Abstract
The thermostabilization of an enzyme while maintaining its activity for industrial or biomedical applications using traditional selection methods can be difficult. We demonstrate a rapid computational approach that identified three mutations within a model enzyme that produce a 10°C increase in apparent Tm and a 30-fold increase in half-life at 50°C, with no reduction in catalytic efficiency. The effects of the mutations were synergistic, giving an increase in excess of the sum of their individual effects. The redesigned enzyme induces an increased, temperature-dependent bacterial growth rate under conditions that require its activity, thereby coupling molecular and metabolic engineering.
Enzymes are the most efficient catalysts of chemical reactions known, enhancing reaction rates by as much as twenty-three orders of magnitude (1, 2). However, there has been little evolutionary pressure for them to become more thermostable than is required by their native environment. Many studies indicate that enzymes (like most proteins) exhibit closely balanced free energy profiles for folding and unfolding, thereby allowing functionally important dynamic motions and appropriate degradation in vivo (3). However, in a laboratory or industrial setting this lack of thermostability can lead to undesirable loss of activity (4).
The physical principles of protein folding that result in a balance of stability and flexibility, while maintaining function, are not perfectly understood and have been difficult to exploit for the development of thermostabilized enzymes (4). For hyperthermophiles, selective pressures have generated proteins with denaturation temperatures upwards of 110° C (5). Their proteins exhibit topologies and stabilizing interactions similar to those from mesophilic and thermophilic organisms (6, 7) leading to diverse hypotheses regarding their relative behaviors (8). However, a key mechanism for thermostabilization appears to be optimization of interactions between amino acids within their core (5), complementing computational design methods which optimize a sequence for a given fold (9-13).
The thermostabilization of an enzyme presents additional challenges for computational protein design methods because the active site substrate geometry and the molecular dynamic behavior during an enzymatic reaction often appear fine-tuned for maximum catalytic efficiency (2, 3). Therefore the design method must be capable of predicting thermostabilizing mutations within a given fold while minimizing any shift in the backbone that might structurally disrupt the active site structure or quench its flexibility.
In the past several years, methods for computational protein structure prediction and design have improved significantly (10, 11, 14). Recently, computational design has been used successfully in thermostabilizing non-catalytic proteins (15-18), redesigning binding pockets (19-23), creating a novel protein fold (24) and designing catalytic activity into a bacterial receptor (25). We use the program RosettaDesign (26), which consists of an energy function for evaluating the fitness of a particular sequence for a given fold and a Metropolis Monte Carlo search algorithm for sampling sequence space. The program requires a backbone structure as input, and generates sequences that are lowest energy for that fold.
We picked the homodimeric hydrolase enzyme yeast cytosine deaminase (yCD), which converts cytosine to uracil, as a target for computational thermostabilization. yCD was chosen because its high-resolution crystal structure is available (27), its catalytic mechanism is well characterized (27), it is thermolabile (28, 29) and it has potential use in anti-tumor suicide gene therapy applications (27, 29-31). As do many commercially useful enzymes, the enzyme displays irreversible unfolding behavior at high temperatures (presumably due to aggregation), rather than the more simple, fully reversible behavior common among model systems for the study of protein folding. The problems inherent in engineering such catalysts have been recently reviewed (4). Computational redesign was used to predict a series of point mutations in the enzyme core that might lead to thermostabilization of the enzyme while maintaining catalytic efficiency. A series of designed enzyme variants were prepared and their folded thermostability, catalytic behavior, ability to complement metabolic cytosine deaminase activity and three-dimensional crystal structures were determined.
The general computational strategy is largely unchanged from that described in (26). An energy function evaluates target sequences threaded onto a template backbone (12, 13, 26, 32). Sequence space is searched with an iterative Metropolis Monte Carlo procedure starting with a random sequence, replacing a single amino acid rotamer with a rotamer from the Dunbrack backbone dependent rotamer library(33) and reevaluating the energy using the energy function. Sequences with lower energy are automatically adopted, while sequences with higher energy are accepted with a probability based on the Metropolis criterion in order to prevent trapping in a local energy minimum. The methodology for computational redesign and all subsequent experiments are provided in supplementary online materials (34).
All residues directly involved in catalysis, those located within 4Å of the active site, and those involved in the dimer interface were held fixed (Figure S1). The remaining 65 residues of the 153 residue monomer were included in the redesign, allowing them to be changed to any amino acid except cysteine. Thirty-three of the 65 residues subjected to redesign (49%) remained wild-type, a result similar to prior applications (18, 26). Sixteen of the point mutations suggested by the program were located on the surface of the protein and were not pursued, while the remainder were in the core. The core mutations could be further subdivided into two localized clusters of interacting residues, as well as four additional isolated point mutations.
Site directed mutagenesis was used to generate each of the two complete clusters of point mutations, and the four individual mutations described above. Cluster 1, consisting of nine simultaneous mutations packed between an α-helix and several β-strands (including replacement of a buried salt-bridge) aggregated at concentrations above 0.4 mg/mL and was not characterized further. Cluster 2, consisting of four mutations packed between two α-helices, remained soluble when concentrated to 20 mg/mL. Individual mutations from this cluster revealed that A23L and I140L were key to the thermostabilization of the enzyme and were included in the final construct described below. Of the remaining four individual mutations, one (V108I) was also incorporated in the thermostabilized triple-mutant enzyme, while the remaining three (W10T, T67E and E69L) were not as well behaved and were not characterized further. Both the double mutant (A23L/I140L) and the final triple mutant (A23L/I140L/V108I) were well behaved during expression and purification, were more thermostable than the wild-type enzyme and fully active.
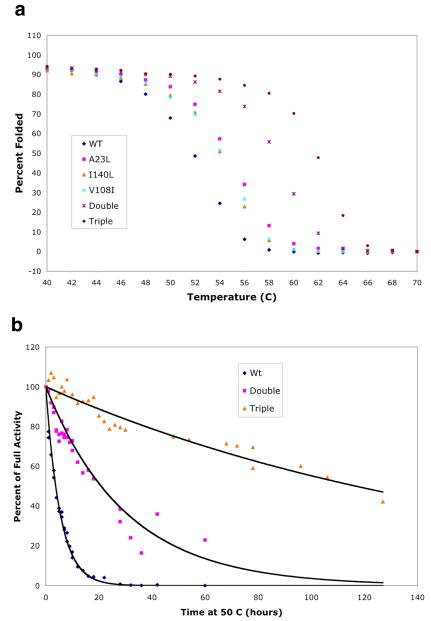
Thermal denaturation experiments were performed on all constructs using CD spectroscopy (Figure 1a). Wild-type yCD and the redesigned mutants displayed largely reversible unfolding behavior over the range of temperatures examined, however at higher temperatures they unfold irreversibly (data not shown). The thermal stability of yCD and mutant constructs was quantified by deriving an apparent Tm from the CD-unfolding curves. This value for the wild-type enzyme was determined to be 52 °C. The isolated single mutations A23L, I140L and V108I each slightly thermostabilized the enzyme, increasing the apparent Tm by approximately 2 °C. However, simultaneous encorporation of all three mutations increased apparent Tm to 62 °C, 10 °C higher than wild-type. Therefore, combination of individual point mutations in a single construct produces a synergistic effect beyond their individual contributions. This result is not simply due to the formation of contacts between redesigned residues, as residue 108 is physically separated from residues 23 and 140.
Figure 1. Thermal denaturation and activity half-life measurements.
A) Temperature melt measuring the change in signal at 220nm over a range of temperatures. All constructs show a folded baseline followed by a sigmodial two-state transition to an unfolded baseline. Only a blow-up of data from 40° to 70° is shown; at lower temperatures the baseline plateaus corresponding to an assignment of 100% folded protein. B) Activity decay at 50 °C. Wild-type yCD and the double and triple mutant constructs were incubated at 50° C and their activity was measured over time as described in methods. The resulting curves gave half-lives for the enzymes at 50° C of 4 hours for WT, 21 hours for the Double mutant and 117 hours for the triple mutant.
The kinetic behavior of the wild-type enzyme and the double and triple mutants were measured at 22 °C to determine the effects of the mutations (Table 1 and Figure S2), as were their relative activities as a function of temperature (Figure S3). At 22 °C, the wild-type enzyme displays a kcat of 160 moles/mol enzyme sec−1, and a Km (cytosine) of 1.98 mM, while the double and triple mutants displayed a slightly reduced Vmax coupled with a reduction in the Michaelis constant Km. The catalytic efficiency of the enzyme mutants (expressed as the ratio kcat/Km) is unchanged relative to the wild-type enzyme. The overall temperature activity profile was broadened, for the redesigned enzyme, with near wild-type activity retained at lower temperatures and higher activity above 50 °C.
Table 1. Kinetic behavior of wild-type and redesigned yCD catalysts.
Both the double and triple mutants displayed a slightly reduced Vmax, coupled with a reduction in the Michaelis constant Km. The catalytic efficiency of the various enzyme mutants (expressed as the ratio kcat/Km) is unchanged relative to the wild-type enzyme.
| WT |
Double Mutant |
Triple Mutant |
|
|---|---|---|---|
| Km (mM) |
1.98 | 1.50 | 1.33 |
| Vmax (M prod / sec) |
0.0016 | 0.0012 | 0.0011 |
| kcat (M prod / M Enz • sec) |
160 | 120 | 110 |
| kcat/Km (1 / sec • M enz) |
8150 | 8190 | 8080 |
The preservation of overall catalytic efficiency (achieved by reducing both kcat and Km, rather than by maximizing overall velocity) and the unusual change in shape and breadth of the enzyme’s thermal profile might suggest that the computational redesign has generated mutations that natural or directed evolution pathways might not select, except perhaps as intermediate species. Therefore, computational strategies for thermostabilization might offer a bonus of selecting mutations that differ in these properties, as compared to selection or redeisgn experiments based on natural selection.
In order to visualize the time-dependent decay of activity at elevated temperatures, wild-type yCD and the double and triple mutants were incubated at 50 °C and the decrease in their relative activity was monitored over time (Figure 1b). The wild-type enzyme showed a rapid loss of activity at 50 °C, with a half-life around 4 hours. The double mutant displayed a half-life around 21 hours while the triple mutant had a half-life at 50 °C of around 117 hours.
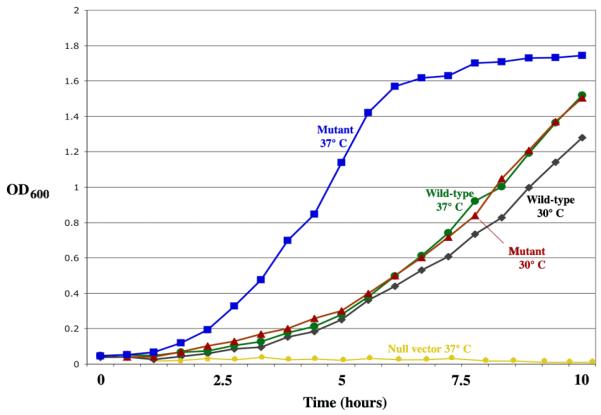
In order to determine the effects of the mutations in vivo, a strain of E.coli dependent on cytosine deaminase function for uracil synthesis was engineered and transformed with both wild-type and mutant yCD reading frames. Doubling times were then measured at 30 and 37 °C on minimal media lacking uracil (Figure 2). The thermostabilized mutant construct induces slightly accelerated growth relative to the wild-type enzyme at 30 °C, and a more significant acceleration at 37 °C. This suggests that the properties of the engineered variants (a reduced KM and thermostabilization) measured in vitro, as described above, correlate with improved enzyme flux in vivo under growth conditions limited by the activity of the enzyme.
Figure 2. In vivo assay for metabolic growth phenotype.
Bacterial growth curves in media conditions requiring cytosine deaminase activity for generation of uracil, as described in methods. Both wild-type and reengineered mutants of yCD complement the bacterial activity; the thermostabilized enzyme variant displays a slight increase in growth rate at 30 °C and a significant increase at 37°C.
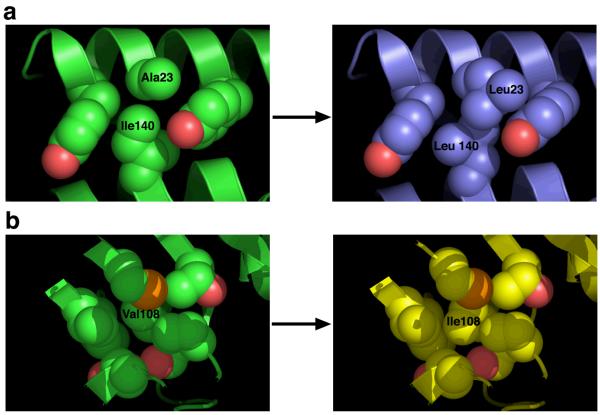
The crystal structures of both the double and triple mutants were solved to 1.9 Å and 1.7 Å respectively (RCSB ID codes 1YSD and 1YSB). The interpretation of density around the redesigned regions of the protein core (in unbiased omit maps) was unambiguous (Figure S4). The rmsd values comparing the wild-type enzyme and both constructs were under 0.5 Å on all common α-carbons and under 0.8 Å on all common atoms. Thus, the redesign and subsequent incorporation of point mutations in the enzyme core had a negligible effect on overall structure of the enzyme, including the active site (Figure S4). The redesigned, mutated residues all appear to pack more tightly in the enzyme core, with more surface area in contact with neighboring residues without altering the nearby side chain rotamers or backbone conformation. Approximately 70 Å2 of additional buried surface area is encorporated as a result of the three mutations (based on an analysis of residue-by-residue packing, using the program NACCESS (49)). The A23L I140L double mutation increased the amount of hydrophobic packing against a neighboring tyrosine ring (Figure 3a), while the addition of V108I in the triple mutant added an additional methyl group to fill a cavity (Figure 3b).
Figure 3. Structural analyses.
A) Van der Waals representation of residues Y19, A23, Y26 and I140 in the wild-type yCD crystal structure (left) and same representation and orientation for the mutant construct with A23L and I140L mutations (right). B) Van der Waals radii representation of the area around V108 in the wild-type structure (left) and similar representation of the area around the V108I mutation in the triple mutant crystal structure.
The stabilized triple mutant was pieced together from part of a cluster of mutations predicted by the program and another single mutation predicted in a separate part of the core. While the degree of thermostabilization produced by these mutations was relatively modest (an increase for Tm of 2 °C for the first change and 4 °C for each subsequent mutation), there is no obvious reason why additional mutations predicted by the program could not be iteratively incorporated into the enzyme core, resulting in a panel of catalysts that display sequential increases in thermal stability.
Not all mutations predicted by the program were equally thermostabilizing. Redesigned involving incorporation or alteration of polar or charged residues in the core (such as replacement of a buried salt-bridge in cluster 1, and individual mutations T67E, E69L or W10T) were less successful than mutations involving substitution of one hydrophobic side chain for another. These latter mutations were predicted and observed to fill cavities within the core with additional van der Waals packing interactions. In future design efforts, selecting mutations of this type in silico may be most successful. Furthermore, modeling of packing forces involving buried polar and charged side chains in the enzyme core is an area for future development in our computational redesign algorithms.
Supplementary Material
Acknowledgments
The authors acknowledge the assistance and advice of the Baker laboratory in running RosettaDesign the FHCRC structural biology program for assistance with data collection, and critiques and suggestions from Roland Strong and Adrian Ferre-D’Amare. Funding was proviced by the NIH to BLS (GM49857 and CA97328), MEB (CA97328 and CA85939), DB (GM59224) and AK (T32-GM08268).
References
- 1.Haldane JBS. In: Enzymes. Longmans G. a. C., editor. MIT Press; Cambridge: 1930. [Google Scholar]
- 2.Kraut DA, Carroll KS, Herschlag D. Annu Rev Biochem. 2003;72:517. doi: 10.1146/annurev.biochem.72.121801.161617. [DOI] [PubMed] [Google Scholar]
- 3.Daniel RM, Dunn RV, Finney JL, Smith JC. Annu Rev Biophys Biomol Struct. 2003;32:69. doi: 10.1146/annurev.biophys.32.110601.142445. [DOI] [PubMed] [Google Scholar]
- 4.Eijsink VG, et al. J Biotechnol. 2004 Sep 30;113:105. doi: 10.1016/j.jbiotec.2004.03.026. [DOI] [PubMed] [Google Scholar]
- 5.Scandurra R, Consalvi V, Chiaraluce R, Politi L, Engel PC. Biochimie. 1998 Nov;80:933. doi: 10.1016/s0300-9084(00)88890-2. [DOI] [PubMed] [Google Scholar]
- 6.Eidsness MK, Richie KA, Burden AE, Kurtz DM, Jr., Scott RA. Biochemistry. 1997 Aug 26;36:10406. doi: 10.1021/bi970110r. [DOI] [PubMed] [Google Scholar]
- 7.Rees DC, Adams MW. Structure. 1995 Mar 15;3:251. doi: 10.1016/s0969-2126(01)00155-1. [DOI] [PubMed] [Google Scholar]
- 8.Sterner R, Liebl W. Crit Rev Biochem Mol Biol. 2001;36:39. doi: 10.1080/20014091074174. [DOI] [PubMed] [Google Scholar]
- 9.Dahiyat BI, Mayo SL. Science. 1997 Oct 3;278:82. doi: 10.1126/science.278.5335.82. [DOI] [PubMed] [Google Scholar]
- 10.Street AG, Mayo SL. Structure Fold Des. 1999 May;7:R105. [Google Scholar]
- 11.Kraemer-Pecore CM, Wollacott AM, Desjarlais JR. Curr Opin Chem Biol. 2001 Dec;5:690. doi: 10.1016/s1367-5931(01)00267-8. [DOI] [PubMed] [Google Scholar]
- 12.Gordon DB, Marshall SA, Mayo SL. Curr Opin Struct Biol. 1999 Aug;9:509. doi: 10.1016/s0959-440x(99)80072-4. [DOI] [PubMed] [Google Scholar]
- 13.Mendes J, Guerois R, Serrano L. Curr Opin Struct Biol. 2002 Aug;12:441. doi: 10.1016/s0959-440x(02)00345-7. [DOI] [PubMed] [Google Scholar]
- 14.Venclovas C, Zemla A, Fidelis K, Moult J. Proteins. 2003;53(Suppl 6):585. doi: 10.1002/prot.10530. [DOI] [PubMed] [Google Scholar]
- 15.Dahiyat BI. Curr Opin Biotechnol. 1999 Aug;10:387. doi: 10.1016/S0958-1669(99)80070-6. [DOI] [PubMed] [Google Scholar]
- 16.Luo P, et al. Protein Sci. 2002 May;11:1218. doi: 10.1110/ps.4580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malakauskas SM, Mayo SL. Nat Struct Biol. 1998 Jun;5:470. doi: 10.1038/nsb0698-470. [DOI] [PubMed] [Google Scholar]
- 18.Dantas G, Kuhlman B, Callender D, Wong M, Baker D. J Mol Biol. 2003 Sep 12;332:449. doi: 10.1016/s0022-2836(03)00888-x. [DOI] [PubMed] [Google Scholar]
- 19.Benson DE, Haddy AE, Hellinga HW. Biochemistry. 2002 Mar 5;41:3262. doi: 10.1021/bi011359i. [DOI] [PubMed] [Google Scholar]
- 20.Reina J, et al. Nat Struct Biol. 2002 Aug;9:621. doi: 10.1038/nsb815. [DOI] [PubMed] [Google Scholar]
- 21.Shifman JM, Mayo SL. J Mol Biol. 2002 Oct 25;323:417. doi: 10.1016/s0022-2836(02)00881-1. [DOI] [PubMed] [Google Scholar]
- 22.Berg DT, et al. Proc Natl Acad Sci U S A. 2003 Apr 15;100:4423. doi: 10.1073/pnas.0736918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Looger LL, Dwyer MA, Smith JJ, Hellinga HW. Nature. 2003 May 8;423:185. doi: 10.1038/nature01556. [DOI] [PubMed] [Google Scholar]
- 24.Kuhlman B, et al. Science. 2003 Nov 21;302:1364. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 25.Dwyer MA, Looger LL, Hellinga HW. Science. 2004 Jun 25;304:1967. doi: 10.1126/science.1098432. [DOI] [PubMed] [Google Scholar]
- 26.Kuhlman B, Baker D. Proc Natl Acad Sci U S A. 2000 Sep 12;97:10383. doi: 10.1073/pnas.97.19.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ireton GC, Black ME, Stoddard BL. Structure (Camb) 2003 Aug;11:961. doi: 10.1016/s0969-2126(03)00153-9. [DOI] [PubMed] [Google Scholar]
- 28.Katsuragi T, Sonoda T, Matsumoto K, Sakai T, Tonomura K. Agric. Biol. Chem. 1989;53:1313. [Google Scholar]
- 29.Kievit E, et al. Cancer Res. 1999 Apr 1;59:1417. [PubMed] [Google Scholar]
- 30.Black ME. Genet Eng (N Y) 2001;23:113. doi: 10.1007/0-306-47572-3_7. [DOI] [PubMed] [Google Scholar]
- 31.Greco O, Dachs GU. J Cell Physiol. 2001 Apr;187:22. doi: 10.1002/1097-4652(2001)9999:9999<::AID-JCP1060>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 32.Lazaridis T, Karplus M. Proteins. 1999 May 1;35:133. doi: 10.1002/(sici)1097-0134(19990501)35:2<133::aid-prot1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 33.Dunbrack RL, Jr., Cohen FE. Protein Sci. 1997 Aug;6:1661. doi: 10.1002/pro.5560060807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Materials and methods provided in Supplementary Online Materials.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.