Abstract
Integrating vectors such as viruses and transposons insert transgenes semi-randomly and can potentially disrupt or deregulate genes. For these techniques to be of therapeutic value, a method for controlling the precise location of insertion is required. The piggyBac (PB) transposase is an efficient gene transfer vector active in a variety of cell types and proven to be amenable to modification. Here we present the design and validation of chimeric PB proteins fused to the Gal4 DNA binding domain with the ability to target transgenes to pre-determined sites. Upstream activating sequence (UAS) Gal4 recognition sites harbored on recipient plasmids were preferentially targeted by the chimeric Gal4–PB transposase in human cells. To analyze the ability of these PB fusion proteins to target chromosomal locations, UAS sites were randomly integrated throughout the genome using the Sleeping Beauty transposon. Both N- and C-terminal Gal4-PB fusion proteins but not native PB were capable of targeting transposition nearby these introduced sites. A genome-wide integration analysis revealed the ability of our fusion constructs to bias 24% of integrations near endogenous Gal4 recognition sequences. This work provides a powerful approach to enhance the properties of the PB system for applications such as genetic engineering and gene therapy.
INTRODUCTION
The ability of integrating vectors to permanently introduce foreign genes into chromosomes has resulted in major advances in the fields of genetic engineering, functional genomics and gene therapy. For these techniques to be of value in the clinical setting it is imperative that insertions occur at known safe loci in order to avoid deregulation of the cell due to deleterious integrations and to control expression of transgenes. Commonly used viral vectors have been shown to preferentially insert their cargo near transcriptional start sites (1–3) and there has been increasing concern for the implications of insertional mutagenesis (4–6). Thus, the safety of insertional therapies would be improved by the ability to target vector integration to a specific genomic safe harbor.
Cys2His2 zinc finger proteins (ZFPs) can bind to specific sequences by inserting an alpha-helix into the major groove of the DNA double helix. These DNA binding domains (DBDs) are specific for 6–18 bp DNA sites and can now be easily designed in a few weeks and be made to target almost any location in the genome (7–11).
By fusing ZFPs to activator or repressor domains, novel zinc finger effectors have been used to up-regulate or down-regulate transcription (12–15). Recently, zinc finger nucleases (ZFNs), chimeric proteins that consist of a ZFP and a Fok1 nuclease domain, have proven to be effective in a variety of applications such as gene disruption, transgene integration and the generation of knockout mice (16–20). By inducing targeted double-stranded breaks (DSBs) and using the host cell’s repair machinery, ZFNs have been used to cause intentional mutations or insert whole genes at the respective targets (21–24). However, the nuclease component of ZFNs can cause off-target cleavage events that result in undesired mutations. Concerns about cyto and genotoxicity remain significant obstacles to be overcome for this to be a safe strategy (16,25–28).
An alternative approach has been to directly fuse DNA integrating enzymes to ZFPs in an attempt to localize activity of the vector to a specific genomic location (29). For example, ZFP-HIV-1 integrase fusions packaged in virions showed promise with preferential targeting in both plasmids and genomic DNA (gDNA), albeit at low levels (30–34). Programmable recombinases using zinc fingers bound to a catalytic domain have been shown to precisely integrate transgenes at pre-determined sites (35–38). However, the catalytic domains for these proteins are sequence-specific, thus targeting is limited to sites containing the required sequences. Steps have been taken to alter the sequence specificity of these catalytic domains in order to allow integration into novel sites (39,40).
Kaminski et al. suggested using transposases fused to a DBD as a method for directing transgene insertion and it was shown previously that modifications to the yeast retrotransposase Ty5 could influence its target selection profile (41–44). Others have shown in Escherichia coli that the ISY100 transposase bound to a ZFP from the mouse transcription factor Zif268 could target transgenes near the expected binding site on recipient plasmids 48% of the time (45). In addition, the prokaryotic mobile element IS30 fused to the Gli1 transcription factor is able to target extrachromosomal plasmids in zebrafish embryos (46).
The Sleeping Beauty (SB) transposase has shown activity in mammalian cells and has been used for diverse non-viral applications (47–49). However, the direct fusion of DBDs to SB has lead to complete or significant reductions in transposase activity (50–52). Despite this, Yant et al. demonstrated DBD–SB fusion protein-mediated transgene targeting at efficiencies of 18–33% to specific sites within plasmids in human cells. However, this group was unable to detect targeted integration when coupling SB with a DBD specific to an endogenous chromosomal location.
The piggyBac (PB), an insect transposase isolated from the moth Trichoplusia Ni, is highly efficient in a broad range of organisms including yeast and mouse, as well as human cell lines and is able to integrate relatively large cassettes of >100 kb. PB inserts transgenes at TTAA tetranucleotide sites and the transposase has been shown to be able to excise the transposon without leaving a DNA footprint (53–64). Previously, we have shown that PB is amenable to a Gal4 DBD fusion with little loss in activity (50). Furthermore, we demonstrated targeting to a plasmid recipient harboring the upstream activating sequence (UAS) Gal4 recognition site in Aedes aegypti embryos (65,66).
A wide range of applications would directly benefit from safe targeted transgenic insertion. We have tested both N- and C-terminal fusions of the classic Gal4 DBD to the PB transposase and assessed for targeted integration in human cells near the UAS DNA binding sequence in a genomic setting. The aim of our experiments was to demonstrate the ability of a chimeric PB transposase fused to a DBD to target the genome. This proof of principle is important for transposition research and will serve as a basis for future improvements that one day may lead to safer transpositional gene therapy treatments in a clinical setting.
MATERIALS AND METHODS
Plasmid development
The mammalian codon biased PB transposase was a gift from Dr Allan Bradley. The backbone for all PB plasmids was the self-inactivating, helper-independent pmGENIE-plasmid system described earlier (58). The Gal4 DBD, (amino acids: KLLSSIEQACDICRLKKLKCSKEKPKCAKCLKNNWEC RYSPKTKRSPLTRAHLTEVESRLERLEQLFLLFPREDLDMILKMDSLQDIKALLTGLFVQDNVNKDAVTDRLASVETDMPLTLRQHRISATSSSEESSNKGQRQLTVS) was introduced via homologous recombination along with the linker: KLGGGAPAVGGGPK (65). The transgene for all pmGENIE constructs was a PCR amplified fragment from pERV3 (Agilent Technologies), including the bacterial and eukaryotic promoter-driven neomycin gene. This fragment was TA-cloned into the pENTR1a vector (Invitrogen) that had been digested with HincII and EcoRI and then t-tailed using a terminal transferase (New England Biolabs). The ligation product, including the neomycin gene, was Gateway (Invitrogen) recombineered between the attR sites in the pmGENIE plasmid transposon.
The CMV-SB11 helper plasmid was a gift from Dr Perry Hackett. To create the SB UAS donor plasmid pT2/HB cam UAS hygro, we used an intermediate plasmid (pELO4) that contains the chloramphenicol gene. The UAS site was isolated from the pGDV1-UAS plasmid by restriction digestion with HindIII and BamHI (65), and cloned into the pELO4 vector adjacent to its chloramphenicol gene. Two identical 650-bp TTAA rich regions, custom synthesized from GenScript and excised from the pUC57 shuttle vector using BamHI or HindIII, were ligated to both sides of the UAS target sequence to make pELO4 cam TTAA-region UAS TTAA-region. The hygromycin gene from pEGSH (Agilent Technologies) was PCR amplified and ligated into the pT2/HB SB donor plasmid with BglII and HindIII and a unique NheI site was introduced with the PCR primers used. The pT2/HB hygro construct was linearized with NheI and a pELO4 PCR product, including cam, UAS and TTAA-rich regions, was added via In-Fusion (Clontech). This plasmid was then used to generate hygromycin resistant cell lines containing the SB transposon and UAS. The UAS-negative control SB donor plasmid was made by PCR amplifying all but the UAS site from the pT2/HB cam UAS hygro construct. This inverse PCR product was re-ligated to form the control plasmid, which retained the TTAA-rich regions but had the UAS removed.
The pT2/HB cam UAS hygro construct was reduced in size by self-ligation using XmnI and SmaI, and used as the recipient in the plasmid into plasmid experiment. In order to eliminate false positives during the plasmid into plasmid assay, the bacterial suicide ccdB gene was added to the backbone of the pmGENIE plasmids by In-Fusion. This strategy prevented recovery of double resistant colonies that contained delivery plasmid backbone resulting from non-transpositional insertion. All restriction enzymes were purchased from New England Biolabs.
Cell transfections
Human embryonic kidney (HEK293) cells were maintained in complete DMEM supplemented with 10% heat inactivated FBS (Invitrogen) and prior to transfection, 0.5 × 105 cells per well were seeded in 12-well plates. To make the SB UAS and SB control cell lines lacking a UAS, 100 ng of CMV-SB11 helper and 200 ng of the appropriate pT2/HB SB donor plasmid were transfected using FuGene6 (Roche Applied Science) into cells at 90% confluency. Stable cell lines were obtained after 30 days of culture in 100 μg/ml HygromycinB (Invitrogen) and were cultured for an additional 3 weeks. We subjected these stable SB transposon containing cell lines to a second round of transfection with 400 ng of helper-independent pmGENIE construct containing the neomycin selection cassette within the transposon. pmGENIE constructs with N- or C-terminal Gal4 fusion or native PB were transfected in triplicates into two independent SB UAS cell lines. Stable G418 resistant lines were obtained after 30 days and equal numbers of cells for each experiment were pelleted and frozen. Genomic DNA was isolated using the DNeasy kit (Qiagen) following the manufacturer’s protocol. A standard colony count assay was performed using all three constructs and a transposase-negative control as described previously (58).
Plasmid into plasmid assay
HEK293 cells were transfected with 750 ng each of UAS recipient plasmid pT2/HB cam UAS hygro and helper-independent pmGENIE delivery plasmids containing either of Gal4–PB fusions or native PB. Cells were grown without selection for 3 days, then pelleted and episomal plasmids were isolated using the Zyppy plasmid miniprep kit (Zymo Research). Lucigen 10G Elite E. coli cells were electroporated with the isolated plasmid DNA and plated on double antibiotic camR/kanR plates for selection. Colonies were screened by duplex PCR to simultaneously confirm the integration of the delivery neomycin transposon, as well as the excision of the transposon from the backbone of the delivery pmGENIE plasmid. Plasmid DNA from positive double antibiotic resistant colonies was purified by miniprep and sequenced using the primer PB 5TRE: ACG GAT TCG CGC TAT TTA GA that extends from the PB transposon into the adjacent sequence. Obtained sequences were aligned to the recipient plasmid pT2/HB cam UAS hygro to determine the insertion site and distance from the UAS site. The efficiency by which PB delivery transposons were integrated into recipient SB plasmids by the respective PB transposase was calculated by dividing the total number of correctly aligned sequences recovered by the percent of colonies screened. The percent of colonies screened was calculated by dividing the total number of colonies PCR screened by the total colony count. (n = 3, mean ± standard deviation) Statistics include two-sided, two-sample Student’s t-test assuming equal variance P = 0.01.
Targeted genomic integration site recovery
Genomic DNA was extracted from pooled clones of stably double-transfected HEK293 cells and nested PCR was performed using forward primers designed to extend from either terminal repeat element (TRE) of the delivery PB transposon, whereas the reverse primers were designed to the SB UAS transposon target. Because of the repetitive nature of the UAS and surrounding sequence we did not obtain PCR products extending through the UAS and therefore designed two sets of reverse primers to identify insertions upstream and downstream of the UAS. See Supplementary Table S1 for primer sequences. Primary PCR products obtained using KOD Xtreme Hot Start DNA Polymerase (Novagen) were diluted 1:100 in H2O and used as template for nested PCR using Easy-A High-Fidelity Polymerase (Agilent Technologies). Amplification products were either first gel purified with Zymoclean Gel DNA Recovery Kit (Zymo Research) or directly TA-cloned into the pGEM-T Easy Vector (Promega). Unique clones were verified by colony PCR, plasmid DNA was purified by miniprep and then sequenced with Sp6 or T7 primers. Sequences were aligned to the SB UAS transposon and distance to the UAS and exact insertion site locations were recorded.
Copy number assay
Monoclonal expansions of double-resistant hygromycin/G418 HEK293 cells that had been first transfected with SB UAS (pT2/HB cam UAS hygro) then with N- or C-terminal Gal4–PB, or PB control pmGENIE plasmids (n = 5 each) were seeded into 96-well plates at low density and visually verified. gDNA from expanded clones was isolated for Southern blot and quantitative PCR (qPCR) copy number assay. In order to standardize the qPCR copy number assay, a Southern blot was performed to identify a known number of insertions for representative PB and SB experiments. Genomic DNA was isolated using the DNeasy kit (Qiagen) following the manufacturer’s protocol. A quantity of 20 µg of each sample was digested overnight using 10 U of HindIII per microgram of gDNA. A quantity of 12 µg of digested gDNA was run for 75 h on a 1% agarose gel at 15 V. The gDNA was then blotted to a Hybond + nylon membrane (GE Healthcare) for 12 h and processed for hybridization. A DIG-labeled probe was generated by PCR amplification using the PCR DIG Probe Synthesis Kit (Roche Applied Science) and the following primers: PB Southern Forward ACGTAAACGGCCACAAGTTC, PB Southern Reverse TGC-TCAGGTAGTGGTTGTCG. SB Southern Forward AACTCGTTTTTCAACTACTCC-ACA, SB Southern Reverse ACTGTCGGGCGTACACAAAT. PCR parameters used: initial denaturation at 94°C for 2 min, 35 cycles of 30 s denaturation at 94°C, 30 s annealing at 56°C and 1 min elongation at 72°C, with a final elongation for 10 min. Hybridization was performed overnight at 55°C using the DIG Easy Hyb Kit, (Roche Applied Science) and were processed according to the manufacturer’s protocol. Chemiluminescent signals were visualized with an LAS-3000 imaging system (Fujifilm).
The qPCR copy number assays were performed by duplex Taqman real-time PCR, where one assay interrogates the transgene copy number, whereas the other assay to Ribonuclease P (RNaseP) serves as a reference. Primers and probes were custom designed (to the 5′TRE for PB and the 3′TRE for SB) or pre-made (RNAseP) and were supplied by Applied Biosystems. The primer and probe sequences are as follows: PB 5TRE Copy Forward GTGACACTTACCGCATTGACAAG, PB 5TRE Copy Reverse GCTGTGCATTTAGGACATCTCAGT, PB Reporter ACGCCTCACGGGAGCTC, SB 3TRE Copy Forward CTCGTTTTTCAACTACTCCACAAATTTCT, SB 3TRE Copy Reverse ACAATTGTTGGAAAAATGACTTGTGTCA, SB ReporterTTTGGCAAGTCAGTTAGGACATCTA. The assays were performed according to the TaqMan copy number assay protocol (Applied Biosystems) using the Step-One-Plus real-time PCR machine in a 20 -µl reaction volume containing 50 ng DNA. A minimum of four replicates per sample was assayed. One sample with known transgene copy number (as determined by Southern blot analysis) was included. The copy number assays were normalized to RNAseP, known to occur in two copies in the genome (Applied Biosystems). The results were analyzed using the software CopyCaller v1.0 (Applied Biosystems).
Western Blotting
Cells were cultured as described. After 48 h of incubation, cells were lysed with lysis buffer supplemented with Set III protease inhibitors (Calbiochem). A quantity of 60 µg of total protein was resolved on a pre-cast SDS–polyacrylamide gel (Bio-Rad). Expression of PB proteins was determined by western blotting using a mouse monoclonal anti-PB non-purified antibody. Binding of primary antibody was detected using an anti-mouse horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology). Bands were visualized with an LAS-3000 imaging system (Fujifilm).
Non-restrictive linear amplification-mediated PCR and 454 sequencing
For the off-target insertion analysis we adapted non-restrictive linear amplification-mediated (LAM) PCR (67). Briefly, 1 µg of gDNA from double-resistant hygromycin/G418 HEK293 cells for both Gal4 fusion samples, as well as PB control (n = 4 each) was used as template for linear PCR using single primers for linear amplification extending from the PB-TREs into flanking genomic sequence. See Supplementary Table S2 for primer sequences. Single stranded linkers were ligated to these linear PCR products and nested PCR was performed to amplify the flanking genomic sequence. GS-FLX sequencing primers were added by PCR and samples were sequenced using a 454 GS-FLX Titanium sequencer (Life Sciences) in accordance with the manufacturer’s protocol by the University of Hawaii Advanced Studies in Genomics, Proteomics and Bioinformatics (ASGPB) unit. Resulting sequences were trimmed and de-multiplexed using CLC Genomics Workbench version 4.7 (CLC Bio). The reads were mapped against the human genome reference, version GRCh37.63, using the short read alignment component (bwa-short) of the Burrows–Wheeler Aligner (68), selecting for reads that align over ∼80% of the sequence with a minimum of 90% similarity. Distances of insertion sites to endogenous Gal4 recognition sites were obtained using custom scripts, which are available upon request. Gal4 recognition sequences were defined as CGGNNNNNNNNNNNCCG and a total of 56 898 sites were identified in the human genome. The position weight matrix for the Gal4 DBD is depicted in Supplementary Table S3. Distances to transcriptional start sites, as well as other annotations were obtained using the Homer bioinformatics tool (69) available online at: http://biowhat.ucsd.edu/homer/ngs/annotation.html. Of the 66 414 integration sites recovered using nrLAM PCR, 7004 sites aligned to unique genomic locations. A list of all insertion sites recovered by non-restrictive (nr) LAM PCR is available upon request.
RESULTS
Chimeric Gal4 PB directs transposition into plasmid targets
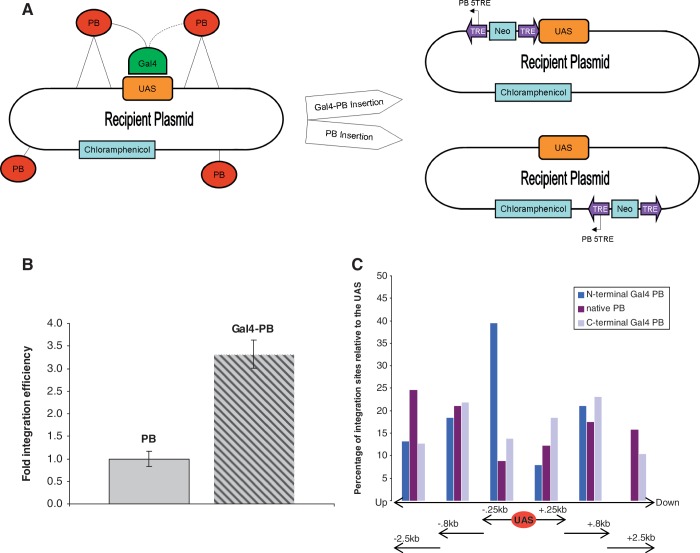
In a previous publication, we described highly efficient PB plasmids that maintained 92% activity of integration after addition of the Gal4 DBD (50). Here, we have tested the hypothesis that by tethering the PB transposase to Gal4, we are able to target integration of transposons near UAS recognition sites in mammalian cells. In the first set of experiments, we used a plasmid into plasmid approach. The recipient plasmid contained a chloramphenicol gene (camR) and a UAS site that consisted of five recognition sequences described earlier (65). The delivery plasmid contained the PB transposase, with or without a Gal4 DBD and a transposon delivery cassette harboring the neomycin (kanR) gene for bacterial selection. Integration of the delivery cassette into the recipient plasmid conferred double resistance to camR/kanR when transformed into E. coli (Figure 1A).
Figure 1.
(A) Schematic for the plasmid into plasmid experiment. Both the delivery plasmid containing the delivery transposon and transposase coding sequence, as well as the recipient plasmid containing the chloramphenicol gene (camR) and the UAS were transfected into HEK293 cells. The tethering of Gal4 to the PB transposase (red circles) is thought to restrict integration to TTAA sites found near the UAS recognition sequence. Native PB proteins are free to integrate throughout the recipient plasmid. Delivery transposons contain the 5′TRE and 3′TRE for PB (purple arrows) and the neomycin gene (Neo) and confer kanR to the recipient plasmid. Recovered camR/kanR plasmids were sequenced with PB 5TRE (black arrows) in order to identify insertion sites. (B) Plasmid into plasmid integration efficiency of PB versus Gal4–PB. Increased total integrations into the recipient plasmid were observed by fusing the Gal4 DBD to PB. (C) Percentages of integration sites recovered at increasing distances from the UAS. N- and C-terminal Gal4 PB integrate closer to the UAS on the recipient plasmid compared with native PB.
The recipient and delivery plasmids were transfected into HEK293 cells and plasmid DNA was isolated 3 days later. Total isolates were electroporated into E. coli and plated on camR/kanR for selection. Colonies were screened by colony PCR to confirm enzymatic excision of the transposon from the delivery plasmid. Plasmid DNA from positive clones was purified and sequenced.
To identify the transposon insertion sites within the recipient plasmid we sequenced out from the delivery cassette into the adjoining plasmid DNA. We flanked both sides of the UAS target with a 650-bp region in which 65 TTAA sites were spaced 10 bp apart. This design would allow us to analyze distance requirements for integration from the UAS. For example, preferential integration at a certain distance from the UAS might be evidence for spatial protein tension during integration or be the result of Gal4 linker length. Only sequences where the PB TRE was immediately followed by a TTAA and consecutively flanked by recipient sequence were considered as verified transpositional insertions. A total of 182 verified sequences were recovered for the Gal4 fusions and native PB experiments. The efficiency of total integrations of the chimeric Gal4 protein was over three times that of native PB control (Figure 1B). This was expected because the Gal4 DBD is thought to bring the PB protein and recipient plasmid together via UAS target binding (Figure 1A). Furthermore, an increase in total integrations is suggestive of targeting to sites nearby the UAS on the recipient plasmid.
By analyzing the distance of integration sites on the recipient plasmid relative to the UAS site, we found that 87% of N-terminal Gal4-PB (nGal4-PB) insertions were located within a region 800 bp up- or downstream of the UAS, whereas only 59% of the PB control insertions fell within this region. Similarly, the C-terminal fused PB (PB-cGal4) also had a significant ability (77%) to integrate near the UAS site (Table 1). More specifically, 47% of the nGal4–PB and 32% of the PB–cGal4 directed integrations were within 250 bp of the UAS compared with 21% for the native PB control. In addition, 39% of these nGal4–PB mediated insertions were detected within 250 bp upstream of the UAS site (Figure 1C). In contrast, the PB–cGal4 sample displayed a more evenly spaced distribution of insertion sites. Native PB frequently integrated farther upstream or downstream from the UAS compared with both fusion proteins (Figure 1C). In summary, this data shows that in our plasmid model the addition of a Gal4 DBD to either N- or C-terminal end of the PB protein confers a propensity for transposition near the UAS site.
Table 1.
Distances of recovered plasmid into plasmid insertions sites from UAS targets
| Plasmid to Plasmid | N-terminal Gal4 PB (%) | Native PB (%) | C-terminal Gal4 PB (%) |
|---|---|---|---|
| insertions < 800 bp from UAS | 87**** | 59 | 77** |
| insertions < 250 bp from UAS | 47*** | 21 | 32* |
Both N- and C-terminal Gal4 PB fusion constructs significantly biased integration near the UAS compared with native PB by the Fisher’s exact test. *P = 0.05, **P = 0.01, ***P = 0.005, ****P = 0.002
Genomic targeting of the chimeric Gal4 PB
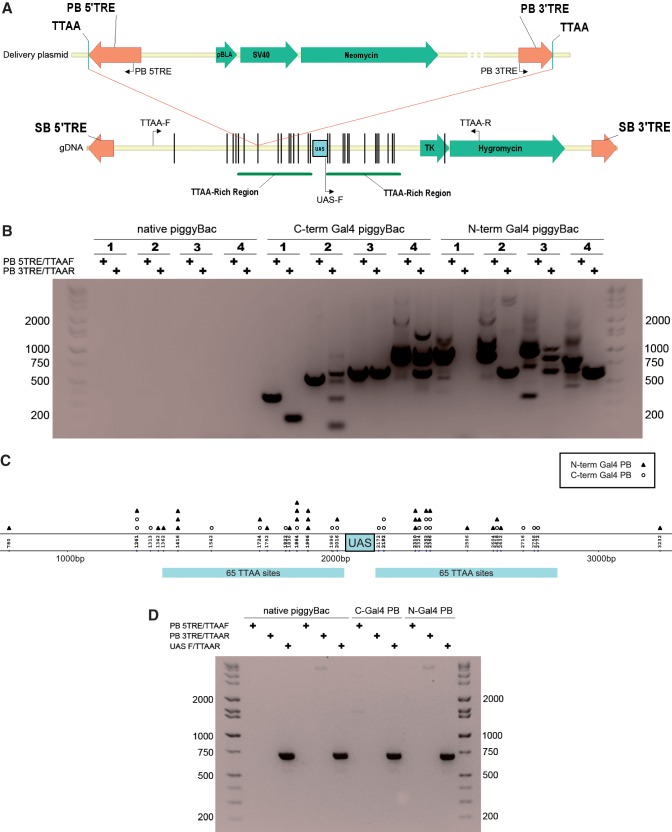
In order to determine whether targeting could be achieved in a genomic setting, we used a recipient transposon containing the UAS and TTAA-rich regions flanked by the TREs of SB (pT2/HB cam UAS hygro). This allowed for random integration of SB transposon targets into the genome of HEK293 cells, which in turn were targeted by the chimeric Gal4-PB. The host repair machinery can be used to uptake fragments of exogenous DNA sequences into cells and integrants can be isolated following selection. However, this process is relatively inefficient and only the sequence for the selection marker is required for cell survival. In order to ensure that an intact sequence was efficiently integrated at a large number of genomic loci, we used the SB system for transposition of the UAS target. By using this approach we avoided the possibility of PB recognizing its own TREs and consequentially excising the target transposon. We used the two plasmid SB approach in which the recipient transposon contained the mammalian selection gene hygromycin and the UAS site flanked by TTAA-rich regions on one plasmid, as well as the SB11 transposase encoded on a second plasmid (Figure 2A).
Figure 2.
(A) Schematic for the genomic DNA targeting experiment. Helper-independent pmGENIE plasmids containing both the delivery transposon, as well as the PB coding sequence were used in both the plasmid into plasmid and genomic targeting experiments. The neomycin gene is driven by both bacterial (pBLA) and eukaryotic (SV40) promoters. Sleeping Beauty and the SB recipient transposons were encoded on different plasmids. The recipient transposon harboring the UAS target was first integrated into gDNA by SB and stable integrants were selected with hygromycin. A second transfection was performed with the pmGENIE delivery plasmid containing both the PB transposase and the PB transposon with the neoR gene. Insertions by the chimeric Gal4–PB transposase that had been directed to the vicinity of the UAS target (red lines) were detected by nested PCR (primers represented by black arrows). Black vertical lines represent actual distances of insertions recovered from the UAS on the SB transposon. UAS-F and TTAA-R served as positive control primers to verify the presence of the SB transposon target. (B) Evidence for genomic targeting and the requirement of Gal4. Genomic DNA from hygroR/G418R cell populations transfected first with SB11 and UAS–SB transposon then next with delivery plasmids containing PB, PB–cGal4 or nGal4–PB was isolated and analyzed by PCR. Shown is a representative gel displaying 4 of 6 independent samples, each of nested products recovered for both Gal4 fusions but not native PB. (C) Schematic map of the UAS–SB target transposon showing integrations of piggyBac donor elements using the Gal4–PB chimeric transposase. Open circles and closed triangles represent insertions by C-terminal and N-terminal Gal4–PB, respectively. The vertical numbers represent the nucleotide location of targeted TTAA sites on the UAS–SB transposon. The UAS was flanked on both sides by 65 TTAA sites spaced 10 bp apart. (D) Evidence for the requirement of the UAS. Stable cells transfected with the UAS-negative SB recipient transposon were re-transfected with PB delivery plasmids. Shown is a control gel displaying PCR products for positive control UAS F/TTAAR but not products from targeting primers for two native PB samples and both Gal4 fusion samples (HI-LO DNA Marker, Bionexus).
HEK293 cells were transfected with both SB plasmids and stable lines were obtained after 4 weeks of culture under hygromycin selection. Two stable polyclonal expansions of HEK293 cell lines harboring the SB UAS transposon were transfected with delivery plasmid expressing N- or C-terminal Gal4-fused PB transposases or the native PB transposase. As with the plasmid into plasmid experiment, these delivery plasmids integrated a PB transposon containing the antibiotic resistance gene neomycin and conferred G418 resistance. Genomic DNA was isolated from 6 replicates each for the 3 experiments. To demonstrate the requirement of the UAS for Gal4-directed targeting, control transfections with a SB recipient transposon plasmid that lacked the UAS target sequence were performed, and hygromycin resistant HEK293 cells were subjected to a second round of transfections with the PB delivery plasmids described above.
In order to detect targeted genomic insertion we used nested PCR with the forward set of primers complementary to the PB transposon and the reverse primers extending from the SB transposon target (Figure 2A and B). As expected, we did not obtain any PCR products from the UAS-negative control samples indicating that neither Gal4 fusion, nor native PB transposase targeted the SB recipient transposon alone (Figure 2D). Furthermore, we did not detect any targeted PCR products for the 6 UAS-positive cell populations that had been transfected with native PB control transposase. In contrast, for all UAS-positive populations tested with a Gal4-PB fusion, nested PCR products were obtained and sequenced (Figure 2B). Within the 8000 bp region that we analyzed, all 49 unique integrations recovered, localized within 1300 bp of the UAS site with the vast majority (96%) found <800 bp up- or down-stream from the UAS (Figure 2C and Table 2). About 95% of sequenced PCR product bands were verified SB transposon insertions; the remaining 5% resulted from non-specific primer binding. Both N- and C-terminal fusions displayed a similar targeting efficiency and the integration profile within the TTAA-rich region flanking the UAS site appeared somewhat random without a predictable integration distance from the UAS. We also identified a number of hot spots where the same insertion site was found across multiple samples; seven sites that were targeted three or more times and for one site, 193 bp upstream of the UAS site, we detected a total of four integrations by both nGal4–PB and PB–cGal4. Furthermore, eight loci shared integrations from both fusion constructs (Figure 2C).
Table 2.
Total genomic insertions into the SB recipient transposon recovered by nested PCR
| HEK 293 Cell Line | N-terminal Gal4 PB | Native PB | N-terminal Gal4 PB |
|---|---|---|---|
| UAS + | 28 | 0 | 21 |
| UAS − | 0 | 0 | 0 |
Transposon copy number and off-target analysis
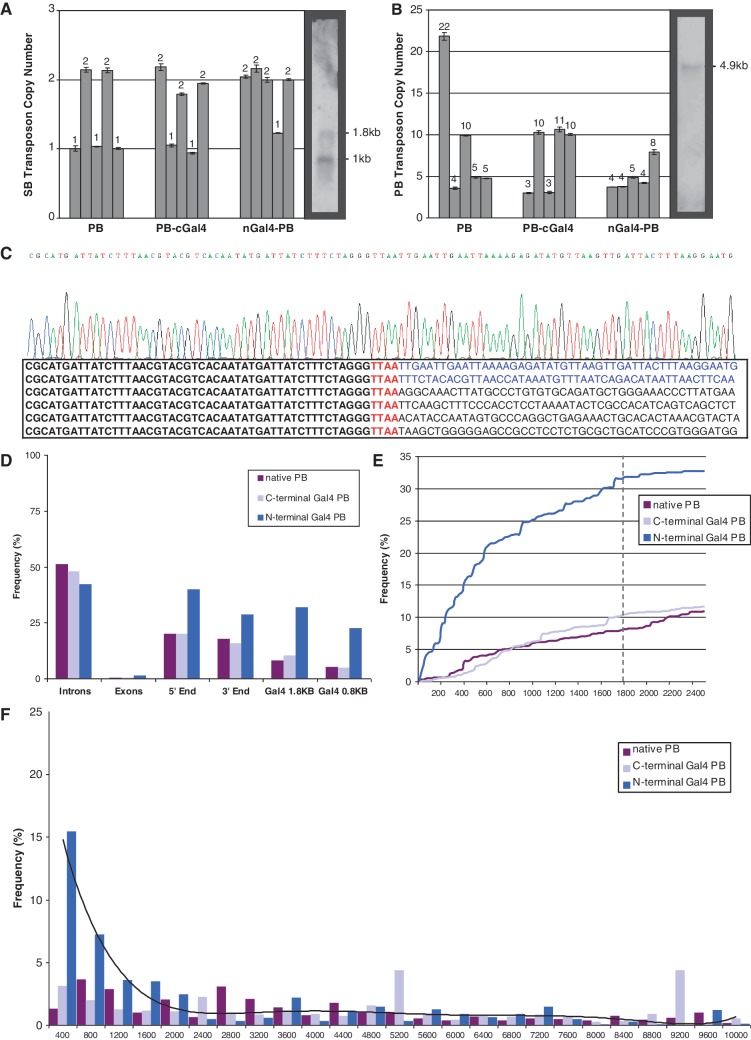
In order to assess the number of possible transposon targets per cell, monoclonal expansions were established from HEK293 cells, transfected with both SB and PB transposons as described above. Individual clones from nGal4–PB, PB–cGal4 and native PB samples were subjected to Southern blot (n = 1) and qPCR (n = 5 each) analysis. All samples tested contained either one or two UAS–SB transposons (Figure 3A). We additionally analyzed the number of PB-mediated integrations per clonal sample by qPCR. Here, the majority contained three to five delivery transposons with an average number of seven integrations per cell (Figure 3B).
Figure 3.
(A and B) Copy number assays for number of SB and PB transposon integrations. The gDNA from 5 single clones each for N- and C-terminal Gal4 fusion and native PB experiments was analyzed by duplex Taqman real-time PCR. The numbers above the bars represent the estimated copy number for each sample. The Southern blot shown on the right of each graph was applied as a standard of known number of transposon integrations and was used to calibrate the qPCR data. (C) Sequences recovered from a representative sample showing the PB TRE on the left in bold, TTAA and flanking sequence on the right. The top 2 lines with flanking sequence in blue show nested PCR products that align to the genomic UAS–SB recipient transposon. The bottom four lines with flanking sequence in black show recovered nrLAM and 454 sequences representing off-target events with alignments to various locations in the human genome. (D) The frequency of insertion sites recovered from nrLAM PCR that land within introns and exons, within a 10 kb window surrounding transcriptional start sites (5′-end) or polyA termination sites (3′-end), and ±1.8 kb and ±0.8 kb of endogenous Gal4 recognition sites. (E) The cumulative percentage of total integrations from 0 to 2400 bp from endogenous recognition sequences. The frequency of insertions for native PB and PB–cGal4 increased linearly. nGal4–PB insertion frequency increased logarithmically until 1800 bp and then increased linearly. (F) Histogram displaying the percentage of total integrations that occurred within 400 bp intervals from 0 to 10 000 bp from endogenous Gal4 recognition sequences. The black line represents the best fit curve for nGal4–PB.
The protein levels of PB, nGal4–PB and PB–cGal4 were determined by western blot (Supplementary Figure S1). Both fusion proteins expressed at similar levels and ran at a higher molecular weight than native PB. The ability to form G418R colonies in a standard colony count assay (58) was used to estimate transposase efficiency. The three constructs produced similar numbers of colonies with a slight reduction in efficiency for nGal4–PB compared with native PB control (Supplementary Figure S2) reconfirming that the Gal4 fusion did not inactivate the PB transposase.
Non-restrictive LAM PCR can be used to amplify genomic sequences flanking known insertion cassettes such as transposons to identify off-target insertion sites. Single-stranded adaptors were ligated onto linear PCR products made from primers designed to extend away from the TREs of the PB delivery transposon. Nested PCR was used to amplify the TRE–gDNA junction and products were subjected to 454 pyrosequencing. Verified insertion sites included TRE sequence followed by the TTAA tetranucleotide followed by genomic sequence (Figure 3C). Four independent transfections for each of the three experiments were pooled and analyzed using published human genome annotations. A total of 66 414 integration sites were recovered from the nrLAM sequencing, many of which were repeated reads indicating good sequencing coverage. In total, 7004 of the integration sites were aligned to unique genomic locations. An off-target analysis was performed on the integration sites identified from the 12 polyclonal samples confirming previous integration profiles for PB reported by us and others (Figure 3D) (53,54,70–74). PB showed a slight preference for integration near the 5′-end (within 10 kb of transcriptional start sites) and 3′-end (within 10 kb of polyA sites) of genes, as well as a preference for introns, possibly due to the large size and number of TTAA sites found within introns. The frequencies of insertions recovered within known genes can be found in Table 3. When compared with viruses such as HIV, PB displayed a more random insertion site distribution and targeted genes much less frequently (71–73). However, PB-mediated insertions into genes were significantly more common when compared with a random insertion pattern.
Table 3.
Frequencies of integration into intragenic regions and transcriptional start sites of RefSeq genes
| Genomic location | Integrations (%) |
||||
|---|---|---|---|---|---|
| Randoma | PB | PB–c Gal4 | nGal4– PB | HIVa,b,c | |
| In RefSeq genes | 33.2 | 60.8 | 57.8 | 61.5 | 83.4 |
| ±5 kb from start site | 5.4 | 19.9 | 19.9 | 39.9 | 11.4 |
Frequency of off-target integrations into genes and regions near transcriptional start sites, recovered from nrLAM PCR, compared with random and viral integration. Results from this study are boldfaced.
aValues from the work of Yant et al. (72).
HIV, human immunodeficiency virus.
Gal4 PB biases integrations near endogenous Gal4 recognition sites
A comparison of off-target integrations for the three constructs revealed that the nGal4–PB sample profile was shifted; we observed an increased number of integrations into exons, polyA sites and transcription start sites, presumably because of altered preference of integration due to Gal4 binding (Figure 3D). To estimate the efficiency of the Gal4–PB fusion protein’s ability to insert near endogenous Gal4 recognition sites, we annotated the 56 898 UAS-like sites found in the human genome and examined insertions that were located in the vicinity of these sites. Gal4 binds tightly to a specific 6 bp binding site defined as CGGNNNNNNNNNNNCCG. Sequence variability or alterations in the number of variable (N) basepairs greatly reduces binding affinity (75). We counted the number of insertions that occurred in 20 bp increments from 0 to 10 000 bp from each Gal4 recognition site. About 32% of nGal4–PB transpositions landed within 1.8 kb of endogenous Gal4 sites compared with 8% for native PB and 23% were within 0.8 kb compared with 5% for native PB (Figure 3D). The cumulative percentage of integrations recovered (Figure 3E) dramatically increases up until 1800 bp for nGal4–PB but not for PB–cGal4 or native PB. A histogram displaying the percentage of total integrations that occurred within 400 bp intervals shows increased insertions recovered in regions up to 1800 bp from Gal4 sites for nGal4–PB but not for PB–cGal4 or native PB (Figure 3F). While PB–cGal4 targeted the exogenous UAS–SB transposon almost as efficiently as nGal4–PB, we were surprised to find no preferential targeting of the endogenous consensus sites by PB–cGal4, which had an overall integration profile that resembled that of native PB.
DISCUSSION
Traditionally, integrating vectors such as viruses have been used to insert transgenes semi-randomly and have led to deleterious effects due to integration at unwanted sites (76,77). To address this problem we have designed novel proteins encompassing the classical Gal4 DBD fused to a PB transposase in an attempt to bias genomic insertion to specific sites within the genome. We have demonstrated targeting to UAS recipient plasmids in human cells using a tethered Gal4–PB and shown that integration preferentially occurs near the UAS recognition sequence (Table 1). Gal4 is a tight binding Zn2/Cys6 zinc finger with a 6 bp binding site that occurs not only in our inserted UAS sites, but also at many endogenous human loci (75). Despite the numerous target sites for Gal4, we were successful in showing that genomic targeting can be achieved near introduced target UAS sites. It is important to assay transpositional events that occur on genomic DNA because histone-associated DNA may influence transposition as compared with naked DNA. Hence, we stably introduced our recipient transposon to ensure that the subtleties of the genomic environment could be accounted for during Gal4-directed transposition.
In our experiments merely integrating SB transposons containing TTAA sites without a UAS was not sufficient for Gal4–PB targeting. Both the UAS recognition sequence, as well as the Gal4 DBD fused to the PB transposase were required for enhanced genomic targeting (Figure 2B and D; Table 2). It should be noted that during the preparation of this publication, a similar methodology was published. In that publication the PB transposase was fused to CHK2-ZFP to direct integration (78). However, of the targeted single clones that were isolated in that study, >20% of negative control clones showed evidence of native PB targeting into SB transposon targets containing the CHK2 recognition sequence. We did not observe such insertions in our negative control experiments, which was expected because it seems unlikely that an unmodified PB would preferentially integrate into one of the <200 TTAA sites found on a SB transposon target given the >10 million possible TTAA sites available in the human genome. Currently, the discrepancy between these studies is unclear.
Given that there were only 1–2 inserted exogenous UAS targets per cell (Figure 3A) but millions of available TTAA sequences throughout each cell’s genome, it is remarkable that we have been able to detect a bias for targeted transgene insertion. In this study, both N- and C-terminal Gal4 fusions but not native PB preferentially integrated within 800 bp of the UAS site. This information could prove important in the future design of alternative DBD–PB fusion proteins. Despite the even distribution of TTAA sites in the regions flanking the UAS site, one every 10 bp, certain hot spots were targeted frequently. A possible explanation for this observation is that the physical structure of the DNA places restraints on some of the potential integrations, whereas other TTAA sites are more readily available for transposition. If indeed this is the case, this phenomenon may explain why the same sites were targeted in repeated transfections with the same plasmid and why the N-terminal fused PB transposase shared hotspots with C-terminal modified PB (Figure 2C).
An extensive off-target analysis using nrLAM PCR and 454 pyrosequencing revealed that fusing the Gal4 DBD to the C-terminal of PB, in contrast to an N-terminal fusion, did not significantly modify PB’s off-target integration profile (Figure 3D). It may be that PB retains its intrinsic ability to bind DNA and is therefore, able to bind to one of the many available TTAA sites within the genome, thereby mediating off-target integration despite its fusion to the Gal4 DBD. While the same mechanism should apply for the N-terminal fusion of Gal4 to PB, we noticed a different integration pattern: in addition to targeting the UAS–SB transposon, nGal4–PB directed integration within 0.8 kb of endogenous genomic Gal4 recognition sites, 23% of the time compared with 5% for native PB controls, and within 1.8 kb of endogenous sites, 32% of the time compared with 8% for controls. Because 8% of integrations occur near Gal4 sites at random, and the percentage of total recovered sites for nGal4–PB is 32%, the difference of 24% represents the percentage of targeted integrations due to the presence of the Gal4 DBD. Given that there was an average of 7 PB insertions per cell (Figure 3B) we estimate that, on average, 1.7 insertions per cell (24% of 7 PB insertions) were targeted to within 1.8 kb of a Gal4 site by nGal4–PB and that 5.3 insertions landed at random TTAA sites. We detected a variable number of PB insertions per cell (Figure 3B) and recognized that it is possible to have cells with or without any targeted insertions and with or without any random off-target insertions. It is entirely possible that the targeting efficiency would be reduced, should the number of available recognition sites be decreased. Because the Gal4-fused PB transposase remains functionally active, there may be a higher probability of encountering and inserting at a random off-target TTAA site before a TTAA site near a single unique recognition sequence. Further experiments are needed to determine the level of influence of the number of possible target sequences in relation to the targeting efficiency of a DBD–PB fusion.
It is not evident to us why nGal4–PB but not PB–cGal4 targeted endogenous sites so much more effectively. The protein levels for the two transposases are comparable (Supplementary Figure S1) and the efficiency of integration for nGal4–PB was not higher than PB–cGal4 (Supplementary Figure S2). It is possible that this phenomenon may be due to spatial/steric interactions between Gal4 and PB. For example, in the N-terminal Gal4–PB configuration the linker and PB transposase extend from the C-terminal side of the Gal4 DBD. This configuration is similar to the orientation of the natural activation domain in the full length wild-type Gal4 protein. However, this model of favorable fusion architecture does not explain why both fusions to the PB transposase apparently target the exogenous UAS on the SB transposon with similar efficiency; neither does it explain why both constructs are able to integrate near the UAS on recipient plasmids. One explanation could be that PB–cGal4 may only efficiently bind naked DNA such as episomal plasmids. Exogenous UAS sites were chromatinized, however, and PB–cGal4 retained the ability to target these sites, indicating that PB–cGal4 can bind DNA associated with histones. An alternative explanation for the difference in exogenous targeting between the N- and C-terminal fusions is that the introduced UAS sequence, like classic UAS sequence arrays, is made up of repeated Gal4 recognition sequences, and that the C-terminal fusion requires a number of sites in tandem for efficient binding. It is possible that, in addition to binding to UAS arrays, the N-terminal fusion also effectively binds endogenous monomeric Gal4 recognition sequences.
Although both chimeric proteins were effective in biasing integration toward exogenous or endogenous Gal4 binding sites, the number of off-target integrations identified in this study remains high and points to the need for a system where the binding of a specific DBD is a prerequisite for PB-mediated transposition. It may be possible to achieve this by mutational molecular evolution in which the activity of the PB protein is made to be dependent on its DBD. Redesign of the dimerization interface of ZFNs has reduced off-target toxicity by reducing binding of the protein dimers in solution (79). Perhaps mutations in PB’s dimerization domain could inhibit activity in solution but retain the ability of the protein to dimerize and integrate, should the dimers unite at a DBD target sequence. It is clear that modifications such as this will be necessary for the chimeric PB strategy to mature into a viable method for genetic engineering and therapy.
Φ-C31 integrase mediates efficient DNA delivery to recipient plasmids using site-specific recombination between its attP and attB recognition sites and to pseudo attP sites of which there is an estimated 370 in the mammalian genome. A study looking at 196 independent genomic integration events revealed that 80% of insertions occurred near Φ-C31 attP sequence motifs, with 7.5% of integrations at a unique site on chromosome 19 (80). Unfortunately, a high proportion of cells expressing Φ-C31 integrase were found to have numerous chromosomal abnormalities including various translocations (80,81).
The adeno-associated virus (AAV) is able to insert its genome into a specific site on chromosome 19 (AAVS1) through the activity of its Rep78 protein. Hybrid adenovirus/AAV vectors have been developed to target transgenes to this site (82). Co-infection of human hematopoietic cells with two helper-dependent adenovirus vectors containing the rep78 gene on one vector and a GFP reporter on the second resulted in 30% of integration into the AAVS1 region (83).
Gersbach et al. (84) recently described zinc-finger recombinase (ZFR) fusion proteins with high specificity in targeting and with few off-target consequences (84). This group was able to efficiently integrate transgenes into a specific target sequence harbored on PB transposons randomly integrated within the genome. The utility of this approach, however, is hindered by the necessity to introduce target sites into the genome of interest due to the fact that the catalytic domain of the integrase retains strict sequence specificity for its native recognition sequence.
The plasmid-based PB system has many advantages over viral techniques including tolerance by the immune system, large cargo capacity, as well as inexpensive and simple preparation. PB was shown to tolerate an assortment of fusion domains (50,55,65) while retaining high integration efficiencies. Unlike recombinase-based approaches, which are restrained by a specific DNA-binding sequence, it should be possible to replace Gal4 with any custom DBD. In order to reduce the number of off-target integrations, a DBD with a unique 18 bp+ recognition sequence could be used to tailor targeting of the chimeric transposase to virtually any pre-defined genomic site.
The need for highly specific endogenous integration is paramount and our future focus is to explore the use of highly specific DBDs to target a natural safe harbor in the human genome. One of the advantages of this strategy would be the ability to isolate single clones with safe single targeted insertions for use in a wide variety of cell replacement therapies. By expanding clones and verifying both the presence of a targeted insertion and the absence of multiple insertions, it would be possible to control gene silencing resulting from position effects and provide a means for avoiding detrimental mutations. The current study has laid the groundwork for using DBD–PB transposase fusion proteins for directed genomic integration. We anticipate that further improvements to this versatile framework will ultimately permit researchers to safely target a genetic cassette to any location within the genome.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3 and Supplementary Figures 1 and 2.
FUNDING
National Institutes of Health (NIH) [5P20RR024206-05, 8P20GM103457-05, R01GM083158-01A1, P20RR018727]. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Joseph Kaminski for critical reading of the manuscript and the Sanger Institute, Cambridge, England for the mouse codon-optimized piggyBac plasmid. We would also like to thank the principal investigator of the University of Hawaii at Manoa’s Bioinformatics Laboratory (BiL Manoa), Dr Poisson Guylaine, for her help in the high-throughput sequence analysis.
REFERENCES
- 1.Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 2.Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, Ecker JR, Bushman FD. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daniel R, Smith JA. Integration site selection by retroviral vectors: molecular mechanism and clinical consequences. Hum. Gene Ther. 2008;19:557–568. doi: 10.1089/hum.2007.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deichmann A, Hacein-Bey-Abina S, Schmidt M, Garrigue A, Brugman MH, Hu J, Glimm H, Gyapay G, Prum B, Fraser CC, et al. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J. Clin. Invest. 2007;117:2225–2232. doi: 10.1172/JCI31659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baum C. Insertional mutagenesis in gene therapy and stem cell biology. Curr. Opin. Hematol. 2007;14:337–342. doi: 10.1097/MOH.0b013e3281900f01. [DOI] [PubMed] [Google Scholar]
- 6.Fehse B, Roeder I. Insertional mutagenesis and clonal dominance: biological and statistical considerations. Gene Ther. 2008;15:143–153. doi: 10.1038/sj.gt.3303052. [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez B, Schwimmer LJ, Fuller RP, Ye Y, Asawapornmongkol L, Barbas CF., 3rd Modular system for the construction of zinc-finger libraries and proteins. Nat. Protoc. 2010;5:791–810. doi: 10.1038/nprot.2010.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. Rapid "open-source" engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Isalan M, Klug A, Choo Y. A rapid, generally applicable method to engineer zinc fingers illustrated by targeting the HIV-1 promoter. Nat. Biotechnol. 2001;19:656–660. doi: 10.1038/90264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Segal DJ, Dreier B, Beerli RR, Barbas CF., 3rd Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5'-GNN-3' DNA target sequences. Proc. Natl Acad. Sci. USA. 1999;96:2758–2763. doi: 10.1073/pnas.96.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mandell JG, Barbas CF., 3rd Zinc Finger Tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006;34:W516–W523. doi: 10.1093/nar/gkl209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beerli RR, Segal DJ, Dreier B, Barbas CF., 3rd Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc. Natl Acad. Sci. USA. 1998;95:14628–14633. doi: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beerli RR, Barbas CF., 3rd Engineering polydactyl zinc-finger transcription factors. Nat. Biotechnol. 2002;20:135–141. doi: 10.1038/nbt0202-135. [DOI] [PubMed] [Google Scholar]
- 14.Blancafort P, Chen EI, Gonzalez B, Bergquist S, Zijlstra A, Guthy D, Brachat A, Brakenhoff RH, Quigley JP, Erdmann D, et al. Genetic reprogramming of tumor cells by zinc finger transcription factors. Proc. Natl Acad. Sci. USA. 2005;102:11716–11721. doi: 10.1073/pnas.0501162102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beltran AS, Russo A, Lara H, Fan C, Lizardi PM, Blancafort P. Suppression of breast tumor growth and metastasis by an engineered transcription factor. PLoS One. 2011;6:e24595. doi: 10.1371/journal.pone.0024595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X, et al. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325:433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moehle EA, Rock JM, Lee YL, Jouvenot Y, DeKelver RC, Gregory PD, Urnov FD, Holmes MC. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc. Natl Acad. Sci. USA. 2007;104:3055–3060. doi: 10.1073/pnas.0611478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu C, Smith T, McNulty J, Rayla AL, Lakshmanan A, Siekmann AF, Buffardi M, Meng X, Shin J, Padmanabhan A, et al. Evaluation and application of modularly assembled zinc-finger nucleases in zebrafish. Development. 2011;138:4555–4564. doi: 10.1242/dev.066779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat. Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 22.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 23.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl Acad. Sci. USA. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolfe SA, Grant RA, Pabo CO. Structure of a designed dimeric zinc finger protein bound to DNA. Biochemistry. 2003;42:13401–13409. doi: 10.1021/bi034830b. [DOI] [PubMed] [Google Scholar]
- 25.Cornu TI, Thibodeau-Beganny S, Guhl E, Alwin S, Eichtinger M, Joung JK, Cathomen T. DNA-binding specificity is a major determinant of the activity and toxicity of zinc-finger nucleases. Mol. Ther. 2008;16:352–358. doi: 10.1038/sj.mt.6300357. [DOI] [PubMed] [Google Scholar]
- 26.Pattanayak V, Ramirez CL, Joung JK, Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat. Methods. 2011;8:765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gabriel R, Lombardo A, Arens A, Miller JC, Genovese P, Kaeppel C, Nowrouzi A, Bartholomae CC, Wang J, Friedman G, et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat. Biotechnol. 2011;29:816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- 28.Gupta A, Meng X, Zhu LJ, Lawson ND, Wolfe SA. Zinc finger protein-dependent and -independent contributions to the in vivo off-target activity of zinc finger nucleases. Nucleic Acids Res. 2011;39:381–392. doi: 10.1093/nar/gkq787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coates CJ, Kaminski JM, Summers JB, Segal DJ, Miller AD, Kolb AF. Site-directed genome modification: derivatives of DNA-modifying enzymes as targeting tools. Trends Biotechnol. 2005;23:407–419. doi: 10.1016/j.tibtech.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 30.Su K, Wang D, Ye J, Kim YC, Chow SA. Site-specific integration of retroviral DNA in human cells using fusion proteins consisting of human immunodeficiency virus type 1 integrase and the designed polydactyl zinc-finger protein E2C. Methods. 2009;47:269–276. doi: 10.1016/j.ymeth.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan W, Dong Z, Wilkinson TA, Barbas CF, 3rd, Chow SA. Human immunodeficiency virus type 1 incorporated with fusion proteins consisting of integrase and the designed polydactyl zinc finger protein E2C can bias integration of viral DNA into a predetermined chromosomal region in human cells. J. Virol. 2006;80:1939–1948. doi: 10.1128/JVI.80.4.1939-1948.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan W, Zhu K, Segal DJ, Barbas CF, 3rd, Chow SA. Fusion proteins consisting of human immunodeficiency virus type 1 integrase and the designed polydactyl zinc finger protein E2C direct integration of viral DNA into specific sites. J. Virol. 2004;78:1301–1313. doi: 10.1128/JVI.78.3.1301-1313.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bushman FD. Tethering human immunodeficiency virus 1 integrase to a DNA site directs integration to nearby sequences. Proc. Natl Acad. Sci. USA. 1994;91:9233–9237. doi: 10.1073/pnas.91.20.9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katz RA, Merkel G, Skalka AM. Targeting of retroviral integrase by fusion to a heterologous DNA binding domain: in vitro activities and incorporation of a fusion protein into viral particles. Virology. 1996;217:178–190. doi: 10.1006/viro.1996.0105. [DOI] [PubMed] [Google Scholar]
- 35.Akopian A, He J, Boocock MR, Stark WM. Chimeric recombinases with designed DNA sequence recognition. Proc. Natl Acad. Sci. USA. 2003;100:8688–8691. doi: 10.1073/pnas.1533177100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bolusani S, Ma CH, Paek A, Konieczka JH, Jayaram M, Voziyanov Y. Evolution of variants of yeast site-specific recombinase Flp that utilize native genomic sequences as recombination target sites. Nucleic Acids Res. 2006;34:5259–5269. doi: 10.1093/nar/gkl548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gordley RM, Gersbach CA, Barbas CF., 3rd Synthesis of programmable integrases. Proc. Natl Acad. Sci. USA. 2009;106:5053–5058. doi: 10.1073/pnas.0812502106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gordley RM, Smith JD, Gräslund T, Barbas CF., 3rd Evolution of programmable zinc finger-recombinases with activity in human cells. J. Mol. Biol. 2007;367:802–813. doi: 10.1016/j.jmb.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 39.Gaj T, Mercer AC, Gersbach CA, Gordley RM, Barbas CF., 3rd Structure-guided reprogramming of serine recombinase DNA sequence specificity. Proc. Natl Acad. Sci. USA. 2011;108:498–503. doi: 10.1073/pnas.1014214108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gersbach CA, Gaj T, Gordley RM, Barbas CF., 3rd Directed evolution of recombinase specificity by split gene reassembly. Nucleic Acids Res. 2010;38:4198–4206. doi: 10.1093/nar/gkq125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu Y, Dai J, Fuerst PG, Voytas DF. Controlling integration specificity of a yeast retrotransposon. Proc. Natl Acad. Sci. USA. 2003;100:5891–5895. doi: 10.1073/pnas.1036705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaminski JM, Huber MR, Summers JB, Ward MB. Design of a nonviral vector for site-selective, efficient integration into the human genome. FASEB J. 2002;16:1242–1247. doi: 10.1096/fj.02-0127hyp. [DOI] [PubMed] [Google Scholar]
- 43.Brady TL, Schmidt CL, Voytas DF. Targeting integration of the Saccharomyces Ty5 retrotransposon. Methods Mol. Biol. 2008;435:153–163. doi: 10.1007/978-1-59745-232-8_11. [DOI] [PubMed] [Google Scholar]
- 44.Kaminski J, Summers JB. Delivering zinc fingers. Nat. Biotechnol. 2003;21:492–493. doi: 10.1038/nbt0503-492b. [DOI] [PubMed] [Google Scholar]
- 45.Feng X, Bednarz AL, Colloms SD. Precise targeted integration by a chimaeric transposase zinc-finger fusion protein. Nucleic Acids Res. 2010;38:1204–1216. doi: 10.1093/nar/gkp1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szabó M, Müller F, Kiss J, Balduf C, Strähle U, Olasz F. Transposition and targeting of the prokaryotic mobile element IS30 in zebrafish. FEBS Lett. 2003;550:46–50. doi: 10.1016/s0014-5793(03)00814-7. [DOI] [PubMed] [Google Scholar]
- 47.Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 48.Ivics Z, Izsvak Z. Non viral gene delivery with the sleeping beauty transposon system. Hum. Gene Ther. 2011;22:1043–1051. doi: 10.1089/hum.2011.143. [DOI] [PubMed] [Google Scholar]
- 49.Izsvák Z, Ivics Z. Sleeping beauty transposition: biology and applications for molecular therapy. Mol. Ther. 2004;9:147–156. doi: 10.1016/j.ymthe.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 50.Wu SC, Meir YJ, Coates CJ, Handler AM, Pelczar P, Moisyadi S, Kaminski JM. piggyBac is a flexible and highly active transposon as compared to sleeping beauty, Tol2, and Mos1 in mammalian cells. Proc. Natl Acad. Sci. USA. 2006;103:15008–15013. doi: 10.1073/pnas.0606979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yant SR, Huang Y, Akache B, Kay MA. Site-directed transposon integration in human cells. Nucleic Acids Res. 2007;35:e50. doi: 10.1093/nar/gkm089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ivics Z, Katzer A, Stüwe EE, Fiedler D, Knespel S, Izsvak Z. Targeted Sleeping Beauty transposition in human cells. Mol. Ther. 2007;15:1137–1144. doi: 10.1038/sj.mt.6300169. [DOI] [PubMed] [Google Scholar]
- 53.Lacoste A, Berenshteyn F, Brivanlou AH. An efficient and reversible transposable system for gene delivery and lineage-specific differentiation in human embryonic stem cells. Cell Stem Cell. 2009;5:332–342. doi: 10.1016/j.stem.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 54.Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 55.Cadiñanos J, Bradley A. Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res. 2007;35:e87. doi: 10.1093/nar/gkm446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yusa K, Rad R, Takeda J, Bradley A. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat. Methods. 2009;6:363–369. doi: 10.1038/nmeth.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mitra R, Fain-Thornton J, Craig NL. piggyBac can bypass DNA synthesis during cut and paste transposition. EMBO J. 2008;27:1097–1109. doi: 10.1038/emboj.2008.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Urschitz J, Kawasumi M, Owens J, Morozumi K, Yamashiro H, Stoytchev I, Marh J, Dee JA, Kawamoto K, Coates CJ, et al. Helper-independent piggyBac plasmids for gene delivery approaches: strategies for avoiding potential genotoxic effects. Proc. Natl Acad. Sci. USA. 2010;107:8117–8122. doi: 10.1073/pnas.1003674107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feschotte C. The piggyBac transposon holds promise for human gene therapy. Proc. Natl Acad. Sci. USA. 2006;103:14981–14982. doi: 10.1073/pnas.0607282103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li MA, Turner DJ, Ning Z, Yusa K, Liang Q, Eckert S, Rad L, Fitzgerald TW, Craig NL, Bradley A. Mobilization of giant piggyBac transposons in the mouse genome. Nucleic Acids Res. 2011;39:e148. doi: 10.1093/nar/gkr764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Claeys Bouuaert C, Chalmers RM. Gene therapy vectors: the prospects and potentials of the cut-and-paste transposons. Genetica. 2010;138:473–484. doi: 10.1007/s10709-009-9391-x. [DOI] [PubMed] [Google Scholar]
- 62.Chen YT, Furushima K, Hou PS, Ku AT, Deng JM, Jang CW, Fang H, Adams HP, Kuo ML, Ho HN, et al. PiggyBac transposon-mediated, reversible gene transfer in human embryonic stem cells. Stem Cells Dev. 2010;19:763–771. doi: 10.1089/scd.2009.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim A, Pyykko I. Size matters: versatile use of PiggyBac transposons as a genetic manipulation tool. Mol. Cell. Biochem. 2011;354:301–309. doi: 10.1007/s11010-011-0832-3. [DOI] [PubMed] [Google Scholar]
- 64.Jang CW, Behringer RR. Transposon-mediated transgenesis in rats. CSH Protoc. 2007;2007 doi: 10.1101/pdb.prot4866. pdb.prot4866. [DOI] [PubMed] [Google Scholar]
- 65.Maragathavally KJ, Kaminski JM, Coates CJ. Chimeric Mos1 and piggyBac transposases result in site-directed integration. FASEB J. 2006;20:1880–1882. doi: 10.1096/fj.05-5485fje. [DOI] [PubMed] [Google Scholar]
- 66.Kolb AF, Coates CJ, Kaminski JM, Summers JB, Miller AD, Segal DJ. Site-directed genome modification: nucleic acid and protein modules for targeted integration and gene correction. Trends Biotechnol. 2005;23:399–406. doi: 10.1016/j.tibtech.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 67.Paruzynski A, Arens A, Gabriel R, Bartholomae CC, Scholz S, Wang W, Wolf S, Glimm H, Schmidt M, von Kalle C. Genome-wide high-throughput integrome analyses by nrLAM-PCR and next-generation sequencing. Nat. Protoc. 2010;5:1379–1395. doi: 10.1038/nprot.2010.87. [DOI] [PubMed] [Google Scholar]
- 68.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang X, Guo H, Tammana S, Jung YC, Mellgren E, Bassi P, Cao Q, Tu ZJ, Kim YC, Ekker SC, et al. Gene transfer efficiency and genome-wide integration profiling of Sleeping Beauty, Tol2, and piggyBac transposons in human primary T cells. Mol. Ther. 2010;18:1803–1813. doi: 10.1038/mt.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Narezkina A, Taganov KD, Litwin S, Stoyanova R, Hayashi J, Seeger C, Skalka AM, Katz RA. Genome-wide analyses of avian sarcoma virus integration sites. J. Virol. 2004;78:11656–11663. doi: 10.1128/JVI.78.21.11656-11663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yant SR, Wu X, Huang Y, Garrison B, Burgess SM, Kay MA. High-resolution genome-wide mapping of transposon integration in mammals. Mol. Cell. Biol. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schröder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 74.Wilson MH, Coates CJ, George AL., Jr PiggyBac transposon-mediated gene transfer in human cells. Mol. Ther. 2007;15:139–145. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- 75.Liang SD, Marmorstein R, Harrison SC, Ptashne M. DNA sequence preferences of GAL4 and PPR1: how a subset of Zn2 Cys6 binuclear cluster proteins recognizes DNA. Mol. Cell. Biol. 1996;16:3773–3780. doi: 10.1128/mcb.16.7.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, Morillon E, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 77.Li Z, Düllmann J, Schiedlmeier B, Schmidt M, von Kalle C, Meyer J, Forster M, Stocking C, Wahlers A, Frank O, et al. Murine leukemia induced by retroviral gene marking. Science. 2002;296:497. doi: 10.1126/science.1068893. [DOI] [PubMed] [Google Scholar]
- 78.Kettlun C, Galvan DL, George AL, Jr, Kaja A, Wilson MH. Manipulating piggyBac transposon chromosomal integration site selection in human cells. Mol. Ther. 2011;19:1636–1644. doi: 10.1038/mt.2011.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Szczepek M, Brondani V, Büchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat. Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 80.Chalberg TW, Portlock JL, Olivares EC, Thyagarajan B, Kirby PJ, Hillman RT, Hoelters J, Calos MP. Integration specificity of phage phiC31 integrase in the human genome. J. Mol. Biol. 2006;357:28–48. doi: 10.1016/j.jmb.2005.11.098. [DOI] [PubMed] [Google Scholar]
- 81.Liu J, Jeppesen I, Nielsen K, Jensen TG. Phi c31 integrase induces chromosomal aberrations in primary human fibroblasts. Gene Ther. 2006;13:1188–1190. doi: 10.1038/sj.gt.3302789. [DOI] [PubMed] [Google Scholar]
- 82.Recchia A, Parks RJ, Lamartina S, Toniatti C, Pieroni L, Palombo F, Ciliberto G, Graham FL, Cortese R, La Monica N, et al. Site-specific integration mediated by a hybrid adenovirus/adeno-associated virus vector. Proc. Natl Acad. Sci. USA. 1999;96:2615–2620. doi: 10.1073/pnas.96.6.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang H, Lieber A. A helper-dependent capsid-modified adenovirus vector expressing adeno-associated virus rep78 mediates site-specific integration of a 27-kilobase transgene cassette. J Virol. 2006;80:11699–11709. doi: 10.1128/JVI.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gersbach CA, Gaj T, Gordley RM, Mercer AC, Barbas CF., 3rd Targeted plasmid integration into the human genome by an engineered zinc-finger recombinase. Nucleic Acids Res. 2011;39:7868–7878. doi: 10.1093/nar/gkr421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.