Abstract
The discovery of translocations that involve one of the genes of the ETS family (ERG, ETV1, ETV4 and ETV5) has been a major advance in understanding the molecular basis of prostate cancer (PC). Each one of these translocations results in deregulated expression of one of the ETS proteins. Here, we focus on the mechanism whereby overexpression of the ETV4 gene mediates oncogenesis in the prostate. By siRNA technology, we show that ETV4 inhibition in the PC3 cancer cell line reduces not only cell mobility and anchorage-independent growth, but also cell proliferation, cell cycle progression and tumor growth in a xenograft model. Conversely, ETV4 overexpression in the nonmalignant human prostate cell line (RWPE) increases anchorage-independent growth, cell mobility and cell proliferation, which is probably mediated by downregulation of p21, producing accelerated progression through the cell cycle. ETV4 overexpression is associated with changes in the pattern of E-cadherin and N-cadherin expression; the cells also become spindle-shaped, and these changes are characteristic of the so-called epithelial to mesenchymal transition (EMT). In RWPE cells overexpressing ETV4 EMT results from a marked increase in EMT-specific transcription factors such as TWIST1, SLUG1, ZEB1 and ZEB2. Thus, whereas ETV4 shares with the other ETS proteins (ERG, ETV5 and ETV1) a major role in invasiveness and cell migration, it emerges as unique in that it increases at the same time also the rate of proliferation of PC cells. Considering the wide spectrum in the clinical course of patients with PC, it may be highly relevant that ETV4 is capable of inducing most and perhaps all of the features that make a tumor aggressive.
Keywords: prostate cancer, ETV4, epithelial to mesenchymal transition, cell cycle, ETS proteins, chromosomal translocations
Introduction
Chromosomal translocations have been regarded for a long time as characteristic of hematological malignancies and sarcomas, as though they were not relevant to epithelial tumors. In recent years, it has become instead clear that chromosomal translocations are frequent in epithelial tumors:1 indeed they are present in 40–80% of prostate cancer (PC) cases. In most cases the translocation brings about the fusion of the androgen-regulated gene, TMPRSS2, with one of the members of the ETS family of transcription factors, the most frequent being ERG.2 In this case, the promoter of TMPRSS2 replaces the ERG promoter with the consequence that ERG expression will become androgen dependent. In some cases, the TMPRSS2 promoter is juxtaposed to one of other ETS family members: ETV1, ETV4, ETV5 and FLI1. Rarely, the translocation juxtaposes one ETS gene to the promoter of a gene highly expressed in the prostate other than TMPRSS2.3 In any case, the common result is that the involved ETS gene will be aberrantly expressed in the prostate.
The role of ERG overexpression has been investigated in various models (including transgenic mice) with somewhat controversial results: some studies suggest that ERG was sufficient for cancer development4, 5 whereas others suggest that additional genetic changes are required:6, 7 either way, it is currently accepted that ERG overexpression has a role in PC pathogenesis. This notion would be greatly strengthened by finding that it applies to other ETS family members involved in translocations in PC. Here, we have focused on one of these, ETV4. ETV4 is overexpressed in a proportion of PC cases: in some cases overexpression is associated with translocations of ETV4 to the promoter of a gene highly expressed in prostate (TMPRSS2, KLK2, DDX5 and CANT1);8, 9, 10 in others it has been observed without any detectable translocation.11, 12 ETV4 is also overexpressed in other types of cancers, including head and neck, lung and breast cancer;13, 14, 15 and its overexpression has been associated with metastasis and with poor prognosis.16 ETV4 expression has been correlated with the activation of cancer-related genes relevant to cell proliferation and to invasiveness.14, 17, 18, 19 On the other hand, some reports have suggested that ETV4 may function as a tumor suppressor.20, 21
Evidence has accumulated over the last years that a major mechanism underlying tumor invasion is that epithelial tumor cells may acquire a mesenchymal phenotype.22, 23 This is similar to a process, crucial for embryonic development, called epithelial to mesenchymal transition (EMT).24 EMT-like processes may have a central role in cancer progression;25 accordingly, the acquisition of an EMT-like phenotype has been associated with the ability of cancer cells to migrate, to invade and to metastasize.26, 27 EMT has been reported in different cancers including prostate28, 29 and it has been also associated with self-renewal of cancer cells.30
By overexpression, as well as by silencing, we have investigated the role of ETV4 in PC. We have found that ETV4 expression promotes proliferation, anchorage-independent growth and tumor growth in a xenograft model. In addition, we have found that ETV4 expression affects cell mobility and favors EMT.
Results
Downregulation of ETV4 inhibits proliferation, anchorage-independent growth and migration in PC cells
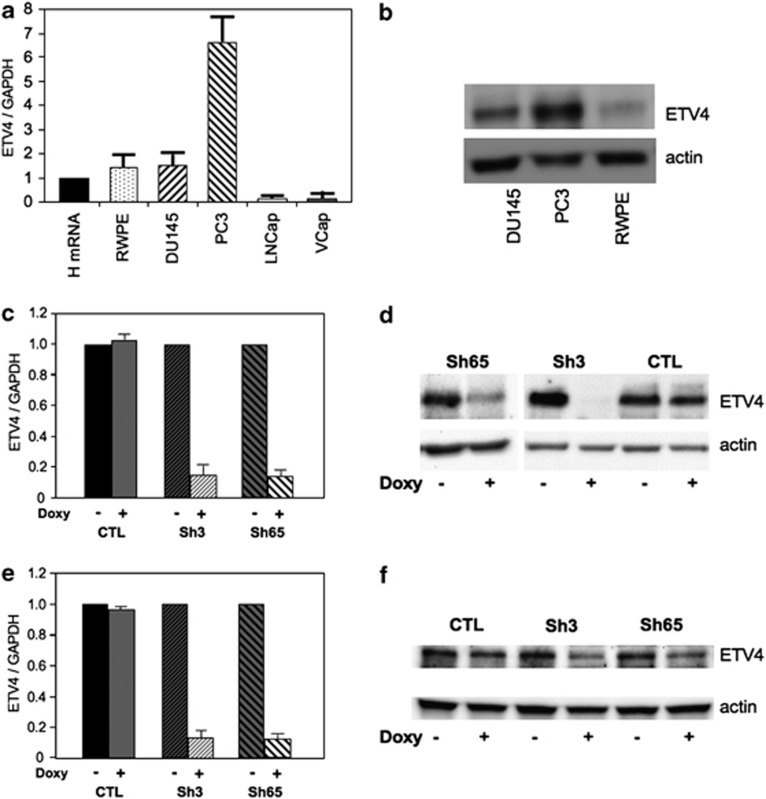
We measured ETV4 expression in normal prostate tissue, in one immortalized but nonmalignant human prostate cell line (RWPE), and in four human PC cell lines (DU145, PC3, LnCap and V-Cap). ETV4 expression was undetectable in LnCap and in V-Cap cells (Figure 1a, Supplementary Figure S1). ETV4 mRNA levels were increased in PC3 (sevenfold) and were similar to normal prostate tissue in RWPE and DU145 (Figure 1a). However, the level of ETV4 protein in PC DU145 cells was about fivefold higher than in the nonmalignant RWPE cells (Figure 1b). Thus, ETV4 is overexpressed in DU145 and in PC3 cells: this overexpression may be causative of their cancer phenotype.
Figure 1.
ETV4 expression levels in human prostate cell lines. (a) ETV4 expression level (normalized to the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH)) in comparison with normal human prostate (HmRNA) was assessed by quantitative real-time (qRT)–PCR in a nonmalignant human prostate cell line (RWPE) and in human cancer prostate cell lines (DU145, PC3, LNCaP and VCap). (b) ETV4 expression level assessed by western blot analysis in human prostate cell lines expressing ETV4 mRNA. (c and e) ETV4 expression level (normalized to the housekeeping gene GAPDH) assessed by qRT-PCR before and after the doxycycline (Doxy) induction of DU145 (c) and PC3 (e) cell lines stably transduced with vectors expressing the indicated shRNA. (d and f) ETV4 expression level assessed by western blot analysis before and after the Doxy induction of DU145 (d) and PC3 (f) cell lines stably transduced with vectors expressing the indicated shRNA. CTL (irrelevant shRNA); sh3 (anti-ETV4 shRNA 3); sh65 (anti-ETV4 shRNA 65); see Materials and methods for details.
ETV4 expression was silenced by two short hairpin RNAs (sh3 and sh65) that, either in a constitutive or in an inducible31 system, produced a marked and durable reduction of ETV4 mRNA levels in both PC3 and DU145 cells (Figures 1c and e). This reduction was confirmed in terms of protein levels (Figures 1d and f). By using doxycycline-inducible stably transduced cell lines, we have reduced the experimental variability of growth assays, because it was possible to test in parallel exactly the same cells with or without ETV4 silencing.
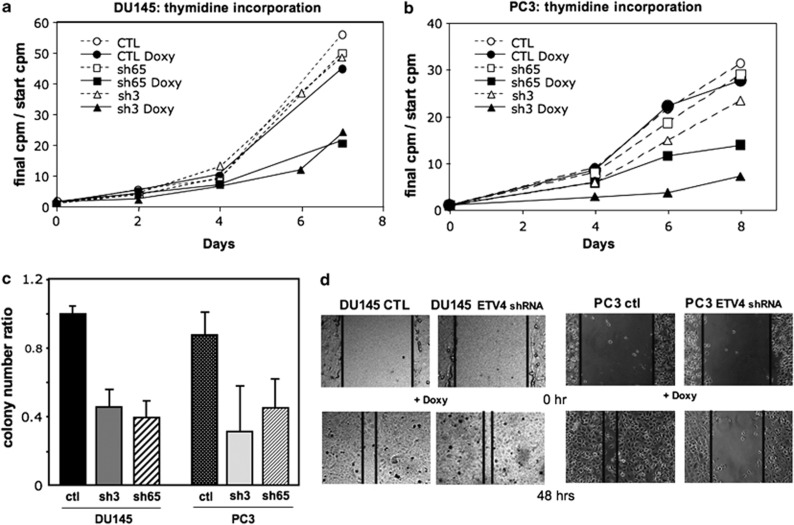
Using this system, we found that after 14 days of ETV4 shRNA induction the number of DU145 and PC3 cells was reduced to 47.6±10.5% and 20.7±5.6%, respectively. In both of these cell lines, a similar reduction in H3 thymidine uptake was also observed upon ETV4 silencing but not after induction of an irrelevant shRNA (Figures 2a and b). Furthermore, ETV4 shRNA induction, obtained with either sh3 or sh65, strongly impaired the ability of PC cell lines to form colonies in soft agar (Figure 2c). This suggests that ETV4 expression is important in determining the proliferation rate and the anchorage-independent growth of both PC3 and DU145 cells. In addition, the fact that similar results were obtained with two different shRNAs supports the notion that their effect is mediated by specific reduction of ETV4 mRNA and not by off-target modulation.
Figure 2.
Effects of ETV4 silencing on the growth of DU145 and PC3 human PC cell lines. (a and b) Doxycycline-induced ETV4 silencing reduces the growth of DU145 (a) and PC3 (b) cell lines. Cell growth has been assessed by H3-thymidine uptake. At each time point, it is represented the ratio between initial and the actual c.p.m. (average of three experiments done in quadruplicate). Circles represent the cell lines stably transduced with a vector expressing an irrelevant shRNA; squares represent the cell lines stably transduced with a vector expressing the anti-ETV4 shRNA 65 (sh65); and triangles represent the cell lines stably transduced with a vector expressing the anti-ETV4 shRNA 3 (sh3). Full symbols connected by continuous lines represent the culture with doxycycline (with reduced ETV4 expression levels). Empty symbols connected by dotted lines represent the culture without doxycycline (normal ETV4 expression levels). (c) Doxycycline-induced ETV4 silencing reduces the substrate-independent growth of DU145 (P<0.01) and PC3 cell lines (P<0.05). Each bar represents the ratio between the number of colonies from plates with doxycycline and the number of colonies from plates without doxycycline. S.d.'s are shown. CTL (irrelevant shRNA); sh3 (anti-ETV4 shRNA 3); sh65 (anti-ETV4 shRNA 65); see Materials and methods for details. (d) Monolayer of stably transduced DU145 (on the left) and PC3 (on the rigth) cells after culture for 5 days with doxycicline was scored and then cultured in medium for the time showed in the picture and in presence of mitomycin C. Cell migration into the wound was examined by phase contrast microscopy (pictures of a representative experiment are shown).
ETS proteins, such as ERG and ETV1, have an important role in cell motility and in the invasive tendency of prostate cell lines;5, 32 and ETV4 itself is important for motility of PC, breast cancer and colon cancer cells.19, 33, 34 Both PC3 and DU145 cells are able to efficiently migrate in the wound-healing assay and to invade matrigel. After doxycycline induction, the migration of PC3 cells containing an inducible ETV4 shRNA was 3.5±0.8-fold less than that of PC3 cells containing an irrelevant shRNA (P<0.01; Figure 2d). In a similar way, doxycycline reduced by 2.9±0.2-fold the number of matrigel invading cells of PC3 containing an inducible ETV4 shRNA, whereas the invading ability of PC3 containing the irrelevant shRNA was only slightly reduced (1.4-fold). In contrast, with DU145 cells ETV4 inhibition did not affect migration (Figure 2d) or invading ability (data not shown). Matrix metalloproteinases (MMPs) have pivotal roles in invasion through the degradation of extracellular matrix; ETV4 is known to regulate some of these MMPs;19, 34, 35 indeed, we found that ETV4 silencing in PC3 cells was associated with reduced expression of MMP1 (0.28±0.2) and MMP3 (0.14±0.1), a slight decrease of MMP9 (0.7±0.2), and no variation of MMP7 (1.0±0.1), PLAU (1.2±0.1) and ADAMs (ADAM9: 1.0±0.2; ADAM10: 1.2±0.3; ADAM17: 1.2±0.1). In addition, there was an increased expression of the metalloproteinase inhibitor TIMP1 (3.24±0.1) but not of TIMP2 (1.0±0.1).36 In the DU145 cells, we did not find any variation of MMPs (MMP1: 1.2±0.1; MMP9: 1.1±0.1), ADAMs (ADAM9: 1.2±0.2; ADAM10: 1.2±0.1; ADAM17: 1.1±0.4) and TIMPs (TIMP1: 0.9±0.1; TIMP2: 1,2±0.2) upon ETV4 silencing.
Overexpression of ETV4 stimulates proliferation, anchorage-independent growth and migration in the nonmalignant RWPE prostate cells
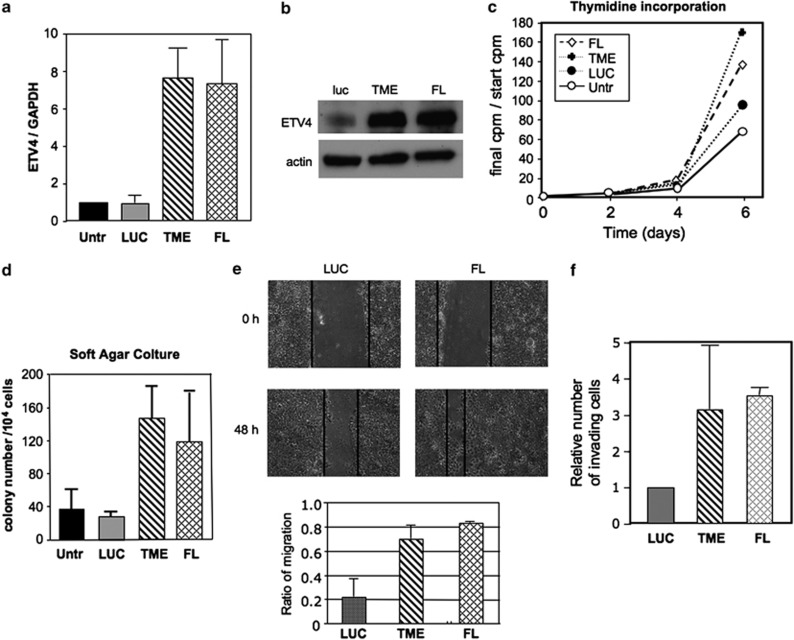
To investigate directly the effect of ETV4 overexpression, we have constructed two expression vectors: one encoding the full length ETV4 cDNA (FL) and one encoding the TMPRSS2–ETV4 (TME) fusion cDNA found in PC patients.8 The nonmalignant RWPE human prostate cell line stably transfected with either the FL or the TME vector showed an increased expression of ETV4 at both mRNA (about 7.5-fold) and protein level (five to sixfold; Figures 3a and b) compared with controls RWPE cells (untransfected or luciferase transfected).
Figure 3.
ETV4 overexpression and its effects on human RWPE. (a) ETV4 expression level (normalized to the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH)) by quantitative real-time (qRT)–PCR in RWPE cells transfected with ETV4 expression vectors FL and TME (see Materials and methods for details) compared with untransfected (untr) and mock-transfected (LUC) RWPE cells. (b) ETV4 expression level assessed by western blot analysis in mock (LUC), FL and TME-transfected RWPE cells. (c) ETV4 overexpression increases the growth of RWPE cell line (assessed by H3-thymidine uptake). At each time point it is represented the ratio between initial and the actual c.p.m. (average of three experiments done in quadruplicate). Empty circle: untransfected RWPE; full circle, mock transfected (LUC) RWPE; diamond, FL-transfected RWPE; and cross, TME-transfected RWPE. (d) ETV4 overexpression increases the substrate-independent growth of RWPE cell lines (UNTR or LUC vs TME or FL RWPE, P<0.05). Each bar represents the average number of colonies (three experiments done in triplicate). S.d.'s are shown. (e) Upper panels: monolayer of mock transfected, FL-transfected RWPE cells was scored and then cultured in medium for 48 h in the presence of mitomycin C. Cell migration into the wound was examined by phase contrast microscopy (pictures of a representative experiment are shown). Lower panel: ETV4 overexpression increases the migration of RWPE cell lines (LUC vs TME-RWPE or FL-RWPE, P<0.03): each bar shows the difference between the width of wound at time 0 and that after 48 h normalized by the width of wound at time 0. Values are expressed as mean+s.d. (three experiments in duplicate). (f) ETV4 overexpression increases the migration through matrigel of RWPE cell lines (LUC vs TME-RWPE or FL-RWPE, P<0.05). Cells that migrated out of the chamber were stained and counted. Each bar shows the number of migrating cells normalized to that of the mock-transfected cells. The mean+s.d. of three experiments done in triplicates is shown.
By H3 thymidine incorporation, we found that RWPE cells transfected with either the FL or the TME vector have an increased rate of proliferation compared with controls (Figure 3c). ETV4 overexpression increased the ability of RWPE cells to form colonies in soft agar (Figure 3d), providing RWPE cells with anchorage-independent growth, an important characteristic of cancer cells. In addition, in wound-healing and in matrigel invasion assays ETV4-transfected RWPE cells showed increased migration (Figure 3e) and invasion ability when compared with controls (Figure 3f). These results confirmed that also in RWPE cells ETV4 has a role in cell proliferation, anchorage-independent growth, migration and invasion ability.
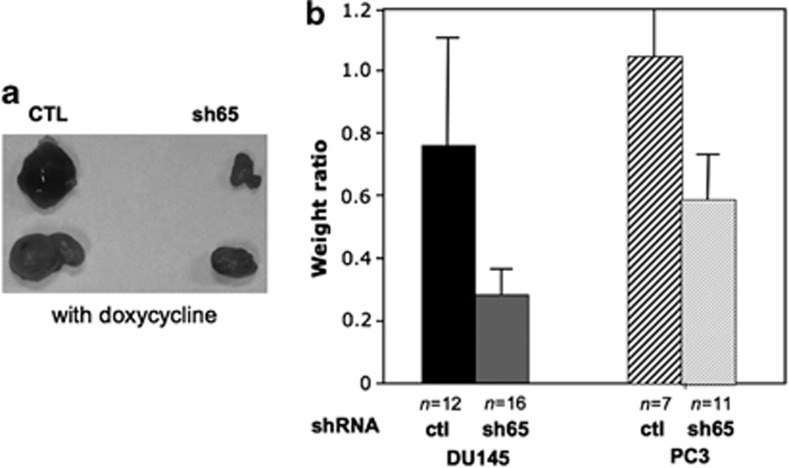
Downregulation of ETV4 inhibits tumor growth in immune-deficient mice
Many types of cancer cells give tumors when injected in immune-deficient mice (xenograft model): this is true for both PC3 and DU145 cells.37, 38 To reduce experimental variability of such experiments, we have used PC3 and DU145 stably transduced with the doxycycline-inducible shRNA against ETV4. Thus, we have injected in each mouse cells derived from exactly the same culture and, after 2 months, we have compared the size of xenografts in mice assigned randomly to drink water with or without doxycycline. The average weight of PC3 xenografts from mice treated with doxycycline was about half of control mice (P<0.04). With DU145 xenografts the effect of ETV4 inhibition was even stronger with a weight reduction of about 70% (P<0.02; Figures 4a and b). In PC3 xenografts harvested from doxycycline-treated mice the ratio at DNA level ETV4-silencing vector/glyceraldehyde-3-phosphate dehydrogenase was much lower (about 4%) than in xenografts from control mice suggesting that in doxycycline-treated mice the tumors originate mainly or entirely from the few untransfected cells included in the initial injection. Thus, ETV4 downregulation reduces the ability of PC3 and in DU145 cells to form tumors in mice, supporting the notion that ETV4 expression is necessary to maintain the cancer status of these human PC cell lines.
Figure 4.
ETV4 silencing reduces the growth of DU145 and PC3 cell in xenograft model. (a) Tumors derived from DU145 cells stably transduced with doxycycline-inducible vectors expressing either an irrelevant shRNA (CTL) or the anti-ETV4 shRNA 65 and injected in athymic nude mice (representative experiment). Tumors have been excised from the immunodeficient mice treated with doxycycline after 8 weeks from the injection. (b) Each bar shows the ratio between the average weight of tumors recovered from doxycycline-treated mice and the average weight of tumors recovered from untreated mice. CTL (irrelevant shRNA); sh65 (anti-ETV4 shRNA 65); see Materials and methods for details.
ETV4 affects cell growth by modulating cell cycle
In principle, cell growth could be increased either by an increased rate of cell proliferation or by a decreased rate of apoptosis. By flow cytometry, we did not observe any significant variation of apoptotic cells percentage neither after ETV4 silencing in PC3 and DU145 cells nor after ETV4 overexpression in RWPE cells (data not shown).
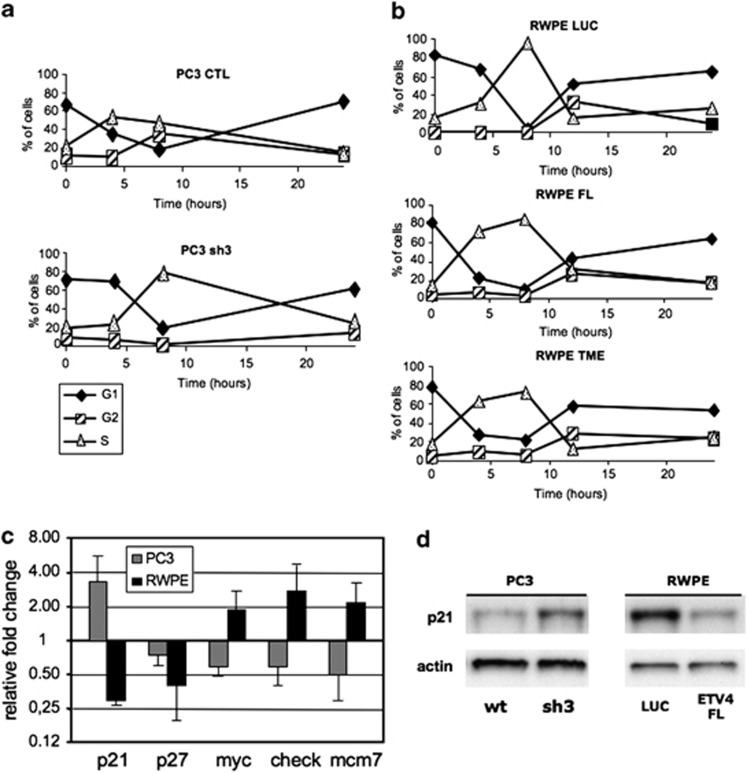
Next, we analyzed cell cycle progression. With ETV4 silenced, 4 h after synchronization with hydroxyurea, the fraction of PC3 cells in S phase was reduced in comparison with controls (Figure 5a), indicating that silencing of ETV4 results in a slower cell cycle progression.
Figure 5.
The effect of ETV4 silencing and of ETV4 overexpression on the cell cycle. (a and b) Cell cycle analysis carried out in PC3 (a) and RWPE (b) cells after treatment with 10 mℳ hydroxyurea for 14 h followed by culture in normal medium. The analysis of the cell cycle phases was performed at 0, 4, 8 and 24 h. The percentage of cells present in each specific phase of the cell cycle at each time point is shown. Filled diamond, cells in G1-phase; empty triangles, cells in S-phase; and dashed square, cells in G2-phase. PC3-CTL (PC3 cells stably transduced with an irrelevant shRNA); PC3-sh3 (PC3 cells stably transduced with anti-ETV4 shRNA 3); RWPE LUC, RWPE TME and RWPE FL (RWPE cells transfected with mock, TME and FL vectors, respectively). (c) Fold changes of expression levels (normalized to the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase) of genes known to be involved in cell cycle in PC3-sh3 (PC3 cells transduced with anti-ETV4 shRNA 3) compared with PC3-CTL (PC3 cells transduced with irrelevant shRNA) and RWPE cells transfected with FL-ETV4 plasmids compared with LUC-transfected RWPE cells. Fold changes are plotted on a Log 2 scale. (d) Expression (western blot analysis) of p21 (Waf1/Cip1) in PC3-sh3 compared with PC3-CTL and RWPE cells transfected with mock and FL-ETV4 plasmids.
An analysis of the cell cycle in RWPE cells overexpressing ETV4 gave a mirror image of the above: that is, compared with controls there was an increase in the fraction of cells in S phase after 4 h after synchronization with hydroxyurea (Figure 5b). We have tested some of the proteins known to be involved in cell cycle regulation.39, 40 In RWPE-overexpressing ETV4, we have found reduced expression of P21 (WAF1/CIP1) and P27, together with a slight increase of MYC, CHECK1 and MCM7 expression. In the ETV4-silenced PC3, we have found increased expression of P21 (Figures 5c and d) and reduced expression of MYC, CHECK1 and MCM7 (Figure 5c). These results suggest a direct or indirect role of ETV4 in cell cycle regulation.
ETV4 overexpression induces EMT in nonmalignant RWPE prostate cells
ETV4 expression in the nonmalignant RWPE cells results in an increase of their ability to migrate and invading that is paralleled by the modulation of the expression of genes involved in this function such as metalloproteinases and their inhibitors. Specifically, we found increased expression of MMP2 (TME-RWPE, 7.4±1.5; FL-RWPE, 41±5.5), MMP3 (TME-RWPE, 2.3±0.2; FL-RWPE, 16±1.2) and a slight reduction of TIMP1 (TME-RWPE, 0.5±0.1; FL-RWPE, 0.8±0.1).
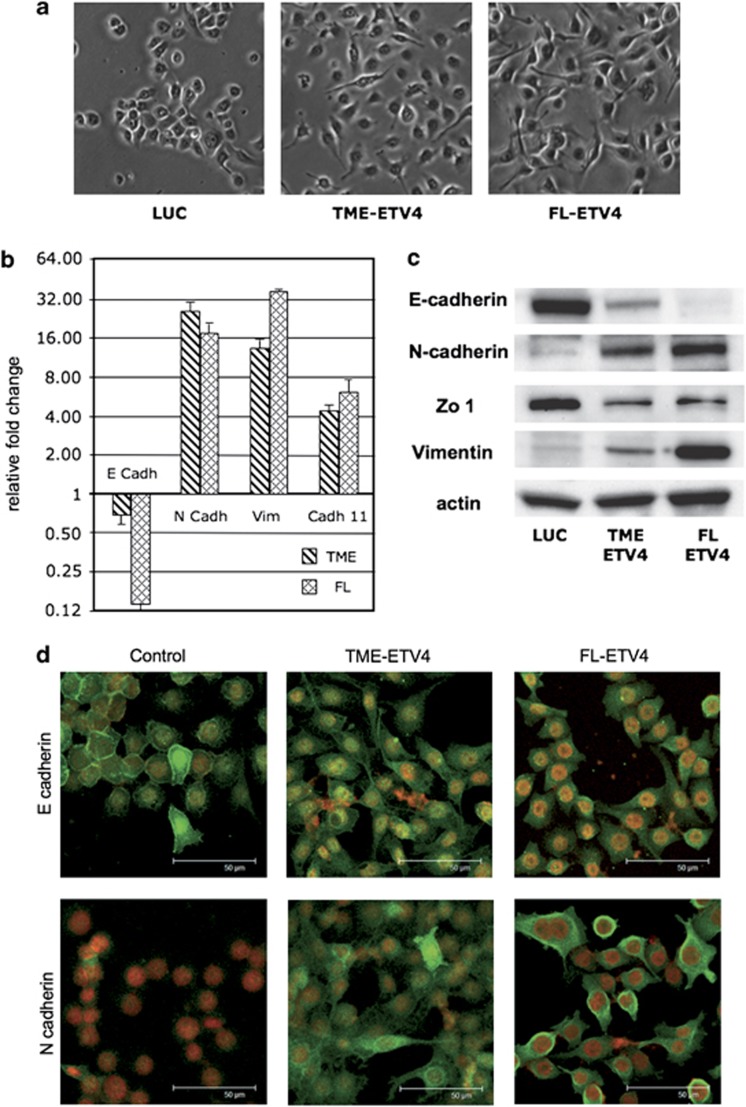
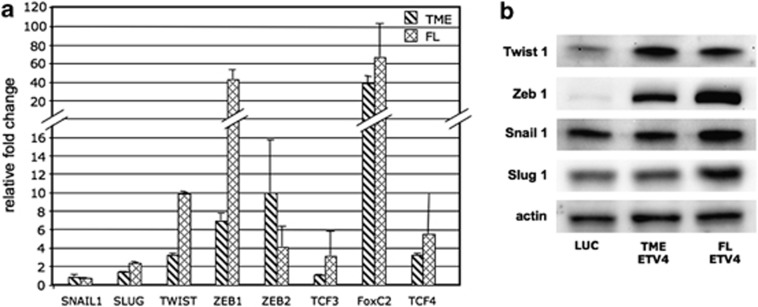
We observed that RWPE cells transfected with ETV4 expression vectors underwent shape changes. Spindle-shaped (mesenchymal-like) cells are rare in control RWPE cells (about 1%); in RWPE transfected with either TME or FL the frequency of spindle-shaped cells increased to 22% and 53%, respectively (Figure 6a). In addition, molecular markers of EMT were also affected. Overexpression of ETV4 was associated with decreased expression of E-cadherin and Zonula-occludens 1 (epithelial markers) and increased expression of vimentin, N-cadherin and cadherin-11 (mesenchymal markers) (Figures 6b and c). In addition, luciferase-transfected RWPE cells do not express N-cadherin, whereas E-cadherin is expressed and localized on the plasma membrane. In keeping with the acquisition of mesenchymal morphology, we found that in RWPE cells overexpressing ETV4, the epithelial marker protein E-cadherin is reduced and shifted from the plasma membrane to the cytoplasm (Figure 6d). In parallel, there is upregulation of the mesenchymal protein N-cadherin, which is now seen in both the cytoplasm and the plasma membrane (Figure 6d). We can therefore presume that the morphological changes of RWPE cells induced by ETV4 expression are mediated by changes in the levels and intracellular localization of E-cadherin and N-cadherin typical of mesenchymal cells. Next, we have tested for proteins that are known regulators of E-cadherin. ETV4-transfected RWPE cells have an increased expression of TWIST1, ZEB1, ZEB2, FOXC2 and TCF4, with only a slight increase of SLUG and TCF3 (Figures 7a and b). Interestingly, there was no variation of SNAIL1 (Figures 7a and b) and no expression of KLF8 and Goosecoid. In addition, we found no variations in the levels of two regulators of ZEB2,41 such as SPINT1 and IL2R (data not shown).
Figure 6.
ETV4 overexpression induces EMT in the RWPE. (a) Representative microphotographs of RWPE cells are shown. Almost all mock-transfected RWPE cells displayed a rounded epithelial cell shape with rare spindle-shaped cells. A large fraction of cells RWPE transfected with FL and TME vectors displayed a spindle-like shape. (b) Fold changes of expression levels (normalized to the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase) of epithelial (E-cadherin) and mesenchymal (N-cadherin, vimentin and cadherin 11) markers in RWPE cells transfected with FL-ETV4 and TME-ETV4 plasmids compared with mock-transfected RWPE cells. Fold changes are plotted on a Log 2 scale. (c) Expression assessed by western blot analysis of epithelial (E-cadherin and zonula occludens) and mesenchymal (N-cadherin and vimentin) markers in RWPE cells transfected with mock (Luc), FL-ETV4 and TME-ETV4 plasmids. (d) Analysis by confocal microscopy of E-cadherin and of N-cadherin in RWPE cells transfected with mock, FL-ETV4 and TME-ETV4 plasmids. The luciferase-transfected RWPE cells do not express the mesenchymal marker protein N-cadherin and the epithelial marker protein E-cadherin is expressed and localized on the plasma membrane. In RWPE cells overexpressing ETV4, either TME or FL, the E-cadherin is reduced and has migrated from the plasma membrane to the cytoplasm; the N-cadherin is upregulated and it appeared in both the cytoplasm and plasma membrane.
Figure 7.
The effect of ETV4 overexpression on the expression of genes involved in EMT in the RWPE. (a) Expression level (normalized to the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase) of transcription factors associated with EMT in RWPE cells transfected with mock, FL and TME plasmids. (b) ETV4 expression level assessed by western blot analysis of some transcription factors (TWIST1, ZEB1, SNAIL1 and SLUG1) associated with EMT in RWPE cells transfected with mock, FL and TME plasmids.
EMT is often associated with upregulation of the WNT pathway, and the overexpression of some components of this pathway has been reported in RWPE cells overexpressing ERG.42 In RWPE cells expressing ETV4, we found a marked increase of FZD4 (TME-RWPE, 7.0±4.0; FL-RWPE, 5.1±0.8) and a slight increase of WNT7A expression (TME-RWPE, 2.4±0.8; FL-RWPE, 1.6±0.40).
Taken together, these data indicate that ETV4 is a powerful inducer of EMT: the increase of several transcription factors relevant to EMT is in keeping with this notion.
Discussion
The discovery of translocations involving ETS proteins, of which TMPRSS2-ERG has been the trailblazer, is a major advance in understanding the molecular basis of PC. Among the ETS proteins, ERG, ETV5 and ETV1 have emerged as major factors in invasiveness and cell migration, but not in fostering cell proliferation;4,5,32 indeed, in a mouse model the prostate-specific overexpression of ERG has proven important in cancer progression, but its ability to initiate cancer is still controversial.4, 5, 6, 7 In contrast, we show here that ETV4, an ETS protein overexpressed in several types of carcinomas (that is, breast, lung and colon) and a partner of TMPRSS2 in chromosomal translocations found in PC, exerts a powerful function not only in invasiveness and in migration, but also in cell proliferation.
For our studies, we have obtained inducible silencing of ETV4 in two PC cell lines that overexpress this protein: PC3 and DU145. Interestingly, in PC3 the high level of ETV4 results from a high level of gene transcript; in DU 145, the ETV4 protein is just as high, despite a level of ETV4 mRNA comparable to that of the nonmalignant prostate cell line RWPE.
ETV4 confers migration and invading ability to PC cells
Both PC3 and DU145 cells are able to efficiently migrate in the wound-healing assay and to invade in the matrigel assay. ETV4 silencing reduces the migration and invasion of PC3 cells33 but not that of DU145 cells. In a variety of tissues ETV4 is a transcriptional activator of several MMPs,35, 43, 44 endopeptidases capable of degrading the extracellular matrix and thus promoting tumor invasion.45 Indeed, when we silenced ETV4 in PC3 cells the expression of MMP1, MMP3 and MMP9 also decreased; however, this was not the case in DU145 cells, suggesting that in these cells the mechanism of MMPs regulation is different. In complementary experiments, ETV4 overexpression confers migration and invading ability to the nonmalignant RWPE cells, and this is probably mediated by increased expression of MMP2 and MMP3.
ETV4 is involved in cell proliferation and in cell cycle regulation
We have confirmed previous evidence that ETV4 silencing reduces the anchorage-independent growth of PC3 cells,33 and we have shown here that this also applied to DU145 cells. In addition, we have shown that ETV4 expression increases anchorage-independent growth of RWPE cells. Whereas these data already suggest that ETV4 overexpression is oncogene-like, we have also uncovered an effect of ETV4 directed onto cell proliferation. Indeed, the progression of PC3 cells through the cell cycle is slowed down by ETV4 silencing; whereas the progression of RWPE cells through the cell cycle is accelerated by ETV4 expression. This effect on the rate of cycling appears to be mediated by or associated with the modulation of the cell cycle genes P21 (WAF1/CIP1), MYC, MCM7 and CHECK1; in RWPE cells also P27 (KIP1) is downregulated. Thus, ETV4 overexpression promotes cell cycle progression of the nonmalignant RWPE prostate cells probably by causing a reduction in P21 expression: accordingly, ETV4 silencing increases the levels of P21 in PC3 cells. These data are strengthened by the fact that for most genes the downregulation in PC3 is the mirror image of the upregulation in RWPE cells. The mechanism by which ETV4 hinders P21 transcription remains to be investigated. As P53 is a major activator of P21 transcription, one might have presumed that the ETV4 effect is mediated by P53.46, 47 However, this effect is seen even in PC3 that do not have P53 cells.48 It is possible that ETV4 downregulates directly P21 gene, resembling the downregulation of the ErbB2 (HER-2/neu) gene reported in breast and ovarian cancer cells as a result of ETV4 binding to the ErbB2 promoter.20, 21 However, it is also possible that part of the effect of ETV4 on P21 is mediated by a slightly increase of the expression of MYC and of its targets,39, 49 which we have observed in prostate cell lines overexpressing ETV4.
ETV4 overexpression activates EMT
The acquisition of invasiveness and metastatic capabilities by epithelial tumor cells is associated with a set of morphological and functional changes that are similar to EMT.23, 25 Various studies have reported about EMT-like process in PC (reviewed in Nauseef and Henry50) and in prostate cell lines after different stimuli (transforming growth factor-β,51, 52 DAB2IP53, 54). Recently, it has been reported that the TMPRSS2-ERG translocation is associated with EMT.41, 42 Here, we have shown that overexpression of ETV4 in RWPE cells induces EMT changes: in fact, not only the cells did acquire a fibroblast-like shape, but this was also associated with an increased expression of mesenchymal markers (vimentin, N-cadherin and cadherin-11) and a decreased expression of epithelial markers (E-cadherin, zonula occludens 1). Specifically, the reduction of the epithelial marker protein E-cadherin paralleled quantitatively an increase in the mesenchymal marker protein N-cadherin on the cell membrane (Figure 6d). Thus, just as in lung cancer cells55 and in ovarian cancer cells,56 ETV4 expression is able to induce EMT transition also in PC cells.
As for the mechanism whereby ETV4 can induce EMT, this is probably mediated by several transcription factors, including members of the Snail family (SNAIL1, SNAIL2/SLUG), of the zinc finger E-box-binding homeobox family (ZEB1 and ZEB2), TCF3 and KLF826, 57, 58 that directly repress E-cadherin transcription, and others that are indirect E-cadherin repressors (the basic helix loop-helix factor TWIST1, Goosecoid, FOXC2 and TCF4).23, 59, 60, 61
Recently, it has been found that ETV4 upregulates TWIST1 in a murine breast cancer cell line62 and SNAIL1 in MCF10a (an immortalized nontumorigenic human breast cell line);63 in addition, a direct correlation between the mRNA levels of ETV4 and SNAIL1 has been observed in breast cancer patients.63 We show here that ETV4 expression in the nonmalignant RWPE prostate cells results in a strong increase of TWIST1, ZEB1 and ZEB2 expression; but, unlike in breast cell lines, not that of SNAIL1: this suggests, that the mechanism whereby ETV4 induces EMT is at least in part tissue specific.
ETV4 resembles ERG in some of the mechanisms leading to EMT, such as activation of the WNT pathway42 and induction of ZEB1 and ZEB2.41 However, ETV4 differs from ERG because it does not affect the expression of ZEB2 regulators. At any rate, although the mechanism through which ETV4 and ERG induce EMT may be somewhat different, it appears that EMT is a final common pathway through which these ETS proteins exert their oncogenic potential in prostate cells.
Conclusion
ETV4 is not alone in causing EMT, but what sets it apart from the other ETS proteins, we have mentioned, is that it exerts also a major effect on the rate of cell proliferation. This finding is novel with respect to prostate, and it confirms what has been observed for other types of cancer.18, 64, 65 Our findings in complementary cellular models of PC strongly support the notion that a certain level of expression of ETV4, by affecting positively both proliferation and invasiveness, is capable of inducing most and possibly all neoplastic features of PC. Most important, these findings have been confirmed in vivo in xenografts. Thus, it is possible that ETV4 overexpression, whether driven by the TMPRSS2 promoter or otherwise, may turn out to be a more powerful oncogene than others. This is of considerable importance, as the correlation between translocations involving ETS genes and the clinical course of PC is still controversial. From this point of view, it is tempting to regard ETV4 as a potential target for new therapeutic approaches.
Materials and methods
Vectors
shRNAs against ETV4 (NM_001986) were designed with the ‘siRNA selection program' (Whitehead Institute: http://jura.wi.mit.edu/siRNAext): GGCGCTTCCCAACTTCATA (sh65) and CCCTGTGTACATATAAATGAA (sh3). These shRNAs and an irrelevant shRNA (GCCTATTTACGCCTGACAA) were cloned in the pLVTHM lentiviral plasmid (gift of D Trono): a vector whose shRNA expression is doxycycline-inducible in cells expressing the modified tetracycline repressor tTR-Krab. The fragment containing the tTR-Krab under the control of the human EF1a promoter, excised from the pLV-tTRKrab plasmid,31 was cloned into pcDNA3 to generate the pcEF-Krab vector. shRNA vectors were used to transduce prostate cells transfected (doxycycline-inducible ETV4 silencing) or not transfected (constitutive ETV4 silencing) with pcEF-Krab.
The FL-ETV4 vector was obtained cloning the human ETV4 cDNA, amplified from PC3 cells, in a pLG4.2 vector (Promega, Madison, WI, USA) derivative containing the EF1a promoter. The TME-ETV4 vector was obtained from the FL-ETV4 replacing the 5′ end of ETV4 with a PCR amplicon in which we have introduced the 5′ portion of TMPRSS2–ETV4 fusion gene.8
Cell lines
Du145, PC3, LnCap and Hek293 (IST Cell Bank and Cell Factory, Genoa, Italy), V-Cap and RWPE (American Type Culture Collection, Manassas, VA, USA) cell lines were cultured according to cell-bank instructions. Serum-free KER medium for RWPE was supplemented with epidermal growth factor and Bovine Pituitary Extract (Gibco, Carlsband, CA, USA).
Du145 and PC3 cells were electroporated with pEF1-Krab plasmid and selected with G418 (Sigma, St Louis, MO, USA). The clones expressing higher Krab levels were stably transduced by calcium phosphate precipitation66 with doxycycline-inducible shRNA pLVTHM vectors. shRNAs expression was induced by doxycycline (0.5 μg/ml, Sigma) for at least 5 days. RWPE cells were stably transfected with FL-ETV4, TME-ETV4 and luciferase-containing vector using Lipofectamine (Invitrogen, Carlsbad, CA, USA) and selected with puromycin (Sigma).
Quantitative real-time PCR
Total RNA was isolated with the RNeasy Mini Kit (Qiagen, Valencia, CA, USA) and reverse transcribed (iScript, Biorad, Hercules, CA, USA). The expression of genes listed in Table 1 was quantified by quantitative real-time PCR (SsoFast EvaGreen Supermix with CFX96 thermocycler: Biorad).
Table 1. Sequences of oligonucleotides used for quantitative RT–PCR.
| Primer name | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| ETV4 | 5′-GCTCGCTGAAGCTCAGGT-3′ | 5′-TCCTTCTTGATCCTGGTGGT-3′ |
| GAPDH | 5′-AACGGATTTGGTCGTATTGGGC-3′ | 5′-TTGATTTTGGAGGGATCTCG-3′ |
| TBP | 5′-CCACAGCTCTTCCACTCACA-3′ | 5′-GCGGTACAATCCCAGAACTC-3′ |
| MMP1 | 5′-AACTCGGCCATTCTCTTGGA-3′ | 5′-TGGCTTGGATGCCATCAAT-3′ |
| MMP2 | 5′-ACGACCGCGACAAGAAGTAT-3′ | 5′-ATTTGTTGCCCAGGAAAGTG-3′ |
| MMP3 | 5′-ATGCAGAAGTTCCTTGGATTGG-3′ | 5′-GATGCCAGGAAAGGTTCTGAAG-3′ |
| MMP7 | 5′-GCTGGCTCATGCCTTTGC-3′ | 5′-TCATGAGTTGCAGCATACAGGAA-3′ |
| MMP9 | 5′-GGGCTCCCGTCCTGCTT-3′ | 5′-CCTCCACTCCTCCCTTTCCT-3′ |
| UPAR | 5′-TCTGCAGGACCACGATCGT-3′ | 5′-TCTTCAAGCCAGTCCGATAGC-3′ |
| UPA | 5′-GATCCCCAGTTTGGCACAAG-3′ | 5′-ACACTCCCGGTGGGAAATC-3′ |
| TIMP1 | 5′-GGGACACCAGAAGTCAACCA-3′ | 5′-GGCTTGGAACCCTTTATACATC-3′ |
| TIMP2 | 5′-CCACCCAGAAGAAGAGCCTGA-3′ | 5′-GTGACCCAGTCCATCCAGAG-3′ |
| ADAM9 | 5′-CAGATGGCAAAAATCAAGCA-3′ | 5′-GATGGGAACTGCTGAGGTTG-3′ |
| ADAM10 | 5′-TCCACAGCCCATTCAGCAA-3′ | 5′-AGGCACTAGGAAGAACCAA-3′ |
| ADAM17 | 5′-TTCACGTTTGCAGTCTCCAA-3′ | 5′-ATGTATCTGTAGAAGCGATGATCTG-3′ |
| ADAMTS1 | 5′-AAGGACAGGTGCAAGCTCAT-3′ | 5′-GAGGTGGAATCTGGGCTACA-3′ |
| P27 | 5′-AATAAGGAAGCGACCTGCAA-3′ | 5′-TTCTGAGGCCAGGCTTCTT-3′ |
| p21 | 5′-CCATGTGGACCTGTCACTGT-3′ | 5′-TGGTAGAAATCTGTCATGCTGGTC-3′ |
| MYC | 5′-TGCAGCTGCTTAGACGCTGG-3′ | 5′-CGAGGTCATAGTTCCTGTTGG-3′ |
| CHEK1 | 5′-TGGTATTGGAATAACTCACAGGG-3′ | 5′-CCGAAATACTGTTGCCAAGCC-3′ |
| MCM7 | 5′-CGAAGCTCTTTGCTGATGCC-3′ | 5′-CCGATGCTCAATGTAAACGTCC-3′ |
| VIM | 5′-CGCCATCAACACCGAGTTC-3′ | 5′-ATCTTATTCTGCTGCTCCAGGAA-3′ |
| E-cadherin | 5′-GAACGCATTGCCACATACAC-3′ | 5′-ATTCGGGCTTGTTGTCATTC-3′ |
| N-cadherin | 5′-TCAGTGGCGGAGATCCTACT-3′ | 5′-GTGCTGAATTCCCTTGGCTA-3′ |
| 11-cadherin | 5′-CAGCCCGATAAGGTATTCCA-3′ | 5′-TGTGGATTTCTGCTGCAAAG-3′ |
| SNAIL1 | 5′-GCGAGCTGCAGGACTCTAAT-3′ | 5′-GGACAGAGTCCCAGATGAGC-3′ |
| SNAIL2 | 5′-GCCTCCAAAAAGCCAAACTA-3′ | 5′-CACAGTGATGGGGCTGTATG-3′ |
| TWIST1 | 5′-GCCGGAGACCTAGATGTCATT-3′ | 5′-CACGCCCTGTTTCTTTGAAT-3′ |
| ZEB1 | 5′-GCCAATAAGCAAACGATTCTG-3′ | 5′-TTTGGCTGGATCACTTTCAAG-3′ |
| ZEB2 | 5′-AAGCCAGGGACAGATCAGC-3′ | 5′-CCACACTCTGTGCATTTGAACT-3′ |
| FOXC2 | 5′-GCCCAGCAGCAAACTTTCC-3′ | 5′-CCGGTGGGAGTTGAACATCT-3′ |
| GSC | 5′-AGCAGCTCGAAGCTCTCGAG-3′ | 5′-CACGTCCGGGTACTTGGTCT-3′ |
| IL1R2 | 5′-CTACGCACCACAGTCAAGGAAG-3′ | 5′-CGTCTGTGCATCCATATTCCC-3′ |
| SPINT | 5′-AGACTACTGCCTCGCATCCAA-3′ | 5′-CAAGCAGCCTCCATAAACGAA-3′ |
| KLF8 | 5′-TCAGAAGGTGGCTCAATGCA-3′ | 5′-CCGAACAGAAGCAGTGACCTG-3′ |
| TCF3 | 5′-AGCTCCTCCTTTGACCCCAG-3′ | 5′-ACTCAGTGAAGTGGGTGCCC-3′ |
| TCF4 | 5′-TGCGATGTTTTCACCTCCTG-3′ | 5′-TGCCAAAGAAGTTGGTCCATT-3′ |
| ZO1 | 5′-GGAGAGGTGTTCCGTGTTGT-3′ | 5′-GGCTAGCTGCTCAGCTCTGT-3′ |
| WNT7A | 5′-TGTGGCTGCGACAAAGAGAA-3′ | 5′-TCCACAAAGACCTTGGCGA-3′ |
| FZD4 | 5′-TTCACACCGCTCATCCAGTA-3′ | 5′-TGCACATTGGCACATAAACA-3′ |
Abbreviations: RT–PCR, real-time–PCR; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Western blot analysis
Proteins, obtained from 70% confluent cells, were separated by either 10 or 15% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Biorad). Proteins were detected by the following primary antibodies: ETV4 monoclonal antibody (Abnova, Atlanta, GA, USA); actin (Sigma-Aldrich, St Louis, MO, USA); vimentin, twist, E-cadherin, N-cadherin and Snail1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA); Zeb1 Zo-1 and Slug (Cell Signaling, Danvers, MA, USA). Horseradish peroxidase-conjugated secondary antibody signals were detected using enhanced chemiluminescence (Pierce, Rockford, IL, USA) and ImageQuant 350 apparatus (GE Healthcare, Pittsburg, PA, USA).
Soft agar assays
Briefly, 1 × 104 and 1 × 105 cells of each cell line were suspended in 4 ml 0.3% Noble agar (BD Biosciences; Bedford, MA, USA) in the appropriate medium (0.5 μg/ml of doxycycline was added for shRNA induction). The 4 ml agar–cell mixture was layered on 4-ml bottom layer of 0.5% agar medium. Weekly, 300 μl of medium (or medium with 0.8 μg/ml doxycycline) has been added to each dish. After 21 days at 37 °C, colonies were stained and counted by microscope. Experiments have been performed three times in triplicate.
Cell cycle analysis
Apoptosis was analyzed using the AnnexinV/PI apoptosis kit (BD Biosciences) on FACScan flow cytometer (BD Bioscience). Cell cycle analysis was performed on synchronized cells (overnight 10 mℳ hydroxyurea) by flow cytometry. At different time points from hydroxyurea removal, cells were washed, fixed with 70% ethanol, centrifuged and suspended in a propidium iodide (50 μg/ml) staining buffer. Cell cycle distribution was analyzed using the ModFit LT software (BD Biosciences).
Migration assay
A wound was created by a yellow tip on cells grown to confluence on 6-well plates; fresh medium (with 0.5 μℳ Mitomycin C to prevent cell proliferation) was added and migration was measured on photographs taken at 0 and 48 h.
Invasion assay
Transwell filters of 8-μm pore size (Corning, Lowell, MA, USA) were coated with 50 μl of 5% matrigel (BD Biosciences); 500 μl of the appropriate medium with 10% fetal bovine serum was loaded in the lower chambers. Single-cell suspensions (1 × 105 PC3/DU145 or 1.5 × 105 RWPE cells/well) in medium without serum (and without supplements for RWPE cells) were loaded in the upper chambers. Filters were incubated 24 h at 37 °C in CO2 incubator and, after removal of cells from the upper surface with a cotton swab, were stained with DIFF-QUICK (Medicult, Firenze, Italy). Invading cells, attached to the lower surface, were counted by microscope.
Mouse xenograft
Athymic nude immunodeficient mice (Harlan, Udine, Italy) were maintained at the CeSAL (University of Florence, Firenze, Italy) and treated according to the European Union guidelines and to protocols approved by the local Animal Ethical Committee. Xenografts were generated by subcutaneous injection of 200 μl of phosphate-buffered saline containing either PC3 (4 × 106) or DU145 (2 × 106) cells. Half of the animals were randomly chosen to receive doxycycline (1 mg/ml in drinking water together with 5% saccarose) to induce shRNA. After 8 weeks tumor weight and volume were measured.
Immunofluorescence
Cells, after overnight growth on sterile coverslips, were fixed with 2% formaldehyde, permeabilized with 0.1% Triton X-100 and blocked in 5% horse serum. Cells were incubated with primary antibodies against E-cadherin and N-cadherin followed by incubation with AlexaFluor488-conjugated goat-anti-rabbit secondary antibodies (Invitrogen) and nuclei counterstained with propidium iodide (Sigma). Immunofluorescence was examined using a Leica SP2-AOBS confocal microscope (Leica Microsystems, Nussloch, Germany).
Statistical analysis
All data are expressed as mean±s.d. Statistical analysis was performed using t-test. Statistical significance was accepted for P<0.05.
Acknowledgments
We are indebted to Lucio Luzzatto for much support, helpful suggestions and fruitful discussions. We thank Paola Chiarugi and Giannino Del Sal for valuable discussions and suggestions. We also thank Tommaso Rondelli for the help in the laboratory, Tommaso Melo for his help with confocal microscopy and Luca Boni for statistical advice. AP is a PhD student at the Department of Biochemistry, University of Florence. This work has been supported by a start-up grant from the Istituto Toscano Tumori.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Oncogenesis website (http://www.nature.com/oncsis).
Supplementary Material
References
- Mitelman F, Johansson B, Mertens F. Fusion genes and rearranged genes as a linear function of chromosome aberrations in cancer. Nat Genet. 2004;36:331–334. doi: 10.1038/ng1335. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- Rubin MA, Maher CA, Chinnaiyan AM. Common gene rearrangements in prostate cancer. J Clin Oncol. 2011;29:3659–3668. doi: 10.1200/JCO.2011.35.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Laxman B, Varambally S, Cao X, Yu J, Helgeson BE, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10:177–188. doi: 10.1593/neo.07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klezovitch O, Risk M, Coleman I, Lucas JM, Null M, True LD, et al. A causal role for ERG in neoplastic transformation of prostate epithelium. Proc Natl Acad Sci USA. 2008;105:2105–2110. doi: 10.1073/pnas.0711711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41:524–526. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Mehra R, Rhodes DR, Smith LR, Roulston D, Helgeson BE, et al. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66:3396–3400. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- Han B, Mehra R, Dhanasekaran SM, Yu J, Menon A, Lonigro RJ, et al. A fluorescence in situ hybridization screen for E26 transformation-specific aberrations: identification of DDX5-ETV4 fusion protein in prostate cancer. Cancer Res. 2008;68:7629–7637. doi: 10.1158/0008-5472.CAN-08-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans KG, Bressers AA, van der Korput HA, Dits NF, Jenster G, Trapman J. Two unique novel prostate-specific and androgen-regulated fusion partners of ETV4 in prostate cancer. Cancer Res. 2008;68:3094–3098. doi: 10.1158/0008-5472.CAN-08-0198. [DOI] [PubMed] [Google Scholar]
- Iljin K, Wolf M, Edgren H, Gupta S, Kilpinen S, Skotheim RI, et al. TMPRSS2 fusions with oncogenic ETS factors in prostate cancer involve unbalanced genomic rearrangements and are associated with HDAC1 and epigenetic reprogramming. Cancer Res. 2006;66:10242–10246. doi: 10.1158/0008-5472.CAN-06-1986. [DOI] [PubMed] [Google Scholar]
- Paulo P, Barros-Silva JD, Ribeiro FR, Ramalho-Carvalho J, Jeronimo C, Henrique R, et al. FLI1 is a novel ETS transcription factor involved in gene fusions in prostate cancer. Genes Chromosomes Cancer. 2012;51:240–249. doi: 10.1002/gcc.20948. [DOI] [PubMed] [Google Scholar]
- Hida K, Shindoh M, Yoshida K, Kudoh A, Furaoka K, Kohgo T, et al. Expression of E1AF, an ets-family transcription factor, is correlated with the invasive phenotype of oral squamous cell carcinoma. Oral Oncol. 1997;33:426–430. doi: 10.1016/s0964-1955(97)00047-x. [DOI] [PubMed] [Google Scholar]
- Hiroumi H, Dosaka-Akita H, Yoshida K, Shindoh M, Ohbuchi T, Fujinaga K, et al. Expression of E1AF/PEA3, an Ets-related transcription factor in human non-small-cell lung cancers: its relevance in cell motility and invasion. Int J Cancer. 2001;93:786–791. doi: 10.1002/ijc.1410. [DOI] [PubMed] [Google Scholar]
- Benz CC, O'Hagan RC, Richter B, Scott GK, Chang CH, Xiong X, et al. HER2/Neu and the Ets transcription activator PEA3 are coordinately upregulated in human breast cancer. Oncogene. 1997;15:1513–1525. doi: 10.1038/sj.onc.1201331. [DOI] [PubMed] [Google Scholar]
- de Launoit Y, Chotteau-Lelievre A, Beaudoin C, Coutte L, Netzer S, Brenner C, et al. The PEA3 group of ETS-related transcription factors. Role in breast cancer metastasis. Adv Exp Med Biol. 2000;480:107–116. doi: 10.1007/0-306-46832-8_13. [DOI] [PubMed] [Google Scholar]
- Discenza MT, Vaz D, Hassell JA, Pelletier J. Activation of the WT1 tumor suppressor gene promoter by Pea3. FEBS Lett. 2004;560:183–191. doi: 10.1016/S0014-5793(04)00104-8. [DOI] [PubMed] [Google Scholar]
- Jiang J, Wei Y, Liu D, Zhou J, Shen J, Chen X, et al. E1AF promotes breast cancer cell cycle progression via upregulation of Cyclin D3 transcription. Biochem Biophys Res Commun. 2007;358:53–58. doi: 10.1016/j.bbrc.2007.04.043. [DOI] [PubMed] [Google Scholar]
- Kaya M, Yoshida K, Higashino F, Mitaka T, Ishii S, Fujinaga K. A single ets-related transcription factor, E1AF, confers invasive phenotype on human cancer cells. Oncogene. 1996;12:221–227. [PubMed] [Google Scholar]
- Xing X, Wang SC, Xia W, Zou Y, Shao R, Kwong KY, et al. The ets protein PEA3 suppresses HER-2/neu overexpression and inhibits tumorigenesis. Nat Med. 2000;6:189–195. doi: 10.1038/72294. [DOI] [PubMed] [Google Scholar]
- Yu Z, Xia W, Wang HY, Wang SC, Pan Y, Kwong KY, et al. Antitumor activity of an Ets protein, PEA3, in breast cancer cell lines MDA-MB-361DYT2 and BT474M1. Mol Carcinog. 2006;45:667–675. doi: 10.1002/mc.20212. [DOI] [PubMed] [Google Scholar]
- Hugo H, Ackland ML, Blick T, Lawrence MG, Clements JA, Williams ED, et al. Epithelial–mesenchymal and mesenchymal–epithelial transitions in carcinoma progression. J Cell Physiol. 2007;213:374–383. doi: 10.1002/jcp.21223. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymkowsky MW, Savagner P. Epithelial-mesenchymal transition: a cancer researcher's conceptual friend and foe. Am J Pathol. 2009;174:1588–1593. doi: 10.2353/ajpath.2009.080545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Portillo F, Cano A. Transcriptional regulation of cadherins during development and carcinogenesis. Int J Dev Biol. 2004;48:365–375. doi: 10.1387/ijdb.041794hp. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- Thompson EW, Newgreen DF, Tarin D.Carcinoma invasion and metastasis: a role for epithelial-mesenchymal transition Cancer Res 2005655991–5995.discussion 5995. [DOI] [PubMed] [Google Scholar]
- Armstrong AJ, Marengo MS, Oltean S, Kemeny G, Bitting RL, Turnbull JD, et al. Circulating tumor cells from patients with advanced prostate and breast cancer display both epithelial and mesenchymal markers. Mol Cancer Res. 2011;9:997–1007. doi: 10.1158/1541-7786.MCR-10-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- Wiznerowicz M, Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J Virol. 2003;77:8957–8961. doi: 10.1128/JVI.77.16.8957-8961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- Hollenhorst PC, Paul L, Ferris MW, Graves BJ. The ETS gene ETV4 is required for anchorage-independent growth and a cell proliferation gene expression program in PC3 prostate cells. Genes Cancer. 2011;1:1044–1052. doi: 10.1177/1947601910395578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi S, Yamamoto H, Min Y, Adachi Y, Itoh F, Imai K. Association of ets-related transcriptional factor E1AF expression with tumour progression and overexpression of MMP-1 and matrilysin in human colorectal cancer. J Pathol. 2003;200:568–576. doi: 10.1002/path.1387. [DOI] [PubMed] [Google Scholar]
- Higashino F, Yoshida K, Noumi T, Seiki M, Fujinaga K. Ets-related protein E1A-F can activate three different matrix metalloproteinase gene promoters. Oncogene. 1995;10:1461–1463. [PubMed] [Google Scholar]
- Bourboulia D, Stetler-Stevenson WG. Matrix metalloproteinases (MMPs) and tissue inhibitors of metalloproteinases (TIMPs): Positive and negative regulators in tumor cell adhesion. Semin Cancer Biol. 2010;20:161–168. doi: 10.1016/j.semcancer.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickey DD, Stone KR, Wunderli H, Mickey GH, Vollmer RT, Paulson DF. Heterotransplantation of a human prostatic adenocarcinoma cell line in nude mice. Cancer Res. 1977;37:4049–4058. [PubMed] [Google Scholar]
- Ware JL, DeLong ER. Influence of tumour size on human prostate tumour metastasis in athymic nude mice. Br J Cancer. 1985;51:419–423. doi: 10.1038/bjc.1985.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold S, Herkert B, Eilers M. Facilitating replication under stress: an oncogenic function of MYC. Nat Rev Cancer. 2009;9:441–444. doi: 10.1038/nrc2640. [DOI] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- Leshem O, Madar S, Kogan-Sakin I, Kamer I, Goldstein I, Brosh R, et al. TMPRSS2/ERG promotes epithelial to mesenchymal transition through the ZEB1/ZEB2 axis in a prostate cancer model. PLoS One. 2011;6:e21650. doi: 10.1371/journal.pone.0021650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Iljin K, Sara H, Mpindi JP, Mirtti T, Vainio P, et al. FZD4 as a mediator of ERG oncogene-induced WNT signaling and epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res. 2011;70:6735–6745. doi: 10.1158/0008-5472.CAN-10-0244. [DOI] [PubMed] [Google Scholar]
- Habelhah H, Okada F, Kobayashi M, Nakai K, Choi S, Hamada J, et al. Increased E1AF expression in mouse fibrosarcoma promotes metastasis through induction of MT1-MMP expression. Oncogene. 1999;18:1771–1776. doi: 10.1038/sj.onc.1202465. [DOI] [PubMed] [Google Scholar]
- Crawford HC, Fingleton B, Gustavson MD, Kurpios N, Wagenaar RA, Hassell JA, et al. The PEA3 subfamily of Ets transcription factors synergizes with beta-catenin-LEF-1 to activate matrilysin transcription in intestinal tumors. Mol Cell Biol. 2001;21:1370–1383. doi: 10.1128/MCB.21.4.1370-1383.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs WB, Carter BS, Ewing CM. Wild-type p53 suppresses growth of human prostate cancer cells containing mutant p53 alleles. Cancer Res. 1991;51:4716–4720. [PubMed] [Google Scholar]
- Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN, et al. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci USA. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nauseef JT, Henry MD. Epithelial-to-mesenchymal transition in prostate cancer: paradigm or puzzle. Nat Rev Urol. 2011;8:428–439. doi: 10.1038/nrurol.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao M, Williams K, Bhowmick NA, Hayward SW. Transforming growth factor-beta promotes invasion in tumorigenic but not in nontumorigenic human prostatic epithelial cells. Cancer Res. 2006;66:8007–8016. doi: 10.1158/0008-5472.CAN-05-4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slabakova E, Pernicova Z, Slavickova E, Starsichova A, Kozubik A, Soucek K. TGF-beta1-induced EMT of non-transformed prostate hyperplasia cells is characterized by early induction of SNAI2/Slug. Prostate. 2011;71:1332–1343. doi: 10.1002/pros.21350. [DOI] [PubMed] [Google Scholar]
- Min J, Zaslavsky A, Fedele G, McLaughlin SK, Reczek EE, De Raedt T, et al. An oncogene-tumor suppressor cascade drives metastatic prostate cancer by coordinately activating Ras and nuclear factor-kappaB. Nat Med. 2010;16:286–294. doi: 10.1038/nm.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D, Gore C, Liu J, Pong RC, Mason R, Hao G, et al. Role of DAB2IP in modulating epithelial-to-mesenchymal transition and prostate cancer metastasis. Proc Natl Acad Sci USA. 2010;107:2485–2490. doi: 10.1073/pnas.0908133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay S, Liu C, Chatterjee A, Hoque MO, Kim MS, Engles J, et al. LKB1/STK11 suppresses cyclooxygenase-2 induction and cellular invasion through PEA3 in lung cancer. Cancer Res. 2006;66:7870–7879. doi: 10.1158/0008-5472.CAN-05-2902. [DOI] [PubMed] [Google Scholar]
- Cowden Dahl KD, Dahl R, Kruichak JN, Hudson LG. The epidermal growth factor receptor responsive miR-125a represses mesenchymal morphology in ovarian cancer cells. Neoplasia. 2009;11:1208–1215. doi: 10.1593/neo.09942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- Moody SE, Perez D, Pan TC, Sarkisian CJ, Portocarrero CP, Sterner CJ, et al. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 2005;8:197–209. doi: 10.1016/j.ccr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Sobrado VR, Moreno-Bueno G, Cubillo E, Holt LJ, Nieto MA, Portillo F, et al. The class I bHLH factors E2-2A and E2-2B regulate EMT. J Cell Sci. 2009;122:1014–1024. doi: 10.1242/jcs.028241. [DOI] [PubMed] [Google Scholar]
- Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Qin L, Liu Z, Chen H, Xu J. The steroid receptor coactivator-1 regulates twist expression and promotes breast cancer metastasis. Cancer Res. 2009;69:3819–3827. doi: 10.1158/0008-5472.CAN-08-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen HF, Chan YK, Grills C, McCrudden CM, Gunasekharan V, Shi Z, et al. Polyomavirus enhancer activator 3 protein promotes breast cancer metastatic progression through Snail-induced epithelial-mesenchymal transition. J Pathol. 2011;224:78–89. doi: 10.1002/path.2859. [DOI] [PubMed] [Google Scholar]
- Moss AC, Lawlor G, Murray D, Tighe D, Madden SF, Mulligan AM, et al. ETV4 and Myeov knockdown impairs colon cancer cell line proliferation and invasion. Biochem Biophys Res Commun. 2006;345:216–221. doi: 10.1016/j.bbrc.2006.04.094. [DOI] [PubMed] [Google Scholar]
- Galang CK, Muller WJ, Foos G, Oshima RG, Hauser CA. Changes in the expression of many Ets family transcription factors and of potential target genes in normal mammary tissue and tumors. J Biol Chem. 2004;279:11281–11292. doi: 10.1074/jbc.M311887200. [DOI] [PubMed] [Google Scholar]
- Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.