Abstract
RNA localization is a mechanism to post-transcriptionally regulate gene expression. Eukaryotic organisms ranging from fungi to mammals localize mRNAs to spatially restrict synthesis of specific proteins to distinct regions of the cytoplasm. In this review, we provide a general summary of RNA localization pathways in Saccharomyces cerevisiae, Xenopus, Drosophila and mammalian neurons.
Introduction
Eukaryotic cells need to temporally and spatially regulate gene expression in response to a variety of cues. While temporal regulation of gene expression can be achieved through transcriptional regulation, the ability of a cell to spatially restrict synthesis of a specific protein within the cytoplasm requires post-transcriptional control. Consequently, eukaryotic cells have developed a variety of mechanisms to spatially sort specific proteins to distinct intracellular locations. Proteins destined for organelles, such as mitochondria and the ER (endoplasmic reticulum), contain signal peptides that direct the protein to the proper location within the cell. Another mechanism to spatially sort proteins is RNA localization. In this process the mRNA encoding the protein is localized to the site where the protein functions. Once the mRNA reaches the proper destination, the mRNA is translated, resulting in the spatial restriction of the corresponding protein. In addition to sorting proteins to distinct regions within the cytoplasm, RNA localization has a role in sorting proteins to the ER and mitochondria [1,2].
There are at least three mechanisms by which an mRNA can be localized: (1) local protection from degradation, (2) diffusion and entrapment by a localized anchor and (3) direct transport by motor proteins on cytoskeletal filaments [3,4]. In Drosophila, Hsp83 mRNA localizes to the posterior pole plasma of the egg using the protection from degradation pathway [5]. In this RNA localization pathway, the RNAs are protected from degradation at the site of localization and are highly susceptible to degradation in other areas of the cell. In contrast, the Xcat-2 mRNA in Xenopus utilizes the diffusion and entrapment mechanism [6]. RNA localization substrates that utilize this pathway randomly diffuse throughout the cytoplasm, and, when the RNA reaches the site of localization, it is captured and retained. In this article, we will focus on direct transport (Figure 1). In this mechanism, the RNA contains a cis-acting localization element, also referred to as a “zip code”. The cis-acting element directs the RNA to the proper intracellular location. RNA-binding proteins recognize a nucleotide sequence and/or structure in the cis-acting localization element. Some RNA-binding proteins identify the RNA localization substrate in the nucleus and escort the RNA to the cytoplasm. Subsequently, the RNA-protein complex interfaces with a molecular motor (myosin, kinesin or dynein), which directly transports the RNA on cytoskeletal components. During transport of the RNA to the site of localization, translation of the RNA is repressed. Once the RNA arrives at its destination, it is hypothesized that the RNA is anchored at the site of localization, and translational repression of the RNA is relieved.
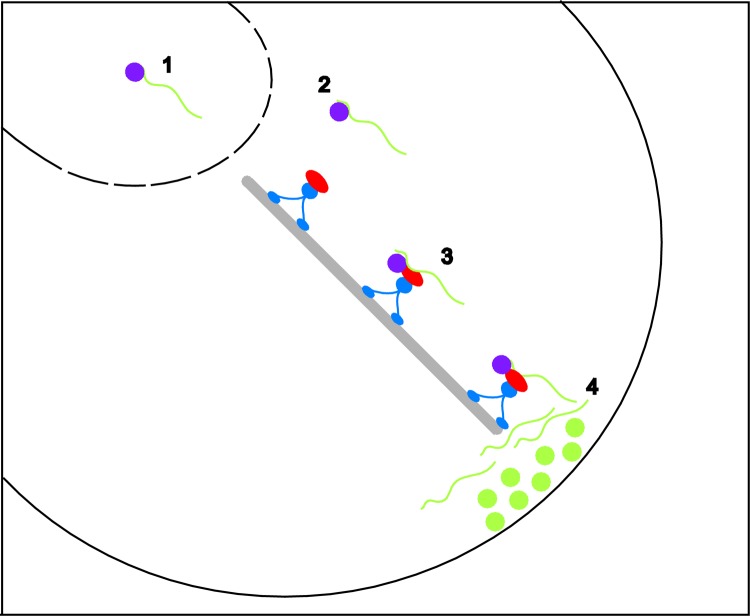
Figure 1. A general pathway for cytoplasmic RNA localization can be broken into distinct steps.
Step 1 – RNA localization substrates (green line) can be identified in the nucleus by RNA-binding proteins (purple ball). Step 2 – the RNA-protein complex is exported from the nucleus to the cytoplasm through the nuclear pore complex. Step 3 – once in the cytoplasm, the RNA can associate with additional proteins (red) that interface the RNA with motor proteins (blue) that transport the RNA along cytoskeletal filaments (gray) to the site of localization. Step 4 – upon reaching the correct destination, translational repression is relieved and the corresponding protein (green ball) is synthesized at the site of action.
ASH1 mRNA localization in S. cerevisiae
In the yeast S. cerevisiae, there are at least 30 mRNAs that are directly transported and localized to the bud tip [7-11], and of these ASH1 mRNA is the most extensively studied. Localization of ASH1 mRNA to the distal tip of the daughter cell (Figure 2) results in the asymmetric sorting of Ash1 to the daughter cell nucleus [12-14]. Ash1 is a transcriptional repressor, and asymmetric sorting of Ash1 results in differential gene expression between mother and daughter cells [15-19].
Figure 2. ASH1 mRNA localization in S. cerevisiae.
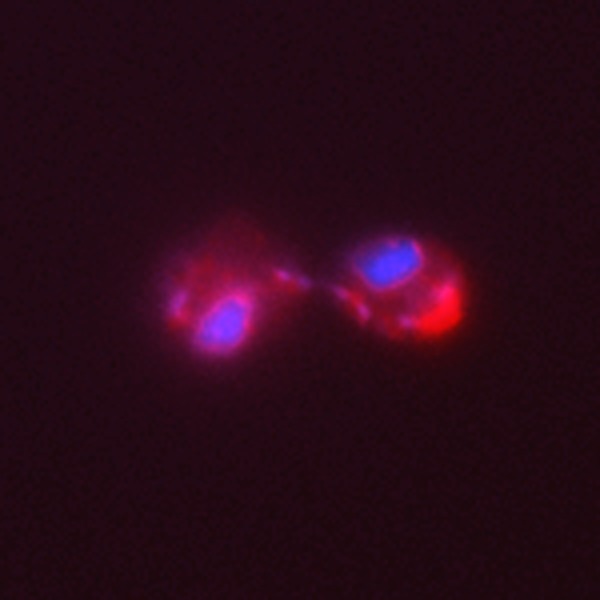
The distribution of ASH1 mRNA is shown in red, and the positions of the nuclei are shown in blue.
Localization of ASH1 mRNA requires four cis-acting localization elements: E1, E2A, E2B and E3 [20,21]. Each of the elements is mechanistically redundant, since each is dependent on the same set of trans-acting factors for mRNA localization to the bud tip [22]. She2 is a unique RNA-binding protein that directly associates with each of the cis-acting localization elements [22-24]. The ASH1 mRNA localization pathway originates in the nucleus where She2 co-transcriptionally identifies RNA localization substrates [25-27]. After exiting the nucleus, She2 forms a transport particle with She3 and the Myo4 motor protein, and the transport particle directly delivers ASH1 mRNA to the bud tip on the actin cytoskeleton [13,14,16,28-32]. Initially, the function for She3 in ASH1 mRNA localization was limited to its ability to simultaneously associate with She2 and Myo4 [32]. However, more recent studies indicate that She3 has additional roles in ASH1 mRNA localization [33,34]. One of these studies provides compelling evidence that She3 directly contacts the cis-acting localization elements [34]. In addition, She2 increases the specificity of She3 for cis-acting localization elements, and She3p increases the specificity of She2 for cis-acting localization elements [34]. Myo4 is a type V non-processive myosin motor protein that directly transports ASH1 mRNA to the bud tip through its association with She3 and the actin cytoskeleton [13,14, 28-31,35,36]. Processive motors are characterized by a single motor molecule possessing the ability to promote continuous transport of a cargo over long distances, while non-processive motors require multiple motor molecules to promote continuous transport over long distances. Given that Myo4 is a non-processive motor, the mechanism by which Myo4 continuously transports ASH1 mRNA is an active area of interest [36]. Recently, a mechanism has been described that results in continuous transport of ASH1 mRNA. In this mechanism, a tetramer of She2 binds to a cis-acting localization element, subsequently recruiting multiple molecules of Myo4, which increases processivity of the transport complex [37-39].
During the transport of ASH1 mRNA from the mother cell to the daughter cell, translation of Ash1 is repressed. The E1, E2A and E2B cis-acting localization elements are located in the ORF (open reading frame), and the position of these cis-acting localization elements participate in reducing translation of Ash1 [12]. Furthermore, Khd1 and Puf6 participate in translational repression of ASH1 mRNA [40,41]. Khd1 is an RNA-binding protein that associates with the E1 element and with the C terminus of the translational initiation factor eIF4G1 [41]. However, the mechanism by which these interactions repress translation of ASH1 mRNA remains elusive. Translational repression of ASH1 mRNA by Khd1 is relieved at the bud tip through the action of Yck1, a protein kinase located in the plasma membrane [41]. Puf6, a member of the PUF family of RNA-binding proteins, represses translation of ASH1 mRNA in a manner similar to Khd1. The E3 cis-acting localization element contains a binding site for Puf6 [40]. Translational repression of ASH1 mRNA by Puf6 occurs through an interaction with the translational initiation factor Fun12, also known as eIF5B [42]. Through an unknown mechanism the interaction between Puf6 and Fun12 prevents conversion of the 43S pre-initiation complex to the 80S initiation complex [42]. Analogous to the mechanism employed by Khd1, translational repression by Puf6 is relieved by phosphorylation at the bud tip through the action of casein kinase II [42].
Xenopus
Vg1 mRNA is localized to the vegetal hemisphere of developing Xenopus oocytes [43]. Spatial restriction of Vg1 protein is necessary for proper specification of endoderm and mesoderm fates during embryogenesis [44-47]. As with ASH1 mRNA, Vg1 mRNA appears to be marked for its unique fate while still in the nucleus. Two proteins, hnRNP (heterogeneous nuclear ribonucleoprotein) I and Vg1RBP/vera, are thought to identify Vg1 mRNA in the nucleus and to accompany the transcript into the cytoplasm during nuclear export [48]. These proteins bind to localization elements present within the 3’UTR (3’untranslated region) of Vg1 mRNA. The hnRNP I protein binds to a localization element termed VM1 (YYUCU) (Vg1 motif 1), whereas Vg1RBP/vera binds to a localization element termed E2 (A/U,YCAC) [49-53]. Both localization elements are present in multiple copies within the 3’UTR of Vg1 mRNA [50, 54-56]. The specific sequence of these elements, rather than secondary structure, is the determining factor for binding by hnRNP I and Vg1RBA/vera [49-52]. Once in the cytoplasm, additional factors, such as Prrp and XStau, are thought to assemble on the mRNP (messenger ribonucleoprotein) [48]. Importantly, XStau is the Xenopus homolog of Staufen, a protein that has been shown to participate in mRNA localization in numerous systems [57-60].
Vg1 mRNA is transported to the vegetal cortex in a microtubule-dependent manner [61]. Recent evidence has implicated two different microtubule plus end-directed motor proteins in the transport step: Kinesin-1 and Kinesin-2 [62,63]. Initial studies described a role for Kinesin-2 in the transport of Vg1 mRNA [62]. Kinesin-2 is a heterotrimeric protein containing two distinct motor subunits and a non-motor accessory protein [64]. Consistent with a role for this motor in the transport of Vg1 mRNA, Kinesin-2 co-localizes with the message at the vegetal cortex of developing oocytes [62]. Furthermore, injecting oocytes with function-blocking antibodies directed against Kinesin-2 disrupts the localization of Vg1 mRNA [62].
Interestingly, XStau was shown to co-immunoprecipitate with Kinesin-1 [65]. Kinesin-1 is also known as conventional Kinesin, and is composed of two identical heavy chains containing the motor domain and two identical light chains [66]. This finding raised an important question: does Kinesin-1 also function in Vg1 mRNA transport? A recent report demonstrated that it does [63]. The two motors do not appear to function in a redundant manner, as blocking the activity of either motor causes defects in the localization of Vg1 mRNA [63]. It is unclear why Vg1 mRNA requires two motors for efficient transport to the vegetal cortex. One possibility is that the motors perform mechanistically distinct roles in the transport of Vg1 mRNA. Another possibility is that the endogenous level of either motor is insufficient for efficient transport. The latter possibility is supported by the finding that over-expression of either Kinesin motor is able to compensate for a deficit in the other [63].
The finding of this requirement for plus end motor proteins in the transport of Vg1 mRNA was somewhat surprising given that the prevailing view at the time held that the cortex of the oocyte was enriched in microtubule minus ends [67]. However, it was observed that a sub-set of plus ends are indeed enriched at the vegetal cortex [63]. Thus, Vg1 mRNA appears to be transported by Kinesin-1 and Kinesin-2 on a specialized population of microtubules. The mechanism by which the motor complexes are able to identify this unique population of microtubules remains elusive.
As with most localized fate determinants, it is important that Vg1 mRNA not be translated until the message is delivered to the correct site within the cell. Consistent with this notion, a region, named the VTE (Vg1 transitional element), in the 3’-untranslated region (UTR) of Vg1 mRNA, has been identified that functions to maintain the mRNA in a translationally repressed state [68,69]. An intriguing feature of the VTE is that it is very AU rich. Recent work demonstrated that the AU-rich binding proteins ElrA and ElrB bind to the VTE and function to repress the translation of Vg1 mRNA [70], but further work is needed to define the mechanism.
Drosophila
In a somewhat Herculean effort, Lécuyer and colleagues performed in situ hybridization against 3300 mRNAs expressed in Drosophila embryos [71]. Surprisingly, approximately 70% of these mRNAs are localized [71], showing that mRNA localization is widely used during Drosophila development for spatial sorting of proteins. Although the mechanism by which these thousands of mRNAs are localized has not been characterized, the localization of bicoid, gurken and oskar mRNA in Drosophila oocytes has been extensively studied [72-75]. In addition, apically localizing mRNAs, such as even skipped, wingless and hairy has also been studied in Drosophila embryos [76-79]. In this review, we will focus our discussion on the mechanism of oskar mRNA localization.
oskar mRNA is localized to the posterior of the oocyte in a microtubule-dependent manner [4,73,74] (Figure 3). The specific localization of oskar mRNA, as well as precise translational regulation of the message, results in restriction of Oskar protein to the posterior of the oocyte [73,80,81], which is essential for establishing the anterior-posterior polarity of the egg [81,82]. The importance of this is illustrated by the demonstration that mis-localization of oskar mRNA results in an oocyte that lacks polarity and an embryo that dies during development [73,81,82].
Figure 3. mRNA localization in Drosophila oocytes.
A schematic representation of the Drosophila egg chamber (top panel) oskar mRNA is transcribed in the nurse cells, transported into the oocyte and localized at the posterior pole (red). The bottom panel shows oskar mRNA (red) localization as detected by in situ hybridization. DAPI stained DNA is shown in blue.
Like many localized mRNAs, oskar is identified as a target for localization while still in the nucleus. Splicing at the first intron position of oskar mRNA appears to be required for its localization [83]. The position of this intron, but not the nucleotide sequence, is the critical factor for localization [83]. The mechanism by which splicing at this position regulates mRNA localization is unclear, but the Exon Junction Complex (EJC) proteins have all been shown to be involved in oskar mRNA localization [84-89]. Although EJC proteins are deposited on all spliced mRNAs, it is possible that they recruit an unknown factor important for oskar mRNA localization when they are present at the position of the first intron. In addition to the presence of this first intron, the 3’UTR of oskar mRNA is also required for localization [90]. Various regions in the 3’UTR of oskar mRNA are thought to mediate distinct steps in its transport from the nurse cells to the posterior of the oocyte [90]. However, the identity of specific proteins that bind to these elements is unknown.
The motor responsible for transporting oskar mRNA to the oocyte posterior appears to be Kinesin-1 [91]. Typically, cargoes that are transported by Kinesin-1 are directly bound by the Kinesin light chain (Klc) adaptor protein [66], and this complex then directly interacts with the motor subunit of the complex, Kinesin heavy chain (Khc) [66]. However, the transport of oskar mRNA does not fit this paradigm. While null mutants in khc result in oskar mRNA delocalization around the oocyte cortex [91,92], null mutants in klc do not have a significant effect on oskar mRNA localization [92]. Thus, oskar mRNA is transported to the oocyte posterior by a Kinesin complex that lacks the canonical light chain adaptor. Consistent with this picture, another study found that the number of oskar mRNP particles displaying fast, directed movement was reduced by five-fold in khc null oocytes [93]. However, a significant number of fast-moving oskar mRNP particles could still be detected in khc null mutant [93]. These results suggest that another, as yet unidentified, motor participates in oskar mRNP transport.
Collectively, these findings raise two very important questions. The first is “How does Khc recognize oskar mRNA as a target for localization?”. One possibility is that an unknown protein links Khc to oskar mRNA. If so, identifying this factor will be a critical next step. The expectation would be that this protein co-localizes with Khc at the posterior of the oocyte, and that mutations in this gene resemble khc null mutants. Another possibility is that a known component of the oskar mRNP directly binds to Khc and serves as the bridge between the motor and the transcript. This scenario does not seem likely because mutants in known components of the oskar mRNP result in mRNA that is either diffusely distributed throughout the oocyte, or in mRNA that is trapped at the anterior of the oocyte. This is in contrast to the khc null phenotype, in which oskar mRNA is found around the oocyte cortex [91,92]. A final possibility is that Khc directly binds to oskar mRNA. At present there are no data to support a direct interaction between Khc and oskar mRNA. Thus, although there is ample evidence implicating Khc in oskar mRNA transport, the mechanism by which this motor binds oskar mRNA is still an open question. The second intriguing question suggested by the live imaging of oskar mRNPs is whether a motor other than Khc also participates in oskar transport.
In addition to transporting oskar mRNA to the posterior pole, spatial restriction of Oskar protein is achieved by numerous mechanisms of translational repression [80,94]. Only once the mRNA is delivered to the posterior is the repression relieved. Additionally, specific mechanisms operate at the posterior of the oocyte to activate translation of oskar [80,94]. A detailed discussion of this topic is beyond the scope of this review. However, some recent findings have shed light on the mechanism by which the translation of oskar mRNA is regulated.
The first factor identified with a role in translational repression of oskar mRNA was Bruno [95]. Bruno was shown to bind to specific BRE (Bruno response element) sequences in the 3’UTR of oskar mRNA [95]. Initially, these elements were thought to function solely in translational repression. However, a recent report from the MacDonald lab demonstrated that one of these BREs also functions in activating the translation of oskar mRNA [96]. The precise mechanism by which Bruno regulates translation is not known. However, Bruno has been shown to associate with the protein Cup [97]. Cup in turn forms a complex with the transition initiation factor eIF4E and prevents it from binding eIF4G [97,98], an interaction which is required for translation initiation. Thus, Bruno and Cup may function together to prevent translation initiation of oskar mRNA. Interestingly, Bruno also appears to package oskar mRNPs into large aggregates called silencing particles, rendering them inaccessible to the translation machinery [99]. Finally, the Drosophila ortholog of hnRNP I/PTB has also been shown to bind oskar mRNA and to package the message into higher-order particles that are translationally silent [100].
RNA localization in neurons
Mature neurons are highly polarized and generally consist of a cell body, a single long axon and several shorter branching dendrites. In developing neurons, the axonal growth cone receives extracellular signals and directs the axon to the proper destination. For a neuron to locate its final destination and properly function, distinct proteins need to be sorted to these various regions of the cell. RNA localization is one mechanism by which proteins are sorted to dendrites, axons and axonal growth cones [101]. Specifically, beta-actin mRNA localizes to the growth cone where its localized translation is required for axon turning in response to guidance cues [102,103].
Beta-actin mRNA contains a 54 nt cis-acting localization element in the 3’UTR, and the cis-acting localization element functions to target beta-actin mRNA to dendrites and axonal growth cones [104-107]. The RNA-binding protein Zbp1 binds the cis-acting localization element and, through an unknown mechanism, helps target beta-actin mRNA to the sites of localization [108-110]. Zbp1 co-transcriptionally identifies beta-actin mRNA in the nucleus of fibroblasts [111], but it is not yet known whether this also happens in neurons. Binding of Zbp1 with beta-actin mRNA in the nucleus is facilitated by another RNA-binding protein, Zbp2 [111]. It has been hypothesized that Zpb2 initially identifies beta-actin mRNA in the nucleus and transfers it to Zbp1 [111]. Subsequently, the Zbp1/beta-actin mRNA complex is exported from the nucleus to the cytoplasm. In neurons, the working hypothesis is that cytoplasmic beta-actin mRNA is interfaced with a molecular motor that directly transports the mRNA on the cytoskeleton to the site of localization. This hypothesis is supported by a number of key observations. First, RNA granules enriched from rat embryonic brains contain numerous proteins and RNAs, including Zbp1, the molecular motor dynein and beta-actin mRNA [112]. Second, in A549 lung carcinoma cells and human dermal fibroblasts, beta-actin mRNA is transported by molecular motors on microtubules [113]. Third, the RNA-binding protein FMRP participates in the transport of MAP1b and CaMKIIα mRNAs to dendrites by associating these RNAs with Kinesin [114]. Consequently, based on these observations, we anticipate that a molecular motor will be identified that functions to directly transport beta-actin mRNA to the site of localization.
Besides participating in the targeting of beta-actin mRNA to the sites of localization, Zbp1 represses its translation [115]. The presence of Zbp1 prevents the formation of the 80S ribosomal initiation complex by an unknown mechanism [115]. Translational repression by Zbp1 is relieved through Src-dependent phosphorylation of Tyr396, which negatively regulates Zbp1 RNA-binding activity [115]. The signalling pathway that leads to the localized synthesis of beta-actin in axonal growth cones has been further elucidated. Axonal growth cones turn in response to brain-derived neurotrophic factor (BDNF). Phosphorylation of Zbp1 by Src family kinases is stimulated by BDNF, and when phosphorylation of Zbp1 is prevented in the presence of BDNF, axonal growth cones are unable to turn, providing additional insight into the mechanism by which local translation of beta-actin contributes to the turning of axonal growth cones [116].
Conclusion
RNA localization was once considered a mechanism employed by eukaryotic cells for the sorting of only a few select proteins. However, it has become increasingly evident that RNA localization is a common mechanism to post-transcriptionally regulate gene expression. A genomic screen of 3370 genes in Drosophila revealed that 71% of the analysed mRNAs are localized to specific regions of the cytoplasm [71]. In this review, we have provided a general overview of the transit of a localization substrate from the site of transcription to the site of localization. We have focused on the cis- and trans-acting factors that have a role in the transport of ASH1, Vg1, oskar and beta-actin mRNAs. We anticipate the discovery of more details concerning how nuclear factors influence RNA localization in the cytoplasm. In addition to transport of the cargo to the site of localization, it is widely hypothesized that, once the RNA reaches the site of localization, it is anchored at that position. Compared with our knowledge of transport, there is very little information concerning the mechanistic details of anchoring, so we look forward to insights into these mechanisms. Besides mechanistic details related to mRNA localization, we expect future developments related to the physiological significance of RNA localization. The genetic advantages in studying lower eukaryotes have provided some understanding of the physiological significance of RNA localization. However, in higher eukaryotic organisms, the physiological significance for RNA localization has been harder to determine, since investigations have been limited to cell culture systems. However, a number of mouse strains have recently been developed to investigate this question [117-119]. Consequently, we can expect advances in understanding the role of RNA localization in human development and disease.
Acknowledgments
Work in the Long laboratory is supported by National Science Foundation Grant 0918446.
Abbreviations
- BDNF
brain-derived neurotrophic factor
- BRE
Bruno response element
- EJC
exon junction complex
- ER
endoplasmic reticulum
- hnRNP
heterogeneous nuclear ribonucleoprotein
- mRNP
messenger ribonucleoprotein
- ORF
open reading frame
- VM1
Vg1 motif 1
- VTE
Vg1 translational element
- 3’UTR
3’untransated region
Competing interests
The authors declare that they have no competing interests.
The electronic version of this article is the complete one and can be found at: http://f1000.com/reports/b/4/16
References
- 1.Kraut-Cohen J, Gerst JE. Addressing mRNAs to the ER: cis sequences act up! Trends in biochemical sciences. 2010;35:459–69. doi: 10.1016/j.tibs.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Margeot A, Garcia M, Wang W, Tetaud E, di Rago JP, Jacq C. Why are many mRNAs translated to the vicinity of mitochondria: a role in protein complex assembly? Gene. 2005;354:64–71. doi: 10.1016/j.gene.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 3.Palacios IM. How does an mRNA find its way? Intracellular localisation of transcripts. Semin Cell Dev Biol. 2007;18:163–70. doi: 10.1016/j.semcdb.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.St Johnston D. Moving messages: the intracellular localization of mRNAs. Nature reviews Molecular cell biology. 2005;6:363–75. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- 5.Ding D, Parkhurst SM, Halsell SR, Lipshitz HD. Dynamic Hsp83 RNA localization during Drosophila oogenesis and embryogenesis. Molecular and cellular biology. 1993;13:3773–81. doi: 10.1128/mcb.13.6.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Roy Long 17 Jul 2012
- 6.Chang P, Torres J, Lewis RA, Mowry KL, Houliston E, King ML. Localization of RNAs to the mitochondrial cloud in Xenopus oocytes through entrapment and association with endoplasmic reticulum. Molecular biology of the cell. 2004;15:4669–81. doi: 10.1091/mbc.E04-03-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Roy Long 17 Jul 2012
- 7.Andoh T, Oshiro Y, Hayashi S, Takeo H, Tani T. Visual screening for localized RNAs in yeast revealed novel RNAs at the bud-tip. Biochem Biophys Res Commun. 2006;351:999–1004. doi: 10.1016/j.bbrc.2006.10.139. [DOI] [PubMed] [Google Scholar]
- 8.Aronov S, Gelin-Licht R, Zipor G, Haim L, Safran E, Gerst JE. mRNAs encoding polarity and exocytosis factors are cotransported with the cortical endoplasmic reticulum to the incipient bud in Saccharomyces cerevisiae. Molecular and cellular biology. 2007;27:3441–55. doi: 10.1128/MCB.01643-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shepard KA, Gerber AP, Jambhekar A, Takizawa PA, Brown PO, Herschlag D, DeRisi JL, Vale RD. Widespread cytoplasmic mRNA transport in yeast: identification of 22 bud-localized transcripts using DNA microarray analysis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:11429–34. doi: 10.1073/pnas.2033246100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takizawa PA, DeRisi JL, Wilhelm JE, Vale RD. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 2000;290:341–4. doi: 10.1126/science.290.5490.341. [DOI] [PubMed] [Google Scholar]
- 11.Oeffinger M, Wei KE, Rogers R, DeGrasse JA, Chait BT, Aitchison JD, Rout MP. Comprehensive analysis of diverse ribonucleoprotein complexes. Nature methods. 2007;4:951–6. doi: 10.1038/nmeth1101. [DOI] [PubMed] [Google Scholar]
- 12.Chartrand P, Meng XH, Huttelmaier S, Donato D, Singer RH. Asymmetric sorting of ash1p in yeast results from inhibition of translation by localization elements in the mRNA. Mol Cell. 2002;10:1319–30. doi: 10.1016/S1097-2765(02)00694-9. [DOI] [PubMed] [Google Scholar]
- 13.Long RM, Singer RH, Meng X, Gonzalez I, Nasmyth K, Jansen RP. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 1997;277:383–7. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- 14.Takizawa PA, Sil A, Swedlow JR, Herskowitz I, Vale RD. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature. 1997;389:90–3. doi: 10.1038/38015. [DOI] [PubMed] [Google Scholar]
- 15.Bobola N, Jansen RP, Shin TH, Nasmyth K. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell. 1996;84:699–709. doi: 10.1016/S0092-8674(00)81048-X. [DOI] [PubMed] [Google Scholar]; F1000 Factor 8Evaluated by Roy Long 17 Jul 2012
- 16.Jansen RP, Dowzer C, Michaelis C, Galova M, Nasmyth K. Mother cell-specific HO expression in budding yeast depends on the unconventional myosin myo4p and other cytoplasmic proteins. Cell. 1996;84:687–97. doi: 10.1016/S0092-8674(00)81047-8. [DOI] [PubMed] [Google Scholar]
- 17.Maxon ME, Herskowitz I. Ash1p is a site-specific DNA-binding protein that actively represses transcription. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:1495–500. doi: 10.1073/pnas.98.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sil A, Herskowitz I. Identification of asymmetrically localized determinant, Ash1p, required for lineage-specific transcription of the yeast HO gene. Cell. 1996;84:711–22. doi: 10.1016/S0092-8674(00)81049-1. [DOI] [PubMed] [Google Scholar]; F1000 Factor 8Evaluated by Roy Long 17 Jul 2012
- 19.Di Talia S, Wang H, Skotheim JM, Rosebrock AP, Futcher B, Cross FR. Daughter-specific transcription factors regulate cell size control in budding yeast. PLoS biology. 2009;7:e1000221. doi: 10.1371/journal.pbio.1000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chartrand P, Meng XH, Singer RH, Long RM. Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Current biology: CB. 1999;9:333–6. doi: 10.1016/S0960-9822(99)80144-4. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez I, Buonomo SB, Nasmyth K, von Ahsen U. ASH1 mRNA localization in yeast involves multiple secondary structural elements and Ash1 protein translation. Current biology: CB. 1999;9:337–40. doi: 10.1016/s0960-9822(99)80145-6. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Roy Long 17 Jul 2012
- 22.Long RM, Gu W, Lorimer E, Singer RH, Chartrand P. She2p is a novel RNA-binding protein that recruits the Myo4p-She3p complex to ASH1 mRNA. The EMBO journal. 2000;19:6592–601. doi: 10.1093/emboj/19.23.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bohl F, Kruse C, Frank A, Ferring D, Jansen RP. She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. The EMBO journal. 2000;19:5514–24. doi: 10.1093/emboj/19.20.5514. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Roy Long 17 Jul 2012
- 24.Niessing D, Huttelmaier S, Zenklusen D, Singer RH, Burley SK. She2p is a novel RNA binding protein with a basic helical hairpin motif. Cell. 2004;119:491–502. doi: 10.1016/j.cell.2004.10.018. [DOI] [PubMed] [Google Scholar]; F1000 Factor 8Evaluated by Roy Long 17 Jul 2012
- 25.Du TG, Jellbauer S, Muller M, Schmid M, Niessing D, Jansen RP. Nuclear transit of the RNA-binding protein She2 is required for translational control of localized ASH1 mRNA. EMBO Rep. 2008;9:781–7. doi: 10.1038/embor.2008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Roy Long 17 Jul 2012
- 26.Shen Z, Paquin N, Forget A, Chartrand P. Nuclear Shuttling of She2p Couples ASH1 mRNA Localization to its Translational Repression by Recruiting Loc1p and Puf6p. Molecular biology of the cell. 2009;20:2265–75. doi: 10.1091/mbc.E08-11-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Roy Long 16 Mar 2009
- 27.Shen Z, St-Denis A, Chartrand P. Cotranscriptional recruitment of She2p by RNA pol II elongation factor Spt4-Spt5/DSIF promotes mRNA localization to the yeast bud. Genes & development. 2010;24:1914–26. doi: 10.1101/gad.1937510. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 8Evaluated by Roy Long 10 Sep 2010
- 28.Beach DL, Salmon ED, Bloom K. Localization and anchoring of mRNA in budding yeast. Current biology: CB. 1999;9:569–78. doi: 10.1016/S0960-9822(99)80260-7. [DOI] [PubMed] [Google Scholar]
- 29.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–45. doi: 10.1016/S1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 30.Munchow S, Sauter C, Jansen RP. Association of the class V myosin Myo4p with a localised messenger RNA in budding yeast depends on She proteins. J Cell Sci. 1999;112:1511–8. doi: 10.1242/jcs.112.10.1511. [DOI] [PubMed] [Google Scholar]
- 31.Takizawa PA, Vale RD. The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5273–8. doi: 10.1073/pnas.080585897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonsalvez GB, Little JL, Long RM. ASH1 mRNA anchoring requires reorganization of the Myo4p-She3p-She2p transport complex. The Journal of biological chemistry. 2004;279:46286–94. doi: 10.1074/jbc.M406086200. [DOI] [PubMed] [Google Scholar]
- 33.Landers SM, Gallas MR, Little J, Long RM. She3p possesses a novel activity required for ASH1 mRNA localization in Saccharomyces cerevisiae. Eukaryot Cell. 2009;8:1072–83. doi: 10.1128/EC.00084-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muller M, Heym RG, Mayer A, Kramer K, Schmid M, Cramer P, Urlaub H, Jansen RP, Niessing D. A cytoplasmic complex mediates specific mRNA recognition and localization in yeast. PLoS biology. 2011;9:e1000611. doi: 10.1371/journal.pbio.1000611. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 9Evaluated by Michael Kiebler and Michael Doyle 01 Aug 2011, Roy Long 17 Jul 2012
- 35.Haarer BK, Petzold A, Lillie SH, Brown SS. Identification of MYO4, a second class V myosin gene in yeast. J Cell Sci. 1994;107:1055–64. doi: 10.1242/jcs.107.4.1055. [DOI] [PubMed] [Google Scholar]
- 36.Reck-Peterson SL, Tyska MJ, Novick PJ, Mooseker MS. The yeast class V myosins, Myo2p and Myo4p, are nonprocessive actin-based motors. The Journal of cell biology. 2001;153:1121–6. doi: 10.1083/jcb.153.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chung S, Takizawa PA. Multiple Myo4 motors enhance ASH1 mRNA transport in Saccharomyces cerevisiae. The Journal of cell biology. 2010;189:755–67. doi: 10.1083/jcb.200912011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krementsova EB, Hodges AR, Bookwalter CS, Sladewski TE, Travaglia M, Sweeney HL, Trybus KM. Two single-headed myosin V motors bound to a tetrameric adapter protein form a processive complex. The Journal of cell biology. 2011;195:631–41. doi: 10.1083/jcb.201106146. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 8Evaluated by Richard Rachubinski and Fred David Mast 23 Nov 2011
- 39.Muller M, Richter K, Heuck A, Kremmer E, Buchner J, Jansen RP, Niessing D. Formation of She2p tetramers is required for mRNA binding, mRNP assembly, and localization. RNA. 2009;15:2002–12. doi: 10.1261/rna.1753309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu W, Deng Y, Zenklusen D, Singer RH. A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes & development. 2004;18:1452–65. doi: 10.1101/gad.1189004. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 7Evaluated by Roy Long 28 Jun 2004, Elizabeth Gavis 02 Aug 2004
- 41.Paquin N, Menade M, Poirier G, Donato D, Drouet E, Chartrand P. Local activation of yeast ASH1 mRNA translation through phosphorylation of Khd1p by the casein kinase Yck1p. Mol Cell. 2007;26:795–809. doi: 10.1016/j.molcel.2007.05.016. [DOI] [PubMed] [Google Scholar]; F1000 Factor 7Evaluated by Linda Huang 29 Jun 2007, Roy Long 09 Jul 2007
- 42.Deng Y, Singer RH, Gu W. Translation of ASH1 mRNA is repressed by Puf6p-Fun12p/eIF5B interaction and released by CK2 phosphorylation. Genes & development. 2008;22:1037–50. doi: 10.1101/gad.1611308. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Roy Long 21 Apr 2008
- 43.Weeks DL, Melton DA. A maternal mRNA localized to the vegetal hemisphere in Xenopus eggs codes for a growth factor related to TGF-beta. Cell. 1987;51:861–7. doi: 10.1016/0092-8674(87)90109-7. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Roy Long 17 Jul 2012
- 44.Dale L, Matthews G, Colman A. Secretion and mesoderm-inducing activity of the TGF-beta-related domain of Xenopus Vg1. The EMBO journal. 1993;12:4471–80. doi: 10.1002/j.1460-2075.1993.tb06136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomsen GH, Melton DA. Processed Vg1 protein is an axial mesoderm inducer in Xenopus. Cell. 1993;74:433–41. doi: 10.1016/0092-8674(93)80045-G. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Roy Long 17 Jul 2012
- 46.Kessler DS, Melton DA. Induction of dorsal mesoderm by soluble, mature Vg1 protein. Development. 1995;121:2155–64. doi: 10.1242/dev.121.7.2155. [DOI] [PubMed] [Google Scholar]
- 47.Joseph EM, Melton DA. Mutant Vg1 ligands disrupt endoderm and mesoderm formation in Xenopus embryos. Development. 1998;125:2677–85. doi: 10.1242/dev.125.14.2677. [DOI] [PubMed] [Google Scholar]
- 48.Kress TL, Yoon YJ, Mowry KL. Nuclear RNP complex assembly initiates cytoplasmic RNA localization. J Cell Biol. 2004;165:203–11. doi: 10.1083/jcb.200309145. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 9Evaluated by Roy Long 04 May 2004, Michael Kiebler 14 Oct 2004
- 49.Cote CA, Gautreau D, Denegre JM, Kress TL, Terry NA, Mowry KL. A Xenopus protein related to hnRNP I has a role in cytoplasmic RNA localization. Mol Cell. 1999;4:431–7. doi: 10.1016/S1097-2765(00)80345-7. [DOI] [PubMed] [Google Scholar]
- 50.Lewis RA, Kress TL, Cote CA, Gautreau D, Rokop ME, Mowry KL. Conserved and clustered RNA recognition sequences are a critical feature of signals directing RNA localization in Xenopus oocytes. Mech Dev. 2004;121:101–9. doi: 10.1016/j.mod.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 51.Deshler JO, Highett MI, Abramson T, Schnapp BJ. A highly conserved RNA-binding protein for cytoplasmic mRNA localization in vertebrates. Current biology: CB. 1998;8:489–96. doi: 10.1016/S0960-9822(98)70200-3. [DOI] [PubMed] [Google Scholar]
- 52.Deshler JO, Highett MI, Schnapp BJ. Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science. 1997;276:1128–31. doi: 10.1126/science.276.5315.1128. [DOI] [PubMed] [Google Scholar]
- 53.Havin L, Git A, Elisha Z, Oberman F, Yaniv K, Schwartz SP, Standart N, Yisraeli JK. RNA-binding protein conserved in both microtubule- and microfilament-based RNA localization. Genes & development. 1998;12:1593–8. doi: 10.1101/gad.12.11.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Betley JN, Frith MC, Graber JH, Choo S, Deshler JO. A ubiquitous and conserved signal for RNA localization in chordates. Curr Biol. 2002;12:1756–61. doi: 10.1016/S0960-9822(02)01220-4. [DOI] [PubMed] [Google Scholar]
- 55.Bubunenko M, Kress TL, Vempati UD, Mowry KL, King ML. A consensus RNA signal that directs germ layer determinants to the vegetal cortex of Xenopus oocytes. Dev Biol. 2002;248:82–92. doi: 10.1006/dbio.2002.0719. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Robert H. Singer 04 Sep 2002
- 56.Kwon S, Abramson T, Munro TP, John CM, Kohrmann M, Schnapp BJ. UUCAC- and vera-dependent localization of VegT RNA in Xenopus oocytes. Curr Biol. 2002;12:558–64. doi: 10.1016/S0960-9822(02)00740-6. [DOI] [PubMed] [Google Scholar]
- 57.Mallardo M, Deitinghoff A, Muller J, Goetze B, Macchi P, Peters C, Kiebler MA. Isolation and characterization of Staufen-containing ribonucleoprotein particles from rat brain. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:2100–5. doi: 10.1073/pnas.0334355100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tang SJ, Meulemans D, Vazquez L, Colaco N, Schuman E. A role for a rat homolog of staufen in the transport of RNA to neuronal dendrites. Neuron. 2001;32:463–75. doi: 10.1016/S0896-6273(01)00493-7. [DOI] [PubMed] [Google Scholar]
- 59.St Johnston D, Beuchle D, Nusslein-Volhard C. Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell. 1991;66:51–63. doi: 10.1016/0092-8674(91)90138-O. [DOI] [PubMed] [Google Scholar]
- 60.Barbee SA, Estes PS, Cziko AM, Hillebrand J, Luedeman RA, Coller JM, Johnson N, Howlett IC, Geng C, Ueda R, Brand AH, Newbury SF, Wilhelm JE, Levine RB, Nakamura A, Parker R, Ramaswami M. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 2006;52:997–1009. doi: 10.1016/j.neuron.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yisraeli JK, Sokol S, Melton DA. A two-step model for the localization of maternal mRNA in Xenopus oocytes: involvement of microtubules and microfilaments in the translocation and anchoring of Vg1 mRNA. Development. 1990;108:289–98. doi: 10.1242/dev.108.2.289. [DOI] [PubMed] [Google Scholar]
- 62.Betley JN, Heinrich B, Vernos I, Sardet C, Prodon F, Deshler JO. Kinesin II mediates Vg1 mRNA transport in Xenopus oocytes. Curr Biol. 2004;14:219–24. doi: 10.1016/j.cub.2004.01.028. [DOI] [PubMed] [Google Scholar]; F1000 Factor 10Evaluated by Ilan Davis 18 Mar 2004, Roy Long 17 Jul 2012
- 63.Messitt TJ, Gagnon JA, Kreiling JA, Pratt CA, Yoon YJ, Mowry KL. Multiple kinesin motors coordinate cytoplasmic RNA transport on a subpopulation of microtubules in Xenopus oocytes. Dev Cell. 2008;15:426–36. doi: 10.1016/j.devcel.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 8Evaluated by Roy Long 17 Jul 2012
- 64.Cole DG, Chinn SW, Wedaman KP, Hall K, Vuong T, Scholey JM. Novel heterotrimeric kinesin-related protein purified from sea urchin eggs. Nature. 1993;366:268–70. doi: 10.1038/366268a0. [DOI] [PubMed] [Google Scholar]
- 65.Yoon YJ, Mowry KL. Xenopus Staufen is a component of a ribonucleoprotein complex containing Vg1 RNA and kinesin. Development. 2004;131:3035–45. doi: 10.1242/dev.01170. [DOI] [PubMed] [Google Scholar]
- 66.Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nature reviews Molecular cell biology. 2009;10:682–96. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- 67.Pfeiffer DC, Gard DL. Microtubules in Xenopus oocytes are oriented with their minus-ends towards the cortex. Cell motility and the cytoskeleton. 1999;44:34–43. doi: 10.1002/(SICI)1097-0169(199909)44:1<34::AID-CM3>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 68.Otero LJ, Devaux A, Standart N. A 250-nucleotide UA-rich element in the 3' untranslated region of Xenopus laevis Vg1 mRNA represses translation both in vivo and in vitro. RNA. 2001;7:1753–67. [PMC free article] [PubMed] [Google Scholar]
- 69.Wilhelm JE, Vale RD, Hegde RS. Coordinate control of translation and localization of Vg1 mRNA in Xenopus oocytes. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:13132–7. doi: 10.1073/pnas.97.24.13132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Colegrove-Otero LJ, Devaux A, Standart N. The Xenopus ELAV protein ElrB represses Vg1 mRNA translation during oogenesis. Molecular and cellular biology. 2005;25:9028–39. doi: 10.1128/MCB.25.20.9028-9039.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Roy Long 17 Jul 2012
- 71.Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–87. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]; F1000 Factor 22Evaluated by Talila Volk 18 Oct 2007, Ken Irvine 22 Oct 2007, Michael Sendtner 30 Oct 2007, Elizabeth Gavis 31 Oct 2007, Roy Long 02 Nov 2007, Paul Lasko 12 Dec 2007, Douglas Turner 10 Mar 2008
- 72.Berleth T, Burri M, Thoma G, Bopp D, Richstein S, Frigerio G, Noll M, Nusslein-Volhard C. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. The EMBO journal. 1988;7:1749–56. doi: 10.1002/j.1460-2075.1988.tb03004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ephrussi A, Dickinson LK, Lehmann R. Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell. 1991;66:37–50. doi: 10.1016/0092-8674(91)90137-N. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Roy Long 17 Jul 2012
- 74.Kim-Ha J, Smith JL, Macdonald PM. oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell. 1991;66:23–35. doi: 10.1016/0092-8674(91)90136-M. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Roy Long 17 Jul 2012
- 75.Neuman-Silberberg FS, Schupbach T. The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGF alpha-like protein. Cell. 1993;75:165–74. [PubMed] [Google Scholar]
- 76.Davis I, Ish-Horowicz D. Apical localization of pair-rule transcripts requires 3' sequences and limits protein diffusion in the Drosophila blastoderm embryo. Cell. 1991;67:927–40. doi: 10.1016/0092-8674(91)90366-7. [DOI] [PubMed] [Google Scholar]
- 77.Simmonds AJ, dosSantos G, Livne-Bar I, Krause HM. Apical localization of wingless transcripts is required for wingless signaling. Cell. 2001;105:197–207. doi: 10.1016/S0092-8674(01)00311-7. [DOI] [PubMed] [Google Scholar]
- 78.Bullock SL, Zicha D, Ish-Horowicz D. The Drosophila hairy RNA localization signal modulates the kinetics of cytoplasmic mRNA transport. EMBO J. 2003;22:2484–94. doi: 10.1093/emboj/cdg230. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Robert H. Singer 04 Jul 2003
- 79.dos Santos G, Simmonds AJ, Krause HM. A stem-loop structure in the wingless transcript defines a consensus motif for apical RNA transport. Development. 2008;135:133–43. doi: 10.1242/dev.014068. [DOI] [PubMed] [Google Scholar]
- 80.Wilhelm JE, Smibert CA. Mechanisms of translational regulation in Drosophila. Biology of the cell / under the auspices of the European Cell Biology Organization. 2005;97:235–52. doi: 10.1042/BC20040097. [DOI] [PubMed] [Google Scholar]
- 81.Smith JL, Wilson JE, Macdonald PM. Overexpression of oskar directs ectopic activation of nanos and presumptive pole cell formation in Drosophila embryos. Cell. 1992;70:849–59. doi: 10.1016/0092-8674(92)90318-7. [DOI] [PubMed] [Google Scholar]
- 82.Ephrussi A, Lehmann R. Induction of germ cell formation by oskar. Nature. 1992;358:387–92. doi: 10.1038/358387a0. [DOI] [PubMed] [Google Scholar]
- 83.Hachet O, Ephrussi A. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature. 2004;428:959–63. doi: 10.1038/nature02521. [DOI] [PubMed] [Google Scholar]; F1000 Factor 13Evaluated by Roy Long 11 May 2004, Lynne Maquat 13 May 2004, Ilan Davis 18 May 2004, Gabriele Varani 20 May 2004, Reinhard Luehrmann 03 Jun 2004
- 84.Hachet O, Ephrussi A. Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Current biology: CB. 2001;11:1666–74. doi: 10.1016/S0960-9822(01)00508-5. [DOI] [PubMed] [Google Scholar]; F1000 Factor 10Evaluated by David Ish-Horowicz 24 Jan 2002, Ilan Davis 27 Feb 2002, Elsebet Lund 11 Mar 2002
- 85.Micklem DR, Dasgupta R, Elliott H, Gergely F, Davidson C, Brand A, Gonzalez-Reyes A, St Johnston D. The mago nashi gene is required for the polarisation of the oocyte and the formation of perpendicular axes in Drosophila. Curr Biol. 1997;7:468–78. doi: 10.1016/S0960-9822(06)00218-1. [DOI] [PubMed] [Google Scholar]
- 86.Mohr SE, Dillon ST, Boswell RE. The RNA-binding protein Tsunagi interacts with Mago Nashi to establish polarity and localize oskar mRNA during Drosophila oogenesis. Genes Dev. 2001;15:2886–99. doi: 10.1101/gad.927001. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by David Ish-Horowicz 24 Jan 2002
- 87.Newmark PA, Mohr SE, Gong L, Boswell RE. mago nashi mediates the posterior follicle cell-to-oocyte signal to organize axis formation in Drosophila. Development. 1997;124:3197–207. doi: 10.1242/dev.124.16.3197. [DOI] [PubMed] [Google Scholar]
- 88.Palacios IM, Gatfield D, St Johnston D, Izaurralde E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004;427:753–7. doi: 10.1038/nature02351. [DOI] [PubMed] [Google Scholar]; F1000 Factor 12Lynne Maquat 02 Mar 2004, Ilan Davis 05 Mar 2004, Miles Wilkinson 15 Apr 2004, Evaluated by Gabriele Varani 11 Aug 2004
- 89.van Eeden FJ, Palacios IM, Petronczki M, Weston MJ, St Johnston D. Barentsz is essential for the posterior localization of oskar mRNA and colocalizes with it to the posterior pole. J Cell Biol. 2001;154:511–23. doi: 10.1083/jcb.200105056. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 8Evaluated by Ilan Davis 27 Feb 2002
- 90.Kim-Ha J, Webster PJ, Smith JL, Macdonald PM. Multiple RNA regulatory elements mediate distinct steps in localization of oskar mRNA. Development. 1993;119:169–78. doi: 10.1242/dev.119.1.169. [DOI] [PubMed] [Google Scholar]
- 91.Brendza RP, Serbus LR, Duffy JB, Saxton WM. A function for kinesin I in the posterior transport of oskar mRNA and Staufen protein. Science. 2000;289:2120–2. doi: 10.1126/science.289.5487.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 8Evaluated by Roy Long 17 Jul 2012
- 92.Palacios IM, St Johnston D. Kinesin light chain-independent function of the Kinesin heavy chain in cytoplasmic streaming and posterior localisation in the Drosophila oocyte. Development. 2002;129:5473–85. doi: 10.1242/dev.00119. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Roy Long 17 Jul 2012
- 93.Zimyanin VL, Belaya K, Pecreaux J, Gilchrist MJ, Clark A, Davis I, St Johnston D. In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell. 2008;134:843–53. doi: 10.1016/j.cell.2008.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 16Michael Kiebler 15 Sep 2008, Anna Akhmanova 24 Sep 2008, Gabriele Varani 28 Evaluated by Nov 2008, Roy Long 17 Jul 2012
- 94.Kugler JM, Lasko P. Localization, anchoring and translational control of oskar, gurken, bicoid and nanos mRNA during Drosophila oogenesis. Fly. 2009;3:15–28. doi: 10.4161/fly.3.1.7751. [DOI] [PubMed] [Google Scholar]
- 95.Kim-Ha J, Kerr K, Macdonald PM. Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell. 1995;81:403–12. doi: 10.1016/0092-8674(95)90393-3. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Roy Long 17 Jul 2012
- 96.Reveal B, Yan N, Snee MJ, Pai CI, Gim Y, Macdonald PM. BREs mediate both repression and activation of oskar mRNA translation and act in trans. Developmental cell. 2010;18:496–502. doi: 10.1016/j.devcel.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 7Evaluated by Phillip Newmark 23 Mar 2010, Paul Lasko 19 Jul 2010
- 97.Nakamura A, Sato K, Hanyu-Nakamura K. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Developmental cell. 2004;6:69–78. doi: 10.1016/S1534-5807(03)00400-3. [DOI] [PubMed] [Google Scholar]; F1000 Factor 8Evaluated by Nancy Standart 09 Feb 2004, Talila Volk 23 Feb 2004, Claudia Bagni 01 Dec 2004
- 98.Wilhelm JE, Hilton M, Amos Q, Henzel WJ. Cup is an eIF4E binding protein required for both the translational repression of oskar and the recruitment of Barentsz. The Journal of cell biology. 2003;163:1197–204. doi: 10.1083/jcb.200309088. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Nancy Standart 09 Feb 2004
- 99.Chekulaeva M, Hentze MW, Ephrussi A. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell. 2006;124:521–33. doi: 10.1016/j.cell.2006.01.031. [DOI] [PubMed] [Google Scholar]; F1000 Factor 7Evaluated by Jeff Coller 16 Feb 2006, Kimberly Mowry 09 Mar 2006
- 100.Besse F, Lopez de Quinto S, Marchand V, Trucco A, Ephrussi A. Drosophila PTB promotes formation of high-order RNP particles and represses oskar translation. Genes & development. 2009;23:195–207. doi: 10.1101/gad.505709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Doyle M, Kiebler MA. Mechanisms of dendritic mRNA transport and its role in synaptic tagging. The EMBO journal. 2011;30:3540–52. doi: 10.1038/emboj.2011.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Leung KM, van Horck FP, Lin AC, Allison R, Standart N, Holt CE. Asymmetrical beta-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nature neuroscience. 2006;9:1247–56. doi: 10.1038/nn1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yao J, Sasaki Y, Wen Z, Bassell GJ, Zheng JQ. An essential role for beta-actin mRNA localization and translation in Ca2+-dependent growth cone guidance. Nature neuroscience. 2006;9:1265–73. doi: 10.1038/nn1773. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Claudia Bagni 20 Oct 2006
- 104.Kislauskis EH, Li Z, Singer RH, Taneja KL. Isoform-specific 3'-untranslated sequences sort alpha-cardiac and beta-cytoplasmic actin messenger RNAs to different cytoplasmic compartments. The Journal of cell biology. 1993;123:165–72. doi: 10.1083/jcb.123.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. The Journal of cell biology. 1994;127:441–51. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tiruchinapalli DM, Oleynikov Y, Kelic S, Shenoy SM, Hartley A, Stanton PK, Singer RH, Bassell GJ. Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and beta-actin mRNA in dendrites and spines of hippocampal neurons. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2003;23:3251–61. doi: 10.1523/JNEUROSCI.23-08-03251.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Roy Long 17 Jul 2012
- 107.Zheng JQ, Kelly TK, Chang B, Ryazantsev S, Rajasekaran AK, Martin KC, Twiss JL. A functional role for intra-axonal protein synthesis during axonal regeneration from adult sensory neurons. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2001;21:9291–303. doi: 10.1523/JNEUROSCI.21-23-09291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 10Evaluated by Clifford J. Woolf 10 Dec 2001, Christine Holt 20 Dec 2001
- 108.Farina KL, Huttelmaier S, Musunuru K, Darnell R, Singer RH. Two ZBP1 KH domains facilitate beta-actin mRNA localization, granule formation, and cytoskeletal attachment. The Journal of cell biology. 2003;160:77–87. doi: 10.1083/jcb.200206003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. Characterization of a beta-actin mRNA zipcode-binding protein. Molecular and cellular biology. 1997;17:2158–65. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Liebelt DA, Dictenberg JB, Singer RH, Bassell GJ. Neurotrophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron. 2001;31:261–75. doi: 10.1016/S0896-6273(01)00357-9. [DOI] [PubMed] [Google Scholar]
- 111.Pan F, Huttelmaier S, Singer RH, Gu W. ZBP2 facilitates binding of ZBP1 to beta-actin mRNA during transcription. Molecular and cellular biology. 2007;27:8340–51. doi: 10.1128/MCB.00972-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Elvira G, Wasiak S, Blandford V, Tong XK, Serrano A, Fan X, del Rayo Sanchez-Carbente M, Servant F, Bell AW, Boismenu D, Lacaille JC, McPherson PS, DesGroseillers L, Sossin WS. Characterization of an RNA granule from developing brain. Molecular & cellular proteomics: MCP. 2006;5:635–51. doi: 10.1074/mcp.M500255-MCP200. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Roy Long 17 Jul 2012
- 113.Lifland AW, Zurla C, Yu J, Santangelo PJ. Dynamics of native beta-actin mRNA transport in the cytoplasm. Traffic. 2011;12:1000–11. doi: 10.1111/j.1600-0854.2011.01209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dictenberg JB, Swanger SA, Antar LN, Singer RH, Bassell GJ. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Developmental cell. 2008;14:926–39. doi: 10.1016/j.devcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Huttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–5. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]; F1000 Factor 11Evaluated by Michael Kiebler 24 Nov 2005, Roy Long 08 Dec 2005, Sui Huang 13 Mar 2006
- 116.Sasaki Y, Welshhans K, Wen Z, Yao J, Xu M, Goshima Y, Zheng JQ, Bassell GJ. Phosphorylation of zipcode binding protein 1 is required for brain-derived neurotrophic factor signaling of local beta-actin synthesis and growth cone turning. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2010;30:9349–58. doi: 10.1523/JNEUROSCI.0499-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Roy Long 17 Jul 2012
- 117.Donnelly CJ, Willis DE, Xu M, Tep C, Jiang C, Yoo S, Schanen NC, Kirn-Safran CB, van Minnen J, English A, Yoon SO, Bassell GJ, Twiss JL. Limited availability of ZBP1 restricts axonal mRNA localization and nerve regeneration capacity. The EMBO journal. 2011;30:4665–77. doi: 10.1038/emboj.2011.347. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Roy Long 17 Jul 2012
- 118.Welshhans K, Bassell GJ. Netrin-1-induced local beta-actin synthesis and growth cone guidance requires zipcode binding protein 1. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2011;31:9800–13. doi: 10.1523/JNEUROSCI.0166-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lionnet T, Czaplinski K, Darzacq X, Shav-Tal Y, Wells AL, Chao JA, Park HY, de Turris V, Lopez-Jones M, Singer RH. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nature methods. 2011;8:165–70. doi: 10.1038/nmeth.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Roy Long 17 Jul 2012