Abstract
Deep sequencing studies frequently identify small RNA fragments of abundant RNAs. These fragments are thought to represent degradation products of their precursors. Using sequencing, computational analysis, and sensitive northern blot assays, we show that constitutively expressed non-coding RNAs such as tRNAs, snoRNAs, rRNAs and snRNAs preferentially produce small 5′ and 3′ end fragments. Similar to that of microRNA processing, these terminal fragments are generated in an asymmetric manner that predominantly favors either the 5′ or 3′ end. Terminal-specific and asymmetric processing of these small RNAs occurs in both mouse and human cells. In addition to the known processing of some 3′ terminal tRNA-derived fragments (tRFs) by the RNase III endonuclease Dicer, we show that several RNase family members can produce tRFs, including Angiogenin that cleaves the TψC loop to generate 3′ tRFs. The 3′ terminal tRFs but not the 5′ tRFs are highly complementary to human endogenous retroviral sequences in the genome. Despite their independence from Dicer processing, these tRFs associate with Ago2 and are capable of down regulating target genes by transcript cleavage in vitro. We suggest that endogenous 3′ tRFs have a role in regulating the unwarranted expression of endogenous viruses through the RNA interference pathway.
INTRODUCTION
Recent applications of deep sequencing methods have led to the identification of a surprising diversity of non-protein-coding RNAs (ncRNAs), including degradation-like small RNA fragments derived from miRNAs (1,2), snoRNAs (3–5) and tRNAs (6–12). Evidence is accumulating that these small RNA fragments are precisely processed to participate in diverse biological processes and are conserved in distantly related species (2,3,5–7,13). The first report of a small snoRNA-derived RNA and its function as a miRNA was reported 3 years ago (3). Since then, ∼10 additional snoRNA-derived small (≥18 nt) RNAs that can function as miRNAs have been described (5). We have also reported a group of human and viral unusually small (∼15 nt) RNAs (usRNAs) that are derived from miRNAs as well as other non-coding regions, and can regulate gene (e.g., RAD21) expression (2). Production of these usRNAs are evolutionarily conserved (13,14) and are associated with hippocampal functions in mice (13).
It is common to find small RNA fragments that match long RNA transcripts such as mRNAs in small RNA sequencing data (8). The presence of such small RNA fragments is generally attributed to the extreme sensitivity of deep sequencers in detecting low-abundance RNAs including degradation products. However, even very low-abundance RNAs, as low as four copies per cell, originating from the Cyclin D1 (CCND1) promoter region are reported to have regulatory functions (15). While many of the RNA fragments in deep-sequencing data frequently correspond to low abundance, potentially non-functional RNA products, RNA fragments found at even greater abundance than most miRNAs are also frequently observed in small RNA sequencing data (2,7). Common sources for such RNA fragments are tRNAs and rRNAs. Since tRNAs and rRNAs constitute most of the cellular RNA, it is reasonable to assume that these RNAs generate much more degradation products than other RNAs, resulting in the preferential detection of their degradation products in deep-sequencing. While the notion of degradation products is appealing, it was recently shown that tRNAs and rRNAs undergo stress-induced cleavage to produce stable RNA products, and this mechanism is conserved from yeast to human cells (11). In human cells, tRNA fragments have been reported to induce transient translational arrest (12). Various tRNA fragments are produced in cells under stress; notably during starvation in Tetrahymena thermophila (9) and serum deprivation in Giardia lambia (10), or during development in Aspergillus fumigatus (16). Stress-induced tRNA cleavage is mediated by Rny1p in yeast (17) and angiogenin (ANG) in humans (12,18). These stress-induced tRNA cleavage products are 30–50 nt in length and seem to form related RNA classes, termed sitRNA (10), tiRNA (12,19) and tRNA halves (11,16,18). An additional class of small RNAs that are ∼20−30 nt long, and are derived from 5′ or 3′ ends of tRNAs or from the genomic region following the 3′ end of tRNAs, broadly termed tRNA fragments (tRFs) were also reported (7). Subsequently, another class of seemingly related tRNA-derived small RNAs (tsRNAs) was found to function similarly to miRNAs. These tsRNAs are processed by either Dicer or RNase Z, depending on the tRF locations within both the mature tRNAs and their precursors (6). In summary, the widespread occurrence of stable small RNAs and their emerging roles in cellular processes indicate that we cannot ignore such molecules as randomly generated degradation products. Systematic analysis of all sequence reads from deep sequencing data may prove useful in defining new biological features and mechanisms relating to both canonical and non-canonical forms of small RNAs.
In this study, we found that the majority of the ∼20 nt long fragments deriving from the mature sequences of the widely expressed tRNAs, rRNAs, snoRNAs and snRNAs, but not mRNAs, are produced in a specific cleavage pattern from the 5′ or 3′ ends. We note that this pattern was previously observed for snoRNAs (3–5), and tRNAs (6–8). Similar to the processing of miRNAs, these terminal RNAs accumulate in the cell in an asymmetric manner that favors the expression of either the 5′ or the 3′ end fragments. The terminal fragments, are largely independent of canonical RNA interference (RNAi) processing machineries, which include Dicer that processes precursor miRNAs (pre-miRNAs), and DGCR8, which partners with Drosha to form the microprocessor complex that processes primary miRNA transcripts to pre-miRNAs. The biogenesis of tRFs is likely to depend on multiple RNase family members, including ANG that cleaves tRNAs within the TψC loop in a sequence-specific manner. Notably, antisense sequences of 3′-terminal tRFs are highly enriched in genomic regions harboring retroviral sequences, and 3′ tRFs are found to cleave target RNA through endogenous association with Ago2.
MATERIALS AND METHODS
Small RNA sequence analysis
Small RNAs reads from the B lymphoma BCP1 cell line were processed and mapped to the transcriptome as reported earlier (2). Small RNA sequences from mouse embryonic stem cells and HEK293 cells were retrieved from NCBI (GSE 12521 and GSE 16579). A total of 522 tRNA genes were curated from gtRNAdb (20), excluding 109 predicted pseudo tRNA genes. Perfect matches between reads and tRNAs were imposed to obtain the most reliable list of tRNA-derived RNAs. To eliminate any bias in our analysis due to terminal CCA motifs that are known to occur in tRNAs, the terminal CCA of tRNAs were masked. For comparison of the abundance of miRNA and different classes of terminal RNA in Dicer or DGCR8 knockout libraries, reads from different libraries were normalized using the total number of mRNA fragments in each library. For normalization, the total number of mRNA fragments (≥15 nt) in each library was used to first divide the read counts of each distinct RNA in that library, and then scaled by multiplying by the total number of mRNA reads in the reference (wild-type) library. To focus on the most reproducible set of tRFs, a threshold of 10 reads/million was used for the analysis of tRFs (Figures 3A, 4B, C and 8B; Supplementary Figure S5).
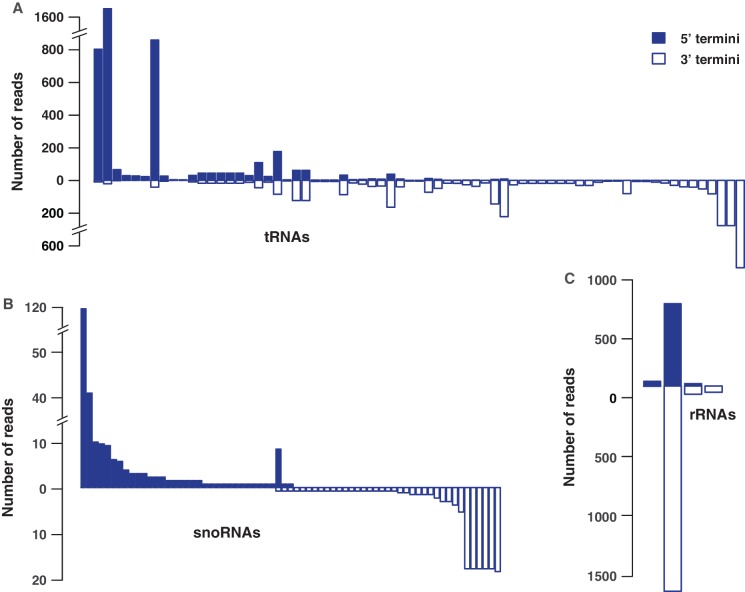
Figure 3.
Terminal small RNA fragments from tRNAs, snoRNAs and rRNAs manifest a bias for either 5′ or 3′ processing. (A) Abundance of 5′ (solid blue) and 3′ (blue) terminal RNAs produced from individual tRNAs (X-axis). For clarity, illustration is limited to non-redundant tRNA sequences producing at least 10 reads (per million) of terminal RNAs. Similar patterns are observed for snoRNAs (B) and rRNAs (C). Profiles of snRNAs are omitted because they yield terminal RNAs exclusively from their 3′ end (Figure 2).
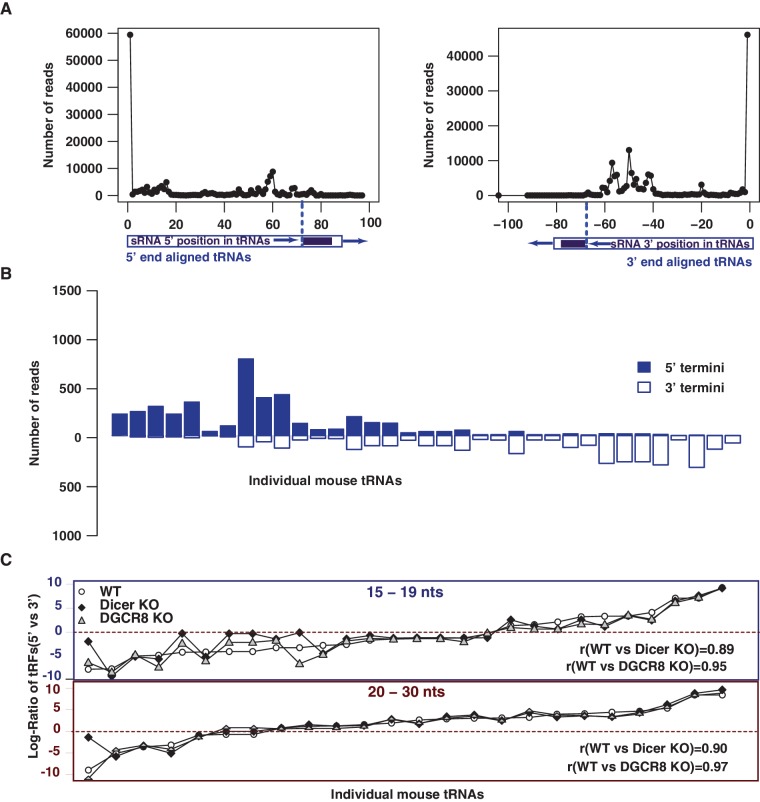
Figure 4.
Processing of 5′–3′ terminal RNA fragments are conserved in mice. (A) Position of the small RNA reads relative to 5′ (left panel) and 3′ (right panel) termini of tRNAs. (B) Similar to human 5′–3′ terminal tRFs, the mouse tRFs preferentially yield either 5′ or 3′ fragments. (C) Asymmetric bias of abundant (≥10 reads/million) 5′ and 3′ tRFs is well preserved across different conditions [wild-type, Dicer/DGCR8 Knock outs (KO)] for each tRNA, with high Pearson correlation coefficients (0.89−0.97).
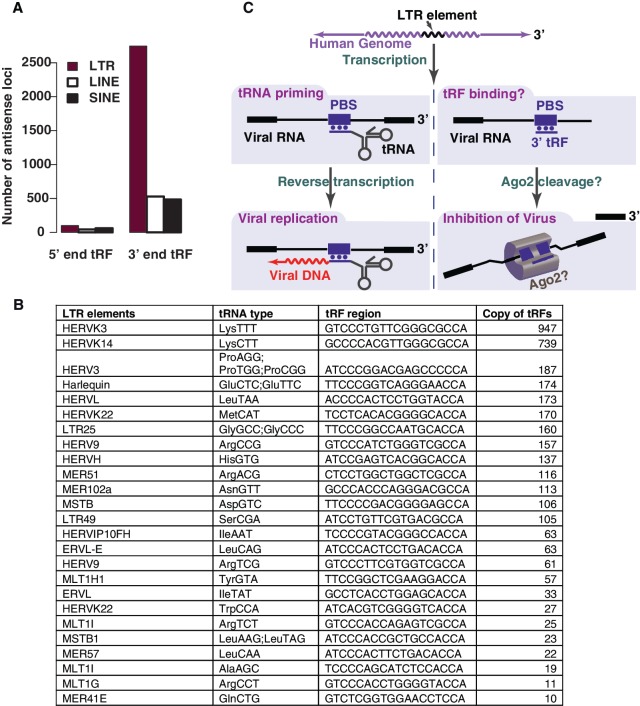
Figure 8.
3′ terminal tRFs might inhibit endogenous retroviral replication. (A) LTRs are highly enriched in complementary sequences of 3′ terminal tRFs. (B) The major tRNA terminal regions that produce abundant tRFs (reads ≥ 10, column 4) and their complementary LTR elements. (C) A potential model for the 3′ tRF pathway. The binding of 3′ tRF to the transcribed viral RNA could recruit double-stranded RNA specific endonucleases such as the highly efficient Ago2, enabling the rapid cleavage of the transcribed endogenous viral RNA.
To detect putative tRNA orthologs, we identified 137 human and mouse tRNA pairs with significantly high similarity (>80%) over >90% of the tRNA sequences. For analysis of retrotransposons, coordinates for human LTR, LINE and SINE elements were downloaded from UCSC genome database (21). To be considered as a potential tRF binding site, we required perfect match of the antisense sequence to the LTR, LINE and SINE elements. To eliminate artifacts due to the different sizes of LTR, LINE and SINE regions, the number of putative binding sites was normalized with respect to the total length of each of those elements. Since the length of the regions spanned by LINE and SINE elements were larger than LTR elements, the appropriate normalization factors of 0.42 and 0.67 were used to multiply the number of binding sites in LINE and SINE elements, respectively. All tRF sequences identified in BCP1 and were at least 10 nt long were used for the analysis. Since tRNAs can share similar terminal sequences, the terminal sequences that match to the most anti-LTR tRFs were selected as representative regions for illustrations.
Northern blot analysis
Northern blots were based on our recently published protocol (22). Briefly, total RNA (10–50 µg) was separated on 18% denaturing polyacrylamide gels and electro-transferred to positively charged nylon membranes from Roche (Roche Applied Science, Indianapolis, IN, USA). LNA–DNA mixed oligonucleotide probes were synthesized by IDT (Integrated DNA Technologies, Inc. Coralville, IA, USA). Probes were labeled with the small molecule Digoxigenin (DIG) using Oligo End Tailing Kit (Roche Applied Science, Indianapolis, IN, USA). Hybridization was carried out overnight in Ultrahyb buffer (Ambion, Austin, TX, USA) at 42°C and washing was performed in two rounds of 0.1 × SSC/0.1%SDS at 60°C. DIG signals were detected with alkaline phosphatase-conjugated anti-DIG antibody (Cat. 11093274910, Roche). Probes used for northern blotting corresponds to (LNAs in bold): 3′ end tRF of tRNA LysTTT (5′-TGGCGCCCGAACAGGA-3′); 5′ end tRF of tRNA LysTTT (5′-CTGAGCTATCCGGGAA-3′); 3′ end tRF of tRNA PheGAA (5′-GTGCCGAAACCCG-3′); 5′ end tRF of tRNA PheGAA (CTCCCAACTGAGCTATTTCG); miR-17 control RNA (5′-CTACCTGCACTGTAAGCACTTTG-3′), 5.8S rRNA (5′-TGATCCACCGCTAAGAGT-3′); U6 snRNA (5′-ATATGGAACGCTTCACGAATT-3′).
For Ago2-association studies, immunoprecipitated RNA and 5 µg total RNA isolated from HEK293 cells using Trifast (Peqlab) was separated by 12% denaturing RNA PAGE and transferred to a nylon membrane (GE Healthcare) by semidry electroblotting. Membranes were cross-linked by 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) chemical crosslink incubating for 1 h at 50°C and hybridized overnight at 50°C with the following probes 5′-TGGTGCCGTGACTCGGA-3′ (tRNA HisGTG), 5′-ATGGTGTCAGGAGTGGGA-3′ (tRNA LeuCAG) and 5′-TCACAAGTTAGGGTCTCAGGGA complementary to miR-125b.
In vitro cleavage assay
Recombinant human ANG (Sigma-Aldrich) was dissolved and diluted to a final concentration of 10 µg/ml in 30 mM HEPES, 30 mM NaCl and 0.01% BSA. The working buffer in absence of ANG was used as the control buffer. RNase A was purchased from Qiagen (Valencia, CA, USA). RNase I and RNase T1 were purchased from Ambion (Carlsbad, CA, USA). Incubations of total RNA were carried out at 37°C for 0, 1, 2 and 4 h. Incubations of in vitro transcribed RNA were carried out at 37°C for 0.5 h. The incubation was stopped by adding Trizol (Invitrogen) into the reaction mixture from which RNA was extracted. To help with RNA precipitation, 20 µg glycogen (Life Technologies, Carlsbad, CA, USA) was added to each sample.
Ago immunoprecipitation and cleavage assay
The construction of human FLAG/HA-Ago1 and FLAG/HA-Ago2, was reported earlier (23). For RNA cleavage assay, a synthetic transcript was generated based PCR amplification and in vitro transcription (T7 RNA polymerase, Fermentas). Primers used were: 5′-TAATACGACTCACTATAGAACAATTGCTTTTACAG-3′ (T7 primer), 5′-ATTTAGGTGACACTA TAGGCATAAAGAATTGAAGA-3′ (SP6 primer). Target sequences were 5′-GAACAATTGCTTTTACAGATGCACATATCGAGGTGAACATCACGTACGTGGTGCCGTGACTCGGATCGGTTGGCAGAAGCTAT-3′ (tRNA HisGTG) and 5′-GAACAATTGCTTTTACAGA TGCACATATCGAGGTGAACATCACGTACGATGGTGTCAGGAGTGGGATCGGTTGGCAGAAGCTAT-3′ (tRNA LeuCAG) and 5′-GGCATAAAGAATTGAAGAGAGTTTTCACTGCA TACGACGATTCTGTGATTTGTATTCAGCCCATATCGTTTCATAGCTTCTGCCAACCGA for the sequence complementary to the oligonucleotide with the specific cleavage sequence. PCR products were purified on denaturing RNA–PAGE followed by ethanol precipitation. For RISC activity assays, substrates were 32P-cap labeled as follows: 1.5 µl (α-32P)-GTP (3000 Ci/mmol), 2 µl 10× buffer (0.4 M Tris pH 8.0, 60 mM MgCl2, 100 mM DTT, 20 mM spermidine), 0.25 µl RNasin (Promega), 1 µl S-adenosyl-Met (500 µM), 1 µl DTT (1 M), 1 µl Guanyltransferase and RNA from in vitro transcription reaction was incubated for 3 h at 37°C. RNA was purified by denaturing RNA–PAGE and recovered by ethanol precipitation. Ago complex-containing anti-FLAG beads were incubated in a reaction containing 5 nM target RNA, 1 mM ATP, 0.2 mM GTP, 10 U/ml RNasin (Promega), 100 mM KCl, 1.5 mM MgCl2 and 0.5 mM DTT for 1.5 h at 30°C. The reaction was stopped by proteinase K treatment and RNA was isolated by phenol/chloroform extraction and ethanol precipitation. Cleavage products were analysed by denaturing RNA–PAGE followed by autoradiography using BioMax MS film (Kodak) and an intensifying screen (Kodak).
Production of wild-type and mutated tRNA molecules
To ensure minimum structural disruptions in mutated tRNA (LysTTT) sequences, the RNAs were folded by Vienna RNA Package (24), and then manually inspected using VARNA (25). Wild-type and mutated tRNA sequences were constructed using mirVana miRNA Probe Construction kit (Ambion, Austin, TX, USA). Briefly, wild-type or mutated tRNA DNA oligos were synthesized (IDT, IA, USA) and annealed to a shorter DNA oligo harboring T7 promoter sequence. The partial double-stranded DNA was filled in by Klenow polymerase and then subjected to in vitro transcription by T7 RNA polymerase. Synthesized RNA was purified by Trizol (Invitrogen), before performing in vitro cleavage assays.
RESULTS
Constitutive ncRNAs but not mRNAs generate termini-specific RNAs
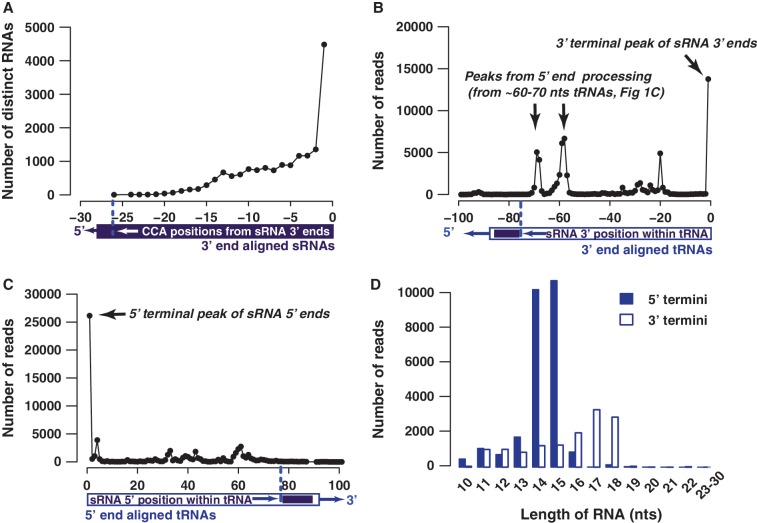
We previously reported on ∼15 nt long unusually small RNAs (usRNAs) and their sub-groups (2), consisting of 3′ end-specific motifs in human and viral genomes. Two of these usRNA subgroups share a CCA element in their 3′ end. To further study RNAs with the terminal CCA motif, we reanalyzed 307 269 RNA sequence reads (10−30 nt) that were identified in the human KSHV-infected primary-effusion lymphoma cell line, BCP1 (26). The CCA motif occurs precisely at the 3′ end in 24 601 reads (4482 distinct sequences, Figure 1A). Terminal, 3′-specific CCA motifs are known to occur in almost all mature tRNAs (∼70−100 nt), which act as sites for amino acid residue attachment and as stabilizers for tRNA-ribosome interactions (27). Since the majority (85.6%) of reads containing terminal CCA motifs consisted of usRNAs or even smaller RNAs that have not been the focus of previous studies (6,7,10–12,16,18,19), we sought to fully characterize the spectra of ∼10−30 base long terminal CCA-containing RNAs. To reduce artifacts from short length reads, the analysis was limited to those reads that perfectly matched tRNA ends. More than half (13 579 of 24 601 reads) of the terminal CCA-containing small RNAs matched tRNAs. We next tested whether tRNAs produce other 10−30 nt long RNAs at other positions in tRNAs in an unbiased manner as expected for a degradation process involving random exo/endo-nuclease cleavage. The majority of tRFs precisely match 5′ or 3′ ends of tRNAs (Figure 1B and C) and these 5′–3′ terminal tRFs matches to 76% (447/522) of human tRNAs. We find a significantly higher proportion of tRFs reads (76%) than previously reported (7) for tRFs (∼55%), likely because gel-based isolation of RNAs >10 nt seem to be necessary to retain the full spectrum of usRNAs (2). Interestingly, 5′ and 3′ tRFs display different size distributions (Figure 1D). The size range of 5′ terminal tRFs peaks at 14 and 15 nt, which corresponds to a smaller size estimate than the previous report of 18−19 nt (7); this difference may be due to our inclusion of a broader range (10−30 nt) of RNA fragments, or is a result of differences between the cell lines in the two studies.
Figure 1.
Characteristics of small RNAs (sRNAs) matching to 5′ and 3′ termini of tRNAs. All positions are determined with respect to either the 5′ or 3′ ends of sRNAs/tRNAs. To determine the positions, the sRNAs (violet) and tRNAs (blue) are aligned by their 5′–3′ ends, as noted by respective illustrations. (A) The CCA motif preferentially occurs at the 3′ termini (position −1) of the sRNAs. To avoid artifacts from single sequences with high number of reads, only distinct sequences that contain CCA were used for the analysis. (B) The visualization of positions of all reads that map to tRNAs indicates that 3′ terminal of tRNAs produce the most number of fragments (3′ terminal peak). The two prominent peaks at ∼−60 and ∼−70 nt are due to 5′ specific processing of tRNAs that are ∼60 and ∼70 nt long, as illustrated in panel C. (C) Similar to tRNA 3′ specific reads, 5′ terminal of tRNAs are also specifically processed to yield RNA fragments (5′ terminal peak). The tRNA position (with respect to its 5′ end) at which the 5′ end of the sRNA reads are matched are used, as depicted by the bottom illustration. (D) Length distribution of 5′ (black) and 3′ (white) terminal RNAs from tRNAs indicate that 5′ terminal RNAs are predominantly 14−15 nt long, while 3′ terminal RNAs seem to manifest a broader range (∼16−18 nt).
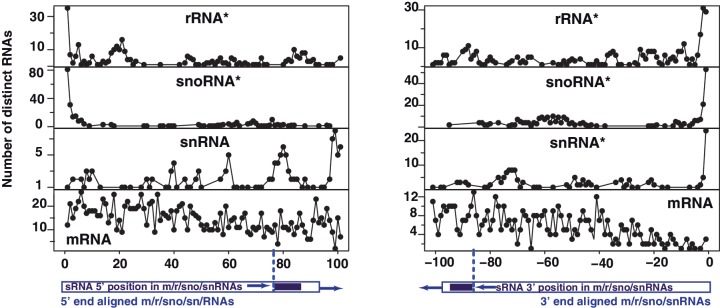
We next examined if other classes of RNAs also generate termini-specific small RNAs. The phenomenon of 5′–3′-specific processing is observed across all major classes of ncRNAs but not mRNAs (Figure 2). Terminal tRFs are the most abundant fragments found in deep sequencing, followed by rRNA-derived RNAs (Supplementary Figure S1; Figure 1B and C). All ncRNAs generate both 5′ and 3′ products except snRNAs, which produce small RNAs exclusively from their 3′ termini. We note that 5′ modifications on mRNAs and snRNAs (28) hinder the cloning of 5′ fragments, which might be the most plausible reason for the absence of 5′ snRNA-derived reads in deep sequencing data. However, despite the large number of mRNAs, very few mRNA-derived fragments are detected at 5′–3′ ends and the proportion of mRNA-derived fragments across all mRNAs is negligible (Figure 2) suggesting that the processing of mRNAs into stable fragments is not a common phenomenon. In comparison to tRNA-derived tRFs with a read number density of ∼1.0 read per base, mRNAs yield a noticeably low read density of 0.0005. Furthermore, there were no reads with purely poly(A) tails of at least 15 nt long sequences, suggesting that end processing of mRNAs is a rare event. Although termini-specific stable fragments of mRNAs are not detected in our analysis, it is important to note that very low abundance (1 copy/10 cells) short RNAs also occur near transcription start sites of mRNAs (29,30). These transcription start site-associated (29) RNAs (TSSa-RNAs) or transcription initiation (30) RNAs (tiRNAs) are distinct from termini-specific RNA fragments, because they map to a wide range of up/down-stream regions near transcription start sites. Thus, small RNAs from ncRNA termini are likely to be produced by a different mechanism than that used for TSSa-RNA and tiRNA generation, as well as mRNA degradation.
Figure 2.
Small terminal RNA fragments are produced preferentially from 5′ and 3′ termini of rRNAs, snoRNAs, snRNA, but not mRNAs. Reads (violet) that map to rRNA, snoRNAs, snRNAs and mRNAs are depicted as in Figure 1; positions are with respect to either the 5′ end (left panel) or the 3′ end (right panel) of the RNAs (blue) as illustrated. Subpanels with the characteristic 5′ or 3′ preferences are highlighted (asterisk) for clarity. For instance, snRNAs yield terminal RNAs exclusively from their 3′ end (right panel).
Asymmetric processing of 5′–3′ terminal fragments
During miRNA biogenesis, each miRNA precursor is processed to generate a small (∼22 nt) duplex, from which one of the strands (miRNA) is preferentially incorporated into Argonaute (AGO) and thus stabilized, while the other strand, termed the passenger strand is often degraded (31,32). Since the 5′ and 3′ ends of both tRNAs (33) and many snoRNAs (28) form RNA duplexes, we speculated that the terminal RNAs may be generated asymmetrically from the 5′ and 3′ ends of the duplex. To explore this possibility, for each tRNA, we compared the number of 5′ terminal RNAs to that of 3′ terminal RNAs. Among the 447 tRNA genes that match terminal tRFs, 335 (75%) tRNAs match both 5′ and 3′ terminal tRFs. Remarkably, 93% of these (313/335) tRNAs manifest considerable (2×) difference between their 5′ and 3′ tRF levels, while nearly half of them (161) yield a difference of 10×. To ensure that these biases are not due to tRNAs that are similar in their sequences, we reanalyzed our data using a non-redundant set of 99 tRNAs that shared <90% sequence identity to reconfirm the observed biases (Figure 3A). To further confirm that the biases are not limited to our BCP1 library and to rule out protocol-based artifacts, we also analyzed the results from an independent library from HEK293 cells (34). We found similar patterns that were additionally confirmed using northern blots for four tRFs derived from two different tRNAs (Supplementary Figure S2). These results support the notion that tRNAs are generally processed into two terminal tRFs but only one fragment is preferentially maintained in cells, reminiscent of miRNA maturation. The bias is stronger for snoRNAs and snRNAs; the 95% (66/69) snoRNAs generate terminal RNAs primarily from either the 5′ or 3′ ends (Figure 3B), while all terminal snRNA fragments are exclusively derived from 3′ ends (Figure 2). Terminal RNAs of rRNAs also seem to manifest a general preference toward 3′ terminal (Figure 3C). Since 5′ and 3′ ends of rRNAs likely do not form intra-molecular base pairs to form a double-stranded structure within the molecule (35), the asymmetry in ncRNA terminal RNAs could be due to stabilization of the 5′ or 3′ fragment that is cleaved off by the processing machinery.
We next sought to find out whether the terminal RNAs and their asymmetric processing bias are characteristics that are evolutionarily conserved. Analysis of small RNA libraries (36) from mouse embryonic stem cells (mESCs) revealed that mESCs contain very abundant terminal RNAs derived precisely from the 5′ and 3′ ends of tRNAs (Figure 4A) that also manifest asymmetry in processing (Figure 4B). Similar to human terminal tRFs, termini-specific processing (Supplementary Figure S3) and asymmetric stabilization (Supplementary Figure S4) are also observed for mouse rRNAs, snoRNAs and snRNAs. Furthermore, comparison of the asymmetric processing bias of individual human tRNAs and their putative mouse orthologs reveal a modest conservation in their processing bias in generating terminal tRFs (Supplementary Figure S5). Intriguingly, the processing biases of 20−30 nt long tRFs are more correlated (r = 0.67) between human and mice than that of the 15 - to 19-nt-long tRFs. Taken together, these data suggest that the mechanism to generate terminal RNAs from various ncRNAs is a conserved process between human and mice.
Terminal small RNAs are primarily independent of Dicer and DGCR8
Since a few Dicer-dependent RNA products from tRNAs (8) and snoRNAs (3) are known, we investigated whether terminal RNAs are commonly processed through the canonical miRNA pathway. Unlike other studies (4,36) that normalized the data sets using RNAs derived from tRNAs and snoRNAs, we used mRNA-derived fragments for normalization. We used a normalization strategy (see ‘Materials and Methods’ section) based on mRNA fragments because of our finding that mRNAs but not tRNAs and snoRNAs, produce fragments that largely resemble random degradation. Since the dependence of shorter tRFs (∼15 nt) on Dicer/DGCR8 has not been investigated yet, we separately analyzed the terminal RNAs of the two different size groups (20−30 and 15−19 nt). As a class, each terminal small RNA group manifest statistically insignificant (P > 0.05) changes after either dicer 1 or dgcr 8 knockouts (Supplementary Figure S6A). However, it is important to note that specific RNAs within tRNAs, snoRNAs and snRNAs do manifest considerable (>2-fold) difference in both Dicer and DGCR8 knockdowns (Supplementary Figure S6B). We next tested whether the observed asymmetric processing of 5′ and 3′ tRFs is maintained during Dicer/DGCR8 deletions. To compare asymmetric processing across different samples, the processing bias was evaluated in each library and for the 20−30 and 15−19 nt size groups (Figure 4C). Consistent with the notion that these RNAs are largely independent of the RNAi pathway, the asymmetry profiles within each group are constant across wild-type and dicer 1/dgcr8 knockout mES cells, yielding Pearson correlation coefficients (r) in the range of 0.89–0.97.
Presence of endogenous Ago2-associated 3′ tRFs that are capable of cleaving target RNAs
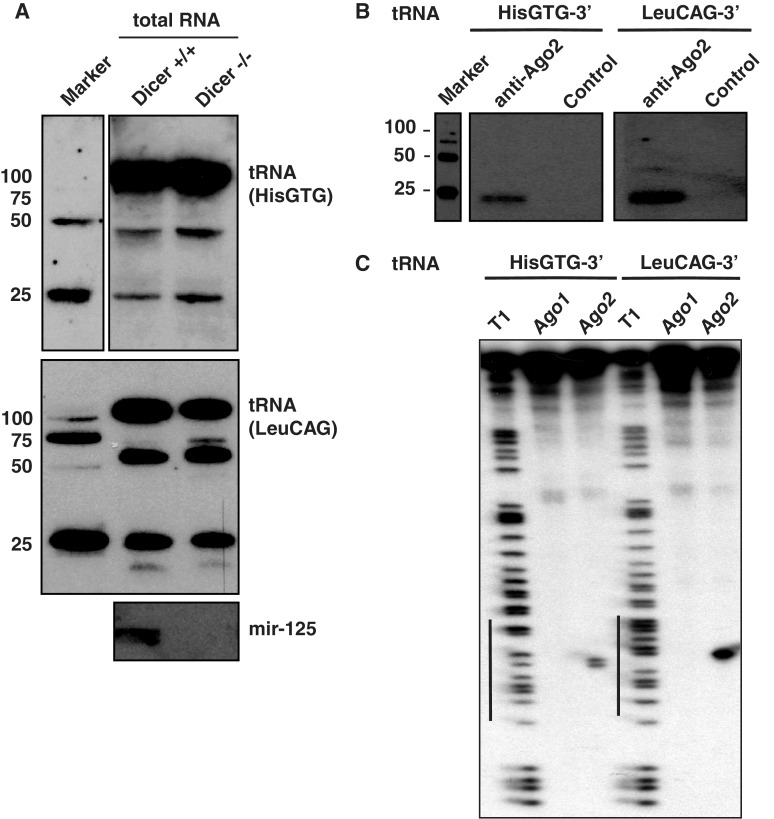
To further investigate processing and function of tRF, we selected two sequences (HisGTG, LeuCAG) for further in vitro studies (Figure 5). We first analyzed possible Dicer requirements for their biogenesis (Figure 5A). Total RNA from wild-type (Dicer+/+) and Dicer knock out (Dicer−/−) mouse embryonic fibroblasts (MEFs) was used for Northern blotting using probes against the HisGTG (Figure 5A, upper panel) and the LeuCAG fragment (lower panel). Indeed, small RNA production from these tRNAs is readily detectable in Dicer-deficient cells, indicating that these tRFs are independent of Dicer processing. To probe into the possibility that some terminal RNAs might use the RNAi pathway to inhibit target transcripts, we further examined whether tRFs associate with Ago2. Endogenous Ago2 was immunoprecipitated using anti-Ago2 antibodies and co-immunoprecipitated RNA was analyzed by Northern blotting using probes specific to HisGTG and LeuCAG (Figure 5B). Both tRFs were detected in anti-Ago2 immunoprecipitates demonstrating that tRFs can be specifically loaded into Ago protein complexes. Although previous studies have indicated that Ago2 co-immunoprecipitates with tRF-like fragments (6,8), it has not been shown yet whether tRFs can cleave target RNAs. Indeed, based on the selection of high abundance of reads in Ago2-associated data, northern blotting assays confirmed that 3′ tRFs from two tRNAs are associated with endogenous Ago2 (Figure 5B). Analysis using Flag-HA-Ago2 complexes also reconfirmed the association of these 3′ tRFs with Ago2 (Figure 5C). Since Ago2, but not Ago1 cleaves target mRNA (37,38), we immunoprecipitated Flag-HA-Ago1 and Flag-HA-Ago2 complexes from transfected HEK293 cells using anti-Flag antibodies (Figure 5C) and incubated them with a 32P-cap labeled artificial target mRNA (∼100 nt) containing a region (17–18 nt) fully complementary to the endogenous 3′ tRFs from the two different tRNAs (LeuCAG and HisGTG). Both endogenous 3′ tRFs directed Ago2-mediated cleavage. In contrast, although both 3′ tRFs are found in the Ago1 sequencing library (3), the catalytically inactive Ago1 (38), does not co-purify with any cleavage products. These results suggest that 3′ tRFs from tRNAs can function in an RNAi-like pathway. Taken together with previous reports using luciferase assays (6), our data support the notion that tRFs have the potential to knock down target genes both at RNA and protein levels.
Figure 5.
Presence of Ago2-associated 3′ tRFs that can cleave target RNA. (A) Total RNA from Dicer knockout mouse cells (Dicer−/−) or wild-type cells (Dicer+/+) were analyzed by northern blotting using probes specific for the 3′ of tRNAs (HisGTG and LeuCAG), and miR-125 b. (B) Endogenous Ago2 was immunoprecipitated from human HEK293 lysates, and the co-immunoprecipitated RNAs were extracted and analyzed by northern blots using probes against the 3′ tRFs (HisGTG and LeuCAG). GST antibody is used as control (C) Immunoprecipitated Flag-HA-Ago1 and Flag-HA-Ago2 were incubated with a 32P-cap-labeled target RNA (∼100 nt long), which contained a perfect complementary sequence to the 3′ termini of tRNAs (HisGTG and LeuCAG). Lanes indicated with T1 represent RNaseT1 digestions of the RNA substrates as ladders. The RNA sequence complementary to tRNA HIS-3′ and tRNA LEU-3′ is indicated by a black bar on the left.
ANG and other RNases can cleave tRNAs to produce terminal tRFs
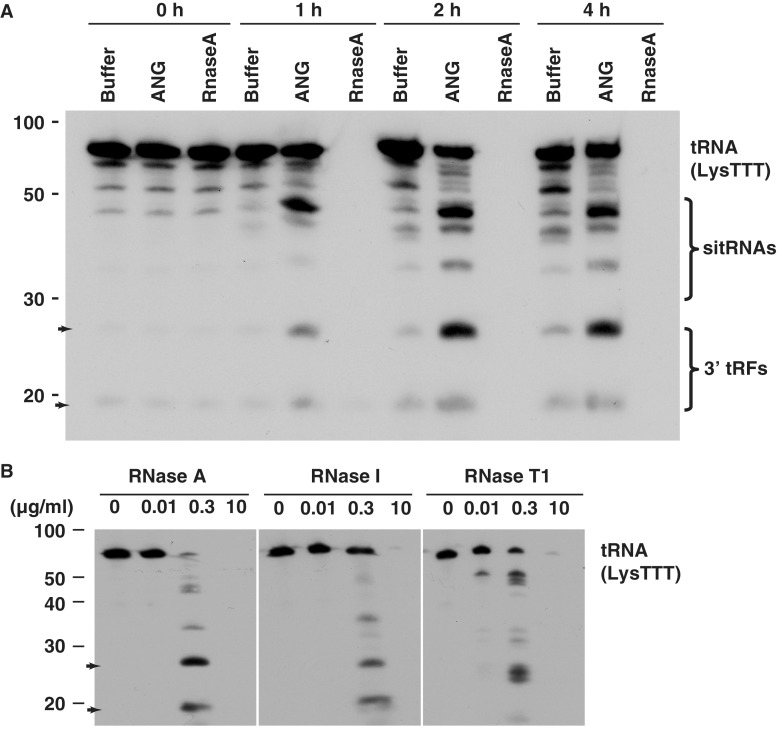
Since tRFs correspond to the most abundant and diverse group of terminal RNAs and are independent of the canonical miRNA processors, we sought to understand their biogenesis. To test potential candidate proteins involved in the production of terminal tRFs, we focused on the ANG nuclease (39), which generates 30−50 nt long sitRNAs from anti-codon loops during stress (12,18) and was reported as a tRNA-specific enzyme (39,40). We treated total RNA from 293 cells in vitro with recombinant ANG and monitored the production of terminal RNA fragments for tRNAs, rRNAs and snRNAs. At the initial time point (0 h), endogenous small RNA species are present in the absence of ANG treatment (Figure 6A and Supplementary Figure S7). Expression of these endogenous RNA species is enhanced after ANG treatment in a time-dependent manner, suggesting ANG is involved in generating a variety of tRNA fragments, the sizes of which correspond to previously reported 30−50 nt sitRNAs (12), as well as the ∼20 nt 5′ (Supplementary Figure S7) and 3′ (Figure 6A) terminal RNAs reported in our study. Processing of rRNA and snRNA, by ANG seems to reflect random hydrolysis, resulting in a series of RNA fragments (Supplementary Figure S8). Since ANG belongs to the super family of RNase A (41), we examined if RNase A produces terminal tRFs at low concentrations, and tested the cleavage pattern of additional RNases. At a much (∼30×) lower concentration (0.3 µg/ml) of RNase A than that was used for ANG, RNase was able to generate both the ∼20 and ∼25 nt tRF bands specifically after 1-h digestion (Figure 6B). Intriguingly, RNase I, a bacterial RNase that can non-specifically digest all di-nucleotide bonds (42), also generated similar tRFs at low concentrations. In contrast, RNase T1, which cleaves only after unpaired G residues (43), generates fragments that are different than ANG, and RNase A/I (Figure 6B). These observations indicate that multiple RNases expressed in a given cell-type are likely responsible for the diverse range of abundant terminal tRFs with varying lengths.
Figure 6.
Multiple RNases generate terminal tRFs from total RNA. (A) ANG (10 µg/ml) generates ∼20 nt long-terminal tRF as well as longer sitRNAs from total RNA extracts. In comparison an equal concentration of RNase A completely degrades the RNA within an hour, while a control lane with solution buffer (Buffer) generate terminal tRFs at much lower levels. (B) In vitro cleavage assay (t = 1 h) using total RNA from HEK293 cells (10 µg/lane) and different RNases at a series of concentrations indicate that at low concentrations, both RNase A and RNase I are able to produce similar terminal tRFs as that of ANG (arrows). In contrast, RNase T1 generates a different set of 3′ tRFs.
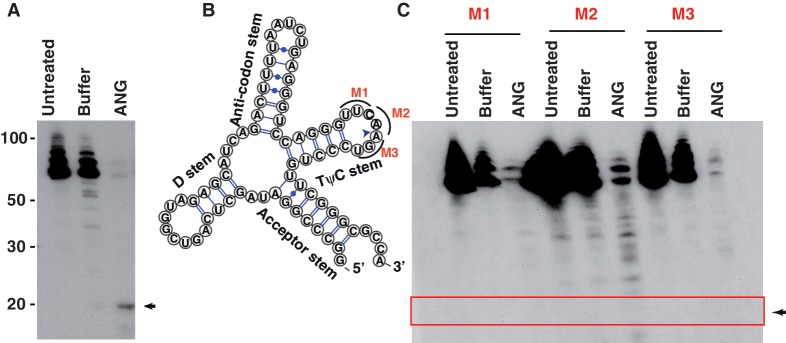
To further analyze the roles of different tRNA regions on tRF production, we designed a series of mutations to disrupt specific functional sub-structures of the tRNA and monitored the tRNA cleavage patterns generated by ANG (Figure 7 and Supplementary Figure S9). The mutations were designed by an in silico screening of base substitutions to yield candidates that preserve secondary structures outside the mutated domains. ANG cleavage of wild-type tRNAs produced ∼20 nt long 3′ terminal tRFs (Figure 7A). Mutations that individually disrupt the acceptor stem, the D stem, the anti-codon stem and the TψC stem were first examined (Supplementary Figure S9A−S9D). None of these mutations interfered with the production of the terminal tRNA fragments by ANG after in vitro cleavage of 0.5 h. Another mutation that completely altered the tRNA clover-leaf structure also did not visibly affect tRF production (Supplementary Figure S9E). However, three different mutants, each involving point mutations around the ANG cleavage site on the TψC loop, resulted in complete disappearance of the terminal RNA bands (Figure 7C). Since the in vitro transcribed tRNAs do not have modified nucleosides, these results also indicate that post-transcriptional modifications of tRNA are not a requirement for terminal RNA processing. In summary, the major determinant in processing of terminal small tRNAs by ANG and possibly related RNases is the TψC loop within tRNA.
Figure 7.
ANG processes tRFs within the TψC loop. (A) ANG can process in vitro transcribed, synthetic tRNA (untreated/buffer/ANG incubations at 0.5 h) to generate terminal tRFs. (B) Secondary structure of the in vitro-transcribed tRNA, highlighting the three mutations in TψC loop (M1−M3) and the tRF cleavage site (arrow). (C) ANG processing of the tRF is impaired by tRNA mutations in TψC loop (M1−M3).
Endogenous retroviral replication loci are preferentially antisense to 3′ terminal tRFs
The binding of tRNAs to retroviral primer binding sites (PBS) facilitates the initiation of retroviral genome replication (44,45), such as that for the human immunodeficiency virus (HIV). Since endogenous human retroviral sequences comprise ∼7% of the human genome (46), it is possible that cells have evolved mechanisms to regulate replication of these retroviral elements, particularly for preventing accidental transcription of deleterious retroviral sequences. We therefore investigated whether the terminal tRFs have the potential to bind to human endogenous retroviruses (HERVs). The majority of HERVs exist in the human genome as long terminal repeat (LTR) retrotransposons (47) which, together with non-viral retrotransposons LINEs (long interspersed nucleotide elements) and SINEs (small interspersed nucleotide elements), form roughly 40% of the mammalian genome (48). Terminal 3′ tRFs are remarkably more complementary to HERV LTRs than to LINEs and SINEs (Figure 8A). Furthermore, even though the number of distinct 3′ and 5′ BCP1 tRFs differ only by a factor of two (1002 versus 507), 3′ tRF sequences are 26-fold (2614 versus 100) more frequently complementary to retrotransposon elements than the 5′ tRFs; corresponding to a normalized enrichment of 13-fold. Similarly, 3′ tRFs are 4- to 6-fold more enriched (normalized) to complementary sites in LINES and SINEs than 5′ tRFs. Analysis of the major LTR elements that are complementary to BCP1 tRFs (Figure 8B), indicate that the top candidate tRF (LysTTT tRNA, 947 reads) is nearly identical to a known antisense DNA oligonucleotide that targets the PBS region of HIV and can inhibit HIV replication (49). Moreover, the two 3′ tRFs (LeuCAG and HisGTG tRNAs) that we found to associate with Ago2 are also capable of degrading their target RNAs, and are complementary to the ERVL-E and HERVH LTR elements (Figure 5C).
DISCUSSION
We present here an in-depth study of terminal-specific small RNAs from rRNAs, snRNAs, snoRNAs, tRNAs and mRNAs that are generally missed in deep sequencing studies because they are commonly filtered out as potential degradation products during bioinformatics analysis. While such strict filtering greatly helps in focusing on the most abundant and likely functional small RNA molecules, emerging studies continue to provide evidence that degradation-like products can have functional impact and are abundant. Most importantly, whether these small RNA products are functional or cellular noise, their study continue to open up novel cellular mechanisms and interesting cellular attributes (1–4,6–13,16,18,19,30,50–53).
Our own detailed analyses of small RNAs derived from various classes of RNAs reveal novel, biologically important features for these small RNAs and support the notion that terminal RNAs are not due to artifacts of cloning methods used in deep sequencing methods. For clarity, we note that the tRFs in this study are derived from mature RNAs and do not include tRFs that could be also processed from mature RNA precursors such as that of pre-tRNAs (6,7). Comparisons across all novel and other previously reported classes of tRNA/snoRNA-derived RNAs, (3,4,6–8), indicate that the production of terminal- and 5′–3′ end specific small RNAs, are common features of constitutively expressed ncRNAs. The distinct size distribution of terminal RNAs, as shown by sequencing and northern blots, seem atypical for RNA turnover products generated by common nuclear proofreading pathways, where mutant or defective RNAs are degraded by nuclear exosomes (50,54), or by 5′–3′ exonucleases Rat1 and Xrn1 (52). The extensive identification of both 5′ and 3′ terminal tRFs with similar size distributions in independent deep-sequencing studies that include Ago bound small RNAs, suggests that tRFs exists due to a yet unknown terminal-specific degradation mechanism or by a processing mechanism that yields functional RNA fragments. The phenomena of terminal RNAs is also present in mice indicative of a conserved terminal RNA processing/degradation mechanism. While our in vitro cleavage assay results suggest that various RNase families including ANG are likely the factors that drive tRF production, the association of these RNAs to AGO as well as the dearth of mRNA-derived tRFs further seem to support the notion that these RNAs could have a biological function, perhaps in a coordinated manner (55).
Why do ncRNAs but not mRNAs manifest terminal-specific processing of small RNAs? Since mRNAs are comprised of many constitutively expressed RNAs, terminal RNA processing is unlikely to be a simple phenomenon for all constitutively expressed RNAs, and provides additional support to the notion that the phenomenon is independent of a random degradation process. We also did not detect a preference for terminal small RNAs among other well-known ncRNAs such as Y-RNAs (56). One possibility is that terminal RNA processing might be a hallmark of RNAs involved in fundamental and ancient core processes such as translation. Consistent with the notion of terminal RNAs as a process that evolved early, we postulate that 3′ tRFs may have evolved as RNAi-based modulators that block the replication of endogenous retroviral sequences (Figure 8C). Indeed, the RNAi pathway evolved as an immune defense mechanism against viruses in basal organisms that do not have protein-based adaptive immune systems (57). While these concepts support the view that terminal RNAs might be limited to core ncRNAs as a pervasive phenomenon, we cannot yet rule out that other classes of RNAs under various environmental conditions (e.g. stress) may also preferentially generate such terminal small RNAs.
Despite the prominent abundance of tRFs, their precise functions remain unknown, yet several observations suggest that the terminal tRFs may have a functional role in the cell. The depletion of a tRF derived from a pre-tRNA is known to increase cell proliferation and elevate the number of cells in G2 phase of the cell cycle (7). Similarly, the longer class of sitRNAs that are generated by ANG can inhibit protein translation (12) to counteract stress, and terminal tRFs likely have a similar function. Indeed, the general role of small RNAs in stress response programs is an emerging theme (58). Clearly, these observations warrant mechanistic studies of the underlying pathways. Our own observations that link 3′ tRFs to HERVs provide a testable hypothesis for a specific cellular role for 3′ terminal tRFs. It is conceivable that in normal cells 3′ tRFs may be able to bind to the PBS region of human endogenous retroviruses, blocking their replication though endonuclease (e.g. AGO2) cleavage of the target transcript. Indeed, we found that 3′ tRFs are endogenously associated with Ago2 and are able to guide Ago2 to cleave the target RNA. Detailed experimental testing of this hypothesis could take several years of research not only because of the complexity of the regulation but also the difficulty in working with endogenous viral products. It also remains difficult to study these RNAs because common technologies such as RT-PCR are either not reliable or laborious (e.g. northern blot) for detecting these RNAs. Our own studies would have been considerably more difficult than if we did not use recently developed northern blot protocols (22) to routinely assay for the expression of these small RNAs. It is likely that emerging third generation sequencing technologies and other microfluidic technologies, combined with multiplexing, will provide methods for cheaper and more robust detection of these difficult-to-study small RNAs that are typically smaller than even miRNAs (59).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–9.
FUNDING
American Cancer Society Research Professorships [RP0909401 to Y.C., RP0909601 to P.S.M.]; American Cancer Society [RSG0905401 to B.J.]; Bavarian Genome Research Network [BayGene to G.M.]; Deutsche Forschungsgemeinschaft [SFB 960 and FOR855 to G.M.]; European Research Council [sRNAs to G.M.]; National Institutes of Health (NIH) [GM079756 to B.J., CA136363 and CA136806 to Y.C./P.S.M.]. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the laboratories of Y.C. and P.S.M., B.J. for helpful discussions and Mr Boles for computer systems administration. C.E thanks Marie Curie Actions for the Intra-European Fellowship.
REFERENCES
- 1.Shi W, Hendrix D, Levine M, Haley B. A distinct class of small RNAs arises from pre-miRNA-proximal regions in a simple chordate. Nat. Struct. Mol. Biol. 2009;16:183–189. doi: 10.1038/nsmb.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Z, Kim SW, Lin Y, Moore PS, Chang Y, John B. Characterization of viral and human RNAs smaller than canonical MicroRNAs. J. Virol. 2009;83:12751–12758. doi: 10.1128/JVI.01325-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ender C, Krek A, Friedländer MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 4.Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brameier M, Herwig A, Reinhardt R, Walter L, Gruber J. Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res. 2011;39:675–686. doi: 10.1093/nar/gkq776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ, Hutvagner G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee SR, Collins K. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J. Biol. Chem. 2005;280:42744–42749. doi: 10.1074/jbc.M510356200. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Luo J, Zhou H, Liao JY, Ma LM, Chen YQ, Qu LH. Stress-induced tRNA-derived RNAs: a novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res. 2008;36:6048–6055. doi: 10.1093/nar/gkn596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smalheiser NR, Lugli G, Thimmapuram J, Cook EH, Larson J. Endogenous siRNAs and noncoding RNA-derived small RNAs are expressed in adult mouse hippocampus and are up-regulated in olfactory discrimination training. RNA. 2011;17:166–181. doi: 10.1261/rna.2123811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hess AM, Prasad AN, Ptitsyn A, Ebel GD, Olson KE, Barbacioru C, Monighetti C, Campbell CL. Small RNA profiling of Dengue virus-mosquito interactions implicates the PIWI RNA pathway in anti-viral defense. BMC Microbiol. 2011;11:45. doi: 10.1186/1471-2180-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jöchl C, Rederstorff M, Hertel J, Stadler PF, Hofacker IL, Schrettl M, Haas H, Huttenhofer A. Small ncRNA transcriptome analysis from Aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis. Nucleic Acids Res. 2008;36:2677–2689. doi: 10.1093/nar/gkn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson DM, Parker R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J. Cell Biol. 2009;185:43–50. doi: 10.1083/jcb.200811119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583:437–442. doi: 10.1016/j.febslet.2008.12.043. [DOI] [PubMed] [Google Scholar]
- 19.Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, Hu GF, Anderson P. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem. 2010;285:10959–10968. doi: 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic. Acids Res. 2009;37:D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karolchik D, Hinrichs AS, Kent WJ. The UCSC Genome Browser. Curr. Protoc. Bioinformatics. 2009 doi: 10.1002/0471250953.bi0104s40. Chapter 1, Unit1 4. [DOI] [PubMed] [Google Scholar]
- 22.Kim SW, Li Z, Moore PS, Monaghan AP, Chang Y, Nichols M, John B. A sensitive non-radioactive northern blot method to detect small RNAs. Nucleic Acids Res. 2010;38:e98. doi: 10.1093/nar/gkp1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Lührmann R, Tuschl T. Identification of novel argonaute-associated proteins. Curr. Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 24.Hofacker IL. RNA secondary structure analysis using the Vienna RNA package. Curr. Protoc. Bioinformatics. 2009 doi: 10.1002/0471250953.bi1202s26. Chapter 12, Unit12 12. [DOI] [PubMed] [Google Scholar]
- 25.Darty K, Denise A, Ponty Y. VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009;25:1974–1975. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boshoff C, Gao SJ, Healy LE, Matthews S, Thomas AJ, Coignet L, Warnke RA, Strauchen JA, Matutes E, Kamel OW, et al. Establishing a KSHV+ cell line (BCP-1) from peripheral blood and characterizing its growth in Nod/SCID mice. Blood. 1998;91:1671–1679. [PubMed] [Google Scholar]
- 27.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 28.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell. Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 29.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, Lassmann T, Forrest AR, Grimmond SM, Schroder K, et al. Tiny RNAs associated with transcription start sites in animals. Nat. Genet. 2009;41:572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 31.Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 32.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 33.Wolin SL, Matera AG. The trials and travels of tRNA. Genes Dev. 1999;13:1–10. doi: 10.1101/gad.13.1.1. [DOI] [PubMed] [Google Scholar]
- 34.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc. Natl Acad. Sci. USA. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 38.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 39.Saxena SK, Rybak SM, Davey RT, Jr, Youle RJ, Ackerman EJ. Angiogenin is a cytotoxic, tRNA-specific ribonuclease in the RNase A superfamily. J. Biol. Chem. 1992;267:21982–21986. [PubMed] [Google Scholar]
- 40.Rybak SM, Vallee BL. Base cleavage specificity of angiogenin with Saccharomyces cerevisiae and Escherichia coli 5S RNAs. Biochemistry. 1988;27:2288–2294. doi: 10.1021/bi00407a007. [DOI] [PubMed] [Google Scholar]
- 41.Leonidas DD, Shapiro R, Allen SC, Subbarao GV, Veluraja K, Acharya KR. Refined crystal structures of native human angiogenin and two active site variants: implications for the unique functional properties of an enzyme involved in neovascularisation during tumour growth. J. Mol. Biol. 1999;285:1209–1233. doi: 10.1006/jmbi.1998.2378. [DOI] [PubMed] [Google Scholar]
- 42.Zhu L, Deutscher MP. The Escherichia coli rna gene encoding RNase I: sequence and unusual promoter structure. Gene. 1992;119:101–106. doi: 10.1016/0378-1119(92)90072-w. [DOI] [PubMed] [Google Scholar]
- 43.Pfeiffer S, Karimi-Nejad Y, Ruterjans H. Limits of NMR structure determination using variable target function calculations: ribonuclease T1, a case study. J. Mol. Biol. 1997;266:400–423. doi: 10.1006/jmbi.1996.0784. [DOI] [PubMed] [Google Scholar]
- 44.Das AT, Klaver B, Berkhout B. Reduced replication of human immunodeficiency virus type 1 mutants that use reverse transcription primers other than the natural tRNA(3Lys) J. Virol. 1995;69:3090–3097. doi: 10.1128/jvi.69.5.3090-3097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Das AT, Berkhout B. Efficient extension of a misaligned tRNA-primer during replication of the HIV-1 retrovirus. Nucleic Acids Res. 1995;23:1319–1326. doi: 10.1093/nar/23.8.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bock M, Stoye JP. Endogenous retroviruses and the human germline. Curr. Opin. Genet Dev. 2000;10:651–655. doi: 10.1016/s0959-437x(00)00138-6. [DOI] [PubMed] [Google Scholar]
- 47.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 48.Goodier JL, Kazazian HH., Jr Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 49.Goodchild J, Agrawal S, Civeira MP, Sarin PS, Sun D, Zamecnik PC. Inhibition of human immunodeficiency virus replication by antisense oligodeoxynucleotides. Proc. Natl Acad. Sci. USA. 1988;85:5507–5511. doi: 10.1073/pnas.85.15.5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suhasini AN, Sirdeshmukh R. Transfer RNA cleavages by onconase reveal unusual cleavage sites. J. Biol. Chem. 2006;281:12201–12209. doi: 10.1074/jbc.M504488200. [DOI] [PubMed] [Google Scholar]
- 52.Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ, Phizicky EM. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5′-3′ exonucleases Rat1 and Xrn1. Genes Dev. 2008;22:1369–1380. doi: 10.1101/gad.1654308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee HC, Chang SS, Choudhary S, Aalto AP, Maiti M, Bamford DH, Liu Y. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature. 2009;459:274–277. doi: 10.1038/nature08041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kadaba S, Wang X, Anderson JT. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA. 2006;12:508–521. doi: 10.1261/rna.2305406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cloonan N, Wani S, Xu Q, Gu J, Lea K, Heater S, Barbacioru C, Steptoe AL, Martin HC, Nourbakhsh E, et al. MicroRNAs and their isomiRs function cooperatively to target common biological pathways. Genome Biology. 2011;12:R126. doi: 10.1186/gb-2011-12-12-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Christov CP, Gardiner TJ, Szüts D, Krude T. Functional requirement of noncoding Y RNAs for human chromosomal DNA replication. Mol. Cell Biol. 2006;26:6993–7004. doi: 10.1128/MCB.01060-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Rij RP, Saleh MC, Berry B, Foo C, Houk A, Antoniewski C, Andino R. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006;20:2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hogg JR, Collins K. Structured non-coding RNAs and the RNP Renaissance. Curr. Opin. Chem. Biol. 2008;12:684–689. doi: 10.1016/j.cbpa.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat. Rev. Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.