thermo-Pierce). Sample protein concentrations were quantified using a Micro BCA Protein Assay Kit (Thermo Scientific). Lysates were separated by SDS–PAGE electrophoresis using NuPAGE (Invitrogen) and transferred to PDVF membrane. Membranes were probed with one or more primary antibodies including: anti-human Blk clone 7A12 (Novus Biologicals), anti-Blk clones 9D10D1 and 9D10B7H6 (Santa Cruz Biotechnology), anti-Blk rabbit polyclonal (Cell Signaling), anti-SDHA rabbit polyclonal (Cell Signaling) and anti-GAPDH clone 1G5 (Sigma) followed by incubation with anti-mouse or anti-rabbit IR Dye 800CW or IR Dye 680 and visualized using an Odyssey Infrared Imaging System (LI-COR Biosciences). The recombinant proteins used were as follows: Recombinant Human Active Fyn A-GST (Sigma), Recombinant Human Active: Lck-GST, Lyn B-GST, Blk-GST (R&D Systems).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institutes of Health (RC2AR059092 to P.K.G and B.D.); The Muriel Fusfeld Foundation through the Lauri Strauss Leukemia Foundation to K.R.S. and P.K.G.; and The Eileen and Mel Ludwig Endowment for Rheumatoid Arthritis to P.K.G.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank all members of the Center for Genomics and Human Genetics, and Diamond lab for technical assistance, secretarial assistance and helpful discussions. We thank the FIMR flow facility personnel and the Chiorazzi lab at FIMR for tonsil. We are especially grateful to Mary Keogh, Cathleen Mason and Margaret DeFranco of the FIMR Tissue bank, and to Christine Metz, for sample acquisition and to the FIMR Biorepository for sample processing. We thank Robert Graham (Genentech) for sharing unpublished data with us.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Pastinen T., Ge B., Hudson T.J. Influence of human genome polymorphism on gene expression. Hum. Mol. Genet. 2006;15:9–16. doi: 10.1093/hmg/ddl044. [DOI] [PubMed] [Google Scholar]
- 2.Murphy A., Chu J.H., Xu M., Carey V.J., Lazarus R., Liu A., Szefler S.J., Strunk R., Demuth K., Castro M., et al. Mapping of numerous disease-associated expression polymorphisms in primary peripheral blood CD4+ lymphocytes. Hum. Mol. Genet. 2010;19:4745–4757. doi: 10.1093/hmg/ddq392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toung J.M., Morley M., Li M., Cheung V.G. RNA-sequence analysis of human B-cells. Genome Res. 2011;21:991–998. doi: 10.1101/gr.116335.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ge B., Pokholok D.K., Kwan T., Grundberg E., Morcos L., Verlaan D.J., Le J., Koka V., Lam K.C., Gagne V., et al. Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nat. Genet. 2009;41:1216–1222. doi: 10.1038/ng.473. [DOI] [PubMed] [Google Scholar]
- 5.Dimas A.S., Deutsch S., Stranger B.E., Montgomery S.B., Borel C., Attar-Cohen H., Ingle C., Beazley C., Gutierrez Arcelus M., Sekowska M., et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gregersen P.K., Amos C.I., Lee A.T., Lu Y., Remmers E.F., Kastner D.L., Seldin M.F., Criswell L.A., Plenge R.M., Holers V.M., et al. REL, encoding a member of the NF-kappaB family of transcription factors, is a newly defined risk locus for rheumatoid arthritis. Nat. Genet. 2009;41:820–823. doi: 10.1038/ng.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hom G., Graham R.R., Modrek B., Taylor K.E., Ortmann W., Garnier S., Lee A.T., Chung S.A., Ferreira R.C., Pant P.V., et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N. Engl. J. Med. 2008;358:900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 8.Freudenberg J., Lee H.S., Han B.G., Shin H.D., Kang Y.M., Sung Y.K., Shim S.C., Choi C.B., Lee A.T., Gregersen P.K., et al. Genome-wide association study of rheumatoid arthritis in Koreans: population-specific loci as well as overlap with European susceptibility loci. Arthritis Rheum. 2011;63:884–893. doi: 10.1002/art.30235. [DOI] [PubMed] [Google Scholar]
- 9.Han J.W., Zheng H.F., Cui Y., Sun L.D., Ye D.Q., Hu Z., Xu J.H., Cai Z.M., Huang W., Zhao G.P., et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat. Genet. 2009;41:1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 10.Gourh P., Agarwal S.K., Martin E., Divecha D., Rueda B., Bunting H., Assassi S., Paz G., Shete S., McNearney T., et al. Association of the C8orf13-BLK region with systemic sclerosis in North-American and European populations. J. Autoimmun. 2010;34:155–162. doi: 10.1016/j.jaut.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nordmark G., Kristjansdottir G., Theander E., Appel S., Eriksson P., Vasaitis L., Kvarnstrom M., Delaleu N., Lundmark P., Lundmark A., et al. Association of EBF1, FAM167A(C8orf13)-BLK and TNFSF4 gene variants with primary Sjogren's syndrome. Genes Immun. 2011;12:100–109. doi: 10.1038/gene.2010.44. [DOI] [PubMed] [Google Scholar]
- 12.Yin H., Borghi M.O., Delgado-Vega A.M., Tincani A., Meroni P.L., Alarcon-Riquelme M.E. Association of STAT4 and BLK, but not BANK1 or IRF5, with primary antiphospholipid syndrome. Arthritis Rheum. 2009;60:2468–2471. doi: 10.1002/art.24701. [DOI] [PubMed] [Google Scholar]
- 13.Dymecki S.M., Niederhuber J.E., Desiderio S.V. Specific expression of a tyrosine kinase gene, blk, in B lymphoid cells. Science. 1990;247:332–336. doi: 10.1126/science.2404338. [DOI] [PubMed] [Google Scholar]
- 14.Borowiec M., Liew C.W., Thompson R., Boonyasrisawat W., Hu J., Mlynarski W.M., El Khattabi I., Kim S.H., Marselli L., Rich S.S., et al. Mutations at the BLK locus linked to maturity onset diabetes of the young and beta-cell dysfunction. Proc. Natl Acad. Sci. USA. 2009;106:14460–14465. doi: 10.1073/pnas.0906474106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Islam K.B., Rabbani H., Larsson C., Sanders R., Smith C.I. Molecular cloning, characterization, and chromosomal localization of a human lymphoid tyrosine kinase related to murine Blk. J. Immunol. 1995;154:1265–1272. [PubMed] [Google Scholar]
- 16.Brunswick M., Burkhardt A., Finkelman F., Bolen J., Mond J.J. Comparison of tyrosine kinase activation by mitogenic and nonmitogenic anti-IgD antibodies. J. Immunol. 1992;149:2249–2254. [PubMed] [Google Scholar]
- 17.Burkhardt A.L., Brunswick M., Bolen J.B., Mond J.J. Anti-immunoglobulin stimulation of B lymphocytes activates src-related protein-tyrosine kinases. Proc. Natl Acad. Sci. USA. 1991;88:7410–7414. doi: 10.1073/pnas.88.16.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saouaf S.J., Mahajan S., Rowley R.B., Kut S.A., Fargnoli J., Burkhardt A.L., Tsukada S., Witte O.N., Bolen J.B. Temporal differences in the activation of three classes of non-transmembrane protein tyrosine kinases following B-cell antigen receptor surface engagement. Proc. Natl Acad. Sci. USA. 1994;91:9524–9528. doi: 10.1073/pnas.91.20.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saouaf S.J., Kut S.A., Fargnoli J., Rowley R.B., Bolen J.B., Mahajan S. Reconstitution of the B cell antigen receptor signaling components in COS cells. J. Biol. Chem. 1995;270:27072–27078. doi: 10.1074/jbc.270.45.27072. [DOI] [PubMed] [Google Scholar]
- 20.Texido G., Su I.H., Mecklenbrauker I., Saijo K., Malek S.N., Desiderio S., Rajewsky K., Tarakhovsky A. The B-cell-specific Src-family kinase Blk is dispensable for B-cell development and activation. Mol. Cell Biol. 2000;20:1227–1233. doi: 10.1128/mcb.20.4.1227-1233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malek S.N., Dordai D.I., Reim J., Dintzis H., Desiderio S. Malignant transformation of early lymphoid progenitors in mice expressing an activated Blk tyrosine kinase. Proc. Natl Acad. Sci. USA. 1998;95:7351–7356. doi: 10.1073/pnas.95.13.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tretter T., Ross A.E., Dordai D.I., Desiderio S. Mimicry of pre-B cell receptor signaling by activation of the tyrosine kinase Blk. J. Exp. Med. 2003;198:1863–1873. doi: 10.1084/jem.20030729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saijo K., Schmedt C., Su I.H., Karasuyama H., Lowell C.A., Reth M., Adachi T., Patke A., Santana A., Tarakhovsky A. Essential role of Src-family protein tyrosine kinases in NF-kappaB activation during B cell development. Nat. Immunol. 2003;4:274–279. doi: 10.1038/ni893. [DOI] [PubMed] [Google Scholar]
- 24.Laird R.M., Laky K., Hayes S.M. Unexpected role for the B cell-specific Src family kinase B lymphoid kinase in the development of IL-17-producing T cells. J. Immunol. 2010;185:6518–6527. doi: 10.4049/jimmunol.1002766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samuelson E.M., Laird R.M., Maue A.C., Rochford R., Hayes S.M. Blk haploinsufficiency impairs the development, but enhances the functional responses, of MZ B cells. Immunol. Cell Biol. 2011 doi: 10.1038/icb.2011.76. doi:10.1038/icb.2011.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orozco G., Eyre S., Hinks A., Bowes J., Morgan A.W., Wilson A.G., Wordsworth P., Steer S., Hocking L., Thomson W., et al. Study of the common genetic background for rheumatoid arthritis and systemic lupus erythematosus. Ann. Rheum. Dis. 2011;70:463–468. doi: 10.1136/ard.2010.137174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graham R.R., Cotsapas C., Davies L., Hackett R., Lessard C.J., Leon J.M., Burtt N.P., Guiducci C., Parkin M., Gates C., et al. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat. Genet. 2008;40:1059–1061. doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee J., Kuchen S., Fischer R., Chang S., Lipsky P.E. Identification and characterization of a human CD5+ pre-naive B cell population. J. Immunol. 2009;182:4116–4126. doi: 10.4049/jimmunol.0803391. [DOI] [PubMed] [Google Scholar]
- 29.Mulhern T.D., Shaw G.L., Morton C.J., Day A.J., Campbell I.D. The SH2 domain from the tyrosine kinase Fyn in complex with a phosphotyrosyl peptide reveals insights into domain stability and binding specificity. Structure. 1997;5:1313–1323. doi: 10.1016/s0969-2126(97)00283-9. [DOI] [PubMed] [Google Scholar]
- 30.Harris D.T., Schumacher M.J., Locascio J., Besencon F.J., Olson G.B., DeLuca D., Shenker L., Bard J., Boyse E.A. Phenotypic and functional immaturity of human umbilical cord blood T lymphocytes. Proc. Natl Acad. Sci. USA. 1992;89:10006–10010. doi: 10.1073/pnas.89.21.10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao X.R., Scott D.W. Antisense oligodeoxynucleotides to the blk tyrosine kinase prevent anti-mu-chain-mediated growth inhibition and apoptosis in a B-cell lymphoma. Proc. Natl Acad. Sci. USA. 1993;90:7946–7950. doi: 10.1073/pnas.90.17.7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menard L., Saadoun D., Isnardi I., Ng Y.S., Meyers G., Massad C., Price C., Abraham C., Motaghedi R., Buckner J.H., et al. The PTPN22 allele encoding an R620W variant interferes with the removal of developing autoreactive B cells in humans. J. Clin. Invest. 2011;121:3635–3644. doi: 10.1172/JCI45790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krejsgaard T., Vetter-Kauczok C.S., Woetmann A., Kneitz H., Eriksen K.W., Lovato P., Zhang Q., Wasik M.A., Geisler C., Ralfkiaer E., et al. Ectopic expression of B-lymphoid kinase in cutaneous T-cell lymphoma. Blood. 2009;113:5896–5904. doi: 10.1182/blood-2008-09-181024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin J., Justement L.B. The MB-1/B29 heterodimer couples the B cell antigen receptor to multiple src family protein tyrosine kinases. J. Immunol. 1992;149:1548–1555. [PubMed] [Google Scholar]
- 35.Pleiman C.M., Clark M.R., Gauen L.K., Winitz S., Coggeshall K.M., Johnson G.L., Shaw A.S., Cambier J.C. Mapping of sites on the Src family protein tyrosine kinases p55blk, p59fyn, and p56lyn which interact with the effector molecules phospholipase C-gamma 2, microtubule-associated protein kinase, GTPase-activating protein, and phosphatidylinositol 3-kinase. Mol. Cell Biol. 1993;13:5877–5887. doi: 10.1128/mcb.13.9.5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deshmukh H.A., Maiti A.K., Kim-Howard X.R., Rojas-Villarraga A., Guthridge J.M., Anaya J.M., Nath S.K. Evaluation of 19 autoimmune disease-associated loci with rheumatoid arthritis in a colombian population: evidence for replication and gene-gene interaction. J. Rheumatol. 2011;38:1866–1870. doi: 10.3899/jrheum.110199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou X.J., Lu X.L., Nath S.K., Lv J.C., Zhu S.N., Yang H.Z., Qin L.X., Zhao M.H., Su Y., Shen N., et al. Gene-gene interaction of BLK, TNFSF4, TRAF1, TNFAIP3, and REL in systemic lupus erythematosus. Arthritis Rheum. 2012;64:222–231. doi: 10.1002/art.33318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castillejo-Lopez C., Delgado-Vega A.M., Wojcik J., Kozyrev S.V., Thavathiru E., Wu Y.Y., Sanchez E., Pollmann D., Lopez-Egido J.R., Fineschi S., et al. Genetic and physical interaction of the B-cell systemic lupus erythematosus-associated genes BANK1 and BLK. Ann. Rheum. Dis. 2012;71:136–142. doi: 10.1136/annrheumdis-2011-200085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yokoyama K., Su Ih I.H., Tezuka T., Yasuda T., Mikoshiba K., Tarakhovsky A., Yamamoto T. BANK regulates BCR-induced calcium mobilization by promoting tyrosine phosphorylation of IP(3) receptor. EMBO J. 2002;21:83–92. doi: 10.1093/emboj/21.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bohnhorst J.O., Bjorgan M.B., Thoen J.E., Natvig J.B., Thompson K.M. Bm1-Bm5 classification of peripheral blood B cells reveals circulating germinal center founder cells in healthy individuals and disturbance in the B cell subpopulations in patients with primary Sjogren's syndrome. J. Immunol. 2001;167:3610–3618. doi: 10.4049/jimmunol.167.7.3610. [DOI] [PubMed] [Google Scholar]
- 41.Pascual V., Liu Y.J., Magalski A., de Bouteiller O., Banchereau J., Capra J.D. Analysis of somatic mutation in five B cell subsets of human tonsil. J. Exp. Med. 1994;180:329–339. doi: 10.1084/jem.180.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.