Abstract
Array comparative genomic hybridization was used to identify copy number alterations in clear cell renal cell carcinoma (ccRCC) patient tumors to identify associations with patient/clinical characteristics. Of 763 ccRCC patients, 412 (54%) provided frozen biopsies. Clones were analyzed for significant copy number differences, adjusting for multiple comparisons and covariates in multivariate analyses. Frequent alterations included losses on: 3p (92.2%), 14q (46.8%), 8p (38.1%), 4q (35.4%), 9p (32.3%), 9q (31.8%), 6q (30.8%), 3q (29.4%), 10q (25.7%), 13q (24.5%), 1p (23.5%) and gains on 5q (60.2%), 7q (39.6%), 7p (30.6%), 5p (26.5%), 20q (25.5%), 12q (24.8%), 12p (22.8%). Stage and grade were associated with 1p, 9p, 9q, 13q and 14q loss and 12q gain. Males had more alterations compared with females, independent of stage and grade. Significant differences in the number/types of alterations were observed by family cancer history, age at diagnosis and smoking status. Von Hippel–Lindau (VHL) gene inactivation was associated with 3p loss (P<E-05), and these cases had fewer alterations than wild-type cases. The fragile site flanking the FHIT locus (3p14.2) represented a unique breakpoint among VHL hypermethylated cases, compared with wild-type cases and those with sequence changes. This is the first study of its size to investigate copy number alterations among cases with extensive patient, clinical/risk factor information. Patients characterized by VHL wild-type gene status (vs sequence alterations) and male (vs female) cases had more copy number alterations regardless of diagnostic stage and grade, which could relate to poor prognosis.
Keywords: renal cancer, epidemiology, VHL
Introduction
Renal cancer incidence is increasing1 and Central/Eastern Europe has the highest rates worldwide.2 Most (∼90%) adult renal cancers are renal cell carcinomas (RCC), most of which are clear cell RCC (ccRCC).3 ccRCC can be further classified by genetic features, most notably by the presence/absence of Von Hippel–Lindau (VHL) gene inactivation, which occurs through three mechanisms: sequence alteration affecting the transcript or protein, transcriptional silencing because of promoter CpG hypermethylation and loss of heterozygosity/copy number of the 3p25-26 locus.4
The VHL protein is involved in tissue-specific responses to oxygen concentration delivery.5, 6 VHL alteration through sequence changes and/or promoter hypermethylation, leading to transcriptional silencing, prevents formation of a protein complex required for hypoxia-inducible factor (HIF) degradation, resulting in excess HIF protein and a gene expression pattern facilitating adaption to oxygen deprivation, delivery and angiogenesis. VHL inactivation subtypes have been associated with improved prognosis and a favorable treatment response in some but not all studies.7, 8, 9, 10, 11 In contrast, VHL wild-type cases may occur through mechanisms independent of VHL inactivation. Generally, VHL inactivation is considered an early event in ccRCC but other unidentified alterations may accompany VHL alteration at diagnosis.
ccRCC subgroups have demonstrated different frequencies and patterns of VHL inactivation associated with known risk factors, which include obesity, hypertension, smoking, certain occupational exposures and a positive family history of kidney cancer.12, 13 High-intake of fruits and vegetables may reduce risk.1
In this study we used array comparative genomic hybridization to evaluate chromosomal alterations among incident ccRCC cases enrolled in a large case–control study of renal cancer. Risk factors, tumor histopathological features and VHL gene inactivation were evaluated for association with the frequency and type of copy number alterations observed.14, 15, 16
Results
Cases with tumor biopsies (N=412) were more likely to be residents from the Czech Republic, have a family cancer history and have a higher education level compared with those without biopsies (N=348) (Supplementary Table 1). Other factors did not differ.
Fraction of genome altered (FGA)
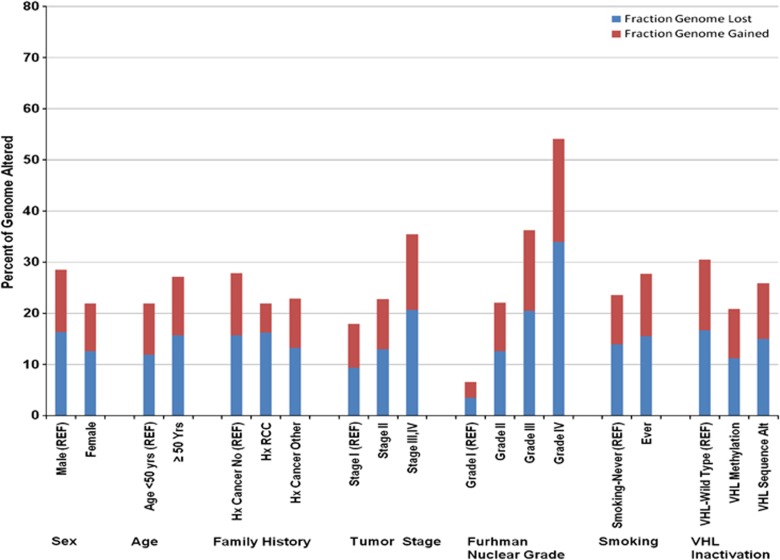
Males had a higher FGA compared with females (P=0.002) (Figure 1). Stage and grade at diagnosis were associated with increasing fraction of the genome lost (FGL), fraction of genome gained (FGG) and total FGA (P<0.0001). Ever smokers had elevated FGG (P=0.05) and FGA (P=0.05) compared with never smokers, but trends with current smoking status were not observed (P=0.62). Individuals without a first degree relative with cancer had a higher FGA compared with those who did (27.9% vs 22.8% P=0.02). A lower FGA was observed in cases with VHL promoter hypermethylation (20.8%, P=0.03) and sequence alteration (25.9%, P=0.11), compared with wild-type cases (30.5%-referent). FGA was not associated with other factors.
Figure 1.
Fraction (%) of the FGL, FGG and FGA among ccRCC case subgroups. P-values between subgroups were as follows: males vs females (P=0.002), age (<50, vs ⩾50 years, P=0.06), any family history of cancer (P=0.02), stage (P<0.00001), grade (P<0.0001), ever vs never smoking (P=0.05), VHL wild-type cases vs those with VHL promoter hypermethylation (P=0.03), VHL wild-type cases vs those with VHL sequence alterations (P=0.17). REF, referent group.
Chromosomal alterations
The exact number and percent of all chromosome p and q arm gains and losses in all ccRCC cases in this study are shown in Supplementary Table 2. Chromosome arms altered in greater than 25% of cases, and those of previous interest were included in multivariate models (Table 1). The strongest association was observed between 3p loss and VHL gene inactivation (P<0.0001). 3p loss was less frequent among VHL wild-type compared with inactivated cases (70.2% vs 95.3%, respectively). Therefore, 29.8% of VHL wild-type and 4.7% of inactivated cases had 3p present. No other factors with the exception of VHL inactivation were associated with 3p loss. Stage was strongly associated with 9p, 9q and 13q (P<0.0001), and to a lesser extent 1p, 10q and 14q losses. Stage was associated with 12p and 20q gains (P=0.001). Grade was strongly associated with 8p and 14q (P=<0.0001) and also 1p, 4q, 6q, 9p, 9q and 13q losses. Gains associated with grade were observed at 5p, 7p, 7q and 12q.
Table 1. Regression analysis of selected chromosomal arm alterations patient/clinical factors.
| Chromosome arm | Risk factors | Odds ratio (95% confidence intervals) | P-value |
|---|---|---|---|
| 1p− | Grade: I, II (REF) vs III, IV | 2.4 (1.3–4.2) | 0.003 |
| Stage: T1 (REF), T2, T3-4 | 1.6 (0.7–3.4); 3.7 (1.6–8.5) | 0.001 | |
| 3p− | VHL gene inactivation: no (REF) vs yes | 1250 (476–3125) | <0.0001 |
| 3q− | BMI: <25 (REF), 25–30, 30+ | 2.5 (1.4–4.6); 1.9 (1.0–3.7) | 0.01 |
| Sex: female (REF) vs male | 2.4 (1.5–4.2) | 0.0004 | |
| 4q− | Grade: I, II (REF), vs III, IV | 2.3 (1.4–3.6) | 0.001 |
| Age: <50 (REF), 50–60, 60+ | 1.6 (0.8–2.9); 3.0 (1.5–5.7) | 0.001 | |
| 5p+ | Grade: I, II (REF), vs III, IV | 1.6 (1.2–3.0) | 0.02 |
| 6q− | Grade: I, II (REF), vs III, IV | 2.1 (1.3–3.3) | 0.004 |
| 7p+ | Grade: I, II (REF), vs III, IV | 2.1 (1.3–3.4) | 0.003 |
| 7q+ | Family history cancer: no (REF) vs yes | 0.6 (0.4–0.9) | 0.02 |
| Grade: I, II (REF), vs III, IV | 2.1 (1.3–3.3) | 0.004 | |
| 8p− | Grade: I, II (REF), vs III, IV | 3.0 (1.8–5.0) | <0.0001 |
| Family history cancer: no (REF) vs yes | 0.6 (0.4–0.9) | 0.01 | |
| 9p− | Grade: I, II (REF), vs III, IV | 2.2 (1.3–3.7) | 0.003 |
| Stage T1 (REF), T2, T3-4 | 3.3 (1.5–7.4); 6.3 (2.6–14.8) | <0.0001 | |
| Age: <50 (REF), 50–60, 60+ | 1.9 (0.9–3.7); 3.1 (1.5–6.3) | 0.006 | |
| Sex: female (REF) vs male | 2.0 (1.2–3.1) | 0.007 | |
| 9q− | Grade: I, II (REF), vs III, IV | 2.5 (1.4–4.2) | 0.001 |
| Stage T1 (REF), T2, T3-4 | 3.9 (1.6–9.2); 7.5 (3.0–18.6) | <0.0001 | |
| Age: <50 (REF), 50–60, 60+ | 1.6 (0.8–3.2); 2.9 (1.4–5.9) | 0.007 | |
| Sex: female (REF) vs male | 2.4 (1.5–4.0) | 0.0006 | |
| 12p+ | Stage T1 (REF), T2, T3-4 | 2.8 (1.2–6.6); 5.0 (2.1–12.2) | 0.001 |
| Sex: female (REF) vs male | 2.1 (1.3–3.7) | 0.005 | |
| 12q+ | Stage T1 (REF), T2, T3-4 | 1.2 (0.9–5.2); 4.8 (2.0–11.9) | 0.001 |
| Grade: I, II (REF), vs III, IV | 2.2 (1.8–4.0) | 0.01 | |
| Sex: female (REF) vs male | 2.3 (1.2–4.4) | 0.01 | |
| 13q− | Stage T1 (REF), T2, T3-4 | 4.4 (1.3–15.1); 13.3(3.8–46.6) | <0.0001 |
| Grade: I, II (REF), vs III, IV | 3.2 (1.9–5.6) | <0.0001 | |
| 14q− | Organic solvents: no (REF) vs yes | 1.9 (1.1–3.2) | 0.03 |
| Sex: female (REF) vs male | 1.9 (1.1–3.1) | 0.03 | |
| Stage T1 (REF), T2, T3-4 | 1.5 (0.8–2.9); 3.1 (1.5–6.7) | 0.006 | |
| Grade: I, II (REF), vs III, IV | 4.4 (2.34–8.2) | <0.0001 | |
| 20q+ | Stage T1 (REF), T2, T3-4 | 1.3 (0.7–2.8); 3.1 (1.5–6.5) | 0.001 |
Abbreviations: BMI, body mass index; VHL, Von Hippel–Lindau gene.
Adjusted for Center, age, sex, stage, grade and VHL gene inactivation.
After adjustment, male cases had more losses of 3q, 9p, 9q, 13q, 14q and gains of 5p, 7q, 12p, 12q, 20q. Most of these alterations were associated with stage and/or grade. Patients 65 years or older had more 4q, 9p and 9q losses compared with younger patients diagnosed before age 50. Individuals that reported a family cancer history had fewer 7q gains and 8p losses, explaining the lower FGA observed. In contrast, there were no chromosome arm alterations associated with smoking status. Interestingly, 5q gain was frequently observed although it was not associated with any of the patient/clinical characteristics or risk factors evaluated. Closer examination revealed that gains occurred distally, specifically at two regions (that is, 5q34–35 (55–58%), 5q31–32 (52–54%) and 5q21–23 (34–49%) (Supplementary Figure 1)).
Loci
Differences in copy number alteration prevalence at specific loci and risk factors were compared using a moderated t-statistic or F statistic with pooled variance, controlling the false discovery rate (FDR). High frequency clonal alterations were sorted initially by FDR-adjusted P-values. Subsequently, significant loci were re-analyzed in adjusted models (Table 2). Grade was associated with loss of large regions on 4q, 9q, 13q, 14q, 18q and gains on 1q, 7p. Stage was associated with large losses on 9p, 9q, 13q, 14q, 17q, 18q and gains on 3q and 7q. Both stage and grade were associated with loss of 1p36; 4q11-12,26; 9p21-22; 9q21-21.3; 14q13-14, 21, 24, 31-32; 18p11.2-12, 18q11-12, 21.3-23, tel; and gains on 3q22-23; 17q24-25; and 20q13. Both stage and grade were associated with losses on 14q23.2, a region harboring the HIF1A gene, and 14q13.2 harboring the PHD1–EGLN3 gene. We examined HIF1A protein expression in tumor tissue microarrays using immunohistochemistry, in relation to 14q and regional (clonal) loss across the HIF1A region (14q23). Although univariate analyses suggested a decrease in expression with 14q and 14q23 loss, neither factor was associated with HIF1A expression after adjustment for stage and grade. Stage and particularly grade remained strongly associated with HIF1A expression, independent of 14q loss (data not shown).
Table 2. Regional clone and copy number alterations and ccRCC tumor stage and grade.
| Chromosome arm | Clone with highest signal | Regions altered | Start (Mb) | End | P-valuea | Grade I, II (%) vs III, IV (%) | ORb | 95% CIb | P-value adjb |
|---|---|---|---|---|---|---|---|---|---|
| Grade | |||||||||
| 1p− | RP11-60J11 (1p36.2) | 1p36 | 7041.726 | 7 232 439 | 0.0041 | 14/33 | 2.94 | (1.66–5.21) | 0.00001 |
| 1q+ | RP11-104A2 (1q32-41) | 1q25, 31-41, 45 | 206 402.345 | 2.07E+08 | 1.58E-05 | 9/31 | 4.42 | (2.34–8.36) | 0.00001 |
| 3q+ | RP11-219P10 | 3q22-23 | 137 606.200 | 1.38E+10 | 0.0037 | 12/25 | 1.96 | (1.07–3.61) | 0.03 |
| 4p− | RP11-176I4 (4p15.1) | 4p14-16 | 33 086.961 | 33 087 410 | 5.85E-05 | 14/33 | 3.10 | (1.73–5.57) | 0.0001 |
| 4q− | RP11-6K22 (4q 26-27) | 4q11-13, 24,26-31,34,35 | 120 282.900 | 1.20E+08 | 7.79E-06 | 13/32 | 3.43 | (1.88–6.26) | 0.0001 |
| 6q− | RP-2202A14 (6q14) | 6q14 | 79 297.260 | 79 474 213 | 0.0034 | 19/37 | 2.09 | (1.22–3.56) | 0.007 |
| 7p+ | RP11-161C7 (7p22.3) | 7p13-14,21-22.3 | 6215.911 | 6 396 692 | 0.0002 | 2543 | 2.27 | (1.36–3.79) | 0.002 |
| 7q+ | RP11-10D8 (7q22.1) | 7q22.1-.3 | 98 067.790 | 98 216 726 | 3.78E-05 | 28/46 | 2.17 | (1.31–3.60) | 0.003 |
| 8p− | RP11-182G2 (8p22) | 8p22-23 | 4457.862 | 4 614 130 | 3.33E-06 | 30/53 | 2.59 | (1.56–4.27) | 0.0001 |
| 9p− | RP11-85J5 (9p21.3) | 9p21-22 | 24 524.208 | 24 524 427 | 1.97E-05 | 26/49 | 2.32 | (1.38–3.92) | 0.002 |
| 9q− | RP11-106O17 (9q22.3) | 9q21-33 | 94 882.444 | 94 882 881 | 8.72E-06 | 25/47 | 2.19 | (1.30–3.68) | 0.003 |
| 11q− | RP11-42L18 (11q22.3-23) | 11q22-23 | 107 422.086 | 1.08E+08 | 2.32E-05 | 8/25 | 3.81 | (1.93–7.53) | 0.0001 |
| 12q+ | RP11-89J2 (12q24.32) | 12q13-14, 24.32 | 124 666.600 | 1.25E+08 | 0.0005 | 17/29 | 1.81 | (1.26–3.68) | 0.04 |
| 13q− | RP11-11C5 (13q21.3) | 13q12-14.3, 21-tel | 72 013.886 | 72 183 774 | 2.34E-06 | 10/41 | 5.36 | (2.94–9.77) | 0.00001 |
| 14q− | RP11-26M6 (14q13) | 14q12-14, 21.3-32.2 | 34 404.822 | 34 591 592 | 2.34E-06 | 34/67 | 3.18 | (1.91–5.28) | 0.00001 |
| 17q+ | RP11-84E24 (17q24-25) | 17q24-25, tel | 67 512.626 | 67 693 011 | 3.78E-05 | 5/22 | 4.50 | (2.16–9.39) | 0.00001 |
| 18p− | CTD-2048F17(18p12) | 18p11.21-11.32,12 | 8396.160 | 8 396 803 | 3.51E-06 | 14/38 | 3.24 | (1.84–5.69) | 0.00001 |
| 18q− | RP11-60P1 (18q21.3) | 18q11-12, 21.3-23, tel | 59 072.800 | 59 251 211 | 3.44E-06 | 16/44 | 3.80 | (2.20–6.55) | 0.00001 |
| 20q+ | CTD2238P18 (20q12) | 20q12,13 | 45 712.070 | 45 864 616 | 0.0001 | 17/37 | 2.28 | (1.32–3.94) | 0.003 |
| Chromosome arm | Clone with highest signal | Regions altered | Start (Mb) | End | P-valuea | Stage I, II (%)vs III, IV (%) | ORb | 95% CI | P-value adjb |
|---|---|---|---|---|---|---|---|---|---|
| Stage | |||||||||
| 1p− | RP11-219F4 (36.2–36.3) | 1p36.2-36.3 | 14 774.025 | 14 774 291 | 0.002 | 17/29 | 2.16 | (1.26–3.72) | 0.005 |
| 2p+ | RP11-57F9 | 2p23 | 28 514.930 | 28 515 223 | 0.0056 | 13/23 | 1.89 | (1.06–3.38) | 0.03 |
| 2q+ | CTD-22500B20 (2q22) | 2q22-24.3 | 158 357.236 | 158 357 713 | 0.0034 | 9/20 | 2.94 | (1.52–5.71) | 0.001 |
| 3q+ | RP11-160A13 (3q23-24) | 22-24, 26-29, tel | 144 154.808 | 144 312 333 | 0.0015 | 12/25 | 1.74 | (0.97–3.11) | 0.06 |
| 4p− | RP11-126C7 (4p13) | 11-12,13 | 38 087.290 | 38 242.92 | 0.0066 | 12/26 | 2.24 | (1.32–3.41) | 0.03 |
| 4q− | RP11-101N17 (4q26) | 4q11-12, 26 | 120 563.175 | 120 720 926 | 0.0003 | 14/29 | 2.42 | (1.35–4.43) | 0.003 |
| 6p− | RP11-284E1 (6p12) | 6p12, 24-25 | 53 823.776 | 53 824 135 | 0.0003 | 14/30 | 2.48 | (1.37–4.42) | 0.001 |
| 7q+ | RP11-38P11 (7q32) | 7q26.2-.3, 32,34,36.3 | 129 557.600 | 129 557 976 | 0.0018 | 26/41 | 1.63 | (1.02–2.63) | 0.04 |
| 9p− | RP11-125K10 (9p24) | 9p21-24, tel | 4819.655 | 4 991 841 | 8.63E-0.05 | 28/49 | 2.17 | (1.33–3.53) | 0.002 |
| 9q− | RP11-8D10 (9q21.3) | 9q12-21.3 | 82 120.447 | 82 209 078 | 8.63E-0.05 | 28/46 | 2.28 | (1.41–3.70) | 0.001 |
| 12p+ | RP11-78F16 (12p11.2) | 12p11.2,12.1-13.1 | 31 959.002 | 32 128 568 | 0.0005 | 20/32 | 2.04 | (1.20–3.45) | 0.008 |
| 12q+ | RP11-89E9 (12q23) | 12q23.3 | 97 222.144 | 97 411 481 | 0.0005 | 18/31 | 2.14 | (1.26–3.64) | 0.005 |
| 13q− | RP11-8C15 (13q12.1) | 13q12.1,12.3-14.3,21.1-tel | 20 985.475 | 20 993 099 | 8.63E-0.05 | 15/38 | 3.72 | (2.11–6.57) | <0.00001 |
| 14q− | RP11-206J4 (14q23) | 14q13-24, 31-32, tel | 63 996.230 | 64 080 508 | 0.0003 | 38/62 | 2.36 | (1.48–3.75) | <0.00001 |
| 16p− | CTD-2032H1 | 16p13 | 3232.028 | 3 296 628 | 0.0104 | 14/28 | 2.01 | (1.28–3.33) | 0.01 |
| 17q+ | RP11-86L18 | 17q12-25.3,36 | 153 645.400 | 153 645 775 | 0.0003 | 27/43 | 1.63 | (1.02–2.60) | 0.04 |
| 18p− | RP11-106J7 (18p11.21-.23) | 18p11.2-12 | 3268.728 | 3 443 000 | 0.0002 | 13/35 | 3.15 | (1.81–5.48) | <0.00001 |
| 18q− | RP11-12J12 (18q22) | 18q11-12, 21-23, tel | 55 424.497 | 55 607 583 | 3.14E-0.06 | 27/43 | 2.84 | (1.68–4.88) | <0.00001 |
| 20q+ | LLNL-255K9 | 20q13.2 | 55 338.240 | 55 352 455 | 0.0005 | 15/34 | 2.24 | (1.31–3.82) | 0.003 |
Abbreviations: ccRCC, clear cell renal cell carcinoma; CI, confidence interval; OR, odds Ratio.
P-value adjusted for multiple comparisons.
OR, 95% CI and P-values from multivariate models(MV) adjusted for age, sex, country stage, grade, smoking status and family history of cancer.
Male cases exhibited losses on 6p22, 6p24–25, large regions of 9q and gains at 1q43-44, 12p12.3, 12q12.4, 24-tel, compared with females (Table 3). These alterations were also associated with stage and grade. Among cases reporting a positive family cancer history, fewer cases demonstrated losses at 4q21-23 (11% vs 24%, P=0.002) and gains at 20q13.2 (13% vs 26%, P=0.002). Former and current smokers exhibited more loss at 3q12 (24% vs 40%, P=0.04) and gain at 16p11.2-13 (22% vs 11%, P=0.003). Interestingly, a significant positive trend was observed with current smoking status and gain at 16p11.2-13.2 (P-trend=0.003). Lastly, copy number alteration of specific clones and VHL inactivation status were evaluated. The lowest FDR-adjusted P-values that remained significant in multivariate analyses were losses observed at clones RP11-245E5 (3p24; P<0.0001) and RP11-180G14 (3p14.3; P<0.0001). RP11-180G14 (3p14.3) is located just distal to CTD-2175D15 (3p14.2), a clone harboring the FHIT locus (Figure 2).
Table 3. Regional clone and copy number alterations and patient/tumor characteristics.
| Sex | Clone with highest signal | Region altered | Start (Mb) | End | P-value adja | Male (%)/female (%) | OR (MV)b,c | 95% CIb | P-valueb |
|---|---|---|---|---|---|---|---|---|---|
| 1q+ | RP11-188A4 (1q43-44) | 1q43-44 | 239 316.500 | 2.39E+08 | 0.002 | 17/9 | 2.56 | (1.43–4.55) | 0.04 |
| 6p− | RP11-52C20 (6p22) | 6p22, 24-25 | 23 943.950 | 24 112 567 | 0.01 | 22/11 | 2.22 | (1.18–4.17) | 0.003 |
| 9q− | RP11-19014 (9q13) | 9q13, 21.1-22, 33 | 70 455.470 | 70 634 947 | 0.003 | 38/21 | 2.13 | (1.30–3.57) | 0.001 |
| 12p+ | RP11-174L5(12p12.3) | 12p12.3 | 14 735.670 | 14 736 047 | 0.001 | 38/19 | 2.44 | (1.45–4.00) | 0.001 |
| 12q+ | RP11-29G23 (12q12.4) | 12q12.4, 24-tel | 43 908.780 | 43 909 199 | 0.02 | 25/14 | 1.92 | (1.11–3.33) | 0.004 |
| Family history | No (%)/yes (%) | ||||||||
| 4q− | RP11-7G22 (4q21.3-22) | 4q21-23 | 84 523.610 | 84 723 458 | 0.002 | 24/11 | 2.70 | (1.45–5.26) | 0.002 |
| 20q+ | RP11-15M15 (20q13.2) | 20q13.2 | 51 109.650 | 51 266 870 | 0.003 | 26/13 | 2.38 | (1.41–4.55) | 0.002 |
| Ever smoker | Never (%)/ever (%) | ||||||||
| 16p+ | RP11-150K5 (16p11.2) | 16p11.2-13.2 | 29 825.190 | 29 825 404 | 0.003 | 11/22 | 2.72 | (1.42–5.21) | 0.003 |
| 3q− | CTD-2014B13 (3q13.2) | 3q12-14 | 100 971.500 | 1.01E+08 | 0.03 | 24/40 | 1.73 | (1.06–2.83) | 0.04 |
| Von Hippel–Lindau gene alteration | Wild-type (%)/altered (%) | ||||||||
| 3p− | RP11-180G14 (3p14.3) | 3p14.3 | 59 538.560 | 59 538 883 | 0.02 | 71/92 | 4.88 | (2.23–10.68) | <0.00001 |
| RP11-245E5 (3p24) | 3p21-25, tel | 22 747.910 | 22 936 416 | 1.97E-07 | 63/95 | 13.48 | (6.01–30.20) | <0.00001 | |
| 20q+ | LLNL-255K9 (20q13.2) | 20q11-13.2 | 55 338.240 | 55 352 455 | 0.03 | 33/20 | 2.08 | (1.04–4.17) | 0.03 |
Abbreviations: CI, confidence interval; MV, multivariate model; OR, Odds Ratio.
P-value adjusted for multiple comparisons.
OR, 95% CI and P-values from MV adjusted for age, sex, country stage, grade, smoking status and family history of cancer.
ORs presented for most prevalent comparison group (underlined).
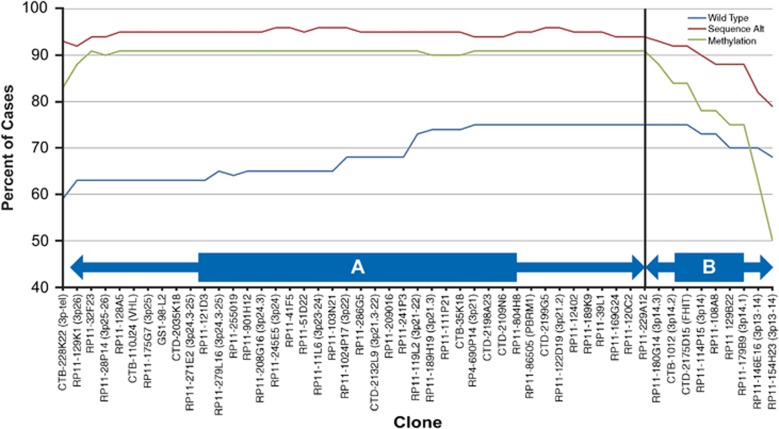
Figure 2.
Frequency of copy number loss of 3p clones among ccRCC cases grouped by absence (wild-type) or presence of VHL gene inactivation (through sequence alteration or promoter hypermethylation). Region A (telomere to RP11-180G14): the frequency of copy number loss of individual 3p clones differed significantly between VHL wild-type and inactivated cases that occurred through both sequence alteration (P<0.00001) and promoter hypermethylation (P=0.03–0.001). Region B (from RP11-180G14to RP11-154H23): the frequency of clonal copy number loss differed significantly between wild-type cases and those with sequence alterations (P=0.003–<0.00001), but was no longer observed with hypermethylated cases (P=0.08–0.59).
To follow-up upon these observations, the prevalence of all 3p losses was compared between groups stratified by VHL inactivation characteristics. Clonal alteration prevalence among wild-type cases were compared separately to cases demonstrating inactivated through VHL promoter hypermethylation or sequence alteration. Loss of these 3p clones was significantly greater among cases with VHL sequence alterations compared with wild-type cases. Losses were significantly more prevalent at the 3p telomere (93% vs 59%, P<0.00001), and clone CTB110-J24 (3p25-26;VHL) (95% vs 62%, P<0.00001) (Region A-Figure 2), and at 3p14.2 (FHIT) (92% vs 75%, P=0.001) but not at regions proximal to the centromere. Loss of 3p loci among VHL hypermethylated cases was similar in prevalence to cases with sequence alterations but significantly higher compared with wild-type cases at the 3p telomere (83%, P=0.03) and the VHL locus (91%, P=0.0006) (Region A-Figure 2), but not significantly different to wild-type cases at loci centromeric to clone RP11-180G14 (3p14.3; Region B-Figure 2). Loss among hypermethylated cases was significantly less prevalent than among cases with sequence alterations at 3p14.2 (84%, P=0.04) and regions proximal to the centromere (50%, P=0.0002). These findings demonstrate differences in 3p loci loss between VHL inactivation subgroups, in particular clones surrounding the fragile FHIT locus. Lastly, VHL wild-type cases were distinguished by having more gains at clone LLNL-255K9 (20q11-12) compared with inactivated cases (33% vs 20%, P=0.03) (Table 3), a clone that was also more prevalent in cases with higher stage.
Amplifications
High-level amplifications were observed at 154 clones among 24 cases (1–28 amplifications/case). The most frequent amplifications were located at clones RP11-28l11 (11p15.2-15.4; N=6), RP11-10M23, RP11-91H14, RP11-121G20 (6p21.1-21.2; N=5) and CTB-136O14 (12q14; N=3). Two cases (one VHL wild-type with 3p present; one VHL inactivated with 3p loss) demonstrated 2q/6p co-amplifications. The 2q overlapping region (∼179.4–199.8 Mb) covers ∼100 genes; the 6p overlapping region (∼39.1–45.7 Mb) included the VEGF (vascular endothelial growth factor) gene.
Discussion
Previous studies have utilized copy number profiling to classify histologically categorized RCC cases into molecularly defined subtypes (Table 4).17, 18, 19, 20, 21, 22 Consistent with previous studies, the most common alterations were loss of 3p, 14q and gain of 5q.17, 18, 19, 20, 21, 22 Two previous comparative genomic hybridization studies examined associations between RCC copy number variation with stage and grade.18, 20 One study (51 cases) reported two clusters defined solely by the alteration subtypes observed.18 In both clusters, 3p loss and 5q gain were frequently observed, however, one cluster, defined by losses on 1p, 4, 9, 13q and 14q, had a higher stage and grade, increased vascular involvement and lower overall survival that were independent of stage and grade.18 Another study (51 cases) reported a difference in genomic copy number and expression profiles between cases with/without biallelic VHL loss.18 A study of 80 cases observed significant associations with stage and loss of 14q and 18p.20 In the current study, both stage and grade were associated with losses on 14q harboring HIF1A (14q23.2) and PHD1-EGLN genes (14q13.2). Recently, genetic and functional studies reported that HIF1A is a target of 14q loss and that HIF1A activity is diminished in 14q deleted RCC cases.21 Loss of these loci being associated with 14q loss was not corroborated in the current study, perhaps because of differences in type of study design and laboratory methodologies. In the current study, loss of specific loci spanning 14q23 was associated with stage and grade, which is consistent with the poor prognosis reported in 14q deleted cases.18
Table 4. List of published comparative genomic hybridization (CGH) studies of kidney and renal cell carcinoma (RCC).
| Manuscript | Sporadic ccRCC Cases | Other tissues evalauted | Method | Factors evaluated | Finding |
|---|---|---|---|---|---|
| Wilhelm et al.22 | 16 | 13 Papillary, 5 chromophobe, | Array CGH-329 clones | RCC subtype segregation | Segregation of kidney tumor histological sub-types by copy number variation (CNV) differences |
| 6 Oncocytoma, 2 normal kidney | ccRCC 3p−, 5q+, and 6q−, 8p−, 9p− or 14q− | ||||
| Toma et al.17 | 22 | 22 Matched normal tissue | Affymetrix 10k chip | Survival | CNV most frequent at 3p−, 5q+, 14q−, 9−,7q+, 6q− |
| qPCR, immunohistochemistry (IHC) | Novel microdeletions at Chr1 , Chr 4, Chr6, Chr8 and Chr 9 | ||||
| Some alterations validated using gene and immunohistochemical (IHC) protein expression assays | |||||
| Arai et al.18 | 51 | NA | array CGH (4361 BACs) | Survival, grade, stage, vascular InvolvementPromoter methylation in 9 genes | CNV most frequent at 3p−, 5q+, 7+Genetic cluster profiles associated with survivial, independent of grade, stage |
| Chen et al.20 | 80 | NA | Illumina 317K SNP array | Stage, grade, telomere length | CNV most frequent at 3p−, 5q+, 8p12-pter, 6q23.3-27, 14q24.1-qter, 9q32-qter, |
| Demographics, smoking, alcohol, medical Hx | 10q22.3-qter, 9p13.3-pter, 4q28.3-qter, 1q25.1-qter, 7q21.13-qter, 8q24.12-qterLoss of heterozygosity (LOH) 9p, 9q, 14q, 18q associated with grade | ||||
| LOH 14q, 18p, 21q associated with stage | |||||
| LOH 2q, 6p, 6q, 9p, 9q, 17p associated with short telomere length | |||||
| Beroukhim et al.19 | 49, 5 Metastases | 36 Tumors/12 VHL patients | Affymetrix 250K StyI | Von Hippel–Lindau (VHL) disease, VHL inactivation, grade, | CNV most frequent at 3p−, 14q−, 5q+CNV higher in VHL WT compared with VHL disease cases |
| Gene expression differences | LOH 1p, 3p (VHL), 4q, 6q, 8p, 9p (CDKN2A, 2B) and 14q | ||||
| Amplification 1q, 2q, 5q, 7q, 8q (MYC), 12p and 20q | |||||
| Shuib et al.21 (data combined with Beroukim et al.) | 22 VHL wild-type (WT) | 57 VHL patients | Affymetrix 250K | VHL disease (germline) | CNV most frequent at 3p−, 14q−, 5q+7p+, 8p− (TNFRSF10C, DUSP4) associated with germline VHL clear cell RCC17p− (TP53), 2p+ (HIF2A) associated with VHL wild-type ccRCC |
| Moore et al., current study | 412 Clear cell RCC | Array CGH (2464 BACs) | VHL inactivation, age, sex, stage, grade | CN loss most frequent at 3p, 14q, 8p, 4q, 9p, 9q, 6q, 3q, 10q, 13q, 1q, | |
| Body mass index, hypertension, family cancer Hx, | CN gain at 5q, 7q, 7p, 5p, 20q, 12q, 12pStage and grade associated with 1p−, 9p−, 9q− 13q−, 14q−, 12q+ | ||||
| Occupational solvent, lead, pesticide, exposures | CNV higher among VHL wild-type compared to hypermethylated casesLOH across 3p varied by VHL inactivation status | ||||
| LINE1 methylation | CNV higher in male compared to females cases, independent of grade, stage | ||||
| CNV higher in cases without positive family Hx cancer | |||||
| Smoking associated with 16p11.2-13.2 gain |
Abbreviations: ccRCC, clear cell renal cell carcinoma; CGH, array comparative genomic hybridization; NA, not applicable.
This is the first study to evaluate 3p loci loss among large numbers of cases that were also defined by VHL status. In multivariate analyses, only VHL gene alteration was associated with 3p loss. Cases with VHL promoter hypermethylation were similar to those with sequence alterations at loci distal to the FHIT locus, and to wild-type cases at loci centromeric to FHIT. This observation suggests that in hypermethylated cases, the FHIT breakpoint could be critical in terms of somatic copy number loss of the chromosome segment distal to 3p14.2. In contrast, the FHIT fragile site was not a significant breakpoint in VHL wild-type or cases with sequence alterations. The FHIT region spans the most common fragile site of the human genome (the FRA3B fragile site) encompassing the previously observed familial kidney cancer breakpoint (t(3;8) p14.2;q24) and a viral integration site.23, 24, 25 Hemizygous, interstitial or terminal 3p deletions involving the FHIT gene and reduced protein expression have been observed in over 90% of ccRCC cases, similar to that observed in the current study (89%).
In addition to heterogeneity across 3p by VHL inactivation subgroups, we found that wild-type tumors had more alterations compared with those with VHL inactivation through promoter hypermethylation; corroborating a previous report that wild-type cases may be less genetically stable than hypermethylated cases, and possibly those with sequence alterations.20 The higher genomic instability among wild-type cases suggests a greater potential for progression, as copy number alterations have been associated with tumor stage, grade and worse prognosis.
Analyses of patient characteristics revealed that male cases had more alterations compared with females. The most prevalent copy number alterations in males occurred independently of stage and grade. This could indicate that incident male ccRCC tumors could be more aggressive compared with female cases, regardless of stage or grade at diagnosis. Male patients might also benefit from more aggressive treatment, follow-up and their genetic alterations could provide clues for targeted therapies. Several target genes on chromosome 9 include CDKN2A and CDKN2B.19, 20 Other risk factors examined did not explain the sex differences observed. Additional characteristics associated with tumor heterogeneity included having a first degree relative with cancer in which fewer copy number alterations were observed compared with those without cancer history. Smoking was associated with higher FGA. Ever smoking was associated with copy number loss of 3q12-14, gain of 16p11.2-13.2 for which a significant, positive trend with current smoking status was found.
As previously reported, 5q gain was highly prevalent and was not associated with of stage, grade, VHL inactivation suggesting that these loci may provide clues to the location of additional genes that are also altered in the early stages of ccRCC development. Two recent studies that employed SNP arrays19, 20 also observed gains at the distal end of 5q. Expression analysis revealed overexpression of 12 of 22 5q genes in tumor compared with normal cortex, including: GNB2L1, MGAT1, RUFY1, RNF130, MAPK9, CANX, CNOT6, SQSTM1, LTC4S, TBC1D9B, HNRPH1, and FLT4.19 A second study reported focal amplification of 5q35.3 containing the SQSTM1 gene that is overexpressed in other cancers.20 The weak correlation between 5q gain and 3p loss across studies suggests that new targets, perhaps functioning through VHL-HIF independent mechanisms, could be identified on 5q.
This study included a large series of histologically-confirmed ccRCC cases with detailed risk factor and patient/clinical information. Our two-step analytical approach, entailing FDR adjustment and subsequent multivariate analyses of significant loci, reduced possible false-positive reporting. Weaknesses included a low sample size for interaction analyses and potential selection bias from exclusion of almost 50% of cases. We also used a relatively low resolution array compared with some studies. However, the current study utilized available high-quality information on each patient and RCC risk factors to an extent not previously evaluated in earlier studies. Results from reports that employed higher resolution techniques will complement our study findings and enable identification of smaller target regions/genes for follow-up.
In summary, we identified novel associations between patient/clinical characteristics and copy number alterations in a well-defined ccRCC case-series. Considerable heterogeneity spanning the FHIT gene locus suggests that this fragile site could be critical in terms of chromosomal loss of regions distal to 3p14.2 in cases with VHL promoter hypermethylation. Our findings also suggest that male tumors may be less genetically stable compared with those in female cases, and may benefit from more aggressive follow-up. Lastly, this study provides evidence that loci on 5q may provide clues to RCC carcinogenesis that could be distinct to the VHL-HIF pathway.
Materials and methods
Study population
Incident cases (ICD-02 code C.64) were participants in a case–control study of RCC conducted in seven centers in four Central and Eastern European countries (Moscow, Russia; Bucharest, Romania; Lodz, Poland; and Prague, Olomouc, Ceske-Budejovice, and Brno, Czech Republic).14, 15 Diagnostic slides were re-reviewed by an expert renal cancer pathologist for standardized classification (MM). Of 1097 cases, 763 were ccRCC, and frozen biopsies were available from 412 (54%) cases. Protocols were approved by ethics committees and institutional review boards of participating centers, the International Agency for Research on Cancer and the US National Cancer Institute. All patients and physicians provided written informed consent. Patients were asked about lifestyle habits, height/weight one year before diagnosis, personal/family medical history and diet.13, 14, 15, 16
Array comparative genomic hybridization
Areas of frozen biopsies containing ⩾70% tumor cells were macrodissected and non-tumor tissue was removed.13 A standard protocol (http://cc.ucsf.edu/people/waldman/Protocols) was used to extract DNA. Arrays included 2464 BACs at approximately Mb intervals along the genome, printed in triplicate.22, 26 DNA samples (500 ng) were labeled as described previously.27
VHL
Inactivating alterations in exons 1–3 of the VHL gene and promoter hypermethylation were assessed as described previously.13 Wild-type tumors included those without sequence alterations affecting the protein coding region, splice junctions or promoter hypermethylation.
Data processing
Data were processed using SPROC to filter data points with low DAPI intensity, reference to DAPI signal intensity and correlation between Cy3 and Cy5 within spots. Ratios that were derived from a single clone or those with a triplicate log2 SD >0.2 were considered missing. Samples were hybridized on two versions of human array (3.1, 3.2). Clone data were combined sharing data from both arrays. Post-processing, 2111 clones remained. Data were mapped to the human DNA sequence May 2004 freeze.
Data were segmented using circular binary segmentation,28 implemented in the DNAcopy package from Bioconductor29 to translate intensity measurements into equal copy number regions. Median absolute deviation, scaled by the factor 1.4826 of the difference between the observed (log2 ratio) and segment values of autosomal clones, were used to estimate sample-specific experimental variation. Clone status outcome assignment (gain/loss/normal) was conducted, applying the merge levels procedure to segment values.30 To identify single technical or biological outliers (that is, high-level amplifications), their presence was allowed within each segment. Outlier clones included those for with an observed value at least four sample-specific median absolute deviation from the segment. High-level amplifications included narrow and single outliers with high copy numbers, as described previously.31
To quantify the total amount of genome altered, each clone was assigned a genomic distance equal to the sum of one half the distance between its center and that of neighboring clones or to the chromosome end for probes with one neighbor.31 FGA was defined as the proportion of the distance of the autosomal clone regions altered to that of the entire genome. FGG or FGL, considered only distances that were gained or lost.
Clone-wise association tests between copy number alterations and clinical variables were based on the segment values with observed values for outlier clones. Moderated t-tests for dichotomous variables and moderated F-statistics32 were used for more than two groups. P-values were adjusted for multiple testing by controlling the FDR.32, 33 Wilcoxon's rank-sum tests were performed to evaluate differential associations of summary genomic events (that is, FGA, FGG, FGL, amplifications and specific alterations) with dichotomous variables. Kruskal–Wallis rank-sum tests were used for events having more than two groups. Statistical analyses of autosomes were performed using R/Bioconductor.29, 34 P-values <0.05 were considered significant.33
Copy number alterations
The prevalence of ccRCC cases with/without frozen biopsies were compared by patient characteristics using χ2 and Fisher's Exact tests. Alterations of chromosome arms were coded as ‘0' (no alterations) and ‘1' (dominant change), defined as gain or loss in >25% of cases. Also included were those previously reported as important alterations in renal cancer. To compare cases with/without p or q arm alterations, cases with gains/losses were considered as dependent variables in stepwise logistic regression models with patient/clinical characteristics, lifestyle and occupational exposure variables. Criteria for inclusion into multivariate models was a P-value <0.20. Study design matching variables (country, sex, age) were included in all models. All variables were fitted in logistic models to obtain odds ratios (OR) and 95% confidence intervals (95% CI) as risk estimates of each alteration. Individual clone copy number differences between groups were initially tested using a moderated t-statistic or F-statistic with pooled variance, adjusted for multiple comparisons controlling the FDR.32 Clones identified through the first screen, observed in >20% of cases, were included in multivariate analyses conducted using SAS 9.1.3 (SAS Institute Inc.) and STATA 10.0 software. Clone-wise tests were conducted in R,34 all were two-sided.
Acknowledgments
This project has been funded in whole or in part with federal funds from the US NCI, NIH, DHHS, Contract HHSN261200800001E. Mention of trade names, commercial products or organizations does not imply endorsement by the US Government.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Oncogenesis website (http://www.nature.com/oncsis)
Supplementary Material
References
- Chow WH, Devesa SS. Contemporary epidemiology of renal cell cancer. Cancer J. 2008;14:288–301. doi: 10.1097/PPO.0b013e3181867628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curado MP, Edwards B, Shin HR, Storm H, Ferlay J, Meanue M, et al. (2008)Cancer Incidence in Five Continents vol. IXIARC Scientific Publications; Lyon, France, 1980. [Google Scholar]
- Linehan WM, Walther MM, Zbar B. The genetic basis of cancer of the kidney. J Urol. 2003;170:2163–2172. doi: 10.1097/01.ju.0000096060.92397.ed. [DOI] [PubMed] [Google Scholar]
- Latif F, Tory K, Gnarra J, Yao M, Duh FM, Orcutt ML, et al. Identification of the von Hippel-Lindau disease tumor-suppressor gene. Science. 1993;260:1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- Maxwell PH, Weisener MS, Chang GW, Clifford SC, Vaux EC, Cockman M, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- Kaelin WG. Molecular basis of the VHL heteditary cancer syndropme. Nat Rev Cancer. 2002;2:673–682. doi: 10.1038/nrc885. [DOI] [PubMed] [Google Scholar]
- Golshayan AR, Brick AJ, Choueiri TK. Predicting outcome to VEGF-targeted therapy in metastatic clear-cell renal cell carcinoma: data from recent studies. Future Oncol. 2008;4:85–92. doi: 10.2217/14796694.4.1.85. [DOI] [PubMed] [Google Scholar]
- Patard JJ, Rioux-Leclercq N, Masson D, Zerrouki S, Jouan F, Collet N, et al. Absence of VHL gene alteration and high VEGF expression are associated with tumour aggressiveness and poor survival of renal-cell carcinoma. Br J Cancer. 2009;101:1417–1424. doi: 10.1038/sj.bjc.6605298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M, Yoshida M, Kishida T, Nakaigawa N, Baba M, Kobayashi K, et al. VHL tumor suppressor gene alterations associated with good prognosis in sporadic clear-cell renal carcinoma. J Natl Cancer Inst. 2002;94:1569–1575. doi: 10.1093/jnci/94.20.1569. [DOI] [PubMed] [Google Scholar]
- Kim JH, Jung CW, Cho YH, Lee J, Lee SH, Kim HY, et al. Somatic VHL alteration and its impact on prognosis in patients with clear cell renal cell carcinoma. Oncol Rep. 2005;13:859–864. [PubMed] [Google Scholar]
- Rini BI, Jaeger E, Weinberg V, Sein N, Chew K, Fong K, et al. Clinical response to therapy targeted at vascular endothelial growth factor in metastatic renal cell carcinoma: impact of patient characteristics and Von Hippel-Lindau gene status. BJU Int. 2006;98:756–762. doi: 10.1111/j.1464-410X.2006.06376.x. [DOI] [PubMed] [Google Scholar]
- Young AC, Craven RA, Cohen D, Taylor C, Booth C, Hamden P, et al. Analysis of VHL gene alterations and their relationship to clinical parameters in sporadic conventional renal cell carcinoma. Clin Cancer Res. 2009;15:7582–7592. doi: 10.1158/1078-0432.CCR-09-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LE, Nickerson ML, Brennan P, Toro JR, Jaeger E, Rinsky J, et al. Association of somatic VHL alteration with risk factors for RCC and germline VHL gene locus polymorphisms. PLoS Genet. 2011;7:E1002312. doi: 10.1371/journal.pgen.1002312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan P, van der Hel O, Moore LE, Zaridze D, Matveev V, Holcatova I, et al. Tobacco smoking, body mass index, hypertension, and kidney cancer risk in central and eastern Europe. Br J Cancer. 2008;99:1912–1915. doi: 10.1038/sj.bjc.6604761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CC, Chow WH, Boffetta P, Moore L, Zaridze D, Moukeria A, et al. Dietary risk factors for kidney cancer in Eastern and Central Europe. Am J Epidemiol. 2007;166:62–70. doi: 10.1093/aje/kwm043. [DOI] [PubMed] [Google Scholar]
- Moore LE, Boffetta P, Karami S, Brennan P, Stewart PS, Hung R, et al. Occupational trichloroethylene exposure and renal carcinoma risk: evidence of genetic susceptibility by reductive metabolism gene variants. Cancer Res. 2010;70:6527–6536. doi: 10.1158/0008-5472.CAN-09-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toma MI, Grosser M, Herr A, Aust DE, Meye A, Hoefling C, et al. Loss of heterozygosity and copy number abnormality in clear cell renal cell carcinoma discovered by high-density affymetrix 10k single nucleotide polymorphism mapping array. Neoplasia. 2008;10:634–642. doi: 10.1593/neo.08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai E, Ushijima S, Tsuda H, Fjimoto H, Hosoda F, Shibata T, et al. Genetic clustering of clear cell renal cell carcinoma based on array-comparative genomic hybridization: its association with DNA methylation alteration and patient outcome. Clin Cancer Res. 2008;14:5531–5539. doi: 10.1158/1078-0432.CCR-08-0443. [DOI] [PubMed] [Google Scholar]
- Beroukhim R, Brunet JP, di Napoli A, Mertz KD, Seeley A, Pires MM, et al. Patterns of gene expression and copy-number alterations in von-Hippel Lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res. 2009;69:4674–4681. doi: 10.1158/0008-5472.CAN-09-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Ye Y, Yang H, Tamboli P, Matin S, Tannir NM, et al. Genome-wide profiling of chromosomal alterations in renal cell carcinoma using high-density single nucleotide polymorphism arrays. Int J Cancer. 2009;125:2342–2348. doi: 10.1002/ijc.24642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuib S, Wei W, Sur H, Morris MR, McMullan D, Ratternberry E, et al. Copy number profiling in von Hippel-Lindau disease renal cell carcinoma. Genes Chromosomes Cancer. 2011;50:479–488. doi: 10.1002/gcc.20865. [DOI] [PubMed] [Google Scholar]
- Wilhelm M, Veltman JA, Olshen AB, Jain AN, Moore DH, Presti JC, et al. Array-based comparative genomic hybridizaiont for the differential diagnosis of renal cell cancer. Cancer Res. 2002;62:957–960. [PubMed] [Google Scholar]
- Wali A. FHIT: Doubts are clear now. Sci World J. 2010;10:1142–1151. doi: 10.1100/tsw.2010.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao GH, Jin F, Klein-Szanto AJP, Goodrow TL, Linehan MW, Yeung RS. The FHIT gene product is highly expressed in the cytoplasm of renal tubular epithelium and is down-regulated in kidney cancers. Am J Pathol. 1997;151:1541–1548. [PMC free article] [PubMed] [Google Scholar]
- Pekarsky Y, Zanesi N, Palamarchuk A, Huebner K, Croce CM. FHIT: from gene discovery to cancer treatment and prevention. Lancet Oncol. 2002;3:748–754. doi: 10.1016/s1470-2045(02)00931-2. [DOI] [PubMed] [Google Scholar]
- Hodgson G, Hager GH, Volik S, Horiono S, Wernick M, Moore D, et al. Genome scanning with array CGH delineates regional alterations in mouse islet carcinomas. Nat Genet. 2001;29:459–464. doi: 10.1038/ng771. [DOI] [PubMed] [Google Scholar]
- Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, et al. High resloultion analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- Gentleman R, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willenbrock H, Fridlyand J. A comparison study: applying segmentation to array CGH data for downstream analyses. Bioinformatics. 2005;21:4084–4091. doi: 10.1093/bioinformatics/bti677. [DOI] [PubMed] [Google Scholar]
- Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Smyth GK.Linear models and empirical Bayes methods for assessing differential expression in microarray experiments Statistical Applications in Genetics and Molecular Biology 20043Article 3. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- R Development Core TeamR A Language and Environment for Statistical Computing R Foundation for Statistical Computing, Vienna, Austria; 2010. ISBN 900051-07-0 URL http://www.R-project.org . [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.