Abstract
We herein report on the formation and high-resolution NMR solution-state structure determination of a G-quadruplex adopted by d[G3ATG3ACACAG4ACG3] comprised of four G-tracts with the third one consisting of four guanines that are intervened with non-G streches of different lengths. A single intramolecular antiparallel (3+1) G-quadruplex exhibits three stacked G-quartets connected with propeller, diagonal and edgewise loops of different lengths. The propeller and edgewise loops are well structured, whereas the longer diagonal loop is more flexible. To the best of our knowledge, this is the first high-resolution G-quadruplex structure where all of the three main loop types are present.
INTRODUCTION
Guanine-rich DNA can form four-stranded structures termed G-quadruplexes (1–3) through stacking of coplanar arrangements of four guanines, G-quartets, which are stabilized with eight Hoogsteen hydrogen bonds and interactions with metal ions that are coordinated in the central cavity. In silico analyses of different genomes showed enrichment of guanine-rich sequences in telomeric and promoter regions (4–7). Formation of G-quadruplexes in promoters of several genes was shown to affect their expression and indicated that G-quadruplexes can act as transcriptional regulators (8). With the aim to down-regulate transcription of proto-oncogenes, a considerable effort has been made to find small ligands that would facilitate formation and stabilize G-quadruplex structures in their promotor regions (9). A very common feature of G-quadruplexes from promoter regions is a longer central loop in comparison to the other two loops that together with G-quartet core constitute the architecture of the intramolecular fold. In addition to being constitutive elements of G-quartets, guanine residues could be involved in loops thus contributing to the variety of their lengths and orientations. It was observed that structures with the longest central loop are favoured (10–13).
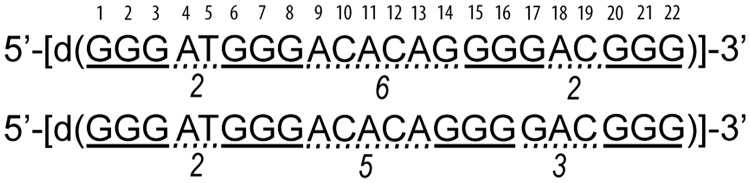
The goal of the current study was to ascertain whether oligonucleotide d[G3ATG3ACACAG4ACG3] comprised of four G-tracts with the third one consisting of four guanines folds into a stable G-quadruplex (refered to as ODN in the present study, Figure 1) and to determine its 3D structure with atomic resolution based on the high-resolution NMR spectra. Structure prediction was intriguing as the third G-tract with four guanine residues increases the possibilities for different topologies and loop lengths of potential G-quadruplex(es). It is of interest to compare fold(s) with the most stable and frequently found G-quadruplexes with respect to strand orientation, loop types and lengths. We were especially interested in the length of the central loop whose structural properties cannot be predicted from sequence alone. In ODN, G14 could be part of the central loop, thus making it 6 nt long. Noteworthy, such topology with the longest possible central loop is expected based on the available data in the literature (8). Alternatively, G17 could be part of the third loop, thus making it 3 nt long, which is predisposed for edgewise orientation. Details of the novel 3D structure would add to architectures of G-quadruplexes and help to understand the principles of folding of these unique DNA structures in general. In addition, it would provide much needed knowledge on the preference of intramolecular topologies for 2:6:2 or 2:5:3 loop arrangements. A few related oligonucleotides of different sequence context and loop lengths were analysed with respect to their ability to fold into a single G-quadruplex structure (Supplementary Table S1).
Figure 1.
The G-rich sequence (designated ODN) investigated in this study can form two different G-quadruplexes that differ in the length of the loops. Guanines that potentially form G-quadruplex core are underlined with full line. Loop-forming residues are underlined with dotted line and the length of the loop is indicated under the loop. The nucleotides in the ODN are numbered.
MATERIALS AND METHODS
Sample preparation
Oligonucleotides were purchased from IDT (Leuven, Belgium) or synthesized on Expedite Nucleic Acid Synthesis 8909 synthesizer (PerSeptive Biosystems) using standard phosphoramidite solid-phase chemistry following the manufacturer's protocol. Oligonucleotides containing partially (6%) 15N site-specifically labelled guanine residues were synthesized to enable unequivocal spectral assignment. Cleavage of protecting groups was carried out in concentrated aqueous ammonia at 55°C overnight. Samples were purified by gel filtration on a Sephadex G25 column, lyophilized and dissolved in modified Britton–Robinson (pH = 7.0), potassium phosphate buffer (pH = 6.8) or H2O containing 10% of 2H2O. pH of the oligonucleotide samples in H2O was adjusted to 6.8 using HCl or LiOH solution. KCl, NaCl or 15NH4Cl were titrated into the samples to final concentrations of 50 and 100 mM. The samples were heated up to 95°C for 3 min and then slowly cooled down to room temperature. Oligonucleotide concentration was between 0.4 and 2.7 mM. The sample in 2H2O was prepared by repeated lyophilization and final dissolution in 2H2O. Concentration of the samples was determined by UV absorption at 260 nm using UV/VIS Spectrophotometer Varian CARY-100 BIO UV–VIS and computer program UV WinLab. Extinction coefficients of 2.26 × 105, 2.34 × 105 and 2.89 × 105 l mol−1 cm−1 for ODN, ODN1 and ODN 2, respectively, were determined by the nearest neighbour method.
Native gel electrophoresis
Electrophoretic separation was run in a temperature-controlled vertical electrophoretic apparatus (Z375039-1EA; Sigma-Aldrich, San Francisco, CA) at 10°C in 25 mM modified Britton–Robinson buffer (pH 7.0) and 50 mM KCl. Polyacrylamide gel concentration was 15%. Prior to being loaded, the DNA samples were heated in the same buffer for 5 min at 98°C and slowly cooled to room temperature within 50 min. AC18 (d(AC)9) and AC36 (d(AC)18) were used as markers for 18 and 36 bases, respectively. DNA was visualized by Stains-all (Sigma-Aldrich) staining.
CD and UV experiments
Samples for circular dichroism (CD) and UV measurements were prepared at 60 and 12 μM oligonucleotide concentrations, respectively, in modified 25 mM Britton–Robinson and 10 mM potassium phosphate buffer containing 50 mM K+. The CD spectra were recorded on an Applied Photophysics Chirascan CD spectrometer at 25°C using a quartz cell with a 0.1 cm path length. The wavelength was varied from 220 to 320 nm. A blank sample containing only modified Britton–Robinson or potassium phosphate buffer was used for baseline correction. Three scans were averaged to obtain a CD spectrum. Thermal stability measurements were performed on a Varian CARY-100 BIO UV–VIS spectrophotometer with the Cary Win UV Thermal program using a 1 cm path-length cell. Samples were heated at 0.1°C min−1 from 10°C to 85°C and absorbance at 295 nm was measured at 0.5°C steps. Measurements were repeated three times. A combination of mineral oil and a fixed cuvette cap was used to prevent evaporation and sample loss due to high temperatures. A stream of nitrogen was applied throughout the measurements to prevent condensation at lower temperatures. Tm was determined from the A295 versus temperature plot.
NMR experiments
NMR spectra were recorded on Varian VNMRS 600 MHz and 800 MHz NMR spectrometers at 25°C using triple resonance probes with inverse detection. The diffusion coefficient was determined using a PFG-STE pulse sequence. Identification of guanine imino protons in partially (6%) 15N site-specifically labelled samples was performed by 1D 15N-filtered HSQC experiments. H1–H8 intra-residual connectivities through the C5 atom were established with the use of a 2D JRHMBC experiment. Standard homonuclear 2D NMR experiments recorded in 2H2O including 2D DQF-COSY, TOCSY (τm of 20 and 80 ms) and NOESY (τm of 80, 150 and 250 ms) were used to assign non-exchangeable protons. Resonances of exchangeable protons were assigned using 2D NOESY (τm of 80, 150 and 250 ms) recorded in H2O. Jump-and-return and DPFGSE pulse sequences were used to suppress the water signal.
Spectra were processed and analysed using programs VNMRJ (Agilent Technologies) and NMRPipe. Cross-peak assignment and integration was achieved using the software SPARKY (UCSF). NOE (Nuclear Overhauser Effect) distance restraints for non-exchangeable protons were obtained from a 2D NOESY spectrum recorded at 25°C in 100% 2H2O with a mixing time of 150 ms. Only non-overlapping cross-peaks were integrated and used for the distance restraints calculation. The average volume of H5–H6 cross-peaks of C10, C12 and C19 was used as the distance reference of 2.45 Å. Cross-peaks were classified as strong (1.8–3.6 Å), medium (2.6–5.0 Å) and weak (3.5–6.5 Å). NOE distance restraints for exchangeable protons were obtained from 2D NOESY spectra recorded at 25°C in 10% 2H2O, 90% H2O with mixing times of 80 and 250 ms. Cross-peaks of medium and weak intensity in 2D NOESY spectrum with a mixing time of 80 ms were classified as strong (1.8–3.6 Å) and medium (2.6–5.0 Å), respectively. Cross-peaks that appeared in 2D NOESY spectrum with a mixing time of 250 ms were classified as weak (3.5–6.5 Å).
Structure calculations
Structure calculations were performed using AMBER 9 software and parmbsc0 force field (14). The initial extended single-stranded DNA structure was obtained using the 3DNA program (15). A total of 50 structures were calculated in 100 ps of NMR restrained simulated annealing (SA) simulations using the generalized Born implicit model to account for solvent effects. The cut-off for non-bonded interactions was 20 Å and the SHAKE algorithm for hydrogen atoms was used with the tolerance of 0.0005 Å. For each SA simulation, a different starting velocity was used. The temperature programme was as follows: after 15 ps at 300 K, the temperature was raised to 1000 K in the next 10 ps and held constant at 1000 K for 30 ps. Temperature was scaled down to 100 K in the next 25 ps and reduced to 0 K in the last 20 ps. Restraints used in the calculation were hydrogen bond and NOE-derived distance restraints (force constant 60 kcal mol−1 Å−2), torsion angle ε, χ and ν2 restraints (force constant 200 kcal mol−1 rad−2) and planarity restraints for G-quartets (force constant 20 kcal mol−1 rad−2). Van der Waals, electrostatic components and force constants for restraints were linearly increased to their final value in the first 35 ps of SA. NOE-derived distance restraints were used after the first 30 ps. Planarity restraints for G-quartets were excluded in the last 20 ps of SA. All structures were minimized with a maximum of 10 000 steps of energy minimization and a family of 10 structures was selected based on the smallest restraints violations and lowest energy.
RESULTS AND DISCUSSION
Formation of a single monomeric G-quadruplex structure by ODN in potassium ion containing solution
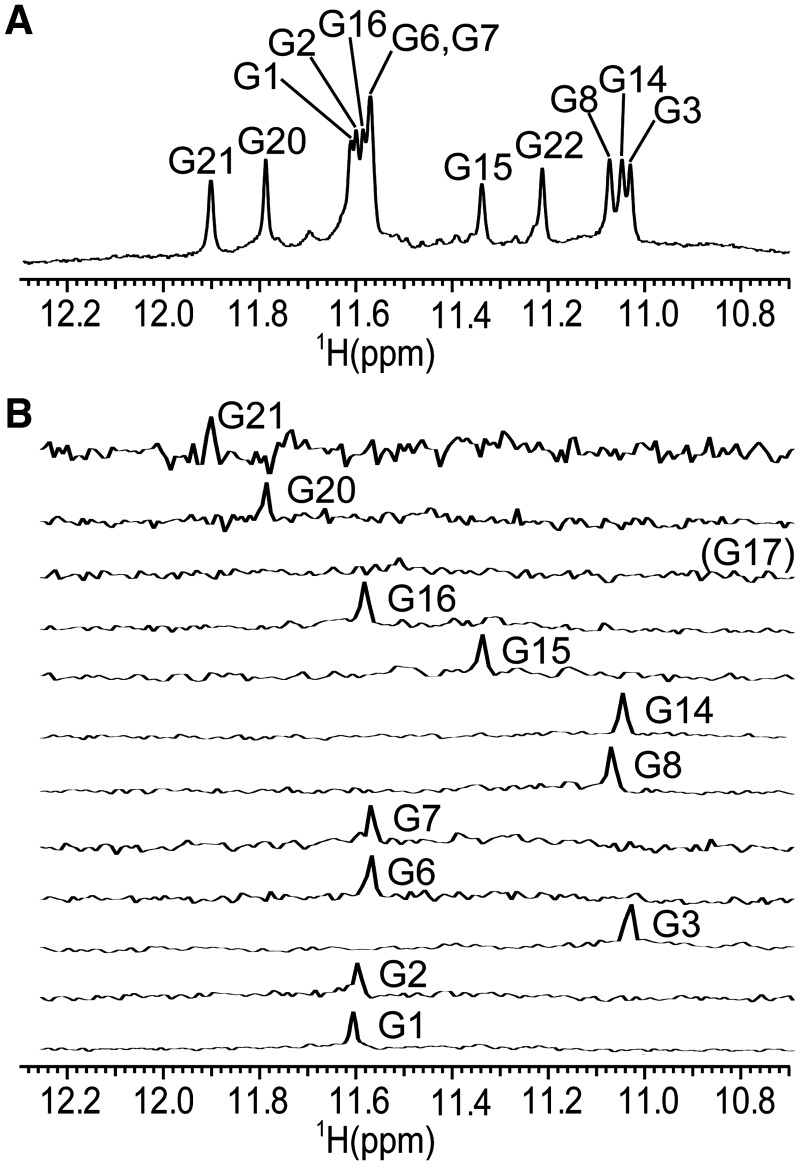
Titration of ODN was performed with additions of 1 M KCl into a buffered aqueous solution of oligonucleotide at 0.4 mM concentration per strand at 25°C. No signals could be observed in the imino region of 1H-NMR spectrum before addition of cations (Supplementary Figure S1A). Increase of the K+ ion concentration to 2.5 mM led to the folding of ODN into a G-quadruplex. The intensity of signals in the imino region increased with increasing K+ concentration (Supplementary Figure S1). Twelve partially resolved signals could be identified in the region from δ 11 to 12 ppm, characteristic of Hoogsteen hydrogen bonds suggesting the formation of a single G-quadruplex structure with three G-quartets (Figure 2A). The translation diffusion coefficient of 1.33 × 10−6 ± 0.04 cm2 s−1 suggested the monomeric nature of a fold. The diffusion coefficient calculated with the program Hydropro (16) on the basis of the molecular structure determined in the current study (vide infra) was 1.37 × 10−6, which is in excellent agreement with the experimentally determined value. UV melting experiments showed one transition between folded and melted states and no hysteresis after several steps of heating and cooling. Tm of 58°C was determined from A295 versus temperature plot (Supplementary Figure S2).
Figure 2.
Assignment of imino proton resonances. (A) The imino region of 1D 1H-NMR spectrum and assignment of guanine residues in G-quartets. (B) 1D 15N filtered HSQC spectra of samples containing partially (6%) 15N residue-specifically labelled oligonucleotides. Spectra were recorded at 800 MHz (1D 1H-NMR spectrum) and 600 MHz (1D 15N filtered HSQC spectra) at 25°C in 10% 2H2O, 50 mM KCl, 10 mM phosphate buffer with pH 6.8. Oligonucleotide concentrations were between 1.5 and 2.7 mM.
1D 1H-NMR spectra in sodium and ammonium ion containing solutions showed many partially resolved signals of low intensity in the imino region denoting the formation of several structures (Supplementary Figure S3). Spectrum of the ODN oligonucleotide in sodium solution exhibits an additional peak around δ 12.8 ppm, indicating the existence of a GC base pair.
In addition a set of related sequences listed in Supplementary Table S1 has been studied. The longest sequence ODN2 consisted of 27 nucleotides and five G-tracts and its first to the fourth G-tract were identical to the ODN, whereas ODN1 comprised of the second to the fifth G-tract of the ODN2. It was of interest to evaluate the effect of the position of the longest stretch of potential loop-forming residues on the polymorphism of the G-quadruplex structures. The slowest electrophoretic mobility exhibited by ODN2 was consistent with its largest length compared to ODN and ODN1 (Supplementary Figure S4). Although five G-tracts in ODN2 imply possibility of formation of several distinct conformations, only one clear band was observed excluding the presence of topologically different molecules. CD spectra of ODN2 (Supplementary Figure S5) suggested anti-parallel fold of the oligonucleotide. In contrast, a smeared band was observed in the PAGE gel for ODN1, indicating formation of multiple structures. Arrangement of signals in CD spectra of ODN1 and their relative intensity was similar to CD spectra of ODN2. Polymorphism of ODN1 was further investigated with NMR spectroscopy. Twelve signals of high intensity and multiple poorly resolved signals of low intensity could be observed in imino region of 1D 1H-NMR spectra (Supplementary Figure S6), which confirmed that ODN1 folds into one prevailing and several minor structures. In concordance with anti-parallel fold of prevailing species determined with CD spectroscopy, six cross-peaks of high intensity were observed in aromatic–anomeric region of 2D NOESY spectra of ODN 1 (data not shown). The fastest electrophoretic mobility was exhibited by ODN, which appeared as one clear band in PAGE gel, which was in complete agreement with imino proton NMR characteristics (vide supra).
Resonance assignment, glycosidic torsion angle determination and sugar conformation
In order to unambiguously assign imino proton resonances and in particular to determine whether G14 or G17 is involved in G-quartet formation, 15N residue-specific partially (6%) labelled oligonucleotides were synthesized and 1D 15N-filtered HSQC spectra were recorded. One signal was observed in the 1D 15N-filtered HSQC spectrum for the imino proton of the isotopically labelled guanine residue in each sample, with the exception of the sample which was labelled at G17 (Figure 2B). G17 is therefore not involved in a G-quartet but is rather part of the third loop. Identified imino signals of 1D 1H-NMR spectrum allowed further assignment of proton resonances by 2D NMR methods.
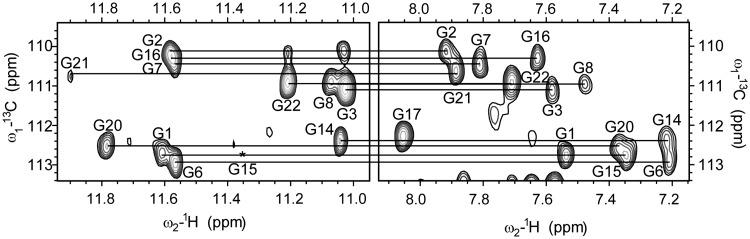
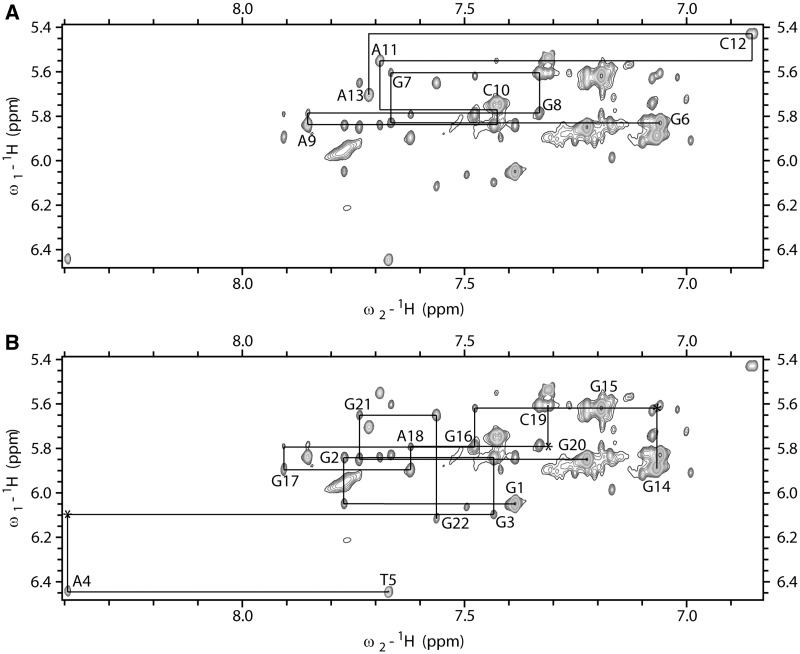
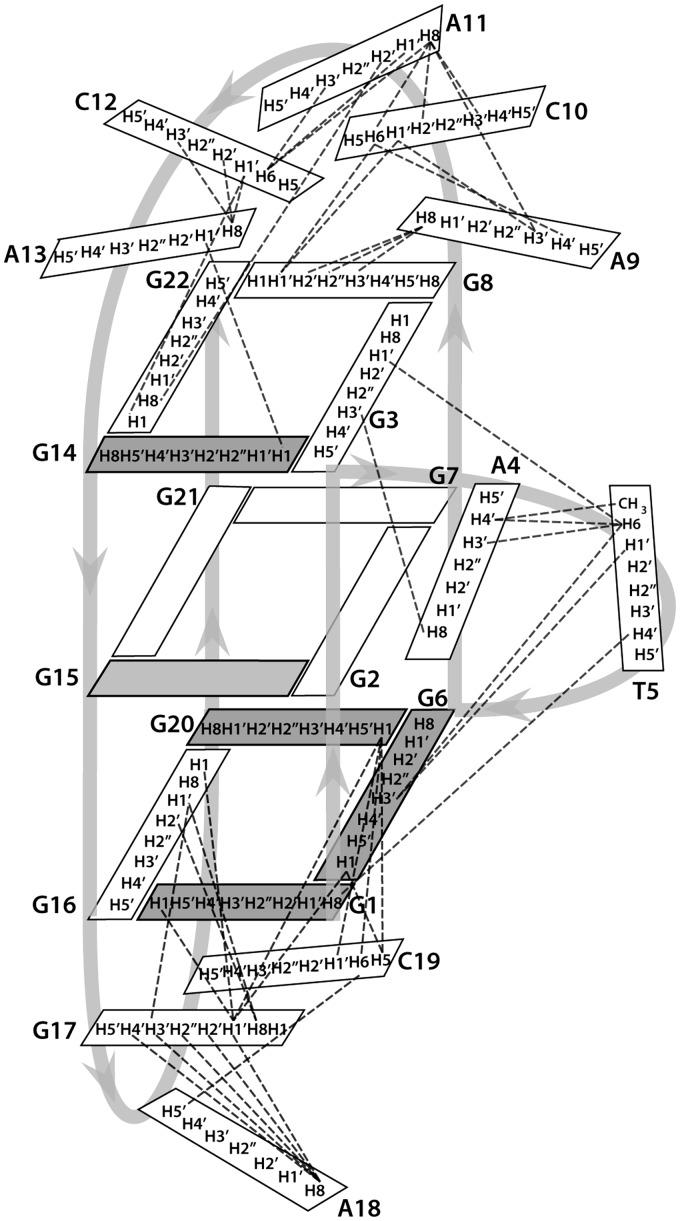
A 2D JRHMBC experiment was performed to correlate guanine H8 and H1 protons within the same guanine base and thus assign H8 protons of guanine residues (Figure 3). H1′ proton resonances were identified with the use of 2D NOESY spectra through correlations with aromatic H6/H8 protons of the same residue. Aromatic H6/H8-sugar H1′ connectivities could be followed along oligonucleotide sequence and were interrupted at the anti-syn steps (Figure 4). H2′/H2″ proton resonances were identified and stereospecifically assigned (17) with the use of 2D DQF-COSY and NOESY spectra. Most of the H3′ and H4″ sugar protons were assigned by 2D TOCSY and DQF-COSY experiments. Signal overlap and close proximity to the diagonal allowed assignment of only a few H5′/H5″ sugar protons, namely of the residues T5, A9 and A11. H5–H6 cross-peaks of C10, C12 and C19 were identified with the help of the 2D TOCSY spectrum with a mixing time of 20 ms. The resonance of a thymine methyl group was assigned through strong intra-residual cross-peak with intra-residual H6 proton in 2D NOESY spectra.
Figure 3.
The imino and aromatic regions of 2D JRHMBC spectrum showing correlations between H8 and the corresponding imino proton with C5 of guanine residues. Missing cross-peak of G15 in the imino region is designated with asterisk. 2D JRHMBC spectrum was recorded at 800 MHz, 25°C in 10% 2H2O, 50 mM KCl, 10 mM phosphate buffer with pH 6.8. Oligonucleotide concentration was 1.7 mM.
Figure 4.
The aromatic–anomeric region of 2D NOESY spectrum (τm = 150 ms) of ODN. The lines connect aromatic H6/H8–sugar H1' NOE cross-peaks. Sequential connectivities for (A) G6-A13 segment and (B) G1-T5 and G14-G22 segments are shown separately to reduce cross-peaks’ overlap. Missing cross-peaks are designated with asterisk. 2D NOESY spectrum was recorded at 800 MHz, 25°C in 10% 2H2O, 50 mM KCl, 10 mM phosphate buffer with pH 6.8. Oligonucleotide concentration was 1.7 mM.
Five strong cross-peaks in H6/H8-H1′ region of the 2D NOESY spectrum acquired at mixing time of 80 ms were assigned to nucleotides with syn conformation of glycosidic torsion angle, namely G1, G6, G14, G15 and G20. Downfield shifts of H2′ and H2″ protons of these residues were consistent with the determined syn glycosidic torsion angle (17).
H1′–H2′ cross-peaks in 2D DQF-COSY were observable for all residues. Approximate JH1′–H2′ values determined on the basis of the sums of JH1′–H2′ and JH1′–H2″ coupling constants (18) were in range from 7 to 10 Hz, which showed that the nucleotides adopted predominantly S-type sugar conformation. In full concordance, H3′–H4′ cross-peaks were of weak intensity for residues G7, G8, A9, A11, C12, G15, G16 and G21, and absent for all the other residues.
Topology of a G-quadruplex
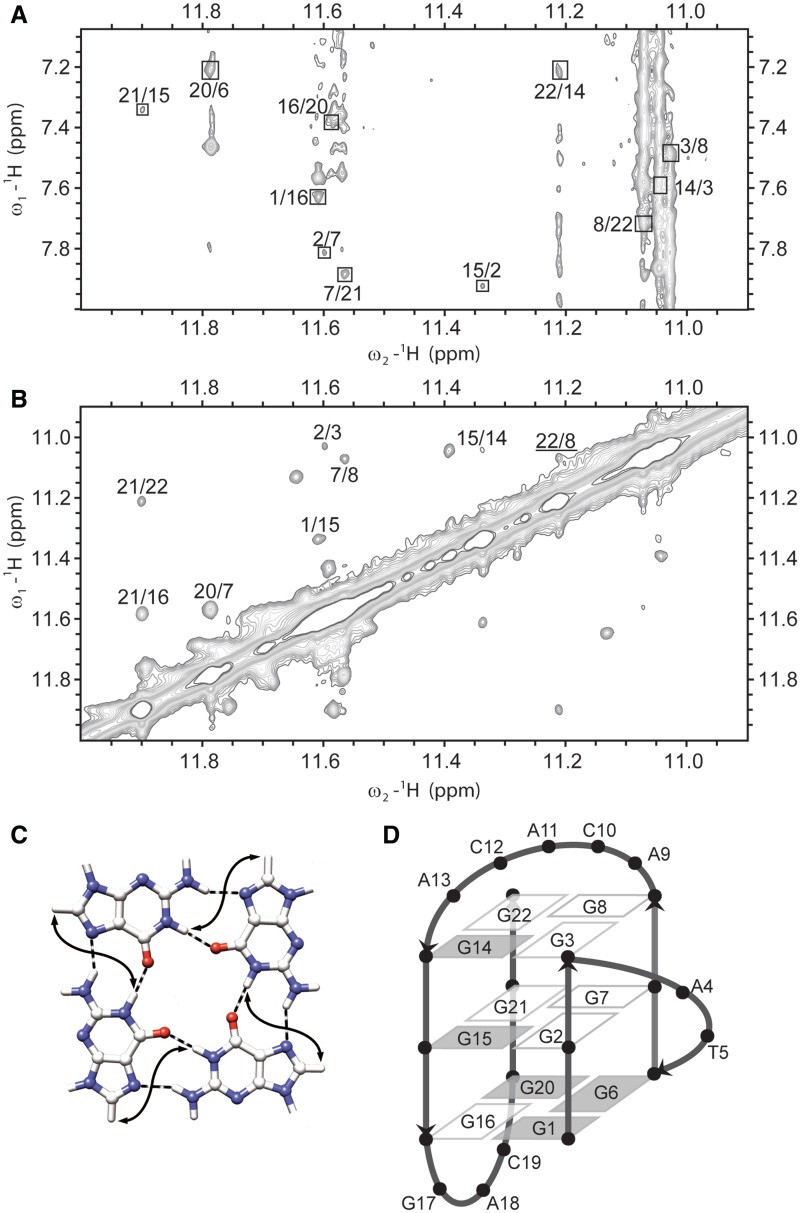
With the assignment of imino–aromatic (Figure 5A) and imino–imino (Figure 5B) regions in 2D NOESY spectrum (τm = 250 ms), we could attribute guanines to individual G-quartets. Cross-peaks correlating H1 and H8 protons of adjoining guanines within a given G-quartet (Figure 5C) helped to determine the direction of hydrogen bonds. Identified G-quartets were G1 → G16 → G20 → G6, G2 → G7 → G21 → G15 and G3 → G8 → G22 → G14, where an arrow indicates hydrogen bond directionality. In full concordance, imino protons of the central G2 → G7 → G21 → G15 quartet showed slow exchange in 2H2O (Supplementary Figure S7).
Figure 5.
(A) The imino-aromatic region of 2D NOESY spectrum (τm = 150 ms). Cross-peaks between H1 and H8 protons of neighbouring guanine residues within the same quartet are designated with squares. (B) The imino–imino region of the same 2D NOESY spectrum. Cross-peak between G8 H1 and G22 H1 protons of neighbouring guanine residues in G-quartet is underlined. 2D NOESY spectrum was recorded at 800 MHz, 25°C in 10% 2H2O, 50 mM KCl, 10 mM phosphate buffer with pH 6.8. Oligonucleotide concentration was 1.7 mM. (C) Presentation of contacts between H1 and H8 protons (full arrows) of neighbouring guanine residues in G-quartet. (D) Schematic presentation of (3+1) topology of G-quadruplex adopted by ODN. Residues with syn and anti conformation along glycosidic bond are represented with grey and white rectangles, respectively.
In the imino–imino region of the 2D NOESY spectra, H1–H1 cross-peaks between protons of neighbouring guanines in individual G-quartet could usually be observed. In the case of ODN, considerable overlap and proximity to the diagonal prevented assignment of most of the H1–H1 cross-peaks of guanine residues within the same G-quartet. In fact, only the G8 H1–G22 H1 cross-peak (underlined in the Figure 5B) could be observed. On the other hand, all cross-peaks except G6 H1–G2 H1 could be assigned to the interaction between imino protons of guanine residues in adjacent G-quartets. The cross-peak of G6 H1–G2 H1 was not visible because of the close proximity to the diagonal.
Imino–imino connectivities between the second (G2–G7–G21–G15) and the third (G3–G8–G22–G14) quartet correlated sequential guanine residues in the DNA strands, for example, H1 of G2 with H1 of G3. NOE interactions between the first (G1–G16–G20–G6) and the second (G2–G7–G21–G15) quartet correlated guanine residues of neighbouring DNA strands, for example, H1 of G1 with H1 of G15. This clockwise twist was observed for all three NOE correlations between the first and the second G-quartets (G1–G15, G16–G21 and G20–G7), which was in agreement with the disposition of glycosidic torsion angles in these two G-quartets. NOE connectivities in imino–imino and imino–aromatic regions of the 2D NOESY spectra together with sequential walk enabled determination of the G-quadruplex topology (Figure 5D). G-quadruplex consists of G1–G3, G6–G8, G14–G16 and G20–G22 G-runs where the first, second and fourth are parallel to one another, while the third is anti-parallel to other strands, thus clearly confirming (3+1) topology. G-runs are connected with propeller, diagonal and edgewise loops of two (AT), five (ACACA) and three (GAC) nucleotides, respectively. The edgewise loop contains the fourth guanine residue that formally belongs to the third G-tract.
CD spectrum offered a complementary assessment of (3+1) G-quadruplex topology and displayed three signals, one negative around 239 nm and two positive around 264 and 287 nm (Supplementary Figure S5). The G-quadruplex exhibits one syn–syn–anti and three syn–anti–anti guanine glycosidic bond angle orientations along DNA strands of the G-quadruplex core. The topology shown in Figure 6 could be assigned to group II according to Webba da Silva et al. (19). CD spectra of G-quadruplexes from this group display the same signals arrangement (− + +) as CD spectrum of the G-quadruplex adopted by ODN, thus offering complementary confirmation of a (3+1) topology.
Figure 6.
Schematic presentation of inter-residue NOE connectivities in loops of a G-quadruplex adopted by ODN. Residues with syn and anti conformation along glycosidic bond are represented with grey and white rectangles, respectively.
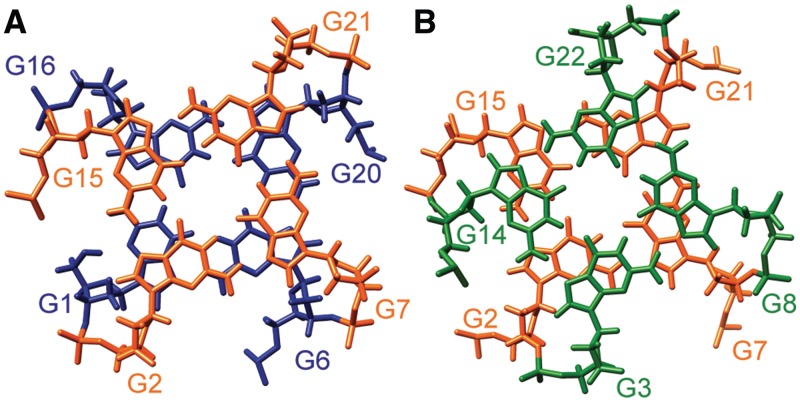
A (3+1)-fold has been observed for bcl2Mid (13) and several telomeric G-quadruplexes (20–24). It was referred to as a robust folding topology that may be used for structure-based drug design (3). Hitherto reported intramolecular (3+1) G-quadruplexes share similar folds with one propeller and two sequential edgewise loops. Recent communication established (3+1) topology with three different types of loops (25). Loop length in intramolecular telomeric G-quadruplexes with four G-tracts is similar [Tetrahymena 4:3:2 (22), human telomeric sequence 3:3:3 (20)], whereas the G-quadruplex adopted by the Bcl-2 promoter region displays a longer central loop compared to the first and the last loop (3:7:1). The fold adopted by ODN exhibits the greatest variability in loop types among known (3+1) G-quadruplexes.
NOE interactions within loop regions
Interestingly, most of the sequential NOE interactions in the loops were visible. In addition, several nucleotides exhibit long-range NOE contacts, indicating that the position of these nucleotides is well defined. In the propeller loop, the T5 residue exhibits NOE interactions with G1, G3 and G6, as well as with A4, positioning T5 in the groove between G1–G3 and G6–G8 strands (Figure 6). In the diagonal loop, there are, apart from sequential connectivities, NOE correlations between A9 and A11, C10 and G8, A11 and G22 as well as C12 and G22. These interactions place C12 over the G3–G8–G22–G14 quartet, where it could get involved in stacking interactions. In the edgewise loop, NOE interactions could be observed between G17 and G6, G16, A18 and G20 as well as between C19 and G6, A18 and G20, positioning G17 and C19 under the G1–G16–G20–G6 quartet where they are involved in base–base stacking interactions (Figure 6). Several residues in the loops, namely A9, A13 and A18, have shown scarce inter-residue NOE contacts.
NMR-restrained structure calculations
The solution structure of the G-quadruplex was calculated using a SA protocol, starting from an extended single-stranded DNA structure. NOE distance restraints were obtained from 2D NOESY spectra acquired with different mixing times. On average 19 NOE-derived distance restraints per residue were used for structure calculations (Supplementary Figure S8). Glycosidic torsion angle χ was restrained to the syn region (0 ± 90°) for residues G1, G6, G14, G15 and G20, based on the intensity of respective H8–H1′ 2D NOESY cross-peaks. For all the other residues, torsion angle χ was restrained to the anti range (180 ± 90°). Torsion angle ε was restrained to the 150°–300° region. The sugar moiety was restrained to the S-type conformation for all residues based on the values of JH1′–H2′ coupling constants. For each G-quartet, eight hydrogen bond restraints were used (Table 1). Planarity restraints for G-quartets were used in the first 80 ps of SA and were excluded in the last 20 ps of the calculation.
Table 1.
Structural statistics
| NMR distance and torsion angle restraints | |
| NOE-derived distance restraints | |
| Total | 316 |
| Intra-residue | 216 |
| Inter-residue | 100 |
| Sequential | 70 |
| Long-range | 30 |
| Hydrogen bond restraints | 24 |
| Torsion angle restraints | 65 |
| G-quartet planarity restraints | 36 |
| Structure statistics | |
| Violations | |
| Mean NOE restraint violation (Å) | 0.079 ± 0.0002 |
| Max. NOE restraint violation (Å) | 0.196 |
| Max. torsion angle restraint violation (°) | 4.765 |
| Deviations from idealized geometry | |
| Bond length (Å) | 0.012 ± 0.0002 |
| Bond angle (°) | 2.525 ± 0.0000 |
| Pairwise heavy atom RMSD (Å) | |
| Overall | 2.63 |
| G-quartets | 0.82 |
| G-quartets and propeller (AT) loop | 0.96 |
| G-quartets and diagonal (ACACA) loop | 2.60 |
| G-quartets and edgewise (GAC) loop | 1.51 |
| G-quartets and A4, T5, G17, C19 | 0.99 |
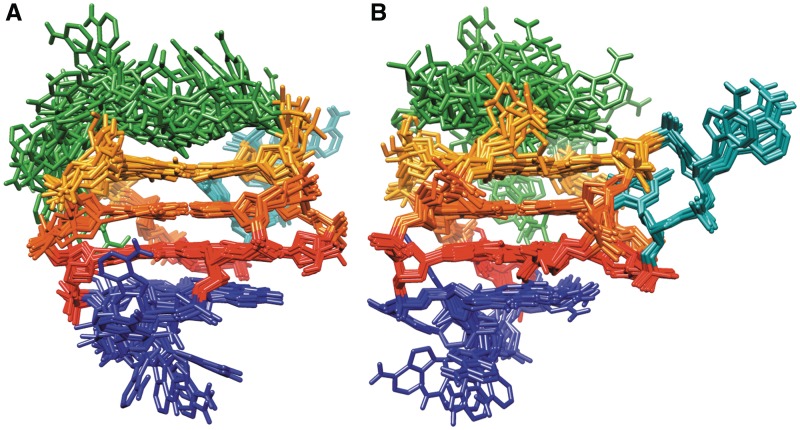
A set of 50 structures was calculated, all of which had NOE distance restraints violations lower than 0.3 Å. Following energy minimization, 10 structures with the lowest energy and the smallest restraint violations were selected (Table 1, Figure 7). A few structures that met energy and restraint violations criteria had G17 and C19 aligned under G1–G16–G20–G6 quartet. Since no experimental NOE contacts between G17 and C19 could be found, those structures were not evaluated further. Final structures are defined by one narrow (between G1–G3 and G14–G16), one wide (between G14–G16 and G20–G22) and two medium grooves (between G1–G3 and G6–G8, and between G6–G8 and G20–G22). The first and the second G-quartets display only partial stacking of guanine bases involving imidazole moieties. NOE cross-peaks among protons in the second and the third G-quartets suggest their favourable stacking interactions (Figure 8). The diagonal loop is the most flexible part of the molecule and several different conformations that are all in agreement with experimentally observed NOE contacts have been found during SA simulation. Interestingly, all conformations of the diagonal loop in the 10 final structures display stacking of the nucleotide bases of loop residues, most commonly between C12–A13, as well as between A9–C10, C10–A11 and A11–C12 pairs. This indicates that the diagonal loop is switching quickly on the NMR time-scale among different conformations that are stabilized with stacking interactions.
Figure 7.
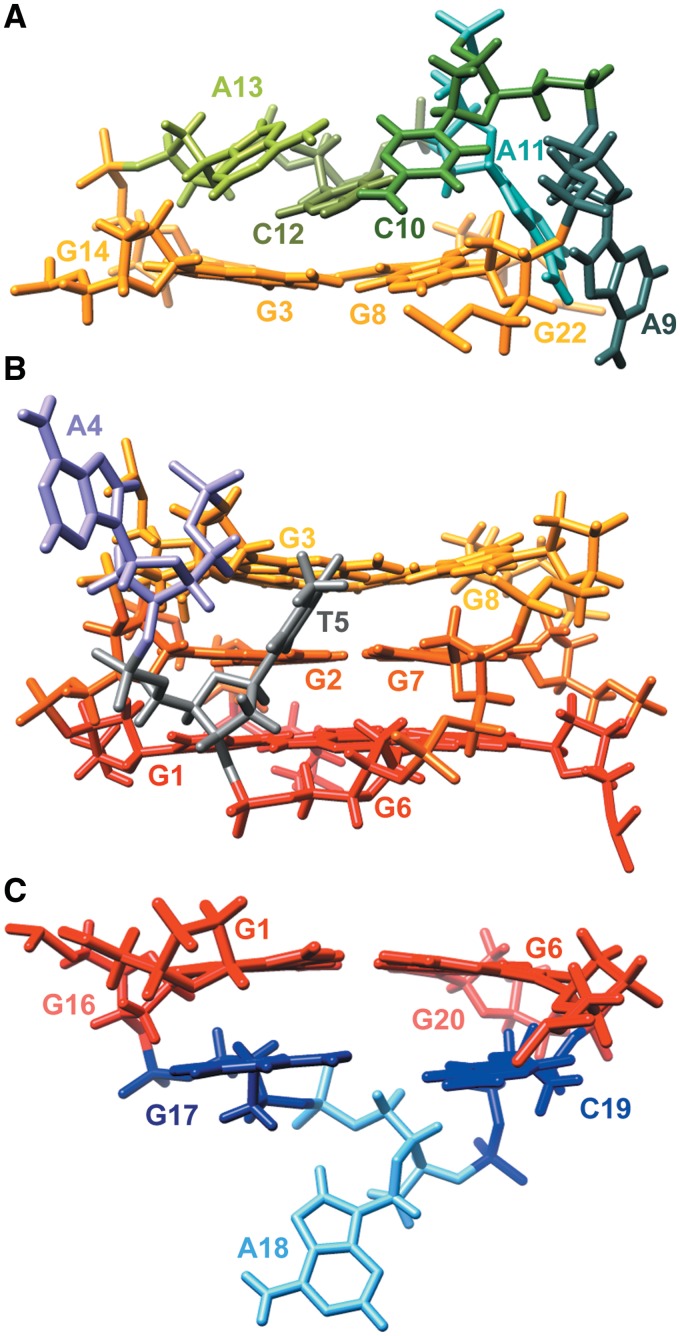
Superimposition of the 10 structures with the lowest energy and the smallest restraints violations. Figures in (A) and (B) are rotated with respect to one another to better represent superimposed structures. G1–G16–G20–G6, G2–G7–G21–G15 and G3–G8–G22–G14 quartets are shown in red, orange and yellow, respectively. Propeller (A4–T5), diagonal (A9–C10–A11–C12–A13) and edgewise (G17–A18–C19) loops are presented in cyan, green and blue, respectively.
Figure 8.
Stacking interactions between (A) G1–G16–G20–G6 (blue) and G2–G7–G21–G15 (orange) quartets, and between (B) G2–G7–G21–G15 (orange) and G3–G8–G22–G14 (green) quartets.
Among five residues (A9–C10–A11–C12–A13) in the diagonal loop, cytosine residues exhibit the highest number of NOE contacts with the G3–G8–G22–G14 quartet. However, only C12 is reasonably well ordered and stacked over the G-quartet (Figure 9A). Several NOE contacts of C12 with A11 and A13 as well as with G14 and G22 are in agreement with the upfield chemical shifts of both aromatic and sugar protons of C12 with respect to C10 and C19. The loop structure shown in Figure 9A shows that C12 is stacked between A13 and the neighbouring G-quartet, and protected from solvent with other residues in the diagonal loop. On the other hand, C10 is considerably more flexible; positioned above G3–G8–G22–G14 quartet in some structures and turned away in others. The most flexible residues in the diagonal loop are A9 and A11. A9 adopts a range of conformations from flipped away from the G-quadruplex to residing in the medium groove formed by G6–G8 and G20–G22 strands. In some structures, A11 is also turned away from the G-quadruplex whereas it is placed between G22 and G8 in the others. The representative structure displaying mean positions of A9 and A11 is shown in Figure 9A. Even though A13 has only few inter-residue NOE contacts, its position is considerably well defined above G3 and G14 where it is also stacked with C12 in most of the structures. This conformation is in agreement with the observation of several NOE contacts between C12 and A13 (C12 H1′/H2′/H3′–A13 H8).
Figure 9.
Position of loop residues in the G-quadruplex adopted by ODN. (A) Diagonal, (B) propeller and (C) edgewise loops in the representative structure, showing stacking of G17 and C19 on G1–G16–G20–G6 quartet, and stacking of C12 on G3–G8–G22–G14 quartet. G1–G16–G20–G6, G2–G7–G21–G15 and G3–G8–G22–G14 quartets are shown in red, orange and yellow, respectively.
The propeller loop spans across the medium groove defined by G1–G3 and G6–G8 strands. Interestingly, T5 is positioned in the centre of the groove, which is in excellent agreement with several of its NOE contacts with G1, G3 and G6 (Figure 9B). Positioning of thymine residues in the grooves of G-quadruplex structures has been observed earlier for two and three-residue propeller loops (22,26). The purine moiety of A4, on the other hand, is turned away from the structure. Protons of A4 exhibit downfield chemical shifts suggesting exposure of this residue to solvent. However, its sugar-phosphate backbone is positioned close to the groove and its flexibility is restricted due to restraints imposed by conformations of the two-residue loop in the propeller-type topology.
The edgewise loop connects anti to syn guanine residues across the wide groove defined by the G14–G16 and G20–G22 strands. Perusal of Figure 7 shows that residues in this loop form a cap under the G1–G16–G20–G6 quartet. Whereas G17 adopts a well-defined structure being stacked on the G1–G16–G20–G6 quartet, C19 is more flexible. The third residue in the edgewise loop, A18, is the least defined residue in this loop and can adopt several conformations. It is flipped away from the G-quadruplex core in most of the structures (Figure 9C).
ODN G-quadruplex in comparison to other G-quadruplexes: loop lengths and stability
ODN could be expected to fold into G-quadruplex structures with different topologies or loop lengths, since its sequence contains four guanine residues in the third G-tract. In principle, any of the four guanine residues in the third G-tract could participate in G-quartet formation. Loop length could consequently vary; loops could be comprised 2, 5 and 3 or 2, 6 and 2 nt. Experimental data, however, demonstrated the presence of only one species with a shorter central loop of 5 nt in the presence of potassium ions. In contrast, other G-quadruplex structures where guanine residues are also found in loops and therefore have potential to adopt structures with different loop lengths (10,13), adopt topologies with one long(er) and two short(er) loops that are more stable than folds with loops of roughly the same length.
It could be hypothesized that in the case of ODN, a longer (central) loop would destabilize the potential G-quadruplex structure. Mergny et al. have compared melting temperatures of G-quadruplexes containing four tracts of three guanines with loops of variable lengths (27). They found a strong inverse correlation between loop length and melting temperature as an indicator of G-quadruplex stability. Increasing the length of any loop has led to a decrease in stability of G-quadruplex(es) and a destabilizing effect has been observed always for loop lengths of ≥6 nt. The differences in melting temperatures could not be attributed to loop lengths alone, though, because G-quadruplex structures were not of uniform topology. Related studies have shown that melting temperatures decrease with increasing length of the third propeller loop, while maintaining the (3+1)-fold of a G-quadruplex and the length of the other two edgewise loops. This effect was considerable when loop length increased from 3 to 9 nt, but hardly noticeable when loop length increased further to 15 and 21 nt (23). We can therefore reason that the hypothetical six-residue loop of G-quadruplex adopted by ODN represents a critical destabilizing effect and compromises the stability of a potential G-quadruplex with 2:6:2 loop lengths.
In comparison to related G-quadruplexes with a long central loop, the structure adopted by ODN exhibits a lower Tm value (58°C), although several of them exhibit a central loop that is longer than the central loop of ODN. A majority of the related G-quadruplexes, however, adopts a parallel orientation of strands or exhibits at least one single-residue propeller loop, both of which were shown to have a stabilizing effect on G-quadruplex structures (27–29). Anti-parallel G-quadruplexes with loops longer than one residue have significantly lower melting temperatures and their values are comparable to the Tm value of the structure adopted by ODN (8,23). It therefore seems that the destabilizing effect of a long(er) central loop can be compensated with the stabilization of a structure with a one-residue loop and parallel topology.
It is interesting to note that the diagonal loop in the G-quadruplex adopted by ODN is not well ordered despite several sequential and long-range NOE contacts. Long loops in G-quadruplexes are often ordered and especially thymine and adenine residues can be included in triads, whereas adenine residues can form pentads, hexads and heptads with guanine residues in G-quartets (3). Such ordered structures additionally stabilize G-quadruplexes. The relatively low melting temperature of the G-quadruplex adopted by ODN may be a reflection of a disordered diagonal loop. However, an even lower melting temperature (53°C) could be observed for the (3+1) wtTel26 G-quadruplex (30), which exhibits ordered T:A:T capping structures and three-residue loops. It therefore seems that the ordering of loop residues alone cannot represent the main contribution to the G-quadruplex stability. However, it appears that correlations of thermodynamic stability are very hard to generalize in terms of sequence details or specific structural elements alone due to polymorphism of G-quadruplex structures.
CONCLUSIONS
The current study established that d[G3ATG3ACACAG4ACG3] in a potassium ion-rich environment folds into an intramolecular monomeric (3+1) G-quadruplex consisting of three stacked G-quartets. The four strands are connected with three different types of loops: propeller, diagonal and edgewise consisting of 2, 5 and 3 nt, respectively. To the best of our knowledge, this is the first high-resolution G-quadruplex structure with all three main known loop types assembled in a single structure.
ACCESSION NUMBERS
Pdb 2LOD, BMRB 18209.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–8.
FUNDING
The Slovenian Research Agency, the Ministry of Higher Education, Science and Technology of the Republic of Slovenia [P1-0242 and J1-4020]; Slovak Grant Agency [1/0153/09 and 1/0504/12]; EAST-NMR FP7 project [contract 228461]; Bio-NMR FP7 project [contract 261863] and European Cooperation in Science and Technology [COST MP0802]. Funding for open access charge: The Slovenian Research Agency (ARRS).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Wong HM, Payet L, Huppert JL. Function and targeting of G-quadruplexes. Curr. Opin. Mol. Ther. 2009;11:146–155. [PubMed] [Google Scholar]
- 2.Huppert JL. Structure, location and interactions of G-quadruplexes. FEBS J. 2010;277:3452–3458. doi: 10.1111/j.1742-4658.2010.07758.x. [DOI] [PubMed] [Google Scholar]
- 3.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5'-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rawal P, Kummarasetti VBR, Ravindran J, Kumar N, Halder K, Sharma R, Mukerji M, Das SK, Chowdhury S. Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 2006;16:644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verma A, Halder K, Halder R, Yadav VK, Rawal P, Thakur RK, Mohd F, Sharma A, Chowdhury S. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J. Med. Chem. 2008;51:5641–5649. doi: 10.1021/jm800448a. [DOI] [PubMed] [Google Scholar]
- 6.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang R, Lin Y, Zhang C. Greglist: a database listing potential G-quadruplex regulated genes. Nucleic Acids Res. 2008;36:372–376. doi: 10.1093/nar/gkm787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kendrick S, Hurley LH. The role of G-quadruplex/i-motif secondary structures as cis-acting regulatory elements. Pure Appl. Chem. 2010;82:1609–1621. doi: 10.1351/PAC-CON-09-09-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balasubramanian S, Hurley LH, Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat. Rev. Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo K, Pourpak A, Beetz-Rogers K, Gokhale V, Sun D, Hurley LH. Formation of pseudosymmetrical G-quadruplex and i-motif structures in the proximal promoter region of the RET oncogene. J. Am. Chem. Soc. 2007;129:10220–10228. doi: 10.1021/ja072185g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun D, Guo K, Rusche JJ, Hurley LH. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Armond R, Wood S, Sun D, Hurley LH, Ebbinghaus SW. Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1α promoter. Biochemistry. 2005;44:16341–16350. doi: 10.1021/bi051618u. [DOI] [PubMed] [Google Scholar]
- 13.Dai J, Chen D, Jones RA, Hurley LH, Yang D. NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region. Nucleic Acids Res. 2006;34:5133–5144. doi: 10.1093/nar/gkl610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pérez A, Marchán I, Svozil D, Sponer J, Cheatham TE, III, Laughton CA, Orozco M. Refinement of the AMBER force field for nucleic acids: improving the description of α/γ conformers. Biophys. J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu X, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De La Torre JG, Huertas ML, Carrasco B. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys. J. 2000;78:719–730. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Webba da Silva M. NMR methods for studying quadruplex nucleic acids. Methods. 2007;43:264–277. doi: 10.1016/j.ymeth.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 18.Plavec J, Tong W, Chattopadhyaya J. How do the gauche and anomeric effects drive the pseudorotation equilibrium of the pentofuranose moiety of nucleosides? J. Am. Chem. Soc. 1993;115:9734–9746. [Google Scholar]
- 19.Karsisiotis AI, Hessari NM, Novellino E, Spada GP, Randazzo A, Webba da Silva M. Topological characterization of nucleic acid G-quadruplexes by UV absorption and circular dichroism. Angew. Chem. Int. Ed. 2011;50:10645–10648. doi: 10.1002/anie.201105193. [DOI] [PubMed] [Google Scholar]
- 20.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang N, Phan AT, Patel DJ. (3+1) Assembly of three human telomeric repeats into an asymmetric dimeric G-quadruplex. J. Am. Chem. Soc. 2005;127:17277–17285. doi: 10.1021/ja0543090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Patel DJ. Solution structure of the Tetrahymena telomeric repeat d(T2G4)4 G-tetraplex. Structure. 1994;2:1141–1156. doi: 10.1016/s0969-2126(94)00117-0. [DOI] [PubMed] [Google Scholar]
- 23.Yue DJE, Lim KW, Phan AT. Formation of (3+1) G-quadruplexes with a long loop by human telomeric DNA spanning five or more repeats. J. Am. Chem. Soc. 2011;133:11462–11465. doi: 10.1021/ja204197d. [DOI] [PubMed] [Google Scholar]
- 24.Crnugelj M, Sket P, Plavec J. Small change in a G-rich sequence, a dramatic change in topology: new dimeric G-quadruplex folding motif with unique loop orientations. J. Am. Chem. Soc. 2003;125:7866–7871. doi: 10.1021/ja0348694. [DOI] [PubMed] [Google Scholar]
- 25.Webba da Silva M, Trajkovski M, Sannohe Y, Ma'ani Hessari N, Sugiyama H, Plavec J. Design of a G-quadruplex topology through glycosidic bond angles. Angew. Chem. Int. Ed. Engl. 2009;48:9167–9170. doi: 10.1002/anie.200902454. [DOI] [PubMed] [Google Scholar]
- 26.Heddi B, Phan AT. Structure of human telomeric DNA in crowded solution. J. Am. Chem. Soc. 2011;133:9824–9833. doi: 10.1021/ja200786q. [DOI] [PubMed] [Google Scholar]
- 27.Guédin A, Gros J, Alberti P, Mergny J. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 2010;38:7858–7868. doi: 10.1093/nar/gkq639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bugaut A, Rodriguez R, Kumari S, Hsu ST, Balasubramanian S. Small molecule-mediated inhibition of translation by targeting a native RNA G-quadruplex. Org. Biomol. Chem. 2010;8:2771–2776. doi: 10.1039/c002418j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang AYQ, Bugaut A, Balasubramanian S. A sequence-independent analysis of the loop length dependence of intramolecular RNA G-quadruplex stability and topology. Biochemistry. 2011;50:7251–7258. doi: 10.1021/bi200805j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dai J, Carver M, Punchihewa C, Jones RA, Yang D. Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007;35:4927–4940. doi: 10.1093/nar/gkm522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.