Abstract
The Sleeping Beauty (SB), piggyBac (PB) and Tol2 transposons are promising instruments for genome engineering. Integration site profiling of SB, PB and Tol2 in human cells showed that PB and Tol2 insertions were enriched in genes, whereas SB insertions were randomly distributed. We aimed to introduce a bias into the target site selection properties of the transposon systems by taking advantage of the locus-specific integration system of adeno-associated virus (AAV). The AAV Rep protein binds to Rep recognition sequences (RRSs) in the human genome, and mediates viral integration into nearby sites. A series of fusion constructs consisting of the N-terminal DNA-binding domain of Rep and the transposases or the N57 domain of SB were generated. A plasmid-based transposition assay showed that Rep/SB yielded a 15-fold enrichment of transposition at a particular site near a targeted RRS. Genome-wide insertion site analysis indicated that an approach based on interactions between the SB transposase and Rep/N57 enriched transgene insertions at RRSs. We also provide evidence of biased insertion of the PB and Tol2 transposons. This study provides a comparative insight into target site selection properties of transposons, as well as proof-of-principle for targeted chromosomal transposition by composite protein–protein and protein–DNA interactions.
INTRODUCTION
The prospect to manipulate the genome of somatic cells of a patient in order to correct a genetic deficiency holds promise for the treatment of numerous inherited and acquired diseases. One major hurdle to overcome is the development of gene therapy vectors that ensure efficient delivery and sustained expression of therapeutic transgenes while minimizing potential side-effects. Retroviral and lentiviral vectors can efficiently deliver transgenes into cells, and have the potential to provide long-term transgene expression by stably integrating into the target cell’s genome (1). However, large-scale surveys on the integration site distribution of HIV-1 revealed a preference for integrations to occur in actively transcribed genes (2). Similar studies showed that the murine leukaemia virus (MLV) has a strong preference for integrating into regions surrounding transcription start sites (3). Thus, the bias in the integration profiles of retroviral gene therapy vectors may result in insertional mutagenesis (4) by activating oncogenes, as observed in clinical trials for SCID-X1 (5,6), X-CGD (7) and WAS (8).
Another promising vector system for gene therapy is based on the adeno-associated virus (AAV). In the absence of a helper virus AAV establishes latency by preferentially integrating its genome locus-specifically into a region on the q arm of human chromosome 19 (19q13.3-qter) termed AAVS1 (9). The only factors needed for targeted integration are the viral cis-acting inverted terminal repeats (ITRs), and the trans-acting viral Rep proteins (Rep). The ITRs have a binding site for the Rep proteins, an imperfect tetrameric GAGC tandem repeat termed Rep-recognition sequence (RRS) (10). Locus-specific integration of the viral genome is determined by DNA sequences present in the AAVS1 locus. A 33-bp sequence encompassing the RRS motif was shown to be necessary and sufficient to mediate targeted integration (11). The viral Rep proteins bind simultaneously to the RRSs in the viral ITRs and in the genomic AAVS1 locus, introduce a nick at the genomic site and integrate the AAV genome through non-homologous recombination (involving partial duplication of the target locus) (Figure 1A) (12). AAV provides several advantages as a gene delivery vehicle. The virus shows no pathogenicity, and is able to efficiently transduce various proliferating and non-proliferating cells (13–17). Serious limitations of AAV for gene therapy are the negative effects of the large Rep proteins on cell viability as they were shown to induce DNA damage, cell-cycle arrest and apoptosis (18). This led to the development of recombinant AAV vectors (rAAV) (19) that lack the Rep genes and therefore persist primarily as episomes in the cell. Nevertheless, rAAV vectors can genomically integrate with a preference for integration into transcription start sites and CpG islands (20). Plasmid-based systems using one plasmid harbouring a gene of interest flanked by AAV ITRs and another expressing the Rep protein have been used to support AAVS1-specific integration (21,22). Other approaches utilized AAV hybrid viruses conditionally expressing the Rep protein (23). AAV-based vectors are involved in diverse gene therapy approaches such as clinical trials for the treatment of hemophilia B (24), rheumatoid arthritis (25) or Parkinson’s disease (26) with modest success and serious complications due to a cytotoxic T lymphocyte response to the AAV capsid (27).
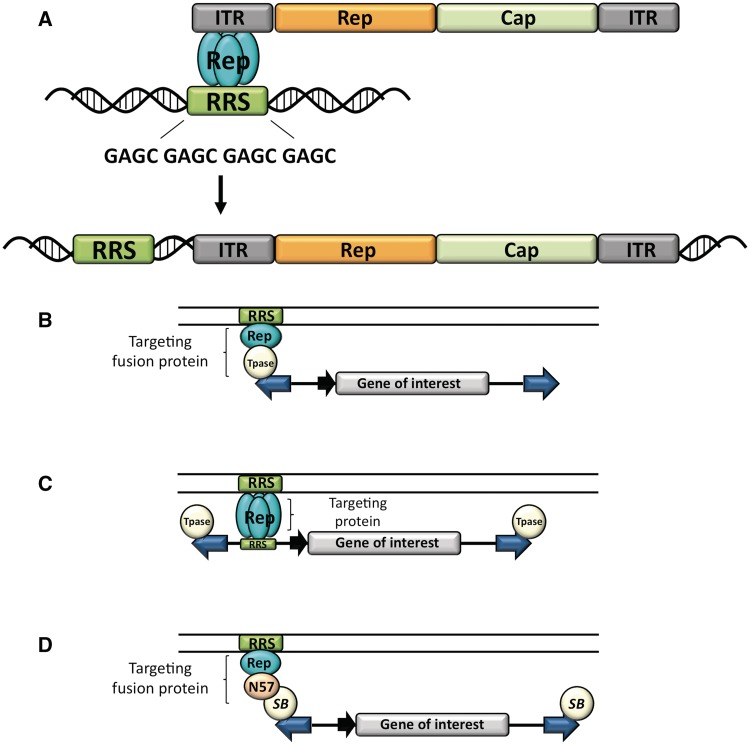
Figure 1.
Experimental strategies for targeting transposition. (A) Site-directed integration of the AAV. The viral Rep proteins (blue ovals) bind to a RRS in the viral ITRs and in the human genome, and mediate site-specific integration of the viral genome adjacent to genomic RRS sites. (B–D) Strategies for transposon targeting. The transposable element is composed of the IRs (blue arrows) flanking a gene of interest (grey box) equipped with a suitable promoter (black arrow). The transposase (Tpase, spheres) binds to the IRs and catalyses transposition. The DNA-binding domain of Rep (blue oval) recognizes an RRS (green box) in the target DNA (parallel lines). (B) Targeting with transposase fusion proteins. Targeting is achieved by fusing the DNA-binding domain of Rep to the transposase. (C) Targeting with proteins that bind the transposon DNA. Targeting is achieved by the DNA-binding domain of Rep that simultaneously interacts with RRS sites present in the transposon and in the target DNA. (D) Targeting with fusion proteins that interact with the transposase. Targeting is achieved by fusing the DNA-binding domain of Rep to the N57 domain (beige oval) that interacts with the SB transposase. Figure redrawn from (71).
DNA transposons are powerful alternatives to viral vectors as well as classical non-viral delivery systems. Transposons are discrete DNA sequences with the ability to move from one genomic location to another via a cut-and-paste mechanism, in which the transposon gets excised from the donor locus and is subsequently reinserted elsewhere in the genome. This transpositional mechanism can be adapted to the crafting of gene delivery vectors, in which transposition out of engineered, plasmid-based vectors forms the basis of stable genomic insertion of gene constructs. Transposon-based vector systems typically consist of two components, the transposon and the transposase. The inverted repeats (IRs) of the transposon that flank a gene of interest to be mobilized contain transposase-binding sites. Transposases typically have two major functional domains: an N-terminal part mediating binding to the transposon IRs, protein multimerization and nuclear transport and a C-terminal catalytic domain responsible for the DNA cleavage and integration reactions required for transposition (28). Virtually any gene of interest can be cloned between the IRs and mobilized by supplying the transposase function. Hence, transposon-based gene therapy vectors offer robust, stable delivery of the desired gene and thus long-term expression. Clinical-grade, plasmid-based transposon vectors can be prepared from well-defined components at much reduced costs and with presumably lower immunogenicity than viral vectors. The three most widely used DNA transposons in vertebrates are the Sleeping Beauty (SB), piggyBac (PB) and Tol2 transposon systems. The SB transposon was resurrected from multiple inactive Tc1/mariner elements found in fish genomes (29). SB transposition exclusively occurs into TA dinucleotides, which are duplicated upon transposon insertion by cellular DNA repair pathways (30). Extensive efforts were made to enhance the transposition efficiency of SB, yielding a set of hyperactive transposase versions with SB100X being the most active transposase to date (31). The SB transposon system has been used to correct several genetic deficiencies in pre-clinical animal models including those for tyrosinemia type I (32), Huntington disease (33), hemophilia A and B (34–38), junctional epidermolysis bullosa (39), mucopolysaccharidosis (40,41), type 1 diabetes (42) and glioblastoma (43,44). In 2008 the National Institute of Health Recombinant DNA Advisory Committee (NIH RAC) approved the first-in-man gene therapy clinical trial that uses transposons. This trial is using the SB transposon/transposase system to generate genetically modified autologous T cells that are transferred into patients with CD19+ B-lymphoid malignancies (45). The PB transposon was first identified in the cabbage looper moth Trichoplusia ni as an active transposon moving from the host genome into a baculovirus genome (46). PB integrates exclusively at TTAA tetranucleotide sequences that are duplicated upon insertion (47). PB elements have been extensively used for germline transgenesis in a wide range of insect species (48), and were also found to efficiently transpose in human and mouse cell lines and in mice in vivo (49). The Tol2 transposon was first identified in the genome of the medaka fish Oryzias latipes, and is the only known naturally occurring active DNA transposon of vertebrate origin (50). Tol2 transposons show no obvious requirements for primary DNA sequence for insertion, and create 8-bp target-site duplications (51). Tol2 is the preferred transposon system for transgenesis and insertional mutagenesis in zebrafish, but also efficiently transposes in cultured mammalian cell lines (52). On the genomic scale, SB transposons exhibit a random integration profile with a target site selection primarily determined by physical properties of the DNA rather than primary DNA sequences (30,53–56). In contrast, PB transposons have a non-random genomic integration profile with a preference for integrating into genes and regions surrounding transcriptional start sites (49,53), whereas Tol2 shows preferences for integrating into transcription units and close to transcription start sites (53). Thus, in addition to its potency in catalysing robust gene transfer in primary cell types, including stem cells (53), a key feature of turning SB to an attractive gene therapy tool is its fairly random integration profile, which might make it safer than integrating viral vectors. Even though SB exhibits a fairly random integration profile, arbitrary integration of a transposon can still be potentially mutagenic, because an internal promoter/enhancer present in the transgene construct could negatively influence the regulation of endogenous genes. Random genomic integration may also limit transgene expression through silencing by position effects. One possibility to alleviate the danger of random transposon integration is targeting of the transposon complex to a specific chromosomal region, where insertion is not predicted to have mutagenic effects.
Attempts to achieve targeted transposition by using hybrid DNA-binding domain-transposase enzymes were undertaken by several research groups. Most of these efforts showed biased transposon insertion into plasmid targets, but no targeted transposition has ever been shown on the genome level. For example, fusions of the bacterial IS30 transposase with the λ repressor and with the DNA-binding domain of the transcription factor Gli1 showed altered insertion profiles in plasmid targets in Escherichia coli and zebrafish embryos (57). A chimeric transposase protein composed of a zinc-finger (ZF) DNA-binding domain from the mouse transcription factor Zif268 fused to the C-terminus of the Synechochystis transposon ISY100 was efficiently targeting transposition into a region adjacent to a Zif268-binding site on a target plasmid (58). Finally, fusions of the polydactyl ZF protein E2C and the SB transposase retained DNA-binding specificity for the E2C-binding site, were capable of mediating bona fide transposition reaction and could direct SB transposition into the vicinity of an E2C-binding site in plasmids, but not in the genome (59).
We performed a large-scale, genome-wide, comparative analysis of target site selection properties of SB, PB and Tol2 transposons, and evaluated the potential of transposase fusion proteins to target transposition in the human genome. We aimed to mimic the site-specific integration ability of AAV and to target transposon integrations near genomic Rep-binding sites. We considered three molecular strategies, all based on the specific RRS DNA-binding activity of the AAV Rep proteins. Based on the binary nature of the transposon vector systems, tethering of the transpositional complex could be achieved by interactions with either the transposase enzyme or the transposon DNA. The first targeting strategy involves a fusion of the site-specific DNA-binding domain of Rep to the transposase polypeptide (Figure 1B). In this setup the transposase fusion protein is predicted to bind site-selectively to a Rep-binding sequence in the genome, thereby tethering the transposition complex to this region and to mediate transposon integration into nearby sites. The second, more indirect, approach was to use the Rep DNA-binding domain that simultaneously binds RRS sites engineered into the transposon vectors and RRS target sites in the human genome, thereby mimicking the locus-specific insertion of AAV (Figure 1C). The third strategy (only applicable to the SB system) relied on non-covalent binding activities of a third protein. This protein combines site-selective DNA-binding functions with the ability to form protein–protein interactions with the SB transposase. The protein acts as a bridge or adapter that is designed to target the transpositional complex to endogenous RRS motifs in the genome through protein–protein interactions with the SB transposase polypeptide (Figure 1D). A series of hybrid proteins consisting of Rep and the SB, PB or Tol2 transposases were engineered, and their activities in the respective transposition reactions were confirmed. We also tested the DNA-binding activities of the Rep/transposase fusions and performed a plasmid-based transposition assay to assess site-directed transposon integration. Finally, we used Illumina sequencing and chromosomal mapping of transposon insertion sites to determine overall target site distribution of the three transposon systems as well as enrichment of insertions close to genomic RRS sequences. We provide proof-of-principle that fusion protein-mediated tethering can effectively redirect transposon insertion site selection in human cells.
MATERIALS AND METHODS
Plasmid constructs
The AAV Rep DNA-binding domain originated from a pcDNA3.1/Myc-His (Invitrogen, CarlSBad, CA) expression plasmid containing the amino-terminal 244 residues of the AAV Rep78 protein (pcDNARepTZ) (60). The SB expression plasmids were produced by PCR amplification of the AAV Rep DNA-binding domain and, if appropriate, together with a GCN4-based wild-type leucine zipper domain (LZ) or an engineered GCN4-based leucine zipper oligomerization domain (TZ). The Rep DNA-binding domain was then ligated in frame to the N-terminus of the hyperactive SB transposase M3a present in a pEGFP-C1 (Clontech) expression vector. The RepTZ/N57 expression plasmid was made by PCR amplification of the 56 amino-acids (excluding the initiator methionine) N-terminal helix–turn–helix domain of the SB transposase and subsequently insertion into the pcDNARepTZ vector creating pcDNARepTZ/N57. The Rep/N57 expression plasmid was constructed by replacing the RepTZ fragment with a Rep fragment in pcDNARepTZ/N57. The effector plasmids were generated by inserting Rep/N57 and RepTZ/N57 into a herpes simplex virus VP16 transcriptional activation domain (AD) containing expression plasmid, yielding Rep/N57-AD and RepTZ/N57-AD. The HA–ADRep/N57 and HA–ADRepTZ/N57 expression plasmids were generated by inserting Rep/N57 and RepTZ/N57 into a pRK5–AD expression vector. The PB fusion proteins were produced by PCR amplification of the PB coding sequence from a PB ORF helper plasmid and subsequent insertion into the Rep expression vector. RepTZL/PB was generated by introducing a double-stranded oligonucleotide coding for a flexible linker (KLGGGAPAVGGGPK) in between the RepTZ and the PB transposase sequence of RepTZ/PB. The Tol2 fusion vector was generated by PCR amplification of the Tol2 transposase coding sequence and subsequent insertion into the pcDNARepTZ expression plasmid. With the incorporation of the double-stranded oligonucleotide flexible linker (KLGGGAPAVGGGPK) in between Rep and the Tol2 transposase the RepL/Tol2 expression plasmid was generated. The SB transposon donor plasmid pTneoRRS was generated by inserting a double-stranded oligonucleotide containing a single repeat of the RRS into the pTneo plasmid (29). In order to generate the pXLSV40neoRRS PB donor plasmid a set of two anti-parallel oligonucleotides containing a single copy of the RRS motif were inserted into a neomycin containing PB transposon vector (53). The pTol2neoRRS donor vector was generated by inserting a double-stranded RRS fragment into the pTol2neo transposon vector (53).
Transposition and western blot analysis
The transposition assay was used to assess the integration capabilities of the fusion transposases. A total of 5 × 105 HeLa cells were transfected with 500 ng of a neomycin containing transposon donor plasmid and 50 ng of a fusion transposase, unmodified transposase or green fluorescent protein (GFP) expression plasmid using the JetPEI™ transfection reagent (Polyplus transfection) according to the manufacturer’s instructions. Two days post-transfection cells were seeded on 10-cm diameter dishes, and selected in DMEM supplemented with 600 µg/ml G418 (Biochrom). Fourteen days later selection was terminated by fixing and staining the neomycin resistant colonies. To determine the relative transposition efficiency, the number of G418-resistant colonies on each plate were counted and compared to the number of G418-resistant colonies obtained with the wild-type transposase. The expression of the fusion proteins was tested by western blot analysis. HeLa cells were transfected with 1 µg of fusion transposases expression plasmids or GFP expression plasmid as a control. Two days later cells were lysed in protein lysis buffer supplemented with protease inhibitor mixture (Roche) and proteins were extracted. Equal concentrations of proteins were separated on a 10% polyacrylamide gel, transferred onto nitrocellulose membranes and incubated with goat polyclonal anti-SB transposase antibody (R&D Systems) or an anti-HA high-affinity antibody (Roche). The bound antibodies were visualized by chemiluminescence (ECL Plus western blotting detection system) and signals were captured on a film.
PCR-based excision assay
The PCR-based excision assay was performed in order to monitor the transposon excision activity of the transposase chimeras in human cells. HeLa cells were transfected with 500 ng of a neomycin resistance gene-containing transposon donor plasmid and 50 ng of a fusion transposase, a wild-type transposase or an expression plasmid for the GFP. Two days post-transfection cells were harvested, low molecular weight DNA was isolated and used as template for nested PCR reactions. Primer pairs flanking the transposon donor sites enable the amplification of transposon footprint products that are generated after transposon excision and repair of the excision site.
Luciferase reporter assay
The RRS-binding capabilities of the transposase fusion proteins were tested in a luciferase-reporter assay. Triplicates of 5 × 105 HeLa cells were each transfected with 50 ng of the pRRStkLuc reporter plasmid, 15 ng of the pcDNARepTZ–AD activator plasmid, 935 ng of fused or unfused transposase expression plasmid and 100 ng of pCMV-βgal (Clonetech) as an internal control for the transfection efficiency. Two days post-transfection, cells were prepared and analysed on a luminometer following the instructions of the Dual-Luciferase® Reporter Assay System protocol (Promega). The measured luciferase activities were related to the total amount of protein in the lysates.
Co-immunoprecipitation
Cultured HeLa cells were plated into 10-cm diameter dishes and transfected with 3 µg of plasmids encoding HA-tagged, untagged or Myc-tagged protein. Forty-eight post-transfection cells were lysed in 1 ml of protein lysis buffer supplemented with protease inhibitor mixture (Roche) and incubated on a shaker at 4°C for 15 min. All of the procedures were done at 4°C. The lysates were pre-clarified with 20 µl of protein G-agarose (Sigma) and subsequently incubated with 40 µl of EZview™ Red Anti-HA Affinity Gel (Sigma). The captured immunocomplexes were treated according to the manufacturers instruction and analysed by immunoblotting using either a polyclonal goat anti-SB transposase antibody (R&D systems), a monoclonal rat anti-HA high affinity antibody (Roche) or a polyclonal rabbit anti-Myc antibody (Abcam).
Inter-plasmid transposition assay
The targeting potential of the fusion proteins was determined by analysing the integration site distribution of a kanamycin-marked transposon into pre-defined target DNA molecules. We transfected 5 × 105 HeLa cells with 200 ng of a kanamycin donor plasmid, 500 ng of either pFVLuc or pFVLuc5xRRS target plasmid and 320 ng of fusion transposase expression plasmid. For the inter-plasmid transposition assay performed with the N57 targeting protein the cells were transfected with 100 ng of the kanamycin-marked donor plasmid, 250 ng of pFVLuc or pFVLuc5xRRS target plasmid, 20 ng of SB helper plasmid and 500 ng of either Rep/N57 or RepTZ/N57 fusion protein expression plasmid. Two days post-transfection the cells were lysed and the low molecular weight DNA was extracted using the Hirt method. The plasmid DNA was electroporated into ElectroMAX DH10B competent E. coli cells (Invitrogen) according to the manufacturer’s instructions. The transformation mix was plated on an LB agar plate containing 100 µg/ml ampicillin and 25 µg/ml kanamycin. Plasmid DNA samples isolated from colonies growing on amp/kan plates were pre-screened by diagnostic restriction digests, and further analysed by DNA sequencing using transposon-specific primers.
Construction of integration site libraries
In order to assess the chromosomal targeting capabilities of the fusion transposases integration site libraries were generated by transfecting 1.5 × 105 HeLa with 90 ng neomycin resistance gene-containing transposon plasmid together with 9 ng of fusion or unfused transposase expression plasmid. In transfection experiments with one of the targeting constructs that do not have transposase capabilities, HeLa cell were transfected with 90 ng of transposon plasmid, 9 ng of a transposase expression plasmid and 450 ng of a targeting fusion protein expression plasmid. Two days post-transfection cells were divided to several 10-cm diameter dishes and set on G418 selection over a period of 2 weeks. At this point, approximately 10 000 G418-resistant colonies were pooled together and used to isolate genomic DNA. The isolated DNA samples were randomly fragmented by sonication and sequences neighboring the insertion sites were amplified by a modified linear amplification-mediated PCR (LAM–PCR) (55,61). Briefly, vector-genome junctions were pre-amplified in a linear PCR with primer specific for the transposon IRs, the PCR products were double-stranded and ligated to linkers with known sequences. The DNA fragments consisting of a linker cassette-, genomic- and transposon-sequences were then subjected to nested PCRs that amplify the vector genome junctions with primer that anneal to the transposon IRs and to the linker. During the amplification we used barcoded primer that allowed us to pool the libraries. Finally, the samples were prepared for high-throughput sequencing by an additional PCR adding the Illumina-specific adaptor sequences required for the solid-phase bridge amplification. The PCR products were purified, pooled and then subjected to high throughput sequencing on the Illumina/Solexa Genome Analyser IIx platform with single end run settings. Primer and linker sequences are specified in Supplementary Methods.
Computational analysis
We used a set of error-correcting barcodes for distinguishing different data sets pooled into a single Illumina Genome Analyser IIx flow cell lane. The 76-bp long sequencing reads were checked for starting exactly with the barcode (4 bp) followed by the adapter sequence (23 bp for SB, 24 bp for PB and 22-bp for Tol2). The adapter for SB ended with ‘TA’ and the adapter for PB with ‘TTAA’ indicating transposase-mediated transposition events as opposed to random integration. The remaining parts of the reads (between 48 and 50 bp) were mapped using Bowtie (62) to the chromosomes of the human genome (NCBI built GRCh37/hg19, February 2009) excluding the Y-chromosome because HeLa is a female cell line. Only reads occurring without mismatches at a single genomic position (exact uniquely mapped reads) were used for the subsequent analysis. Multiple reads mapping to the same genomic position and strand were treated as a single integration site. To improve the data quality, we discarded all sites supported by <10 reads. In case of the SB data sets, we only accepted sites having ‘TA’ adjacent to the integration site. In case of the PB data sets, we only accepted sites having ‘TTAA’ adjacent to the integration site. For the statistical analysis, we generated three control sets, each containing 10 000 sites, which were randomly selected from inside of contigs on human chromosomes excluding the Y-chromosome. For the SB control set, only control sites adjacent to ‘TA’ were selected, and for the PB control set, only control sites adjacent to ‘TTAA’ were selected. Databases of genomic regions (RefSeq) were downloaded from UCSC (http://genome.ucsc.edu). We used the H3K27me3 domains determined by Cuddapah et al. (63) (GEO accession number GSM325898). Regions enriched for H3K4me1 and H3K4me3 were determined as follows: The raw ChIP-Seq reads by Robertson et al. (64) (http://www.bcgsc.ca/data/histone-modification) were mapped to the human genome using Bowtie, then peaks were called using MACS (65), and H3K4me1/3 domains are then defined as 5-kb windows around the centers of the peaks. The search for RRS motifs was done by a custom C++ program.
RESULTS
Comparative analysis of target site selection of SB, PB and Tol2 transposons in human cells
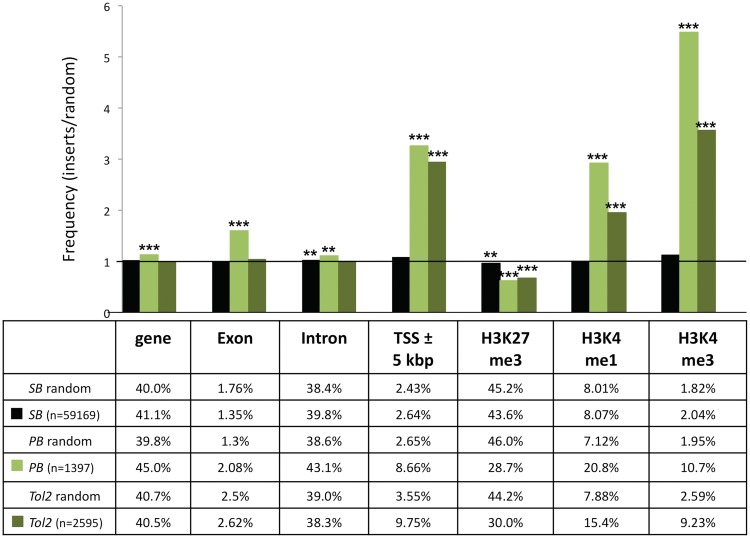
We generated integration site libraries for the SB, PB and Tol2 transposons by transfecting cultured HeLa cells with neo-marked transposon donor plasmids and transposase expression plasmids. Approximately 10 000 antibiotic-resistant colonies were pooled together and used to extract genomic DNA. To recover genomic DNA flanking the transposon integration sites, we performed a modified LAM–PCR (55,61). In total we analysed 59 169 SB, 1397 PB and 2595 Tol2 genomic integration sites. Integration sites were mapped onto the human genome, and local features at the integration sites were quantified. The three integration site data sets were analysed in relation to various chromosomal features, and compared to 10 000 computer-simulated random integrations. The random control sites were matched to the experimental sites with respect to the integration frequencies into genes, exons, introns, regions around transcription start sites (5-kb regions surrounding transcriptional start sites; 5-kb ± TSS), and sites of histone modifications. We concentrated on analysing transposon integration frequencies with regard to H3K4 mono-and trimethylation, a marker for active promoter (H3K4me3) and enhancer regions (H3K4me1) associated with open chromatin (66), and trimethylated H3 K27 (H3K27me3), a marker for condensed chromatin regions associated with gene repression (67). The bioinformatic analysis revealed a random integration profile for SB with no apparent bias for integrating into genes or near transcriptional regulatory regions of genes (Figure 2). In contrast, PB showed a non-random integration profile with significant bias towards integrating into genes, transcription start sites and their upstream regions (Figure 2). The Tol2 transposon showed an intermediate integration profile with no bias for inserting into genes, but preferential integration into transcription start sites and transcriptional regulatory regions (Figure 2). The biased integration of PB and Tol2 transposons into transcriptional start sites correlated with significant enrichment in chromatin regions characterized by H3K4me3 and H3K4me1 associated with open chromatin (Figure 2). In sum, the three transposon systems displayed different preferences for local features of the target DNA, with SB being the closest to showing a fully random integration profile in the human genome.
Figure 2.
Comparison of integration site selection of the SB, PB and Tol2 transposon systems. The values are presented as the percentage of integration events divided by the percentage of random events. The horizontal line at value 1 represents the expected random distribution. The statistical significance of differences from the random control is shown by asterisks above the bars *P ≤ 0.05, **P ≤ 0.01 and ***P ≤ 0.001 (using Fisher’s exact test). Depicted are the integration frequencies within genes, exons, introns, into 5-kb windows surrounding transcription start sites (TSS ± 5-kb) and near sites of the H3K27me3, H3K4me1 and H3K4me3 histone modifications.
Design and transpositional activities of Rep/SB fusion proteins
The subsequent aim of the study was to introduce a measurable bias into the target site selection properties of the transposon systems by targeting transposon integrations to predetermined chromosomal regions. We chose to investigate target-selected transposon insertion by mimicking the site-specific integration mechanism of AAV. All targeting strategies were based on the specific binding of the AAV Rep proteins to a 16-bp imperfect GAGC repeating motif called RRS. The fusion transposases comprised the 244 amino acid N-terminal DNA-binding domain of the AAV Rep protein, sufficient for specific binding to the RRS sequence (60), and hereafter designated as Rep.
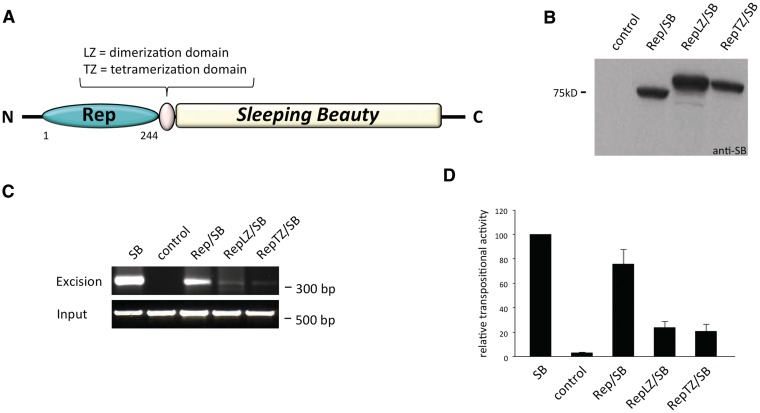
Figure 3A shows a sketch of the Rep/SB fusion protein design. The monomeric form of the Rep protein does not efficiently bind to DNA, whereas addition of oligomerization motifs that induce the formation of multimers were previously found to strongly enhance RRS-binding activity (60). Fusion proteins containing the SB transposase fused to Rep either directly or with a dimerization domain of the yeast transcription factor GCN4 (LZ) or with a mutant GCN4 leucine zipper (TZ) predicted to assemble into a parallel tetramer (68) were engineered. All fusion constructs were cloned under the transcriptional control of the cytomegalovirus (CMV) promoter and expression was confirmed by western blot analysis (Figure 3B). The excision activities of the chimeric transposases were tested in a plasmid-based excision assay, during which the transposase excision activity is assayed by polymerase chain reaction (PCR)-amplification of the footprint that is left behind after transposon excision from a donor plasmid (69). All three SB fusion transposases showed detectable excision activity. Even though the PCR assay is not designed to be quantitative, it is useful for probing the overall potencies of transposases in the excision reaction. Indeed, whereas the Rep/SB transposase showed a transposon excision activity close to that of the wild-type, unfused SB transposase, RepLZ/SB and RepTZ/SB displayed only weak excision activities (Figure 3C). The relative activities of genomic integration catalysed by each fusion protein were assessed by a standard colony-formation assay that is based on acquisition of an antibiotic-resistant phenotype as a result of stable genomic insertion of genetically tagged transposon constructs. In line with the excision data, the Rep/SB fusion retained a high transpositional activity of ∼80% of the wild-type level, whereas the transpositional activities of RepLZ/SB and RepTZ/SB were at only 20% of the wild-type level (Figure 3D).
Figure 3.
Design, expression and transpositional activities of Rep/SB fusion proteins. (A) Schematic representation of Rep/SB fusion transposases consisting of the 244 amino acid N-terminal DNA-binding domain of the AAV Rep protein fused to the SB transposase. The RepLZ/SB and RepTZ/SB fusions contain a GCN4 wild-type (LZ) or modified leucine zipper (TZ) linker domain. (B) Western blot analysis of chimeric transposase proteins detected with a polyclonal antibody against the SB transposase (anti-SB). The position of a molecular size marker is indicated on the left. (C) Excision activities of chimeric transposases. HeLa cells were co-transfected with a neomycin-marked (neoR) transposon plasmid together with an expression plasmid producing either a transposase fusion, unfused transposase as positive control (SB) or the GFP as negative control (control). The agarose gels show the electrophoretic separation of the 300-bp PCR product diagnostic of transposon excision. The lower gel photograph shows PCR amplification products of a segment of the neoR-marked transposon plasmids that served as input controls. The positions of the size markers are indicated on the right. (D) Transpositional activities of chimeric transposases. HeLa cells were co-transfected with a transposon donor plasmid carrying a selectable neoR gene together with either a transposase-expressing helper plasmid or with a plasmid expressing GFP (control), and cells were placed under G418 selection. Resistant colonies were fixed, stained and counted. The ratio of colony numbers in the presence versus in the absence of the transposase is a measure of transposition efficiency. Average relative transpositional activities obtained from three independent transfections are shown (mean ± SD).
RRS-specific DNA-binding of the Rep/SB fusion proteins
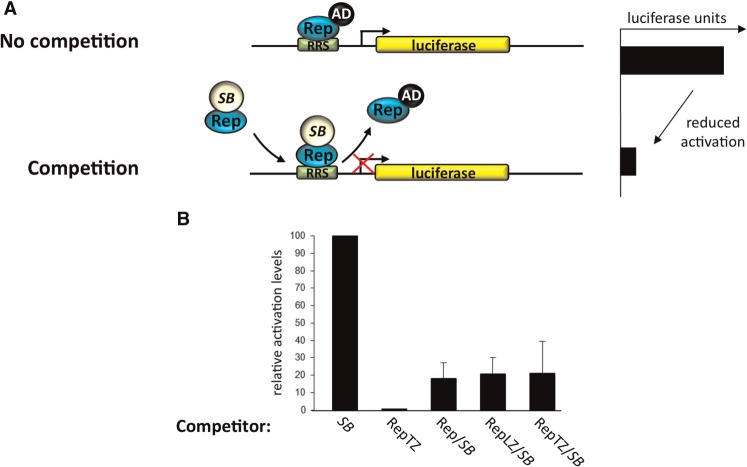
The transposon excision and re-integration assays demonstrated that the SB fusion proteins retained the ability to catalyse authentic DNA transposition in human cells, albeit at reduced frequencies. Thus, by definition, the transposase hybrid proteins were still able to bind the transposon end sequences despite the presence of the large Rep DNA-binding domain at their N-termini. The abilities of the Rep/SB fusion proteins to recognize and specifically bind to RRS sequences were examined in a mammalian one-hybrid reporter assay that measures the abilities of the SB fusion proteins to reduce the level of luciferase reporter gene activation via competitive DNA-binding (Figure 4A).
Figure 4.
DNA-binding activities of transposase fusion proteins. (A) Schematic representation of the competition assay performed to monitor the RRS-specific DNA-binding activity of transposase fusion proteins in human cells. HeLa cells were transfected with a reporter plasmid containing a luciferase gene downstream of an RRS site and a minimal thymidine kinase promoter together with limiting amounts of a plasmid expressing the RepTZ-AD activator protein. Cells also received an excess of plasmids encoding the transposase fusion proteins, the RepTZ protein as positive and the respective unmodified transposase as negative control. The chimeric transposase proteins and the control proteins were tested for their abilities to compete in protein binding at the target site, and to thereby reduce the level of luciferase trans-activation in the cell. (B) Luciferase reporter assays performed with the SB fusion transposases. Each graph displays luciferase activation levels relative to transfection with the respective unmodified transposase. Bars represent the average (mean ± SD) obtained from three independent transfection experiments.
Cultured HeLa cells were transfected with a luciferase reporter plasmid having the RRS motif cloned in front of a minimal promoter (pRRStkLuc) together with limiting amounts of activator plasmids expressing Rep fused to the herpes simplex virus VP16 transcriptional activation domain (RepTZ–AD) and an excess of plasmids expressing the corresponding SB fusion transposases. The unfused SB protein had no RRS-binding activity, and therefore did not compete with RepTZ–AD, whereas the addition of excess amounts of RepTZ (lacking AD) showed significant competition in RRS binding, and decreased luciferase activation to background levels (Figure 4B). Upon expression of either Rep/SB, RepLZ/SB or RepTZ/SB a reduction of ∼80% in reporter gene activation level was detected, showing that all SB transposase fusion proteins were able to compete with the Rep protein in RRS-binding (Figure 4B). Apparently, and unexpectedly, the presence of the LZ and TZ multimerization domains in the RepLZ/SB or RepTZ/SB fusions did not have a major effect on the RRS-binding activities of the Rep/transposase fusions. First, it could be that the transposase partner in the fusions had a negative effect on the functions of the LZ and TZ multimerization domains. Second, because Rep does not bind the RRS as a monomer, it is conceivable that multimerization of Rep/SB was driven by the SB transposase that is believed to function as a tetramer (28), which in turn favoured binding to the RRS. Taken together, all fusion transposases were competent in transposition as well as in binding to the RRS sequence, suggesting that these fusion proteins might prove useful in redirecting transposon insertions in human cells.
DNA–protein and protein–protein interactions of Rep fusions with the N57 domain of the SB transposase
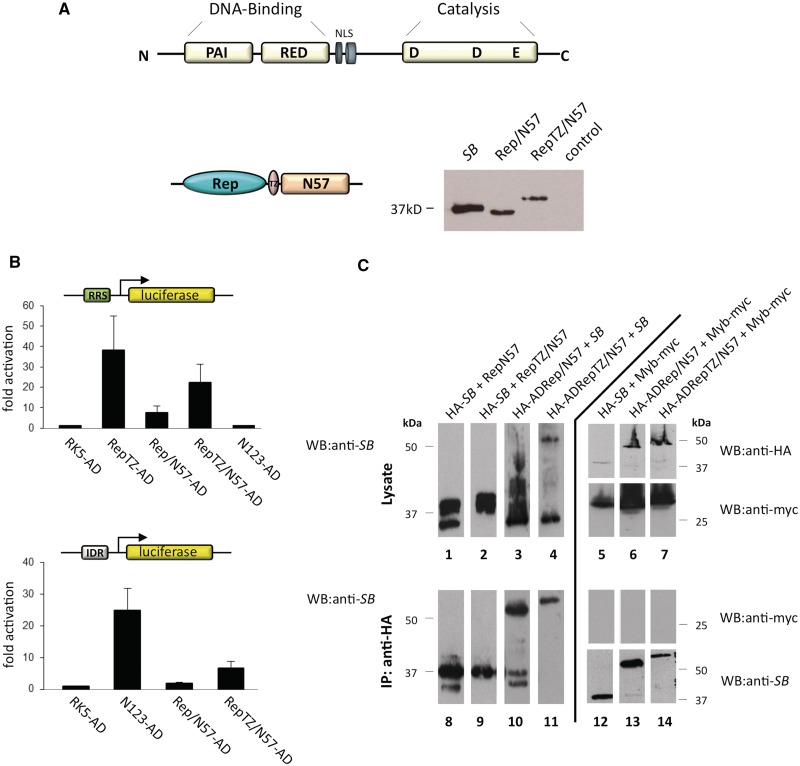
Several, naturally occurring transposable elements evolved strategies to target their insertion into defined sites using protein–protein interactions between a transposon-encoded factor and a cellular DNA-binding factor (70). Adapting such a strategy for targeting transposition requires the identification of proteins that interact with the transposase. Such an interacting protein was previously reported for the SB transposase. The DNA-binding region of the SB transposase consists of two helix–turn–helix subdomains (PAI + RED = PAIRED) (Figure 5A). The N-terminal 57 amino acids (N57) of the transposase encompassing the PAI subdomain of the PAIRED complex mediates specific interactions with the transposase-binding sites within the transposon IRs as well as protein–protein interactions between transposase subunits (28). Furthermore, a fusion protein consisting of N57 and the tetracycline repressor was previously shown to efficiently target SB integration in ∼10% of transposant cells into a chromosomally integrated tetracycline response element in cultured human cells (71).
Figure 5.
Design, expression and binding activities of the chimeric N57 fusion proteins. (A) Structure of the SB transposase. The transposase has an N-terminal paired-like DNA-binding domain consisting of two helix–turn–helix motifs, PAI and RED, and a nuclear localization signal (NLS) followed by the catalytic domain responsible for the DNA cleavage and joining reactions characterized by the conserved DDE signature. The targeting fusion proteins consist of the AAV Rep DNA-binding domain (Rep) and the N-terminal interaction domain of the SB transposase (N57). RepTZ/N57 additionally contained the modified leucine zipper oligomerization domain (TZ). Expression of the N57 fusion proteins was examined by western blot experiments using a polyclonal antibody against the SB transposase. The position of the molecular size marker is indicated on the left. (B) One-hybrid assays for monitoring RRS- and IR-binding of N57 fusion proteins. The reporter plasmid for Rep binding contained a luciferase gene downstream of an RRS motif, whereas the reporter plasmid for IR binding contained a luciferase gene downstream of a 5-mer repeat of the SB transposase-binding site. Each graph displays luciferase activity relative to transfection with the empty AD vector (pRK5–AD). Bars represent the average (mean ± SD) obtained from three independent transfection experiments. (C) Physical interactions between the SB transposase and the N57 fusion proteins. Interactions between the SB transposase and the Rep/N57 or RepTZ/N57 fusion proteins were examined by co-immunoprecipitation experiments. The HA-tagged proteins were precipitated with an anti-HA antibody and the immunoprecipitated proteins (IPs) were analysed for the presence of the Rep/N57 and RepTZ/N57 or the SB transposase proteins by western blot (WB) analysis using a polyclonal anti-SB antibody. Transfection of Myb-myc served as negative control, for which lysates and IPs were analysed by WB with an anti-myc antibody. The positions of the molecular size markers are indicated on the right.
We generated fusion transposases consisting of N57 and Rep with or without TZ, and confirmed their expression by western blotting (Figure 5A). Active tethering of the integration complex through N57 fusion proteins requires RRS-specific-binding activity of the Rep domain as well as the N57 domain mediating either specific binding to the transposon IRs or protein–protein interaction with the SB transposase. The DNA-binding activities of the N57 fusion proteins were tested in a one-hybrid luciferase assay using two different reporters. HeLa cells were co-transfected with the RRS-containing reporter plasmid pRRStkLuc and an AD-containing expression plasmid. The expression of RepTZ–AD resulted in significant (>35-fold) luciferase gene activation as compared to values obtained with AD domain only (Figure 5B, upper panel). Upon expression of Rep/N57 and RepTZ/N57, a 7-fold and a 22-fold increase in luciferase gene activation was observed, respectively (Figure 5B, upper panel). We included the entire N-terminal DNA-binding domain including the multimerization domain of the SB transposase consisting of 123 amino acids fused to AD (N123-AD) as a further negative control in the experiments. Similar to AD domain only, N123-AD showed background levels of luciferase transactivation on the pRRStkLuc reporter (Figure 5B, upper panel), indicating that the transposase domain does not interact with the pRRStkLuc plasmid in a non-specific manner. Binding activities of the N57 fusion proteins to the IRs of the SB transposon were tested with co-transfection of a reporter construct having five SB transposase-binding sites cloned in front of a minimal reporter (pIDRLuc) together with either of the N57 effector constructs (Figure 5B, lower panel). The strongest luciferase activation was observed with N123-AD, consistent with previous studies (Figure 5B, lower panel) (59). Expression of Rep/N57 and RepTZ/N57 led to an increase in luciferase activation of ∼1.8-fold and 6-fold relative to the background level, respectively (Figure 5B, lower panel). In sum, based on efficient activation of reporter gene expression, we conclude that both chimeric proteins were able to bind to the RRS as well as to the transposon IRs.
Since targeting of the transposition complex could not only be achieved by binding to the transposon DNA but also through interaction with the SB transposase protein, we examined possible physical interactions between N57 fusion proteins and the SB transposase by co-immunoprecipitation experiments. This assay was performed in HeLa cells by co-expressing a haemagglutinin-tagged SB transposase (HA–SB) together with either of the N57 fusion proteins. The SB transposase and associated proteins were immunoprecipitated using an anti-HA antibody, and subsequently subjected to immunoblotting with a polyclonal anti-SB antibody to examine the presence of the N57 chimeras. Hybridization of the anti-SB antibody to the HA-tagged SB as well as to the Rep/N57 and RepTZ/N57 validated their expression in co-transfected HeLa cells (Figure 5C, lanes 1 and 2). Immunoprecipitation with an anti-HA antibody showed co-precipitation of SB with Rep/N57 (Figure 5C, lane 8), but not with RepTZ/N57 (Figure 5C, lane 9). These experiments indicated a protein–protein interaction between the SB transposase and the Rep/N57 fusion protein but not between SB and RepTZ/N57. The HA–SB and RepTZ/N57 proteins have similar molecular weights, and thus were difficult to separate by gel electrophoresis. We therefore repeated the co-immunoprecipitation assays with untagged SB transposase and HA-tagged fusion proteins containing N57 fused to a VP16 AD (HA–ADRep/N57 and HA–ADRepTZ/N57). As before, immunoprecipitation experiments revealed co-precipitation of SB with Rep/N57 (Figure 5C, lane 10), but not with RepTZ/N57 (Figure 5C, lane 11). To ensure that the detected interactions were specific, we performed co-immunoprecipitations with protein lysates from cells transfected with either SB, Rep/N57 or RepTZ/N57 and an unrelated Myc-tagged protein of the Harbinger transposon system (Myb-myc) (72). As expected, immunoprecipitation with an anti-HA antibody revealed the presence of proteins interacting with an anti-SB antibody and the absence of proteins interacting with an anti-myc antibody (Figure 5C, lanes 12–14).
In sum, the one-hybrid DNA-binding assays showed that both N57 chimeras retained their abilities to bind to the RRS motif and to the transposon IRs. The co-immunoprecipitation assays showed protein–protein interactions between SB and Rep/N57 but not between the SB transposase and the RepTZ/N57 fusion protein.
Inter-plasmid targeted transposition
We next tested whether DNA tethering mediated by Rep could alter transposase target site selection. For this we made use of a well-established inter-plasmid transposition assay (54). The assay involves the co-delivery of three different plasmid molecules: (i) a donor plasmid containing a kanamycin-marked (KanR) transposon, (ii) an ampicillin-resistant (AmpR) target plasmid with or without RRS repeats and (iii) a chloramphenicol-resistant helper plasmid expressing the transposase. All three plasmids were co-delivered to HeLa cells and allowed to undergo transposition. The transposase fusions excised the transposons and re-inserted them into the target plasmids resulting in AmpR/KanR double-resistant plasmid molecules, which were isolated and sequenced to determine the integration sites in the target plasmids.
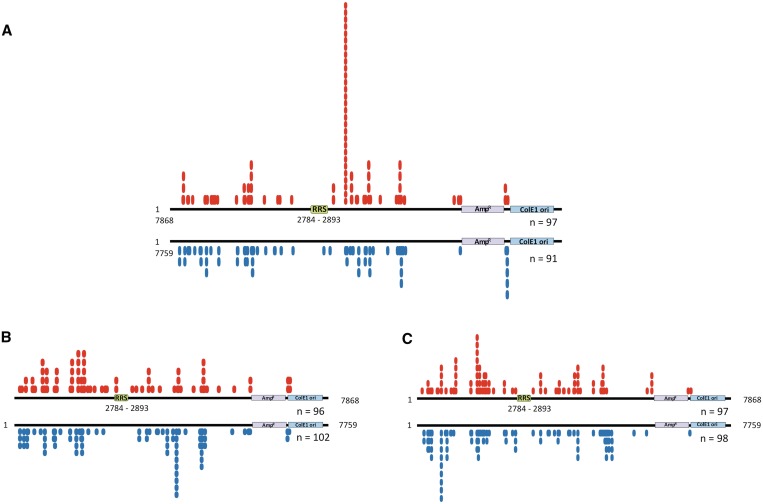
Figure 6 summarizes the results of the inter-plasmid transposition assays performed with the components of the SB transposon system and the Rep/SB fusion transposase as well as the chimeric Rep/N57 and RepTZ/N57 constructs. We mapped 97 Rep/SB-mediated transposition events into the pFVLuc5xRRS target plasmid and 91 integrations into the control plasmid lacking the RRS sites. The target plasmid contained 486 potential TA target sites outside of sequences that are essential for plasmid maintenance. 81 of these sites were used by the Rep/SB transposase. The control experiment revealed an essentially random integration of the donor element at multiple TA target sites, whereas in the presence of the RRS motif Rep/SB-mediated transposition primarily occurred into one particular TA dinucleotide at plasmid position 3583, 700-bp downstream of the RRS (Figure 6A). In total, 29 out of 97 mapped insertions occurred at this TA dinucleotide, representing a 15-fold increase in integration frequency at this particular site as compared to the control (P < 0.001 using Student’s t-test). All of the Rep/SB mediated transposition events into this specific target site were in a 5′–3′ orientation with respect to the RRS motif. The N57 domain of the SB transposase alone is not able to promote the transposition process. For this, we added the wild-type SB transposase promoting excision and insertion of the transposon donor element to the experimental setup. We mapped 97 insertions that occurred into the pFVLuc5xRRS target plasmid in the presence of the Rep/N57 fusion protein, and 98 insertions were recovered from the control experiment (Figure 6B). In total, the 195 transposon integrations occurred into 104 TA dinucleotides distributed over the whole plasmid. We observed some preferred transposon integration sites represented by multiple insertions into particular TA dinucleotides. However, this preference for particular TA sites in the RRS-containing target plasmid was not different from that observed in the control target plasmid (Figure 6B). The inter-plasmid transposition assay with RepTZ/N57 fusion protein generated 96 transposition events in the pFVLuc5xRRS and 102 events in the control plasmid, with integrations having occurred into 81 TA sites. The target site distribution in the RRS-containing target plasmid was almost identical to that observed in the control plasmid lacking the RRS motif (Figure 6C).
Figure 6.
Inter-plasmid transposition assay for investigating site-directed transposition. HeLa cells were transfected with a transposase-encoding plasmid together with a donor plasmid encoding a kanamycin-marked (KanR) transposon and an ampicillin-resistant (AmpR) target plasmid containing five tandem RRS, and allowed to undergo transposition resulting in AmpR/KanR plasmids. Two days post-transfection low-molecular weight plasmid DNA were isolated and transformed into DH10B E. coli bacteria. AmpR/KanR bacterial colonies were amplified, plasmid DNA isolated and the locations of transposon insertion sites were determined by DNA sequencing. The schematic maps of the target plasmids show the locations of the RRS, the AmpR and the ColE1 origin of replication (ColE1ori). The numbers (n) of recovered and mapped integration sites are indicated below the schemes. The red circles represent insertions of the SB donor element into the pFVLuc5xRRS target plasmid. The blue circles indicate integration into the pFVLuc control target plasmid lacking the RRS motif. (A) Distribution of Rep/SB-mediated transposon integration sites. (B) Distribution of SB-mediated transposon integration sites in the presence of the Rep/N57 fusion protein. (C) Distribution of SB-mediated transposon integration sites in the presence of the RepTZ/N57 fusion protein.
In conclusion, the results of the plasmid-based integration assays indicate that the Rep/SB fusion efficiently targeted transposon insertions into a particular TA site, whereas preferred integration into particular TA dinucleotides observed in the presence of the Rep/N57 and RepTZ/N57 fusion proteins were presumably independent from the presence of the targeted RRS sequence in the plasmid.
Biased insertion profile of SB mediated by Rep in human cells
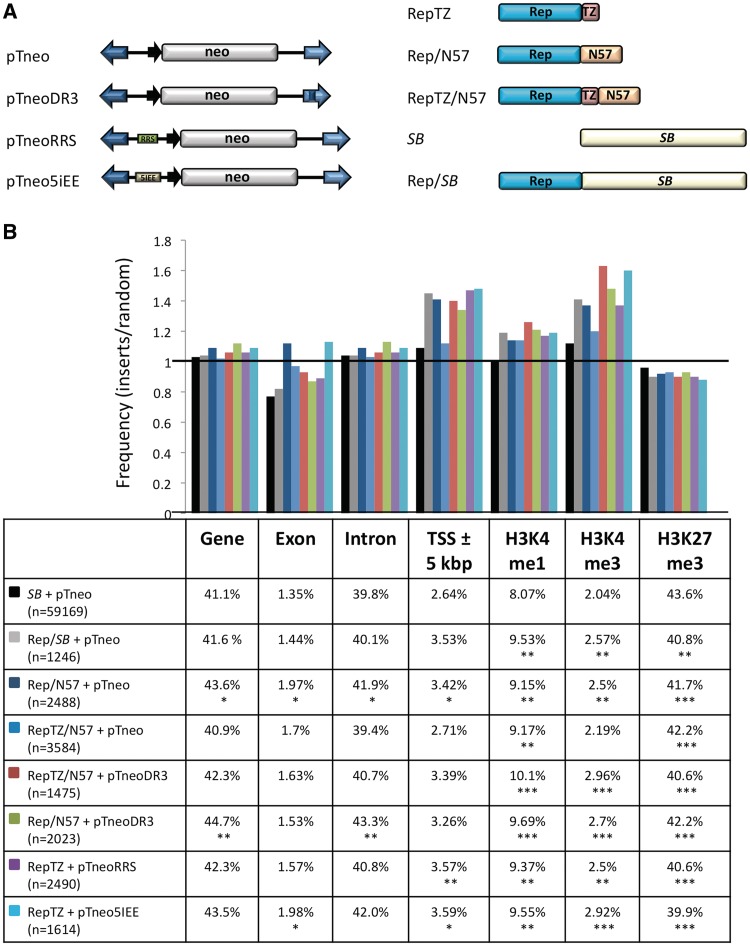
Next we addressed the question whether our Rep-based fusion proteins could target transposon integrations to genomic sites bound by Rep. We also considered to test a targeting strategy that employs modifications of the transposon molecule (Figure 1C). We inserted Rep-binding sequences into the transposon vector (Figure 7A), and used these modified transposons together with an unmodified transposase and the RRS-binding protein RepTZ to target transposition near endogenous Rep-binding sequences. We hypothesized that similar to the process during site-selective integration of AAV, the RepTZ protein could simultaneously bind to the RRS sequence in the transposon and to an RRS site in the genome, thereby guiding the whole transpositional complex into the vicinity of endogenous RRS sites, at which the unmodified transposase could then perform the excision and re-integration of the transposon. On the basis of the neomycin-marked SB transposon pTneo, we designed transposon versions with a single RRS motif (pTneoRRS, Figure 7A) or a 138-bp p5 integration efficiency element (p5IEE) known to enhance Rep-mediated site-specific integration of AAV (pTneop5IEE, Figure 7A) (73). We tested this hypothesis not only with the RepTZ protein, but also with the Rep/N57 and RepTZ/N57 fusion proteins using SB transposons containing additional N57-binding sites in the 3′-IR (pTneoDR3, Figure 7A). All transposon vectors were tested in transposition and excision assays and proved to be fully active (data not shown). There were no differences observed between transposition efficiencies of the original (without RRS) and modified (with RRS) transposons, indicating that the binding of the RepTZ protein did not interfere with transposition.
Figure 7.
Comparison of the integration profiles of unfused SB transposase and Rep/SB chimeras. (A) Schematic representation of the transposon vectors and targeting fusion transposases. (B) Genomic target site distributions. The values are presented as the percentage of integration events divided by the percentage of random events. Random integration has a value of 1 and is marked by a horizontal line. Depicted are the integration frequencies within genes, exons, introns, into 5-kb windows surrounding transcription start sites (TSS ± 5-kb) and near sites of the H3K27me3, H3K4me1 and H3K4me3 histone modifications. The table below the diagram summarizes the percentage of integrations next to the respective genomic annotation. The statistical significance of differences from the unfused SB transposase is shown by asterisks above the bars *P ≤ 0.05, **P ≤ 0.01 and ***P ≤ 0.001.
We generated large numbers of transposon insertion in HeLa cells transfected with the different transposon and transposase components depicted in Figure 7A, recovered the insertions with LAM–PCR as above, mapped the insertions onto the human genome, and quantified local features at the integration sites. The integration site data sets were analysed in relation to integration frequencies into genes, exons, introns, regions around transcription start sites (5-kb regions surrounding transcriptional start sites; 5 kb± TSS), and sites of histone modifications (Figure 7B). Figure 7B presents the frequency of transposon integrations near the indicated genomic features divided by the respective frequencies in the computer-generated random control dataset. As compared to the wild-type SB transposon system Rep/N57 mediated transposition revealed a modest enrichment of integrations into genes (exon and introns) and into regions surrounding transcriptional start sites (Figure 7B). Enhanced targeting of transcriptional start sites was also observed in the presence of the RepTZ protein (Figure 7B). Notably, all targeting datasets showed a preference for integrating next to open chromatin regions characterized by H3K4me1 and H3K4me3 marks, whereas condensed chromatin regions characterized by H3K27me3 marks were avoided (Figure 7B).
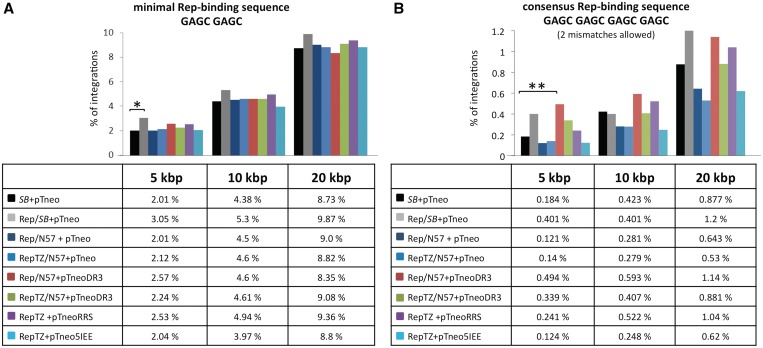
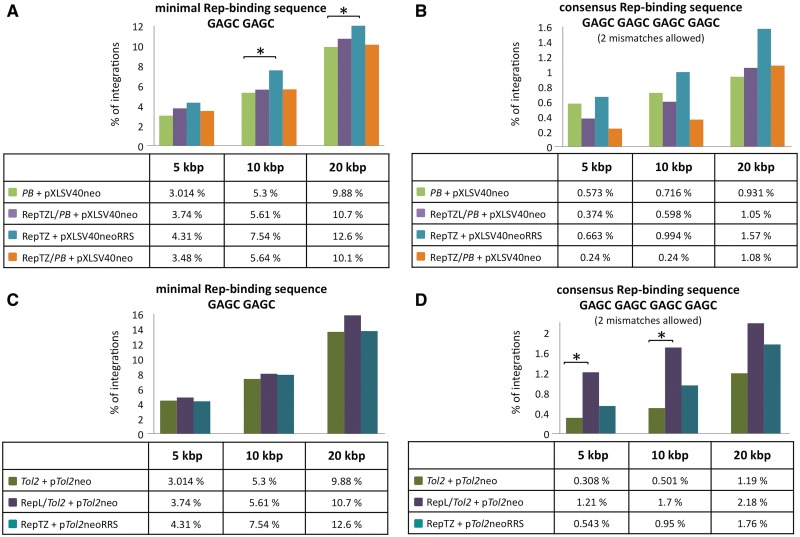
We next tested whether the Rep-based fusion proteins could redirect transposon integration into the vicinity of genomic Rep-binding sites. Since the AAV Rep proteins bind to combinations of two to four GAGC repeats (74,75), we adjusted two different potential Rep-binding site settings: a minimal GAGC GAGC Rep-binding motif (15 726 sites per human genome) and a consensus GAGC GAGC GAGC GAGC Rep-binding motif allowing up to two random mismatches (2134 sites per human genome). All potential Rep-binding sites in the human genome were mapped, and the numbers of integrations that occurred in 5-, 10- and 20-kb windows surrounding the binding sites were determined. The bioinformatic calculations with GAGC GAGC as a minimal Rep-binding motif revealed a modest (P ≤ 0.05) increase of SB transposon integrations into a 5-kb interval in the presence of the Rep/SB fusion transposase (Figure 8A). Our analyses also revealed a statistically significant (P ≤ 0.01), 2.7-fold enrichment of SB transposition events into a 5-kb window around the consensus (GAGC)4 Rep-binding sequence in the presence of the Rep/N57 targeting protein and the modified pTneoDR3 transposon (Figure 8B). The significance of this enrichment disappeared with extended mapping windows (Figure 8B), indicating that the enrichment around the RRSs was indeed dependent on physical interaction of Rep with the consensus binding sequence. Notably, the enrichment was only observed with pTneoDR3 containing an additional N57-binding site within the right IR of the transposon, and not with the canonical pTneo transposon (Figure 8B), suggesting a crucial contribution of N57 binding at the transposon vector to the observed effect. Given the bias of Rep/SB and Rep/N57 to mediate insertions nearby TSSs and into open chromatin regions characterized by H3K4me1 and H3K4me3 marks (Figure 7B) as well as around RSSs (Figure 8) a bioinformatics approach was used to investigate the extent of overlap between the distribution of RSSs and TSSs as well as H3K4me1/3 sites in the human genome. The analysis revealed that the Rep-binding sites are indeed quite enriched in TSS and H3K4me1/3 chromosomal regions (Supplementary Figure S1). This overlap therefore cross-validates the data presented in Figures 7 and 8. In conclusion, we found that the most significant bias towards transposon insertion around genomic RRS sites required the Rep/N57 protein and an engineered transposon with additional N57-binding sites, suggesting the role of multiple protein–DNA interactions in guiding transposon insertions to predetermined sites in the human genome.
Figure 8.
Mapping of SB transposon integrations in proximity to chromosomal Rep-binding motifs. Transposon insertions were recovered from HeLa cells transfected with the components of the targeting system. Sequences neighbouring the insertion sites were analysed with respect to proximity to Rep-binding sites. Calculations were performed with the minimal Rep-binding sequence GAGC GAGC (A) and the consensus Rep-binding sequence GAGC GAGC GAGC GAGC with up to two mismatches (B). Distances were categorized in 5-, 10- and 20-kb windows around putative Rep-binding sites and the percentage of insertions obtained in the presence and absence of the targeting fusion proteins in each category are shown. The percentage of integrations that occurred in the designated windows is represented as bars. The results of statistical tests comparing the distribution of integration sites generated by the targeting proteins to those generated by unfused SB are summarized as asterisks on top of each bar. P-values > 0.05 were not considered statistically significant. *P-values < 0.05 were considered significant and **P-values < 0.01 were considered highly significant.
Activities of PB and Tol2 transposons
We used the same experimental strategy as above to test the targeting capabilities of the two other vertebrate transposon systems: PB and Tol2. In accordance with previous findings suggesting that the PB transposase tolerates protein additions (76), all Rep/PB fusion transposases retained efficient transpositional activities (Supplementary Figure S2), and efficient RRS-binding activity (Supplementary Figure S3A). A modified PB transposon carrying an RRS sequence together with the RepTZ protein displayed a weak enrichment of transposition events into relatively large (>10-kb) windows around the minimal Rep-binding motif (Figure 9A), but none of the Rep/PB chimeras was able to redirect PB transposon insertion into regions adjacent to genomic consensus Rep-binding sites (Figure 9B).
Figure 9.
Mapping of PB and Tol2 transposon integrations in proximity to chromosomal Rep-binding motifs. Transposon insertions (A and B, PB; C and D, Tol2) were recovered from HeLa cells transfected with components of the Rep-based targeting systems. Sequences neighbouring the insertion sites were analysed with respect to proximity to the next Rep-binding site. Calculations were performed with the following putative Rep-binding sites: (A and C) minimal Rep-binding site GAGC GAGC and (B and D) consensus Rep-binding site GAGC GAGC GAGC GAGC with up to two mismatches. Distances were categorized in 5-, 10- and 20-kb windows around putative Rep-binding sites and the percentage of insertions obtained in the presence and absence of the targeting fusion proteins in each category are shown. The levels of significance are marked by asterisks. P-values > 0.05 were not considered statistically significant. P-values < 0.05 (*) were considered significant. The PB data set was composed of 1397 integration sites generated with the unmodified PB transposase (PB + pXLSV40neo), 834 integration sites generated with the RepTZ/PB chimeric transposase (RepTZ/PB + pXLSV40neo) and 1207 integration sites generated with the RRS-containing PB transposon in the presence of the Rep DNA-binding protein (RepTZ + pXLSV40neoRRS). The Tol2 integration site analyses comprised 2595 integrations generated with the unmodified Tol2 transposase (Tol2 + pTol2neo), 412 integrations generated with the chimeric RepL/Tol2 transposases and 737 integrations generated in the presence of the Rep DNA-binding protein and a RRS-containing Tol2 transposon (Rep/TZ + pTol2neoRRS).
The two Rep/Tol2 fusions showed very weak transpositional activities (<10% of the activity of unfused Tol2 transposase (Supplementary Figure S4), but displayed binding to the RRS motif (Supplementary Figure S3B). There was no significant enrichment around the minimal Rep-binding motif seen in any of the datasets (Figure 9C). However, with the RepL/Tol2 fusion transposase we observed a ∼4-fold increase of Tol2 transposon integrations into 5- and 10-kb windows around consensus Rep-binding motifs (Figure 9D). Thus, these alternative transposon systems can also be potentially engineered for targeted transposition in particular applications.
DISCUSSION
Recent developments in transposon-based technologies underscore the emerging potential of transposons in gene therapy applications and induced pluripotent stem cell generation for regenerative medicine. The SB transposon system demonstrated effective gene delivery and sustained transgene expression in a variety of vertebrate organisms including humans. SB combines the integrating abilities of viral gene therapy vectors needed for stable and long-lasting transgene expression with the advantageous properties of easy production, simpler handling and potentially safer chromosomal integration profiles. Studies on site-directed integration using fusion transposases may lead to a new approach for inserting exogenous genes at specific sites and improve the therapeutic application of transposon-derived gene therapy vectors. The integration of a transposon to pre-defined sites in the genome would ensure appropriate expression of the transgene (lack of position effects) and simultaneously prevent hazardous effects to the organism due to insertional mutagenesis of cellular genes (lack of genotoxicity).
We performed a large-scale, genome-wide integration site analysis evaluating the preferences of the SB transposon and two other transposon systems, PB and Tol2, for their propensities to integrate into genes, transcription start sites and near histone modifications. The bioinformatic analysis revealed distinct differences between the three transposon systems (Figure 2). SB had a random integration profile with no apparent bias for integrating into genes or near transcriptional regulatory regions of genes. By contrast, the PB system showed a non-random integration profile with statistically significant bias towards integrating into genes, transcription start sites and their upstream regions. The Tol2 transposon showed an intermediate integration profile with no bias for inserting into genes, but preferential integration into transcription start sites and transcriptional regulatory regions. Tol2 and PB integrations were particularly favoured near transcription start sites and near transcription-associated histone modifications, including monomethyltated H3K4 (a marker for enhancer regions) and trimethylated H3K4 (associated with promoters of active genes), but disfavoured in regions rich in H3K27me3 (a histone modification typically associated with transcriptionally repressed heterochromatin). These data suggest that Tol2 and PB integration preferences resemble the target site preferences of retroviral vectors in that HIV- and MLV-derived vectors strongly favour integration into active genes and near transcription start sites, respectively (77). These results are in agreement with a survey of SB, PB and Tol2 transposon integration sites in primary T cells showing a random distribution of SB integrations and a favoured integration of PB and Tol2 near transcriptional start sites, CpG islands and DNaseI hypersensitive sites (78). Our analysis suggests that the biased integration site distribution of PB and Tol2 transposons might increase the likelihood for insertional mutagenesis potentially activating proto-oncogenes or disrupting tumour-suppressor genes: two effects that need to be avoided in gene therapeutic approaches. In contrast, SB integration sites appear to be random compared to Tol2 and PB, and thus SB likely represents a safer gene transfer vehicle for application in human gene therapy. However, random integration of transposon DNA into host chromosomes still carries a genotoxic risk that needs to be avoided or minimized for gene-delivery purposes.
In this study we explored the molecular components of site-selective integration of AAV in order to manipulate integration site selection of SB, PB and Tol2 in a proof-of-concept experimental setup (Figure 1). We evaluated three molecular strategies to target transposon integrations, all making use of the specific binding of the AAV Rep protein to a RRS consisting of GAGC repeats. We incorporated the components of the site-selective integration machinery of AAV into the transpositional systems by (i) fusing the N-terminal DNA-binding domain of Rep to the transposase, (ii) introducing the RRS-binding site into the transposon DNA and (iii) fusing the Rep DNA-binding domain to a protein domain that interacts with the transposon or the transposase. The rationale behind using only the DNA-binding domain of Rep was that full-length Rep proteins are cytotoxic and were shown to induce chromosomal rearrangements (18), and that in the context of a transpositional mechanism only the DNA-binding function of Rep was required.
We have assessed the feasibility of directly fusing the DNA-binding domain of the AAV Rep protein to the transposases, and tested oligomerization motifs known to promote the multimerization of the Rep proteins, which is needed for efficient binding to the RRS (60). We tested the transposon excision and integration activities of the engineered transposases, and found that all retained excision and integration capabilities able to mediate bona fide transposition reactions. Importantly, both the SB transposase (Figure 3D) as well as the PB transposase (Supplementary Figure S2D) in fusion with an N-terminally attached Rep DNA-binding domain retained up to 80% transpositional activity of that of the unfused respective transposases. In contrast, fusion of Rep to the Tol2 transposase severely inhibited transposition (Supplementary Figure S4D), suggesting considerable difference in sensitivity of transposases to protein fusions. We performed a plasmid-based transposition assay to evaluate the potential of the SB-based fusion transposases to target pre-defined DNA sites. When compared to integrations mediated into the control plasmid, the Rep/SB fusion transposase showed evidence of directed transposition events into a TA dinucleotide 700-bp downstream of the RRS, representing 30% of all insertions and a 15-fold enrichment of insertion at that particular site (Figure 6). In a 2-kb window surrounding the integration hot spot there were 119 TA sites available for integration. The pFVLuc plasmid used in this study is a well-studied target for SB transposition, and was used to analyse the target-site preferences of the SB transposon and to develop an automated method that can generate profiles of predicted integration events (54). According to this, we deliberately placed the RRS motif into a region predicted to be disfavoured for SB insertions in the hope that this would decrease background (non-targeted) integrations within this region. Indeed, the targeted TA dinucleotide at position 3583 is a true cold spot for SB transposition as determined by comparison with the SB integration distribution in pFVLuc. It is reasonable to propose that the Rep/SB transposase is structurally constrained after binding to the RRS motif, and is therefore forced to mediate insertion into one particular TA site. Furthermore, it is conceivable that binding of the Rep domain to the RRS sequence places the transposon/transposase complex at some distance from this region, leading to transposon insertion into a TA dinucleotide 700-bp away. Evidence for this comes from the observation that all insertions into this particular TA dinucleotide were at the same orientation with respect to the RRS motif. Importantly, a bias in insertion site selection was also seen in our genome-wide analysis: a slight, ∼1.5-fold enrichment of integration into 5-kb chromosomal regions surrounding minimal Rep-binding sites was evident with Rep/SB. The Rep/PB transposase fusions did not show evidence of a biased integration profile (Figure 9A,B), whereas the Rep domain fused to the Tol2 transposase was apparently quite efficient in resulting a ∼4-fold enrichment of insertions near genomic consensus RSS sites (Figure 9D).
The potential of targeting transposon integration using fusion proteins with dual DNA-binding activities had been assessed by engineering RRS sequences into benign sites within the transposons. The efficiency of targeting transposition events into endogenous chromosomal RRS sequences were tested by employing a targeting fusion protein containing the Rep DNA-binding domain linked to a tetramerization domain (RepTZ) known to bind efficiently to the RRS motif (60). This approach was supposed to mimic the natural mechanism of AAV integration, during which the full-length Rep protein binds to RRS sequences present in the virus and in the chromosome, thereby bringing both components in close vicinity and mediating viral integration into nearby regions (Figure 1A). Bridging DNA molecules by proteins was also proposed to target some P element transposon vectors in Drosophila. P element vectors containing regulatory sequences from the engrailed gene showed some insertional specificity by frequently integrating near the endogenous parental gene (79). With this strategy we did not observe a statistically relevant insertional bias next to genomic RRS motifs. It is possible that the transposon molecules bound by the RepTZ proteins accumulate at the RRS site, whereas the majority of unbound transposon molecules got inserted by the unmodified transposase at numerous other sites independent from the RRS sequence.
Several naturally occurring transposable elements have evolved strategies for targeted insertion into defined chromosomal sites or regions based on protein–protein interactions between a transposon-encoded factor and a cellular DNA-binding factor. One such example is the bacterial transposon Tn7. The Tn7 element integrates specifically into the attTn7 site in the E. coli chromosome. The targeted integration of the element is mediated by a subunit of the Tn7 transposase named TnsD, which specifically binds to the attTn7 site, and thereby recruits a second transposase subunit called TnsC. The TnsC subunit in turn interacts with the TnsAB transposase, which then promotes the insertion of Tn7 at attTn7 (80). Another example is represented by the yeast Ty retrotransposons. The targeting mechanisms of the Ty1, Ty3 and Ty5 retrotransposons are based on tethering the integration complexes through interactions with proteins bound at the respective preferential insertion sites. Interactions of the retrotransposon-encoded integrase with components of transcription factor III mediate targeted integration of Ty1 and Ty3 into regions upstream of genes transcribed by RNA polymerase III (81). Targeting of the Ty5 element to heterochromatin is mediated by the interaction of the retrotransposon-encoded integrase and the heterochromatin protein Sir4p (82). The integration of the human immunodeficiency virus is particularly favoured in active transcription units. This integration site-selection is controlled by the cellular transcriptional co-activator lens epithelium-derived growth factor/p75 (LEDGF/p75), which binds to the lentiviral integrase and thereby targets integration into active transcription units (83). Fusions of full-length LEDGF/p75 or the LEDGF/p75 integrase binding domain to the DNA-binding domain of phage λ repressor protein showed increased integration near λ repressor-binding sites (84). In a recent study, replacement of the LEDGF/p75 chromatin interaction-binding domain with CBX1, a protein binding to di- and trimethylated H3K9 histone molecules associated with pericentric heterochromatin and intergenic regions, was shown to direct lentiviral integration away from genes near regions bound by the CBX1 protein (85). An approach based on protein–protein interactions between the SB transposase and an engineered targeting protein consisting of the N57 domain and the tetracycline repressor resulted in targeted integration into a chromosomal TRE region in ∼10% of all cells containing transposon insertions (71).
We adapted this strategy by co-expressing the SB transposase with a targeting fusion protein consisting of the Rep DNA-binding domain and the N57 subdomain of the SB transposase, which specifies binding of the transposase to the IR sequences as well as protein–protein interactions between transposase molecules. We designed a direct N57 fusion and one with a separating tetramerization domain. Co-expression of Rep/N57 or RepTZ/N57 with the full-length transposase did not impair transposition. Both fusion proteins showed efficient binding to the RRS sequence, but showed differences in binding to the transposon/transposase (Figure 5). RepTZ/N57 was efficiently binding to the transposon IRs but failed to show protein–protein interaction with the SB transposase, whereas Rep/N57 showed only modest binding to the transposon IRs but robust protein–protein interaction with the SB transposase, suggesting that the multimerization state of N57 fundamentally affects its capacity to engage either in DNA-binding or protein-binding. The results further indicate that the intrinsic multimerization domain of the SB transposase encompassed by the N57 domain is sufficient to mediate tetramerization of the Rep DNA-binding domain crucial for efficient binding to the RRS sequence, whereas the introduction of additional tetramerization domains interfered with the protein–protein interaction function of the transposase. Analysis of insertion sites obtained in the presence of the Rep/N57 fusion protein revealed a statistically significant enrichment of transposition events into 5-kb windows surrounding genomic Rep-binding consensus sequences. This indicates that the Rep/N57 fusion protein was able to direct a relevant number of transposition complexes to regions adjacent to Rep-binding sequences occurring with a frequency of only 2134 sites per human genome. A total of 0.184% of all integrations mediated by the unmodified SB transposase occurred in this targeting window, whereas in the presence of Rep/N57 this number raised to 0.494% of all integrations. Notably, this effect was observed with a modified SB transposon containing an additional N57-binding site in the 3′-IR (pTneoDR3) and not with the unmodified original transposon (pTneo). These findings suggest that the additional binding site in the transposon IR had an enhancing effect on tethering the transpositional complex, and provide an indication for the composite effect of protein–protein and protein–DNA interactions. Furthermore, we observed an increase in transposon integrations into genes and transcription start sites in the presence of Rep/N57 and RepTZ. This indicates that Rep/N57 and to some extent also RepTZ had an effect on the integration site selection of SB, either mediated by protein–protein or protein–DNA interactions.
We queried the integration site libraries for insertions in the vicinity of the endogenous AAVS1 locus on chromosome 19. Of all integration events analysed we found in total only three insertion events that could be mapped into a 20-kb window surrounding the endogenous RRS site in the AAVS1 locus (data not shown). One insertion was mediated by the unmodified SB transposase in the presence of the Rep/N57 fusion, consistent with the proposed targeting mechanism. However, the two other integrations were generated by the unmodified SB transposase alone. Thus, it is clear that the AAVS1 locus was not targeted in these experiments, and we speculate that this might be associated with the relative GC-richness of the AAVS1 locus, which thereby makes this locus relatively unattractive for SB insertions. This prompted us to investigate the GC-content in 5-kb windows around the RRS in AAVS1, all other RRSs elsewhere in the genome and RSSs that were targeted in our datasets. Indeed, GC content in the AAVS1 site is considerably higher (59%) than in genomic regions directly flanking other RSSs (48%) and those RRS sites that were targeted in our experiments (45%) (Supplementary Figure S5), consistent with the hypothesis that local DNA composition in the vicinity of a targeted site may have fundamental impact on the ability of engineered transposases to interact with that site. Indeed, a recent study showed that ZF/PB fusions were unable to redirect PB transposons into an endogenous site in the human genome, and that only engineered, artificial target sequences containing a ZF-binding site flanked by numerous TTAA sites supported a modest (∼2-fold) increase in targeted events (86). In conclusion, our data suggest that targeted transposition is likely to be best achieved by a DNA-binding domain fused to a transposase/transposon-binding protein that would tether the transposition complex into a desired locus through protein–protein and additional protein–DNA interactions. The advantage of such an approach is that unlike with direct transposase fusions, the transposase polypeptide does not have to be modified. Therefore, potential negative effects on transposase activity are eliminated. Furthermore, this strategy resembles naturally occurring insertion targeting hypothesizing that the most promising approach for artificially targeting integration is to mimic nature.
The human genome contains 3.5 billion base pairs, which complicates efforts for targeted integration. Any attempt to target specific sites must face an overwhelming excess of non-specific competitor DNA. All transposases used in this study were fully functional in target DNA binding and capture. It is very likely that the intrinsic target DNA-binding domain of the transposase quickly binds to random target DNA, and completes the transposition reaction rather than dissociating prior to integration. This non-specific binding of the transposases to human chromosomal DNA obviously competes with specific binding to a desired target sequence, thereby limiting the probabilities of targeted transposition events. In this context it is important to note that targeted AAV integration is not as specific for AAVS1 as previously assumed. It was recently found that Rep targets AAV to integrate into numerous sites in the human genome showing similarity to RRS (87). Thus, in light of the presence of numerous sites in the genome competent in Rep binding, the level of enrichment in transposon integration near such sites observed in our study is promising. Clearly, an important challenge in designing sequence-specific transposase fusions will be the reduction of non-specific DNA-binding. The ultimate goal would be the design of transposase mutants deficient in target DNA binding but proficient in catalysis. Moreover, the DNA-binding domain in a fusion transposase could be further developed. Approaches exploiting DNA sequence-specific ZF (88) and TALE (89) DNA-binding domains to target a genomic sequence of choice represent promising alternatives.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5 and Supplementary Methods.
FUNDING
EU FP7 [PERSIST, grant number 222878]; Deutsche Forschungsgemeinschaft ‘Mechanisms of gene vector entry and persistence’ [SPP1230, IV 21/4-2]; Bundesministerium für Bildung und Forschung [InTherGD, 01GU0815]. Funding for the open access charge: Institutional funds.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank S. Yant for kindly providing SB-based plasmids for luciferase transactivation experiments with the N123 domain.
REFERENCES
- 1.Maier P, von Kalle C, Laufs S. Retroviral vectors for gene therapy. Future Microbiol. 2010;5:1507–1523. doi: 10.2217/fmb.10.100. [DOI] [PubMed] [Google Scholar]
- 2.Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 3.Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 4.Baum C, von Kalle C, Staal FJ, Li Z, Fehse B, Schmidt M, Weerkamp F, Karlsson S, Wagemaker G, Williams DA. Chance or necessity? Insertional mutagenesis in gene therapy and its consequences. Mol. Ther. 2004;9:5–13. doi: 10.1016/j.ymthe.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 5.Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, Clappier E, Caccavelli L, Delabesse E, Beldjord K, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H, Brugman MH, Pike-Overzet K, Chatters SJ, de Ridder D, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J. Clin. Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stein S, Ott MG, Schultze-Strasser S, Jauch A, Burwinkel B, Kinner A, Schmidt M, Kramer A, Schwable J, Glimm H, et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat. Med. 2010;16:198–204. doi: 10.1038/nm.2088. [DOI] [PubMed] [Google Scholar]
- 8.Boztug K, Schmidt M, Schwarzer A, Banerjee PP, Diez IA, Dewey RA, Bohm M, Nowrouzi A, Ball CR, Glimm H, et al. Stem-cell gene therapy for the Wiskott-Aldrich syndrome. N. Engl. J. Med. 2010;363:1918–1927. doi: 10.1056/NEJMoa1003548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kotin RM, Siniscalco M, Samulski RJ, Zhu XD, Hunter L, Laughlin CA, McLaughlin S, Muzyczka N, Rocchi M, Berns KI. Site-specific integration by adeno-associated virus. Proc. Natl Acad. Sci. USA. 1990;87:2211–2215. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCarty DM, Ryan JH, Zolotukhin S, Zhou X, Muzyczka N. Interaction of the adeno-associated virus Rep protein with a sequence within the A palindrome of the viral terminal repeat. J. Virol. 1994;68:4998–5006. doi: 10.1128/jvi.68.8.4998-5006.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giraud C, Winocour E, Berns KI. Site-specific integration by adeno-associated virus is directed by a cellular DNA sequence. Proc. Natl Acad. Sci. USA. 1994;91:10039–10043. doi: 10.1073/pnas.91.21.10039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henckaerts E, Dutheil N, Zeltner N, Kattman S, Kohlbrenner E, Ward P, Clement N, Rebollo P, Kennedy M, Keller GM, et al. Site-specific integration of adeno-associated virus involves partial duplication of the target locus. Proc. Natl Acad. Sci. USA. 2009;106:7571–7576. doi: 10.1073/pnas.0806821106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flotte TR, Afione SA, Zeitlin PL. Adeno-associated virus vector gene expression occurs in nondividing cells in the absence of vector DNA integration. Am. J. Respir. Cell. Mol. Biol. 1994;11:517–521. doi: 10.1165/ajrcmb.11.5.7946381. [DOI] [PubMed] [Google Scholar]
- 14.Kaplitt MG, Leone P, Samulski RJ, Xiao X, Pfaff DW, O'Malley KL, During MJ. Long-term gene expression and phenotypic correction using adeno-associated virus vectors in the mammalian brain. Nat. Genet. 1994;8:148–154. doi: 10.1038/ng1094-148. [DOI] [PubMed] [Google Scholar]
- 15.Russell DW, Miller AD, Alexander IE. Adeno-associated virus vectors preferentially transduce cells in S phase. Proc. Natl Acad. Sci. USA. 1994;91:8915–8919. doi: 10.1073/pnas.91.19.8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samulski RJ. Adeno-associated virus: integration at a specific chromosomal locus. Curr. Opin. Genet. Dev. 1993;3:74–80. doi: 10.1016/s0959-437x(05)80344-2. [DOI] [PubMed] [Google Scholar]
- 17.Xiao W, Berta SC, Lu MM, Moscioni AD, Tazelaar J, Wilson JM. Adeno-associated virus as a vector for liver-directed gene therapy. J. Virol. 1998;72:10222–10226. doi: 10.1128/jvi.72.12.10222-10226.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berthet C, Raj K, Saudan P, Beard P. How adeno-associated virus Rep78 protein arrests cells completely in S phase. Proc. Natl Acad. Sci. USA. 2005;102:13634–13639. doi: 10.1073/pnas.0504583102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bueler H. Adeno-associated viral vectors for gene transfer and gene therapy. Biol. Chem. 1999;380:613–622. doi: 10.1515/BC.1999.078. [DOI] [PubMed] [Google Scholar]
- 20.Nakai H, Montini E, Fuess S, Storm TA, Grompe M, Kay MA. AAV serotype 2 vectors preferentially integrate into active genes in mice. Nat. Genet. 2003;34:297–302. doi: 10.1038/ng1179. [DOI] [PubMed] [Google Scholar]
- 21.Kogure K, Urabe M, Mizukami H, Kume A, Sato Y, Monahan J, Ozawa K. Targeted integration of foreign DNA into a defined locus on chromosome 19 in K562 cells using AAV-derived components. Int. J. Hematol. 2001;73:469–475. doi: 10.1007/BF02994009. [DOI] [PubMed] [Google Scholar]
- 22.Recchia A, Perani L, Sartori D, Olgiati C, Mavilio F. Site-specific integration of functional transgenes into the human genome by adeno/AAV hybrid vectors. Mol. Ther. 2004;10:660–670. doi: 10.1016/j.ymthe.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 23.Ueno T, Matsumura H, Tanaka K, Iwasaki T, Ueno M, Fujinaga K, Asada K, Kato I. Site-specific integration of a transgene mediated by a hybrid adenovirus/adeno-associated virus vector using the Cre/loxP-expression-switching system. Biochem. Biophys. Res. Commun. 2000;273:473–478. doi: 10.1006/bbrc.2000.2972. [DOI] [PubMed] [Google Scholar]
- 24.Herzog RW, Hagstrom JN, Kung SH, Tai SJ, Wilson JM, Fisher KJ, High KA. Stable gene transfer and expression of human blood coagulation factor IX after intramuscular injection of recombinant adeno-associated virus. Proc. Natl Acad. Sci. USA. 1997;94:5804–5809. doi: 10.1073/pnas.94.11.5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams DA. RAC reviews serious adverse event associated with AAV therapy trial. Mol. Ther. 2007;15:2053–2054. doi: 10.1038/sj.mt.6300352. [DOI] [PubMed] [Google Scholar]
- 26.Kaplitt MG, Feigin A, Tang C, Fitzsimons HL, Mattis P, Lawlor PA, Bland RJ, Young D, Strybing K, Eidelberg D, et al. Safety and tolerability of gene therapy with an adeno-associated virus (AAV) borne GAD gene for Parkinson's disease: an open label, phase I trial. Lancet. 2007;369:2097–2105. doi: 10.1016/S0140-6736(07)60982-9. [DOI] [PubMed] [Google Scholar]
- 27.Mingozzi F, Maus MV, Hui DJ, Sabatino DE, Murphy SL, Rasko JE, Ragni MV, Manno CS, Sommer J, Jiang H, et al. CD8(+) T-cell responses to adeno-associated virus capsid in humans. Nat. Med. 2007;13:419–422. doi: 10.1038/nm1549. [DOI] [PubMed] [Google Scholar]
- 28.Izsvak Z, Khare D, Behlke J, Heinemann U, Plasterk RH, Ivics Z. Involvement of a bifunctional, paired-like DNA-binding domain and a transpositional enhancer in Sleeping Beauty transposition. J. Biol. Chem. 2002;277:34581–34588. doi: 10.1074/jbc.M204001200. [DOI] [PubMed] [Google Scholar]
- 29.Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 30.Vigdal TJ, Kaufman CD, Izsvak Z, Voytas DF, Ivics Z. Common physical properties of DNA affecting target site selection of sleeping beauty and other Tc1/mariner transposable elements. J. Mol. Biol. 2002;323:441–452. doi: 10.1016/s0022-2836(02)00991-9. [DOI] [PubMed] [Google Scholar]
- 31.Mates L, Chuah MK, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, Grzela DP, Schmitt A, Becker K, Matrai J, et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 32.Montini E, Held PK, Noll M, Morcinek N, Al-Dhalimy M, Finegold M, Yant SR, Kay MA, Grompe M. In vivo correction of murine tyrosinemia type I by DNA-mediated transposition. Mol. Ther. 2002;6:759–769. doi: 10.1006/mthe.2002.0812. [DOI] [PubMed] [Google Scholar]
- 33.Chen ZJ, Kren BT, Wong PY, Low WC, Steer CJ. Sleeping Beauty-mediated down-regulation of huntingtin expression by RNA interference. Biochem. Biophys. Res. Commun. 2005;329:646–652. doi: 10.1016/j.bbrc.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 34.Hausl MA, Zhang W, Muther N, Rauschhuber C, Franck HG, Merricks EP, Nichols TC, Kay MA, Ehrhardt A. Hyperactive sleeping beauty transposase enables persistent phenotypic correction in mice and a canine model for hemophilia B. Mol. Ther. 2010;18:1896–1906. doi: 10.1038/mt.2010.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kren BT, Unger GM, Sjeklocha L, Trossen AA, Korman V, Diethelm-Okita BM, Reding MT, Steer CJ. Nanocapsule-delivered Sleeping Beauty mediates therapeutic Factor VIII expression in liver sinusoidal endothelial cells of hemophilia A mice. J. Clin. Invest. 2009;119:2086–2099. doi: 10.1172/JCI34332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu L, Mah C, Fletcher BS. Sustained FVIII expression and phenotypic correction of Hemophilia A in neonatal mice using an endothelial-targeted Sleeping Beauty transposon. Mol. Ther. 2006;13:1006–1015. doi: 10.1016/j.ymthe.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 37.Ohlfest JR, Frandsen JL, Fritz S, Lobitz PD, Perkinson SG, Clark KJ, Nelsestuen G, Key NS, McIvor RS, Hackett PB, et al. Phenotypic correction and long-term expression of factor VIII in hemophilic mice by immunotolerization and nonviral gene transfer using the Sleeping Beauty transposon system. Blood. 2005;105:2691–2698. doi: 10.1182/blood-2004-09-3496. [DOI] [PubMed] [Google Scholar]
- 38.Yant SR, Meuse L, Chiu W, Ivics Z, Izsvak Z, Kay MA. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nat. Genet. 2000;25:35–41. doi: 10.1038/75568. [DOI] [PubMed] [Google Scholar]
- 39.Ortiz-Urda S, Thyagarajan B, Keene DR, Lin Q, Fang M, Calos MP, Khavari PA. Stable nonviral genetic correction of inherited human skin disease. Nat. Med. 2002;8:1166–1170. doi: 10.1038/nm766. [DOI] [PubMed] [Google Scholar]
- 40.Aronovich EL, Bell JB, Belur LR, Gunther R, Koniar B, Erickson DC, Schachern PA, Matise I, McIvor RS, Whitley CB, et al. Prolonged expression of a lysosomal enzyme in mouse liver after Sleeping Beauty transposon-mediated gene delivery: implications for non-viral gene therapy of mucopolysaccharidoses. J. Gene. Med. 2007;9:403–415. doi: 10.1002/jgm.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aronovich EL, Bell JB, Khan SA, Belur LR, Gunther R, Koniar B, Schachern PA, Parker JB, Carlson CS, Whitley CB, et al. Systemic correction of storage disease in MPS I NOD/SCID mice using the sleeping beauty transposon system. Mol. Ther. 2009;17:1136–1144. doi: 10.1038/mt.2009.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He CX, Shi D, Wu WJ, Ding YF, Feng DM, Lu B, Chen HM, Yao JH, Shen Q, Lu DR, et al. Insulin expression in livers of diabetic mice mediated by hydrodynamics-based administration. World J. Gastroenterol. 2004;10:567–572. doi: 10.3748/wjg.v10.i4.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohlfest JR, Demorest ZL, Motooka Y, Vengco I, Oh S, Chen E, Scappaticci FA, Saplis RJ, Ekker SC, Low WC, et al. Combinatorial antiangiogenic gene therapy by nonviral gene transfer using the sleeping beauty transposon causes tumor regression and improves survival in mice bearing intracranial human glioblastoma. Mol. Ther. 2005;12:778–788. doi: 10.1016/j.ymthe.2005.07.689. [DOI] [PubMed] [Google Scholar]
- 44.Peng PD, Cohen CJ, Yang S, Hsu C, Jones S, Zhao Y, Zheng Z, Rosenberg SA, Morgan RA. Efficient nonviral Sleeping Beauty transposon-based TCR gene transfer to peripheral blood lymphocytes confers antigen-specific antitumor reactivity. Gene Ther. 2009;16:1042–1049. doi: 10.1038/gt.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh H, Manuri PR, Olivares S, Dara N, Dawson MJ, Huls H, Hackett PB, Kohn DB, Shpall EJ, Champlin RE, et al. Redirecting specificity of T-cell populations for CD19 using the Sleeping Beauty system. Cancer Res. 2008;68:2961–2971. doi: 10.1158/0008-5472.CAN-07-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fraser MJ, Smith G, Summers M. Acquisition of host cell DNA sequences by baculoviruses: relationship between host DNA insertions and FP mutants of Autographa californica and Galleria mellonella nuclear polyhedrosis viruses. J. Virol. 1983;47:287–300. doi: 10.1128/jvi.47.2.287-300.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fraser MJ, Ciszczon T, Elick T, Bauser C. Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect Mol. Biol. 1996;5:141–151. doi: 10.1111/j.1365-2583.1996.tb00048.x. [DOI] [PubMed] [Google Scholar]
- 48.Handler AM. Use of the piggyBac transposon for germ-line transformation of insects. Insect Biochem. Mol. Biol. 2002;32:1211–1220. doi: 10.1016/s0965-1748(02)00084-x. [DOI] [PubMed] [Google Scholar]
- 49.Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 50.Koga A, Inagaki H, Bessho Y, Hori H. Insertion of a novel transposable element in the tyrosinase gene is responsible for an albino mutation in the medaka fish, Oryzias latipes. Mol. General Genet. 1995;249:400–405. doi: 10.1007/BF00287101. [DOI] [PubMed] [Google Scholar]
- 51.Kawakami K, Shima A, Kawakami N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc. Natl Acad. Sci. USA. 2000;97:11403–11408. doi: 10.1073/pnas.97.21.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawakami K. Tol2: a versatile gene transfer vector in vertebrates. Genome Biol. 2007;8(Suppl 1):S7. doi: 10.1186/gb-2007-8-s1-s7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grabundzija I, Irgang M, Mates L, Belay E, Matrai J, Gogol-Doring A, Kawakami K, Chen W, Ruiz P, Chuah MK, et al. Comparative analysis of transposable element vector systems in human cells. Mol. Ther. 2010;18:1200–1209. doi: 10.1038/mt.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu G, Geurts AM, Yae K, Srinivasan AR, Fahrenkrug SC, Largaespada DA, Takeda J, Horie K, Olson WK, Hackett PB. Target-site preferences of Sleeping Beauty transposons. J. Mol. Biol. 2005;346:161–173. doi: 10.1016/j.jmb.2004.09.086. [DOI] [PubMed] [Google Scholar]
- 55.Moldt B, Miskey C, Staunstrup NH, Gogol-Doring A, Bak RO, Sharma N, Mates L, Izsvak Z, Chen W, Ivics Z, et al. Comparative genomic integration profiling of Sleeping Beauty transposons mobilized with high efficacy from integrase-defective lentiviral vectors in primary human cells. Mol. Ther. 2011;19:1499–1510. doi: 10.1038/mt.2011.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yant SR, Wu X, Huang Y, Garrison B, Burgess SM, Kay MA. High-resolution genome-wide mapping of transposon integration in mammals. Mol. Cell. Biol. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Szabo M, Muller F, Kiss J, Balduf C, Strahle U, Olasz F. Transposition and targeting of the prokaryotic mobile element IS30 in zebrafish. FEBS Lett. 2003;550:46–50. doi: 10.1016/s0014-5793(03)00814-7. [DOI] [PubMed] [Google Scholar]
- 58.Feng X, Bednarz AL, Colloms SD. Precise targeted integration by a chimaeric transposase zinc-finger fusion protein. Nucleic Acids Res. 2009;38:1204–1216. doi: 10.1093/nar/gkp1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yant SR, Huang Y, Akache B, Kay MA. Site-directed transposon integration in human cells. Nucleic Acids Res. 2007;35:e50. doi: 10.1093/nar/gkm089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cathomen T, Collete D, Weitzman MD. A chimeric protein containing the N terminus of the adeno-associated virus Rep protein recognizes its target site in an in vivo assay. J. Virol. 2000;74:2372–2382. doi: 10.1128/jvi.74.5.2372-2382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmidt M, Schwarzwaelder K, Bartholomae C, Zaoui K, Ball C, Pilz I, Braun S, Glimm H, von Kalle C. High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR) Nat. Methods. 2007;4:1051–1057. doi: 10.1038/nmeth1103. [DOI] [PubMed] [Google Scholar]
- 62.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robertson AG, Bilenky M, Tam A, Zhao Y, Zeng T, Thiessen N, Cezard T, Fejes AP, Wederell ED, Cullum R, et al. Genome-wide relationship between histone H3 lysine 4 mono- and tri-methylation and transcription factor binding. Genome Res. 2008;18:1906–1917. doi: 10.1101/gr.078519.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 67.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 68.Waterman MJ, Waterman JL, Halazonetis TD. An engineered four-stranded coiled coil substitutes for the tetramerization domain of wild-type p53 and alleviates transdominant inhibition by tumor-derived p53 mutants. Cancer Res. 1996;56:158–163. [PubMed] [Google Scholar]
- 69.Izsvak Z, Stuwe EE, Fiedler D, Katzer A, Jeggo PA, Ivics Z. Healing the wounds inflicted by sleeping beauty transposition by double-strand break repair in mammalian somatic cells. Mol. Cell. 2004;13:279–290. doi: 10.1016/s1097-2765(03)00524-0. [DOI] [PubMed] [Google Scholar]
- 70.Voigt K, Izsvak Z, Ivics Z. Targeted gene insertion for molecular medicine. J. Mol. Med. 2008;86:1205–1219. doi: 10.1007/s00109-008-0381-8. [DOI] [PubMed] [Google Scholar]
- 71.Ivics Z, Katzer A, Stuwe EE, Fiedler D, Knespel S, Izsvak Z. Targeted Sleeping Beauty transposition in human cells. Mol. Ther. 2007;15:1137–1144. doi: 10.1038/sj.mt.6300169. [DOI] [PubMed] [Google Scholar]
- 72.Sinzelle L, Kapitonov VV, Grzela DP, Jursch T, Jurka J, Izsvak Z, Ivics Z. Transposition of a reconstructed Harbinger element in human cells and functional homology with two transposon-derived cellular genes. Proc. Natl Acad. Sci. USA. 2008;105:4715–4720. doi: 10.1073/pnas.0707746105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Philpott NJ, Gomos J, Berns KI, Falck-Pedersen E. A p5 integration efficiency element mediates Rep-dependent integration into AAVS1 at chromosome 19. Proc. Natl Acad. Sci. USA. 2002;99:12381–12385. doi: 10.1073/pnas.182430299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ryan JH, Zolotukhin S, Muzyczka N. Sequence requirements for binding of Rep68 to the adeno-associated virus terminal repeats. J. Virol. 1996;70:1542–1553. doi: 10.1128/jvi.70.3.1542-1553.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wonderling RS, Owens RA. Binding sites for adeno-associated virus Rep proteins within the human genome. J. Virol. 1997;71:2528–2534. doi: 10.1128/jvi.71.3.2528-2534.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu SC, Meir YJ, Coates CJ, Handler AM, Pelczar P, Moisyadi S, Kaminski JM. piggyBac is a flexible and highly active transposon as compared to sleeping beauty, Tol2, and Mos1 in mammalian cells. Proc. Natl Acad. Sci. USA. 2006;103:15008–15013. doi: 10.1073/pnas.0606979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, Ecker JR, Bushman FD. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang X, Guo H, Tammana S, Jung YC, Mellgren E, Bassi P, Cao Q, Tu ZJ, Kim YC, Ekker SC, et al. Gene transfer efficiency and genome-wide integration profiling of Sleeping Beauty, Tol2, and PiggyBac transposons in human primary T cells. Mol. Ther. 2010;18:1803–1813. doi: 10.1038/mt.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kassis JA, Noll E, VanSickle EP, Odenwald WF, Perrimon N. Altering the insertional specificity of a Drosophila transposable element. Proc. Natl Acad. Sci. USA. 1992;89:1919–1923. doi: 10.1073/pnas.89.5.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peters JE, Craig NL. Tn7: smarter than we thought. Nat. Rev. Mol. Cell. Biol. 2001;2:806–814. doi: 10.1038/35099006. [DOI] [PubMed] [Google Scholar]
- 81.Kirchner J, Connolly CM, Sandmeyer SB. Requirement of RNA polymerase III transcription factors for in vitro position-specific integration of a retroviruslike element. Science. 1995;267:1488–1491. doi: 10.1126/science.7878467. [DOI] [PubMed] [Google Scholar]
- 82.Xie W, Gai X, Zhu Y, Zappulla DC, Sternglanz R, Voytas DF. Targeting of the yeast Ty5 retrotransposon to silent chromatin is mediated by interactions between integrase and Sir4p. Mol. Cell. Biol. 2001;21:6606–6614. doi: 10.1128/MCB.21.19.6606-6614.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 84.Ciuffi A, Diamond TL, Hwang Y, Marshall HM, Bushman FD. Modulating target site selection during human immunodeficiency virus DNA integration in vitro with an engineered tethering factor. Hum. Gene Ther. 2006;17:960–967. doi: 10.1089/hum.2006.17.960. [DOI] [PubMed] [Google Scholar]
- 85.Gijsbers R, Ronen K, Vets S, Malani N, De Rijck J, McNeely M, Bushman FD, Debyser Z. LEDGF hybrids efficiently retarget lentiviral integration into heterochromatin. Mol. Ther. 2010;18:552–560. doi: 10.1038/mt.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kettlun C, Galvan DL, George AL, Jr, Kaja A, Wilson MH. Manipulating piggyBac transposon chromosomal integration site selection in human cells. Mol. Ther. 2011;19:1636–1644. doi: 10.1038/mt.2011.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huser D, Gogol-Doring A, Lutter T, Weger S, Winter K, Hammer EM, Cathomen T, Reinert K, Heilbronn R. Integration preferences of wildtype AAV-2 for consensus rep-binding sites at numerous loci in the human genome. PLoS Pathog. 2010;6:e1000985. doi: 10.1371/journal.ppat.1000985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Feng X, Bednarz AL, Colloms SD. Precise targeted integration by a chimaeric transposase zinc-finger fusion protein. Nucleic Acids Res. 2010;38:1204–1216. doi: 10.1093/nar/gkp1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.