Abstract
Endometrial stromal sarcomas are rare malignancies, accounting for less than 10% of uterine sarcomas. The most characteristic chromosomal aberration of this tumor type is the translocation t(7;17)(p15-p21;q12-q21) leading to the fusion of two zinc finger genes, JAZF1 and SUZ12. Recently, the presence of the neoplastic JAZF1/SUZ12 fusion transcript was reported in normal cells of human endometrium. One of the positive samples for the JAZF1/SUZ12 transcript was the immortalized T HESCs cell line. This cell line was derived from the stromal cells obtained from an adult female with myomas and immortalized by transfection of a human telomerase gene. Since T HESCs has a normal karyotype and no fusion of the two genes occurs at the genomic level, the JAZF1/SUZ12 transcript was proposed to be generated by regulated trans-splicing between precursor RNAs for JAZF1 and SUZ12. However, no confirmatory reports currently exist. To determine whether the results could be reproduced, the T HESCs cell line was subjected to three different RT-PCR amplifications for the JAZF1/SUZ12 fusion transcript. RT-PCR assays did not amplify JUZF1/SUZ12 cDNA fragments in the T HESCs cell line, whereas the same assays easily generated JUZF1/SUZ12-amplified transcripts in an endometrial stromal cell sarcoma carrying the t(7;17) chromosomal aberration. Thus, the presence, if any, of a JUZF1/SUZ12 chimeric transcript in the immortalized normal T HESCs is not a constant, reproducible result.
Keywords: endometrial stromal sarcoma, cell line, non-neoplastic, T HESCs immortalized, fusion transcript, JAZF1/JJAZ1, trans-splicing
Introduction
Endometrial stromal sarcomas (ESS) are rare malignancies, accounting for less than 10% of uterine sarcomas (1). The most characteristic chromosomal aberration of this tumor type is the translocation t(7;17)(p15-p21;q12-q21) leading to the fusion of two zinc finger genes, JAZF1 (juxtaposed with another zinc finger) and SUZ12 (also known as JJAZ1) (2). Recently, Li et al (3) reported the presence of the neoplastic JAZF1/SUZ12 fusion transcript in normal cells of human endometrium. One of the samples found to be positive for the JAZF1/SUZ12 transcript was the immortalized T HESCs cell line. This cell line was derived from the stromal cells obtained from an adult female with myomas and immortalized by transfection of a human telomerase gene (4). Since this cell line has a normal karyotype and no fusion of the two genes occurs at the genomic level, it was proposed that the JAZF1/SUZ12 transcript in the T HESCs cells is generated from regulated trans-splicing between precursor RNAs for JAZF1 and SUZ12 (3). However, no confirmatory reports exist thus far. To determine whether the results obtained by Li et al (3) could be reproduced, the T HESCs cell line was subjected to various RT-PCR for the JAZF1/SUZ12 fusion transcript.
Materials and methods
Materials
The cell line T HESCs was an American Type Culture Collection (ATCC) culture (ATCC-CRL-4003; Lot No. 3857441) purchased from LGC Promochem (http://www.lgcpromochem-atcc.com/) and cultured according to ATCC recommendations. Thus, the cells were grown in DMEM/F12 medium with 3.1 g/l glucose, 1 mM sodium pyruvate and 1.2 g/l sodium bicarbonate, without phenol red (Invitrogen Cat no. 11039). The medium was supplemented with 1% ITS + Premix (BD Cat no. 354352) and 10% fetal bovine serum (Invitrogen Cat no. 10106-151). Prior to the molecular investigation, a cytogenetic analysis verified the previously published karyotype of 46, XX (3,4). An endometrial stromal sarcoma carrying the t(7;17)(p15;q12) was used as a positive control for all analyses.
Methods
Total RNA was extracted from the cell line using the RNeasy micro kit (Qiagen, Hilden, Germany) and from the tumor biopsy with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. cDNA was synthesized using 2.5 μg of total RNA in a 20-μl reaction mixture containing 50 mM Tris-HCl pH 8.3 (at 25°C), 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, 1 mM each dNTP, 48 units RNA inhibitor (RNA guard; GE Healthcare, Uppsala, Sweden), 25 μM random hexamers and 200 units M-MLV Reverse Transcriptase (Invitrogen). The reaction was carried out at 37°C for 60 min, heated for 5 min at 65°C and then maintained at 4°C until analysis. cDNA quality was checked using ABL1-specific primers (5).
Assays
For the amplification of the JAZF1/SUZ12 fusion transcript, the assays used were: i) the method described by Micci et al (6). Briefly, PCR amplification was performed using 1 μl of the cDNA as a template in 50-μl reaction volume containing 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 1.25 mM MgCl2, 0.2 mM of each dNTP, 1 unit Platinum Taq polymerase (Invitrogen) and 0.5 μM of each of the primers JAZF1-357F CCACAGCAGTGGAAGCCTTA and JJAZ1-843R CCGGGT TTTGTTTGATTGAGG. To increase the sensitivity of the assay, 1 μl of the first PCR was re-amplified using the inner primers JAZF1-390F CCCACCCATCACCCCCTCCT and JJAZ1-827R AGGTCAGGAATCAAAGGCACCTGC in 50-μl reaction volume with the same composition as above. Both the first and second PCR amplifications were run on a PCT-200 DNA Engine (MJ Research, Waltham, MA, USA) with the following cycling profile: an initial denaturation at 94°C for 5 min, followed by 30 cycles of 1 min at 94°C, 1 min at 60°C and 1 min at 72°C and a final extension for 10 min at 72°C.
ii) A one-step PCR using the primers described by Li et al (3). The 50-μl reaction had the same composition as the one mentioned above, except that the forward primer Fusion-286F AAGATTCAGCCGAAGCTCTCG and the reverse primer Fusion-541R TGTTTGTTCTGGAGTTTCGATGAGACA were used.
iii) The method described by Li et al (3). For the detection of the JAZF1/SUZ12 fusion transcript, PCR was performed with Platinum Taq DNA Polymerase High Fidelity (Invitrogen). The 50-μl reaction volume contained 1X High Fidelity PCR buffer, 0.2 mM of each dNTP, 2 mM MgSO4, 0.2 μM of each of the forward and reverse primers, 1 unit Platinum Taq DNA Polymerase High Fidelity and 1 μl of the cDNA. Four primer combinations were used: i) The human-JAZF1-284 primer AGAAGATTCAGCCGAAGCTC with Fusion-541R; ii) the Rhesus-JAZF1-284 primer AGAAGATTCAGCCGAAGCTG with Fusion-541R; iii) the human-JAZF1-286F primer AAGATTCAGCCGAAGCTCTCG with Fusion-541R; and iv) the Rhesus-JAZF1-286F AAGATTCAGCCGAAGCTGTCA with Fusion-541R. The above amplifications were performed using the touchdown protocol described by Li et al (3). The cycle profile was an initial denaturation step at 94°C for 30 sec followed by 2 cycles of 30 sec at 94°C, 30 sec at 66°C and 1 min at 68°C, 2 cycles of 30 sec at 94°C, 30 sec at 64°C and 1 min at 68°C, 4 cycles of 30 sec at 94°C, 30 sec at 62°C and 1 min at 68°C and 37 cycles 30 sec at 94°C, 30 sec at 60°C and 1 min at 68°C.
Amplification of the wild-type and SUZ12 transcripts
For the detection of the wild-type normal JAZF1 and SUZ12 transcripts, PCR was performed with Platinum Taq DNA Polymerase High Fidelity (Invitrogen). The 50-μl reaction volume contained 1X High Fidelity PCR buffer, 0.2 mM of each dNTP, 2 mM MgSO4, 0.2 μM of each of the forward and reverse primers, 1 unit Platinum Taq DNA Polymerase High Fidelity and 1 μl of the cDNA. The JAZF1 wild-type transcript was amplified using the primers JAZF1-12F GGCTCTCGATGTAGCACCATGACAG and JAZF1-776R GCTGGTGAGGATTTCTTGGCACAG. The SUZ12 transcript was amplified using the primers JJAZ1-329F CTGTGGAGGGGGTGGCAGTTACTC and JJAZ1-827R AGGTCAGGAATCAAAGGCACCTGC.
The cycle profile was an initial denaturation step at 94°C for 30 sec followed by 35 cycles of 30 sec at 94°C, 30 sec at 60°C and 1 min at 68°C and a final extension for 5 min at 68°C.
For sequence analyses, the RT-PCR-amplified fragments were run on 1.5% agarose gels, purified using the Qiagen gel extraction kit (Qiagen) and directly sequenced using the dideoxy procedure with an ABI Prism BigDye terminator v1.1 cycle sequencing kit (PE Applied Biosystems, Foster City, CA, USA) on the Applied Biosystems Model 3100-Avant DNA sequencing system. The BLAST software (http://www.ncbi.nlm.nih.gov/BLAST/) was used for the computer analysis of the sequence data. For comparison, the reference sequences with accession numbers NM_175061 (JAZF1) and NM_015355 (SUZ12) were used.
Results
RT-PCR analysis of the T HESCs cell line for the detection of JAZF1/SUZ12 chimeric transcript
RT-PCR with the JAZF1-357F forward and JJAZ1-843R reverse primers amplified a 480-bp cDNA fragment from endometrial stromal sarcoma carrying the t(7;17)(p15;q12), but not from the immortalized normal T HESCs cell line (Fig. 1). To verify the presence of the JAZF1/SUZ12 chimeric transcript, the amplified products were analyzed by direct sequencing (Fig. 1). The fusion was identical to that previously reported, i.e., junction of exon 3 of JAZF1 with exon 2 of SUZ12 (2,6). Similarly, nested PCR with the inner primer combination JAZF1-390F/JJAZ1-827R amplified JAZF1/SUZ12 cDNA fragments from endometrial stromal sarcoma, but not from the T HESCs cell line (Fig. 1). The same results were obtained in PCR with the primer set Fusion-286F/Fusion-541R. To elucidate, the JAZF1/SUZ12 chimeric transcript was amplified from the endometrial stromal sarcoma cDNA, but no JAZF1/SUZ12 was found in the T HESCs cell line (Fig. 1).
Figure 1.
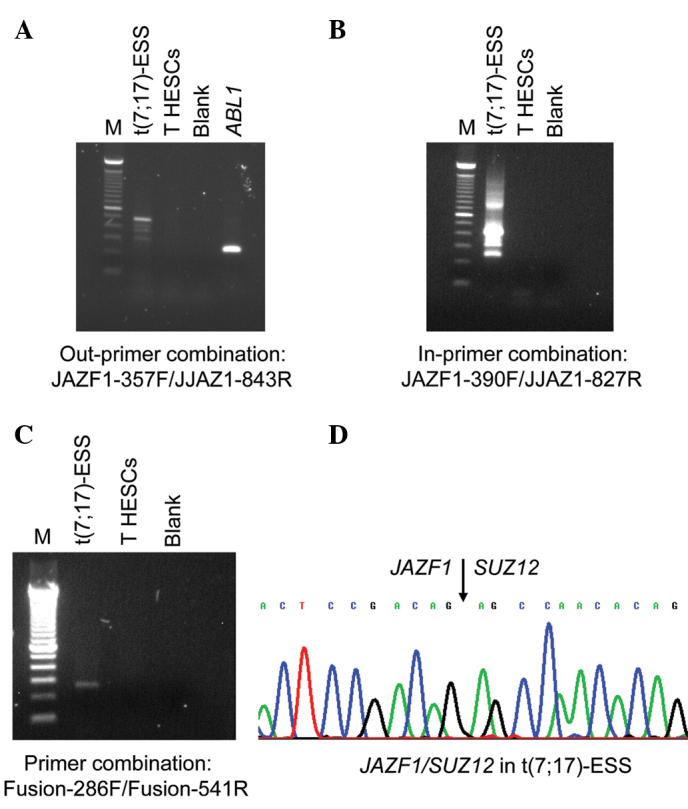
RT-PCR analysis of the T HESCs cell line for the detection of the JAZF1/SUZ12 chimeric transcript. (A) PCR amplification using the out-primer combination JAZF1-357/JJAZ1-843R. (B) The first PCR-amplified products (1 μl) were used as a template in a second nested PCR with the primers JAZF1-390F/JJAZ1-827R. (C) PCR amplification with the primer combination Fusion-286F/Fusion-541R. JAZF1/SUZ12 chimeric transcripts were not amplified in the T HESCs cell line, while they were found in an endometrial stromal sarcoma carrying the t(7;17) chromosomal aberration [t(7;17)-ESS]. Amplification of a cDNA fragment for the ABL1 gene in the T HESCs was used as an internal control. For the out-primer combination, blank is no RNA in the cDNA synthesis. For nested PCR, no first round PCR amplified products in the second PCR. M, 100-bp DNA ladder. (D) Partial chromatogram showing the fusion of JAZF1 and SUZ12 in the JAZF1/SUZ12 chimeric transcript found in the t(7;17)-ESS.
The quality of the cDNA synthesis was examined by amplification of a cDNA fragment of the ABL1 gene, used as an internal control. A 300-bp ABL1 cDNA fragment was amplified in the T HESCs cell line, indicating the good quality of the synthesized cDNA.
Using Platinum Taq DNA Polymerase High Fidelity, JAZF1/SUZ12 chimeric transcripts were amplified with the human-JAZF1-284/Fusion-541R and human-JAZF1-286F/Fusion-541R primer combinations in the positive endometrial stromal sarcoma sample, but not in the T HESCs cell line (Fig. 2A). The Rhesus-JAZF1-284/Fusion-541R and the Rhesus-JAZF1-286F/Fusion-541R did not amplify any cDNA fragments (Fig. 2B). RT-PCR analysis showed that in the T HESCs cell line, both the normal JAZF1 and SUZ12 are expressed. Subsequently, the primers JAZF1-12F and JAZF1-776R amplified a 788-bp cDNA fragment, which corresponds to the entire open reading of JAZF1. Additionally, the primer set JJAZ1-329F and JJAZ1-827R generated a 522-bp SUZ12 cDNA fragment that contained part of exon 1, exons 2-6 and part of exon 7 (Fig. 2).
Figure 2.
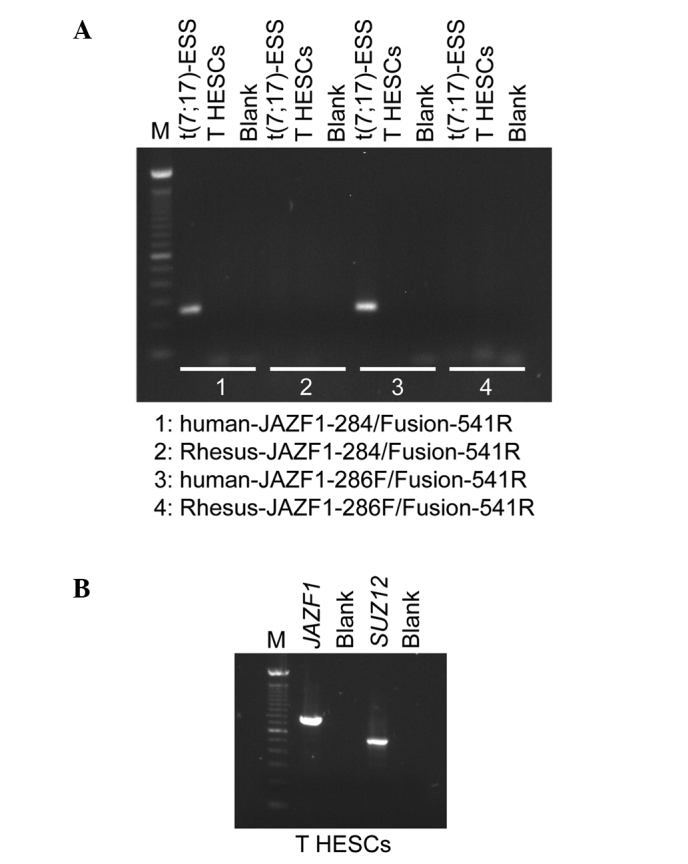
RT-PCR analysis of the T HESCs cell line. (A) For the detection of the JAZF1/SUZ12 chimeric transcript, 1 μl cDNA was used as a template in PCR amplification with the primer combinations: human-JAZF1-284/Fusion-541R, Rhesus-JAZF1-284/Fusion-541R, human-JAZF1-286F/Fusion-541R and Rhesus-JAZF1-286F/Fusion-541R, the enzyme Platinum Taq DNA Polymerase High Fidelity and a touchdown PCR cycling protocol. Using the human-JAZF1-284/Fusion-541R and human-JAZF1-286F/Fusion-541R, JAZF1/SUZ12 chimeric transcripts were amplified in an endometrial stromal sarcoma carrying the t(7;17) chromosomal aberration [t(7;17)-ESS], whereas no JAZF1/SUZ12 chimeric transcripts were amplified in the T HESCs cell line. The Rhesus-JAZF1-284/Fusion-541R and Rhesus-JAZF1-286F/Fusion-541R did not generate any PCR products. (B) RT-PCR analysis showed that in the T HESCs cell line, both the normal JAZF1 and SUZ12 genes are expressed. The primer set for JAZF1 amplified a cDNA fragment that corresponds to the entire open reading of JAZF1. The amplified cDNA fragment of SUZ12 contained part of exon 1, exons 2–6 and part of exon 7. M, 100-bp DNA ladder. Blank, no RNA in the cDNA synthesis.
Discussion
The present results do not support the previous finding by Li et al (3), that is the presence of a JUZF1/SUZ12 chimeric transcript in the immortalized normal T HESCs cell line derived from endometrial stromal cells. Various RT-PCR assays did not amplify JUZF1/SUZ12 cDNA fragments in the T HESCs cell line, whereas the same assays easily generated JUZF1/SUZ12-amplified transcripts in an endometrial stromal cell sarcoma carrying the t(7;17) chromosomal aberration (Figs. 1 and 2). The discrepancy between the present and the previous findings (3) may be due to a different group of cells examined, different culturing conditions or difference in the PCR routine. The formation of chimeric cDNA fragments has been reported as artefacts of the PCR (7,8). The commercially available AMV and MMLV reverse transcriptase have the ability to switch from one template to another resulting in recombinant DNA artefacts (9,10). In addition, thermostable polymerases generate artificial recombinant fragments by template switching (11–14). Thus, any amplified PCR product deviated from the anticipated ones should be considered with great caution.
The inherent RT-PCR risk of cross contamination or plasmid contamination may be another explanation for the difference between the findings of the present study and the results of Li et al (3). In their original study, in which the fusion JAZF1/SUZ12 was reported for the first time, Koontz et al examined 10 normal endometria. None of the endometria had the JAZF1/SUZ12 fusion. (2). In another study, Hrzenjak et al examined 10 additional normal endometria, but again, none of the endometria had the JAZF1/SUZ12 fusion (15).
In conclusion, the presence, if any, of a JUZF1/SUZ12 chimeric transcript, in human normal endometrial stromal cells is not a constant, reproducible result.
Acknowledgements
This study was supported by The Swedish Childhood Cancer Foundation.
References
- 1.Baker P, Oliva E. Endometrial stromal tumours of the uterus: a practical approach using conventional morphology and ancillary techniques. J Clin Pathol. 2007;60:235–243. doi: 10.1136/jcp.2005.031203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koontz JI, Soreng AL, Nucci M, et al. Frequent fusion of the JAZF1 and JJAZ1 genes in endometrial stromal tumors. Proc Natl Acad Sci USA. 2001;98:6348–6353. doi: 10.1073/pnas.101132598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li H, Wang J, Mor G, Sklar J. A neoplastic gene fusion mimics trans-splicing of RNAs in normal human cells. Science. 2008;321:1357–1361. doi: 10.1126/science.1156725. [DOI] [PubMed] [Google Scholar]
- 4.Krikun G, Mor G, Alvero A, et al. A novel immortalized human endometrial stromal cell line with normal progestational response. Endocrinology. 2004;145:2291–2296. doi: 10.1210/en.2003-1606. [DOI] [PubMed] [Google Scholar]
- 5.Panagopoulos I, Mertens F, Domanski HA, et al. No EWS/FLI1 fusion transcripts in giant-cell tumors of bone. Int J Cancer. 2001;93:769–772. doi: 10.1002/ijc.1415. [DOI] [PubMed] [Google Scholar]
- 6.Micci F, Walter CU, Teixeira MR, et al. Cytogenetic and molecular genetic analyses of endometrial stromal sarcoma: nonrandom involvement of chromosome arms 6p and 7p and confirmation of JAZF1/JJAZ1 gene fusion in t(7;17) Cancer Genet Cytogenet. 2003;144:119–124. doi: 10.1016/s0165-4608(03)00025-6. [DOI] [PubMed] [Google Scholar]
- 7.Brakenhoff RH, Schoenmakers JG, Lubsen NH. Chimeric cDNA clones: a novel PCR artifact. Nucleic Acids Res. 1991;19:1949. doi: 10.1093/nar/19.8.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeng XC, Wang SX. Evidence that BmTXK beta-BmKCT cDNA from Chinese scorpion Buthus martensii Karsch is an artifact generated in the reverse transcription process. FEBS Lett. 2002;520:183–185. doi: 10.1016/s0014-5793(02)02812-0. [DOI] [PubMed] [Google Scholar]
- 9.Ouhammouch M, Brody EN. Temperature-dependent template switching during in vitro cDNA synthesis by the AMV-reverse transcriptase. Nucleic Acids Res. 1992;20:5443–5450. doi: 10.1093/nar/20.20.5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaphiropoulos PG. Template switching generated during reverse transcription? FEBS Lett. 2002;527:326. doi: 10.1016/s0014-5793(02)03239-8. [DOI] [PubMed] [Google Scholar]
- 11.Judo MS, Wedel AB, Wilson C. Stimulation and suppression of PCR-mediated recombination. Nucleic Acids Res. 1998;26:1819–1825. doi: 10.1093/nar/26.7.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Odelberg SJ, Weiss RB, Hata A, White R. Template-switching during DNA synthesis by Thermus aquaticus DNA polymerase I. Nucleic Acids Res. 1995;23:2049–2057. doi: 10.1093/nar/23.11.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shammas FV, Heikkila R, Osland A. Fluorescence-based method for measuring and determining the mechanisms of recombination in quantitative PCR. Clin Chim Acta. 2001;304:19–28. doi: 10.1016/s0009-8981(00)00374-0. [DOI] [PubMed] [Google Scholar]
- 14.Zaphiropoulos PG. Non-homologous recombination mediated by Thermus aquaticus DNA polymerase I. Evidence supporting a copy choice mechanism. Nucleic Acids Res. 1998;26:2843–2848. doi: 10.1093/nar/26.12.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hrzenjak A, Moinfar F, Tavassoli FA, et al. JAZF1/JJAZ1 gene fusion in endometrial stromal sarcomas: molecular analysis by reverse transcriptase-polymerase chain reaction optimized for paraffin-embedded tissue. J Mol Diagn. 2005;7:388–395. doi: 10.1016/S1525-1578(10)60568-5. [DOI] [PMC free article] [PubMed] [Google Scholar]


