Abstract
Although protein expression is regulated both temporally and spatially, most proteins have an intrinsic, “typical” range of functionally effective abundance levels. These extend from a few molecules per cell for signaling proteins, to millions of molecules for structural proteins. When addressing fundamental questions related to protein evolution, translation and folding, but also in routine laboratory work, a simple rough estimate of the average wild type abundance of each detectable protein in an organism is often desirable. Here, we introduce a meta-resource dedicated to integrating information on absolute protein abundance levels; we place particular emphasis on deep coverage, consistent post-processing and comparability across different organisms. Publicly available experimental data are mapped onto a common namespace and, in the case of tandem mass spectrometry data, re-processed using a standardized spectral counting pipeline. By aggregating and averaging over the various samples, conditions and cell-types, the resulting integrated data set achieves increased coverage and a high dynamic range. We score and rank each contributing, individual data set by assessing its consistency against externally provided protein-network information, and demonstrate that our weighted integration exhibits more consistency than the data sets individually. The current PaxDb-release 2.1 (at http://pax-db.org/) presents whole-organism data as well as tissue-resolved data, and covers 85,000 proteins in 12 model organisms. All values can be seamlessly compared across organisms via pre-computed orthology relationships.
The state-of-the-art concerning systematic studies of the proteome is progressing quickly, from initially rather qualitative protein identifications to more precise and quantitative global measurements. A variety of experimental techniques are currently being employed for genome-wide proteome quantification (1–4), ranging from affinity-based and biophysical methods to the large array of mass spectrometry-based quantification techniques. Because the expressed proteome constitutes the “business end” of a cell, such measurements are arguably among the most biologically meaningful functional genomics data sets; they support multiple application scenarios, including genome annotation (5, 6), biomarker discovery (7, 8), posttranslational modification detection (9), and even environmental studies (10, 11).
Several databases and repositories already exist that are dedicated to mass spectrometry proteomics data (12–17), each serving a somewhat different purpose and aiming for various levels of reprocessing and meta-annotation. Because of the cutting edge development of proteomics technology, and because of the wide spectrum of different application scenarios and experimental protocols, the challenges met by these repositories are far greater than is the case for other types of data (such as DNA or transcriptomics data). At one end of the tool spectrum is Tranche/ProteomeCommons (16), a distributed data sharing facility uniquely positioned to handle the very large files that hold the primary experimental data as well as the downstream analysis results. Next, PRIDE is a repository largely dedicated to provide the “submitter's view” on the data (13). It already provides much more meta-information, by requesting the submitter to follow controlled vocabularies and standards for submission, and by providing file formats, converters and associated tools. Finally, GPMDB, PeptideAtlas and MOPED are repositories that have the additional aim of re-processing submitted raw data in a consistent way (14, 15, 17). The latter resources are based on the assumption that proteomics experiments are often made more valuable by subsequent re-analyses with updated search and statistics algorithms.
The data sets and experiments that are currently stored in these various repositories span a wide range of organisms, sample materials and preparation protocols, reflecting the diverse research motivations behind the various proteomics projects. Some submissions are focused on specific subcellular organelles, some on cultured cells or specific body fluids (e.g. human plasma) and yet others on normal or diseased tissues (e.g. tumors). The necessity to systematically annotate the meta-information describing each sample is a unique challenge for the database repositories, although the infrastructure for this has much improved recently (notably by introducing standards and controlled vocabularies at the Proteomics Standards Initiative (18), and at PRIDE (13)). Nevertheless, the metadata is often not provided by the submitters, so it remains challenging to achieve straightforward integration of the various data submissions into higher levels of organization (such as “combine all data on nuclear preparations” or “aggregate data from all mouse tissues but not cell lines”). Yet, such integration is often highly desirable: With respect to the proteome as a whole, many current data sets still suffer from under-sampling and have relatively high levels of technical noise (particularly for low-abundance proteins). Hence, integration of data from several experiments/data sets would offer the opportunity to reduce technical and biological noise, and to increase proteome coverage. Furthermore, many of the currently available proteomics data sets were not originally intended as proteome quantifications, but merely as qualitative descriptions of detectable proteins. Nevertheless, these data sets often hold quantifiable information as well (mainly through spectral counting (19–21)), which is again best used by aggregating over a large number of samples.
Of all the conceivable levels at which proteome quantifications might be aggregated (e.g. by tissue, by organelle, by cell-type, by technology, etc), our initial focus here is on two levels: a) the organism-wide average, and b) the organ/tissue-wide average. The organism-wide average is the level at which the data is most easily compared across the entire tree of life. It is also the level at which the largest data sets are available, because it applies to single-celled organisms and because even in multicellular organisms, many experimental efforts are directly targeting the whole organism by design. Often, an organism-wide average is meaningful in and of itself, particularly for evolutionary studies. The typical expression level of a protein—determined largely by its function—has a surprisingly large impact on its evolutionary trajectory, controlling the extent of purifying selection on its amino acid sequence (22), its codon usage (23), translation regime (24), folding accuracy (25, 26), and its genomic organization (27, 28), among others. Because all the expression states of a given protein are controlled by the same fixed gene locus, an organism-wide average offers a good approximation of the evolutionary influence of protein abundance felt by that locus.
The second level of our data aggregation is the organ/tissue level; it mainly applies to larger, well-studied model organisms. Here, proteomics data availability is still somewhat more limited, but on the other hand, tissue data is more immediately biologically relevant. Where gaps still exist in tissue proteomics, these can to some extent be complemented by extensive transcriptomics collections that are also available at tissue resolution.
As a meta-resource, another important focus for PaxDb is to provide an intuitive user experience, for example by including concise tables and visuals, and by directly integrating accessory information. This allows seamless ad hoc browsing and queries of the database by non-expert users in proteomics, and brings together disparate aspects of biology for high-throughput analysis. The resource provides functional information on each protein, including sequence features, protein domains, functional annotations and 3D protein structure data. Importantly, PaxDb also provides pre-computed orthology relationships at various hierarchy levels in the tree of life. This is particularly useful for global comparative analysis across different organism groups. It allows an instant view on any protein family of interest in, say, eukaryotes—detailing the various expression states of the family members in each of the organisms and tissues that have been assessed so far.
EXPERIMENTAL PROCEDURES
Data Sources
Because PaxDb is a meta-resource, it draws exclusively from published experiments and from the tedious work done at the primary proteomics data repositories. PaxDb itself does not accept any author submissions of experimental data. For the current release 2.1, we imported 81 quantitative proteomics data sets—each addressing either a particular organ/tissue, or the entire detectable proteome, in one of 12 model organisms. The data sets were either curated directly from published literature (29–52) (whereby the actual data files were often retrieved via PRIDE), or were downloaded from PeptideAtlas (15, 53), taking advantage of PeptideAtlas “Builds.” The PeptideAtlas Builds are the result of aggregations over multiple proteomics data sets stored in PeptideAltas, and the data has been reprocessed there via standardized database searches and peptide scoring (for some organisms, we average over two available builds to strike a balance between coverage and stringency). From the PeptideAtlas builds, we analyzed the actual spectral count information—i.e. which peptides have been identified, and how often, over the full build. This part of PaxDb's data import is entirely based, and dependent, on the original scoring and quality cutoffs implemented at PeptideAtlas (15, 53).
Identifier Remapping and Spectral Counting
In PaxDb, each protein abundance data set is remapped to an up-to-date, consistently annotated version of the respective model-organism genome/proteome. The reference genomes are imported from the STRING database (54), which in its current version holds more than 1000 completely sequenced genomes. Using the built-in synonym tables of STRING, source identifiers are first mapped to their respective genome loci, and from there to a single, “canonical” protein encoded at each locus. By design, PaxDb aggregates any splicing-specific abundance information at the gene locus level; splice-form-specific abundance information is currently not stored (because it is often under-sampled and not very informative at the high level of integration that PaxDb provides).
In the case of identified peptide sequences reported from MS-MS spectra searches, we remap each peptide to the corresponding protein, based on sequence matches. Importantly, any peptide that cannot be mapped unequivocally to a single locus, even after collapsing alternative splice isoforms, is assigned “fractionally” to all matching loci. This effectively averages peptide counts over recently duplicated gene paralogs; note that such paralogs often have at least broadly similar molecular functions and are further aggregated in PaxDb, for example when comparing between organisms based on orthologous groups.
Protein Abundance Values
Next, we convert the information in each data set into protein abundance estimates, using a consistent expression unit. For this, instead of using “molar concentration” or “molecules per cell,” we express all abundances in “parts per million” (ppm), i.e. each protein entity is enumerated relative to all other protein molecules in the sample. In our view, this has the advantage of being independent of cell-size and other factors; furthermore, this definition can encompass arbitrary extracellular structures, volumes or dilutions. Abundances in “ppm” are essentially a way of describing each protein with reference to the entire expressed proteome and in particular to the most abundant proteins therein; the latter are usually confined to the translation apparatus and to a few core proteins in metabolism or cell structure maintenance. A further advantage of this way of counting is that it is easily comparable between tissues and cell culture samples, and of course also between different model organisms with vastly distinct cell sizes and tissue structures.
In the case of biochemical, biophysical or label-free-MS quantifications, we compute the ppm values directly, by re-scaling the author-provided abundance estimates by their sum-total. In the case of spectral counting data, we estimate abundance values as described earlier (34, 55). Briefly, we first weigh each expected peptide in a protein by its estimated likelihood of detection based on its length (we have shown this likelihood to be at present relatively uniform across the diverse organisms and mass spectrometers (55)). We then compute the actual peptide coverage of each protein (ambiguous peptides are counted fractionally for each matching protein), and normalize those counts by the expected peptide coverage in the protein. Lastly, all spectral counts of an organism are added up, and rescaled by their sum-total.
Data Set Scoring
For many organisms, benchmarking information on the proteome-wide abundance of proteins is not available. In addition, the limited congruence between independent data sets targeting the same organism (55) suggest a considerable amount of technical and biological noise. For PaxDb, we have developed an indirect and somewhat approximate way of estimating data quality. It is based on the assumption that proteins that contribute jointly to a shared function (such as members of a protein complex) should tend to have roughly similar protein abundance levels. We therefore systematically compute abundance ratios for pairs of proteins that are known to be functionally connected (i.e. those having an interaction score of at least 0.900 in the STRING database (54)). For a given data set, the median of these ratios is a rough quality/consistency indicator—the closer the median is to 1.0, the better the data set consistency. To convert this into an easily understandable and consistent score, we also compute the median for the same data set after shuffling its abundance values; this is done several hundred times, and the actual median is compared with the distribution of randomized medians in a Z-score setup. We term this metric the “interaction consistency score.” In our view, this indirect way of scoring the quality of a data set has several advantages: a) it is applicable to every model organism, provided that it is represented with protein-protein interactions in the STRING database, b) the score is not based on a small number of reference proteins only, but instead based on a large fraction of all the proteins in a data set, c) regulated expression changes do not affect the score as long as all interacting proteins are regulated together, and d) by relying on functional protein-protein interactions, we choose a reference that is very distinct from abundance measurement, and thus presumably shares very few technical biases with it.
Integrated Data Set
Based on the individual interaction consistency scores of each contributing data set in an organism, an integrated data set is computed that corresponds to the weighted average of the data sets. Within a given data set, any proteins that are reported to be not detectable are assigned an abundance of zero. For the weighted average, the decision on what weight to give to each data set is taken manually (for some data sets it can also be zero). First, the best-scoring data set is given a weight of 1.0, and then for the second-best data set a weight is chosen that maximizes the score for the resulting weighted combination. This is repeated until the addition of another data set no longer increases the overall score of the integrated data set. Occasionally, the addition of a data set would not raise the overall score, but would bring in additional proteins and thus increase the overall coverage. In this case, it is included if its quality is deemed acceptable. In general, the designation of the weights is necessarily somewhat arbitrary - we would like to stress that they should not be taken as a statement on data quality.
Hierarchical Orthology
To aid the user in comparing protein abundances across diverse organisms, we precompute and store all detectable orthology relationships between the proteins of the various organisms in PaxDb. This is implemented via a “group-orthology” framework, a concept used in a number of resources such as COG (56), eggNOG (57), or OrthoDB (58). Group orthology has the advantage of bundling paralogous, recently duplicated genes into the same group, and thus providing a natural aggregator over broadly similar functions in diverse organisms. Because the definition of orthology and paralogy depends on the choice of the last common ancestor under consideration, we provide orthology relations at various levels of resolution; a given human protein, for example, can be viewed in the context of its orthologs in primates only, in metazoa, in eukaryotes, or in all living organisms. The further back in time this choice goes, the less specific the orthology relations become, but the more general is the function captured. For PaxDb, we compute orthologous groups using the eggNOG pipeline (57), which we tailor specifically to the organisms contained in PaxDb. Within each orthologous group, PaxDb provides the integrated abundance estimate of each protein in each organism for easy comparison, but it also provides the sum over all paralogous protein members within each organism (and hence the total abundance of a given gene family in that organism).
Tissue Ontology Terms
For each protein abundance data set, PaxDb attempts to provide a standardized ontology reference to formally describe its tissue/sample origin. Because there is currently no single, unified provider for anatomical ontologies across organisms, we employ several ontology frameworks, most of which are cross-referenced by the “Uberon” project (59). Importantly, PaxDb also connects these ontology terms across organisms where applicable (e.g. by declaring that “mouse brain” is a concept that can be meaningfully compared with “human brain”). To systematically build these references across organisms is daunting, but for the limited number of tissues currently covered by proteomics data, this can be done manually.
Database Implementation
PaxDb is based on a semi-automated import pipeline that can be executed repeatedly for each new release. The data is stored in a document-oriented database backend (http://www.mongodb.org/), and is served via a web-frontend based on Java and the Google Web Toolkit. All data is freely available via downloadable flat-files under a Creative Commons license.
RESULTS AND DISCUSSION
The PaxDb database (“Protein Abundances Across Organisms”) currently covers 12 model species from all three domains of life, ranging from a single-celled archaeon to complex eukaryotes. For each of these organisms, PaxDb aims to provide individual (tissue-resolved) data sets, but in addition also a single, consolidated abundance estimate of all detectable proteins. This latter estimate is meant to be an organism-wide average of protein expression, aggregating over all available data sets (from various environmental conditions and developmental stages). Where applicable, consolidated averages are provided for specific tissues as well, i.e. wherever several independent data sets are available for a given tissue. Fig. 1 outlines the basic flowchart for each new release of PaxDb (the current release version is 2.1). Apart from the final, aggregated averages, each imported data set is also made available, as is—after re-mapping onto a common, up-to-date version of the respective model organism genome. All abundance data is presented in the same numerical framework, i.e. expressing average steady-state protein abundances in molecular counts, normalized to “parts per million” (ppm; see section “Experimental Procedures” above). Apart from these abundance estimates, each protein is presented together with accessory information regarding the annotated function, sequence and structural information, and within a network context of known or predicted functional interaction partners. All of this latter, additional information is imported from the STRING database (54), with which PaxDb shares the protein name-space and all functional annotation information. Apart from the protein-centered information, PaxDb also contains summary metrics describing each data set, such as its abundance distribution over the entire proteome. Furthermore, all proteins are grouped into families of orthologs (“orthologous groups”), which enables a direct comparison of abundance estimates across organisms.
Fig. 1.
PaxDb overview. For each release of PaxDb, protein abundance information is imported from a number of sources, including proteomics repositories and published studies. All data is preprocessed and, in the case of raw MS/MS data, protein abundances are recalculated by spectral counting. Additional information is imported from the STRING database. The representation of data is structured in three different views: 1) information about a single protein, 2) abundance tables for all detectable proteins in an organism, and 3) a summary page for every organism, listing available data sets. Where several data sets exist for one organism, PaxDb also provides a weighted-average integrated data set that is more comprehensive and has less noise than the single data sets.
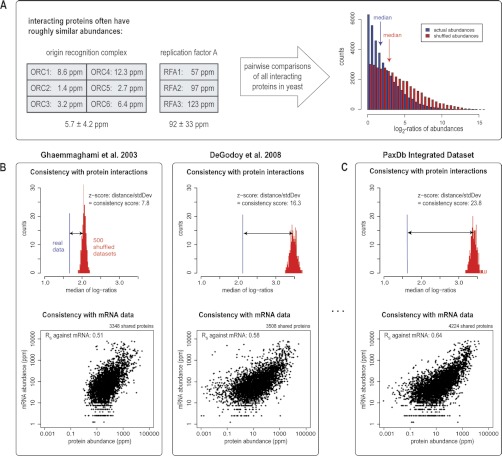
Because gold-standard benchmark/reference information on protein abundance is often not available, gauging the quality of the individual data sets in PaxDb is far from trivial. Here, we employ two distinct, indirect strategies for obtaining a rough estimate on data quality: one is based on protein-protein interaction information, and the other is based on abundance measurements of mRNA molecules. The use of protein-protein interaction data is based on the assumption that interacting proteins should on average have roughly similar steady-state abundances (see also above, “Experimental Procedures”). As shown in Fig. 2, this assumption is an oversimplifying approximation at best: the median abundance ratio of two interacting proteins currently stands at about 3:1 in yeast. Indeed, interacting proteins do not necessarily have to be expressed at similar levels, especially in the case of transient or regulated interactions. However, the observed 3:1 ratio is much less than the abundance ratio of 10:1 that can be expected by chance (assessed from repeatedly shuffling the abundance data), hence it does provide an intrinsic quality estimator. We express this measure as a Z-score-distance from the random distribution, and designate it as the “interaction consistency score.” Likewise, the use of mRNA abundances as a quality measure is also based on a simplifying assumption: that the average steady-state abundance of an mRNA species should be a rough predictor of the steady-state abundance of its encoded protein. Of course, this does not necessarily have to hold for any given transcript/protein pair, but the overall correlation has been shown to be highly significant, and to improve recently as measurement accuracies improved as well (60). Similar to the interaction test described above, this mRNA correlation test has the advantage of including the majority of proteins in a sample, and of sharing very little systematic technical biases with protein quantifications in general. Using yeast as an example, Fig. 2 shows that our scoring indeed reveals strong differences between the various protein abundance data sets (see also supplemental Figs. S1 and S2). Importantly, the data consolidation performed at PaxDb (i.e. a weighted average over the contributing data sets) does score highest on both measures, achieving an interaction consistency score of about 23.8 and a Spearman rank correlation against mRNA of about 0.64 in yeast (Fig. 2, supplemental Fig. S3). Relative to each other, both of our consistency measures exhibit a reasonably good correlation (RS = 0.74, p value < 0.0005, Supplemental Fig. S4A), and thus allow us to provide at least an initial, rough estimate of data quality/consistency.
Fig. 2.
Data set quality. The quality scoring system in PaxDb is based on the assumption that interacting proteins should have roughly similar abundances. A, left: two examples of protein complexes in S. cerevisiae, both involved in replication but with different abundance levels. Right: abundance ratios of all interacting protein pairs, plotted as histograms. Shuffled data (in red) shows a shift toward higher ratios compared with actual data (in blue). B, For two different data sets, both the median of the actual ratios (in blue) and the distribution of medians obtained from 500 × shuffling of abundances (in red) are shown. The PaxDb score corresponds to the z-transformed distance. The same two data sets are plotted against RNAseq data below. C, The consolidated data set, with a higher PaxDb score, also correlates better with mRNA abundances, and covers more proteins. Yeast mRNA quantification data is from ref (62).
The website of PaxDb (Figs. 3 and 4; and http://pax-db.org/) has been designed for intuitive and fast access, allowing both the ad-hoc query of a protein family of interest, as well as browsing and comparing entire data sets. Protein queries are resolved against a large collection of identifier name-spaces, and multiple proteins can be requested simultaneously in one query. In addition to searching for known identifiers, the user queries are also searched against the annotations of all proteins in PaxDb, using a fast full-text search. For each organism in PaxDb, a distinct summary page provides information on data provenance, its coverage and estimated quality (Fig. 3). This page also provides the distribution of abundance values of each data set as a histogram, as well as listing the most abundant proteins in the organism. Users can open and browse entire data sets from this page, and from these data set tables they can proceed directly to the protein details page and, in the case of PeptideAtlas data, also directly back to the underlying peptide information via deep links.
Fig. 3.
The organism page. This page provides general information about the selected organism, such as the number of data sets and the proteome coverage; it also provides a histogram showing the distribution of protein abundances in the “integrated” data set. The bottom panel lists all available data sets for the selected species. On each page in PaxDb, the very top panel contains a search box that allows the user to search for both protein names and annotations.
Fig. 4.
The protein page. This page displays information on a single protein. A short description of the protein is followed by a table of abundances in all available data sets, along with the corresponding abundance ranks. In the panel below, the abundances of orthologs in other species are displayed. The resolution of orthologs can be filtered via a drop-down menu. The bottom panel contains additional information imported from other sources, such as domain structure, interaction partners, as well as links to relevant databases (truncated here).
The PaxDb protein pages (Fig. 4) finally constitute the core of the information provided. The protein in question is first identified and briefly described in terms of its functional role, as annotated at UniProt (61) and/or at dedicated model organism databases. Its estimated abundance values are then listed, for each available data set separately, including rank and quantile information. In the case of the integrated data sets, which constitute PaxDb's “best estimates,” the relative position of the protein in the entire detectable proteome is then visualized in the abundance histogram. Next, the protein is shown in the context of all family members in other organisms, for which abundance information is available. Users can select the tissue to be shown, as well as control the phylogenetic depth of the orthologs; the abundance of paralogs that have diverged because the last common ancestor is shown separately, but also added up within each organism. This view directly allows the assessment of the gene family in question throughout evolution. Finally, further accessory information on the protein is presented, including interaction partners, sequence domains and structural information, if available. The latter information is cross-linked directly to the relevant data providers.
As of the current release 2.1 of PaxDb, the coverage and generality of the abundance estimates are still very much limited by data availability. In some organisms, the organism-wide average is affected by relatively large sampling biases (notably in the data on human proteins, which has a strong overrepresentation of blood serum samples). Furthermore, membrane proteins and other “difficult” subsets may be systematically underrepresented. Nevertheless, even at this early stage, the quantitative makeup of the core proteome is coming into view. For example, the abundance correlation of the eukaryotic core proteome, when comparing animals (human, fly, worm) with other eukaryotes (fungi, plants), is now standing at RS = 0.80 (55), which is remarkable given the technical difficulties still associated with whole-proteome quantification. This correlation is likely to soon rise even further, given the growth and increased quality of proteomics measurements. Looking ahead, PaxDb will continue to focus on quantification—based on mass spectrometry data (including the growing label-free approaches), but also based on biochemistry or molecular biology approaches. Future releases of PaxDb will also take advantage of the expected increase in meta-information, and will provide aggregation and quantification at more levels of interest, such as for intracellular organelles or specific cell-lines - all in the context of seamless cross-species comparisons via orthologs.
Footnotes
* Work on PaxDb has been funded by the Swiss National Science foundation, by the SystemsX.ch initiative and by the University of Zurich through its Research Priority Program “Systems Biology and Functional Genomics”.
 This article contains supplemental Figs. S1 to S4.
This article contains supplemental Figs. S1 to S4.
REFERENCES
- 1. Vaudel M., Sickmann A., Martens L. (2010) Peptide and protein quantification: a map of the minefield. Proteomics 10, 650–670 [DOI] [PubMed] [Google Scholar]
- 2. Wang D., Bodovitz S. (2010) Single cell analysis: the new frontier in ‘omics’. Trends Biotechnol. 28, 281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nilsson T., Mann M., Aebersold R., Yates J. R., 3rd, Bairoch A., Bergeron J. J. (2010) Mass spectrometry in high-throughput proteomics: ready for the big time. Nat. Methods 7, 681–685 [DOI] [PubMed] [Google Scholar]
- 4. Rees J., Lilley K. (2011) Enabling technologies for yeast proteome analysis. Methods Mol Biol. 759, 149–178 [DOI] [PubMed] [Google Scholar]
- 5. Castellana N., Bafna V. (2010) Proteogenomics to discover the full coding content of genomes: a computational perspective. J. Proteomics 73, 2124–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Krug K., Nahnsen S., Macek B. (2011) Mass spectrometry at the interface of proteomics and genomics. Mol. Biosyst 7, 284–291 [DOI] [PubMed] [Google Scholar]
- 7. Surinova S., Schiess R., Huttenhain R., Cerciello F., Wollscheid B., Aebersold R. (2011) On the development of plasma protein biomarkers. J. Proteome Res. 10, 5–16 [DOI] [PubMed] [Google Scholar]
- 8. Rifai N., Gillette M. A., Carr S. A. (2006) Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat. Biotechnol. 24, 971–983 [DOI] [PubMed] [Google Scholar]
- 9. Zhao Y., Jensen O. N. (2009) Modification-specific proteomics: strategies for characterization of post-translational modifications using enrichment techniques. Proteomics 9, 4632–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. VerBerkmoes N. C., Denef V. J., Hettich R. L., Banfield J. F. (2009) Systems biology: Functional analysis of natural microbial consortia using community proteomics. Nat. Rev. Microbiol. 7, 196–205 [DOI] [PubMed] [Google Scholar]
- 11. Keller M., Hettich R. (2009) Environmental proteomics: a paradigm shift in characterizing microbial activities at the molecular level. Microbiol. Mol. Biol. Rev. 73, 62–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vizcaino J. A., Foster J. M., Martens L. (2010) Proteomics data repositories: providing a safe haven for your data and acting as a springboard for further research. J. Proteomics 73, 2136–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vizcaino J. A., Côté R., Reisinger F., Barsnes H., Foster J. M., Rameseder J., Hermjakob H., Martens L. (2010) The Proteomics Identifications database: 2010 update. Nucleic Acids Res. 38, D736–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Craig R., Cortens J. P., Beavis R. C. (2004) Open source system for analyzing, validating, and storing protein identification data. J. Proteome Res. 3, 1234–1242 [DOI] [PubMed] [Google Scholar]
- 15. Deutsch E. W., Lam H., Aebersold R. (2008) PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 9, 429–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Smith B. E., Hill J. A., Gjukich M. A., Andrews P. C. (2011) Tranche distributed repository and ProteomeCommons.org. Methods Mol. Biol. 696, 123–145 [DOI] [PubMed] [Google Scholar]
- 17. Kolker E., Higdon R., Haynes W., Welch D., Broomall W., Lancet D., Stanberry L., Kolker N. (2012) MOPED: model organism protein expression database. Nucleic Acids Res. 40, D1093–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Orchard S., Hermjakob H. (2011) Data standardization by the HUPO-PSI: how has the community benefitted? Methods Mol. Biol. 696: p. 149–60 [DOI] [PubMed] [Google Scholar]
- 19. Liu H., Sadygov R. G., Yates J. R., 3rd, (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 20. Braisted J. C., Kuntumalla S., Vogel C., Marcotte E. M., Rodrigues A. R., Wang R., Huang S. T., Ferlanti E. S., Saeed A. I., Fleischmann R. D., Peterson S. N., Pieper, (2008) The APEX Quantitative Proteomics Tool: generating protein quantitation estimates from LC-MS/MS proteomics results. BMC Bioinformatics 9, 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ishihama Y., Oda Y., Tabata T., Sato T., Nagasu T., Rappsilber J., Mann M. (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics 4, 1265–1272 [DOI] [PubMed] [Google Scholar]
- 22. Drummond D. A., Bloom J. D., Adami C., Wilke C. O., Arnold F. H. (2005) Why highly expressed proteins evolve slowly. Proc. Natl. Acad. Sci. U.S.A. 102, 14338–14343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sharp P. M., Emery L. R., Zeng K. (2010) Forces that influence the evolution of codon bias. Philos. Trans. R. Soc. Lond. B 365, 1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tuller T., Carmi A., Vestsigian K., Navon S., Dorfan Y., Zaborske J., Pan T., Dahan O., Furman I., Pilpel Y. (2010) An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell 141, 344–354 [DOI] [PubMed] [Google Scholar]
- 25. Drummond D. A., Wilke C. O. (2008) Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 134, 341–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Powers E. T., Balch W. E. (2008) Costly mistakes: translational infidelity and protein homeostasis. Cell 134, 204–206 [DOI] [PubMed] [Google Scholar]
- 27. Castillo-Davis C. I., Mekhedov S. L., Hartl D. L., Koonin E. V., Kondrashov F. A., (2002) Selection for short introns in highly expressed genes. Nat. Genet. 31, 415–418 [DOI] [PubMed] [Google Scholar]
- 28. Zaslaver A., Baugh L. R., Sternberg P. W. (2011) Metazoan operons accelerate recovery from growth-arrested states. Cell 145, 981–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Castellana N. E., Payne S. H., Shen Z., Stanke M., Bafna V., Briggs S. P. (2008) Discovery and revision of Arabidopsis genes by proteogenomics. Proc. Natl. Acad. Sci. U.S.A. 105, 21034–21038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Baerenfaller K., Grossmann J., Grobei M. A., Hull R., Hirsch-Hoffmann M., Yalovsky S., Zimmermann P., Grossniklaus U., Gruissem W., Baginsky S. (2008) Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320, 938–941 [DOI] [PubMed] [Google Scholar]
- 31. Newman J. R., Ghaemmaghami S., Ihmels J., Breslow D. K., Noble M., DeRisi J. L., Weissman J. S. (2006) Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441, 840–846 [DOI] [PubMed] [Google Scholar]
- 32. Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Global analysis of protein expression in yeast. Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 33. de Godoy L. M., Olsen J. V., Cox J., Nielsen M. L., Hubner N. C., Fröhlich F., Walther T. C., Mann M. (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254 [DOI] [PubMed] [Google Scholar]
- 34. Schrimpf S. P., Weiss M., Reiter L., Ahrens C. H., Jovanovic M., Malmstrom J., Brunner E., Mohanty S., Lercher M. J., Hunziker P. E., Aebersold R., von Mering C., Hengartner M. O. (2009) Comparative functional analysis of the Caenorhabditis elegans and Drosophila melanogaster proteomes. PLoS Biol. 7, e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Brunner E., Ahrens C. H., Mohanty S., Baetschmann H., Loevenich S., Potthast F., Deutsch E. W., Panse C., de Lichtenberg U., Rinner O., Lee H., Pedrioli P. G., Malmstrom J., Koehler K., Schrimpf S., Krijgsveld J., Kregenow F., Heck A. J., Hafen E., Schlapbach R., Aebersold R. (2007) A high-quality catalog of the Drosophila melanogaster proteome. Nat. Biotechnol. 25, 576–583 [DOI] [PubMed] [Google Scholar]
- 36. Kuntumalla S., Braisted J. C., Huang S. T., Parmar P. P., Clark D. J., Alami H., Zhang Q., Donohue-Rolfe A., Tzipori S., Fleischmann R. D., Peterson S. N., Pieper R. (2009) Comparison of two label-free global quantitation methods, APEX and 2D gel electrophoresis, applied to the Shigella dysenteriae proteome. Proteome Sci. 7, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Malmström J., Beck M., Schmidt A., Lange V., Deutsch E. W., Aebersold R. (2009) Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature 460, 762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Taniguchi Y., Choi P. J., Li G. W., Chen H., Babu M., Hearn J., Emili A., Xie X. S. (2010) Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329, 533–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lewis N. E., Hixson K. K., Conrad T. M., Lerman J. A., Charusanti P., Polpitiya A. D., Adkins J. N., Schramm G., Purvine S. O., Lopez-Ferrer D., Weitz K. K., Eils R., Konig R., Smith R. D., Palsson B. O. (2010) Omic data from evolved E. coli are consistent with computed optimal growth from genome-scale models. Mol. Syst. Bio. 6, 390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kuhner S., van Noort V., Betts M. J., Leo-Macias A., Batisse C., Rode M., Yamada T., Maier T., Bader S., Beltran-Alvarez P., Castano-Diez D., Chen W. H., Devos D., Guell M., Norambuena T., Racke I., Rybin V., Schmidt A., Yus E., Aebersold R., Herrmann R., Bottcher B., Frangakis A. S., Russell R. B., Serrano L., Bork P., Gavin A. C. (2009) Proteome organization in a genome-reduced bacterium. Science 326, 1235–1240 [DOI] [PubMed] [Google Scholar]
- 41. Lu P., Vogel C., Wang R., Yao X., Marcotte E. M. (2007) Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 25, 117–124 [DOI] [PubMed] [Google Scholar]
- 42. Kislinger T., Cox B., Kannan A., Chung C., Hu P., Ignatchenko A., Scott M. S., Gramolini A. O., Morris Q., Hallett M. T., Rossant J., Hughes T. R., Frey B., Emili A. (2006) Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell 125, 173–186 [DOI] [PubMed] [Google Scholar]
- 43. Huttlin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Beausoleil S. A., Villen J., Haas W., Sowa M. E., Gygi S. P. (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Krüger M., Moser M., Ussar S., Thievessen I., Luber C. A,., Forner F., Schmidt S., Zanivan S., Fassler R., Mann M. (2008) SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell 134, 353–364 [DOI] [PubMed] [Google Scholar]
- 45. Martens L., Müller M., Stephan C., Hamacher M., Reidegeld K. A., Meyer H. E., Bluggel M., Vandekerckhove J., Gevaert K., Apweiler R. (2006) A comparison of the HUPO Brain Proteome Project pilot with other proteomics studies. Proteomics 6, 5076–5086 [DOI] [PubMed] [Google Scholar]
- 46. Wang H., Qian W. J., Chin M. H., Petyuk V. A., Barry R. C., Liu T., Gritsenko M. A., Mottaz H. M., Moore R. J., Camp D. G., Ii, Khan A. H., Smith D. J., Smith R. D. (2006) Characterization of the mouse brain proteome using global proteomic analysis complemented with cysteinyl-peptide enrichment. J. Proteome Res. 5, 361–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Waanders L. F., Chwalek K., Monetti M., Kumar C., Lammert E., Mann M. (2009) Quantitative proteomic analysis of single pancreatic islets. Proc. Natl. Acad. Sci. U.S.A. 106, 18902–18907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Guo X., Shen J., Xia Z., Zhang R., Zhang P., Zhao C., Xing J., Chen L., Chen W., Lin M., Huo R., Su B., Zhou Z., Sha J. (2010) Proteomic analysis of proteins involved in spermiogenesis in mouse. J. Proteome Res. 9, 1246–1256 [DOI] [PubMed] [Google Scholar]
- 49. Aye T. T., Scholten A., Taouatas N., Varro A., Van Veen T. A., Vos M. A., Heck A. J. (2010) Proteome-wide protein concentrations in the human heart. Mol. Biosyst. 6, 1917–1927 [DOI] [PubMed] [Google Scholar]
- 50. Kline K. G., Frewen B., Bristow M. R., Maccoss M. J., Wu C. C. (2008) High quality catalog of proteotypic peptides from human heart. J. Proteome Res. 7, 5055–5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Abdul-Salam V. B., Wharton J., Cupitt J., Berryman M., Edwards R. J., Wilkins M. R. (2010) Proteomic analysis of lung tissues from patients with pulmonary arterial hypertension. Circulation 122, 2058–2067 [DOI] [PubMed] [Google Scholar]
- 52. Grobei M. A., Qeli E., Brunner E., Rehrauer H., Zhang R., Roschitzki B., Basler K., Ahrens C. H., Grossniklaus U. (2009) Deterministic protein inference for shotgun proteomics data provides new insights into Arabidopsis pollen development and function. Genome Res. 19, 1786–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Deutsch E. W., (2010) The PeptideAtlas Project. Methods Mol. Biol. 604, 285–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., Jensen L. J., von Mering C. (2011) The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39, D561–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Weiss M., Schrimpf S., Hengartner M. O., Lercher M. J., von Mering C. (2010) Shotgun proteomics data from multiple organisms reveals remarkable quantitative conservation of the eukaryotic core proteome. Proteomics 10, 1297–1306 [DOI] [PubMed] [Google Scholar]
- 56. Tatusov R. L., Fedorova N. D., Jackson J. D., Jacobs A. R., Kiryutin B., Koonin E. V., Krylov D. M., Mazumder R., Mekhedov S. L., Nikolskaya A. N., Rao B. S., Smirnov S., Sverdlov A. V., Vasudevan S., Wolf Y. I., Yin J. J., Natale D. A. (2003) The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Muller J., Szklarczyk D., Julien P., Letunic I., Roth A., Kuhn M., Powell S., von Mering C., Doerks T., Jensen L. J., Bork P. (2010) eggNOG v2.0: extending the evolutionary genealogy of genes with enhanced non-supervised orthologous groups, species and functional annotations. Nucleic Acids Res. 38, D190–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Waterhouse R. M., Zdobnov E. M., Tegenfeldt F., Li J., Kriventseva E. V. (2011) OrthoDB: the hierarchical catalog of eukaryotic orthologs in 2011. Nucleic Acids Res. 39, D283–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mungall C. J., Torniai C., Gkoutos G. V., Lewis S. E., Haendel M. A. (2012) Uberon, an integrative multi-species anatomy ontology. Genome Biol. 13, R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tuller T., Kupiec M., Ruppin E. (2007) Determinants of protein abundance and translation efficiency in S. cerevisiae. PLoS Comput. Biol. 3, e248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Apweiler R., Martin M. J., O'Donovan C., Magrane M., Alam-Faruque Y., Antunes R., Barrell D., Bely B., Bingley M., Binns D., Bower L., Browne P., Chan W. M., Dimmer E., Eberhardt R., Fazzini F., Fedotov A., Foulger R., Garavelli J., Castro L. G., Huntley R., Jacobsen J., Kleen M., Laiho K., Legge D., Lin Q., Liu W., Luo J., Orchard S., Patient S., Pichler K., Poggioli D., Pontikos N., Pruess M., Rosanoff S., Sawford T., Sehra H., Turner E., Corbett M., Donnelly M., van Rensburg P., Xenarios I., Bougueleret L., Auchincloss A., Argoud-Puy G., Axelsen K., Bairoch A., Baratin D., Blatter M. C., Boeckmann B., Bolleman J., Bollondi L., Boutet E., Quintaje S. B., Breuza L., Bridge A., deCastro E., Coudert E., Cusin I., Doche M., Dornevil D., Duvaud S., Estreicher A., Famiglietti L., Feuermann M., Gehant S., Ferro S., Gasteiger E., Gateau A., Gerritsen V., Gos A., Gruaz-Gumowski N., Hinz U., Hulo C., Hulo N., James J., Jimenez S., Jungo F., Kappler T., Keller G., Lara V., Lemercier P., Lieberherr D., Martin X., Masson P., Moinat M., Morgat A., Paesano S., Pedruzzi I., Pilbout S., Poux S., Pozzato M., Redaschi N., Rivoire C., Roechert B., Schneider M., Sigrist C., Sonesson K., Staehli S., Stanley E., Stutz A., Sundaram S., Tognolli M., Verbregue L., Veuthey A. L., Wu C. H., Arighi C. N., Arminski L., Barker W. C., Chen C., Chen Y., Dubey P., Huang H., Mazumder R., McGarvey P., Natale D. A., Natarajan T. G., Nchoutmboube J., Roberts N. V., Suzek B. E., Ugochukwu U., Vinayaka C. R., Wang Q., Wang Y., Yeh L. S., Zhang J. (2011) Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 39, D214–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nagalakshmi U., Wang Z., Waern K., Shou C., Raha D., Gerstein M., Snyder M. (2008) The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320, 1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]