Abstract
The human cytomegalovirus (HCMV) is suspected to increase tumour malignancy by infection of cancer and/or stroma cells (oncomodulation). So far, oncomodulatory mechanisms have been attributed to the presence of HCMV and direct action of its gene products on cancer cells. Here, we investigated whether the prolonged presence of HCMV can result in the irreversible selection of a cancer cell population with increased malignancy. The neuroblastoma cell line UKF-NB-4 was long-term (200 passages) infected with the HCMV strain Hi91 (UKF-NB-4Hi) before virus eradication using ganciclovir (UKF-NB-4HiGCV). Global gene expression profiling of UKF-NB-4, UKF-NB-4Hi and UKF-NB-4HiGCV cells and subsequent bioinformatic signal transduction pathway analysis revealed clear differences between UKF-NB-4 and UKF-NB-4Hi, as well as between UKF-NB-4 and UKF-NB-4HiGCV cells, but only minor differences between UKF-NB-4Hi and UKF-NB-4HiGCV cells. Investigation of the expression of a subset of five genes in different chronically HCMV-infected cell lines before and after virus eradication suggested that long-term HCMV infection reproducibly causes specific changes. Array comparative genomic hybridisation showed virtually the same genomic differences for the comparisons UKF-NB-4Hi/UKF-NB-4 and UKF-NB-4HiGCV/UKF-NB-4. UKF-NB-4Hi cells are characterised by an increased invasive potential compared with UKF-NB-4 cells. This phenotype was completely retained in UKF-NB-4HiGCV cells. Moreover, there was a substantial overlap in the signal transduction pathways that differed significantly between UKF-NB-4Hi/UKF-NB-4HiGCV and UKF-NB-4 cells and those differentially regulated between tumour tissues from neuroblastoma patients with favourable or poor outcome. In conclusion, we present the first experimental evidence that long-term HCMV infection can result in the selection of tumour cell populations with enhanced malignancy.
Keywords: human cytomegalovirus, neuroblastoma, oncomodulation, long-term infection, cancer cell malignancy
Introduction
Human cytomegalovirus (HCMV), a ubiquitous herpes virus, infection leads (in immunocompetent individuals usually symptomless) to a life-long persistence after primary infection. About 50 to 100% of the general adult population is infected. HCMV routinely reactivates, but this is usually controlled by the host immune response in healthy people. In immuno-compromised individuals such as recipients of organ transplants or AIDS patients, HCMV is a major pathogen.1, 2
HCMV has been suspected to have a role in cancer diseases for decades, based on seroepidemiological evidence and on the detection of viral DNA, messenger RNA (mRNA) and/or antigens in tumour tissues.2, 3 However, in contrast to viruses from different RNA virus (retrovirus, flavivirus) and DNA virus (hepadna virus, herpes virus, papova virus) families that are known to be oncogenic in humans,4 HCMV is not considered a tumour virus because of a lack of proven transformation potential in human cells.2 With our studies about the influence of HCMV on cancer cells,5, 6, 7 we introduced the concept of oncomodulation, meaning that HCMV may infect cancer cells and/or stromal cells in established tumours and increase tumour malignancy also in the absence of transformation potential.1, 2, 3, 6, 8, 9 In the meantime, oncomodulatory effects exerted by HCMV or single cytomegalovirus proteins have been reported in vitro and in vivo by numerous groups.2, 3, 8, 9, 10, 11 Moreover, application of sensitive (although not yet indisputably accepted) pathological methods applied by numerous independent research groups indicated the presence of HCMV and/or virus constituents in cancers from different cancer entities.1, 2, 3, 12, 13, 14, 15, 16, 17 In glioblastomas, the presence of HCMV was correlated with higher disease stage and worse outcome.1, 2, 3, 12, 18, 19 In addition, expression of HCMV proteins appeared to promote oncogenic signalling events.2, 3, 12, 20, 21
Neuroblastoma, a paediatric cancer entity, has been associated with increased HCMV antibody titres and HCMV immediate-early antigen (IEA) expression in a fraction of tumours.2, 3, 22, 23 After primary HCMV infection of different neuroblastoma cell lines, a balance is established between virus production and cell division.6, 7, 24, 25 Chronically HCMV-infected neuroblastoma cells show a more malignant phenotype indicated by properties such as increased invasive potential, metastasis formation in nude mice and resistance to chemotherapy.2, 6, 7, 24
So far, HCMV-induced oncomodulatory effects were attributed to the presence of HCMV and direct action of its gene products, 2, 3, 10, 24 and therefore suspected to be reversible after virus eradication. Here, we investigated the effects of long-term HCMV strain Hi91 infection on UKF-NB-4 neuroblastoma cells. Long-term HCMV-infected (UKF-NB-4Hi) cells showed a very close relationship with ganciclovir-cured UKF-NB-4Hi (UKF-NB-4HiGCV) cells at the level of gene expression and genomic copy number changes, whereas substantial differences were detected between UKF-NB-4Hi/UKF-NB-4HiGCV cells and parental UKF-NB-4 cells. Moreover, UKF-NB-4HiGCV showed the same increased invasive potential as UKF-NB-4Hi cells compared with UKF-NB-4. Bioinformatics signal transduction pathway analysis suggested a substantial overlap in pathways differentially regulated between UKF-NB-4Hi/UKF-NB-4HiGCV cells and UKF-NB-4 cells, as well as between tumour tissues from neuroblastoma patients with poor or favourable outcome. These data indicate that the long-term presence of HCMV can result in the irreversible selection of a cancer cell population with increased malignancy. Investigation of the expression of a subset of five genes in additional long-term HCMV-infected neuroblastoma cells and their cidofovir- or ganciclovir-cured sub-lines suggested that long-term HCMV infection of different neuroblastoma cells reproducibly results in characteristic changes.
Results
Establishment of chronically HCMV-infected neuroblastoma cells and virus eradication
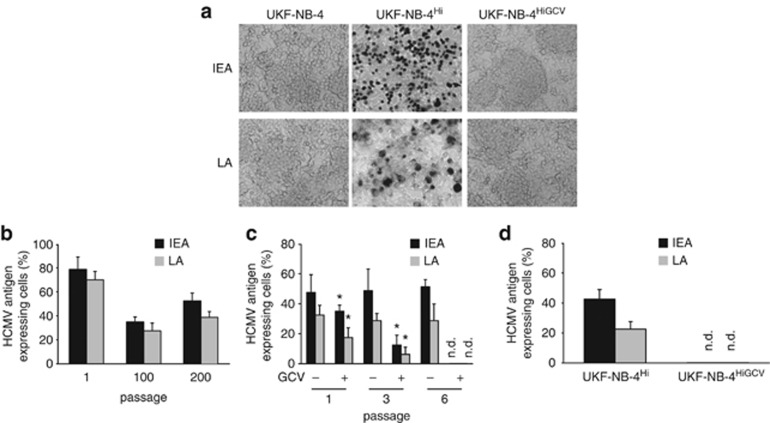
UKF-NB-4 cells, derived from bone marrow metastases of a patient harbouring a MYCN-amplified stage IV neuroblastoma,26 were infected once with the HCMV strain Hi9127 at MOI 10 and then subcultured without further addition of virus (UKF-NB-4Hi). Non-infected UKF-NB-4 cells were passaged in parallel as control. After primary infection, about 80% of UKF-NB-4 cells were HCMV infected (Figure 1). Five days after infection, the amount of viable cells was about 20%, as indicated by trypan blue staining (Supplementary Figure 1). After 200 passages, HCMV IEA and late antigen expression remained detectable in UKF-NB-4Hi cells, resulting in about 30–60% infected cells (Figure 1). Trypan blue staining indicated 70–80% viable cells (5 days after passaging of cells) (Supplementary Figure 1). Virus titres were 8.0 × 102 TCID50 (tissue culture infectious dose)/ml at passage 1 (determined 5 days after primary infection), 4.5 × 102 TCID50/ml at passage 100 and 1.4 × 103 TCID50/ml at passage 200 (both detected 5 days after passaging). HCMV DNA copy numbers were 6.3 × 105/106 cells at passage 1 (determined 5 days after primary infection), 9.8 × 104/106 cells at passage 100 and 9.1 × 105/106 cells at passage 200 (both detected 5 days after passaging).
Figure 1.
Establishment of UKF-NB-4Hi and UKF-NB-4HiGCV cells. UKF-NB-4 cells were infected once with HCMV strain Hi at MOI 10 and then subcultured without further addition of virus (UKF-NB-4Hi). After 200 passages, UKF-NB-4Hi cells were treated for six passages with 20 μM ganciclovir (GCV) until no HCMV IEA or late antigen (LA) expression were detectable anymore. (a) Representative pictures showing immunostaining of UKF-NB-4, UKF-NB-4Hi or UKF-NB-4HiGCV cells for IEA or LA. (b) Fraction of HCMV antigen-expressing cells in UKF-NB-4Hi cells at different passages after initial infection. (c) HCMV antigen expression in GCV-treated UKF-NB-4Hi cells at different passages in comparison with non-treated UKF-NB-4Hi cells. Values represent mean±s.d. from three independent experiments. *P<0.05 relative to non-treated UKF-NB-4Hi cells. (d) HCMV antigen expression in GCV-cured UKF-NB-4Hi (UKF-NB-4HiGCV) cells after cultivation for 10 passages in the absence of GCV in comparison with non-treated UKF-NB-4Hi cells. n.d., not detectable.
Thereafter, UKF-NB-4Hi cells were treated with 20 μM ganciclovir (a concentration that did not affect the viability of UKF-NB-4 or UKF-NB-4Hi cells) for six passages until no HCMV gene expression was detected anymore (UKF-NB-4HiGCV; Figure 1). Further cultivation of UKF-NB-4HiGCV cells for 10 passages did not result in recurrence of HCMV gene expression (Figure 1). The absence of HCMV from UKF-NB-4HiGCV cells was confirmed by virus yield assay and quantitative PCR detecting HCMV genome (Supplementary Figure 2).
Comparison of global gene expression in UKF-NB-4, UKF-NB-4Hi and UKF-NB-4HiGCV cells
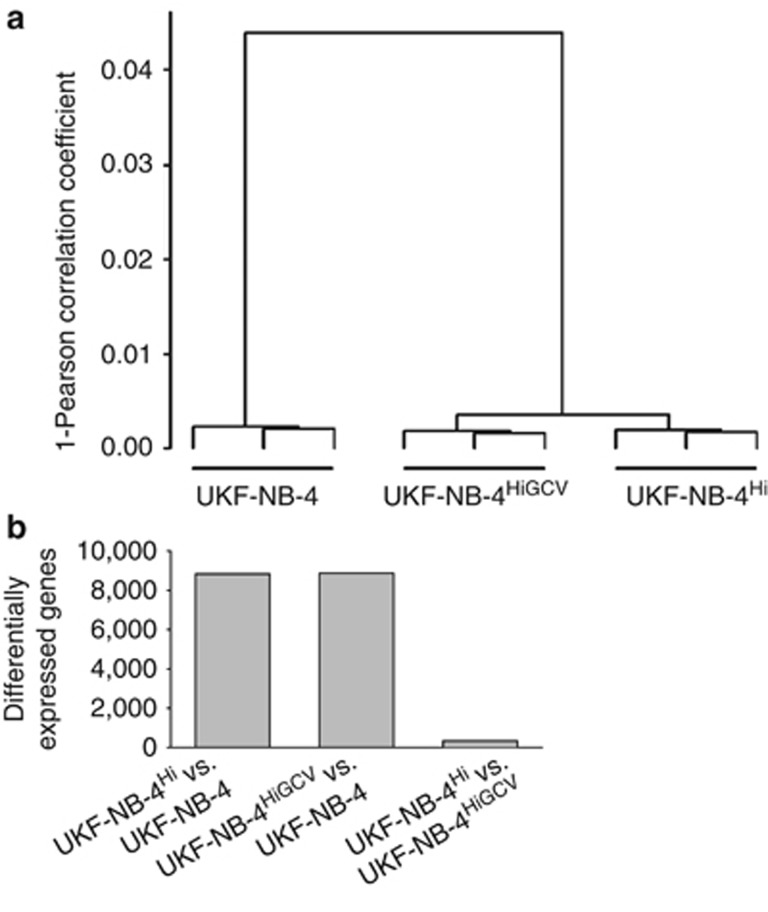
Triplicates of UKF-NB-4, UKF-NB-4Hi and UKF-NB-4HiGCV cells were analysed for global cellular gene expression at the mRNA level using gene microarray (Affymetrix HGU133plus2, Santa Clara, CA, USA). Correlation analysis revealed a very high similarity between gene expression profiles in UKF-NB-4Hi and UKF-NB-4HiGCV cells but clear difference between these two cell lines and UKF-NB-4 (Figure 2a). More than 8000 genes were significantly differentially expressed (false discovery rate (FDR)<0.05, corrected for multiple testing) between UKF-NB-4Hi and UKF-NB-4 cells or UKF-NB-4HiGCV and UKF-NB-4 cells, whereas only about 300 genes were differentially expressed between UKF-NB-4Hi and UKF-NB-4HiGCV cells (Figure 2b, Supplementary Table 1). Further analysis showed that nearly 90% of the genes significantly differentially expressed between UKF-NB-4Hi and UKF-NB-4 cells and between UKF-NB-4HiGCV and UKF-NB-4 cells were changed consistently in both UKF-NB-4Hi and UKF-NB-4HiGCV cells (that is, were both overexpressed or both were underexpressed compared with UKF-NB-4; Supplementary Table 2). These data indicate that both UKF-NB-4Hi and UKF-NB-4HiGCV strongly differ from UKF-NB-4 cells by means of gene expression, whereas gene expression signatures of UKF-NB-4Hi and UKF-NB-4HiGCV were very similar.
Figure 2.
Comparison of global gene expression in UKF-NB-4, UKF-NB-4Hi and UKF-NB-4HiGCV cells by gene microarray. (a) Hierarchical cluster analysis based on the Pearson correlation coefficient. (b) Numbers of genes significantly differentially expressed (FDR<0.05) between the investigated cell lines.
Correlation analysis of UKF-NB-4 cells stored at the beginning of the experiments in parallel with primary infection (UKF-NB-40) and UKF-NB-4 cells that had been cultivated in parallel with the UKF-NB-4Hi cells for 200 passages (UKF-NB-4200) indicated a close relationship between these two passages of the UKF-NB-4 cell line but a substantial difference to UKF-NB-4Hi cells (Supplementary Figure 3).
Bioinformatic signal transduction pathway analysis of global gene expression data
Bioinformatic signal transduction pathway analysis, performed using the PANTHER database (www.pantherdb.org), showed that the top five differentially regulated pathways for the comparisons UKF-NB-4Hi vs UKF-NB-4 and UKF-NB-4HiGCV vs UKF-NB-4 were the same, although the order differed (Table 1). In total, 13 pathways were significantly differentially regulated in both comparisons (P-value<0.05, corrected for multiple testing; Table 1), whereas only three pathways were significantly differentially regulated between UKF-NB-4Hi and UKF-NB-4 cells but not between UKF-NB-4HiGCV and UKF-NB-4 cells (Supplementary Table 3). Four pathways were significantly differentially regulated between UKF-NB-4HiGCV and UKF-NB-4 cells but not between UKF-NB-4Hi and UKF-NB-4 cells (Supplementary Table 4). No significant differentially regulated pathways were detected between UKF-NB-4Hi and UKF-NB-4HiGCV, confirming the high similarity of these cells at the level of gene expression.
Table 1. Signal transduction pathways that are significantly differentially regulated (P<0.05, corrected for multiple testing) in UKF-NB-4Hi vs UKF-NB-4 and UKF-NB-4HiGCV vs UKF-NB-4 cells as identified by PANTHER pathway analysis.
| Pathway (number of genes annotated in the pathway) | UKF-NB-4Hi vs UKF-NB-4 (ranka/P-value/genes affectedb) | UKF-NB-4HiGCV vs UKF-NB-4 (ranka/P-value/genes affectedb) |
|---|---|---|
| Wnt signalling (348) | 1/1.76 × 10−10/191 | 1/3.32 × 10−10/190 |
| Ras pathway (91) | 2/1.57 × 10−6/64 | 3/8.14 × 10−6/62 |
| Ubiquitin proteasome pathway (89) | 3/3.69 × 10−6/62 | 4/8.48 × 10−6/61 |
| Cadherin signalling pathway (168) | 4/1.12 × 10−5/96 | 2/6.21 × 10−6/97 |
| Parkinson disease (106) | 5/7.04 × 10−5/66 | 5/1.72 × 10−5/68 |
| Angiogenesis (229) | 6/3.38 × 10−4/115 | 12/8.50 × 10−3/108 |
| Integrin signalling pathway (227) | 7/6.04 × 10−4/113 | 9/2.51 × 10−3/110 |
| EGF receptor signalling pathway (150) | 8/8.68 × 10−4/81 | 6/8.91 × 10−4/81 |
| PDGF signalling pathway (187) | 9/8.69 × 10−4/97 | 11/6.33 × 10−3/93 |
| p53 pathway (136) | 11/2.94 × 10−3/73 | 7/9.63 × 10−4/75 |
| Huntington disease (172) | 12/1.57 × 10−3/87 | 8/4.23 × 10−3/89 |
| p53 pathways feedback loops 2 (66) | 13/1.02 × 10−2/41 | 13/1.00 × 10−2/41 |
| Cytoskeletal regulation by Rho GTPase (111) | 16/3.93 × 1−2/58 | 16/3.86 × 10−2/58 |
Abbreviations: EGF, epidermal growth factor; PDGF, platelet-derived growth factor.
Pathways were ranked by their P-values with the lowest P-value being rank 1.
Pathway genes differentially expressed between the data sets (FDR<0.05). In total, 25 431 genes were annotated in the PANTHER reference list. 8832 genes were differentially expressed between UKF-NB-4Hi and UKF-NB-4, 8873 between UKF-NB-4HiGCV and UKF-NB-4.
Gross genomic differences of UKF-NB-4Hi and UKF-NB-4HiGCV cells compared with UKF-NB-4 cells by array comparative genomic hybridization (CGH)
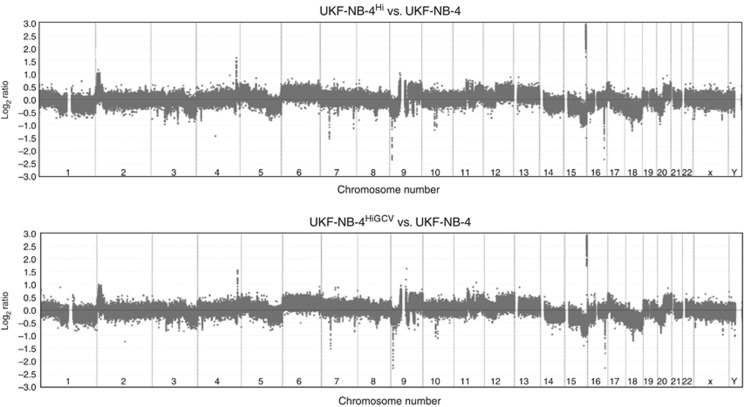
Array CGH is a technology developed to detect genomic copy number variations. UKF-NB-4 cells were compared with UKF-NB-4Hi cells or UKF-NB-4HiGCV cells by array CGH in order to investigate whether genomic differences underlie the observed changes in gene expression. Results revealed virtually identical array CGH profiles in DNA copy number for both the comparisons UKF-NB-4Hi vs UKF-NB-4 and UKF-NB-4HiGCV vs UKF-NB-4 (Figure 3). There is evidence of aneuploidy, chromosome segment gain and loss and individual loci loss/gain. Specifically, we detected aneuploidy (gain) of chromosomes 6, 13 and 20, with loss of 14, 15 and 18. Structural gains were observed at terminal 2p, the whole of 9q (with a small portion of 9p), 12q, terminal 17p and 20q. Losses are detected of chromosome 1q (and about a third of proximal 1p), parts of chromosome 3, terminal 5q, a central portion of 8q, most of terminal 9p, terminal 15q (in addition to the aforementioned aneuploidy) and 20p. Reciprocal gains and losses of p and q arms on chromosomes 9 and 20 (and possibly chromosome 12) may indicate the presence of isochromosomes. Single locus gains (gene amplifications) were apparent on chromosomes 4q and 15q, with losses on 7p, 7q, 9p, 10 (near the centromere), 15q and 16q.
Figure 3.
Array CGH of all 24 human chromosomes resulting from comparisons of UKF-NB-4Hi cells (upper plot) and UKF-NB-4HiGCV cells (bottom plot), both compared with UKF-NB-4 cells. Results suggest that both UKF-NB-4Hi and UKF-NB-4HiGCV have very similar karyotypes: whole-chromosome gain and loss (e.g., on chromosomes 6 and 18), losses and gains of chromosome segments, e.g., terminal 2p and 12q, as well as amplification and loss on individual loci.
UKF-NB-4, UKF-NB-4Hi or UKF-NB-4HiGCV adhesion to and transmigration through endothelial monolayers
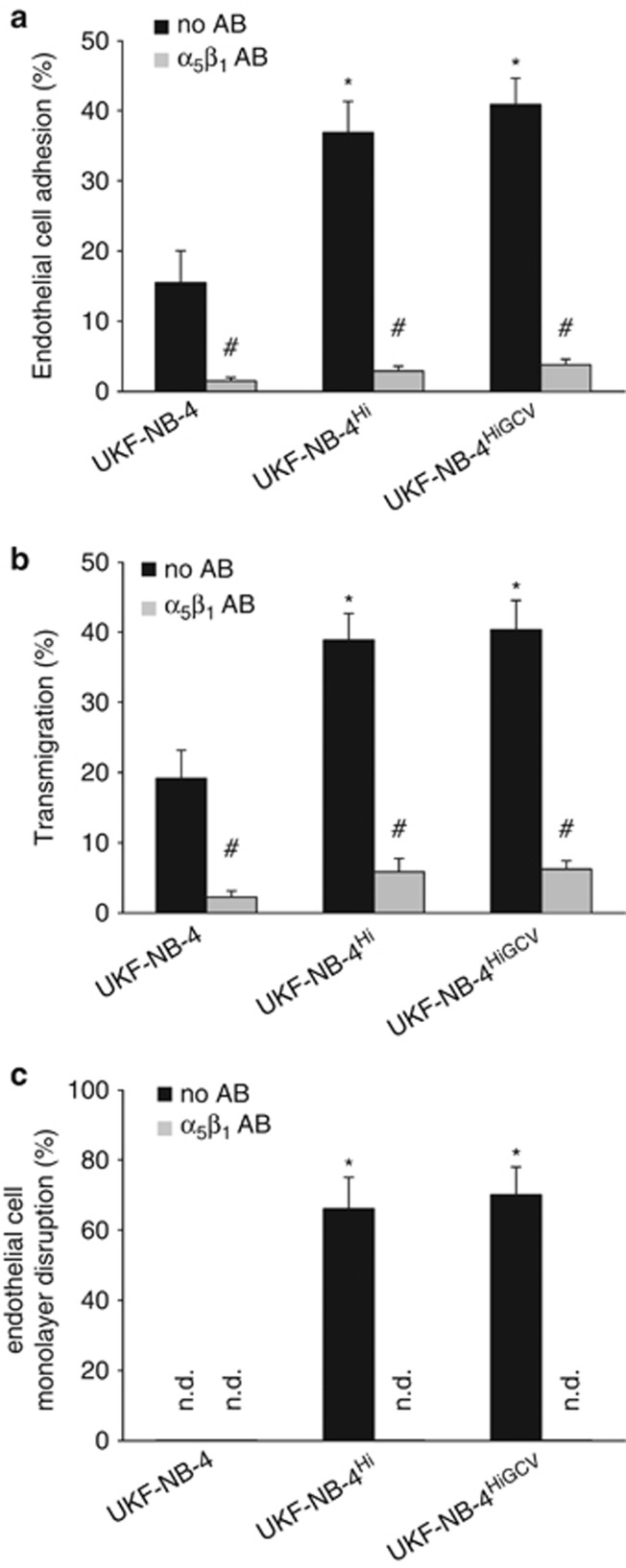
UKF-NB-4, UKF-NB-4Hi and UKF-NB-4HiGCV were investigated for cancer cell adhesion to and migration through human umbilical vein endothelial cell (HUVEC) monolayers. These events are considered to be important during metastasis formation. HCMV-infected neuroblastoma cells had been shown earlier to exhibit a stronger invasive potential than non-infected cells.28 UKF-NB-4Hi and UKF-NB-4HiGCV cells both showed a strongly increased adhesion to endothelial cells (Figure 4a), as well as enhanced numbers of transmigrating cells (Figure 4b) when compared with UKF-NB-4. Moreover, UKF-NB-4Hi and UKF-NB-4HiGCV cells (but not UKF-NB-4 cells) disrupted endothelial cell monolayer integrity, resulting in focal plaques (that is, round openings that are not associated with endothelial cell HCMV infection or death as described28) (Figure 4c). Previous results had shown that interaction of UKF-NB-4 cells with endothelial cells depended on α5β1 integrin binding.28 Here, adhesion of UKF-NB-4, UKF-NB-4Hi and UKF-NB-4HiGCV cells to and transmigration through endothelial cell monolayers could be inhibited by an antibody blocking α5β1 integrin (Figures 4a and b). Moreover, antibody-mediated inhibition of α5β1 integrin binding also interfered with endothelial cell monolayer disruption by UKF-NB-4Hi or UKF-NB-4HiGCV cells.
Figure 4.
Adhesion of UKF-NB-4, UKF-NB-4Hi and UKF-NB-4HiGCV cells to endothelial cell monolayers, transmigration and endothelial cell monolayer distruction. (a) Fractions of cells that adhered to HUVEC monolayers (in the presence or absence of an α5β1-blocking antibody (a, b)) were expressed as the percentage of the total number of input cells. (b) Fractions of cells that transmigrated through HUVEC monolayers (in the presence or absence of an α5β1-blocking antibody (a, b)) were expressed as the percentage of the total number of adherent cells. (c) Focal endothelial cell monolayer disruption (in the presence or absence of an α5β1-blocking antibody (a, b)) was expressed as the percentage of the cell-free area. Values represent mean±s.d. from three independent experiments. *P<0.05 relative to UKF-NB-4; #P<0.05 relative to the corresponding cell line in the absence of α5β1-blocking antibody; n.d., not detectable; AB, antibody.
These findings suggest that the enhanced malignant properties of UKF-NB-4Hi cells are at least partly retained in UKF-NB-4HiGCV cells after and despite virus eradication.
Signal transduction pathways differentially regulated between UKF-NB-4 and UKF-NB-4Hi/UKF-NB-4HiGCV cells and between tumours from neuroblastoma patients with favourable or non-favourable outcome.
Next, signal transduction pathways differentially regulated between UKF-NB-4 and UKF-NB-4Hi/UKF-NB-4HiGCV cells were analysed in the context of two gene expression data sets derived from neuroblastoma patients. The first data set (Oberthuer et al.) was derived from a study comparing gene expression signatures between N-myc-amplified neuroblastoma tissues from patients with favourable outcome (event-free survival) or poor outcome (death or relapse of disease) (www.ebi.ac.uk/arrayexpress; accession E-TABM-38; Oberfhuer et al.29). The second data set (Asgharzadeh et al.) investigated gene expression in non-N-myc-amplified neuroblastoma tissues from patients with favourable outcome (no relapse) or poor outcome (relapse of disease) (www.ncbi.nlm.nih.gov/geo/; accession number GSE344630). In the Oberthuer et al. data set, 21 out of the 153 PANTHER pathways were significantly differentially regulated (P<0.05, corrected for multiple testing) between favourable outcome and poor outcome patients. Nine of the 13 pathways that had been found to be differentially regulated between UKF-NB-4 and UKF-NB-4Hi/UKF-NB-4HiGCV were also significantly differentially regulated between favourable and poor outcome samples in this data set (Table 2). In the Asgharzadeh et al. data set, 32 out of the 153 pathways were significantly differentially regulated, and 12 of the 13 pathways differentially regulated between UKF-NB-4 and UKF-NB-4Hi/UKF-NB-4HiGCV were also significantly differentially regulated between favourable and poor outcome patients (Table 2).
Table 2. Analysis of signalling pathways significantly differentially expressed (P<0.05, corrected for multiple testing) between UKF-NB-4Hi and UKF-NB-4HiGCV cells and UKF-NB-4 in data sets of neuroblastoma patients27, 28 PANTHER pathway analysis.
| Pathway | Oberthuer et al.27,a | Asgharzadeh et al.28,b |
|---|---|---|
| |
Death/relapse from disease vs no relapse rankc (P-value) |
Relapse vs no relapse rankc (P-value) |
| Wnt signalling | 14 (2.33 × 10−3) | 1 (4.13 × 10−14) |
| Ras pathway | 4 (6.70 × 10−7) | 14 (1.58 × 10−4) |
| Ubiquitin proteasome pathway | 44 (4.82 × 10−1) | 15 (1.84 × 10−4) |
| Cadherin signalling pathway | 76 (1.00 × 100) | 23 (4.44 × 10−3) |
| Parkinson disease | 9 (2.49 × 10−4) | 27 (1.49 × 10−2) |
| Angiogenesis | 1 (5.60 × 10−13) | 2 (1.31 × 10−9) |
| Integrin signalling pathway | 10 (2.67 × 10−4) | 11 (1.63 × 10−5) |
| EGF receptor signalling pathway | 11 (5.11 × 10−4) | 6 (4.90 × 10−7) |
| PDGF signalling pathway | 3 (4.53 × 10−8) | 4 (2.74 × 10−8) |
| p53 pathway | 5 (5.57 × 10−6) | 8 (8.60 × 10−7) |
| Huntington disease | 42 (4.34 × 10−1) | 3 (2.15 × 10−8) |
| p53 pathways feedback loops 2 | 6 (6.63 × 10−6) | 39 (1.18 × 10−1) |
| Cytoskeletal regulation by Rho GTPase | 86 (1.00 × 100) | 18 (4.98 × 10−4) |
Abbreviations: EGF, epidermal growth factor; PDGF, platelet-derived growth factor.
21 out of 153 pathways were significantly differentially regulated (P<0.05).
32 out of 153 pathways were significantly differentially regulated (P<0.05).
Pathways were ranked according to their P-values for the comparison of non-favourable vs favourable outcome with the lowest P-value being rank 1.
Moreover, 7 of the 13 pathways (Angiogenesis, platelet-derived growth factor signalling pathway, Ras pathway, p53 pathway, p53 pathway feedback loops 2, Parkinson disease, Integrin signalling pathway) belonged to the 10 most strongly differentially regulated pathways of the Oberthuer et al. data set. Six of the 13 pathways (Wnt signalling pathway, Angiogenesis, Huntington disease, platelet-derived growth factor signalling pathway, epidermal growth factor receptor signalling pathway, p53 pathway) belonged to the 10 most strongly differentially regulated pathways of the Asgharzadeh et al. data set.
Discussion
Oncomodulation is a concept postulating that HCMV may infect cancer cells and/or stromal cells in established tumours and increase tumour malignancy also in the absence or independently of transformation potential.2, 3, 6 To study long-term effects of HCMV on cancer cells, chronically HCMV-infected neuroblastoma cell cultures were established.6, 7, 24, 25 Here, we show that long-term HCMV infection of UKF-NB-4 neuroblastoma cells results in the establishment of a cell population that is clearly distinct from the parental UKF-NB-4 cell line regarding global gene expression and genomic imbalances. Eradication of HCMV from chronically HCMV strain Hi91-infected UKF-NB-4 (UKF-NB-4Hi) cells causes only minor effects on gene expression in the resulting UKF-NB-4HiGCV cell line as indicated by comparison of differentially expressed genes, as well as by bioinformatics signal transduction pathway analyses. Genomic imbalances are virtually the same between UKF-NB-4Hi and UKF-NB-4 cells and between UKF-NB-4HiGCV and UKF-NB-4 cells as indicated by array CGH.
UKF-NB-4Hi cells show an increased invasive phenotype indicating enhanced tumour cell malignancy compared with UKF-NB-4 cells. This more invasive phenotype is completely sustained in ganciclovir-cured UKF-NB-4HiGCV cells. Thirteen signal transduction pathways are significantly differentially regulated between UKF-NB-4 cells and both UKF-NB-4Hi and UKF-NB-4HiGCV cells. The majority of these pathways are also significantly differentially regulated between tissues from neuroblastoma patients with favourable outcome and those from patients with unfavourable disease course (9 out of 13 in the Oberthuer et al. data set,29 and 12 out of 13 in the Asgharzadeh et al. data set30). This demonstrates that long-term HCMV infection of neuroblastoma cells can result in the establishment of a novel cell population with different malignant properties compared with the parental cell line. Moreover, malignant features, at least in part, no longer depend on the presence of HCMV or its gene products after long-term HCMV infection.
To enhance the confidence that the observed effects are associated with the presence of HCMV and to receive some more information about the specificity of the HCMV-induced selection process, five genes were selected that had been shown earlier to be significantly differentially expressed in various chronically HCMV-infected neuroblastoma cell lines relative to the corresponding non-infected cell lines: DHFR (encoding for dihydrofolate reductase), ENPP2 (encoding for autotaxin), KIAA0101 (encoding for a protein that is also known as PCNA-associated factor/p15(PAF)), NDRG1 (encoding for N-myc downstream regulated 1) and TMPRSS15 (also known as PRSS7, encoding for a protein known as enterokinase or transmembrane protease, serine 15).25 These genes were also significantly differentially regulated between UKF-NB-4 and both UKF-NB-4Hi and UKF-NB-4HiGCV cells (as indicated by gene expression microarray data and confirmed by real-time PCR, Supplementary Table 5, Supplementary Figure 4), and between MHH-NB-11 cells chronically infected with HCMV strain AD169 (MHH-NB-11AD169,25) and its sub-lines in which the virus was eradicated by the addition of ganciclovir (MHH-NB-11AD169GCV) or cidofovir (MHH-NB-11AD169CDF) compared with parental MHH-NB-11 cells (Supplementary Figure 5).
Acute infection of UKF-NB-4 cells with HCMV strain Hi91 did not affect the expression of DHFR, ENPP2, KIAA0101, NDRG1 or TMPRSS15. Moreover, chronic infection of UKF-NB-4 cells with varicella zoster virus strains did not influence the expression of these genes (data not shown), indicating that the expression of these genes is not commonly influenced by the long-term presence of different members of the herpes virus family (or a consequence of genetic drift). Together, these findings support the idea that the specific adaptation and selection processes during chronic HCMV infection reproducibly result in novel cell populations with characteristics clearly distinct from the parental cell lines.
Various processes might be involved in the observed establishment of a novel, more malignant neuroblastoma cell population due to long-term HCMV infection. In fully permissive normal cells such as human fibroblasts, HCMV results in cell cycle arrest and subsequent cell lysis.2, 31 However, (at least a fraction of) chronically HCMV-infected cell cultures must escape from HCMV-induced cell cycle block and death. In this context, cell cycle deregulation is regarded as ‘hallmark' of cancer, 32 and HCMV may not be able to induce an effective cell cycle block in certain cancer cells.2 HCMV antigen-positive glioblastoma cells were shown to divide, resulting in two HCMV antigen-expressing progeny cells.33 In sarcoma or glioblastoma cells, virus variants arose that replicated more slowly and produced lower virus yields than the original strain used for primary infection.2, 34, 35 Therefore, changes of virus properties may contribute to the selection of a more malignant cell fraction in our model, although HCMV strain Hi91 reisolated from UKF-NB-4Hi cells showed virtually the same growth kinetics after acute infection in UKF-NB-4 cells and human foreskin fibroblasts as the original virus used for primary infection.
Stress conditions activate cellular survival signalling pathways, resulting in a condition thought to favour adaptation processes in malignant cells.36 In this context, HCMV is known to exert pro-inflammatory effects and to activate crucial events in survival signalling, such as NFκB, PI3K-Akt, MAPKs and/or JNK, and inhibit apoptosis by effects on p53 and p73, as well as by enhancing expression of anti-apoptotic proteins such as bcl-2.1, 2, 37 Moreover, HCMV is suspected to promote chromosomal instability in infected cells.2, 38 Stable expression of the HCMV UL76 protein was sufficient to induce chromosomal aberrations in glioblastoma cells.39 Indeed, the long-term presence of HCMV had resulted in chromosomal imbalances in UKF-NB-4Hi cells in our experiments. As HCMV is known to also modify epigenetic features such as histone acetylation of infected cells,19, 40, 41, 42 epigenetic mechanisms are also likely to contribute.
Finally, cancer cells are characterised by chromosomal instability, resulting in clonal evolution, that is, the permanent appearance and disappearance of novel (sub-)clones (characterised by various mutations and/or genomic aberrations) that compete for space and resources.32, 43 Therefore, HCMV may also exert a selection pressure on clones initially present in a cancer cell population or on clones that develop in the presence, but without the involvement, of HCMV. Five metaphases from UKF-NB-4, UKF-NB-4Hi or UKF-NB-4HiGCV cells were investigated by spectral karyotyping. Apparently, six abnormalities are consistently found in cells from all three cell lines, suggesting the cells to be of a common origin (Supplementary Table 6). Nevertheless, the long-term presence of HCMV may exert a selection pressure that favours the growth of (a) specific cell clone(s).
In conclusion, we present evidence that long-term HCMV infection of neuroblastoma cells can lead to the selection of a novel cell population with increased malignant properties. It is the first report demonstrating that HCMV infection can irreversibly modulate the malignant properties of cancer cells in such a way that the HCMV-induced enhanced cancer cell malignancy is retained (at least in part) even when the virus is eradicated from the cancer cells. This indicates that HCMV can act as a selection agent in cancer cells. Therefore, our findings represent a profound extension to the concept of oncomodulation2, 3, 6 and a novel way by which viruses may influence cancer diseases.
Materials and methods
Immunostaining for HCMV antigen expression
As described previously,44 cells producing HCMV-specific antigens were detected by immunoperoxidase staining using monoclonal antibodies directed against the UL123-coded 72-kDa IEA1 (DuPont, Bad Homburg, Germany) or late antigen gB (kindly provided by K Radsak, Institut für Virologie, Marburg, Germany).
Virus propagation
HCMV Strain Hi91 was isolated from the urine of an AIDS patient with HCMV retinitis.27 Virus stocks were prepared in human foreskin fibroblasts maintained in minimal essential medium with 4% foetal calf serum. The titres were determined by plaque titration as described previously.44
Virus yield assay
The amount of infectious virus was determined by virus yield assay in a single-cycle assay format using human foreskin fibroblasts as described before.44 Virus titres were expressed as 50% of tissue culture infectious dose (TCID50) 120 h post infection.
Cells
UKF-NB-4 cells derived from bone marrow metastases of a patient harbouring a MYCN-amplified stage IV neuroblastoma were used.26 Cells were grown at 37 °C in Iscove's modified Dulbecco's medium supplemented with 10% heat-inactivated foetal calf serum and containing 100 IU/ml of penicillin and 100 μg/ml streptomycin. HUVECs were cultivated as described before.44
Global gene expression analysis
Triplicates of UKF-NB-4, UKF-NB-4Hi and UKF-NB-4HiGCV cells were analysed for global cellular gene expression at the mRNA level using GeneChip HGU133 Plus 2.0 (Affymetrix) by the Fraunhofer Institut für Zelltherapie und Immunologie (Leipzig, Germany). mRNA was isolated using the RNeasy kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Expression data were processed using the R/bioconductor packages ‘gcrma' and ‘limma' (www.r-project.org/; www.bioconductor.org/) in order to detect fold changes and FDRs.45 FDRs were corrected for multiple testing after Benjamini and Hochberg.46 Hierarchical clustering analysis was performed and heatmaps were visualised using R (www.r-project.org).
Signal transduction pathway bioinformatics
Statistical analysis to identify significant expression changes was focusing on a pathway analysis using the PANTHER database, which identifies global patterns in expression.47 For each expert-curated pathway in the database, potential differential expression was determined by a binomial test48 with subsequent Bonferroni correction for multiple testing, using the PANTHER human gene reference list matching our microarrays and lists of differentially expressed genes that passed a FDR threshold of 0.05.
Array CGH
DNA was isolated from UKF-NB-4, UKF-NB-4Hi and UKF-NB-4HiGCV cell lines using the DNAeasy Blood & Tissue kit (Qiagen) following the manufacturer's instructions. Using DNA from cell line UKF-NB-4 as reference cell lines, UKF-NB-4Hi and UKF-NB-4HiGCV were profiled for DNA copy number changes by applying the Nimblegen oligonucleotide microarray platform. Hybridisations on Nimblegen HG18_WG_CGH_v2D_HX1 microarrays and microarray data processing were carried out at Source BioScience imaGenes (Berlin, Germany). The chips used contain 2.5 mio oligonucleotides tiling the human genome at an average probe spacing of 1.2 kb. Microarray images were processed in the NimbleScan 2 software (Roche NimbleGen, Madison, WI, USA).
Adhesion and transmigration assay
Neuroblastoma cell adhesion to and transmigration through HUVEC monolayers was determined as previously shown.28 HUVECs were pretreated with 3-aminopropyl-triethoxy-silan (2% Sigma-Aldrich Chemie GmbH, Munich, Germany). Neuroblastoma cells were added. After 4 h, non-adherent neuroblastoma cells were washed off. Further, cells were fixed with 1% (w/v) glutaraldehyde (Merck KGaA, Darmstadt, Germany). Adherent cells were counted in six different fields (5 × 0.25 mm2) using phase contrast microscopy. Transmigrating neuroblastoma cells were detected using a reflection interference contrast microscope with a Phloem apparatus (Nikon, Düsseldorf, Germany).
HUVEC monolayer disruption was detected and quantified, as described earlier28 after 24 h of cocultivation of HUVEC monolayers with UKF-NB-4, UKF-NB-4Hi or UKF-NB-4HiGCV cells.
For blocking experiments, the α5β1 antibody VLA-5 (clone SAM-1, Hycultec GmbH, Beutelsbach, Germany) was used.
Quantitative real-time PCR
Total RNA was isolated from cell cultures using TRI reagent (Sigma-Aldrich). Quantitative real-time reverse transcriptase PCR (qRT–PCR) for viral mRNA was performed as described previously.44 Cellular mRNA was detected using TaqMan Gene Expression Assays (Applied Biosystems, Darmstadt, Germany).
DNA was purified using the DNeasy Blood & Tissue Kit (Qiagen). Viral DNA quantification was carried out by real-time PCR as described before.44
Acknowledgments
We thank Gesa Meincke for technical support. The work was supported by the charity Hilfe für krebskranke Kinder Frankfurt e.V. and its trust Frankfurter Stiftung für krebskranke Kinder. (MM, FR, JC, JC Jr).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Oncogenesis website (http://www.nature.com/oncsis)
Supplementary Material
References
- Söderberg-Nauclér C. HCMV microinfections in inflammatory diseases and cancer. J Clin Virol. 2008;41:218–223. doi: 10.1016/j.jcv.2007.11.009. [DOI] [PubMed] [Google Scholar]
- Michaelis M, Doerr HW, Cinatl J. The story of human cytomegalovirus and cancer: increasing evidence and open questions. Neoplasia. 2009;11:1–9. doi: 10.1593/neo.81178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis M, Baumgarten P, Mittelbronn M, Hernáiz Driever P, Doerr HW, Cinatl J. Oncomodulation by human cytomegalovirus: Novel clinical findings open new roads. Med Microbiol Immunol. 2011;200:1–5. doi: 10.1007/s00430-010-0177-7. [DOI] [PubMed] [Google Scholar]
- Moore PS, Chang Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat Rev Cancer. 2010;10:878–889. doi: 10.1038/nrc2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinatl J, Cinatl J, Radsak K, Rabenau H, Weber B, Novak M, et al. Replication of human cytomegalovirus in a rhabdomyosarcoma cell line depends on the state of differentiation of the cells. Arch Virol. 1994;138:391–401. doi: 10.1007/BF01379143. [DOI] [PubMed] [Google Scholar]
- Cinatl J, Cinatl J, Vogel JU, Rabenau H, Kornhuber B, Doerr HW. Modulatory effects of human cytomegalovirus infection on malignant properties of cancer cells. Intervirology. 1996;39:259–269. doi: 10.1159/000150527. [DOI] [PubMed] [Google Scholar]
- Cinatl J, Vogel JU, Cinatl J, Weber B, Rabenau H, Novak M, et al. Long-term productive human cytomegalovirus infection of a human neuroblastoma cell line. Int J Cancer. 1996;65:90–96. doi: 10.1002/(SICI)1097-0215(19960103)65:1<90::AID-IJC16>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Barami K. Oncomodulatory mechanisms of human cytomegalovirus in gliomas. J Clin Neurosci. 2010;17:819–823. doi: 10.1016/j.jocn.2009.10.040. [DOI] [PubMed] [Google Scholar]
- Kofman A, Marcinkiewicz L, Dupart E, Lyshchev A, Martynov B, Ryndin A, et al. The roles of viruses in brain tumor initiation and oncomodulation. J Neurooncol. 2011;105:451–466. doi: 10.1007/s11060-011-0658-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maussang D, Verzijl D, van Walsum M, Leurs R, Holl J, Pleskoff O, et al. Human cytomegalovirus-encoded chemokine receptor US28 promotes tumorigenesis. Proc Natl Acad Sci USA. 2006;103:13068–13073. doi: 10.1073/pnas.0604433103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongers G, Maussang D, Muniz LR, Noriega VM, Fraile-Ramos A, Barker N, et al. The cytomegalovirus-encoded chemokine receptor US28 promotes intestinal neoplasia in transgenic mice. J Clin Invest. 2010;120:3969–3978. doi: 10.1172/JCI42563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soroceanu L, Cobbs CS. Is HCMV a tumor promotor. Virus Res. 2011;157:193–203. doi: 10.1016/j.virusres.2010.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkins LE, Matlaf LA, Soroceanu L, Klemm K, Britt WJ, Wang W, et al. Detection of human cytomegalovirus in normal and neoplastic breast epithelium. Herpesviridae. 2010;1:8. doi: 10.1186/2042-4280-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziurzynski K, Wei J, Qiao W, Hatiboglu MA, Kong LY, Wu A, et al. Glioma-associated cytomegalovirus mediates subversion of the monocyte lineage to a tumor propagating phenotype. Clin Cancer Res. 2011;17:4642–4649. doi: 10.1158/1078-0432.CCR-11-0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Zhu H, Shenk T. Human cytomagalovirus IE1 and IE2 proteins are mutagenic and mediate “hit-and-run” oncogenic transformation in cooperation with the adenovirus E1A proteins. Proc Natl Acad Sci USA. 1997;94:3341–3345. doi: 10.1073/pnas.94.7.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baryawno N, Rahbar A, Wolmer-Solberg N, Taher C, Odeberg J, Darabi A, et al. Detection of human cytomegalovirus in medulloblastomas reveals a potential therapeutic target. J Clin Invest. 2011;121:4043–4055. doi: 10.1172/JCI57147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan P, Clark PA, Kuo JS, Salamat MS, Kalejta RF. Significant association of multiple human cytomegalovirus genomic loci with glioblastoma multiforme samples. J Virol. 2012;86:854–864. doi: 10.1128/JVI.06097-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheurer ME, Bondy ML, Aldape KD, Albrecht T, El-Zein R. Detection of human cytomegalovirus in different histological types of gliomas. Acta Neuropathol. 2008;116:79–86. doi: 10.1007/s00401-008-0359-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saddawi-Konefka R, Crawford JR. Chronic viral infection and primary central nervous system malignancy. J Neuroimmune Pharmacol. 2010;5:387–403. doi: 10.1007/s11481-010-9204-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strååt K, Liu C, Rahbar A, Zhu Q, Liu L, Wolmer-Solberg N, et al. Activation of telomerase by human cytomegalovirus. J Natl Cancer Inst. 2009;101:488–497. doi: 10.1093/jnci/djp031. [DOI] [PubMed] [Google Scholar]
- Slinger E, Maussang D, Schreiber A, Siderius M, Rahbar A, Fraile-Ramos A, et al. HCMV-encoded chemokine receptor US28 mediates proliferative signaling through the IL-6-STAT3 axis. Sci Signal. 2010;3:ra58. doi: 10.1126/scisignal.2001180. [DOI] [PubMed] [Google Scholar]
- Wertheim P, Voute PA. Neuroblastoma Wilms' tumor, and cytomegalovirus. J Natl Cancer Inst. 1976;57:701–703. doi: 10.1093/jnci/57.3.701. [DOI] [PubMed] [Google Scholar]
- Nigro G, Schiavetti A, Booth JC, Clerico A, Dominici C, Krzysztofiak A, et al. Cytomegalovirus-associated stage 4S neuroblastoma relapsed stage 4. Med Pediatr Oncol. 1995;24:200–203. doi: 10.1002/mpo.2950240311. [DOI] [PubMed] [Google Scholar]
- Cinatl J, Cinatl J, Vogel JU, Kotchetkov R, Driever PH, Kabickova H, et al. Persistent human cytomegalovirus infection induces drug resistance and alteration of programmed cell death in human neuroblastoma cells. Cancer Res. 1998;58:367–372. [PubMed] [Google Scholar]
- Hoever G, Vogel JU, Lukashenko P, Hofmann WK, Komor M, Doerr HW, et al. Impact of persistent cytomegalovirus infection on human neuroblastoma cell gene expression. Biochem Biophys Res Commun. 2005;326:395–401. doi: 10.1016/j.bbrc.2004.11.042. [DOI] [PubMed] [Google Scholar]
- Cinatl J, Cinatl J, Kotchetkov R, Vogel JU, Woodcock BG, Matousek J, et al. Bovine seminal ribonuclease selectively kills human multidrug-resistant neuroblastoma cells via induction of apoptosis. Int J Oncol. 1999;15:1001–1009. doi: 10.3892/ijo.15.5.1001. [DOI] [PubMed] [Google Scholar]
- Cinatl J, Margraf S, Vogel JU, Scholz M, Cinatl J, Doerr HW. Human cytomegalovirus circumvents NF-kappa B dependence in retinal pigment epithelial cells. J Immunol. 2001;167:1900–1908. doi: 10.4049/jimmunol.167.4.1900. [DOI] [PubMed] [Google Scholar]
- Scholz M, Blaheta RA, Wittig B, Cinatl J, Vogel JU, Doerr HW, et al. Cytomegalovirus-infected neuroblastoma cells exhibit augmented invasiveness mediated by beta1alpha5 integrin (VLA-5) Tissue Antigens. 2000;55:412–421. doi: 10.1034/j.1399-0039.2000.550503.x. [DOI] [PubMed] [Google Scholar]
- Oberthuer A, Berthold F, Warnat P, Hero B, Kahlert Y, Spitz R, et al. Customized oligonucleotide microarray gene expression-based classification of neuroblastoma patients outperforms current clinical risk stratification. J Clin Oncol. 2006;24:5070–5078. doi: 10.1200/JCO.2006.06.1879. [DOI] [PubMed] [Google Scholar]
- Asgharzadeh S, Pique-Regi R, Sposto R, Wang H, Yang Y, Shimada H, et al. Prognostic significance of gene expression profiles of metastatic neuroblastomas lacking MYCN gene amplification. J Natl Cancer Inst. 2006;98:1193–1203. doi: 10.1093/jnci/djj330. [DOI] [PubMed] [Google Scholar]
- Sanchez V, Spector DH. Subversion of cell cycle regulatory pathways. Curr Top Microbiol Immunol. 2008;325:243–262. doi: 10.1007/978-3-540-77349-8_14. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Luo MH, Fortunato EA. Long-term infection and shedding of human cytomegalovirus in T98G glioblastoma cells. J Virol. 2007;81:10424–10436. doi: 10.1128/JVI.00866-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T. A variant of human cytomegalovirus derived from a persistently infected culture. Virology. 1984;137:191–194. doi: 10.1016/0042-6822(84)90023-0. [DOI] [PubMed] [Google Scholar]
- Ogura T, Tanaka J, Kamiya S, Sato H, Ogura H, Hatano M. Human cytomegalovirus persistent infection in a human central nervous system cell line: production of a variant virus with different growth characteristics. J Gen Virol. 1986;67:2605–2616. doi: 10.1099/0022-1317-67-12-2605. [DOI] [PubMed] [Google Scholar]
- Lambert G, Estévez-Salmeron L, Oh S, Liao D, Emerson BM, Tlsty TD, et al. An analogy between the evolution of drug resistance in bacterial communities and malignant tissues. Nat Rev Cancer. 2011;11:375–382. doi: 10.1038/nrc3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblach T, Grandel B, Seiler J, Nevels M, Paulus C. Human cytomegalovirus IE1 protein elicits a type ii interferon-like host cell response that depends on activated STAT1 but not interferon-γ. PLoS Pathog. 2011;7:e1002016. doi: 10.1371/journal.ppat.1002016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortunato EA, Spector DH. Viral induction of site-specific chromosome damage. Rev Med Virol. 2003;13:21–37. doi: 10.1002/rmv.368. [DOI] [PubMed] [Google Scholar]
- Siew VK, Duh CY, Wang SK. Human cytomegalovirus UL76 induces chromosome aberrations. J Biomed Sci. 2009;16:107. doi: 10.1186/1423-0127-16-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron C, Col E, Khochbin S. The viral control of cellular acetylation signaling. Bioessays. 2003;25:58–65. doi: 10.1002/bies.10202. [DOI] [PubMed] [Google Scholar]
- Nevels M, Paulus C, Shenk T. Human cytomegalovirus immediate-early 1 protein facilitates viral replication by antagonizing histone deacetylation. Proc Natl Acad Sci U S A. 2004;101:17234–17239. doi: 10.1073/pnas.0407933101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinatl J, Nevels M, Paulus C, Michaelis M. Activation of telomerase in glioma cells by human cytomegalovirus: another piece of the puzzle. J Natl Cancer Inst. 2009;101:441–443. doi: 10.1093/jnci/djp047. [DOI] [PubMed] [Google Scholar]
- Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis M, Paulus C, Löschmann N, Dauth S, Stange E, Doerr HW, et al. The multi-targeted kinase inhibitor sorafenib inhibits human cytomegalovirus replication. Cell Mol Life Sci. 2011;68:1079–1090. doi: 10.1007/s00018-010-0510-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK.Linear models and empirical Bayes methods for assessing dierential expression in microarray experiments Statist Appl Genet Mol Biol 20043(Article 3). [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- Mi H, Guo N, Kejariwal A, Thomas PD. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007;35:D247–D252. doi: 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho RJ, Campbell MJ. Transcription genomes, function. Trends Genet. 2000;16:409–415. doi: 10.1016/s0168-9525(00)02065-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.