Abstract
Streptomyces sp. KY40-1, a strain isolated from the Kentucky Appalachian foothills, is the producer of moromycins A (18) and B (19). Further investigations of this strain led to the isolation and structure elucidation of the five new saquayamycins G – K (1–5), along with known compounds. Two of the new compounds bear the unusual aminosugar rednose, which was found here for the first time in angucyclines. The different attachment positions of this aminosugar in these two compounds indicates a high acceptor substrate flexibility of the responsible glycosyl transferase or alternatively the involvement of multiple glycosyl transferases. The cytotoxic activity of the isolated compounds was determined using human prostate cancer (PC-3) and non-small cell lung cancer (H460) cell lines. Cell viability assays showed that saquayamycins J (4), K (5), A (7) and B (8) were most active in PC3 cells, with saquayamycin B (8) showing the highest activity (GI50 = 0.0075 μM). The aminosugar-containing saquayamycins H (2) and saquayamycin B (8) showed the highest activity against H460 cells, with a GI50 = 3.3 and 3.9 μM, respectively. The results presented here provide more insights into the structure-activity relationship (SAR) of saquayamycins with respect to the nature, number and linkage of sugar residues.
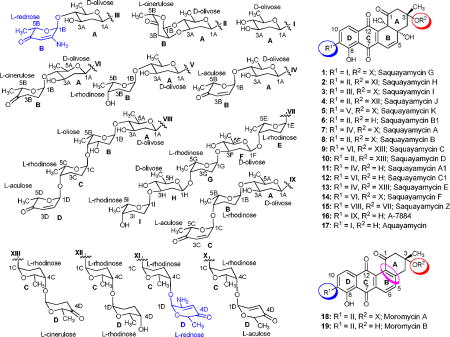
The angucycline group of antibiotics is one of the largest groups of polycyclic aromatic polyketides, rich in chemical scaffolds and biological activities, predominantly anticancer and antibacterial.1–3 Saquayamycins,4–8 urdamycins,9–17 and landomycins,18–24 are well known angucycline antitumor antibiotics. The structures of both saquayamycins and urdamycins contain the same aquayamycin (17)25–27 as aglycone with the C-glycosidic sugar D-olivose attached at C-9 of the angucycline chromophore. So far, seven saquayamycin analogues were reported from Streptomyces spp. Saquayamycins differ from urdamycins by their saccharide patterns, which are attached at C-9 and C-3 positions in the saquayamycins, but at C-9- and C-12b-positions in urdamycins. Saquayamycins A-D (7–10) were first isolated from Streptomyces nodosus MH190-16F3 and were reported as platelet aggregation inhibitors.4 Saquayamycins A-B (7–8) contain three different O-glycosidically linked deoxysugars, L-rhodinose, L-aculose, and L-cinerulose (only in saquayamycin B). Saquayamycin A (7) was reported to be unstable to acid, and even contact with silica gel led to its conversion to saquayamycin B (8). Saquayamycins A1 (11), B1 (6), and C1 (12) were generated by partial acid hydrolysis of saquayamycins A-C (7–9), respectively.4 The two isomers, saquayamycins E (13) and F (14) were produced by Actinomyces strain MK290-AF1 and reported to inhibit the FPTase from bovine brain with IC50 values of 1.8 and 2.0 μM, respectively.5 They differ slightly from the saquayamycins A (7) and C (9) with respect to their sugar moieties. The saquayamycin analogue A-7884 (16) was isolated from the Streptomyces sp. #AM1699; it has a trisaccharide side chain connected at C-9, which contains an L-rhodinose sugar moiety between the first sugar, a C-glycosidic bound D-olivose, and L-aculose of saquayamycin A1 (11).28 Compound 16 showed inhibitory activity in the inducible nitric oxide synthase (iNOS) assay with IC50 values of 43.5 μM, better than saquayamycin A1 (11; IC50 values of 101.2 μM).28 Recently, the largest saquayamycin analogue, saquayamycin Z (15), was reported from Micromonospora sp. strain Tü6368.6 Saquayamycin Z (15) contains tetra- and pentasaccharide side chains linked at C-3- and C-9-positions of the benz[a]anthracene core. The tetrasaccharide side chain of saquayamycin Z (15) is striking due to the presence of an L-oliose, which is not a usual sugar constituent of the angucyclines.6, 29 Very recently, Ren et al., reported three novel members of the angucycline family named N05WA963 A, B and D, with the same aglycones as in the moromycins A (18) and B (19), except for an additional methoxy group attached at C-5.30 N05WA963 A, B and D were reported to have antiproliferative effects on a panel of cancer cell lines such as SW620 (colon cancer), K-562 (chronic myelogeneous leukemia), MDA-MB-231 (estrogen receptor negative breast cancer), YES-4 (esophageal cancer), T-98 and U251SP (both glioblastomae).
To study the structure-activity relationship (SAR) of this saquayamycin group of antibiotics, we looked for further saquayamycin analogues produced by Streptomyces sp. K40-1, the strain earlier reported as the producer of moromycins A-B (18–19).7 The saccharide attachments in moromycins A (18) and B (19) are like those of saquayamycins B (8) and B1 (6), respectively, however, their tetracyclic angucyclinone core has an aromatic ring B, and thus no angular hydroxy groups at C-4a and C-12b positions.
We found five new metabolites, designated as saquayamycins G-K (1–5), produced by repeated fermentations of the same strain, along with saquayamycin B1 (6), which was previously not described as a natural product, and known saquayamycins. Two of the new angucyclines, saquayamycins H (2) and I (3), bear the unusual aminosugar rednose, which was found for the first time in an angucycline compound. Aminosugar-containing angucyclines are very rare, and previously only three examples had been reported, namely the marmycins A and B,31, 32 and mayamycin.33 The marmycins contain an unusual branched and doubly (C- and N)-linked aminosugar, 3-epi-, 4-epi-vancosamine, and mayamycin contains a unique C-glycosidically linked aminosugar, N-demethylangolosamine, attached at the C-5-position of the benz[a]anthracenone core.
Results and Discussion
Cultivation, Isolation and Structure Investigation
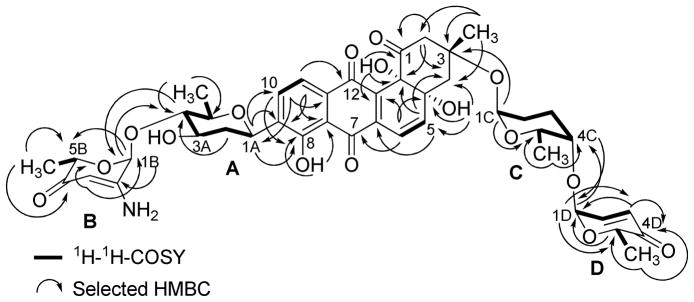
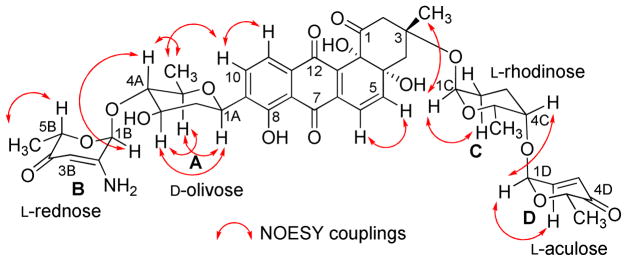
2.3 g of crude extract was obtained from a four day culture of Streptomyces sp KY40-1. On the basis of TLC, UV and HPLC-MS (Figure S5) of the crude extract, several angucyclines containing aquayamycin (17) were detected.34 Fractionation of the extract (2.30 g), using various chromatographic techniques (Figure S1) led to the isolation of six new saquayamycins G-K (1–5) and B1 (6). For comparison reasons we report here also NMR assignments and NOESY correlations of saquayamycins A-B (7–8) (Figure S3–S6), which were previously incompletely reported.7
Saquayamycin G (1)
Compound 1 is an orange-red solid with a molecular formula of C37H41O14, indicating that one sugar molecule was missing compared to saquayamycins A (7) and B (8). The 1H NMR spectrum of 1 displayed the same aromatic pattern as aquayamycin (17), with sugar substitutions at the C-3- and C-9-positions. The aliphatic region between δ 1.37–1.26 revealed three methyl protons as doublets indicating three 6-deoxysugar moieties. This was confirmed by the presence of the three anomeric protons at δ 5.24 (d, 3.5 Hz), 5.22 (br d, 5.5 Hz), and 4.87 (brd, 11.0 Hz), consistent with one β-D- and two α-L-glycosidically linked sugar moieties. The 13C NMR/HSQC spectra of 1 established aquayamycin (17) as the aglycone. The last two carbonyls correspond to a quinone system, with one carbonyl chelated with a peri-hydroxy group. In the sp3 region, two anomeric carbon signals (δ 95.5 and 92.6) were observed along with seven methine, three quaternary, five methylene, and four methyl signals. The 1H and 13C NMR data of 1 are closely related to saquayamycins A (7) and B (8) with Δm/z =110, indicating the missing sugar B (L-cinerulose or L-aculose) or sugar D (L-aculose) in compounds 7 and 8, respectively.
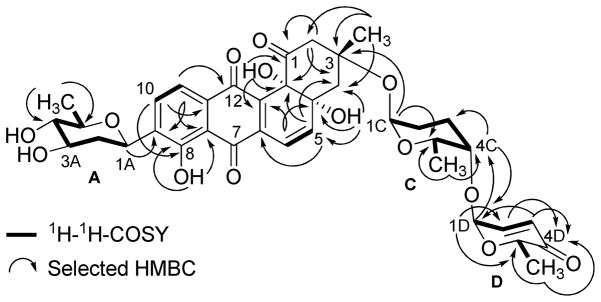
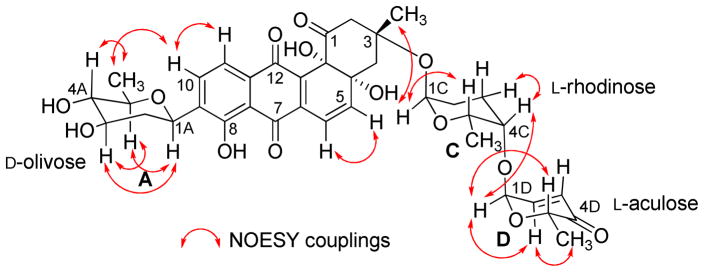
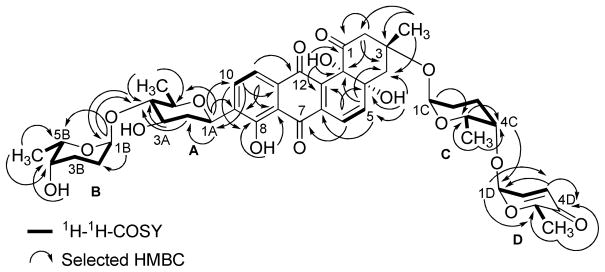
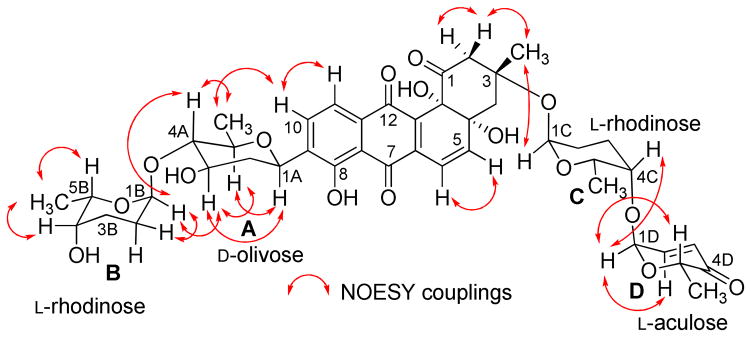
The HMBC and 1H-1H COSY correlations of 1 (Figure 1) revealed two partial structures, the aquayamycin aglycone (17) and a disaccharide system. The attachment of the disaccharide at the usual C-3-position was confirmed by a 3JC-H long range coupling between the anomeric proton of α-L-rhodinose (δH 5.22) and C-3 (δC 82.7) of the aglycone. All the three sugars A, C and D showed the same signal patterns and connectivity as previously found in saquayamycins A (7) and B (8). The oxygenated carbon C-4A (δC 78.2) of the D-olivose moiety of compound 1 appeared upfield compared to the same carbon of saquayamycin A (7; C-4A, δC = 89.3, table 4), indicating a free OH group at C-4A of the β-D-olivose sugar moiety. The couplings and chemical shifts were in full agreement with structure 1 (Figure 1). The relative configuration of the sugar residues was further confirmed by NOESY experiments (Figure 2), determining structure 1 as 3-α-L-rhodinosyl-4-1-α-L-aculosyl-aquayamycin, which was subsequently named saquayamycin G.
Figure 1.
1H-1H-COSY (bold lines) and selected HMBC long range couplings (→) of saquayamycin G (1).
Table 4.
13C NMR (125 MHz) assignments of Saquayamycins A (7), J (4) and K (5) in CDCl3, δ in ppm relative to TMS.
| Position | Saq. A (7) | Saq. J (4) | Saq. K (5) |
|---|---|---|---|
|
| |||
| δC, mult. a) | δC, mult. a) | δC, mult. a) | |
| 1 | 205.1, C | 205.0, C | 205.1, C |
| 2 | 50.3, CH2 | 50.4, CH2 | 50.4, CH2 |
| 3 | 82.7, C | 82.6, C | 82.8, C |
| 3-CH3 | 25.6, CH3 | 25.6, CH3 | 25.6, CH3 |
| 4 | 44.6, CH2 | 44.7, CH2 | 44.7, CH2 |
| 4a | 80.1, C | 80.1, C | 80.2, C |
| 5 | 145.7, CH | 145.8, CH | 145.7, CH |
| 6 | 117.6, CH | 117.6, CH | 117.7, CH |
| 6a | 138.9, C | 138.9, C | 139.0, C |
| 7 | 188.3, C | 188.5, C | 188.4, C |
| 7a | 114.1, C | 114.2, C | 114.1, C |
| 8 | 158.1, C | 158.1, C | 158.3, C |
| 9 | 138.3, C | 138.0, C | 138.8, C |
| 10 | 133.7, CH | 133.8, CH | 133.8, CH |
| 11 | 119.8, CH | 119.8, CH | 119.9, CH |
| 11a | 130.6, C | 130.7, C | 130.6, C |
| 12 | 182.4, C | 182.4, C | 182.5, C |
| 12a | 138.9, C | 139.1, C | 139.0, C |
| 12b | 77.6, C | 77.6, C | 77.6, C |
| Sugar A, α-D-olivose | |||
| 1A | 71.2, CH | 71.6, CH | 71.2, CH |
| 2A | 39.0, CH2 | 36.9, CH2 | 38.9, CH2 |
| 3A | 71.4, CH | 76.9, CH | 71.6, CH |
| 4A | 89.3, CH | 74.6, CH | 89.2, CH |
| 5A | 74.5,CH | 74.7, CH | 74.7, CH |
| 6A | 18.5, CH3 | 17.6, CH3 | 18.7, CH3 |
| Sugar B, α-L-aculose or α-L-cinerulose or α-L-rhodinose | |||
| 1B | 95.2, CH | 91.5, CH | 98.9, CH |
| 2B | 142.4, CH | 71.3, CH | 27.3, CH2 |
| 3B | 127.4, CH | 40.1, CH2 | 30.1, CH2 |
| 4B | 195.4, C | 208.0, C | 71.7, CH |
| 5B | 71.6, CH | 77.9, CH | 71.5, CH |
| 6B | 15.3, CH3 | 16.3, CH3 | 18.0, CH3 |
| Sugar C, α-L-rhodinose | |||
| 1C | 92.5, CH | 92.8, CH | 92.6, CH |
| 2C | 24.8, CH2 | 25.0, CH2 | 24.9, CH2 |
| 3C | 24.7, CH2 | 24.7, CH2 | 24.7, CH2 |
| 4C | 76.3, CH | 74.4, CH | 76.4, CH |
| 5C | 67.1, CH | 67.6, CH | 67.1, CH |
| 6C | 17.3, CH3 | 17.4, CH3 | 17.4, CH3 |
| Sugar D, α-L-aculose or α-L-rhodinose | |||
| 1D | 95.4, CH | 98.8, CH | 95.5, CH |
| 2D | 143.3, CH | 29.9, CH2 | 143.3, CH |
| 3D | 127.4, CH | 27.9, CH2 | 127.5, CH |
| 4D | 197.0, C | 72.3, CH | 197.1, CH |
| 5D | 70.7, CH | 70.4, CH | 70.8, CH |
| 6D | 15.3, CH3 | 18.0, CH3 | 15.4, CH3 |
| - | - | - | - |
See also Figures S32–37, S40–45 and S54–59.
Figure 2.
Selected NOESY correlations (↔) in saquayamycin G (1).
Saquayamycin H (2)
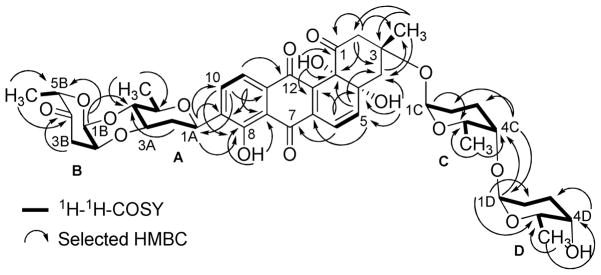
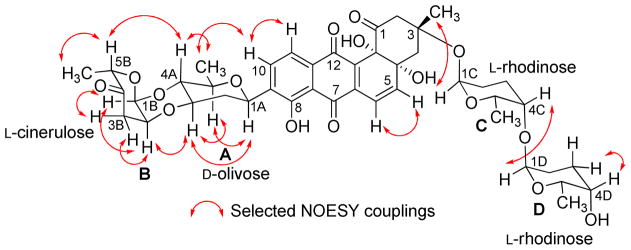
Compound 2 was obtained from fraction FIV as an orange-red solid (Figure S1). It has similar physicochemical properties as saquayamycins G (1), A (7) and B (8), showing an orange fluorescence under long UV (365 nm) light, and blue-violet coloration with 2N NaOH. The molecular formula C43H49NO16 of compound 2 was established by ESI HRMS, indicating a nitrogen-containing compound with Δm/z =15 higher than saquayamycins A (7) and B (8).
The 1H NMR of compound 2 was very similar to that of saquayamycin B (8) with the exception that one of the two doublet olefinic protons of the L-aculose moiety was missing. Instead, one singlet proton at δ 5.19 along with a broad signal with an integration of 2H at δ 4.78 was present. The 13C NMR/HSQC of compound 2 displayed 43 carbon atoms as in saquayamycin B (8) with highly similar chemical shifts. The sole difference in the 13C NMR spectrum was that the two methine carbons at C-2D (δC 143.3, Cq) and C-3D (δC 127.5, CH) of the L-aculose moiety of saquayamycin B (8) were shifted to δ 159.1 (Cq) and δ 97.4 (CH), respectively. The down- field chemical shift of the quaternary carbon δ 159.1 in compound 2 indicated its connection to a heteroatom, which turned out to be NH2 in this case, as shown from the broad signal in the 1H NMR spectrum, in the β-position of the carbonyl.
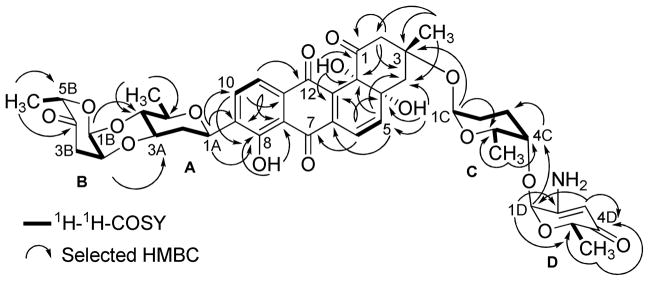
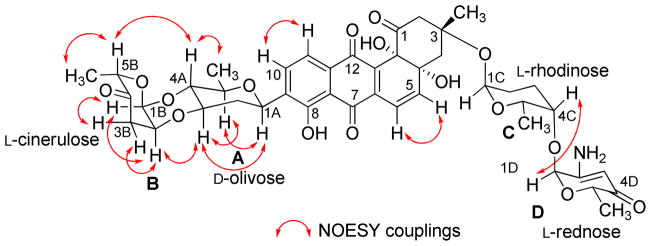
The full NMR assignments for compound 2 were deduced from the 1H-1H COSY, HSQC and HMBC experiments (Figure 3 and Tables 1–2) indicating the presence of the rare aminosugar rednose, which was connected at C-4C of the α-L-rhodinose moiety instead of the α-L-aculose moiety found in the same position in saquayamycins A (7) and B (8). Based on NOESY experiments (Figure 4), coupling constants and comparison with saquayamycin B (8), compound 2 was established as 4A-α-L-cinerulosyl-3-α-L-rhodinosyl- 4C-1D-α-L-rednosyl-aquayamycin, and consequently named saquayamycin H. The unusual trideoxy-keto-aminosugar rednose was previously reported in two anthracycline type compounds, CG21-C35 and rudolphomycin 36 (hence the name).
Figure 3.
1H-1H-COSY (bold lines) and selected HMBC long range couplings (→) of saquayamycin H (2).
Table 1.
1H NMR (500 MHz) assignments of saquayamycins B (8), G (1), H (2) and I (3), δ in ppm relative to TMS (multiplicity, J/Hz).
| Position | Saquayamycin B (8) a) | Saquayamycin G (1) a) | Saquayamycin H (2) a) | Saquayamycin I (3) a) |
|---|---|---|---|---|
|
| ||||
| δH (CDCl3) | δH (CDCl3) | δH (CDCl3) | δH (Acetone-d6) | |
| 2 | 2.48, d (13.5, Ha), 3.15, dd (13.0, 3.0, He) | 2.49, d (13.5, Ha), 3.16, dd (13.5, 3.0, He) | 2.48, d (13.5, Ha), 3.16, dd (13.5, 3.0, He) | 2.89, brd (13.0, Ha), 2.96, brd (13.0, He) |
| 3-CH3 | 1.37, s | 1.38, s | 1.37, s | 1.39, s |
| 4 | 1.81, d (15.5, Ha), 2.25, dd (15.5, 3.0, He) | 1.81, d (15.0, Ha), 2.26, dd (15.0, 3.0, He) | 1.84, d (15.0, Ha), 2.25, dd (15.5, 3.0, He) | 2.22, d (15.5, Ha), 2.31, dd (15.5, 2.0, He) |
| 4a-OH | 4.31, brs | 4.32, brs | 4.35, brs | 4.31, brs |
| 5 | 6.42, d (10.0) | 6.42, d (9.5) | 6.42, d (10.0) | 6.43, d (9.5) |
| 6 | 6.87, d (9.5) | 6.88, brd (10.5) | 6.87, d (9.5) | 6.82, d (9.5) |
| 8-OH | 12.27, s | 12.28, s | 12.26, s | 12.32, s |
| 10 | 7.85, d (7.5) | 7.85, d (8.0) | 7.85, d (7.5) | 7.90, d (7.5) |
| 11 | 7.57, d (7.5) | 7.58, d (7.5) | 7.57, d (7.5) | 7.57, d (8.0) |
| 12b-OH | 4.56, brs | 4.59, brs | 4.56, brs | 4.62, brs |
| Sugar A, β-D-olivose | ||||
| 1A | 4.93, brd (9.5) | 4.87, brd (11.0) | 4.93, brd (9.5) | 4.91, brd (11.0) |
| 2A | 1.45, m (Ha), 2.41, ddd (12.5, 4.0, 2.5, He) | 1.40 m (Ha), 2.44 m (He) | 1.45, ddd (14.0, 11.5, 3.0, Ha), 2.41 (ddd, 12.5, 2.5, 1.5, He) | 1.40, m (Ha), 2.47 brdd (12.5, 4.5) |
| 3A | 3.78, m | 3.80, brm | 3.78, m | 3.95, m |
| 3A-OH | - | 3.46, brs | - | 4.75, brs |
| 4A | 3.45, dd (9.0, 9.0) | 3.16, dd (10.5, 9.0) | 3.45, dd (9.0, 9.0) | 3.40, dd (9.0, 9.5) |
| 4A-OH | - | n.o.b) | - | - |
| 5A | 3.54, dq (9.0, 6.0) | 3.48, m | 3.54, dq (9.0, 6.0) | 3.63, m |
| 6A | 1.36, d (6.5) | 1.37, brd (7.5) | 1.36, d (6.0) | 1.43, d (6.0) |
| Sugar B, α-L-cinerulose or α-L-rednose | ||||
| 1B | 5.15, d (2.5) | - | 5.15, d (3.0) | 5.50 (s) |
| 2B | 4.31, brs | - | 4.32, m | - |
| 2B-NH2 | - | - | - | 6.38, brs |
| 3B | 2.60, dd (17.5, 3.0, Ha), 2.64, dd (17.5, 3.0, He) | - | 2.60, dd (17.5, 3.0, Ha), 2.64, dd (17.5, 3.0, He) | 5.10, s |
| 5B | 4.69, q (7.0) | - | 4.69, q (6.5) | 4.63, q (6.5) |
| 6B | 1.35, d (7.0) | - | 1.34, d (6.5) | 1.31, d (6.5) |
| Sugar C, α-L-rhodinose | ||||
| 1C | 5.22, brd (4.0) | 5.22, brd (5.5) | 5.22, d (2.5) | 5.24, brs |
| 2C | 1.44, m (Ha), 2.01, m (He) | 1.90, m (Ha), 2.00, m (He) | 1.90, m (Ha), 2.00, m (He) | 1.96–1.88, m (Ha) 1.96–1.88, m (He) |
| 3C | 1.88, m (Ha) 1.88, m (He) | 1.45, m (Ha), 1.85, m (He) | 1.45, m (Ha), 1.85, m (He) | 1.43, m (Ha), 1.82, m (He) |
| 4C | 3.66, brs | 3.67, brs | 3.69, brs | 3.73, brs |
| 5C | 4.21, brm | 4.22, m | 4.21, brq (6.5) | 4.20, m |
| 6C | 1.26, d (6.5) | 1.26, d (6.5) | 1.30, d (7.0) | 1.22, d (6.5) |
| Sugar D, α-L-aculose or α-L-rednose | ||||
| 1D | 5.24, d (3.5) | 5.24, d (3.5) | 5.17, s | 5.38, d (3.5) |
| 2D | 6.86, dd (10.5, 3.5) | 6.85, dd (10.0, 3.5) | - | 7.07, dd (10.0, 3.5) |
| 2D-NH2 | - | - | 4.78, brs | - |
| 3D | 6.01, d (10.5) | 6.07, d (10.5) | 5.19, s | 6.05, d (10.0) |
| 5D | 4.51, q (6.5) | 4.51, q (7.0) | 4.32, m | 4.58, q (6.5) |
| 6D | 1.34, d (6.5) | 1.35, d (7.0) | 1.34, d (6.5) | 1.28, d (7.0) |
See also Figures S12–17, S20–31 and S60–65,
not observed.
Table 2.
13C NMR (125 MHz) assignments of saquayamycins B (8), G (1), H (2), and I (3), δ in ppm relative to TMS.
| Position | Saq. B (8)a,b) | Saq. G (1) a,b) | Saq. H (2) a,b) | Saq. I (3) a,c) | Saq. B1 (6) a,b) |
|---|---|---|---|---|---|
|
| |||||
| δC, mult. | δC, mult. | δC, mult. | δC, mult. | δC, mult. | |
| 1 | 205.0, C | 205.1, C | 205.2, C | 205.8, C | 205.1, C |
| 2 | 50.4, CH2 | 50.4, CH2 | 50.2, CH2 | 51.1, CH2 | 52.9, CH2 |
| 3 | 82.7, C | 82.7, C | 82.8, C | 83.3, C | 76.3, C |
| 3-CH3 | 25.6, CH3 | 25.6, CH3 | 25.6, CH3 | 26.1, CH3 | 30.5, CH3 |
| 4 | 44.7, CH2 | 44.7, CH2 | 44.8, CH2 | 43.9, CH2 | 43.4, CH2 |
| 4a | 80.1, C | 80.2, C | 80.2, C | 80.9, C | 80.7, C |
| 5 | 145.8, CH | 145.7, CH | 145.8, CH | 147.2, CH | 144.5, CH |
| 6 | 117.6, CH | 117.7, CH | 117.7, CH | 117.5, CH | 117.7, CH |
| 6a | 138.9, C | 139.0, C | 140.0, C | 139.4, C | 139.0, C |
| 7 | 188.4, C | 188.4, C | 188.4, C | 190.1, C | 188.2, C |
| 7a | 114.1, C | 114.1, C | 114.1, C | 115.2, C | 114.2, C |
| 8 | 158.1, C | 158.2, C | 158.1, C | 158.4, C | 158.2, C |
| 9 | 138.0, C | 138.6, C | 138.1, C | 138.8, C | 138.2, C |
| 10 | 133.8, CH | 133.9, CH | 133.8, CH | 134.1, CH | 134.0, CH |
| 11 | 119.9, CH | 119.9, CH | 119.8, CH | 119.7, CH | 120.0, CH |
| 11a | 130.7, C | 130.6, C | 130.7, C | 132.0, C | 130.6, C |
| 12 | 182.4, C | 182.4, C | 182.4, C | 183.2, C | 182.2, C |
| 12a | 139.0, C | 139.0, C | 140.0, C | 140.4, C | 138.3, C |
| 12b | 77.6, C | 77.6, C | 77.6, C | 78.2, C | 76.3, C |
| Sugar A, β-D-olivose | |||||
| 1A | 71.6, CH | 71.4, CH | 71.6, CH | 71.9, CH | 71.6, CH |
| 2A | 36.9, CH2 | 39.5, CH2 | 36.9, CH2 | 40.5, CH2 | 36.9, CH2 |
| 3A | 76.8, CH | 73.1, CH | 76.8, CH | 71.9, CH | 76.9, CH |
| 4A | 74.6, CH | 78.2, CH | 74.6, CH | 88.4, CH | 74.7, CH |
| 5A | 74.7, CH | 76.1, CH | 74.7, CH | 75.5, CH | 74.8, CH |
| 6A | 17.6, CH3 | 18.3, CH3 | 17.6, CH3 | 19.1, CH3 | 17.7, CH3 |
| Sugar B, α-L-cinerulose or α-L-rednose | |||||
| 1B | 91.5, CH | - | 91.5, CH | 96.6, CH | 91.6, CH |
| 2B | 71.3, CH | - | 71.3, CH | 161.1, C | 71.3, CH |
| 3B | 40.1, CH2 | - | 40.2, CH2 | 95.6, CH | 40.2, CH2 |
| 4B | 208.0, C | - | 208.1, C | 193.2, C | 208.0, C |
| 5B | 77.9, CH | - | 77.9, CH | 71.0, CH | 78.0, CH |
| 6B | 16.3, CH3 | - | 16.4, CH3 | 16.6, CH3 | 16.4, CH3 |
| Sugar C, α-L-rhodinose | |||||
| 1C | 92.6, CH | 92.6, CH | 92.6, CH | 92.9, CH | - |
| 2C | 24.9, CH2 | 24.9, CH2 | 24.9, CH2 | 25.2, CH2 | - |
| 3C | 24.7, CH2 | 24.7, CH2 | 24.8, CH2 | 25.6, CH2 | - |
| 4C | 76.3, CH | 76.4, CH | 76.7, CH | 77.2, CH | - |
| 5C | 67.1, CH | 67.1, CH | 67.2, CH | 67.4, CH | - |
| 6C | 17.3, CH3 | 17.4, CH3 | 17.7, CH3 | 17.6, CH3 | - |
| Sugar D, α-L-aculose or α-L-rednose | |||||
| 1D | 95.5, CH | 95.5, CH | 96.7, CH | 96.1, CH | - |
| 2D | 143.3, CH | 143.3, CH | 159.1, C | 145.2, CH | - |
| 3D | 127.5, CH | 127.5, CH | 97.4, CH | 127.3, CH | - |
| 4D | 197.0, C | 197.1, C | 195.2, C | 197.3, C | - |
| 5D | 70.8, CH | 70.8, CH | 70.5, CH | 71.1, CH | - |
| 6D | 15.4, CH3 | 15.4, CH3 | 16.0, CH3 | 15.5, CH3 | - |
Figure 4.
Selected NOESY correlations (↔) in saquayamycin H (2).
Saquayamycin I (3)
Compound 3 was obtained as an orange-red solid from the same fraction as saquayamycin H (2), with a slightly higher polarity. Compound 3 is an isomer of saquayamycin H (2) with the same molecular formula of C43H48NO16. The UV spectra of compound 3 showed the band at 218 nm, characteristic of the L-rednose sugar moiety (Figure S2).
The only difference in the 1H and 13C NMR spectra between compounds 2 and 3 was that the ABX signals (CH-2B and CH2-3B) of the L-cinerulose sugar moiety in 2 were missing in 3. Instead, two CH groups of the L-aculose moiety in 3 at δ 7.07 (dd, 10.0, 3.5 Hz; δC 145.2) and δ 6.05 (d, 10.0 Hz; δC 127.3) were observed instead.
The full assignment of compound 3 was deduced from the 1H-1H COSY, HSQC and HMBC experiments (Figure 5 and Tables 1–2) indicating the attachment of the L-rednose moiety at C-4A of the β-D-olivose moiety (sugar A). This was confirmed by long range coupling of the proton at δ 5.50 (H-1B, δC 96.6) to C-4A (δC 88.4) and and from H-4A (δH 3.40) to C-1B (δC 96.6). Thus, the rednose moiety was found in compound 3 in a different position than in saquayamycin H (2), and the same O-glycosidic linked disaccharide chain (3-α-L-rhodinosyl-4-1-α-L-aculose) as found in compounds 1, 7, and 8 is connected at C-3 of the aquayamycin aglycone. The relative stereochemistry of the aglycone of compound 3 was deduced from NOESY experiments (Figure 6), coupling constants and by comparison with the related saquayamycins A (7) and B (8) and H (2). The relative configuration of the sugar residues was established to be 4A-α-L-rednosyl-3-α-L-rhodinosyl-4C-1D-α-L-aculosyl-aquayamycin, confirming structure 3 as indicated in Figures 5 and 6, which was subsequently named saquayamycin I. The attachment of the L-rednose moiety in two different positions in saquayamycins H (2) and I (3) indicates acceptor substrate flexibility of the aminosugar glycosyltransferase or the involvement of two different enzymes.
Figure 5.
1H-1H-COSY (bold lines) and selected HMBC long range couplings (→) of saquayamycin I (3).
Figure 6.
Selected NOESY correlations (↔) in saquayamycin I (3).
Saquayamycin J (4)
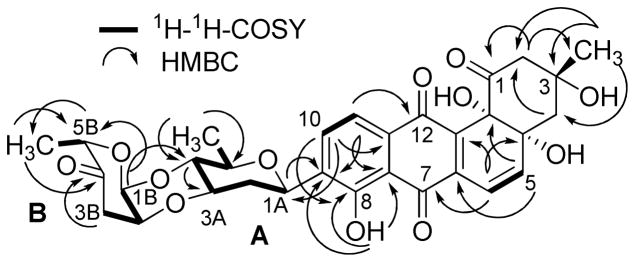
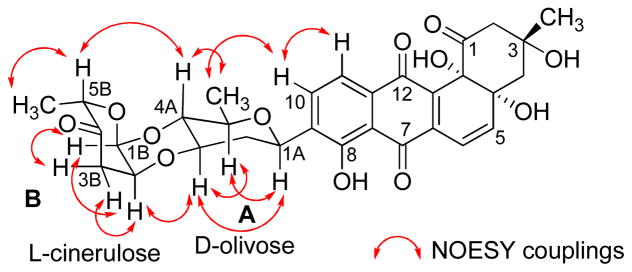
Compound 4 is a yellow solid with similar physicochemical properties and staining to those of the earlier isolated saquayamycins. Its molecular weight was deduced by ESIMS and (−)HRESIMS, establishing its molecular formula to be C43H52O16, 4 amu higher than saquayamycins A (7) and B (8), attributed to the reduction of two double bonds or ring openings. The NMR spectrum showed the same aromatic pattern of the aquayamycin system and the characteristic ABX proton signals CH-2B and CH2-3B along with the carbonyl of the L-cinerulose sugar moiety (sugar B, δC 208.0), confirming the existence of the same sugar residues A and B in compound 4 as in saquayamycin B (8). Comparing the NMR data with those of saquayamycin B (8) showed the absence of the olefinic proton signals of the L-aculose sugar moiety (2D and 3D) and the carbonyl carbon C-4D. Instead, two methylene groups and one oxygenated carbon at δC 29.9, 27.9 and 70.4, respectively, were observed. The 4 amu difference between saquayamycin B (8) and compound 4 was attributed to the reducing of C-2D and C-3D double bonds along with the reduction of the carbonyl group at C-4D. The OH group at C-4D could be axial or equatorial. The structure of 4 was further confirmed by 2D NMR experiments (Figures 7 and 8), exhibiting the same sugar residues A (D-olivose), B (L-cinerulose) and C (L-rhodinose) and connectivity as found in 8 (Figures 7 and 8). However, based on coupling constants and NOESY correlations the fourth sugar, residue D, was deduced to be L-rhodinose instead of L-aculose, which was typically found in the same position, e.g., in compounds 1, 3, 7 and 8. Hence the new compound 4 was determine to be 4A-α-L-cinerulosyl-3-α-L-rhodinosyl-4C-1D-α-L-rhodinosyl-aquayamycin and named saquayamycin J. This is the first angucycline with a disaccharide side chain consisting of the same sugar building blocks (α-L-rhodinose) attached to C-3
Figure 7.
1H-1H-COSY (bold lines) and selected HMBC (→) correlations of saquayamycin J (4).
Figure 8.
Selected NOESY correlations (↔) in saquayamycin J (4).
Saquayamycin K (5)
Compound 5 was isolated as a yellow-orange solid. The two isomers 4 and 5 were separated by HPLC followed by PTLC (see Figure S8). The (−)-HRESI MS suggested the molecular formula C43H52O16 for 5 with a Δm/z 4 amu higher than saquayamycin A (7), corresponding to two less double bond equivalents. Compound 5 has the same mass as saquayamycin J (4). The 1H and 13C NMR spectrum of compound 5 was similar to those of saquayamycins A (7) and J (4), except in the olefinic region, one set of double bond protons of an L-aculose moiety were observed, not two as in 7. This indicates that one of the L-aculose moieties of saquayamycin A (7) was reduced to an L-rhodinose moiety to give compound 5. The overall structure was investigated by 2D NMR studies (Figures 9 and 10; Tables 3 and 4) to determine, at which positions the L-aculose and L-rhodinose moieties were located. In the HMBC spectrum, a 3J coupling was observed from the L-rhodinose anomeric proton at δ 4.87 (δC 98.9) to C-4A (δC 89.2) and from H-4A (δ 3.01) to C-1B (δC 98.9) fixing the linkage of the L-rhodinose sugar moiety at C-4A of the D-olivose (sugar A). The structure of 5 and its configuration was finally determined to be 4A-α-L-rhodinosyl-3-α-L-rhodinosyl-4C-1D-α-Laculosyl- aquayamycin (Figures 9 and 10), and subsequently named saquayamycin K. An α-L-rhodinosyl sugar moiety (B) was reported in the same position in A-7884 (16), where it is inserted between a D-olivose and an L-aculose residue. The fact that a second α-L-rhodinose moiety was found in two different positions in the isomers 4 and 5 indicates again either a high flexibility of the corresponding glycosyltransferase or two different glycosyltransferases, similar to the glycosyltransferase responsible for the L-rednose transfer in saquamycins H (2) and I (3).
Figure 9.
1H-1H-COSY (bold lines) and selected HMBC long range couplings (→) of saquayamycin K (5).
Figure 10.
Selected NOESY correlations (↔) in Saquayamycin K (5).
Table 3.
1H NMR assignments (500 MHz, CDCl3) of saquayamycins A (7), J (4), K (5) and B1 (6), δ in ppm relative to TMS (multiplicity, J/Hz).
| Position | Saquayamycin A (7)a) | Saquayamycin J (4)a) | Saquayamycin K (5)a) | Saquayamycin B1 (6)a) |
|---|---|---|---|---|
|
| ||||
| δH | δH | δH | δH | |
| 2 | 2.47, d (13.5, Ha), 3.13, dd (13.5, 2.5, He) | 2.47, d (13.0, Ha), 3.16, dd (13.0, 3.0, He) | 2.49, d (13.5, Ha), 3.16, dd (13.5, 3.0, He) | 2.60, d (13.5, Ha), 2.93, dd (12.5, 2.5, He) |
| 3-CH3 | 1.36, s | 1.38, s | 1.38, s | 1.27, s |
| 3-OH | - | - | - | 3.90, brs b) |
| 4 | 1.81, d (15.0, Ha), 2.24, dd (15.5, 3.0, He) | 1.81, d (15.5, Ha), 2.24, dd (15.0, 2.5, He) | 1.82, d (15.5, Ha), 2.26, dd (15.5, 3.0, He) | 1.82, d (15.0, Ha) 2.25, dd (14.0, 2.0, He) |
| 4a-OH | 4.30, brs | 4.38, brs | 4.31, brs | 3.54, brs b) |
| 5 | 6.40, d (10.0) | 6.42, d (10.0) | 6.42, d (10.0) | 6.38, d (10.0) |
| 6 | 6.86, d (9.5) | 6.87, d (9.5) | 6.89, d (10.0) | 6.87, d (9.5) |
| 8-OH | 12.25, s | 12.27, s | 12.26, s | 12.26, s |
| 10 | 7.83, d (8.0) | 7.85, d (8.0) | 7.85, d (8.0) | 7.87, d (7.5) |
| 11 | 7.56, d (7.5) | 7.57, d (8.0) | 7.58, d (7.5) | 7.59, d (8.0) |
| 12b-OH | 4.56, brs | 4.56, brs | 4.57, brs | 4.98, brs b) |
| Sugar A, β-D-olivose | ||||
| 1A | 4.84, brd (11.0) | 4.93, brd (9.5) | 4.84, brd (10.0) | 4.94, brd (10.0) |
| 2A | 1.45, m (Ha), 2.48, m (He) | 1.41, m (Ha), 2.41, ddd, 12.5, 4.5, 2.0, He) | 1.33 m (Ha), 2.49 m (He) | 1.40, m (Ha), 2.42, ddd (12.5, 4.5, 2.0, He) |
| 3A | 3.86, m | 3.78, m | 3.79, m | 3.79, m |
| 3A-OH | 4.23, brs | - | 3.46, brs | - |
| 4A | 3.17, dd (9.0, 8.5) | 3.45, dd (9.0, 9.0) | 3.01, dd (8.5, 8.5) | 3.46, dd (9.0, 9.0) |
| 5A | 3.53, m | 3.54, m | 3.53, m | 3.55, m |
| 6A | 1.35, d (6.0) | 1.37, d (6.0) | 1.33, d (6.5) | 1.38, d (6.0) |
| Sugar B, α-L-aculose or α-L-cinerulose or α-L-rhodinose | ||||
| 1B | 5.34, d (3.5) | 5.15, d (3.0) | 4.87, brs | 5.16, d (3.0) |
| 2B | 6.81, dd (10.0, 3.5) | 4.31, brm | 1.90–1.80, m (Ha, He) | 4.32, brq (3.0) |
| 3B | 6.10, d (10.5) | 2.60, dd (17.5, 3.0, Ha), 2.65, dd (17.5, 3.5, He) | 1.20, m (Ha), 1.90, m (He) | 2.60, dd (16.5, 3.0, Ha), 2.66, dd (17.5, 3.5, He) |
| 4B | - | - | 3.32, brm | - |
| 4B-OH | - | - | 4.93, brs | - |
| 5B | 4.72, q (6.5) | 4.69, q (7.0) | 3.84, m | 4.70, q (7.0) |
| 6B | 1.39, d (7.0) | 1.34, d (7.0) | 1.29, d (6.0) | 1.35, d (6.5) |
| Sugar C, α-L-rhodinose | ||||
| 1C | 5.21, brs | 5.23, brd (2.5) | 5.23, brs | - |
| 2C | 1.45, m (Ha), 1.88, m (He) | 1.42, m (Ha), 1.80, m (He) | 1.45, m (Ha), 1.90, m (He) | - |
| 3C | 1.99–1.88, m (Ha, He) | 1.76, m (Ha, He) | 2.01–1.90, m (Ha, He) | - |
| 4C | 3.65, brs | 3.54, brm | 3.67, brs | - |
| 5C | 4.18, m | 4.15, m | 4.22, m | - |
| 6C | 1.24, d (6.0) | 1.20, d (6.0) | 1.26, d (7.0) | - |
| Sugar D, α-L-aculose or α-L-rhodinose | ||||
| 1D | 5.22, d (3.5) | 4.76, brd (2.5) | 5.24, d (3.5) | - |
| 2D | 6.85, dd (9.5, 3.5) | 1.70, m (Ha), 1.95, m (He) | 6.86, dd (10.0, 3.5) | - |
| 3D | 6.04, d (10.0) | 1.80, m (Ha, He) | 6.07, d (10.5) | - |
| 4D | - | 3.26, brm | - | - |
| 4D-OH | - | 5.27, brs | - | - |
| 5D | 4.50, q (7.0) | 3.61, m | 4.52, q (6.5) | - |
| 6D | 1.32, d (7.0) | 1.21, d (6.0) | 1.34, d (7.0) | - |
See also Figures S32–37, S40–45 and S48–59,
Assignments may be interchangeable among the signals.
Saquayamycin B1 (6)
Compound 6 was isolated as yellow solid, after several chromatographic purifications of the main fraction FI. The (−)HRESI MS-derived molecular formula C31H32O12, 224 amu less than saquayamycins A (7) and B (8), indicates two missing sugar residues. The UV data and the proton NMR spectrum of compound 6 revealed its similarity to saquayamycin B (8), except the protons and carbon atoms of the sugar residues C (L-rhodinose) and D (L-aculose) were absent (Tables 1 and 3). The chemical shift of C-3 was upfield (δ 76.3; in saquayamycin B, δC 82.7), due to the attachement of a free hydroxyl group. Thus, compound 6 was identified as saquayamycin B1, which had been previously reported together with saquayamycins A1 (11), and C1 (12), obtained through acid hydrolysis of saquayamycins B (8), A (7), and C (9), respectively,4, 5 although not as a natural product. The reported NMR data of saquayamycin B1 were identical to those from our isolated compound 6. We further confirmed the structure of 6 as saquayamycin B1 by full NMR assignments using 2D NMR experiments (Figure 11). Based on NOESY experiments (Figure 12), coupling constants and comparison with the reported data of saquayamycin B1,4 compound 6 was shown to have the same configuration as saquayamycin B1.
Figure 11.
1H-1H-COSY (bold lines) and selected HMBC (→) correlations of saquayamycin B1 (6).
Figure 12.
Selected NOESY (↔) correlations in saquayamycin B1 (6).
Cytotoxicity assays
The cytotoxic activity of saquayamycins G-K (1–5) and B1 (6), saquayamycins A and B (7 and 8) in comparsion with landomycin A22, 37–39 was determined using PC3 (Prostate cancer) and H460 (non-small cell lung cancer) cell lines (Figures S66 and S67, and Table 5). The results indicate that the cytotoxic activity of the molecules is altered corresponding to their substitution and connectivity patterns (Table 5). The compounds with ether linkage between C-3A and C-2B of the sugar D-olivose (A) and L-cinerulose (B) sugar moieties showed slightly better activity than the opened one, indicating that the rigidity of these two doubly-linked sugar moieties may play an important role in improving the activity. Notably, compounds with fewer sugar residues are less active. The the α,β-conjugated double bond (Michael acceptor) in the L-aculose moiety of saquayamycin B (8) is advantageous compared to the saturated n L-rhodinose sugar moiety in the same position of saquayamycin J (4). In the H460 cell line, saquayamycin H (2) with the rare L-rednose sugar moiety was slightly more active than saquayamycin B (8) which has the L-aculose moiety in the same position, suggesting a potential improvement of activity through the amino group located at β-position of the L-aculose moiety. Currently, we are investigating the molecular mechanism by which saquayamycins exert their cytotoxic effects on both PC3 and H460 cells.
Table 5.
Cytotoxic activity of the new angucyclines 1–6, compared to the previously known saquayamycins A (7) and B (8) and with the angucyclin lead compound landomycin A37–39 (GI50 values, μM).
| No. | Name and Structure | PC3 cells - 48hr | H460 cells - 48hr | ||
|---|---|---|---|---|---|
| GI50 (μM) | 95% Confidence Intervals | GI50 (μM) | 95% Confidence Intervals | ||
| 1: | Saquayamycin G (R1 = I, R2 = X) | 0.5535 | 0.4879 – 0.6280 | 6.718 | 4.771 – 9.459 |
| 2: | Saquayamycin H (R1 = II, R2 = XI) | 0.7898 | 0.7354 – 0.8482 | 3.302 | 2.102 – 5.188 |
| 3: | Saquayamycin I (R1 = III, R2 = X) | 0.9149 | 0.8087 – 1.035 | 7.943 | 5.557 – 11.35 |
| 4: | Saquayamycin J (R1 = II, R2 = XII) | 0.1791 | 0.1425 – 0.2251 | 5.694 | 4.284 – 7.567 |
| 5: | Saquayamycin K (R1 = V, R2 = X) | 0.1478 | 0.09491 – 0.2303 | 7.281 | 5.519 – 9.605 |
| 6: | Saquayamycin B1 (R1 = II, R2 = H) | 1.759 | 0.2774 – 11.16 | 13.20 | 7.965 – 21.87 |
| 7: | Saquayamycin A (R1 = IV, R2 = X) | 0.1057 | 0.09697 – 0.1153 | 6.195 | 4.617 – 8.312 |
| 8: | Saquayamycin B (R1 = II, R2 = X) | 0.07454 | 0.06720 – 0.08267 | 3.929 | 2.895 – 5.333 |
| - | Landomycin A | 0.5505 | 0.4982 – 0.6081 | 4.109 | 2.548 – 6.626 |
Experimental Section
General Experimental Procedures
UV spectra were recorded on a Shimadzu UV-1800 (Model TCC-240A) UV spectrometer. NMR spectra were measured on Varian Vnmr 500 (1H, 500 MHz; 13C, 125 MHz) spectrometer. ESIMS was recorded on a Finnigan LCQ ion trap mass spectrometer. HRMS was recorded by ESIMS on an Agilent LC/MSD TOF (Resolution: 10,000; 3 ppm mass accuracy; inlet systems: Agilent Technologies 1200 Series LC pumps) Mass Spectrometer (Manufacturer: Agilent Palo Alto, CA, USA). HPLC purifications were carried out using a Symmetry Prep C18 7μm column (7.8 × 300 mm) on a binary LC system (Solvent A: 0.2% aq. formic acid, solvent B: acetonitrile; flow rate: 2.0 mL min−1; 0–15 min, 75-0% A (linear gradient), 15–20 min 0% A and 100% B, 20 – 22 min 0 – 75% A (linear gradient), 22 – 27 min 75% A). HPLC-MS analyses were carried out using a Symmetry Anal C18 5μm column (4.6 × 250 mm) on a binary LC system. Flash chromatography was carried out on silica gel MN 60 (140–270 mesh ASTM). Rf values were measured on Polygram SIL G/UV254 (Macherey-Nagel & Co.). Size exclusion chromatography was performed using Sephadex LH-20 (GE Healthcare).
Cell Viability Assay
Prostate cancer cell line PC3 and non-small cell lung cancer cell line H460 were used to determine the cytotoxicity of saquayamycins G-K (1–5), B1 (6), A (7), B (8) and landomycin A. Cells were plated in 96 well plates at a density of 5 × 103 cells per well in 100 μL of full growth media and allowed to adhere overnight. The following day, media was replaced with 100 μL fresh media containing DMSO control or serial concentrations of the test compounds and plates were incubated for 48 h at 37 °C. At the end of the incubation period, 10 μL of Resazurin (Sigma-Aldrich, St. Louis, MO) was added to each well and further incubated for 3 hr at 37 °C. Cell viability was determined by measuring the fluorescence at 560 nm excitation wavelength and 590 nm emission wavelength using a Molecular Devices Spectramax M5 plate reader. Fluorescence was measured at the time of treatment (time zero) and subtracted from the fluorescence values obtained after 48 h treatment. Fifty percent growth inhibition (GI50) was measured using non linear regression analysis and by fitting a sigmoidal dose response curve to the data using Graphpad Prism (Graph Pad software Inc., Version 5.0). The well-studied cytostatic angucycline lead compound landomycin A was used as a positive control standard.37–39 Experiments were performed in four replicates.
SG-Medium
Glucose (20.0 g), yeast extract (5.0 g), Soytone (10.0 g), CoCl2·6H2O (1.0 mg) and calcium carbonate (2.0 g) were dissolved in 1 L of demineralized water. The suspension (pH 7.2) was sterilized by autoclaving for 33 min at 121 °C.
M2-Agar
Glucose (4.0 g), yeast extract (4.0 g), malt extract (10.0 g) and agar (15.0 g) were dissolved in 1 L of demineralized water, then sterilized for 33 min at 121 °C. The medium was adjusted to pH 7.2 with 2 N NaOH before sterilization.
Fermentation, Extraction and Isolation
Streptomyces sp. KY40-1 (originally isolated, purified and taxonomically identified by M. K. Kharel from a soil sample collected from the foothills of the Appalachian mountains near Cave Run Lake, Kentucky, USA, maintained as glycerol spore suspension at −80 °C and named as KY002)7 was cultivated on M2-agar plates at 28 °C for 3 days. With pieces of well-grown agar cultures of Streptomyces sp. KY40-1, a 250 mL Erlenmeyer flask pre-culture containing 100 mL of SG-medium, was inoculated and cultivated at 28 °C (250 rpm) for three days. The obtained 100 mL pre-culture was used to inoculate 60 250 mL Erlenmeyer flasks (each with 1 mL preculture), each containing 100 mL of SG-medium and incubated at 28 °C and 250 rpm. After 4 days the culture broth was harvested. The obtained reddish-brown culture broth was mixed with Celite and filtered affording the mycelium and water phases. The mycelium was extracted with EtOAc (4 × 300 mL), sonicated and filtered, while the water phase was extracted with EtOAc (3 × 2 L). Extracts were combined and evaporated to dryness under vacuum at 35 °C, giving 2.30 g of a dark-red solid crude extract, whose chromatographic purification yielded known metabolites and six new congeners (1–6): The crude extract (2.30 g) was chromatographed on silica gel (column 2 × 50 cm) using a stepwise MeOH-CH2Cl2 gradient (0–100% MeOH) and monitoring by TLC to yield fractions FI (1.0 g, yellow-orange solid), FII (0.1 g, orange solid), FIII (0.2 g, orange-red solid), FIV (0.15 g, orange-red solid), FV (0.1 g, orange solid), FVI (0.95 g, orange solid), and FVII (0.1 g, yellow- orange solid) (see also supporting information Figure S1). Purification of fraction FI using HPLC (SymmetryPrep™ C18 7μm, 7.8 × 300 mm column; MeCN-H2O, 0.2% formic acid; flow rate 2.0 mL min−1) afforded saquayamycin A (7; orange-red solid, 26.3 mg) and saquayamycin B (8; yellow-orange powder, 95.3 mg) along with the sub-fraction FIC. Further fractionation and purification of the sub-fraction FIC using PTLC (see Figure S1, for the PTLC eluting system conditions) and Sephadex LH-20 (2 × 50 cm, 50 % MeOH-CH2Cl2) yielded saquayamycins B1 (6; yellow solid, 2.3 mg), J (4; yellow-orange solid, 10.7 mg) and K (5; yellow-orange solid, 8.3 mg), and further separation and purification of fractions FIII and FIV following figure S1 gave the saquayamycins G (1; orange-red solid, 10.5 mg), H (2; orange-red solid, 8.3 mg), and I (3; orange-red solid, 9.2 mg) along with 4′,7-dihydroxyisoflavanone (white solid, 8.7 mg). Fractions FII, FV-FVII were excluded based on the TLC and HPLC-MS analysis, since no major products were isolated in enough amounts for NMR characterization (Figure S1).
Saquayamycin G (1)
Orange-red solid; Rf 0.56 (silica gel, MeOH/CH2Cl2 7:93 v/v), blue-violet coloration with 2N NaOH; UV/vis (MeOH) λmax (log ε) 218 (4.50), 330 sh (4.05), 433 (4.14), 468 sh (4.11) nm; 1H NMR (CDCl3, 500 MHz), see Table 1; 13C NMR (CDCl3, 125 MHz), see Table 2; (−)-APCI MS m/z 709 [M−H]−; (−)-ESI MS m/z 709 [M−H]−; (+)-ESI MS m/z 711 [M+H]+; (−)-HRESIMS m/z 709.2501 [M−H]− (calcd for C37H41O14, 709.2502) and m/z 691.2393 [M–H2O–H]− (calcd for C37H39O13, 691.2396).
Saquayamycin H (2)
Orange-red solid; Orange fluorescence under long UV (365 nm); Rf 0.47 (silica gel, MeOH/CH2Cl2 7:93 v/v); Blue-violet coloration with 2N NaOH; UV/vis (MeOH) λmax (log ε) 218 (4.66), 281 (4.59), 330 sh (4.14), 433 (4.27), 468 sh (4.22) nm; 1H NMR (CDCl3, 500 MHz), see Table 1; 13C NMR (CDCl3, 125 MHz), see Table 2; (−)-APCI MS m/z 834 [M−H]−; (−)-ESI MS m/z 834 [M−H]−; (+)-ESI MS m/z 836 [M+H]+; (−)-HRESIMS m/z 834.2968 [M−H]− (calcd for C43H48NO16, 834.2978) and m/z 816.2862 [M–H2O–H]− (calcd for C43H46NO15, 816.2872).
Saquayamycin I (3)
Orange-red solid; Orange fluorescence under long UV (365 nm); Rf 0.29 (silica gel, MeOH/CH2Cl2 7:93 v/v); Blue-violet coloration with 2N NaOH; UV/vis (MeOH) λmax (log ε) 218 (4.65), 281 (4.50), 329 sh (4.15), 435 (4.25), 468 sh (4.20) nm; 1H NMR (Acetone-d6, 500 MHz), see Table 1; 13C NMR (Acetone-d6, 125 MHz), see Table 2; (−)-APCI MS m/z 834 [M−H]−; (−)-ESI MS m/z 834 [M−H]−; (+)-ESI MS m/z 836 [M+H]+; (−)-HRESIMS m/z 834.2968 [M−H]− (calcd for C43H48NO16, 834.2978) and m/z 816.2862 [M–H2O–H]− (calcd for C43H46NO15, 816.2872).
Saquayamycin J (4)
Yellow-orange solid; UV absorbing (254 nm), Orange-red fluorescence under long UV (365 nm); Rf 0.41 (silica gel, MeOH/CH2Cl2 7:93 v/v), 0.17 (silica gel, 70% EtOAc/n-Hexane); Blue-violet coloration with 2N NaOH; UV/vis (MeOH) λmax (log ε) 218 (4.61), 328 sh (4.15), 431 (4.25), 468 sh (4.20) nm; 1H NMR (CDCl3, 500 MHz), see Table 3; 13C NMR (CDCl3, 125 MHz), see Table 4; (−)-APCI MS m/z 823 [M−H]−; (−)-ESI MS m/z 823 [M−H]−; (+)-ESI MS m/z 825 [M+H]+; (−)-HRESIMS m/z 823.3189 [M−H]− (calcd for C43H51O16, 823.3182), and m/z 805.3085 [M–H2O–H]− (calcd for C43H49O15, 805.3077).
Saquayamycin K (5)
Yellow-orange solid; UV absorbing (254 nm), Orange-red fluorescence under long UV (365 nm); Rf 0.42 (silica gel, MeOH/CH2Cl2 7:93 v/v), 0.21 (silica gel, 70% EtOAc/n-Hexane); Blue-violet coloration with 2N NaOH; UV/vis (MeOH) λmax (log ε) 218 (4.66), 328 sh (4.15), 434 (4.26), 468 sh (4.21) nm; 1H NMR (CDCl3, 500 MHz), see Table 3; 13C NMR (CDCl3, 125 MHz), see Table 4; (−)-APCI MS m/z 823 [M−H]−; (−)-ESI MS m/z 823 [M−H]−; (+)-ESI MS m/z 825 [M+H]+; (−)-HRESIMS m/z 823.3189 [M−H]− (calcd for C43H51O16, 823.3182), and m/z 805.3085 [M–H2O–H]− (calcd for C43H49O15, 805.3077).
Saquayamycin B1 (6)
Yellow solid; UV absorbing (254 nm); Rf 0.50 (silica gel, MeOH/CH2Cl2 7:93 v/v); Blue-violet coloration with 2N NaOH; UV/vis (MeOH) λmax (log ε) 218 (4.23), 328 sh (3.88), 429 (3.98), 468 sh (3.94) nm; 1H NMR (CDCl3, 500 MHz), see Table 3; 13C NMR (CDCl3, 125 MHz), see Table 2; (−)-APCI MS m/z 595 [M−H]−; (−)-ESI MS m/z 595 [M−H]−; (+)-ESI MS m/z 597 [M+H]+; (−)-HRESIMS m/z 595.1821 [M−H]− (calcd for C31H31O12, 595.1821), m/z 577.1724 [M–H2O–H]− (calcd for C31H29O11, 577.1715), m/z 559.1604 [M–2H2O–H]− (calcd for C31H27O10, 559.1609), and m/z 1191.3703 [2M–H]− (calcd for C62H63O24, 1191.3714).
Saquayamycin A (7)
Orange-red solid; UV absorbing (254 nm); Orange-red fluorescence under long UV (365 nm); Rf 0.69 (silica gel, MeOH/CH2Cl2 7:93 v/v); Blue-violet coloration with 2N NaOH; 1H NMR (CDCl3, 500 MHz), see Table 3; 13C NMR (CDCl3, 125 MHz), see Table 4; (−)-APCI MS m/z 819 [M−H]−.
Saquayamycin B (8)
Yellow-orange powder; UV absorbing (254 nm); Orange-red fluorescence under long UV (365 nm); Rf 0.70 (silica gel, MeOH/CH2Cl2 7:93 v/v); Blue-violet coloration with 2N NaOH; UV/vis (MeOH) λmax (log ε) 218 (4.50), 330 sh (4.12), 428 (4.24), 468 sh (4.20) nm; 1H NMR (CDCl3, 500 MHz), see Table 1; 13C NMR (CDCl3, 125 MHz), see Table 2; (−)-APCI MS m/z 819 [M−H]−.
Supplementary Material
Acknowledgments
We thank the mass spectrometry facility of the Biotechnology center of the University of Wisconsin for the HRESI mass spectra. This work was supported by grants CA 102102 and CA 091901of the National Institutes of Health to J.R.
Footnotes
ASSOCIATED CONTENT: HPLC analysis chromatogram of the crude extract obtained from Streptomyces sp. KY40-1; workup procedure scheme; HRMS, NMR and UV spectra of angucyclines (1–8); TLC of strain extract, cultivation figures and PTLC isolation of saquayamycins J (4), and K (5). This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES AND NOTES
- 1.Krohn K, Rohr J. Topics Curr Chem. 1997;188:127–195. [Google Scholar]
- 2.Rohr J, Thiericke R. Nat Prod Rep. 1992;9:103–137. doi: 10.1039/np9920900103. [DOI] [PubMed] [Google Scholar]
- 3.Kharel MK, Pahari P, Shepherd MD, Tibrewal N, Nybo SE, Shaaban KA, Rohr J. Nat Prod Rep. 2012;29:264–325. doi: 10.1039/c1np00068c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uchida T, Imoto M, Watanabe Y, Miura K, Dobashi T, Matsuda N, Sawa T, Naganawa H, Hamada M, Takeuchi T. J Antibiot. 1985;38:1171–1181. doi: 10.7164/antibiotics.38.1171. [DOI] [PubMed] [Google Scholar]
- 5.Sekizawa R, Iinuma H, Naganawa H, Hamada M, Takeuchi T, Yamaizumi J, Umezawa K. J Antibiot. 1996;49:487–490. doi: 10.7164/antibiotics.49.487. [DOI] [PubMed] [Google Scholar]
- 6.Ströch K, Zeeck A, Antal N, Fiedler HP. J Antibiot. 2005;58:103–110. doi: 10.1038/ja.2005.13. [DOI] [PubMed] [Google Scholar]
- 7.Abdelfattah MS, Kharel MK, Hitron JA, Baig I, Rohr J. J Nat Prod. 2008;71:1569–1573. doi: 10.1021/np800281f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henkel T, Zeeck A. J Antibiot. 1990;43:830–837. doi: 10.7164/antibiotics.43.830. [DOI] [PubMed] [Google Scholar]
- 9.Rohr J, Zeeck A. J Antibiot. 1987;40:459–467. doi: 10.7164/antibiotics.40.459. [DOI] [PubMed] [Google Scholar]
- 10.Rohr J, Zeeck A, Floss HG. J Antibiot. 1988;41:126–129. doi: 10.7164/antibiotics.41.126. [DOI] [PubMed] [Google Scholar]
- 11.Rohr J, Beale JM, Floss HG. J Antibiot. 1989;42:1151–1157. doi: 10.7164/antibiotics.42.1151. [DOI] [PubMed] [Google Scholar]
- 12.Rohr J. J Chem Soc, Chem Commun. 1990:113–114. [Google Scholar]
- 13.Rohr J. Angew Chem Int Ed Engl. 1990;29:1051–1053. [Google Scholar]
- 14.Rohr J. J Antibiot. 1989;42:1482–1488. doi: 10.7164/antibiotics.42.1482. [DOI] [PubMed] [Google Scholar]
- 15.Rix U, Remsing LL, Hoffmeister D, Bechthold A, Rohr J. ChemBioChem. 2003;4:109–111. doi: 10.1002/cbic.200390002. [DOI] [PubMed] [Google Scholar]
- 16.Rohr J. J Chem Soc, Chem Commun. 1989:492–493. [Google Scholar]
- 17.Fedoryshyn M, Nur-e-Alam M, Zhu L, Luzhetskyy A, Rohr J, Bechthold A. J Biotechnol. 2007;130:32–38. doi: 10.1016/j.jbiotec.2007.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henkel T, Rohr J, Beale JM, Schwenen L. J Antibiot. 1990;43:492–503. doi: 10.7164/antibiotics.43.492. [DOI] [PubMed] [Google Scholar]
- 19.Weber S, Zolke C, Rohr J, Beale JM. J Org Chem. 1994;59:4211–4214. [Google Scholar]
- 20.Zhu L, Luzhetskyy A, Luzhetska M, Mattingly C, Adams V, Bechthold A, Rohr J. ChemBioChem. 2007;8:83–88. doi: 10.1002/cbic.200600360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou M, O’Doherty GA. Org Lett. 2008;10:2283–2286. doi: 10.1021/ol800697k. [DOI] [PubMed] [Google Scholar]
- 22.Ostash B, Korynevska A, Stoika R, Fedorenko V. Mini Rev Med Chem. 2009;9:1040–1051. doi: 10.2174/138955709788922593. [DOI] [PubMed] [Google Scholar]
- 23.Shaaban KA, Srinivasan S, Kumar R, Damodaran C, Rohr J. J Nat Prod. 2011;74:2–11. doi: 10.1021/np100469y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaaban KA, Stamatkin C, Damodaran C, Rohr J. J Antibiot. 2011;64:141–150. doi: 10.1038/ja.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sezaki M, Kondo S, Maeda K, Umezawa H, Ono M. Tetrahedron. 1970;26:5171–90. doi: 10.1016/s0040-4020(01)98726-5. [DOI] [PubMed] [Google Scholar]
- 26.Ohta K, Kamiya K. J Chem Soc Chem Commun. 1981:154–155. [Google Scholar]
- 27.Ohta K, Mizuta E, Okazaki H, Kishi T. Chem Pharm Bull. 1984;32:4350–4359. [Google Scholar]
- 28.Alvi KA, Baker DD, Stienecker V, Hosken M, Nair BG. J Antibiot. 2000;53:496–501. [PubMed] [Google Scholar]
- 29.Erb A, Luzhetskyy A, Hardter U, Bechthold A. ChemBioChem. 2009;10:1392–1401. doi: 10.1002/cbic.200900054. [DOI] [PubMed] [Google Scholar]
- 30.Ren X, Lu X, Ke A, Zheng Z, Lin J, Hao W, Zhu J, Fan Y, Ding Y, Jiang Q, Zhang H. J Antibiot. 2011;64:339–343. doi: 10.1038/ja.2011.4. [DOI] [PubMed] [Google Scholar]
- 31.Ding C, Tu S, Li F, Wang Y, Yao Q, Hu W, Xie H, Meng L, Zhang A. J Org Chem. 2009;74:6111–6119. doi: 10.1021/jo9011078. [DOI] [PubMed] [Google Scholar]
- 32.Martin GDA, Tan LT, Jensen PR, Dimayuga RE, Fairchild CR, Raventos-Suarez C, Fenical W. J Nat Prod. 2007;70:1406–1409. doi: 10.1021/np060621r. [DOI] [PubMed] [Google Scholar]
- 33.Schneemann I, Kajahn I, Ohlendorf B, Zinecker H, Erhard A, Nagel K, Wiese J, Imhoff JF. J Nat Prod. 2010;73:1309–1312. doi: 10.1021/np100135b. [DOI] [PubMed] [Google Scholar]
- 34.Sezaki M, Hara T, Ayukawa S, Takeuchi T, Okami Y. J Antibiot. 1968;21:91–97. doi: 10.7164/antibiotics.21.91. [DOI] [PubMed] [Google Scholar]
- 35.Johdo O, Yoshioka T, Naganawa H, Takeuchi T, Yoshimoto A. J Antibiot. 1996;49:669–675. doi: 10.7164/antibiotics.49.669. [DOI] [PubMed] [Google Scholar]
- 36.Nettleton DE, Jr, Balitz DM, Doyle TW, Bradner WT, Johnson DL, O’Herron FA, Schreiber RH, Coon AB, Moseley JE, Myllymaki RW. J Nat Prod. 1980;43:242–258. doi: 10.1021/np50008a003. [DOI] [PubMed] [Google Scholar]
- 37.Depenbrock H, Bornschlegl S, Peter R, Rohr J, Schmid P, Schweighart P, Block T, Rastetter J, Hanauske AR. Ann Hematol. 1996;73(Supl II):A80/316. [Google Scholar]
- 38.Korynevska A, Heffeter P, Matselyukh B, Elbling L, Micksche M, Stoika R, Berger W. Biochem Pharmacol. 2007;74:1713–1726. doi: 10.1016/j.bcp.2007.08.026. [DOI] [PubMed] [Google Scholar]
- 39.Crow RT, Rosenbaum B, Smith R, 3rd, Guo Y, Ramos KS, Sulikowski GA. Bioorg Med Chem Lett. 1999;9:1663–1666. doi: 10.1016/s0960-894x(99)00261-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.