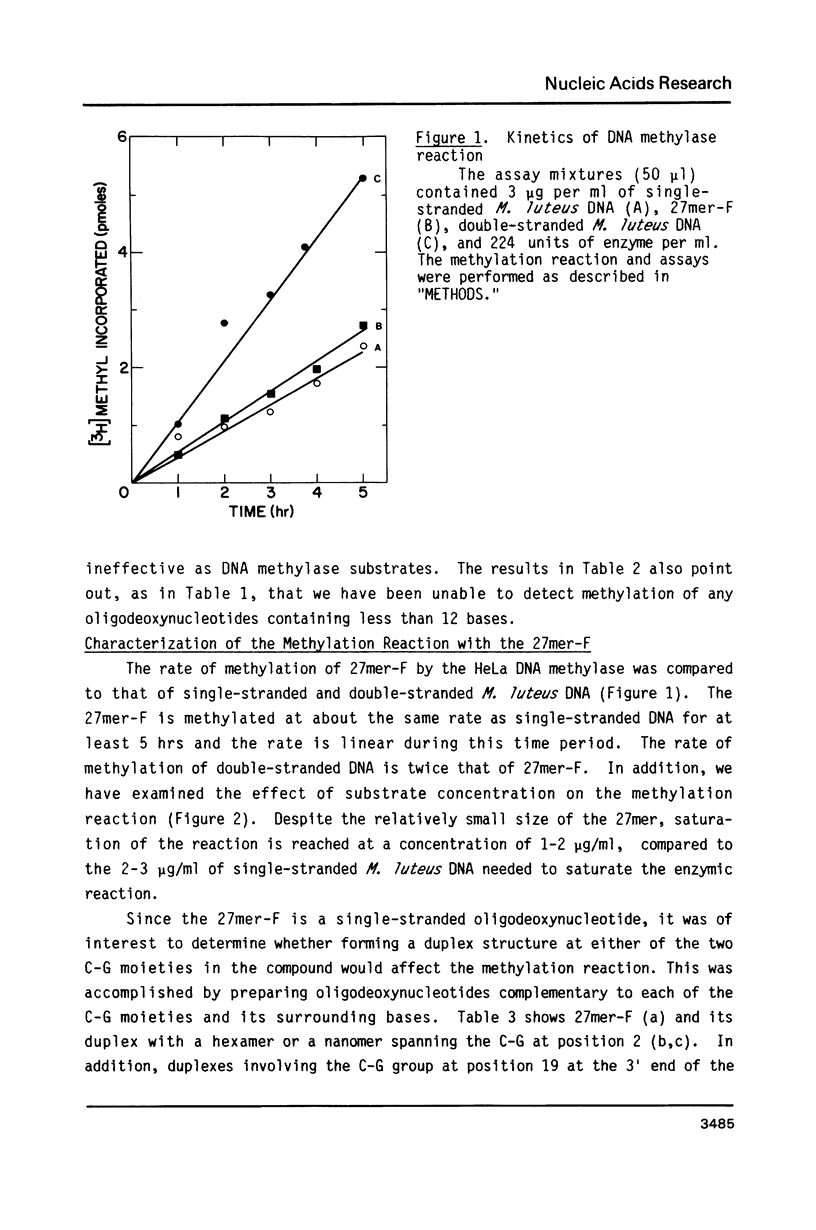
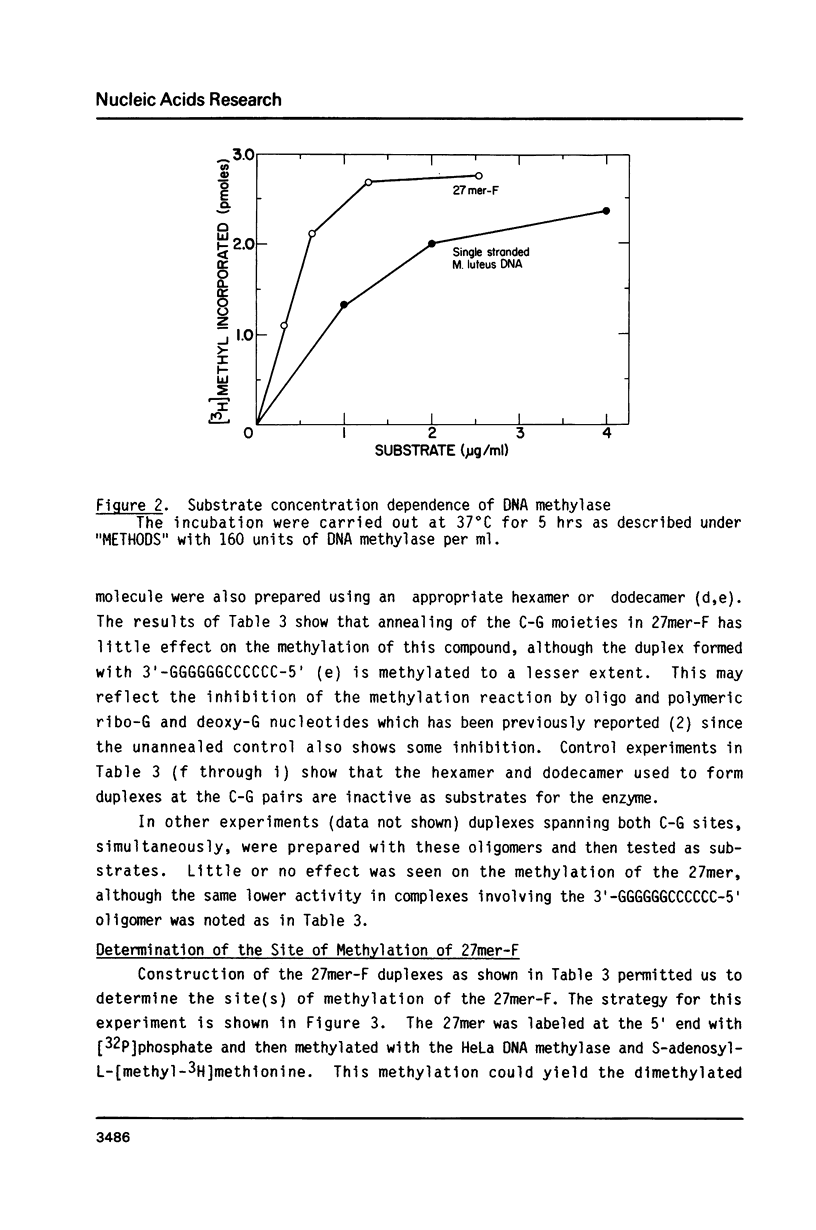
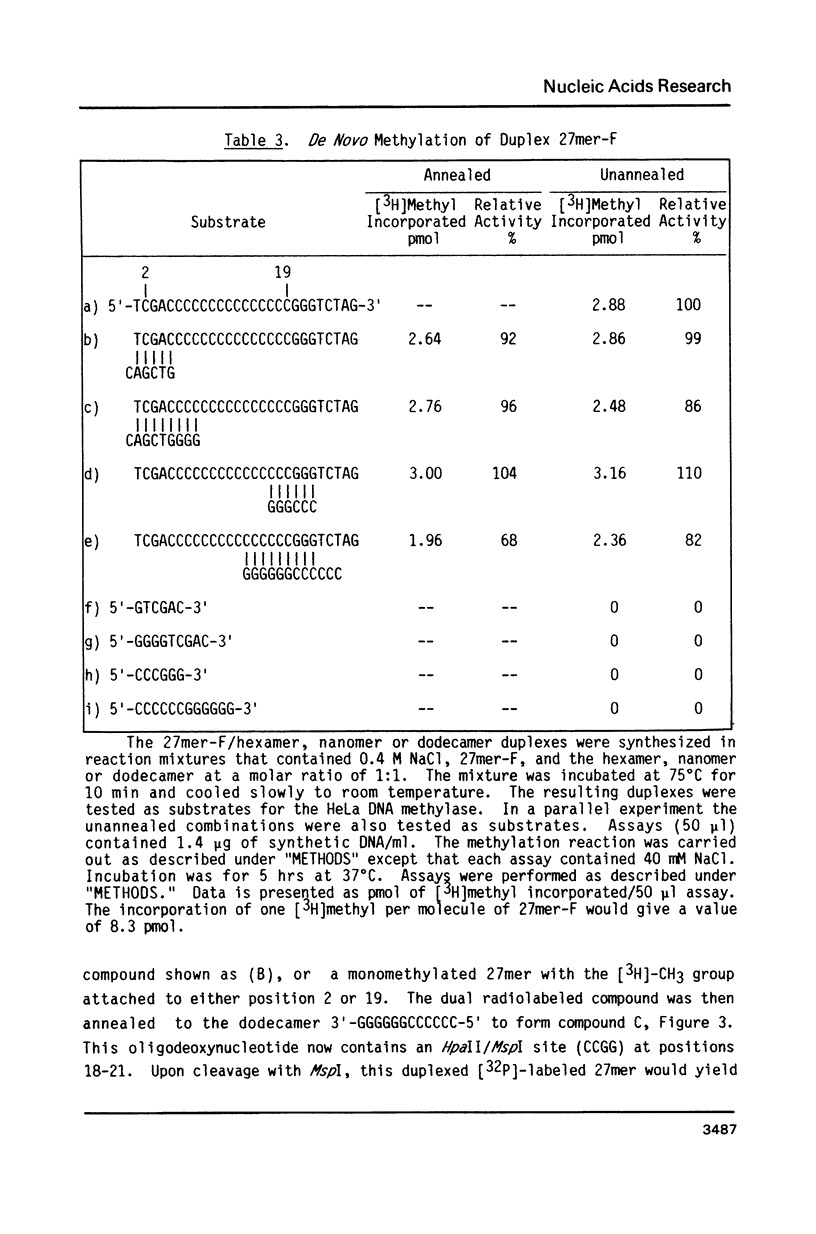
Abstract
Synthetic single-stranded oligodeoxynucleotides of known sequence have been used as in vitro substrates for a partially purified HeLa cell DNA methylase. Although most oligonucleotides tested cannot be used by the HeLa DNA methylase in vitro, we have found a unique 27mer, containing 2 C-G pairs, that is an excellent substrate for the enzyme. Analysis of the methylation of the 27mer, its derivatives and other oligomer substrates reveal that the HeLa DNA methylase does not significantly methylate an oligomer which contains just one C-G pair. In addition, only one of the two C-G pairs in the 27mer is methylated and this methylation is abolished if the other C-G pair is converted to a C-A pair. Furthermore, the HeLa enzyme apparently cannot methylate C-G pairs located in compounds containing a high A + T content. The most efficient methylation occurs with multiple separated C-G pairs in a compound with a high G + C content (greater than 65%). The results suggest that clustering of C-G pairs in regions of the DNA high in G + C content may be the preferred site for DNA methylation in vivo.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bird A. P. DNA methylation--how important in gene control? Nature. 1984 Feb 9;307(5951):503–504. doi: 10.1038/307503a0. [DOI] [PubMed] [Google Scholar]
- Bird A. P., Taggart M. H. Variable patterns of total DNA and rDNA methylation in animals. Nucleic Acids Res. 1980 Apr 11;8(7):1485–1497. doi: 10.1093/nar/8.7.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A., Taggart M., Frommer M., Miller O. J., Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985 Jan;40(1):91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- Bolden A., Ward C., Siedlecki J. A., Weissbach A. DNA methylation. Inhibition of de novo and maintenance methylation in vitro by RNA and synthetic polynucleotides. J Biol Chem. 1984 Oct 25;259(20):12437–12443. [PubMed] [Google Scholar]
- Busslinger M., Hurst J., Flavell R. A. DNA methylation and the regulation of globin gene expression. Cell. 1983 Aug;34(1):197–206. doi: 10.1016/0092-8674(83)90150-2. [DOI] [PubMed] [Google Scholar]
- Christman J. K. DNA methylation in friend erythroleukemia cells: the effects of chemically induced differentiation and of treatment with inhibitors of DNA methylation. Curr Top Microbiol Immunol. 1984;108:49–78. doi: 10.1007/978-3-642-69370-0_5. [DOI] [PubMed] [Google Scholar]
- DOSKOCIL J., SORM F. Distribution of 5-methylcytosine in pyrimidine sequences of deoxyribonucleic acids. Biochim Biophys Acta. 1962 Jun 11;55:953–959. doi: 10.1016/0006-3002(62)90909-5. [DOI] [PubMed] [Google Scholar]
- Doerfler W. DNA methylation and gene activity. Annu Rev Biochem. 1983;52:93–124. doi: 10.1146/annurev.bi.52.070183.000521. [DOI] [PubMed] [Google Scholar]
- Drahovský D., Morris N. R. Mechanism of action of rat liver DNA methylase. I. Interaction with double-stranded methyl-acceptor DNA. J Mol Biol. 1971 May 14;57(3):475–489. doi: 10.1016/0022-2836(71)90104-5. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Gama-Sosa M. A., Huang L. H., Midgett R. M., Kuo K. C., McCune R. A., Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982 Apr 24;10(8):2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld G., Nickol J., Behe M., McGhee J., Jackson D. Methylation and chromatin structure. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):577–584. doi: 10.1101/sqb.1983.047.01.068. [DOI] [PubMed] [Google Scholar]
- Geiser M., Mattaj I. W., Wilks A. F., Seldran M., Jost J. P. Structure and sequence of the promoter area and of a 5' upstream demethylation site of the estrogen-regulated chicken vitellogenin ii gene. J Biol Chem. 1983 Jul 25;258(14):9024–9030. [PubMed] [Google Scholar]
- Gruenbaum Y., Cedar H., Razin A. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature. 1982 Feb 18;295(5850):620–622. doi: 10.1038/295620a0. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y., Stein R., Cedar H., Razin A. Methylation of CpG sequences in eukaryotic DNA. FEBS Lett. 1981 Feb 9;124(1):67–71. doi: 10.1016/0014-5793(81)80055-5. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., Grünwald S., Boehm T. L., Drahovsky D. Isolation and characterization of DNA cytosine 5-methyltransferase from human placenta. Biochim Biophys Acta. 1983 Aug 2;740(3):323–330. doi: 10.1016/0167-4781(83)90141-0. [DOI] [PubMed] [Google Scholar]
- Razin A., Friedman J. DNA methylation and its possible biological roles. Prog Nucleic Acid Res Mol Biol. 1981;25:33–52. doi: 10.1016/s0079-6603(08)60482-1. [DOI] [PubMed] [Google Scholar]
- Roy P. H., Weissbach A. DNA methylase from HeLa cell nuclei. Nucleic Acids Res. 1975 Oct;2(10):1669–1684. doi: 10.1093/nar/2.10.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H., Sager R. Tissue specificity and clustering of methylated cystosines in bovine satellite I DNA. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3584–3588. doi: 10.1073/pnas.79.11.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tykocinski M. L., Max E. E. CG dinucleotide clusters in MHC genes and in 5' demethylated genes. Nucleic Acids Res. 1984 May 25;12(10):4385–4396. doi: 10.1093/nar/12.10.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]