Abstract
Cy3B is an extremely bright and stable fluorescent dye, which is only available for coupling to nucleic acids post-synthetically. This severely limits its use in the fields of genomics, biology and nanotechnology. We have optimized the synthesis of Cy3B, and for the first time produced a diverse range of Cy3B monomers for use in solid-phase oligonucleotide synthesis. This molecular toolkit includes phosphoramidite monomers with Cy3B linked to deoxyribose, to the 5-position of thymine, and to a hexynyl linker, in addition to an oligonucleotide synthesis resin in which Cy3B is linked to deoxyribose. These monomers have been used to incorporate single and multiple Cy3B units into oligonucleotides internally and at both termini. Cy3B Taqman probes, Scorpions and HyBeacons have been synthesized and used successfully in mutation detection, and a dual Cy3B Molecular Beacon was synthesized and found to be superior to the corresponding Cy3B/DABCYL Beacon. Attachment of Cy3, Cy3B and Cy5 to the 5-position of thymidine by an ethynyl linker enabled the synthesis of an oligonucleotide FRET system. The rigid linker between the dye and nucleobase minimizes dye–dye and dye–DNA interactions and reduces fluorescence quenching. These reagents open up new future applications of Cy3B, including more sensitive single-molecule and cell-imaging studies.
INTRODUCTION
Cy-Dyes are a group of highly fluorescent molecules that cover a wide spectral range. They are important fluorophores which are used in many DNA-related applications. Despite their great utility, most Cy-Dyes are vulnerable to cis/trans isomerization about the polymethine linker which leads to loss of fluorescence upon excitation, particularly at elevated temperatures (1). In contrast to other Cy-Dyes, Cy3B is conformationally locked; it is therefore not prone to photo-isomerization and has superior fluorescence properties. However, it is not available as a phosphoramidite monomer, only as an N-hydroxysuccinimide (NHS) ester or maleimide derivative. Consequently it has to be introduced into oligonucleotides post-synthetically. This makes it difficult to synthesize oligonucleotides containing Cy3B together with other dyes that are added after oligonucleotide synthesis, and limits its range of applications. Post-synthetic labelling of one oligonucleotide with different dyes could be achieved by using two orthogonal methods e.g. click chemistry (2–4) in combination with amide bond formation. However, this approach is complex, time consuming and relatively low yielding. It would therefore be advantageous to have recourse to a range of Cy3B phosphoramidite monomers and resins for incorporation of this very bright fluorophore into DNA during solid-phase synthesis.
When designing phosphoramidite monomers for attaching Cy-Dyes to oligonucleotides the nature of the linker warrants careful consideration. The commonly used linkers are long and unstructured, allowing the fluorophore to sample a large volume of space, thereby introducing imprecision into fluorescence resonance energy transfer (FRET) distance measurements. There is a particular problem at short dye–dye distances where the linker length can be comparable to the dye separation (5). In this situation contact quenching can occur, and paradoxically oligonucleotides with several fluorophores have low fluorescence. The imprecision in FRET distance measurement can be partially overcome if the Cy-Dye is added at the 5′-end of the oligonucleotides. In this case the dye stacks on the end of the DNA duplex and is held in a rigid position (6,7). However, a significant proportion of the dye molecules remain unstacked (1), so this is an inadequate method of immobilizing Cy-Dyes for FRET studies. For quantitative biophysical applications an elegant solution has been developed in which a fluorophore is integrated into the structure of a nucleobase which is held firmly in place by base pairing (8–10). Several such fluorescent nucleobases have been studied and are proving to be useful in nucleic acid research (11). However, their applications are somewhat restricted by their low extinction coefficients and poor quantum yields, resulting in weak fluorescence, in stark contrast to the Cy-Dyes.
Integration of the fluorophore into the structure of the nucleobase is not a feasible approach for Cy-Dyes due to their large size and the complex synthesis that would be required. Therefore alternative methods are needed to restrict the mobility of Cy-Dyes in DNA. In this context, Balasubramanian et al. have devised a method to attach Cy-Dyes to the 5-position of thymine bases in DNA via a rigid ethynyl linkage (12). They achieved this by reacting oligonucleotides containing 5-ethynyl-dU with iodo-derivatives of Cy3 and Cy5 under Sonogashira conditions on the DNA synthesis support. In DNA duplexes the rigid linker restricts the mobility of the dye so that it samples a reduced volume of space, minimizing dye/DNA interactions and potentially reducing fluorescence quenching caused by interactions with the nucleobases (Figure 1B). The above approach to oligonucleotide labelling is ingenious but it is complicated and precludes the synthesis of oligonucleotides containing mixed internal Cy-Dyes. A simpler and more efficient strategy would be to synthesize the equivalent rigid Cy-Dye dT phosphoramidite monomers and directly incorporate them into oligonucleotides during automated solid-phase synthesis. This would allow the efficient synthesis of oligonucleotides containing multiple and mixed Cy-Dyes.
Figure 1.
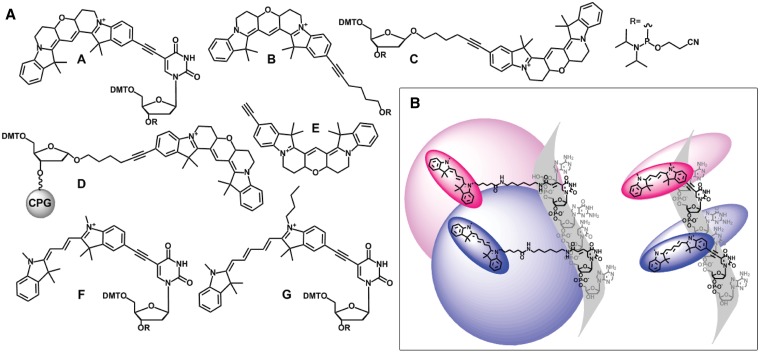
(A) Cy-Dye molecular toolkit; Cy3BdT phosphoramidite A; 5-(hexyn-1-ol)-6-Cy3B phosphoramidite for 5′-addition B; β-Cy3BdR phosphoramidite C; α-Cy3BdR oligonucleotide synthesis resin for 3′-addition D; 5-ethynyl-Cy3B E; Cy3dT phosphoramidite F; Cy5dT phosphoramidite G. The CydT and β-Cy3BdR phosphoramidite monomers can be added either internally or at the 5′-end of oligonucleotides but the latter will not form a base pair. 5-Ethynyl-Cy3B can be used for modification of oligonucleotides by click chemistry. (B) Flexible dye linker (left) and short rigid dye linker (right) in the context of a DNA helix cross-section (only labelled strand shown). Blue shading (Cy5) and pink shading (Cy3) suggests the volume of space that can be sampled by the dyes when the bases are paired. A short rigid linker restricts the dye and prevents dye–DNA interactions.
We have achieved these objectives and now describe the synthesis of the requisite fluorescent phosphoramidites and other monomers for the site-specific incorporation of Cy3, Cy5 and Cy3B analogues into DNA (Figure 1A). The design of an efficient synthesis of the Cy3B dye facilitated the development of a range of Cy3B phosphoramidite monomers and a DNA synthesis resin. This ‘molecular toolkit’ provides a flexible approach for incorporation of Cy3B into DNA by solid-phase phosphoramidite chemistry and enables the synthesis of fluorescent oligonucleotides that were previously unobtainable. Oligonucleotides containing multiple additions of these fluorophores can be readily prepared, and the biophysical properties of fluorogenic DNA probes labelled with these dyes have been determined. The synthesis of oligonucleotides containing the highly fluorescent Cy3B dye is now greatly simplified, extending the potential applications of this important fluorophore.
MATERIALS AND METHODS
Methods for the synthesis of the Cy-Dye monomers, oligonucleotide synthesis, UV and fluorescence protocols and analytical data on synthetic oligonucleotides are described in the Supplementary Methods.
Polymerase chain reaction protocols
Polymerase chain reaction (PCR) mixtures were prepared under sterile conditions using a LabCaire PCR workstation and pipetted into 0.2 ml low-profile white 8-tube strips with optically clear lids (BioRad). All samples were made up to a total volume of 20 µl using a Precision HRM Mastermix (Primer Design, 0.25 U Taq DNA polymerase, 2.5 mM Mg2+, 100 µM of each dNTP) and 1 pg of template [ODN-1 wild-type template (wt), ODN-2 mutant template (mt)]. Template was replaced with sterile water in the negative control samples. Reactions were performed using a BioRad CFX96 Real-Time PCR Detection System with CFX Manager software, monitoring in Channel 3 (excitation range 560–590 nm, detector range 610–650 nm), and all samples were run in triplicate. Primer/probe concentration and thermal protocols are detailed below.
For the Molecular Beacon probes, asymmetric PCR was performed using 0.05 µM forward primer (ODN-3), 0.5 µM reverse primer (ODN-4) and 0.15 µM probe (ODN-8/ODN-9). The thermal protocol was initiated with 8 min at 95°C followed by 50 cycles of 95°C (15 s), 58°C (15 s) and 72°C (15 s). For the Taqman probe, symmetric PCR was performed using 0.5 µM forward primer (ODN-5), 0.5 µM reverse primer (ODN-4) and 0.15 µM probe (ODN-6). The thermal protocol was initiated with 8 min at 95°C followed by 40 cycles of 95°C (15 s), 55°C (15 s) and 68°C (30 s). For the Scorpion probe, symmetric PCR was performed using 0.5 µM forward primer (ODN-3) and 0.5 µM Scorpion probe (ODN-10). The thermal protocol was initiated with 8 min at 95°C followed by 40 cycles of 95°C (15 s), 55°C (15 s) and 61°C (30 s). For the HyBeacon probes asymmetric PCR was performed using 0.5 µM forward primer (ODN-3), 0.05 µM reverse primer (ODN-4) and 0.15 µM probe (ODN-7). The thermal protocol was initiated with 8 min at 95°C followed by 50 cycles of 95°C (15 s), 50°C (15 s) and 72°C (15 s) followed by a melt from 35 to 80°C on the RotorGene3000 (0.5°C increments, 5 s per step).
RESULTS AND DISCUSSION
Synthesis of Cy-Dye monomers, solid supports and oligonucleotides
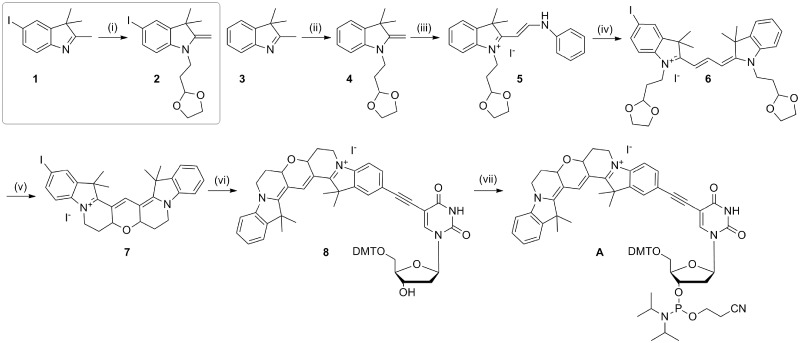
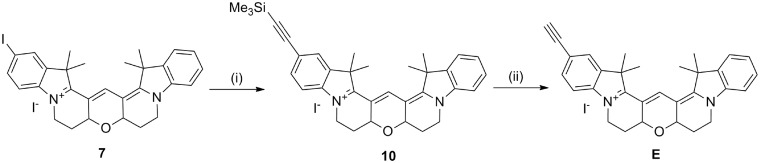
The synthesis of 5-iodo-Cy3B (7; Scheme 1) was adapted from the procedure described in the patent literature for the synthesis of Cy3B (13), which involves the addition of a 3,3-diethyloxypropyl group at the 1N position of the indole ring. This product was very unstable, so instead the ethyldioxolane group was used. The resulting indoles with protected aldehydes (2 and 4, Scheme 1) were used immediately after synthesis to prevent deprotection of the dioxolane and cyclization of the indole. The protection was sufficiently robust during the subsequent reactions to enable the synthesis of the hemicyanine dye (5, Scheme 1) and the cyanine dye (6, Scheme 1). The aldehyde groups were deprotected using aqueous sulphuric acid in chloroform allowing rapid tandem cyclization to produce 5-iodo-Cy3B in high yield (96%).
Scheme 1.
Synthesis of Cy3BdT phosphoramidite. Reagents and conditions: (i) Bromoethyl-1,3-dioxolane, KI, MeCN, reflux, 45 h, 29%; (ii) bromoethyl-1,3-dioxolane, KI, MeCN, reflux, 45 h, 56%; (iii) N,N′-diphenylformamidine, EtOH, triethylorthoformate, 97°C, 16 h, 65%; (iv) 2, pyridine, Ac2O, 50°C, 24 h, 79%; (v) CHCl3, 50% aqueous H2SO4, room temperature, 20 min, 96%; (vi) 5-ethynyl-dU, CuI, Pd(PPh3)4, Et3N, DMF, room temperature, 1.5 h, 42%; (vii) 2-cyanoethyl-N,N-diisopropyl-chlorophosphoramidite, DIPEA, DCM, room temperature, 45 min, 63%.
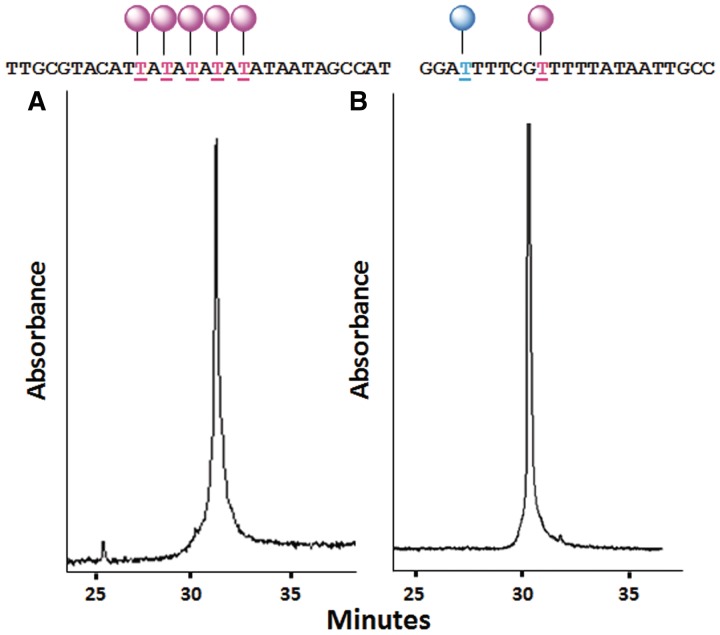
The syntheses of 5-iodo-Cy3 (21, Supplementary Scheme S1) and 5-iodo-Cy5 (23, Supplementary Scheme S2) were carried out using a combination of literature sources (12,14–16). The iodo derivatives of Cy3B, Cy3 and Cy5 were coupled to 5-ethynyl-dU (17) in moderate yield by palladium cross-coupling chemistry to give the corresponding Cy3BdT, Cy3dT and Cy5dT derivatives which were converted to phosphoramidite monomers (A, Scheme 1, F, Supplementary Scheme S1, G, Supplementary Scheme S2). Due to their high polarity and affinity for silica gel, it was not feasible to purify the monomers by chromatography. Instead, they were purified by repeated precipitation using hexane and dichloromethane. The phosphitylation reactions were monitored by thin layer chromatography (TLC) to ensure no more phosphitylation reagent was added than absolutely necessary, and under these conditions precipitation was an effective means of purification. Incorporation of the CydT monomers into oligonucleotides during solid-phase synthesis was achieved in high yield (≥98.0% by trityl analysis) and multiple additions were straightforward, as demonstrated by the synthesis of an oligonucleotide containing five additions of Cy3dT (Figure 2A). In addition, oligonucleotides containing both Cy3dT and Cy5dT were readily prepared (Figure 2B). All oligonucleotides were purified by high performance liquid chromatography (HPLC), analysed by capillary gel electrophoresis and characterized by mass spectrometry.
Figure 2.
Capillary electrophoresis analysis (monitored at 260 nm). (A) An oligonucleotide containing five additions of Cy3dT (ODN-38). (B) An oligonucleotide containing both Cy3dT and Cy5dT (ODN-16).
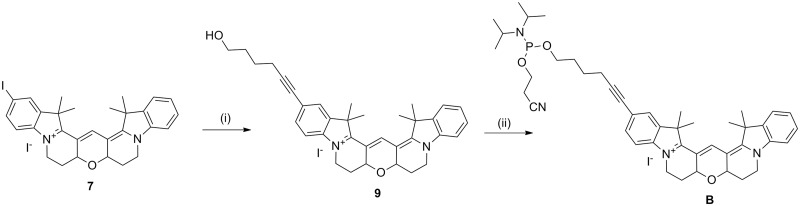
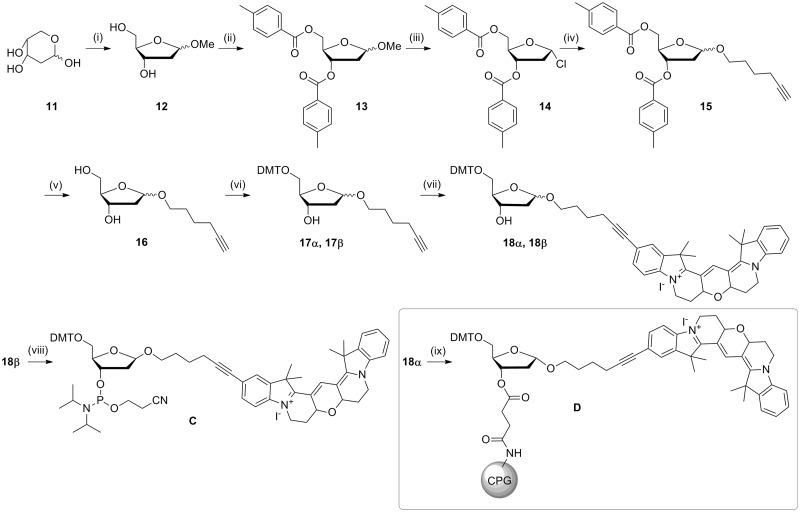
To expand the Cy3B toolkit, a monomer for the 5′-addition of Cy3B was synthesized by reacting iodo-Cy3B with 5-hexyn-1-ol then converting the product to the phosphoramidite (B, Scheme 2). Iodo-Cy3B was also used as an intermediate in the synthesis of 5-ethynyl-Cy3B (E, Scheme 3), which was used for the incorporation of Cy3B into DNA by reverse click chemistry (18). The Cy3B compounds were found to have a very high affinity for silica and so special care was taken during purification; methanolic ammonia was used in the mobile phase to prevent retention of the compounds on the column. To complete the series of Cy3B labelling reagents, Cy3BdR (dR = deoxyribose) was synthesized by reacting iodo-Cy3B with 1′-hexynyl deoxyribose (Scheme 4). Phosphitylation of the β-anomer provided a phosphoramidite monomer for internal or end-addition (C), and attachment of the α-anomer to a solid support gave α-Cy3BdR resin D, which provided a means of adding Cy3B to the 3′-end of oligonucleotides. The above monomers and solid supports enable the facile incorporation of Cy-Dyes into oligonucleotides at any internal or terminal position during oligonucleotide synthesis.
Scheme 2.
Synthesis of 5-(hexyn-1-ol)-6-Cy3B phosphoramidite. Reagents and conditions: (i) 5-hexyn-1-ol, CuI, Pd(PPh3)4, DMF, Et3N, room temperature, 5 h, 82%; (ii) 2-cyanoethyl-N,N-diisopropyl-chlorophosphoramidite, DIPEA, DCM, room temperature, 45 min, 93%.
Scheme 3.
Synthesis of 5-ethynyl-Cy3B. Reagents and conditions: (i) trimethylsilylacetylene, CuI, Pd(PPh3)4, Et3N, DMF, 16 h, room temperature, 71%; (ii) TBAF, THF, 5 min, room temperature, 75%.
Scheme 4.
Synthesis of β-Cy3BdR phosphoramidite and α-Cy3BdR solid support. Reagents and conditions; (i) MeOH, 1% methanolic HCl, room temperature, 30 min; (ii) pyridine, p-toluoyl-chloride, 0°C then room temperature, 16 h; (iii) acetic acid, saturated HCl in acetic acid, acetyl chloride, 0°C then room temperature, 64% over three steps; (iv) THF, DCM, DMAP, 5-hexyn-1-ol, room temperature, 3.5 h, 53%; (v) methanolic ammonia, 45°C, 16 h, 82%; (vi) DMTCl, pyridine, DMAP, 1 h, room temperature, α 47%, β 41% (total 88%); (vii) 7, CuI, Pd(PPh3)4, Et3N, DMF, 52 h, room temperature, α 66%, β 73%; (viii) 2-cyanoethyl-N,N-diisopropyl-chlorophosphoramidite, DIPEA, DCM, room temperature, 45 min, 94%; (ix) succinylated amino-link resin, N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride, DMAP, Et3N.
Properties of oligonucleotides containing Cy-Dyes
The effects of Cy3dT, Cy5dT and Cy3BdT on DNA duplex stability were evaluated by UV melting using a series of 23- to 25-mer oligonucleotides. Each dye slightly destabilized the duplex; Cy3BdT (−3.4°C), Cy5dT (−3.1°C) and Cy3dT (−2.4°C), as is often observed for nucleobases labelled with fluorophores (Supplementary Table S1, sequences in Supplementary Table S2). Nevertheless, these results demonstrate that the Cy-dT monomers can be used as thymidine analogues without greatly perturbing the DNA duplex. This was confirmed by comparing the CD spectra of oligonucleotides containing Cy-Dyes with their unmodified counterparts (Supplementary Figure S1).
The fluorescence quantum yields of the Cy3B monomers were, as expected, significantly higher than Cy3, Cy5 or the corresponding Cy3dT and Cy5dT monomers (0.5 and 0.8 for Cy3BdT and β-Cy3BdR, respectively, compared to 0.15 and 0.28 for Cy3 and Cy5 NHS esters and 0.05 and 0.06 for Cy3dT and Cy5dT, respectively, Supplementary Table S3). Cy3B is not normally vulnerable to the photo-induced cis/trans isomerization or the temperature-dependent decrease in fluorescence exhibited by Cy3 and Cy5 because the rigid cyclized polymethine linker prevents rotation (1,19–23). To demonstrate this holds true for our derivatives, Cy3dT, Cy5dT and Cy3B oligonucleotides ODN-20, ODN-21, ODN-23, ODN-11 (Supplementary Table S4) were subjected to fluorescence melting. The oligonucleotides containing Cy3dT (ODN-20) and Cy5dT (ODN-21) exhibited significant temperature dependence, whereas the Cy3B oligonucleotides (ODN-23, ODN-11) showed virtually no reduction in fluorescence with increased temperature (Supplementary Figure S2). The above results indicate that oligonucleotides containing the new Cy3BdT monomers will be particularly suitable in biochemical and biological applications which require high sensitivity, and in studies that are carried out at elevated temperatures (e.g. high-resolution DNA melting).
In a preliminary study, duplexes containing Cy3dT and Cy5dT in complementary strands showed the expected inverse correlation between FRET efficiency and the distance between the dyes (Supplementary Figure S4, Supplementary Table S5). The dyes are located in the major groove on the periphery of the duplex (5-position of thymine) which means that increasing the base pair separation between Cy-Dye labelled bases along the helical axis sometimes brings the dyes closer together about the helix axis (Supplementary Figure S5), an effect also seen by Clegg et al. (24). This effect is unavoidable for any dye that is attached to a DNA base. The rigidity of the ethynyl linker imposes another important restriction on dye mobility; FRET measurements at shorter dye-acceptor distances are possible as the dyes cannot reach each other to give contact quenching.
As indicated by Seidel et al. (5), short linkers are required for measuring short distances in DNA, because the length of long linkers is comparable to the absolute distances between donor and acceptor dyes. They also note that κ2-related uncertainties for short linkers are outweighed by better defined dye positions. Hence, although rotation about the short linker of the Cy-dT monomers imposes uncertainty in the κ2 value, this is compensated by the precise dye positioning along the helical axis. This is clearly a complicated issue made more complex by the dynamics of the DNA duplex, and a detailed study will be undertaken by us in due course.
Studies on fluorogenic PCR probes containing Cy3B monomers
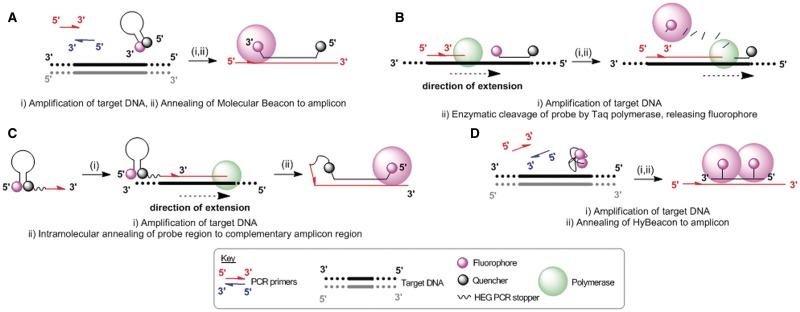
Several fluorogenic PCR probes (Figure 3) containing Cy3B were synthesized; a Taqman probe (25,26), a HyBeacon (27–30), a Molecular Beacon (31–36) and a Scorpion probe (37–39); along with synthetic templates and primers for the cystic fibrosis transmembrane conductance regulatory (CFTR) gene (R516G mutation) (ODN-1 wt, ODN-2 mt, ODN-3 forward primer, ODN-4 reverse primer, Table 1).
Figure 3.
The mechanism of action of (A) Molecular Beacon probes, (B) Taqman probes, (C) Scorpion probes and (D) HyBeacon probes.
Table 1.
Oligonucleotide sequences used in the PCR study
| ODNs | Sequences | ODN descriptions |
|---|---|---|
| ODN-1 | TCTCAGTTTTCCTGGATTATGCCTGGCACCATTAAAGAAAATATCATCTTTGGTG TTTCCTATGATGAATATAGATACAGAAGCGTCATCAAAGCATGCCAACTAGAA GAGGTAAGAAACTATGTGAAAACTTTTTGA | Wild-type template |
| ODN-2 | TCTCAGTTTTCCTGGATTATGCCTGGCACCATTAAAGAAAATATCATCTTTGGTGT TTCCTATGATGAATATGGATACAGAAGCGTCATCAAAGCATGCCAACTAGAAGA GGTAAGAAACTATGTGAAAACTTTTTGA | Mutant template |
| ODN-3 | CAGTTTTCCTGGATTATGCC | Forward primer |
| ODN-4 | CAAAAAGTTTTCACATAGTTTCTT | Reverse primer |
| ODN-5 | GGCACCATTAAAGAAAATATCA | Reverse Taqman primer |
| ODN-6 | hTTCCTATGAqGAATATAGATACAGAAGCGp | Taqman probe |
| ODN-7 | CGCTTCbGTATCbATATTCATCp | HyBeacon |
| ODN-8 | sCCTAGCATGATGAATATAGATACAGAAGCGTCGCTAGGd | Dye-quencher |
| Molecular Beacon | ||
| ODN-9 | sCCTAGCATGATGAATATAGATACAGAAGCGTCGCTAGGr | Dual-fluorophore |
| Molecular Beacon | ||
| ODN-10 | sCCGCGGGATGAATATAGATACAGAAGCGCCGCGGQHTCTTCTAGTTGGCATGCT | Scorpion probe |
Key; b = Cy3BdT phosphoramidite; p = propanol resin; h = 5-(hexyn-1-ol)-6-Cy3B phosphoramidite; d = DABCYL resin; Q = DABCYL-dT phosphoramidite; r = α-Cy3BdR resin; s = β-Cy3BdR phosphoramidite; H = Hexaethylene glycol; q = BHQ2-dT phosphoramidite. PCR templates (ODN-1 wt, ODN-2 mt) show the mutation site underlined. PCR primers (ODN-3 forward, ODN-4 reverse and ODN-5 reverse primer for Taqman probe).
Molecular Beacon probes containing β-Cy3BdR phosphoramidite and α-Cy3BdR resin
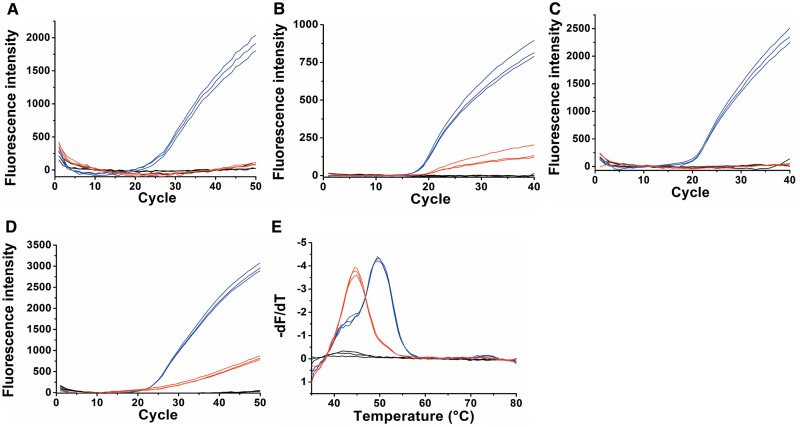
Molecular Beacons are optical switches that have very low fluorescence in the closed form and highly intense fluorescence in the hybridized form. Cy3B Molecular Beacons were designed to probe the antisense strand (instead of the sense strand ODN-1) in order to achieve a good wt/mt discrimination by forming an AC mismatch which is particularly unstable at neutral pH and above, instead of a relatively stable TG mismatch (40–42). Two Molecular Beacon probes were evaluated. Firstly, a conventional Molecular Beacon was designed with a 3′-DABCYL quencher and 5′-β-Cy3BdR fluorophore (ODN-8). It has been established (43) that DABCYL is a suitable contact quencher for Cy3B with minimal unwanted FRET quenching in the open Beacon. Secondly, a dual-fluorophore Molecular Beacon (ODN-9) was synthesized using the α-Cy3BdR solid support at the 3′-end and the β-Cy3BdR phosphoramidite monomer at the 5′-end. Dual-fluorophore Molecular Beacons have been described previously (44). In the closed form the two dyes stack to form a quenched dimer, whereas in the hybridized form they give increased fluorescence compared to standard dye-quencher Molecular Beacons. Variations on the dual-fluorophore Molecular Beacon have been studied using several chromophores (45–47) but it remains a challenge to obtain good closed-Beacon quenching yet retain high fluorescence in the hybridized form. It was hoped that both these requirements could be fulfilled using Cy3B dual-fluorophore Molecular Beacons. Asymmetric real-time PCR using both Beacons gave successful wt/mt discrimination using ODN-8 (Supplementary Figure S3) and ODN-9 (Figure 4A). In both probe formats the wild-type template had a cycle-threshold (CT) value of 20 whereas the mutant-type template showed no real-time fluorescence accumulation. This is because the duplexes formed between the mutant template and the Molecular Beacon probes are unstable due to the mismatch, so are not formed at the monitoring temperature during the PCR (58°C). The dual-fluorophore Molecular Beacon gave a far superior real-time amplification than the traditional Molecular Beacon giving almost double the fluorescence signal.
Figure 4.
Wild-type template (blue), mutant template (red) and negative control (black). Samples performed in triplicate. (A) Fluorescence accumulation during real-time PCR using the dual-fluorophore Molecular Beacon (ODN-9). (B) Fluorescence accumulation of real-time PCR using a Taqman probe containing Cy3B and BHQ2 (ODN-6). (C) Fluorescence accumulation of real-time PCR using a Scorpion probe containing Cy3B and DABCYL (ODN-10). (D) Fluorescence accumulation of real-time PCR using a HyBeacon probe containing Cy3BdT (ODN-7). (E) The derivative of the fluorescence melting curve, giving Tm values for the wild-type (50°C) and mutant (45°C).
Taqman probe containing 5′-Cy3B phosphoramidite
A Taqman probe with 5′-Cy3B (5-(hexyn-1-ol)-6-Cy3B phosphoramidite) and internal BHQ2 was synthesized (ODN-6, Table 1). BHQ2 was chosen as the FRET quencher due to its strong absorbance at the emission wavelength of Cy3B (BHQ2 abs max 580 nm). In a conventional Taqman probe (normally ∼20 bases in length) the fluorophore is located at the 5′-end so that it is cleaved efficiently by the DNA polymerase, and the quencher is located at the 3′-end (25). A longer probe (29 bases) was designed for the AT-rich CFTR R516G locus to ensure that the probe was annealed to the template at the PCR extension temperature. The quencher was located centrally within the probe to allow for efficient FRET quenching of the fluorophore. Symmetric real-time PCR was carried out to give successful discrimination between the wild-type and mutant templates (Figure 4B). The wild-type had a CT value of 16 indicating that probe cleavage during the extension step was very efficient, whereas the mutant had a CT value of 19; in this case the probe-template duplex is unstable and so the probe was cleaved at a far slower rate. The ratio of fluorescence intensity (∼6.5:1 wt:mt) is also a clear indicator of good wt/mt discrimination.
Scorpion probe containing β-Cy3BdR phosphoramidite monomer
A Scorpion probe was synthesized containing β-Cy3BdR and a DABCYL quencher (ODN-10) (Table 1). Symmetric real-time PCR was carried out and excellent wt/mt discrimination was obtained (Figure 4C). The wild-type template had a CT value of 18 and the final fluorescence intensity was high, indicating efficient probe-template duplex formation. This was anticipated, as Scorpion probe-amplicon binding is kinetically favoured over re-annealing of the amplicon, and is thermodynamically favoured over re-folding into a hairpin structure, thereby providing a rapid and robust signalling mechanism (37,38).
HyBeacon probe containing Cy3BdT phosphoramidite monomer
HyBeacon probes can be used to monitor the PCR in real-time, but their major benefit stems from post-amplification melting which gives precise discrimination between wild-type and mutant targets (matched and mismatched). A HyBeacon probe was synthesized containing two Cy3BdT units (ODN-7, Table 1). Asymmetric PCR was carried out followed by post-PCR fluorescence melting and the results are shown in Figure 4. The wild-type has a CT value of 22 while the mutant has a CT value of 25 (Figure 4D). In addition, the ratio of fluorescence intensity is ∼4:1 (wt:mt) as a result of the mutant duplex being less stable at the monitoring temperature. The derivative of a post-amplification melting curve (Figure 4E) gives clearly defined Tm values of 50°C for the wt and 45°C for the mutant, making it possible to unambiguously discriminate between them.
CONCLUSIONS
A new range of Cy-Dye phosphoramidite monomers have been synthesized and incorporated into synthetic oligonucleotides. The phosphoramidite method of insertion during DNA synthesis allows for efficient multiple and mixed dye additions and provides a convenient and efficient site-specific method of inserting cyanine dyes into oligonucleotides. A Cy3B molecular toolkit has been produced based on an efficient synthesis of iodo and ethynyl derivatives of the Cy3B dye. These new Cy3B phosphoramidite monomers and solid supports offer major advantages over the commercially available post-synthetic Cy3B labelling reagents, as they provide a convenient way to incorporate single or multiple Cy3B units at any oligonucleotide position in high yield, as demonstrated by the synthesis of oligonucleotides labelled internally and at both ends. As anticipated, Cy3B oligonucleotides have superior fluorescence properties compared to Cy3 and Cy5 analogues. The synthesis of a Taqman probe, a HyBeacon probe, a Molecular Beacon and a Scorpion probe containing various Cy3B monomers demonstrates the utility of the Cy3B molecular toolkit and a dual-fluorophore Molecular Beacon containing a Cy3B monomer at both termini was found to be superior to a corresponding Cy3B/DABCYL Molecular Beacon. A preliminary study indicated that the rigid ethynyl Cy-dT monomers can be used to measure FRET within an oligonucleotide, and detailed studies of this type will be undertaken in the future. It is envisaged that oligonucleotide probes containing this new range of Cy-Dye monomers could be used for single-molecule and cell-imaging studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Schemes 1 and 2, Supplementary Tables 1–5, Supplementary Figures 1–5, Supplementary Methods (synthesis of the Cy-Dye monomers, methods for oligonucleotide synthesis, purification and biophysical studies) and Supplementary References [12–18,48–62].
FUNDING
The UK Biotechnology and Biological Sciences Research Council and ATDBio via a BBSRC Ph.D. studentship (to L.H.) and a CASE studentship (to M.G.). Funding for open access charge: University of Southampton.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank ATDBio for oligonucleotide synthesis and Dr Rob Powell (Primer Design) and Jane Theaker (Qiagen) for assistance with probe design.
REFERENCES
- 1.Sanborn ME, Connolly BK, Gurunathan K, Levitus M. Fluorescence properties and photophysics of the sulfoindocyanine Cy3 linked covalently to DNA. J. Phys. Chem. B. 2007;111:11064–11074. doi: 10.1021/jp072912u. [DOI] [PubMed] [Google Scholar]
- 2.El-Sagheer AH, Brown T. Click chemistry with DNA. Chem. Soc. Rev. 2010;39:1388–1405. doi: 10.1039/b901971p. [DOI] [PubMed] [Google Scholar]
- 3.Gierlich J, Burley GA, Gramlich PM, Hammond DM, Carell T. Click chemistry as a reliable method for the high-density postsynthetic functionalization of alkyne-modified DNA. Org. Lett. 2006;8:3639–3642. doi: 10.1021/ol0610946. [DOI] [PubMed] [Google Scholar]
- 4.Seela F, Sirivolu VR. Nucleosides and oligonucleotides with diynyl side chains: the Huisgen-Sharpless cycloaddition “click reaction” performed on DNA and their constituents. Nucleosides Nucleotides Nucleic Acids. 2007;26:597–601. doi: 10.1080/15257770701490308. [DOI] [PubMed] [Google Scholar]
- 5.Sindbert S, Kalinin S, Nguyen H, Kienzler A, Clima L, Bannwarth W, Appel B, Müller S, Seidel CAM. Accurate distance determination of nucleic acids via For̈ster resonance energy transfer: implications of dye linker length and rigidity. J. Am. Chem. Soc. 2011;133:2463–2480. doi: 10.1021/ja105725e. [DOI] [PubMed] [Google Scholar]
- 6.Iqbal A, Arslan S, Okumus B, Wilson TJ, Giraud G, Norman DG, Ha T, Lilley DMJ. Orientation dependence in fluorescent energy transfer between Cy3 and Cy5 terminally attached to double-stranded nucleic acids. Proc. Natl Acad. Sci. USA. 2008;105:11176–11181. doi: 10.1073/pnas.0801707105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ouellet J, Schorr S, Iqbal A, Wilson TJ, Lilley DMJ. Orientation of cyanine fluorophores terminally attached to DNA via long, flexible tethers. Biophys. J. 2011;101:1148–1154. doi: 10.1016/j.bpj.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Börjesson K, Preus S, El-Sagheer AH, Brown T, Albinsson B, Wilhelmsson LM. Nucleic acid base analog FRET-pair facilitating detailed structural measurements in nucleic acid containing systems. J. Am. Chem. Soc. 2009;131:4288–4293. doi: 10.1021/ja806944w. [DOI] [PubMed] [Google Scholar]
- 9.Hudson RHE, Ghorbani-Choghamarani A. Oligodeoxynucleotides incorporating structurally simple 5-alkynyl-2'-deoxyuridines fluorometrically respond to hybridization. Org. Biomol. Chem. 2007;5:1845–1848. doi: 10.1039/b705805e. [DOI] [PubMed] [Google Scholar]
- 10.Xie Y, Maxson T, Tor Y. Fluorescent nucleoside analogue displays enhanced emission upon pairing with guanine. Org. Biomol. Chem. 2010;8:5053–5055. doi: 10.1039/c0ob00413h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilhelmsson LM. Fluorescent nucleic acid base analogues. Q. Rev. Biophys. 2010;43:159–183. doi: 10.1017/S0033583510000090. [DOI] [PubMed] [Google Scholar]
- 12.Fegan A, Shirude PS, Balasubramanian S. Rigid cyanine dye nucleic acid labels. Chem. Commun. 2008:2004–2006. doi: 10.1039/b801629a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waggoner AS, Mujumdar R. Rigidized trimethine cyanine dyes. U.S. Patent No. 2001 6133445A. [Google Scholar]
- 14.Zimmermann T, Hennig L. Ring transformations of heterocyclic compounds. XXII. Pyrido[1,2-a]indolium salts from 2-methyl-3H-indoles by pyrylium mediated three carbon annelation. J. Heterocycl. Chem. 2002;39:263–269. [Google Scholar]
- 15.Tomasulo M, Sortino S, Raymo FM. Bichromophoric photochromes based on the opening and closing of a single oxazine ring. J. Org. Chem. 2008;73:118–126. doi: 10.1021/jo7017119. [DOI] [PubMed] [Google Scholar]
- 16.Mason SJ, Hake JL, Nairne J, Cummins WJ, Balasubramanian S. Solid-phase methods for the synthesis of cyanine dyes. J. Org. Chem. 2005;70:2939–2949. doi: 10.1021/jo0479415. [DOI] [PubMed] [Google Scholar]
- 17.Graham D, Parkinson JA, Brown T. DNA duplexes stabilized by modified monomer residues: synthesis and stability. J. Chem. Soc.-Perkin Trans. I. 1998:1131–1138. [Google Scholar]
- 18.Gerowska M, Hall LM, Richardson JA, Shelbourne M, Brown T. Efficient reverse click labeling of azide oligonucleotides with multiple alkynyl Cy-Dyes applied to the synthesis of HyBeacon probes for genetic analysis. Tetrahedron. 2012;68:857–864. [Google Scholar]
- 19.Aramendia PF, Negri RM, Sanroman E. Temperature-dependence of fluorescence and photoisomerisation in symmetrical carbocyanines - influence of medium viscosity and molecular-structure. J. Phys. Chem. 1994;98:3165–3173. [Google Scholar]
- 20.Chibisov AK, Zakharova GV, Goerner H, Sogulyaev YA, Mushkalo IL, Tolmachev AI. Photorelaxation processes in covalently-linked indocarbocyanine and thiacarbocyanine dyes. J. Phys. Chem. 1995;99:886–893. [Google Scholar]
- 21.Chibisov AK, Zakharova GV, Goerner H. Effects of substituents in the polymethine chain on the photoprocesses in indodicarbocyanine dyes. J. Chem. Soc.-Faraday Trans. 1996;92:4917–4925. [Google Scholar]
- 22.Harvey BJ, Levitus M. Nucleobase-specific enhancement of Cy3 fluorescence. J. Fluoresc. 2009;19:443–448. doi: 10.1007/s10895-008-0431-1. [DOI] [PubMed] [Google Scholar]
- 23.Levitus M, Ranjit S. Cyanine dyes in biophysical research: the photophysics of polymethine fluorescent dyes in biomolecular environments. Q. Rev. Biophys. 2011;44:123–151. doi: 10.1017/S0033583510000247. [DOI] [PubMed] [Google Scholar]
- 24.Clegg RM, Murchie AIH, Zechel A, Lilley DMJ. Observing the helical geometry of double-stranded DNA in solution by fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA. 1993;90:2994–2998. doi: 10.1073/pnas.90.7.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Flood SJA, Marmaro J, Giusti W, Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridisation. PCR Methods Appl. 1995;4:357–362. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- 26.Holland PM, Abramson RD, Watson R, Gelfand DH. Detection of specific polymerase chain-reaction product by utilising the 5’-3’ exonuclease activity of thermus-aquaticus DNA-polymerase. Proc. Natl Acad. Sci. USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.French DJ, Archard CL, Andersen MT, McDowell DG. Ultra-rapid DNA analysis using HyBeacon probes and direct PCR amplification from saliva. Mol. Cell. Probes. 2002;16:319–326. doi: 10.1006/mcpr.2002.0425. [DOI] [PubMed] [Google Scholar]
- 28.French DJ, Archard CL, Brown T, McDowell DG. HyBeacon (TM) probes: a new tool for DNA sequence detection and allele discrimination. Mol. Cell. Probes. 2001;15:363–374. doi: 10.1006/mcpr.2001.0384. [DOI] [PubMed] [Google Scholar]
- 29.Dobson N, McDowell DG, French DJ, Brown LJ, Mellor JM, Brown T. Synthesis of HyBeacons and dual-labelled probes containing 2'-fluorescent groups for use in genetic analysis. Chem. Commun. 2003:1234–1235. doi: 10.1039/b302855k. [DOI] [PubMed] [Google Scholar]
- 30.Richardson JA, Gerowska M, Shelbourne M, French D, Brown T. Six-colour HyBeacon probes for multiplex genetic analysis. ChemBioChem. 2010;11:2530–2533. doi: 10.1002/cbic.201000623. [DOI] [PubMed] [Google Scholar]
- 31.Piatek AS, Tyagi S, Pol AC, Telenti A, Miller LP, Kramer FR, Alland D. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat. Biotechnol. 1998;16:359–363. doi: 10.1038/nbt0498-359. [DOI] [PubMed] [Google Scholar]
- 32.Tyagi S, Bratu DP, Kramer FR. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 33.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 34.Marras SAE, Tyagi S, Kramer FR. Real-time assays with molecular beacons and other fluorescent nucleic acid hybridization probes. Clin. Chim. Acta. 2006;363:48–60. doi: 10.1016/j.cccn.2005.04.037. [DOI] [PubMed] [Google Scholar]
- 35.Vet JAM, Majithia AR, Marras SAE, Tyagi S, Dube S, Poiesz BJ, Kramer FR. Multiplex detection of four pathogenic retroviruses using molecular beacons. Proc. Natl Acad. Sci. USA. 1999;96:6394–6399. doi: 10.1073/pnas.96.11.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marras SAE, Kramer FR, Tyagi S. Multiplex detection of single-nucleotide variations using molecular beacons. Genet. Anal. -Biomol. Eng. 1999;14:151–156. doi: 10.1016/s1050-3862(98)00018-7. [DOI] [PubMed] [Google Scholar]
- 37.Whitcombe D, Theaker J, Guy SP, Brown T, Little S. Detection of PCR products using self-probing amplicons and fluorescence. Nat. Biotechnol. 1999;17:804–807. doi: 10.1038/11751. [DOI] [PubMed] [Google Scholar]
- 38.Thelwell N, Millington S, Solinas A, Booth J, Brown T. Mode of action and application of Scorpion primers to mutation detection. Nucleic Acids Res. 2000;28:3752–3761. doi: 10.1093/nar/28.19.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Solinas A, Brown LJ, McKeen C, Mellor JM, Nicol JTG, Thelwell N, Brown T. Duplex Scorpion primers in SNP analysis and FRET applications. Nucleic Acids Res. 2001;29:E96. doi: 10.1093/nar/29.20.e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ikuta S, Takagi K, Wallace RB, Itakura K. Dissociation kinetics of 19 base paired oligonucleotide-DNA duplexes containing different single mismatched base pairs. Nucleic Acids Res. 1987;15:797–811. doi: 10.1093/nar/15.2.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ke SH, Wartell RM. Influence of nearest neighbour sequence on the stability of base pair mismatches in long DNA; determination by temperature-gradient gel electrophoresis. Nucleic Acids Res. 1993;21:5137–5143. doi: 10.1093/nar/21.22.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allawi HT, SantaLucia J. Nearest-neighbor thermodynamics of internal A.C mismatches in DNA: sequence dependence and pH effects. Biochemistry. 1998;37:9435–9444. doi: 10.1021/bi9803729. [DOI] [PubMed] [Google Scholar]
- 43.Marras SAE, Kramer FR, Tyagi S. Efficiencies of fluorescence resonance energy transfer and contact-mediated quenching in oligonucleotide probes. Nucleic Acids Res. 2002;30:e122. doi: 10.1093/nar/gnf121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Venkatesan N, Seo YJ, Kim BH. Quencher-free molecular beacons: a new strategy in fluorescence based nucleic acid analysis. Chem. Soc. Rev. 2008;37:648–663. doi: 10.1039/b705468h. [DOI] [PubMed] [Google Scholar]
- 45.Bernacchi S, Mely Y. Exciton interaction in molecular beacons: a sensitive sensor for short range modifications of the nucleic acid structure. Nucleic Acids Res. 2001;29:e62. doi: 10.1093/nar/29.13.e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conley NR, Pomerantz AK, Wang H, Twieg RJ, Moerner WE. Bulk and single-molecule characterization of an improved molecular beacon utilizing H-dimer excitonic behavior. J. Phys. Chem. B. 2007;111:7929–7931. doi: 10.1021/jp073310d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nesterova IV, Erdem SS, Pakhomov S, Hammer RP, Soper SA. Phthalocyanine dimerization-based molecular beacons using Near-IR fluorescence. J. Am. Chem. Soc. 2009;131:2432–2433. doi: 10.1021/ja8088247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gottlieb HE, Kotlyar V, Nudelman A. NMR chemical shifts of common laboratory solvents as trace impurities. J. Org. Chem. 1997;62:7512–7515. doi: 10.1021/jo971176v. [DOI] [PubMed] [Google Scholar]
- 49.Booth J, Brown T, Vadhia SJ, Lack O, Cummins WJ, Trent JO, Lane AN. Determining the origin of the stabilization of DNA by 5-aminopropynylation of pyrimidines. Biochemistry. 2005;44:4710–4719. doi: 10.1021/bi047561d. [DOI] [PubMed] [Google Scholar]
- 50.Sonogashira K, Tohda Y, Hagihara N. Convenient synthesis of acetylenes - catalytic substitutions of acetylenic hydrogen with bromoalkenes, iodoarenes, and bromopyridines. Tetrahedron Lett. 1975;16:4467–4470. [Google Scholar]
- 51.Hobbs FW. Palladium-catalyzed synthesis of alkynylamino nucleosides - a universal linker for nucleic acids. J. Org. Chem. 1989;54:3420–3422. [Google Scholar]
- 52.Crisp GT, Flynn BL. Palladium-catalyzed coupling of terminal alkynes with 5-(trifluoromethanesulfonyloxy)pyrimidine nucleosides. J. Org. Chem. 1993;58:6614–6619. [Google Scholar]
- 53.Cai C, Vasella A. Oligosaccharide analogs of polysaccharides.3. A new protecting group for alkynes - orthogonally protected dialkynes. Helv. Chim. Acta. 1995;78:732–757. [Google Scholar]
- 54.El-Sagheer AH, Brown T. Synthesis of alkyne- and azide-modified oligonucleotides and their cyclization by the CuAAC (click) reaction. Curr. Protoc. Nucleic Acid Chem. 2008;35:4.33.1–4.33.21. doi: 10.1002/0471142700.nc0433s35. [DOI] [PubMed] [Google Scholar]
- 55.Kocalka P, El-Sagheer AH, Brown T. Rapid and efficient DNA strand cross-linking by click chemistry. ChemBioChem. 2008;9:1280–1285. doi: 10.1002/cbic.200800006. [DOI] [PubMed] [Google Scholar]
- 56.Deglane G, Morvan F, Debart F, Vasseur JJ. 5-Propynylamino alpha-deoxyuridine promotes DNA duplex stabilization of anionic and neutral but not cationic alpha-oligonucleotides. Bioorg. Med. Chem. Lett. 2007;17:951–954. doi: 10.1016/j.bmcl.2006.11.052. [DOI] [PubMed] [Google Scholar]
- 57.Rolland V, Kotera M, Lhomme J. Convenient preparation of 2-deoxy-3,5-di-O-p-toluoyl-alpha-D-erythro-pentofuranosyl chloride. Synth. Commun. 1997;27:3505–3511. [Google Scholar]
- 58.Morvan F, Rayner B, Leonetti JP, Imbach JL. alpha-DNA. VII. Solid-phase synthesis of alpha-anomeric oligodeoxyribonucleotides. Nucleic Acids Res. 1988;16:833–847. doi: 10.1093/nar/16.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Damha MJ, Giannaris PA, Zabarylo SV. An improved procedure for derivatization of controlled-pore glass-beads for solid-phase oligonucleotide synthesis. Nucleic Acids Res. 1990;18:3813–3821. doi: 10.1093/nar/18.13.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.El-Sagheer AH, Brown T. New strategy for the synthesis of chemically modified RNA constructs exemplified by hairpin and hammerhead ribozymes. Proc. Natl Acad. Sci. USA. 2010;107:15329–15334. doi: 10.1073/pnas.1006447107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karstens T, Kobs K. Rhodamine-B and rhodamine-101 as reference substances for fluorescence quantum yield measurements. J. Phys. Chem. 1980;84:1871–1872. [Google Scholar]
- 62.Langley GJ, Herniman JM, Davies NL, Brown T. Simplified sample preparation for the analysis of oligonucleotides by matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 1999;13:1717–1723. doi: 10.1002/(SICI)1097-0231(19990915)13:17<1717::AID-RCM704>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.